Abstract
While the long non-coding RNAs (ncRNAs) constitute a large portion of the mammalian transcriptome, their biological functions has remained elusive. A few long ncRNAs that have been studied in any detail silence gene expression in processes such as X-inactivation and imprinting. We used a GENCODE annotation of the human genome to characterize over a thousand long ncRNAs that are expressed in multiple cell lines. Unexpectedly, we found an enhancer-like function for a set of these long ncRNAs in human cell lines. Depletion of a number of ncRNAs led to increased expression of their neighboring protein-coding genes, including the master regulator of hematopoiesis, SCL (also called TAL1), Snai1 and Snai2. Using heterologous transcription assays we demonstrated a requirement for the ncRNAs in mediating such enhancement of gene expression. These results reveal an unanticipated role for a class of long ncRNAs in activation of critical regulators of development and differentiation.
Recent technological advances have allowed the analysis of the human and mouse transcriptomes with an unprecedented resolution. These experiments indicate that a major portion of the genome is being transcribed and that protein-coding sequences only account for a minority of cellular transcriptional output (Bertone et al., 2004; Birney et al., 2007; Cheng et al., 2005; Kapranov et al., 2007). Discovery of RNA interference (RNAi) (Fire et al., 1998) in C. elegans and the identification of a new class of small RNAs known as microRNAs (Lee et al., 1993; Wightman et al., 1993) led to a greater appreciation of RNA’s role in regulation of gene expression. MicroRNAs are endogenously expressed non-coding transcripts that silence gene expression by targeting specific mRNAs on the basis of sequence recognition (Carthew and Sontheimer, 2009). Over 1,000 microRNA loci are estimated to be functional in humans, modulating roughly 30 percent of protein-coding genes (Berezikov and Plasterk, 2005).
While microRNAs represent a minority of the non-coding transcriptome, the tangle of long and short non-coding transcripts is much more intricate, and is likely to contain as yet unidentified classes of molecules forming transcriptional regulatory networks (Efroni et al., 2008; Kapranov et al., 2007). Long ncRNAs are transcripts longer than 100 nts which in most cases mirror the features of protein-coding genes without containing a functional open reading frame (ORF). Long ncRNAs have been implicated as principal players in imprinting and X-inactivation. The imprinting phenomenon dictates the repression of a particular allele, depending on its paternal or maternal origin. Many clusters of imprinted genes contain ncRNAs, and some of them have been implicated in the transcriptional silencing (Yang and Kuroda, 2007). Similarly, the X-chromosome inactivation relies on the expression of a long ncRNA named Xist, which is thought to recruit, in a cis-specific manner, protein complexes establishing repressive epigenetic marks that encompass the chromosome (Heard and Disteche, 2006). There is also a report indicating that a long ncRNA expressed from the HOXC locus may affect the expression of genes in the HOXD locus which is located on a different chromosome (Rinn et al., 2007). More recently, a set of long ncRNAs has been identified in mouse, through the analysis of the chromatin signatures (Guttman et al., 2009). There has also been reports of divergent transcription of short RNAs flanking transcriptional start sites of the active promoters (Core et al., 2008; Preker et al., 2008; Seila et al., 2008).
In search of a function for long ncRNAs, we used the GENCODE annotation (Harrow et al., 2006) of the human genome. To simplify our search we subtracted transcripts overlapping the protein-coding genes. Moreover, we filtered out the transcripts that may correspond to promoters of protein-coding genes and the transcripts that belong to known classes of ncRNAs. We identified 3,019 putative long ncRNAs that display differential patterns of expression. Functional knock-down of multiple ncRNAs revealed their positive influence on the neighbouring protein-coding genes. Furthermore, detailed functional analysis of a long ncRNA adjacent to the Snai1 locus using reporter assays demonstrated a role for this ncRNA in an RNA-dependent potentiation of gene expression. Our studies suggest a role for a class of long ncRNAs in positive regulation of protein-coding genes.
Results
Non-coding RNAs are expressed and respond to cellular differentiating signals
To assign a function to uncharacterized human long ncRNAs, we identified unique long non-coding transcripts using the annotation of the human genome provided by the GENCODE (Harrow et al., 2006) and performed by human and vertebrate analysis and annotation (HAVANA) group at Sanger Institute. Such genomic annotation is being produced in the framework of the ENCODE project (Birney et al., 2007). At the time of our analysis, the GENCODE annotation encompassed about one third of the human genome. Such an annotation relies on the human expert curation of all available experimental data on transcriptional evidence, such as cloned cDNA sequences, spliced RNAs and ESTs mapped on to the human genome.
We focused on ncRNAs that do not overlap the protein-coding genes in order to simplify the interpretation of our functional analysis of ncRNAs. This included the subtraction of all transcripts mapping to exons, introns and the antisense transcripts overlapping the protein-coding genes. We also excluded transcripts within 1 kb of the first and the last exons as to avoid promoter and 3′-associated transcripts (Fejes-Toth K, 2009; Kapranov et al., 2007), that display a complicated pattern of short transcripts (Core et al., 2008; Preker et al., 2008; Seila et al., 2008). Furthermore, we excluded all known non-coding transcripts from our list of putative long ncRNAs. This analysis resulted in 3,019 ncRNAs, which are annotated by HAVANA to have no coding potential, expressed from 2,286 unique loci (some loci display multiple alternative spliced transcripts) of the human genome (Materials and Methods, Table S1). The average size of the non-coding transcripts is about 800 nts with a range from 100 nts to 9,100 nts. Interestingly, the long ncRNAs display a simpler transcription unit than that of protein-coding genes (Figure S1A). Nearly 50% of our long ncRNAs contain a single intron in their primary transcript (Figure S1A). Moreover, analysis of their chromatin signatures indicated similarities with protein-coding genes. Transcriptionally active ncRNAs display histone H3K4 trimethylation at their 5′-end (Figure S1B) and histone H3K36 trimethylation in the body of the gene (Figure S1C).
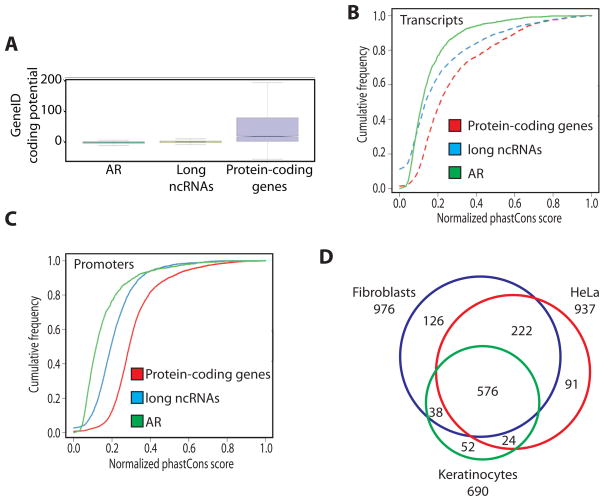
Analysis of protein coding potential of the ncRNAs using GeneID (Blanco et al., 2007; Parra et al., 2000) shows ncRNAs coding potential comparable to that of ancestral repeats (Lunter et al., 2006), supporting the HAVANA annotation of these transcripts as non-coding (Figure 1A). Moreover, comparison of ncRNAs with protein-coding genes and control sequences corresponding to ancestral repeats (Lunter et al., 2006) reveals that ncRNA sequence conservation is lower than that of protein-coding genes, but higher than that of ancestral repeats (Figure 1B). A similar case is seen with the promoter regions (Figure 1C). These results are in concordance with previous observations in the mouse genome (Guttman et al., 2009; Ponting et al., 2009).
Figure 1. Identification of novel long ncRNAs in human annotated by GENCODE.
(A) Analysis of coding potential using Gene ID for ancestral repeats (AR), long ncRNAs annotated by GENCODE and protein-coding genes.
(B) Conservation of the genomic transcript sequences for AR, long ncRNAs, protein-coding genes, and (C) of their promoters.
(D) Expression analysis of 3,019 long ncRNA in human fibroblasts, HeLa cells and primary human keratinocytes, showing numbers for transcripts detected in each cell line and the overlaps between cell lines. All microarray experiments have been done in four replicates. See also Figure S1 and Tables S1 and S2.
Next we used custom-made microarrays (Materials and Methods) which were designed to include an average of six probes (non-repetitive sequences) against each ncRNA transcript to detect their expression. We analyzed the expression pattern of ncRNAs using three different human cell lines (Figure 1D). Overall, we detected 1,167 ncRNAs expressed in at least one of the three cell types and 576 transcripts common among the three cell types (Figure 1D). We validated the expression of 16 ncRNAs that mapped to the 1 percent of the human genome investigated by the original ENCODE study (Birney et al., 2007) using quantitative polymerase chain reaction (qPCR) in three different cell lines (Table S2). Furthermore, we could find evidence for expression of 80% of our non-coding transcripts in at least one human tissue in a recent high throughput sequencing of the human transcriptome (Wang et al., 2008).
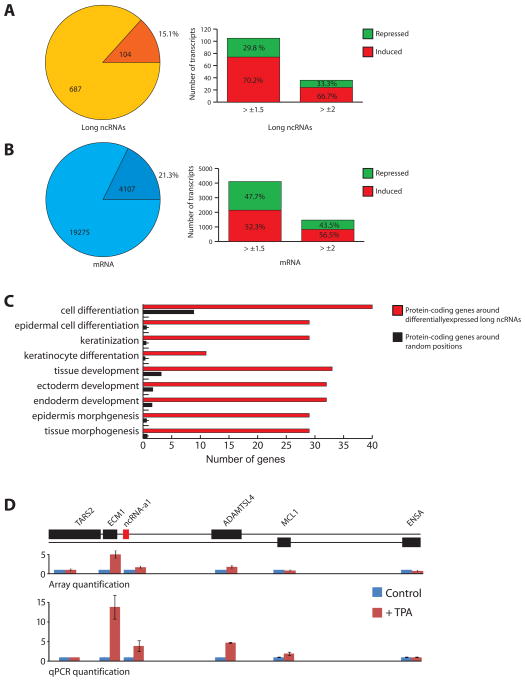
To assess whether ncRNAs respond to cellular differentiating signals, we induced the differentiation of human primary keratinocytes using 12-O-tetradecanoylphobol 13-acetate (TPA). We monitored the expression of ncRNAs using custom microarrays. Expression of protein-coding genes was monitored using conventional Agilent arrays containing nearly all human mRNAs. We prepared RNA from human primary keratinocytes before and following treatment with TPA. As shown in Figure 2A and Table S3, we could detect 687 ncRNAs in keratinocytes, where 104 (or 15.1 percent) respond to TPA treatment by over 1.5 fold. Similarly, 21.3 percent of protein coding-genes display a change in expression of over 1.5 fold (Figure 2B). While around half of the TPA-regulated protein-coding genes increase and a similar proportion decrease their expression following differentiation, 70 percent of the TPA-regulated ncRNAs increase their expression whereas only 30 percent show a decrease (Figures 2A and B). Furthermore, analysis of the protein-coding genes in the 500 Kb window surrounding the TPA-regulated ncRNAs indicates a significant enrichment in genes involved in differentiation and morphogenesis (Figure 2C). An example of such change in expression of an important gene involved in extra-cellular matrix is shown in Figure 2D. Extracellular Matrix Protein 1 (ECM1) gene and an ncRNA adjacent to it displayed a 5 and 1.7 fold induction following TPA treatment, respectively. (Figure 2D, upper panel). qPCR analysis shows the TPA-mediated induction of ECM1 and the ncRNA as 14 and 4 fold, respectively (Figure 2D, bottom panel). Taken together, we found that many of the GENCODE annotated transcripts are expressed in multiple cell lines and that they display gene expression responsiveness to differentiation signals.
Figure 2. Long ncRNAs display responsiveness to differentiation signals in human primary keratinocytes.
(A and B) Distribution of differentially expressed transcripts (dark colors) following TPA treatment for long ncRNAs (A), and mRNAs (B). Lighter colors show total number of transcripts, darker colors and percentage show number of differentially expressed transcripts. Bar-plots show number and fractions of transcripts induced (red) or repressed (green) at different fold-change cut-offs.
(C) Gene onthology analysis of genes flanking the differentially expressed long ncRNAs (red) compared to genes flanking random positions (black).
(D) Graphic representation of a locus with induction of the long ncRNA ncRNA-a1 and the adjacent ECM1 gene, with expression values from microarrays (upper panel) and qPCR quantification of transcripts (lower panel). Microarray experiments and qPCR validation are done in four replicates. Data shown are mean +/− S.D. See also Figure S2 and Table S3.
Non-coding RNAs display a transcriptional activator function
To assess the function of our set of long ncRNAs, we reasoned that similar to long ncRNAs function at the imprinting loci, our collection of ncRNAs may act to regulate their neighboring genes. To test this hypothesis, we used RNA interference to deplete a set of ncRNAs. We initially chose ncRNAs that showed a differential expression following keratinocyte differentiation. However, to obtain a reproducible knock-down we had to use cell lines that are permissive to transfection by siRNAs. We used five different cell lines for our analyses in which the candidate ncRNAs display a detectable expression (Figure 3).
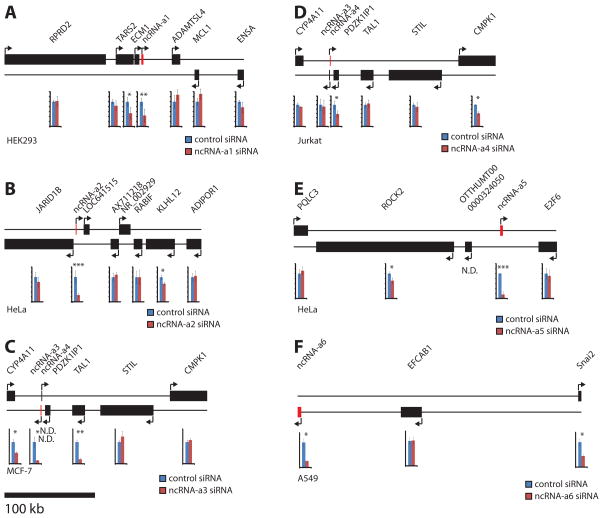
Figure 3. Stimulation of gene expression by activating RNAs.
The thick black line representing each gene shows the span of the genomic region including exons and introns. The targeted activating RNAs are shown in red. Bar-plots show RNA levels as determined by triplicate qPCR experiments and represent at least three independent experiments.
(A) ncRNA-a1 locus in HEK293 cells.
(B) ncRNA-a2 locus in HeLa cells.
(C) ncRNA-a3 locus in MCF-7 cells.
(D) ncRNA-a4 locus in Jurkat cells.
(E) ncRNA-a5 locus in HeLa cells.
(F) ncRNA-a6 locus in A549 cells. All values are relative to GAPDH expression and relative to control siRNA transfected cells set to an average value of 1. Scale bar is 100 kb and applies to all figure panels. Error bars show +/− S.E.M. * p < 0.05, ** p < 0.01, *** p < 0.001 by two-tailed Student’s T-test. See also Figure S3 and Table S4. The results represent at least six independent experiments. See also Figure S3 and Table S4
We validated the expression of our experimental set of ncRNAs and the absence of protein-coding potential using rapid amplification of 5′ and 3′ complementary DNA ends (5′ and 3′ RACE), PCR and in vitro translation (Figure S3). These experiments confirmed the expression of ncRNAs and showed that they do not yield a product in an in vitro translation assay (Figures S3A and S3B), supporting the non-coding annotation of our set of ncRNAs. In two cases, the ncRNAs adjacent to Snai2 and TAL1 loci, we found evidence of a longer ncRNA transcript than that annotated by HAVANA (Figure S3).
We began by examining small interfering RNAs (siRNAs) against the ncRNA next to ECM1 in order to assess its functional role following its depletion (for reasons that will follow this class of RNA is designated as non-coding RNA-activating1 through 7, ncRNA-a1-7). HEK293 cells were used for these experiments because of the ease of functional knock-down and the detectable amounts of ncRNA-a1 and ECM1 in this cell line. We compared the results obtained using two siRNAs against ncRNA-a1 to data obtained following the transfection of two control siRNAs (for the visual simplicity only one siRNA is shown (Figure 3A), the values for both siRNAs can be seen in Table S4). The two siRNAs produced comparable results. We interrogated a 300 Kb window around the ncRNA-a1 containing six protein-coding genes using qPCR.
Surprisingly, unlike the silencing action of long ncRNAs in imprinting and X-inactivation, depletion of ncRNA-a1 adjacent to ECM1 resulted in a concomitant decrease in expression of the neighboring ECM1 gene (Figure 3A). This effect was specific, as we did not detect any change in the other protein-coding genes surrounding ncRNA-a1 (Figure 3A). To ascertain that ncRNA-a1 is not a component of the ECM1 3′ untranslated region, we used primer pairs spanning the ECM1 and ncRNA-a1 genes. We were not able to detect a transcript comprised of the two genes in HEK293 cells, supporting the contention that the two transcripts are independent transcriptional units (Figure S2A). Furthermore, published ChIP experiments (Euskirchen et al., 2007) show the presence of RNA polymerase II and trimethyl H3K4 peaks at the transcription start site of ncRNA-a1 in several cell lines, further attesting to an independent transcriptional start site for ncRNA-a1. Moreover, knocking down the ECM1 gene did not affect the expression level of ncRNA-a1 or any of the other protein-coding genes analyzed in the locus, further supporting the independence of ECM1 transcript from that of ncRNA-a1 (Figure S2B).
Next we analyzed ncRNA-a2 flanking the histone demethylase JARID1B/KDAM 5B which also shows increased expression following keratinocyte differentiation. These experiments were performed in HeLa cells as they showed detectable expression of ncRNA-a2. Interestingly, while depletion of ncRNA-a2 did not change JARID1B/KDAM 5B levels, the KLHL12, a gene known for its negative regulation of the Wnt-beta catenin pathway, on the opposite strand displayed a significant reduction (Figure 3B). Although the decrease in KLHL12 was small (about 20 percent), no other protein-coding gene in the locus displayed a difference in expression (Figure 3B).
To extend our findings and to determine whether regulation of neighboring protein-coding genes is a common function of ncRNAs, we interrogated the ncRNA-a3 flanking the stem cell leukemia gene (SCL, also called TAL1). SCL is a basic helix-loop-helix protein which serves as the master regulator of hematopoiesis (Lecuyer and Hoang, 2004). This locus contains two ncRNAs on different strands of DNA. We used MCF-7 cells to assess the depletion of ncRNA-a3, since the expression of ncRNA-a3 and TAL1 could be readily detected in these cells. However, neither PDZK1IP1 nor ncRNA-a4 could be detected by qPCR in MCF-7 cells. Depletion of ncRNA-a3 resulted in a specific and potent reduction of TAL1 expression (Figure 3C). While depletion of ncRNA-a3 did not affect either STIL or CMPK1 genes, a significant reduction in CYP4A11 gene on the opposite strand of the DNA was detected (Figure 3C).
We next turned our attention to ncRNA-a4 which was not expressed at a detectable level in MCF7 cells. We could reliably detect ncRNA-a4 in Jurkat cells. While we could not efficiently knock-down ncRNA-a3 in Jurkat cells, siRNAs specific to ncRNA-a4 reproducibly reduced its levels by about 50 percent (Figure 3D). Importantly, reduced levels of ncRNA-a4 resulted in a consistent and significant decrease in the level of the gene CMPK1 which is over 150 kb downstream of ncRNA-a4 (Figure 3D). We do not detect any changes in the other protein-coding genes surrounding ncRNA-a4. Next we depleted ncRNA-a5 which is adjacent to the E2F6 gene, an important component of a polycomb-like complex (Ogawa et al., 2002). Knock-down of ncRNA-a5 did not affect the E2F6 gene. However, depletion of ncRNA-a5 resulted in a specific reduction in ROCK2 expression levels in HeLa cells, which is located upstream of ncRNA-a5 (Figure 3E).
Finally, we examined the Snai1 and Snai2 loci in A549 cells (Figure 3F and Figure 4). The Snail family of transcription factors are implicated in the differentiation of epithelia cells into mesenchymal cells (epithelial-mesenchymal transition) during embryonic development (Barrallo-Gimeno and Nieto, 2005; Savagner, 2001). Snai2 shows a significant reduction in expression when the adjacent ncRNA-a6 is depleted, an effect that is not seen on EFCAB1, the only other protein-coding gene within 300 kb of the ncRNA-a6 (Fig. 3F). In total, we have examined 12 loci where we were able to efficiently knock-down the ncRNAs using siRNAs (Table S5). We were able to show that in 7 cases, the ncRNA acts to potentiate the expression of a protein-coding gene within 300 kb of the ncRNA. It is possible that the remaining ncRNAs which did not display a positive effect on the neighboring genes within the 300 kb window, exert their action over longer distances which was not assessed in our analysis. Taken together, our results indicate that a subset of ncRNAs has activating functions and therefore we have named them ncRNA-activator (ncRNA-a) followed by a number to distinguish each activating long ncRNA.
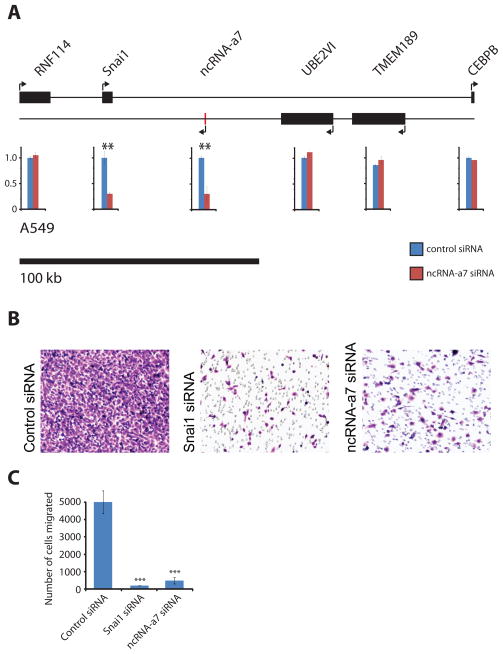
Figure 4. Knock-down of ncRNA-a7 specifically targets Snai1 expression.
(A) As in Figure 3, the ncRNA-a7 locus is depicted showing effects on RNA levels for the surrounding genes with and without knock-down of ncRNA-a7. The results represent at least six independent experiments.
(B) Migration assay of A549 cells with control (right panel) or ncRNA-a7 (left panel) siRNA transfections.
(C) Quantification of the data shown in (B).
All experiments are done in three replicates, and are shown as mean +/− S.E.M. ** p < 0.01, *** p < 0.001 by two-tailed Student’s T-test. See also Figure S4 and Table S5.
ncRNA-a7 is a regulator of Snai1
As mentioned above, Snai1 is a member of the Snail zinc-finger family, which comprises transcription factors with diverse functions in development and disease (Barrallo-Gimeno and Nieto, 2005; Nieto, 2002). The Snail gene family is conserved among species from Drosophila to human and has been shown to function as mesodermal determinant genes (Barrallo-Gimeno and Nieto, 2005; Nieto, 2002). Snail genes are the regulators of cell adhesion, migration and epithelial-mesenchymal transition (EMT) (Barrallo-Gimeno and Nieto, 2005; Nieto, 2002). Analysis of the ncRNA close to the Snai1 gene provided us with an opportunity to combine our gene expression analysis with analysis of changes in cellular migration. Knock-down of ncRNA-a7 resulted in a specific reduction in Snai1 levels (Figure 4A). The expression of the four other protein-coding genes in this locus does not change following the depletion of ncRNA-a7. Concomitantly, knock-down of ncRNA-a7 has a significant phenotypic effect in cell migration assays, reducing the number of migrating cells to about 10 percent of that of the control (Figure 4B–C), consistent with the phenotypic changes following the depletion of Snai1 (Figure 4B–C).
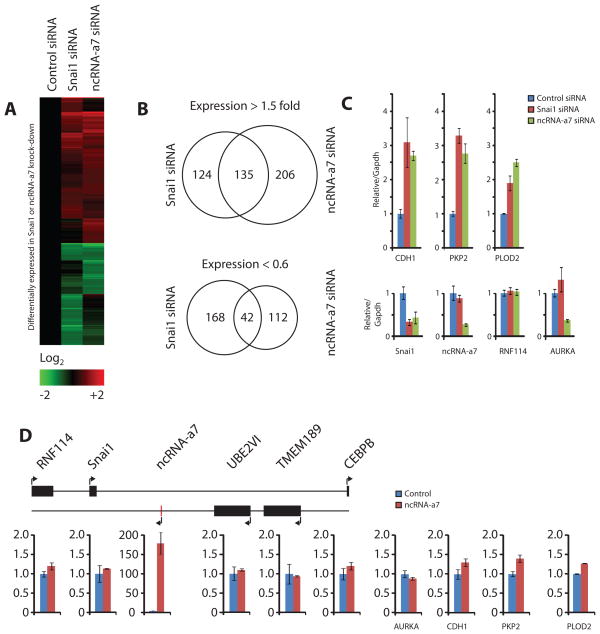
Since the knock-down of ncRNA-a7 or Snai1 had similar consequences on cellular migration, we assessed their depletion on gene expression in A549 cells using Agilent arrays. We could not detect the basal level of Snai1 on the array, while Snai1 was readily detectable using quantitative PCR. Interestingly, depletion of Snai1 or ncRNA-a7 resulted in similar changes in gene expression profiles (Figure 5A and Tables S6). Not only did we observe a similar trend in genes that were affected upon the knockdown of either gene but also a significant number of genes that were up-regulated were in common in both treatments (Figure 5A and B). Since Snai1 is a known transcriptional repressor, depletion of Snai1 or ncRNA-a7 should result in an up-regulation of Snai1 target genes. Indeed, a number of genes that were commonly up-regulated were direct targets of Snai1 (Figure 5C, upper panel) (De Craene et al., 2005). Depletion of either ncRNA-a7 or Snai1 also resulted in down regulation of a set of genes with a partial overlap between the genes down-regulated following the two treatments (Figure 5B). Interestingly, Aurora-kinase A a gene that is 6 MB down-stream of ncRNA-a7 was specifically down-regulated following the depletion of ncRNA-a7, suggesting a long range effect for ncRNA-a7 (Figure 5C). Taken together, these results indicate that while the depletion of ncRNA-a7 partially mimic the gene expression profile observed following Snai1 depletion, there are a number of gene expression changes resulting from the ncRNA-a7 depletion that occur independently of changes in Snai1. Therefore, it is likely that depletion of ncRNA-a7 may have other effects on gene expression which may be mediated through other targets in trans.
Figure 5. Microarray analysis of Snai1 and ncRNA-a7 knock-down.
Snai1 or ncRNA-a7 were knocked down using siRNA in A549 cells and the isolated RNA analyzed on microarrays in duplicate experiments. (A) All genes differentially expressed (>1.5 fold or <0.6 fold compared to control) in either Snai1 or ncRNA-a7 knock-down, or both, are shown clustered in a heat map according to expression profile. Numbers are log(2) transformed and color-scale is shown below the heat map. (B) Analysis of genes showing upregulation (>1.5 fold) or downregulation (<0.6 fold) in both Snai1 and ncRNA-a7 knock-down. Numbers represent number of genes regulated in the indicated condition. (C) Validation of microarray data by qPCR, and (D) analysis of the Snai1 locus and targets of Snai1 upon over-expression of ncRNA-a7. ncRNA-a7 was over-expressed from a vector in A549 cells and expression of select genes were measured by qPCR. Y-axes show expression value relative to GAPDH of the indicated gene. Values are normalized to those of control siRNA transfected cells, set to 1. ** p < 0.01, *** p < 0.001 by one-tailed Student’s T-test. See also Table S6.
To specifically address whether ncRNA-a7 may exert its effects in trans, we assessed the gene expression changes in Snai1 locus as well as some of the targets that were changed by depletion of ncRNA-a7 or Snai1 following the over-expression of ncRNA-a7 (Figure 5D). Overall, we did not observe changes in gene expression for any of the ncRNA-a7 targets following its over-expression (Figure 5D, ncRNA-a7 was over-expressed 150 fold). While these results suggest that ncRNA-a7 exerts its local gene expression changes in cis, it is likely that other targets may be influenced in trans. Taken together, these experiments reveal a role for ncRNA-a in positive regulation of expression of neighboring protein-coding genes and show that this effect is not specific to any one locus and may represent a general function for ncRNAs in mammalian cells.
ncRNA-activation of gene expression of a heterologous reporter
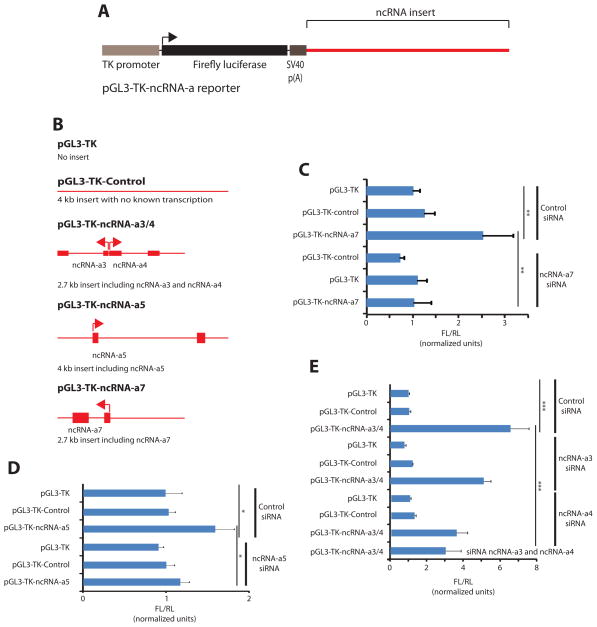
Previous studies have shown that distal activating sequences/enhancers can stimulate transcription when placed adjacent to a heterologous promoter, a methodology widely used to validate potential enhancers (Banerji et al., 1983; Banerji et al., 1981; Gillies et al., 1983; Heintzman et al., 2009; Kong et al., 1997). To functionally dissect the influence of the ncRNA-activation on the expression of an adjacent gene, we constructed vectors with inserts containing either ncRNA-a3 and -a4 from a bidirectional promoter, ncRNA-a5 or ncRNA-a7, and placed them down-stream of Firefly luciferase driven by a thymidine kinase (TK) promoter in a reporter vector ((pGL3-TK-ncRNA-a), (Figure 6A). We included 1–1.5 kb upstream of the ncRNA-as to contain their endogenous promoters and 500 bps downstream in the reporter vector. We also produced a control vector (pGL3-TK-control) in which 4 kb of DNA without transcriptional potential was cloned down-stream of Firefly luciferase similar to the ncRNA-activation reporters (Figure 6B). A vector containing Renilla luciferase was used to control for transfection efficiency. Importantly, inclusion of either of the three ncRNA-a inserts result in an enhancement of transcription ranging from 2 to 7 folds (Figure 6C–E). This effect is specific as pGL3-TK-control vector do not enhance the basal TK promoter activity (Figure 6C–E). To demonstrate that the observed potentiation of gene expression is mediated through the action of ncRNA-a, we knocked down the ncRNA-a in question for each reporter construct using specific siRNAs (Figure 6C–E). Interestingly while depletion of ncRNA-a7 and ncRNA-a5 completely abolished the increased transcription, depletion of ncRNA-a3 and/or ncRNA-a4 resulted in a partial decrease in transcriptional enhancement (Figure 6C–E). These results suggest that while ncRNA-a play a major role in transcriptional activation, other DNA elements in the cloned ncRNAa-3/4 region may also contribute to increased transcription.
Figure 6. ncRNA-activators potentiate transcription of a reporter gene.
(A) ncRNA-a 3/4, 5 and 7 were cloned and inserted downstream of luciferase driven by a TK-promoter in a reporter plasmid as shown. (B) Graphical representation of the inserts in the various vectors used. The pGL3-TK-Control vector contains an insert of approximately 4 kb containing no annotated evidence of transcription. The depicted inserts show exons and transcriptional direction of the ncRNA-a. (C–E) Luciferase reporter assays. The Firefly luciferase vectors were co-transfected with a Renilla luciferase vector (pRL-TK) for transfection control. (C) The vector containing ncRNA-a3 and ncRNA-a4 from a bidirectional promoter, with control siRNA or siRNAs towards either of the two ncRNA-a, or both. (D) Reporter with ncRNA-5, and (E) the reporter with the ncRNA-a7 inserted downstream of luciferase. X-axes show relative Firefly (FL) to Renilla (RL) luciferase activity. Co-transfected siRNAs are indicated to the right of the bars. All data shown are from six independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001 by one-tailed Student’s T-test.
Dissection of the ncRNA-a7 in a reporter construct
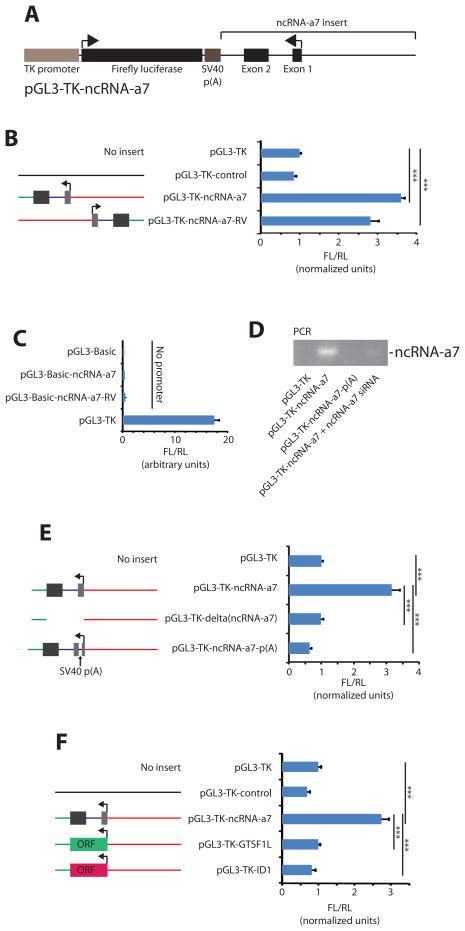
An important property of enhancing sequences is their orientation independence (Imperiale and Nevins, 1984; Khoury and Gruss, 1983; Kong et al., 1997). We designed reporter constructs (Figure 7A) in which the ncRNA-a7 sequence is reversed (pGL3-TK-ncRNA-a7-RV) in order to assess its orientation independence. The ncRNA-a7-RV construct displayed a similar transcriptional enhancing activity as the construct containing the ncRNA-a7 insert in its endogenous orientation with respect to the regulated gene (Figure 7B).
Figure 7. RNA-dependent activation of a reporter gene by ncNRA-a7.
(A) Properties of the ncRNA-a7 containing luciferase reporter vector. (B, C, E and F) Luciferase reporter assays. The Firefly luciferase vectors were co-transfected with a Renilla luciferase vector (pRL-TK) for transfection control. (D) Semiquantitative PCR of ncRNA-a7. (B) Reporter experiments with the ncRNA-a7 insert reversed as indicated in the left panel. (C) The TK-promoter driving luciferase expression was deleted from the construct and expression values are shown relative to the pGL3-TK control plasmid as a reference. (E) Truncated reporter constructs containing the ncRNA-a7 promoter and downstream sequences, but not the ncRNA-a7 sequence (pGL3-TK-delta(ncRNA-a7)), or one with a poly(A) signal in the beginning of the ncRNA-a7 to induce premature polyadenylation (pGL3-TK.ncRNA-a7-p(A)). See also (D) for analysis of expression from these plasmids. (F) Protein coding sequences were inserted in place of ncRNA-a7 downstream of the ncRNA-a7 promoter. Full-length GTSF1L or ID1 sequences are used. X-axes show relative Firefly (FL) to Renilla (RL) luciferase activity. All data shown are from six independent experiments. *** p < 0.001 by one-tailed Student’s T-test.
To show that luciferase expression requires a promoter and that ncRNA-7a cannot act as a proximal promoter, we deleted the TK promoter from the reporter vectors. As shown in Figure 7C, ncRNA-a7 cannot drive transcription of the Firefly luciferase in the absence of a proximal TK promoter. These experiments demonstrate that sequences corresponding to ncRNA-a7 transcription unit can function to activate expression of a heterologous promoter in an orientation-independent manner, but cannot act as a promoter itself.
To further verify that ncRNA-a7 is the active component of the transcriptional enhancement, we constructed two reporters in which ncRNA-a7 sequences are either deleted or shortened by placing a strong polyadenylation signal within the ncRNA-a genomic sequence but close to the transcriptional start site, to induce premature polyadenylation (Figure 7D–E). Both modifications result in loss of the increased gene expression (Figure 7E) compared to constructs where ncRNA-a7 is expressed. Finally, to assess whether the RNA corresponding to ncRNA-7a is critical for increased gene expression, we developed constructs where DNA sequences corresponding to two different protein-coding genes were positioned in the place of ncRNA-a7 (Figure 7F), keeping the endogenous ncRNA-a7 promoter. Neither of these constructs displayed an increased gene expression compared to that of the control constructs (Figure 7F). Taken together, these experiments demonstrate that the potentiation of gene expression is signaled by the ncRNA-a and is not merely the result of the transcription of the ncRNA.
Discussion
We used the annotation of the human genome performed by GENCODE to arrive at a collection of long ncRNAs that are expressed from loci independent of those of protein-coding genes or previously described non-coding RNAs. GENCODE annotation encompasses both protein-coding and non-coding transcripts and relies on experimental data obtained through the analysis of cDNAs, ESTs and spliced RNAs. Our collection of ~3,000 transcripts correspond to the manual curation of about a 1/3 of the human genome. Analysis of the GENCODE data indicates that nearly all of their non-coding annotated transcripts are spliced (Figure S1A).
Importantly, the median distance of an ncRNA transcript to a protein-coding gene is over a 100 kb making it an unlikely scenario for the ncRNA to be an extension of protein-coding transcripts (Figure S2C and D). Moreover, transcriptionally active ncRNAs display similar chromatin modifications seen with expressed protein-coding genes (Figure S1B and C). Furthermore, the analyzed ncRNAs display RNA pol(II), p300 and CBP occupancy at levels similar to those of the surrounding protein coding genes, consistent with their transcriptional independence (Figure S4). Although our analysis is focused on understanding the function of a set of ncRNAs annotated by GENCODE, the human transcriptome includes other forms of ncRNAs with important regulatory functions that have not been included in our study. These include the anti-sense transcripts arising from protein-coding genes, precursors of microRNAs as well as a wealth of unspliced transcripts described in multiple studies (Guttman et al., 2009; Kapranov et al., 2007; Rinn et al., 2007).
Taken together, the novelty of our work lies in the following. First, we show that at multiple loci of the human genome depletion of a long ncRNA leads to a specific decrease in the expression of neighboring protein-coding genes. Previous studies analyzing the function of long ncRNAs in X-inactivation or the imprinting phenomenon point to their role in silencing of gene expression (Mattick, 2009). Second, we show that the enhancement of gene expression by ncRNAs is not cell specific as we observe the effect in five different cell lines. Third, this enhancement of gene expression is mediated through RNA, as depletion of such activating ncRNAs abrogate increased transcription of the neighboring genes. Fourth, through the use of heterologous reporter assays, we suggest that activating ncRNAs mediate this RNA-dependent transcriptional responsiveness in cis. Fifth, we show that similar to classically defined distal activating sequences, ncRNA-mediated activation of gene expression is orientation independent. Sixth, we present evidence that similar to defined activating sequences, ncRNAs cannot drive transcription in the absence of a proximal promoter. Finally, we demonstrate that the activation of gene expression in the heterologous reporter system is mediated through RNA as multiple approaches depleting the RNA levels lead to abrogation of the stimulatory response. Therefore, we have uncovered a new biological function in positive regulation of gene expression for a class of ncRNAs in human cells.
There are previous reports of individual ncRNAs having a positive effect on gene expression. The ~3.8 kb Evf-2 ncRNA was shown to form a complex with the homeodomain-containing protein Dlx2 and lead to transcriptional enhancement (Feng et al., 2006). Similarly, the ncRNA HSR1 (heat-shock RNA-1) forms a complex with HSF1 (heat shock transcription factor 1), resulting in induction of heat shock proteins during the cellular heat shock response (Shamovsky et al., 2006) and an isoform of ncRNA SRA (steroid receptor RNA activator) functions to co-activate steroid receptor responsiveness (Lanz et al., 1999). Our findings that activating ncRNAs positively regulate gene expression extend these previous studies and demonstrate that the activation of gene expression by long ncRNA may be a general function of a class of long ncRNAs. Moreover, whether ncRNA effects seen in our study are mediated through association with specific transcriptional activators is not known. However, this is a likely scenario given previous examples of an RNA-mediated responsiveness. Other possibilities include a formation of an RNA-DNA hybrid at the loci of the ncRNA or the protein-coding gene which may result in enhanced binding of the sequence specific DNA binding proteins or chromatin modifying complexes.
A recent study uncovers a set of bi-directional transcripts (termed eRNA) that are derived from sites in the human genome that show occupancy by CBP, RNA polymerase II and are decorated by monomethyl Histone H3 lysine 4 (H3K4) (Kim et al., 2010). Moreover, they show that the expression of such transcripts is correlated with their nearest protein-coding genes. There are fundamental differences between their collection of ~2,000 transcripts and our GENCODE set of transcripts. First, while all their eRNAs are bidirectional, only about one percent of our ncRNAs show evidence of bi-directionality (see the example shown in the TAL1 locus). Second, our analysis of the histone modifications of a subset of ncRNAs that are expressed in lymph (Barski et al., 2007) indicates the presence of H3K4 trimethylation at the transcriptional start sites and H3K36 trimethylation at the body of the gene (Figure S1B and C). This is in stark contrast to eRNA loci where there is an absence of H3K4 trimethyl marks and the predominant chromatin signature is the monomethyl H3K4. Third, eRNAs are reported to be predominantly not polyadenylated. The majority of our collection of ncRNAs show evidence of polyadenylation as they were amplified using oligo-dT-primed reactions and furthermore 41 percent display the presence of a canonical polyadenylation site. Analysis of the protein-coding transcripts revealed that a similar proportion (52 percent) to that of our ncRNAs contain the canonical polyadenylation sites. Finally, while we show that a set of our ncRNAs function to enhance gene expression, there is no evidence provided for eRNAs exerting a biological function. While we believe that eRNAs designate a different class of ncRNAs than ncRNA-a described in our study, it is temping to speculate that many of the ncRNA-a and their promoters may correspond to mammalian enhancers or polycomb/trithorax response elements (PRE/TREs). In such a scenario, binding of polycomb or trithorax proteins to proximal promoters of ncRNA-a will regulate the expression of ncRNA-a which in turn impact the expression of the protein-coding gene at the distance.
Another set of recently published ncRNAs were termed long intervening non-coding RNA or lincRNAs (Guttman et al., 2009). The comparison of our ncRNAs and the lincRNAs show that about 13 percent of the ncRNAs defined by ENCODE overlap the broad regions encoding a set of recently identified human lincRNAs (Khalil et al., 2009). The overlap between our ncRNAs and lincRNAs are even smaller (~4%) if one considers only the exons corresponding to lincRNAs. Importantly, the studies with lincRNAs did not reveal any transcriptional effects in neighboring genes (Khalil et al., 2009). Therefore, it is likely that lincRNAs describe a distinct set of ncRNAs compared to those annotated by GENCODE. Similar to the diverse functions for proteins, ncRNAs such as lincRNAs may play other roles in regulating gene expression.
The GENCODE annotation used in this study encompasses only a third of the human genome. Therefore, the number of ncRNAs in human cells is likely to grow and may equal or even surpass the number of protein-coding genes. Our considerations for selection of ncRNAs excluded all ncRNAs associated with protein-coding genes and their promoters, as well as known ncRNAs. Therefore, the repertoire of the non-coding transcripts in human cells contains many more transcripts than those cataloged in this study. Specifically, there have been reports of pervasive amount of anti-sense transcription as well as transcription mapping to promoter regions of protein-coding genes (Core et al., 2008; Denoeud et al., 2007; Kapranov et al., 2007; Preker et al., 2008; Seila et al., 2008). Whether such transcripts will have biological functions similar to those described for activating ncRNAs in our study is not known. However, it is clear that future genome-wide genetic analysis of ncRNAs in mammalian cells will begin to shed light on different classes of the ncRNAs.
The precise mechanism by which our ncRNAs function to enhance gene expression is not known. We envision a mechanism by which ncRNAs by virtue of sequence or structural homology targets the neighboring protein-coding genes to bring about increased expression. Our experimental evidence using a heterologous promoter point to the mechanism of action for activating ncRNAs operating in cis. However, genome-wide analysis following depletion of ncRNA-a7 suggested changes in gene expression that may not be related to the action of ncRNA-a7 on its local environment and may be a result of wider trans-mediated effects of ncRNA-a7. Such regulatory functions of ncRNAs could be achieved through an RNA-mediated recruitment of a transcriptional activator, displacement of a transcriptional repressor, recruitment of a basal transcription factor or a chromatin-remodeling factor. While we favor a transcriptional based mechanism for ncRNA-activation, effects on RNA stability cannot be excluded. Taken together, the next few years will bring about new prospects for the long ncRNAs as central players in gene expression.
Experimental procedures
Extracting long ncRNA data
The HAVANA annotation12 has been downloaded using the DAS server26 provided by the Sanger institute (version July, 16th 2008). We removed all annotated biotypes or functional elements belonging to specific categories such as pseudogenes or protein-coding genes. We excluded all transcripts overlapping with known protein coding loci annotated by HAVANA, RefSeq or UCSC27. Transcripts falling into a 1kb window of any protein-coding gene were also removed. Finally, we excluded all transcripts covered by known non-coding RNAs such as miRNAs (miRbase version 11.0 April 2008).
To estimate the evolutionary constraints among mammalian sequences we constructed the cumulative distribution of PhastCons28 scores for ancestral repeats (ARs), RefSeq genes and long ncRNAs. The cumulative distributions of these transcripts or repeats are plotted using a log-scale on the y-axis.
Cell culture and siRNA transfections
Human primary keratinocytes from four different biological donors were grown in Keratinocyte medium (KFSM, Invitrogen). Differentiation was induced by 2.5 ng/ml 12-O-tetradecanoylphorbol-13-acetate (TPA) during 48h.
HEK293, A549, HeLa and MCF-7 cells were cultured in complete DMEM media (GIBCO) containing 10 % FBS, and 1X Anti/Anti (GIBCO). Jurkat cells were cultured in complete RPMI media (GIBCO) containing 10 % FBS and 1X Anti/Anti (GIBCO). Migration assays were performed as previously described(Gumireddy et al., 2009).
For transfections of 293, HeLa, A549 and MCF-7 cells we used Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations and an siRNA concentration of 50 nM. Jurkat cells were transfected using HiPerFect (Qiagen) according to the manufacturer’s recommendations and an siRNA concentration of 100 nM.
RNA purification, cDNA synthesis and quantitative PCR
Cells were harvested and resuspended in TRIzol (Invitrogen) and RNA extracted according to the manufacturer’s protocol. cDNA synthesis was done using MultiScribe reverse transcriptase and random primers from Applied Biosystems. Quantitative PCR was done using SybrGreen reaction mix (Applied Biosystems) and an HT7900 sequence detection system (Applied Biosystems). For all quantitative PCR reactions Gapdh was measured for an internal control and used to normalize the data. Primer sequences are as follows:
Cloning of pGL3-TK reporters and luciferase assay
pGL3-Basic was digested with BglII and HindIII and the TK promoter from pRL-TK was inserted into these sites. Inserts were amplified from genomic DNA and cloned into the BamHI and SalI sites 5′ to the luciferase gene. Luciferase assays were performed in 96-well white plates using Dual-Glo (Promega) according to the manufacturer’s protocol.
Microarrays
Custom-made microarrays (Agilent) were designed based on the library of 3,019 long ncRNA sequences, with on average six probes targeting each transcript. Human whole genome mRNA arrays were from Agilent (G4112F). Total RNA samples were converted to cDNA using oligo-dT primers. Labeling of the cDNA and hybridization to the microarrays were performed according to Agilent standard dye swap protocols. Data analysis was done using the AFM 4.0 software. All microarrays were done in 4 biological replicates.
Supplementary Material
Acknowledgments
Thanks to the HAVANA team for use of their genome annotation. We also thank the CRG Genomic Facility and the Functional Genomics Core Facility at Wistar and UPenn for expertise in Solexa sequencing and microarray analysis. We thank Dr. Ken Zaret for helpful discussions. UAO is supported by a grant from the Danish Research Council, MB is supported by an HFSPO fellowship. AG was supported by a fellowship from the American Italian Cancer Foundation. RG was supported through Spanish ministry and NIH and RS was supported by a grant from NIH, GM 079091.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Plasterk RH. Camels and zebrafish, viruses and cancer: a microRNA update. Hum Mol Genet. 2005;14(Spec No 2):R183–190. doi: 10.1093/hmg/ddi271. [DOI] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco E, Parra G, Guigo R. Using geneid to identify genes. Curr Protoc Bioinformatics. 2007;Chapter 4(Unit 4):3. doi: 10.1002/0471250953.bi0403s18. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene B, Gilbert B, Stove C, Bruyneel E, van Roy F, Berx G. The transcription factor snail induces tumor cell invasion through modulation of the epithelial cell differentiation program. Cancer Res. 2005;65:6237–6244. doi: 10.1158/0008-5472.CAN-04-3545. [DOI] [PubMed] [Google Scholar]
- Denoeud F, Kapranov P, Ucla C, Frankish A, Castelo R, Drenkow J, Lagarde J, Alioto T, Manzano C, Chrast J, et al. Prominent use of distal 5′ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res. 2007;17:746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen GM, Rozowsky JS, Wei CL, Lee WH, Zhang ZD, Hartman S, Emanuelsson O, Stolc V, Weissman S, Gerstein MB, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes-Toth KSV, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, Foissac S, Willingham AT, Duttagupta R, Dumais E, Gingeras TR. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gillies SD, Morrison SL, Oi VT, Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983;33:717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Gumireddy K, Li A, Gimotty PA, Klein-Szanto AJ, Showe LC, Katsaros D, Coukos G, Zhang L, Huang Q. KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nat Cell Biol. 2009;11:1297–1304. doi: 10.1038/ncb1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl 1):S4, 1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiale MJ, Nevins JR. Adenovirus 5 E2 transcription unit: an E1A-inducible promoter with an essential element that functions independently of position or orientation. Mol Cell Biol. 1984;4:875–882. doi: 10.1128/mcb.4.5.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G, Gruss P. Enhancer elements. Cell. 1983;33:313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010 doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong S, Bohl D, Li C, Tuan D. Transcription of the HS2 enhancer toward a cis-linked gene is independent of the orientation, position, and distance of the enhancer relative to the gene. Mol Cell Biol. 1997;17:3955–3965. doi: 10.1128/mcb.17.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O’Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Hoang T. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp Hematol. 2004;32:11–24. doi: 10.1016/j.exphem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lunter G, Ponting CP, Hein J. Genome-wide identification of human functional DNA using a neutral indel model. PLoS Comput Biol. 2006;2:e5. doi: 10.1371/journal.pcbi.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Parra G, Blanco E, Guigo R. GeneID in Drosophila. Genome Res. 2000;10:511–515. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savagner P. Leaving the neighborhood: molecular mechanisms involved during epithelial-mesenchymal transition. Bioessays. 2001;23:912–923. doi: 10.1002/bies.1132. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Yang PK, Kuroda MI. Noncoding RNAs and intranuclear positioning in monoallelic gene expression. Cell. 2007;128:777–786. doi: 10.1016/j.cell.2007.01.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.