Abstract
Transcription by RNA polymerase (RNAP) is interrupted by pauses that play diverse regulatory roles. Although individual pauses have been studied in vitro, the determinants of pauses in vivo and their distribution throughout the bacterial genome remain unknown. Using nascent transcript sequencing we identify a 16-nt consensus pause sequence in E. coli that accounts for known regulatory pause sites as well as ~20,000 new in vivo pause sites. In vitro single-molecule and ensemble analyses demonstrate that these pauses result from RNAP/nucleic-acid interactions that inhibit next-nucleotide addition. The consensus sequence also leads to pausing by RNAPs from diverse lineages and is enriched at translation start sites in both E. coli and B. subtilis. Our results thus reveal a conserved mechanism unifying known and newly identified pause events.
Transcriptional pausing by RNA polymerase (RNAP) is an important feature of gene regulation that facilitates RNA folding (1), factor recruitment (2), transcription termination (3), and synchronization with translation in prokaryotes (4, 5). Previously characterized regulatory pauses (6) represent a very small and biased fraction of potential pause sites in the bacterial genome. Furthermore, it remains unknown whether most pauses identified by in vitro studies significantly affect transcription in vivo. To study transcriptional pausing in vivo, we adapted a high-throughput approach to isolate and sequence nascent elongating transcripts (NET-seq) (7). Escherichia coli nascent transcripts were captured by immunoprecipitating FLAG-tagged RNAP molecules, converted to DNA, and sequenced to a depth of ~30 million reads per sample (figs. S1 to S3 and table S1 and S2). Each sequencing read was mapped to a single site corresponding to the 3′ end of the nascent transcript (Fig. 1A), allowing us to define RNAP locations along ~2,000 genes with single nucleotide resolution (table S2).
Fig. 1. Bacterial NET-seq provides a genome-wide view of transcription dynamics.
(A) Nascent RNA is isolated from bacteria and converted to a DNA library sequenced with deep coverage. Reads are aligned to the reference genome and mapped according to their 3′ end, which corresponds to the RNAP active site. (B) An example of RNAP density in the his leader region (hisL) shows a peak at a single site which matches the previously mapped regulatory pause position (underlined sequence). (C) Biological replicates along the ribosomal L10 protein subunit (rplJ). (D) Histogram of pause frequency for highly transcribed genes (n = 1984, gene average >1 read/bp) within the protein coding sequence.
The number of mapped reads at each genomic position is proportional to the number of RNAP molecules at that position. We observed well-defined single-nucleotide peaks within transcribed regions at known regulatory pause sites, including sites that synchronize transcription with translation, mediate RNA folding, or recruit transcription factors (Fig. 1B and fig. S4A–E). NET-seq profiles also revealed a large number of other highly reproducible peaks in RNAP density throughout the genome (example gene in Fig. 1C). In total, we identified ~20,000 previously undocumented pause sites across well-transcribed genes, representing an average frequency of 1 per 100 bp (Fig. 1D). Thus, known regulatory pause sites represent a tiny fraction of actual pause positions.
We found that in vivo pause propensity depended strongly on the sequence identity at the 3′-end of the transcript (87% of paused transcripts end with either cytosine or uracil), as well as on the identity of the incoming NTP substrate (70% of pause sites occur prior to GTP addition) (Fig. 2A). Sequence dependence extends outside the RNAP active site to 11 nucleotides (nt) upstream and 5 nt downstream of the pause position, consistent with the extent of core nucleic-acid contacts made within the elongation complex (8). To determine the contribution of each base to pause duration, we used the density of reads in the NET-seq profile to calculate the relative dwell time of RNAP at each well-transcribed position in the genome. Modeling the addition of the next nucleotide as a process with a single activation barrier, we calculated the effective energetic barrier to nucleotide addition as the logarithm of the RNAP occupancy signal (supplementary materials). We used these values to determine the sequence dependence of this barrier for all positions within 15 bases of the transcript 3′ end. The resulting plot provides an energetic view of sequence-dependent pausing, in which peaks indicate bases that increase the relative RNAP dwell time (Fig. 2B). These observations implicate a 16-nt consensus pause sequence whose prominent features include GG at the upstream edge of RNA:DNA hybrid and TG or CG at the location of the 3′-end of the nascent transcript and incoming NTP (Fig. 2A).
Fig. 2. Transcriptional pauses are driven by RNAP-nucleic acid interactions.
(A) Sequences corresponding to peaks in RNAP density were aligned at their 3′ end to generate a consensus pause sequence, the length of which matches the size of the transcription bubble (shown below). (B) Relative energetic contribution of neighboring bases as they impact in vivo pause dynamics (mean ± SD). The 16-nt consensus pause sequence, represented by peaks in energy, is shown above. (C) Cumulative distribution function for the energetics of both pause and non-pause sequences. (D) Experimental geometry for the single-molecule pausing assay and representative records of transcription by individual RNAP molecules in GTP-limiting conditions. Long pauses at GTP-coding positions (gray lines) provide register with the template DNA. (E) In vitro pause energetics calculated from the single-molecule data (mean ± SD, see supplement for SD estimation). (F) In vitro pause energetics are well-correlated with in vivo pause energetics determined by NET-seq (Pearson r = 0.6, two-tailed p-value = 9.8×10−17). Each point corresponds to a given nucleotide at a specific scaffold position (unlabeled).
We used the energetic profile as a metric to determine whether most in vivo pauses could be explained by the consensus pause sequence. The energetics of nucleotide addition (Fig. 2B) allowed us to compute the propensity for pausing at every well-transcribed position by summing the energetic contribution of each base from position −1 to −11. The predicted energies were grouped into two categories: sequences for which pausing was observed, and sequences for which pausing was undetectable. A cumulative histogram of the energetics for the two populations shows that pause-associated sequences were well-separated in sequence space from non-pause sequences (Fig. 2C). Using a receiver-operating characteristic (ROC) analysis, we determined the optimal threshold for distinguishing these two populations (fig. S5), and found that the majority of pause sequences lay above the threshold (Fig. 2C). Furthermore, the same threshold correctly classified the group of “canonical” regulatory pauses previously identified in E. coli, suggesting that this seemingly disparate group of pause sequences derive from a single consensus sequence. Intriguingly, the HIV-1 TAR pause element, which affects mammalian RNAPII (9), resembles our consensus sequence (Fig. 2C).
To understand the minimal requirements for pausing, we modified a high-resolution optical-trapping technique to measure sequence-resolved nucleotide addition by individual RNAP molecules in vitro (10, 11). By limiting the concentration of GTP, which is the nucleotide most frequently associated with pausing in vivo, its addition became rate limiting for elongation, allowing us to determine the absolute alignment of single-molecule records with the transcribed sequence. In this fashion, we measured the nucleotide addition rate for E. coli RNAP at over 300 unique positions in a segment of the E. coli rpoB gene (Fig. 2D). These position-specific rates, which ranged over ~2–3 orders of magnitude, yielded activation energy barriers well-correlated to those computed from NET-seq (Fig. 2E–F). Moreover, they are qualitatively consistent with an in vitro consensus proposed previously from a small set of pause-inducing elements (12). This agreement suggests that interactions of RNAP with the DNA template and nascent transcript are sufficient for pausing in vivo, and that these interactions largely dictate genome-wide pause patterns.
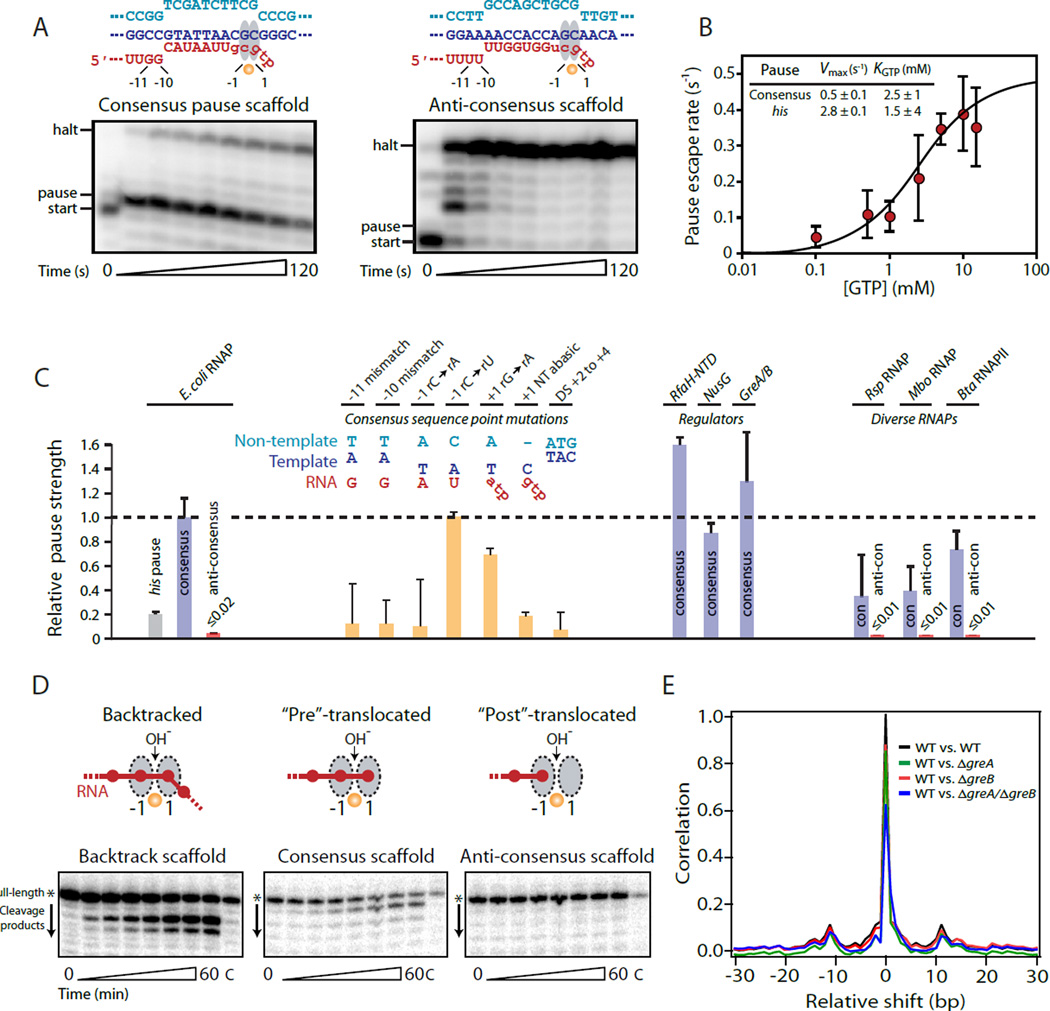
To probe individual elements of the consensus pause sequence, we reconstituted transcription complexes on a series of short, artificial nucleic-acid scaffolds. These scaffolds encoded either the consensus pause or an anti-consensus pause, in which the nucleotide at each position from −11 to +5 (excepting the highly conserved −1/+1 active-site positions) was altered to be the nucleotide predicted to cause the shortest dwell time (Fig. 3A). Strong pausing was observed at the expected position on the short consensus scaffold (Fig. 3A), and also on a template with the same consensus sequence embedded in a long DNA template (fig. S6). The consensus pause was roughly 5-fold stronger than the his pause (τ = 2 s at saturating GTP, Fig. 3B), despite the fact that the his pause is stabilized by a nascent RNA hairpin. Pausing was undetectable at the equivalent position on the anti-consensus scaffold (Fig. 3A). Thus, sequence elements upstream and downstream of the RNAP active site, although less enriched in our analysis, are essential for generating a pause signal. Consistent with prior proposals that discrete pause elements act together to form a multipartite pause signal (13), substitutions that disrupt RNA:DNA base-pairing at the −11 or −10 positions, remove the +1 non-template strand base, or alter the downstream DNA at positions +2 to +4 were found to reduce pause strength significantly (Fig. 3C, see fig. S7 for additional analysis of sequence dependence).
Fig. 3. Pause consensus sequence leads to a long-lived, non-backtracked pause in vitro.
(A) Purified E. coli RNAP was reconstituted on a nucleic-acid scaffold containing either the consensus pause sequence or an anti-consensus sequence. RNA nucleotides in lower case were added after initial reconstitution by extension with α-32P-labeled or unlabeled NTPs. Full sequences are shown in fig. S7. A strong pause is observed at the predicted position on the consensus pause scaffold, but does not occur on the anti-consensus scaffold. (B) Consensus pause escape rate (SD of ≥3 replicates) as a function of GTP concentration reveals a maximal escape rate ~5 times slower than the his pause. (C) Relative pause strengths for variants of the consensus pause (yellow), in the presence of transcription regulators, or with diverse RNAPs (SD of ≥3 replicates). (D) RNAP active site-catalyzed hydrolytic cleavage of nascent RNA in complexes reconstituted with a 3′ mismatch forcing a backtracked register (left), at the pause site on the consensus pause scaffold (middle), and at the equivalent position on the anti-consensus scaffold (right). (E) Mean cross-correlation between NET-seq profiles for WT E. coli and ΔgreA (green), ΔgreB (red), or ΔgreA/ΔgreB (blue) strains for well transcribed genes (n = 1240, gene average >1 read/bp for each sample). The mean autocorrelation for the WT strain is shown for comparison (black).
RNA polymerase has the ability to “backtrack”, shifting the transcript 3′ end downstream from the −1/+1 positions of the active site into the NTP-entry pore. Backtracking is resolved by cleavage of 2 or more nucleotides from the RNA, generating a new 3′ end in the active site. To determine whether RNAP backtracked at the consensus pause, we tested for transcript cleavage at the active site. Pause complexes reconstituted on the consensus scaffold cleaved only a single nucleotide, consistent with no backtracking, clearly different from the 2-nt cleavage observed with complexes prepared with an obligately backtracked scaffold, and also from complexes prepared with an anti-consensus scaffold (Fig. 3D). GreA, a cleavage factor in E. coli known to relieve backtracking, stimulated a 2-nt cleavage of the RNA at the consensus pause, but failed to reduce the pause dwell time (Fig. 3C and fig. S8), suggesting that the consensus pause sequence leads to a predominantly pre-translocated register which may be poised to backtrack, but that such backtracking does not principally determine the barrier to pause escape. It is likely that variations of the consensus sequence may lead to pauses that backtrack more readily. The observed pause profiles in vivo were unaffected by the deletion of GreA and GreB (Fig. 3E), suggesting that most transcriptional pauses in E. coli lead to an elemental non-backtracked pause state (12, 14).
The consensus pause sequence is conserved across diverse lineages, as demonstrated in vitro using RNAPs derived from Rhodobacter sphaeroides (Rsp), Mycobacteria bovis (Mbo), and Thermus thermophilus (Tth), which paused on the consensus template, but not on the anti-consensus template (Fig. 3C and fig. S9–S10). Mammalian RNAPII (B. taurus, Bta) also responded to the consensus sequence (Fig. 3C), but exhibited a somewhat different pattern, involving pausing at the consensus position and even stronger pausing 1 nt downstream (fig. S11). Addition of the cleavage factor TFIIS converted the downstream pause to a strong pause at the consensus position, suggesting the consensus pause leads to backtracking by RNAPII. This result is consistent with other evidence indicating a greater proclivity for eukaryotic RNAPII to backtrack as compared to bacterial RNAP (15).
The average RNAP density across all genes exhibited a sharp peak within the start codon (Fig. 4A), at the juxtaposition of the ribosome-binding sequence (RBS; AGGAGG) and the ATG start codon, which are separated by an average spacing of 10 nt in E. coli (16) and consequently define the ends of a consensus pause sequence (Fig. 4B). Indeed, RBS substitutions abolished the start-codon pause for the lacZ gene in vivo (Fig. S12). Similar to E. coli, we observed frequent pausing throughout the genome of the Gram-positive bacterium Bacillus subtilis, with a consensus pause sequence characterized by −11G/−10G and a −1 pyrimidine, but with A rather than G as the preferred +1 nt (fig. S13A–B). Start-codon pausing also occurred in B. subtilis, just prior to the A of the ATG codon, placing it 2 nt earlier than the E. coli start-codon pause (Fig. 4C). The B. subtilis RBS, which generates the −11G/−10G of the start-codon consensus pause, is, on average, 2 nt further upstream from the ATG codon than in E. coli (Fig. 4D) (16). Thus, the change in the consensus pause sequence in B. subtilis may reflect an evolved alteration that compensates for the 2-nt upstream shift of the RBS relative to the start codon (Fig. 4D).
Fig. 4. Consensus pause sequence is enriched at translation start sites.
(A) Average RNAP density for well-transcribed genes in E. coli. The predicted RNAP density calculated using pause energetics (Fig. 2B) shows a peak at the same position in the start codon. (B) Alignment of sequences surrounding translation start sites in E. coli reveals a sequence that resembles the pause consensus. (C) Average RNAP density for well-transcribed genes in B. subtilis shows a peak 2 nt prior to the center of the start codon. This peak is predicted by the in vivo pause energetics (fig. S13B). (D) Alignment of sequences surrounding translation start sites in B. subtilis shows a 2 nt increase in the average RBS-to-start codon separation compared to E. coli, while the separation between consensus pause features remains unchanged.
In addition to start-codon pausing, RNAP also exhibits a pronounced tendency to pause within the first 100 nt of expressed genes, even though consensus pause sequences are not statistically over-represented within these regions (Fig. 4A, compare RNAP density to predicted density). This 5′-proximal RNAP pausing may be increased until a ribosome can initiate translation and inhibit pausing during coupled transcription-translation (4, 5) (Fig. 4A), which likely explains the promoter-proximal build-up of E. coli RNAP previously observed by ChIP (17).
We have defined a consensus pause sequence that temporarily halts transcription at over 20,000 unique sites in E. coli. Pauses are overrepresented at ATG translation start codons, and this could direct folding of the 5′-UTR into structures that preserve accessibility of the RBS once transcription resumes (fig. S14), consistent with the known ability of paused RNAP to influence nascent RNA folding (1) and the correlation between RBS accessibility and the rate of translation initiation (18, 19). The enhanced pausing downstream of the start codon (in the first 100 nt of genes) may also help preserve the unstructured RBS by limiting synthesis of additional RNA until translation starts. More generally, the conservation of pause sequences across diverse lineages suggest that consensus-sequence pausing may have evolved early in primitive organisms, and was subsequently co-opted to control transcription in a variety of regulatory contexts, accounting for the diverse functions of transcriptional pausing observed today.
Supplementary Material
Acknowledgments
We thank O. Brandman, V. Chu, D. Larson, G. Li, C. McLean, and E. Simmons for critical reading of the manuscript, and J. Lund and E. Chow for assistance with sequencing. This research was supported by the Center for RNA Systems Biology (JSW), the Howard Hughes Medical Institute (JSW), a Ruth L. Kirschstein National Research Service Award (MHL, JMP), and grants from the NIH to CAG, SMB, WJG, and RL. All data are deposited in Gene Expression Omnibus (accession number GSE56720). Jonathan Weissman and Stirling Churchman have submitted a patent on the NET-seq technology."
Footnotes
Supplementary Materials:
Materials and Methods
Supplementary Text
Figs. S1 to S14
Tables S1 to S4
References (20–57)
References
- 1.Pan T, Sosnick T. RNA folding during transcription. Annu. Rev. Biophys. Biomol. Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 2.Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 3.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol. Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 4.Landick R, Carey J, Yanofsky C. Detection of transcription-pausing in vivo in the trp operon leader region. Proc. Natl. Acad. Sci. U. S. A. 1987;84:1507–1511. doi: 10.1073/pnas.84.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem. Soc. Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 7.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 9.Palangat M, Meier TI, Keene RG, Landick R. Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Mol. Cell. 1998;1:1033–1042. doi: 10.1016/s1097-2765(00)80103-3. [DOI] [PubMed] [Google Scholar]
- 10.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenleaf WJ, Block SM. Single-molecule, motion-based DNA sequencing using RNA polymerase. Science. 2006;313:801. doi: 10.1126/science.1130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herbert KM, et al. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan CL, Wang D, Landick R. Multiple interactions stabilize a single paused transcription intermediate in which hairpin to 3’ end spacing distinguishes pause and termination pathways. J. Mol. Biol. 1997;268:54–68. doi: 10.1006/jmbi.1997.0935. [DOI] [PubMed] [Google Scholar]
- 14.Weixlbaumer A, Leon K, Landick R, Darst SA. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–441. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starmer J, Stomp A, Vouk M, Bitzer D. Predicting Shine–Dalgarno Sequence Locations Exposes Genome Annotation Errors. PLoS Comput Biol. 2006;2:e57. doi: 10.1371/journal.pcbi.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mooney RA, et al. Regulator trafficking on bacterial transcription units in vivo. Mol. Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodman DB, Church GM, Kosuri S. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013;342:475–479. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- 19.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.