Abstract
Glutamine synthetase (GS) plays a central role in plant nitrogen metabolism. Plant GS occurs as a number of isoenzymes present in either the cytosol (GS1) or chloroplast/plastid (GS2). There are several reports of improved performance in transgenic plants overexpressing GS1 transgenes driven by the constitutive CaMV35S promoter. Improvement has been attributed to the GS1 transgene product functioning to enhance re-assimilation of NH4+ released by photorespiration or protein degradation. In this paper, alfalfa and tobacco transformants expressing a soybean gene driven by a photosynthetic cell-specific promoter have been compared to transformants with the same transgene driven by the stronger CaMV35S promoter. The two classes of alfalfa and tobacco transformants showed differences in the level of GS1 transcript and GS1 protein accumulation, but the difference in the total GS activity was small. The discrepancy in the transgene expression level and GS activity has been attributed to posttranslational regulation at the level of holoprotein stability. Both classes of transformants exhibited similar level of improvement in soluble protein and in the rates of photosynthesis and photorespiration. The data supports the hypothesis that GS1 made in the mesophyll cells is involved in the re-assimilation of NH4+ released via photorespiration and/or protein degradation.
Keywords: Glutamine synthetase, mesophyll-specific overexpression, nitrogen assimilation, photorespiratory ammonium, transgenic alfalfa
1. Introduction
Alfalfa (Medicago sativa) is one of the most widely grown forage crop. Although closely associated with dairy production, many other livestock industries rely extensively on alfalfa as an important source of nutrition, and as such merits efforts to improve yields [1, 2]. A factor limiting crop yields and plant growth is the availability of the macronutrient nitrogen (N). Plants primarily acquire N from the soil in the form of nitrate (NO3−) which is sequentially reduced to ammonium (NH4+) in the leaves by the enzymes, nitrate reductase and nitrite reductase. Alfalfa and other legumes can also form symbiotic relationships with nitrogen fixing bacteria that reduce atmospheric dinitrogen (N2) to NH4+ in exchange for carbon (C) and energy [3]. While primary metabolism plays an important role in N nutrition, NH4+ can also be released by various secondary metabolic reactions within the plant such as photorespiration, protein turnover and synthesis of phenylpropanoids [3, 4].
The NH4+ taken up or released by the above processes is assimilated by glutamine synthetase (GS; EC 6.3.1.2), which catalyzes the ATP dependent condensation of NH4+ and glutamate (Glu) to yield glutamine (Gln). This is the first step in the incorporation of inorganic N into an organic N compound usable by plants. Glutamine synthetase is, therefore, the key enzyme of N assimilation [5]. Based on electron microscopic observations, plant GS, was originally thought to be an octamer, however, more recently GS has been shown to be decameric [6] with a native molecular weight of 440 kDa. Glutamine synthetase occurs as a number of isoenzyme forms and is present in either the cytosol (GS1) or chloroplast/plastid (GS2). GS1 isoforms are usually encoded by multigene families [7, 8]. Most of the terrestrial plants have single genes encoding GS2 [9]. However, some species may have multiple genes for GS2 [7, 10]. The two isoforms are differentially expressed and regulated throughout the growth and developmental stages in a plant’s life cycle [4, 7, 8]. Both GS isoforms play important roles in N metabolism. The predominant isoform in the leaves is GS2, which functions in the assimilation of NH4+ released from the reduction of NO3− and the re-assimilation of photorespiratory NH4+ [9, 11]. Cytosolic glutamine synthetase is expressed primarily in the roots and nodules. In the roots, GS1 functions to assimilate NH4+ directly taken up from the soil or produced from the reduction of NO3−, while in the nodules, GS1 assimilates NH4+ produced by N2 fixation [12, 13, 14]. In addition, GS1 plays an important role in the remobilization of N during senescence, herbicide treatments, bacterial infections, and water stress [8, 15, 16]. The exclusive location of GS1 in the vasculature of shoots and leaves, also suggests that GS1 may be involved in the synthesis of Gln for transport [8, 17]. While there is ample evidence for transcriptional regulation of GS genes, there is also evidence for regulation at the level of transcript stability and translation initiation [18], posttranslational modification of the protein [19, 20, 21] and at the level of enzyme stability [22].
The coincidence of the positions of the QTLs for yield components and the genes for cytosolic GS in maize, rice, wheat and Arabidopsis genomes suggest that GS1 may represent a key component of nitrogen use efficiency and yield [23, 24, 25, 26]. This has prompted us and others in the field, to make attempts to alter GS1 activity in plants with the purpose of enhancing nitrogen use efficiency and plant performance [27, 28, 29, 30]. Some of these transformants showed an increase in GS1 polypeptide and GS activity and an improvement in plant growth. For example, Lotus japonicus transformants with an alfalfa GS1 gene (MsGS100) driven by the constitutive cauliflower mosaic virus (CaMV35S) promoter exhibited an increase in total protein and chlorophyll content, as well as an increase in total amino acid content in the leaves and stems [27]. Due to the significant increase in the level of Asn/Asp in the stem of the transformants, the authors attributed the increase in plant growth seen in the L. japonicus transformants to improved transport of assimilated NH4+. Other authors, however, have attributed improved performance in GS1 overexpressing plants to the presence of GS1 in the photosynthetic cells, functioning to enhance re-assimilation of NH4+ released during photorespiration and/or senescence [28, 29, 31]. The levels of NH4+ released during photorespiration are very high and re-assimilation of this NH4+ is crucial for nitrogen-use efficiency [32, 33]. Moreover, as the leaves age, the roles of the two GS isoforms switch and GS1 becomes the predominant isoform which functions in the remobilization of N during senescence [8, 15, 34]. With the high levels of NH4+ produced in the leaf during these processes, increased levels of GS1 in the mesophyll cells may improve plant performance by re-assimilating NH4+ that may normally be lost.
The objective of this paper is to determine how alfalfa will respond to mesophyll-specific overexpression in comparison to constitutive overexpression of GS1. We have also included tobacco in this study to determine how alfalfa compares to tobacco with regards to overexpression of GS1. We have used the soybean GS1 (Gmglnβ1) gene as the transgene and since previous studies showed that the 3’UTR of the Gmglnβ1 gene plays a key role in regulating both transcript stability and translation initiation [18], we have used the Gmglnβ1 gene without its 3’UTR as the GS1 transgene. By taking a molecular, biochemical and physiological approach in the analysis of the two classes of transformants, this study casts light on how post-translational regulatory events affect the expression of the GS1 transgene and its eventual repercussions on physiolgoical parameters.
2. Materials and Methods
2.1. Gene constructs
Standard procedures were used for all recombinant DNA manipulations [35]. The alfalfa RUB promoter was isolated from a pGEM-T Easy vector (Promega, Milwaukee, WI) as a 5’ Xba I and 3’ Bam HI fragment [36] and directionally cloned into the pBI101 vector (Clontech BD Biosciences, San Jose, CA; NCBI accession no. U12639) in front of the E. coli GUS coding region (uidA) producing the RUB-uidA-NOS3’ gene construct. Cloning and isolation of the Glycine max (soybean) Gmglnβ1 cDNA (NCBI accession no. AF301590) has been previously reported [37]. Since previous studies showed that the 3’UTR of the Gmglnβ1 gene plays a key role in regulating both transcript stability and translation initiation [18], we used the Gmglnβ1 gene without its 3’UTR in our gene constructs. Removal of a 254 bp fragment containing most of the 3’ UTR from Gmglnβ1 was as previously described [18]. The Gmglnβ1 cDNA without its 3’ UTR was released as a 5’ Cla I and 3’ Sst I fragment (1121 bp) and directionally cloned into the binary vector pBI101, replacing the uidA gene between the alfalfa RUB promoter and the NOS 3’ terminator to produce the RUB-Gmglnβ1-NOS3’ construct.
The RUB-Gmglnβ1-NOS3’ and CaMV35S-Gmglnβ1-NOS3’ gene constructs were used in the transformation of alfalfa (Medicago sativa cv. Regen-SY) and tobacco (Nicotiana tabaccum var. Xanthi nc) as previously described [18]. The gene constructs were mobilized from E. coli (strain XL1-blue) into Agrobacterium tumefaciens receptor strain LBA4404. An Agrobacterium strain with the 35S-uidA-NOS3 was kindly provided by Dr. John Kemp (New Mexico State University).
2.2. Plant material
The RUB-Gmglnβ1 and 35S-Gmglnβ1 transgenic plants were selected on media containing Kanamycin [25 µg/ml]. The insertion of the transgene was confirmed by performing PCR on the genomic DNA using primers specific for the Gmglnβ1 gene and NPTII gene (selectable marker in the TDNA construct). All transformants and control plants have an identical background since all transformations were performed on the same clonal material, except for the presence and position of the transgene in the genome. Independent transgenic lines were selected from several independent primary transformants. Clonal material was regenerated via tissue culture and maintained under greenhouse conditions. The plants were watered alternately with water and 0.5 × N free Hoagland solution and were fed 5 mM ammonium nitrate twice a week.
Plant tissue used for analysis was harvested generally at the same time of the day and used for RNA, protein, and enzyme activity analysis. For analysis of net photosynthetic rates, chlorophyll content and protein content, plants were transferred to fresh soil and grown with no nitrogen for 14 days. Experiments described in this paper were each performed three times and only results from representative experiments are shown here.
2.3. RNA analysis
Total RNA was isolated from leaves by LiCl precipitation, fractionated in 1.35% agarose/formaldehyde gels and blotted onto nitrocellulose. Filters were pre-hybridized overnight followed by hybridization to32P-labeled DNA probes for 20–24h in hybridization buffer (50% formamide, 5× SSC, 5× Denhardt’s solution, 50 mM sodium phosphate (pH 7.0), 0.1 % SDS, 0.1 mg ml−1 denatured salmon sperm DNA). The32P-labeled DNA probes were prepared from plasmid inserts by random primer extension using the Prime-a-Gene system (Promega, Milwaukee, WI). Following hybridization, the filters were washed at 54° C, three times with 2× SSC, 0.5% SDS (w/v), followed by two washes with 0.5× SSC, 0.5% (w/v) SDS for 20 min each and exposed to X-ray film. The hybridization signals were digitized by scanner and quantified using KODAK 1D analysis software (Kodak Scientific Imaging Systems, Rochester, NY).
2.4. Protein extraction and GS enzyme activity assay
All procedures were carried out at 4°C. The different tissues were ground in liquid nitrogen with 15% (w/w) insoluble polyvinylpolypyrolidone and homogenized with two (roots and stems of alfalfa and leaves of tobacco) or five (leaves of alfalfa) volumes of cold extraction buffer: 50 mM Tris–HCl pH 8.0, 20% glycerol (v/v), 5% ethylene glycol (v/v), 1 mM MgCl2, 1 mM DTT, 1 mM EDTA and a protease inhibitors cocktail (Roche Applied Science, Indianapolis, IN). The homogenate was centrifuged for 15 min at 20,000g and desalted in Sephadex G25 columns against desalting buffer [62.5 mM Tris–HCl pH 6.8, 20% glycerol (v/v), 5% ethylene glycol (v/v), 1 mM DTT, 1 mM EDTA, and protease inhibitors]. Protein concentration was measured by the Bradford protein assay (Bio-Rad, Hercules, CA) using bovine serum albumin as the protein standard. The activity of GS was measured spectrophotometrically at 500 ηm by the transferase assay [38]. Transferase units were calculated from a standard curve of γ-glutamyl hydroxamate. Activity values are reported as µmol of γ-glutamyl hydroxamate produced min−1 mg−1 protein at 30°C. The GS activity data are averages of at least three independent experiments.
2.5. Polyacrylamide gel electrophoresis (PAGE)
Two different PAGE systems were utilized: SDS–PAGE using 12% acrylamide (w/v) mini-gels and a non-denaturing (native) PAGE performed in 7.5% acrylamide (w/v) slab gels, in which the pH of the resolving gel was 8.15. The amount of protein loaded per lane was 5 µg for denaturing gels and 25 µg for the non-denatured protein samples. Denatured proteins were fractionated at 100 V for 2 h. For the native gels the proteins were fractionated at 145 V for 16 h at 4°C.
2.6. Immunodetection of GS
For immunoblot analysis, proteins fractionated on acrylamide gels were electroblotted onto nitrocellulose. Glutamine synthetase was detected using an anti-GS antibody raised in rabbits against a recombinant soybean Gmglnβ1 protein expressed in E. coli using a glutathione-S-transferase expression system (GE Healthcare, Piscataway, NJ). The antibodies were made monospecific for GS (GS1 and GS2) by affinity adsorption to purified plant GS protein. Protein bands on gels which were immunoreactive with the GS antibodies were visualized with an alkaline phosphatase-linked secondary antibody using nitroblue tetrazolium and 5-bromo-4-chloro-3-indoyl-phosphate as substrates. The immunoreactive bands were quantified with the KODAK 1D image analysis software (Kodak Scientific Imaging Systems, Rochester, NY). Experiments were repeated at least three times. Only representative experiments are presented.
2.7. Gas Exchange (Net Photosynthesis) measurements
Measurements of net photosynthetic rates (Pnet) on the most recent fully expanded trifoliate leaves of alfalfa and on the second or third leaf of tobacco were performed with an infrared gas analyzer-based photosynthesis system (LI-6400, Li-Cor Inc., Lincoln, Nebraska) using a Li-Cor 6400-05 conifer chamber with an external light source and a Li-Cor 6400-02B LED light source. A photo flux of ~1300 µmols m−2 s−1 (alfalfa) and 1000 µmols m−2 s−1 (tobacco) was maintained. The Pnet measurements were taken at ambient CO2 (flow rate of 400 µmol s−1 and internal CO2 of 400 µmol mol−1) and at low CO2 (flow rate of 40 µmol s−1 and internal CO2 of 40 µmol mol−1) conditions. Leaf areas of alfalfa trifoliates were measured using a Li-Cor LI3000 portable area meter (Li-Cor Inc., Lincoln, Nebraska). Three measurements were done on each of the 4–5 replicates of the controls and the transformants and average values of net photosynthesis per leaf area ± SE for each set of plants are presented. Experiments were repeated at least three times.
2.8. Chlorophyll and protein content
Leaf tissue from alfalfa and tobacco were rapidly weighed (~100 mg) and ground to a powder in liquid nitrogen. Tissue was homogenized in 100% acetone (1 ml) and centrifuged at 10,000 × g. Supernatant was transferred and the insoluble pellets were re-extracted with 100% acetone (1 ml). Acetone extracts were kept at 4° and minimum light exposure was maintained. Chlorophyll a and b content was measured spectrophotometrically (chl a: 664 nm; chl b: 647 nm) and concentrations of total chlorophyll content were calculated using equations derived from predetermined extinction coefficients [39]. Experiments were repeated three times on replicates and average values (mg g−1 FW−1) ± SE are presented.
For protein content determination, 5 leaflets of alfalfa and 5 leaf discs of tobacco, controls and transformants, were freeze dried for two days in a lyophilizer (Virtis, Gardiner, NY) and tissue was measured for dry weights. Total soluble protein was extracted and concentrations calculated as described above in the section on protein extraction and GS activity assay. Experiments were done in replicate and were repeated at least 3 times. Average values ±SE are presented as mg protein g−1 DW−1.
2.9. Histochemical localization of GUS activity
Histochemical localization of GUS activity was performed using 5-bromo-3 indolyl β-DGluronic acid (X-Gluc) as a chromogenic substrate. Hand sections were placed directly into reaction mixture consisting of 1 mM X-Gluc dissolved in 50 mM sodium phosphate buffer (pH 7.0) and allowed to incubate in the dark for 12 h at 37°C. Stained tissue was rinsed with water and incubated in 70% ethanol to remove chlorophyll. Stained tissue was mounted on glass slides and covered with 50% glycerol. Pictures of leaf tissue were taken at 40× magnification using a Zeiss Axiophot Photomicroscope (Carl Zeiss, Germany).
3. Results
3.1. RuBisco promoter is active in the photosynthetic cells while the CaMV35S promoter is active in the mesophyll cells and the vascular bundles of both the leaves and the stem
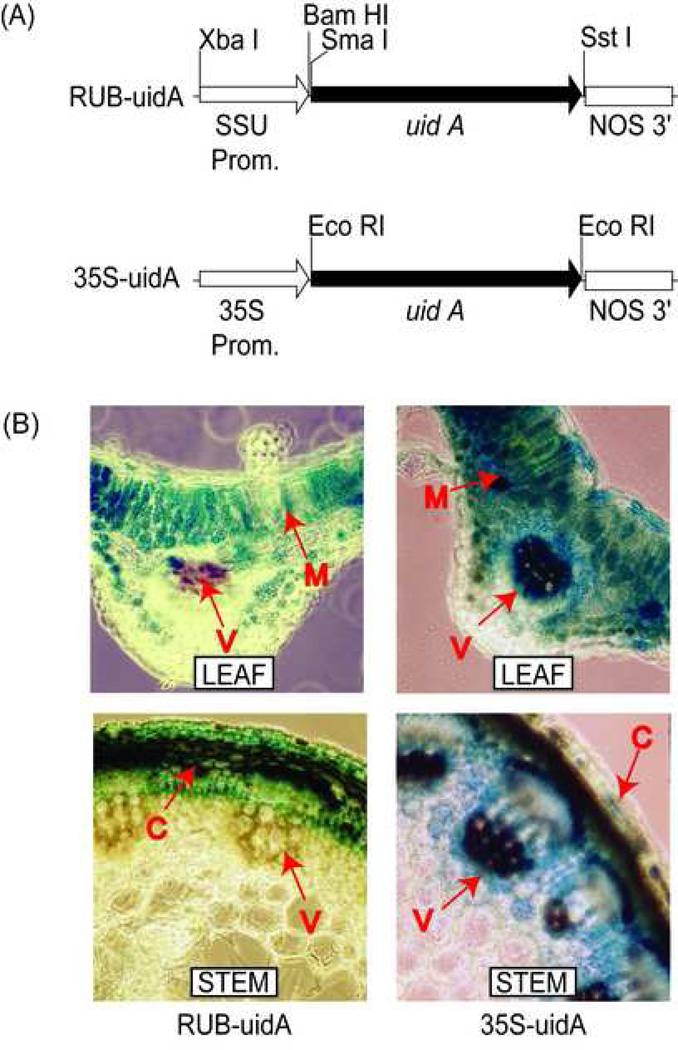
The major objective of this paper is to determine how alfalfa plants will respond to mesophyll-specific overexpression in comparison to constitutive overexpression of GS1. To accomplish overexpression of GS1 in a mesophyll specific manner, we used the RuBisco SSU (RUB) promoter from alfalfa [36]. This promoter, along with the cauliflower mosaic virus 35S (CaMV35S) constitutive promoter, was placed in front of the GUS (uidA) coding region (Fig. 1A). The two constructs were introduced into alfalfa via an Agrobacterium tumefaciens mediated plant transformation. Hand sections of leaf and stem tissue from several RUB- and CaMV35S-GUS transformants were histochemically stained for GUS activity to test the site of promoter function in alfalfa. Representative pictures of the stained leaf and stem sections are shown in Fig. 1B. The RUB-GUS alfalfa transformants showed GUS activity primarily in the palisade layers and at lower levels in the spongy mesophyll. Leaf tissue of CaMV35S-GUS alfalfa transformants showed GUS staining in the palisade layers and spongy mesophyll as well as in the vasculature. Both sets of transformants showed no GUS activity in the epidermis or parenchyma cells below the vasculature. The leaves from the CaMV35S-GUS transformants showed much higher level of GUS staining in the mesophyll cells compared to the RUB-GUS transformants. The stems of the RUB-GUS transformants showed GUS staining in the outer cortical cells, while the CaMV35S-GUS transformants showed heavy staining in both the cortical cells and the vasculature. Overall, the results showed stronger expression of the GUS reporter gene in different tissues of the CaMV35S-GUS transformants, including the leaf mesophyll, stem cortical cells and the vascular bundles. The alfalfa RUB promoter, however, showed weaker strength compared to the CaMV35S promoter and conferred only mesophyllspecific expression in the leaves, and cortex-specific expression in the stem. Thus driving GS1 gene with the RUB promoter would only target GS1 overexpression in the photosynthetic cells.
Fig. 1.
Localization of RUB- and CaMV35S- promoter function in leaves and stem of alfalfa. (A) Maps of gene constructs used to produce the RUB-uidA-NOS and CaMV35S-uidA-NOS alfalfa transformants. The pertinent restriction enzyme sites used for cloning purposes is shown. (B) Analysis of GUS expression in leaf and stem sections of RUB-uidA-NOS and 35S-uidA-NOS alfalfa transformants. Location of GUS activity is indicated by blue precipitate. V, vasculature; M, mesophyll cells; C, cortical cells.
3.2. Alfalfa transformants with the two different GS1 gene constructs: CaMV35S-Gmglnβ1 and RUB-Gmglnβ1 were identified for further analysis
Once the site of promoter function was confirmed, GS1 gene constructs were produced by replacing the GUS coding region from the constructs by a cDNA encoding for a soybean GS1 (Gmglnβ1) cDNA. Since our initial studies have shown that the 3’UTR of the Gmglnβ1 gene plays a key role in regulating transcript stability and translation initiation, we used the gene without its 3’UTR in our gene constructs [18]. The gene devoid of sequences determining posttranscriptional regulation was engineered behind the RUB promoter and the CaMV35S promoter (Fig. 2A). These constructs were introduced into alfalfa via A. tumefaciens mediated plant transformation. Putative RUB- and CaMV35S-Gmglnβ1 alfalfa transformants were selected by their ability to grow and root on selection media containing Kanamycin, and further identified by PCR using primer sets specific for the NPTII gene. All the putative transformants (14) tested positive for the NPTII gene and after initial examination of the transformants by western blot analysis for GS (data not shown), three independent transformants representing each construct were randomly selected and propagated clonally for further analysis.
Fig. 2.
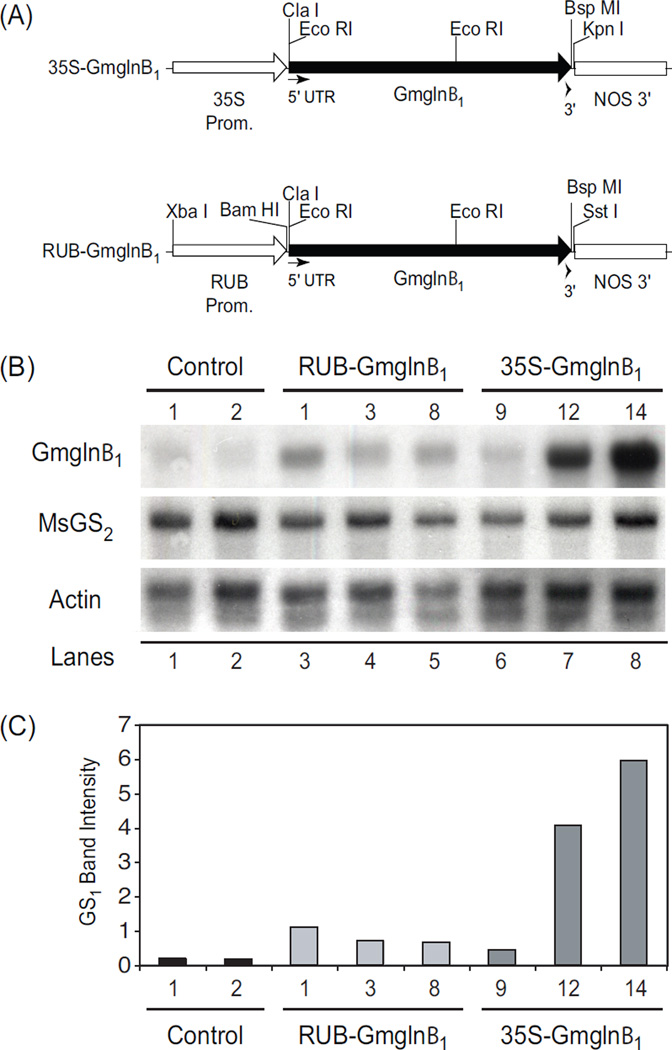
Analysis of GS transcript in the leaves of alfalfa transformants.
(A) Gene constructs used to produce the RUB-Gmglnβ1 and CaMV35S-Gmglnβ1 alfalfa transformants. Positions of relevant restriction enzyme sites is indicated. (B) Northern blot analysis of leaf RNA (10 µg) extracted from green house grown alfalfa control plants, RUBGmglnβ1 and CaMV35S-Gmglnβ1 alfalfa transformants. The blot was probed with 32P-labeled fragments of the soybean GS1 transgene (Gmglnβ1), alfalfa GS2 (MsGS2), and Actin genes. (C) Autoradiographs were quantified using a KODAK 1D image analysis software. Band intensities of Gmglnβ1 and MsGS2 standardized to Actin band intensity is plotted.
3.3. The CaMV35S-Gmglnβ1 transformants showed much higher level of transgene transcript accumulation in the leaves compared to the levels in the leaves of the RUB-Gmglnβ1 transformants
To determine how the two promoters (CaMV35S and RUB) affect the accumulation of the transgene transcript, total leaf RNA isolated from the control plants and the two sets of transformants was subjected to northern blot analysis using the Gmglnβ1 coding region, alfalfa GS2 gene and the actin gene as probes. The actin probe was used to determine RNA loads. The hybridization signals obtained with the different probes were digitized using a scanner and quantified using a Kodak 1D image analysis software. Several clones of the two classes of independent transformants were analyzed and only a representative blot is shown here (Fig. 2B). The values of hybridization signals corresponding to Gmglnβ1 and MsGS2 were standardized against the hybridization signals of actin (Fig. 2C).
The Gmglnβ1 coding region probe showed low level of hybridization to RNA from control plants probably representing the hybridization signal to the endogenous GS1 transcript since the coding region of the different GS1 genes share very high sequence similarity. The hybridization signal with the Gmglnβ1 gene was considerably higher for the transformants compared to the control plants, the signal being much higher in two of the CaMV35S-Gmglnβ1 transformants (#12 and #14) compared to the RUB-Gmglnβ1 transformants (Fig. 2B, C). The CaMV35S-Gmglnβ1 transformant #9 showed hybridization signal lower than seen with all the other transformants and this probably can be ascribed to position effect. The blots were also hybridized with cDNA of alfalfa GS2 to determine if expression of the GS1 transgene had any effect on the expression of the endogenous GS2 gene. The level of GS2 transcript accumulation did not show any difference between the two sets of transformants and the control plants (Fig. 2). Similarly, the expression of the GS1 transgene did not affect the steady state level of the endogenous GS1 gene transcript (data not shown). The results suggest that the steady level of the Gmglnβ1 transcript was 3-fold higher in the CaMV35S-Gmglnβ1 transformants compared to the RUB-Gmglnβ1 transformants, probably indicative of both promoter strength and the ubiquitous expression pattern of the CaMV35S promoter compared to the mesophyll-specific expression of the RUB promoter (Fig. 1B).
3.4. The CaMV35S-Gmglnβ1 transformants showed many fold higher level accumulation of the transgene protein in the leaves compared to the level in the leaves of the RUB-Gmglnβ1 transformants
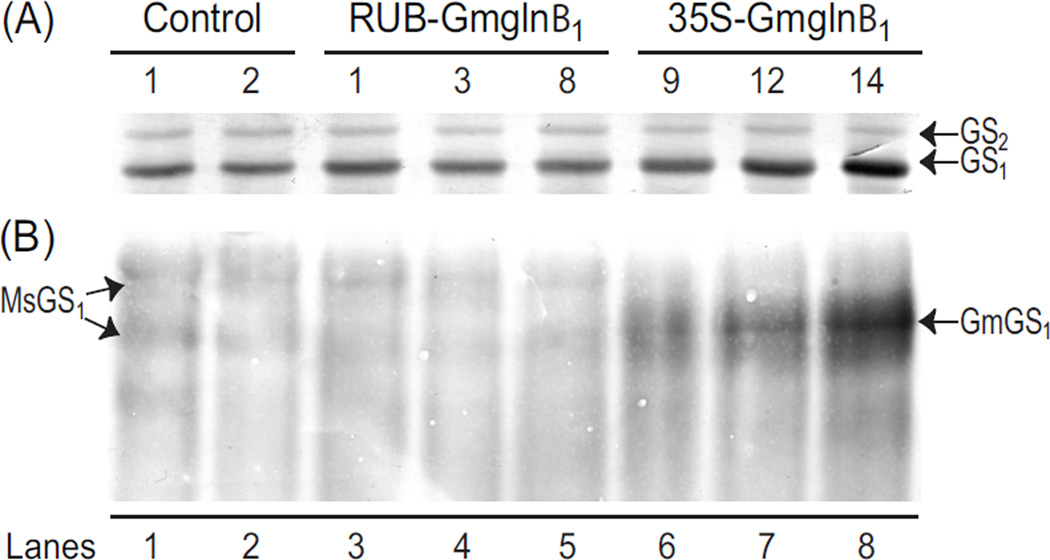
To determine whether RUB- and CaMV35S-Gmglnβ1 alfalfa transformants were accumulating GS1 polypeptide corresponding to the transgene transcript level, the total soluble protein from the leaves of the same alfalfa controls and promoter-Gmglnβ1 transformants used for RNA analysis, was subjected to SDS-PAGE followed by western analysis using antibodies raised against GS1 polypeptide. As seen in a representative blot in Fig. 3A, two immunoreactive bands were observed in the protein extracts from the leaves of control plants and the transformants when subjected to SDS-PAGE with the slower migrating band corresponding to GS2 and the faster migrating band corresponding to GS1. While no differences were observed in the level of GS2 polypeptide accumulation between the controls and the transformants, a substantial increase in the accumulation of GS1 polypeptide was seen in both sets of transformants. As with Gmglnβ1 transcript accumulation, except for the transformant #9, the CaMV35S-Gmglnβ1 transformants accumulated higher levels of GS1 polypeptides than the RUB-Gmglnβ1 transformants. The increase in the GS1 polypeptide level in the leaves of the transformants compared to control plants can be attributed to the transgene product.
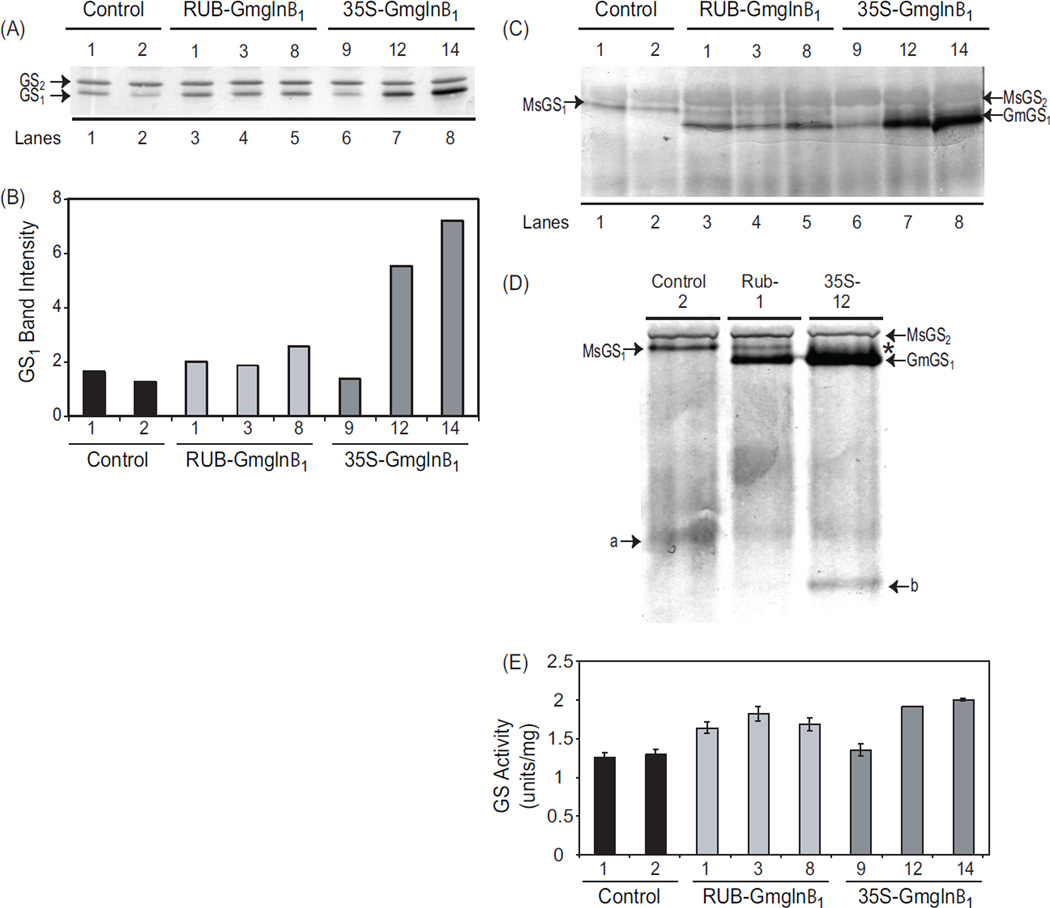
Fig. 3.
Analysis of GS polypeptide, holoenzyme and GS activity in the leaves of RUB-Gmglnβ1 and CaMV35S-Gmglnβ1 alfalfa transformants. (A) Five µg of protein from leaf extracts of alfalfa control plants and independent transformants with the RUB-Gmglnβ1 and CaMV35SGmglnβ1 gene constructs was subjected to SDS PAGE followed by western blot analysis using anti-GS antibodies. Immunoreactive bands corresponding to GS1 (39–40 kd) and GS2 (42–43 kd) polypeptides are shown. (B) Band intensities of the GS1 band from panel A is plotted. (C) Twenty five µg of of leaf extracts from the same plants as in panel A were subjected to native gel electrophoresis followed by western blot analysis using anti-GS antibodies. Immunoreactive bands corresponding to the endogenous GS2 (MsGS2) and GS1 (MsGS1) and the transgene product (GmGS1) are designated. (D) Close-up of a immunoreactive native gel profile of leaf protein from control, RUB-Gmglnβ1 (#1) and CaMV35S-Gmglnβ1 (#12) transformants showing the bands corresponding to the endogenous MsGS2 and MsGS1, and the transgene product GmGS1. Asterisk represents the broad immunoreactive band probably representing a mixture of heteromers made of the endogenous GS1 and the transgene product. Bands a and b represent the turnover products of the MsGS and GmGS1 holoproteins respectively. (E) GS activity was measured in the leaf protein samples used for western analysis in panel A. Average values ±SE of three experiments were plotted as µmol γ-glutamyl hydroxamate produced per minute per mg of protein at 30° C.
3.5. The transgene product assembles into novel holoprotein(s) in the leaves of both the CaMV35S-Gmglnβ1 and the RUB-Gmglnβ1 transformants
While SDS-PAGE followed by western analysis showed higher level of GS1 polypeptide accumulation in the leaf tissues of the transformants compared to the control plants (Figure 3A, B), it did not distinguish between the endogenous GS1 polypeptide and the transgene product. To differentiate between the endogenous GS1 polypeptide and the transgene polypeptide, the same protein extract used for SDS-PAGE was subjected to native gel electrophoresis followed by western blot analysis (Fig. 3C). Non-denaturing (native) gel electrophoresis, besides determining if the transgene GS1 polypeptides assemble into a holoprotein, can also differentiate between the holoproteins made of the endogenous GS polypeptides and those made of the transgene product based on their migration pattern on native gels. Moreover, heteromeric holoprotein formed of endogenous and transgene polypeptides can also be identified.
As seen in Fig. 3C, the native gel immunoblot showed two immunoreactive bands in the controls corresponding to the endogenous GS enzymes: GS1 (MsGS1) and GS2 (MsGS2). GS2 corresponds with the broad, more lightly immunostained and slower migrating band and GS1 corresponds with the more distinct and faster migrating band. The alfalfa GS2 holoprotein shows lower affinity for the GS1 antibody and thus the western blot does not reflect the actual abundance of GS2 and GS1 proteins. A novel immunoreactive band (GmGS1) migrating faster than the endogenous GS1 holoprotein was seen only in the transformants, the level being higher in the CaMV35S-Gmglnβ1 transformants except transformant #9. This novel immunoreactive band, most likely represents a holoprotein which contains the Gmglnβ1 polypeptide. While the RUB-Gmglnβ1 transformants still maintained the distinct endogenous GS1 holoprotein band (MsGS1), though at lower level compared to control, the CaMV35S-Gmglnβ1 transformants did not. This is clearly seen in sample #9 (lane 6, Fig. 3C) and in the close up picture of the immunoblot of the three samples (Fig. 3D). In the CaMV35S-Gmglnβ1 transformants, the novel immunoreactive band appears more diffused represented with an asterick in Fig. 3D. This broad banding pattern seen in the CaMV35S-Gmglnβ1 transformants could be attributed to the higher level accumulation of the holoproteins corresponding to the transgene product. The absence of the band corresponding to the endogenous MsGS1 band in the 35S transformants (Fig. 3C and D), suggests the possible mixing of the endogenous GS1 and Gmglnβ1 gene product. The native gel also showed a faster migrating immunoreactive band (Fig. 3D, a) in all the lanes probably representing the turnover product of the endogenous GS holoprotein and an additional band (Fig. 3D, b) in the CaMV35S-Gmglnβ1 transformants probably representing the turnover product of the holoprotein made of the transgene product. Since the CaMV35S-promoter is active in both the mesophyll cells and the vasculature, while the RUB promoter functions only in the mesophyll cells, the mixing of endogenous and the transgene polypeptide in the CaMV35S-Gmglnβ1 transformants and not in the RUB-Gmglnβ1 transformants, points to the normal location of GS1 in the vasculature of alfalfa leaves. The results suggest that the CaMV35S-Gmglnβ1 transformants accumulate the major amount of transgene GS1 holoprotein in the mesophyll cells as in the case of the RUB-Gmglnβ1 transformants, but also accumulate some amount of GS1 heteromers containing the transgene product in the vasculature.
3.6. The two classes of transformants show higher level of GS activity in the leaves compared to control but not at a level corresponding to the increase in polypeptide or transcript level
The stability of GS holoprotein has been shown to be regulated by the occupancy of the substrate at its active site [22] and as such, just the assembly of a holoprotein is not always reflective of its enzymatic activity. GS activity (the total amount resulting from both the GS2 and GS1 activities) was measured on the same extracts that were used for western analysis and as seen in Fig. 3E, the transformants showed higher GS activity compared to the controls but the increase did not show a direct correspondence with the increase in the GS1 protein levels. The CaMV35S-Gmglnβ1 transformants, while showing a ~ 3-fold higher level of GS1 protein accumulation showed only a slight increase in activity over the RUB-Gmglnβ1 transformants. The CaMV35S-Gmglnβ1 transformant #9, showed a lower level of activity compared to transformants #12 and #14, in keeping with the low level of expression of the transgene, however, the level of activity is not directly proportional to the amount of GS1 protein made. The data suggest that there is an increase in GS1 activity in the mesophyll cells in the leaves of the transformants when compared to control plants and the CaMV35S-Gmglnβ1 (transformants #12 and #14) have higher activity compared to the RUB-Gmglnβ1 transformants. The CaMV35S-Gmglnβ1 transformants, in addition, probably also show increased activity in the vasculature of the leaves.
3.7. Only the CaMV35S-Gmglnβ1 transformants showed increased GS activity in the stem and roots
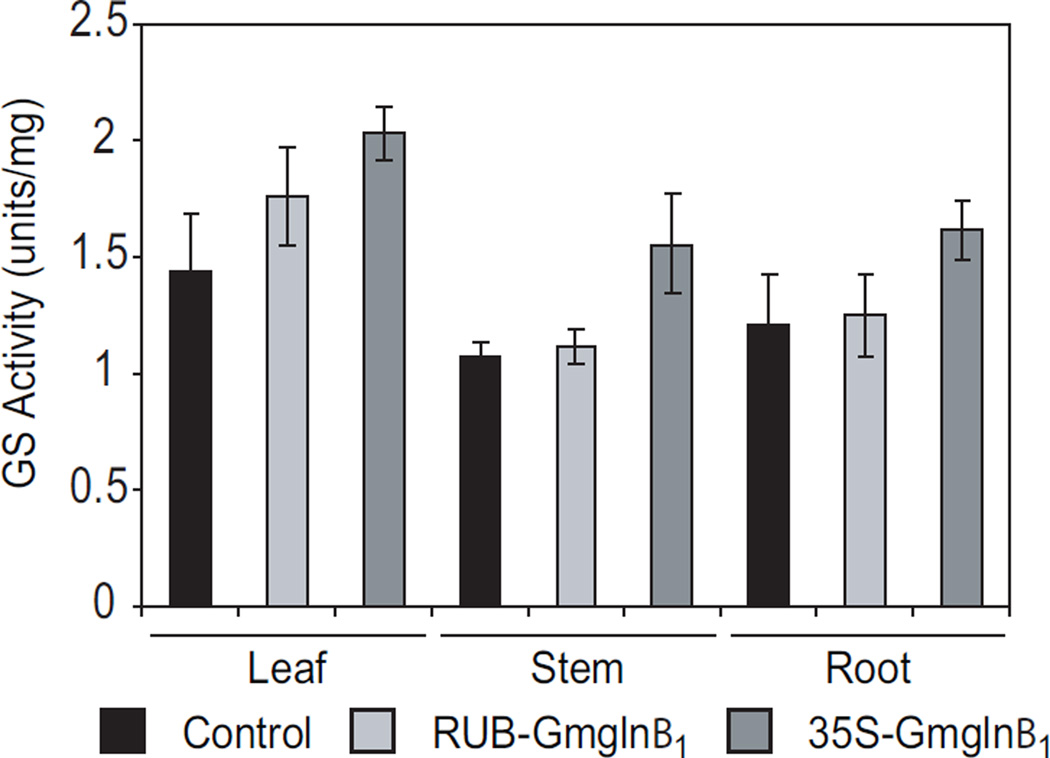
The CaMV35S promoter is known to be expressed in a constitutive manner and the RUB promoter in a photosynthetic cell-specific manner. Localization of GUS activity in the stems of CaMV35S-GUS and RUB-GUS transformants, showed intense staining in the cortex in both classes of transformants but in addition, the CaMV35S transformants also showed strong staining in the vascular bundles (Fig. 1B). To determine if promoter function in the stem and roots translates into GS activity, the leaves, stem and roots of control plants and the two classes of transformants were assayed for GS activity. As seen in Fig. 4, while the leaves of the RUBGmglnβ1 plants showed about a 30% increase in GS activity, the GS activity in the stem and roots was the same as in control plants. The CaMV35S-Gmglnβ1 transformants, on the other hand, showed higher level of GS activity in all the organs, leaves (~44%), stem (~40%) and the roots (~33%) compared to control plants. Because both the RUB and CaMV35S promoters are functional in the cortical cells of the stem (Fig. 1B), the absence of any increase in GS activity in the stem of the RUB-Gmglnβ1 transformants but an increase in the CaMV35S-Gmglnβ1 transformants, would suggest that GS1 protein made in the stem cortical cells is probably not stable but the GS1 made in the vasculature of the stem is stable. This was confirmed by subjecting stem protein from control plants and the two classes of transformants to native gel electrophoresis followed by western blot analysis.
Fig. 4.
GS activity measurements in the leaf, stem and root samples of control, RUB-Gmglnβ1 and CaMV35S-Gmglnβ1 alfalfa transformants. Protein samples isolated from the leaves, stem and roots of control plants and several clonally propagated RUB-Gmglnβ1 (#1 and #3) and CAMV35S-Gmglnβ1 (#12 and #14), were assayed for GS activity by transferase reaction. Values represent the average ±SE.
Stem protein extracts from the same plants as those used for analysis of leaf proteins, was subjected to SDS-PAGE and native gel electrophoresis followed by western blot analysis using the GS antibodies. As seen in Fig. 5, GS1 is the major isoform in the stem and GS2 is the minor isoform. While the GS2 isoform was maintained at the same level in the stem of both the controls and transformants, only the stem proteins from the CaMV35S-Gmglnβ1 transformants showed a considerable increase in the level of the GS1 protein compared to the controls. The immunoreactive profile of the stem proteins subjected to native gel electrophoresis showed the same profile for both the controls and the RUB-Gmglnβ1 transformants while the CaMV35S-Gmglnβ1 transformants showed a broad immunoreactive band, probably representing multiple holoproteins made up of the endogenous and transgene GS1 polypeptides. Since the RUB promoter is functional in the cortical cells of the stem, the absence of a novel GS holoprotein in the stem of RUB-Gmglnβ1 transformants would suggest the GS1 transgene product does not accumulate as a holoprotein in the cortical cells of the stem. The presence of the novel GS holoprotein in the stem of the CaMV35S-Gmglnβ1 transformants suggests that the transgene product and the endogenous GS1 polypeptides assemble into holoproteins in the vasculature of the stem where they are stable. Furthermore, we also conclude that while the CaMV35S-Gmglnβ1 transformants have increased GS1 activity in the leaves, stem and roots, only the leaves of the RUB-Gmglnβ1 transformants have increased GS1 activity (Fig. 4).
Fig. 5.
Analysis of GS polypeptides and holoproteins in the stem of RUB-Gmglnβ1 and CaMV35S-Gmglnβ1 alfalfa transformants. Five µg (denatured) or 25 µg (non-denatured) protein from stem extracts of alfalfa control plants and independent transformants with the RUBGmglnβ1 and CaMV35S-Gmglnβ1 gene constructs (same as used in Fig. 3) were subjected to either SDS PAGE or native gel electrophoresis followed by western blot analysis using anti-GS antibodies. Immunoreactive bands corresponding to GS1 (39–40 kd) and GS2 (42–43 kd) polypeptides, and native GS bands corresponding to the endogenous GS1 (MsGS1) and the transgene product (Gmglnβ1) are designated.
3.8. CaMV35S- and RUB- Gmglnβ1 tobacco transformants showed similar trends in the expression pattern of Gmglnβ1
Since there are several reports dealing with the constitutive overexpression of GS1 in tobacco, we also undertook to compare the physiological outcome of mesophyll-specific overexpression of GS1 to constitutive overexpression of GS1 in tobacco. Thus tobacco transformants overexpressing Gmglnβ1 in a mesophyll-specific and constitutive manner were produced, using the same constructs as in Fig. 2A. The site of both CaMV35S and the RUB promoter function in tobacco was confirmed by localizing GUS activity in tobacco transformants containing the same constructs as in Fig. 1A (data not shown).
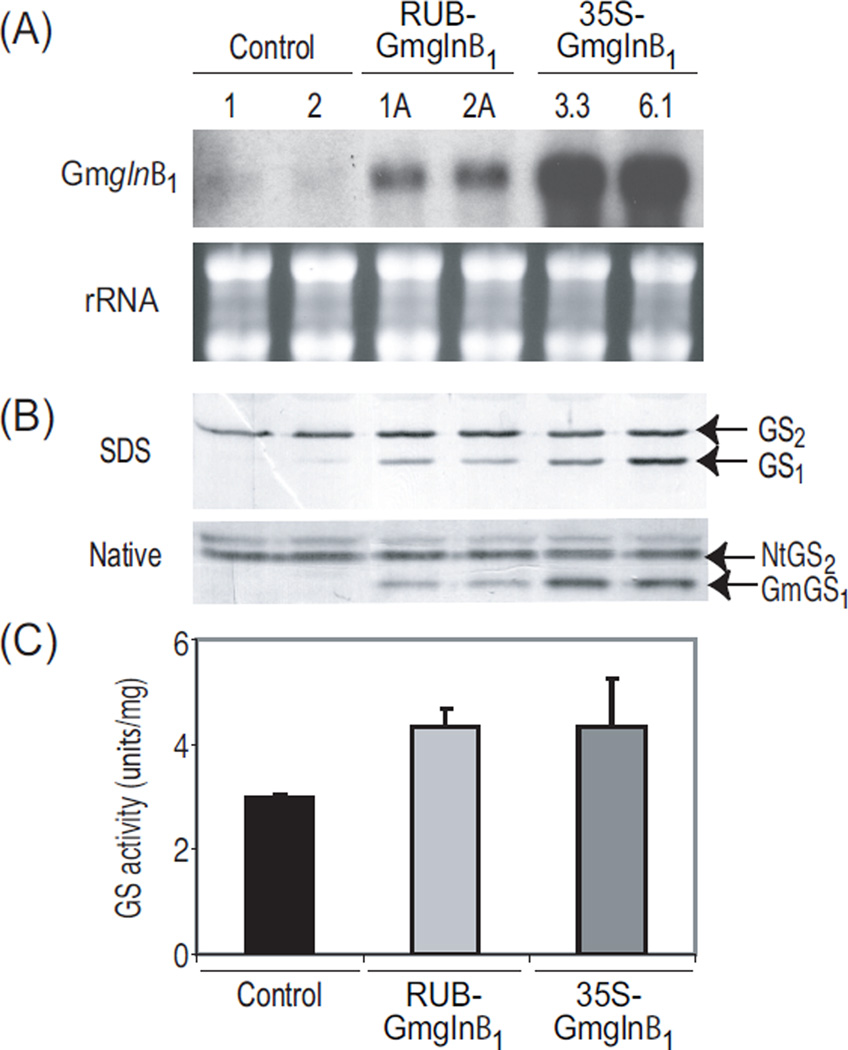
Northern blot analysis, using the Gmglnβ1 gene as a probe, was performed on total RNA extracted from the leaves of tobacco controls and promoter-Gmglnβ1 transformants (Fig. 6A). Transcript accumulation of Gmglnβ1 in both sets of tobacco transformants were analogous to the trend seen in the alfalfa transformants, again with the CaMV35S-Gmglnβ1 tobacco transformants accumulating higher level of Gmglnβ1 transcript compared to the RUB-Gmglnβ1 transformants. Slight cross hybridization of the endogenous GS1 RNA to the Gmglnβ1 probe was seen in the controls.
Fig. 6.
Analysis of transcript and protein corresponding to the Gmglnβ1 transgene in tobacco transformants. Tobacco RUB-Gmglnβ1 and CaMV35S-Gmglnβ1 transformants were produced using the same constructs shown in Fig. 2A. (A) Northern blot analysis of leaf RNA (10 µg) extracted from green house grown tobacco control plants and the two classes of Gmglnβ1 transformants. Transcript was detected using a probe for the soybean GS1 transgene (Gmglnβ1) and EtBr stained ribosomal rRNA is shown to confirm equal loading. (B) Total soluble protein extracted from the same leaf tissue was fractionated by SDS- and native-PAGE (5 µg and 25 µg, respectively) followed by western blot analysis using anti-GS antibodies. Immunoreactive bands corresponding to GS1 (39–40 kD) and GS2 (42–43 kD) polypeptides and the native GS bands corresponding to the endogenous GS2 (NtGS2) and the transgene product (GmGS1) are designated. (C) Transferase assay was performed on the same protein extracts as in panel B. Average values for GS activity were plotted as µmol γ-glutamyl hydroxamate produced per minute per mg of protein at 30° C. Experiments were repeated at least three times.
Total soluble protein from the leaves of the same tobacco controls and promoter-Gmglnβ1 transformants used in the analysis of Gmglnβ1 transcript accumulation was extracted and analyzed for GS activity and GS proteins. Leaf protein extract was subjected to SDS-PAGE and native gel electrophoresis followed by western blot analysis using antibodies specific to GS. The level of GS1 polypeptide accumulation in the CaMV35S- and RUB-Gmglnβ1 tobacco transformants was substantially higher than the levels of GS1 polypeptide in the control plants (Fig. 6B). In contrast to what is seen in alfalfa controls, no GS1 polypeptide was detectable in the tobacco controls. There was no difference between tobacco controls and the Gmglnβ1 transformants in the level of GS2 protein accumulation.
The pattern of accumulation of GS holoproteins followed the same pattern as the GS polypeptides (Fig. 6B). The highest levels of GS1 holoenzyme accumulation was in the CaMV35S-Gmglnβ1 tobacco transformants. The leaves of tobacco controls did not show a discernible immunoreactive band that could be ascribed to an endogenous GS1 holoprotein. The immunoreactive band corresponding to the Gmglnβ1 transgene product was distinct and defined in both classes of transformants. The Gmglnβ1 tobacco transformants did not show a diffused broad band corresponding to heteromers formed between the transgene product and the endogenous GS1 polypeptide as seen in the alfalfa CaMV35S-Gmglnβ1 transformants. GS2 also appeared as a more defined and distinct band. The nature of the immunoreactive band migrating slower than the major GS2 oligomer is not known. We rule out the possibility that it represents an oligomer of the unprocessed GS2 polypeptide since an immunoreactive band corresponding to the GS2 precursor was not seen on SDS-PAGE. The band could represent a decameric oligomer of GS2 [6]. The level of GS2 holoenzyme accumulation was not affected in the two sets of transformants.
Leaf protein extracts were assayed for GS activity and the average values ±SE are shown in Fig. 6C. Both sets of transformants showed an increase in GS activity when compared to controls. While CaMV35S-Gmglnβ1 tobacco transformants had significantly higher level of Gmglnβ1 transcript, GS1 polypeptide and GmGS1 oligomers in the leaves than the RUB-Gmglnβ1 tobacco transformants, the level of GS activity was the same as in the leaves of the RUBGmglnβ1 transformants. This was also seen in the alfalfa transformants and may be attributed to regulation at the level of enzyme stability or metabolite availability.
3.9. Physiological analysis of the two sets of alfalfa and tobacco transformants
The alfalfa and tobacco transformed with the RUB- and CaMV35S-Gmglnβ1 constructs were analyzed for chlorophyll content, net photosynthetic rates and protein content to assess the effect of increased GS1 on plant performance. Clones of independent alfalfa transformants (RUB-Gmglnβ1 #1 and #3; CaMV35S-Gmglnβ1 #12 and #14) produced via tissue culture were transferred to soil and allowed to grow under greenhouse conditions. In the case of tobacco, independent transformants were selfed and seeds were used to produce an F1 generation for both classes of transformants. The F1 seeds were grown in soil under greenhouse conditions (7–8 weeks) for physiological studies. Net photosynthetic rates were measured under ambient and low CO2 conditions, followed by harvesting of leaf tissue that was used for measuring chlorophyll and protein content in both alfalfa and tobacco plants.
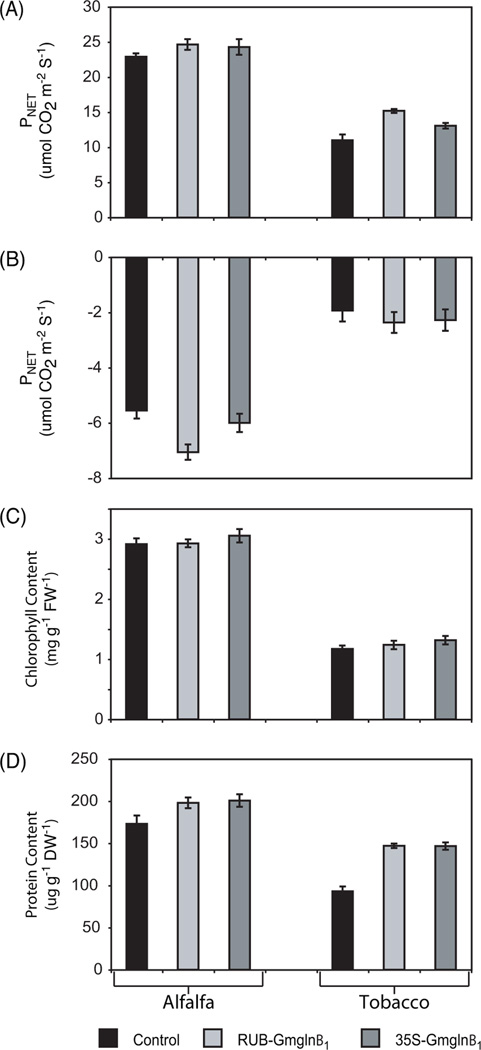
Under ambient CO2 conditions (400 µmol mol−1 CO2), both RUB-Gmglnβ1 and CaMV35S-Gmglnβ1 alfalfa transformants showed an increase in net photosynthetic rates (8% and 6%, respectively) when compared to controls (Fig. 7A). The tobacco RUB- and CaMV 35SGmglnβ1 transformants also showed a larger increase in net photosynthetic rates (38% and 19%, respectively) compared to control plants. To determine the photorespiratory capacity of the transformants, we measured net photosynthesis under low CO2 levels (40 µmol mol−1 CO2), which is conducive to photorespiratory conditions. Under these conditions, the RUB-Gmglnβ1 alfalfa and tobacco transformants showed the lowest levels of net photosynthesis when compared to controls (approximately a 28% and 23% drop, respectively) while CaMV35S-Gmglnβ1 alfalfa and tobacco transformants showed only a 8% and 18% drop in net photosynthetic rates respectively (Fig. 7B). A drop in net photosynthesis is attributable to the CO2 released during photorespiration. Under ambient CO2 conditions, the alfalfa plants in general showed higher rates of net photosynthesis compared to tobacco and similarly under low CO2, the alfalfa plants showed a larger drop in net photosynthesis compared to tobacco.
Fig. 7.
Net photosynthesis, total protein content, and chlorophyll content measurements in the leaves of two classes of alfalfa and tobacco transformants. Net photosynthesis measurements (µmol CO2 m−2 s−1) taken under (A) ambient CO2 (Ci = 400 µmol mol−1) and (B) low CO2 (Ci = 40 µmol mol−1) conditions using the Li-Cor LI 6400. (C) Total chlorophyll content (mg g−1 FW−1) and (D) total protein content (mg g−1 DW−1) was measured spectrophometrically as described in materials and methods. Average values ± SE are presented.
As seen in Fig. 7C, the chlorophyll content showed no dramatic increase in the alfalfa RUB- and CaMV35S-Gmglnβ1 transformants (0.45 and 5%, respectively), while the tobacco transformants showed a 6% (RUB-Gmglnβ1) and 12% (CaMV35S-Gmglnβ1) increase over control. Total soluble protein concentrations, extracted from lyophilized leaf tissue of RUB- and CaMV35S-Gmglnβ1 alfalfa and tobacco transformants, were measured and plotted as mg g−1 DW−1 (Fig. 7D). The RUB- and CaMV35S-Gmglnβ1 alfalfa transformants showed approximately a 14% and 16% increase, respectively, in soluble protein concentrations when compared to controls. Both sets of tobacco transformants showed a considerably higher increase in soluble protein (57%) when compared to controls. The alfalfa plants showed higher level of protein and chlorophyll content per unit tissue dry weight compared to tobacco.
While no sytematic analysis of plant performance was done, in general, both sets of transformants appeared bigger only in the young stages. The 35S-Gmglnβ1 alfalfa transformants showed some signs of premature senescence.
4. Discussion
Genetic approaches have identified the co-localization of QTLs for GS1 and QTLs for yield parameters in several plant species [23, 24, 25, 26], and since GS plays such a central role in N assimilation, it is logical to postulate that enhanced GS activity would lead to improved plant performance. However, the results obtained with the overexpression of GS1 genes in different plants has not been consistent [27, 28, 29, 37, 40, 42, 41, 42, 43, 44]. This inconsistency can be attributed to the posttranscriptional regulatory attributes of the GS1 transgene [18], the source of N and the plant species. In cases where constitutive overexpression of GS1 has been associated with improvement in plant performance, it has been suggested that overexpression of GS1 in the leaves may provide an additional or alternative route for re-assimilation of NH4+ released from photorespiration [27, 28, 29]. However, constitutive overexpression of GS1 does not unambiguously prove that the improved performance of the plant is due to the increased reassimilation of photorespiratory NH4+ [27, 28, 29], since increased GS1 activity in other organs could affect other functions that have a role in promoting plant performance like improvement in N transport in the vasculature. In this paper, we have used the RUB promoter to drive the GS1 gene to ensure that the transgene is expressed only in the cells where photorespiratory NH4+ is produced. Thus any response in plant performance to the photosynthetic cell-specific expression of the GS1 transgene could be ascribed to improved re-assimilation of photorespiratory NH4+. We, however, cannot rule out the possibility of protein degradation as a source of NH4+ for assimilation by the transgene product in the mesophyll cells. The CaMV35S-Gmglnβ1 transformants are included in this study to compare the effects of constitutive expression of Gmglnβ1 to the photosynthetic cell-specific exression of Gmglnβ1 We have also compared the performance of the alfalfa transformants with tobacco transformants expressing GS1 in a mesophyll-specific manner, since there are several reports of improved performance in tobacco resulting from the constitutive overexpression of GS1 [28, 29, 42].
Comparison of the site of GUS localization in the two classes of transformants: 35S-uidA and RUB-uidA, clearly showed that RUB-promoter activity is specifically restricted to the palisade cells and some spongy mesophyll in the leaves, in contrast to the CaMV35S promoter, whose activity extends to both the mesophyll and the vasculature (Fig. 1B). Moreover, based on the intensity of GUS activity, the CaMV35S promoter appeared to have more activity in the leaf mesophyll cells compared to the RUB promoter. The RUB promoter showed transcriptional activity in the cortical cells of the stem while the CaMV35S promoter, besides showing activity in the cortical cells, also showed intense activity in the vasculature of the stem. In earlier studies, we had shown that CaMV35S promoter is also functional only in the vasculature of the roots [37]. Since GS1 is known to be regulated at steps beyond transcription [18, 22], localization of promoter function and strength is not always indicative of the site and level of accumulation of the GS1 protein. In our earlier studies, we had determined that the 3’UTR of the Gmglnβ1 gene has a role in posttranscriptional regulation [18], and as such, we have in this study, eliminated any constraints in expression at the level of transcript turnover or translation initiation by removing the 3’UTR from the Gmglnβ1 gene [18]. However, the expression of the Gmglnβ1 could still be regulated by posttranslational modification and/or at the level of holoprotein assembly and/or protein turnover, which is partly regulated by the availability of the substrate [22].
Native gel electrophoresis of proteins extracted from the leaves of control alfalfa plants followed by western analysis with GS antibodies, showed the presence of immunoreactive bands representing the endogenous GS2 and GS1 holoproteins. Besides the two endogenous immunoreactive protein bands on native gels, the RUB-Gmglnβ1 transformants showed an additional faster migrating immunoreactive band. Since the band representing the endogenous GS1 is maintained in the RUB-Gmglnβ1 transformants, we postulate that this novel immunoreactive band in these transformants represents the holoprotein made exclusively of the transgene product. The band corresponding to the endogenous GS1 is not seen in the CaMV35SGmglnβ1 transformants. However, the band comigrating with the novel holoprotein in the RUBGmglnβ1 transformants, appeared more diffused in the CaMV35S-Gmglnβ1 transformants, probably representing holoproteins made of different combinations of the endogenous GS1 polypeptide and the transgene product. The absence of the band corresponding with the endogenous GS1 holoprotein in the CaMV35S-Gmglnβ1 transformants strongly suggest that some of the transgene product combines with the endogenous GS1 polypeptide to form heteromeric holoprotein, implying that some of the transgene GS1 polypeptides are being made in the same cells where the endogenous GS1 is being made. Since the CaMV35S promoter is highly active in the vasculature (Fig. 1B), and GS1 in the leaves is normally made in the vasculature [7, 17], we would conclude that the endogenous and transgene GS1 subunits mix together to form heteromeric holoproteins in the vasculature. It is important to point out that even though the CaMV35S-Gmglnβ1 transformants showed 3-fold higher level of GS1 protein than the RUB-Gmglnβ1 transformants, the enzyme transferase activity showed only a small increase. The discrepancy between the protein content and enzyme activity can be attributed to differences in the kinetic properties of the different holoproteins and probably to some kind posttranslational regulation [19, 20]. The presence of the novel lower molecular weight GS immunoreactive protein band in the 35S-Gmglnβ1 transformants and not in the RUB-Gmglnβ1 transformants is indicative of higher holoprotein turnover in the former, suggesting that there is also regulation at the level GS holoprotein stability [22]. Any holoprotein made in excess of the substrate is probably disassembled and eventually turned over.
While the RUB promoter is active in the cortical cells of the stem, no GS holoprotein made of the transgene polypeptide accumulates in the stem. The stem of these transformants also do not exhibit any increase in GS activity when compared to control plants. In contrast, the CaMV35S-Gmglnβ1 transformants show a broad immunoreactive band, probably representing the different holoproteins made of different combinations of the endogenous and transgene GS1 polypeptides in the vasculature. Moreover, the stem of CaMV35S-Gmglnβ1 transformants also exhibit higher level of GS activity compared to control plants. Taken together, the results suggest that in the stem, GS1 holoprotein made in the vasculature is stable and active while the GS1 holoprotein made in the cortical cells is unstable.
It is interesting to point out that both the tobacco and alfalfa transformants with the RUBGmglnβ1 exhibited an improvement in certain physiolgical parameters, measured as total protein content and rates of photosynthesis when compared to control plants. This report of improved performance in the transformants overexpressing the GS1 transgene only in the cells involved in photorespiration would strongly support the idea that expression of GS1 in the mesophyll cells provides an alternate route to GS2 for the assimilation of photorespiratory NH4+. The photorespiratory NH4+ produced by the lysine decarboxylase in the mitochondria is either assimilated by a mitochondrial targeted GS2 [9] or has to enter the chloroplast via the cytoplasm to be assimilated by the chloroplastic located GS2. The forced expression of GS1 in the cytoplasm of the photorespiratory cells may give the NH4+ produced during photorespiration an alternate and more accessible avenue for re-assimilation, resulting in less NH4+ lost en route to the chloroplast. Elevated photorespiratory rates in the transformants compared to control plants would suggest that GS may be critical in linking photosynthesis with photorespiration as suggested in earlier studies [11, 28]. Moreover, the increased GS1 activity in the mesophyll cells could also be involved in the re-assimilation of NH4+ produced by catabolic pathways associated with senescence. Overall, the tobacco transformants exhibited better performance compared to the alfalfa transformants, emphasizing the differences in response to GS overexpression attributable to the different plant models.
It is interesting to note that the CaMV35S-Gmglnβ1 transformants, in spite of exhibiting higher levels of transgene expression, showed less or the same level of response in performance as the RUB-Gmglnβ1 transformants. The comparable performance of the CaMV35S-Gmglnβ1 transformants to the RUB-Gmglnβ1 transformants, would suggest that the improvement in plant performance in both sets of transformants is solely due to increased GS1 activity in the leaf mesophyll cells. The higher level expression of the transgene in the mesophyll cells of the CaMV35S-Gmglnβ1 transformants as evidenced by transcript and protein level, does not translate directly into a corresponding increase in overall GS actvity (Fig. 3D). Moreover, since the GS catalyzed reaction involves ATP consumption, the uncontrolled GS activity, as may occur when the GS1 gene is driven by the strong constitutive CaMV35S promoter, could deplete the cells of ATP and C skeletons. Therefore, using the RUB promoter, as shown here, may avoid negative effects of overexpressing Gmglnβ1 in a constitutive manner and at very high levels as the RUB promoter drives expression only in the mesophyll cells, is light inducible and is a weaker promoter than the 35S promoter. The mesophyll cells being the site of C fixation, ATP synthesis and production of photorespiratory NH4+, is the ideal site for increased GS activity with regards to availability of substrate and energy. Thus increased GS activity in the mesophyll at optimal level, does not affect the C/N ratio or the energy status of the plant and is yet able to re-assimilate NH4+ released during photorespiration and/or protein turnover. The higher level improvement in performance of plants transformed with CaMV35S-GS1 gene constructs in earlier studies [27, 28, 29, 30, 43], compared to the performance of CaMV35SGmglnβ1 transformants in this study can probably be ascribed to the gene construct in this study, being devoid of its 3’UTR. We had previously shown that the Gmglnβ1 gene is regulated by its 3’UTR at the level of transcript stability and translation, the process being mediated by the N or the C/N status of the cells [18]. In the absence of the regulatory 3’UTR, the Gmglnβ1 driven by the CaMV35S promoter probably loses that control and functions in an unregulated manner resulting in the depletion of ATP and substrates required for the normal growth of the plant.
Our results suggest that even modest increases in GS1 activity in the leaf mesophyll cells of both alfalfa and tobacco has some beneficial effect on plant performance. It will be interesting to see if higher increases in GS1 activity would have a more profound effect on plant growth. However, because of the multistep regulation of GS, it is a difficult endeavor to increase GS activity at will by just manipulating the promoter driving the GS transgene. Moreover, even our efforts to curtail posttranscriptional regulation by removing the 3’UTR of the GS1 gene, failed. GS function is regulated by the C/N ratio and since it works in conjunction with other key enzymes involved in C metabolism, our efforts to increase GS activity in a predictable manner may involve manipulating levels of other key enzymes along with GS.
Acknowledgements
This work was supported by the National Institutes of Health (Grant Nos. GMO-8136-26, GMO-61222 and GMO-7667-25) and by the Agricultural Experiment Station at New Mexico State University. We would also like to thank the NSF program AGEP for fellowship support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Mark Seger, Email: mseger@nmsu.edu.
Jose Luis Ortega, Email: jortegac@nmsu.edu.
Suman Bagga, Email: sbagga@nmsu.edu.
Champa-Sengupta Gopalan, Email: csgopala@nmsu.edu.
References
- 1.Lamb JSF, Sheaffer CC, Rhodes LH, Sulc RM, Undersander DJ, Brummer EC. Five decades of alfalfa cultivar improvement: Impact on forage yield, persistence, and nutritive value. Crop Sci. 2006;46:902–909. [Google Scholar]
- 2.Putnam D, Russelle M, Orloff S, Kuhn J, Fitzhugh L, Godfrey L, Kiess A, Long R. Alfalfa, wildlife and the environment—the importance and benefits of alfalfa in the 21st century. California Alfalfa and Forage Assoc. 2001:1–24. [Google Scholar]
- 3.Lea PJ, Ireland RJ. Nitrogen metabolism in higher plants. In: Singh BK, editor. Plant amino acids: biochemistry and biotechnology. New York: Marcel Dekker, Inc.; 1999. pp. 1–50. [Google Scholar]
- 4.Miflin BJ, Habash DZ. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 2002;53:979–987. doi: 10.1093/jexbot/53.370.979. [DOI] [PubMed] [Google Scholar]
- 5.Coruzzi G, Last R. Amino acids. In: Buchanan B, Gruissem W, Jones R, editors. Biochemistry and molecular biology of plants: American Society of Plant Physiologists. 2000. pp. 358–412. [Google Scholar]
- 6.Unno H, Uchida T, Sugawara H, Kurisu G, Sugiyama T, Yamaya T, Sakakibara H, Hase T, Kusunoki M. Atomic structure of plant glutamine synthetase: a key enzyme for plant productivity. J. Biol. Chem. 2006;281:29287–29296. doi: 10.1074/jbc.M601497200. [DOI] [PubMed] [Google Scholar]
- 7.Bernard SM, Moller ALB, Dionisio G, Kichey T, Jahn TP, Dubois F, Baudo M, Lopes MS, Tercé-Laforgue T, Foyer CH, Parry MAJ, Forde BG, Araus JL, Hirel B, Schjoerring JK, Habash DZ. Gene expression, cellular localization and function of glutamine synthetase isozymes in wheat (Triticum aestivum L.) Plant Mol. Biol. 2008;67:89–105. doi: 10.1007/s11103-008-9303-y. [DOI] [PubMed] [Google Scholar]
- 8.Morey KJ, Ortega JL, Sengupta-Gopalan C. Cytosolic glutamine synthetase in soybean is encoded by a multigene family and the members are regulated in an organ-specific and developmental manner. Plant Physiol. 2002;128:182–193. [PMC free article] [PubMed] [Google Scholar]
- 9.Taira M, Valtersson U, Burkhardt B, Ludwig RA. Arabidopsis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts. Plant Cell. 2004;16:2048–2058. doi: 10.1105/tpc.104.022046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zozaya-Hinchiffle M, Potenza C, Ortega JL, Sengupta-Gopalan C. Nitrogen and metabolic regulation of the expression of plastidic glutamine synthetase in alfalfa (Medicago sativa) Plant Sci. 2005;168:10141–1052. [Google Scholar]
- 11.Kosaki A, Takeba G. Photorespiration protects C3 plants from photooxidation. Nature. 1996;384:557–560. [Google Scholar]
- 12.Kiyomiya S, Nakanishi H, Uchida H, Tsuji A, Nishiyama S, Futatsubashi M, Tsukada H, Ishioka NS, Watanabe S, Ito T, Mizuniwa C, Osa A, Matsuhashi S, Hashimoto S, Sekine T, Mori S. Real time visualization of 13N-translocation in rice under different environmental conditions using positron emitting tracer imaging system. Plant Physiol. 2001;125:1743–1753. doi: 10.1104/pp.125.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Márquez AJ, Betti M, Garcia-Calderón M, Pal’ove-Balang P, Diaz P, Monza J. Nitrate assimilation in Lotus japonicus. J. Ex. Bot. 2005;56:1741–1749. doi: 10.1093/jxb/eri171. [DOI] [PubMed] [Google Scholar]
- 14.Carvalho H, Lescure N, de Billy F, Chabaud M, Lima L, Salema R, Cullimore J. Cellular expression and regulation of the Medicago truncatula cytosolic glutamine synthetase genes in root nodules. Plant Mol. Biol. 2000;42:741–756. doi: 10.1023/a:1006304003770. [DOI] [PubMed] [Google Scholar]
- 15.Brugiere N, Dubois F, Masclaux C, Sangwan RS, Hirel B. Immunolocalization of glutamine synthetase in senescing tobacco (Nicotiana tabacum L.) leaves suggests that ammonia assimilation is progressively shifted to the mesophyll cytosol. Planta. 2000;211:519–527. doi: 10.1007/s004250000309. [DOI] [PubMed] [Google Scholar]
- 16.Perez-Garcia A, Pereira S, Pissarra J, Garca Gutierrez A, Cazorla FM, Salema R, de Vicente A, Canovas FM. Cytosolic localization in tomato mesophyll cells of a novel glutamine synthetase induced in response to bacterial infection or phosphinothricin treatment. Planta. 1998;206:426–434. [Google Scholar]
- 17.Edwards J, Walker E, Coruzzi G. Cell-specific expression in transgenic plants reveals nonoverlapping roles for chloroplast and cytosolic glutamine synthetase. Proc. Natl. Acad. Sci. 1990;87:3459–3463. doi: 10.1073/pnas.87.9.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortega JL, Moguel-Esponda S, Potenza C, Conklin CF, Quintana A, Sengupta-Gopalan C. The 3’ untranslated region of a soybean cytosolic glutamine synthetase (GS1) affects transcript stability and protein accumulation in transgenic alfalfa. Plant J. 2006;45:832–846. doi: 10.1111/j.1365-313X.2005.02644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finneman J, Schjoerring JK. Post-translational regulation of cytosolic glutamine synthetase by reversible phosphorylation and 14-3-3 protein interaction. Plant J. 2000;24:171–181. doi: 10.1046/j.1365-313x.2000.00863.x. [DOI] [PubMed] [Google Scholar]
- 20.Lima L, Seabra A, Melo P, Cullimore J, Carvalho H. Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula. J. Exp. Bot. 2006;57:2751–2761. doi: 10.1093/jxb/erl036. [DOI] [PubMed] [Google Scholar]
- 21.Riedel J, Tischner R, Mäck G. The chloroplastic glutamine synthetase (GS2) of tobacco is phosphorylated and associated with 14-3-3 proteins inside the chloroplast. Planta. 2001;213:396–401. doi: 10.1007/s004250000509. [DOI] [PubMed] [Google Scholar]
- 22.Ortega JL, Roche D, Sengupta-Gopalan C. Oxidative turnover of soybean root glutamine synthetase: in vitro and in vivo studies. Plant Physiol. 1999;119:1483–1496. doi: 10.1104/pp.119.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirel B, Bertin P, Quillere I, Bourdoncle W, Attagnant C, Dellay C, Gouy A, Cadiou S, Retailliau C, Falque M, Gallais A. Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol. 2001;125:1258–1270. doi: 10.1104/pp.125.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obara M, Sato T, Sasaki S, Kashiba K, Nakamura I, Ebitani T, Yano M, Yamaya T. Identification and characterization of QTL on chromosome 2 for cytosolic glutamine synthetase content and panicle number in rice (Oryza sativa L.) Theor. Appl. Genet. 2004;110:1–11. doi: 10.1007/s00122-004-1828-0. [DOI] [PubMed] [Google Scholar]
- 25.Habash DZ, Bernard S, Schondelmaier J, Weyen J, Quarrie SA. A genetic study of nitrogen use in hexaploid wheat in relation to N utilization, development and yield. Theor. Appl. Genet. 2007;114:403–419. doi: 10.1007/s00122-006-0429-5. [DOI] [PubMed] [Google Scholar]
- 26.Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F. Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol. 2003;131:345–358. doi: 10.1104/pp.102.010785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortega JL, Temple SJ, Bagga S, Ghoshroy S, Sengupta-Gopalan C. Biochemical and molecular characterization of transgenic Lotus japonicus plants constitutively over-expressing a cytosolic glutamine synthetase gene. Planta. 2004;219:807–818. doi: 10.1007/s00425-004-1292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira IC, Brears T, Knight TJ, Clark A, Coruzzi GM. Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light, and photorespiration. Plant Physiol. 2002;129:1170–1180. doi: 10.1104/pp.020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuentes SI, Allen DJ, Ortiz-Lopez A, Hernandez G. Over-expression of cytosolic glutamine synthetase increases photosynthesis and growth at low nitrogen concentrations. J. Exp. Bot. 2001;52:1071–1081. doi: 10.1093/jexbot/52.358.1071. [DOI] [PubMed] [Google Scholar]
- 30.Gallardo F, Fu J, Cantón FR, García-Gutierrez A, Cánovas FM, Kirby EG. Expression of a conifer glutamine synthetase gene in transgenic poplar. Planta. 1999;210:19–26. doi: 10.1007/s004250050649. [DOI] [PubMed] [Google Scholar]
- 31.Habash DZ, Massiah AJ, Rong HL, Wallsgrove RM, Leigh RA. The role of cytosolic glutamine synthetase in wheat. Ann. Appl. Biol. 2001;138:83–89. [Google Scholar]
- 32.Artus NN, Somerville SC, Somerville CR. The biochemistry and cell biology of photorespiration. C.R.C. Crit. Rev. Plant Sci. 1987;4:121–147. [Google Scholar]
- 33.Orea A, Pajuelo P, Pajuelo E, Quidiello C, Romero JM, Marquez AJ. Isolation of photorespiratory mutants from Lotus japonicus deficient in glutamine synthetase. Physiol. Plant. 2002;115:352–361. doi: 10.1034/j.1399-3054.2002.1150304.x. [DOI] [PubMed] [Google Scholar]
- 34.Masclaux C, Valadier MH, Brugiere N, Morot-Gaudry JF, Hirel B. Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta. 2000;211:510–518. doi: 10.1007/s004250000310. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Russell D. Molecular cloning: a laboratory manual: Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 36.Potenza C, Aleman L, Sengupta-Gopalan C. Targeting transgene expression in research, agriculture and environmental applications promoters used in plant transformation. In Vitro Cell Dev. Biol. Plant. 2004;132:1–22. [Google Scholar]
- 37.Ortega JL, Temple SJ, Sengupta-Gopalan C. Constitutive overexpression of cytosolic glutamine synthetase (GS1) gene in transgenic alfalfa demonstrates that GS1 may be regulated at the level of RNA stability and protein turnover. Plant Physiol. 2001;126:109–121. doi: 10.1104/pp.126.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferguson AR, Sims AP. Inactivation in vivo of glutamine synthetase and NAD-specific glutamate dehydrogenase its role in the regulation of glutamine synthesis in yeasts. J.Gen. Microbiol. 1971;69:423–427. doi: 10.1099/00221287-69-3-423. [DOI] [PubMed] [Google Scholar]
- 39.Porra R, Thompson W, Kreidmann P. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extract with four different solvents: verifications of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochem. Biophys. Acta. 1989;975:348–394. [Google Scholar]
- 40.Good AG, Shrawat AK, Muench DG. Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 2004;9:597–605. doi: 10.1016/j.tplants.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Vincent R, Fraisier V, Chaillou S, Limami MA, Deleens E, Phillipson B, Douat C, Boutin JP, Hirel B. Overexpression of a soybean gene encoding cytosolic glutamine synthetase in shoots of transgenic Lotus corniculatus L. plants triggers changes in ammonium assimilation and plant development. Planta. 1997;201:424–433. doi: 10.1007/s004250050085. [DOI] [PubMed] [Google Scholar]
- 42.Temple SJ, Knight TJ, Unkefer PJ, Sengupta-Gopalan C. Moduation of glutamine synthetase gene expression in tobacco by the introduction of an alfalfa glutamine synthetase gene in sense and antisense orientation molecular and biochemical analysis. Mol. Genet. 1993;236:315–325. doi: 10.1007/BF00277128. [DOI] [PubMed] [Google Scholar]
- 43.Fei H, Chaillou S, Hirel B, Polowick P, Mahon JD, Vessey JK. Effects of the overexpression of a soybean cytosolic glutamine synthetase gene (GS15) linked to organ-specific promoters on growth and nitrogen accumulation of pea plants supplied with ammonium. Plant Physiol. Biochem. 2006;44:543–550. doi: 10.1016/j.plaphy.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Martin A, Lee J, Kichey T, Gerentes D, Zivy M, Tatout C, Dubois F, Balliau T, Valot B, Davanture M, Tercé-Laforgue T, Quilleré I, Coque M, Gallais A, Gonzalez-Moro MB, Bethencourt L, Habash DZ, Lea PJ, Charcosset A, Perez P, Murigneux A, Sakakibara H, Edwards KJ, Hirel B. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell. 2006;18:3252–3274. doi: 10.1105/tpc.106.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]