Abstract
Studies of metagenomics and the human microbiome will tremendously expand our knowledge of the composition of microbial communities in the human body. As our understanding of microbial variation and corresponding genetic parameters is refined, this information can be applied to rational remodeling or “tailoring” of human-associated microbial communities and their associated functions. Physiologic features such as the development of innate and adaptive immunity, relative susceptibilities to infections, immune tolerance, bioavailability of nutrients, and intestinal barrier function may be modified by changing the composition and functions of the microbial communities. The specialty of gastroenterology will be affected profoundly by the ability to modify the gastrointestinal microbiota through the rational deployment of antibiotics, probiotics, and prebiotics. Antibiotics might be used to remove or suppress undesirable components of the human microbiome. Probiotics can introduce missing microbial components with known beneficial functions for the human host. Prebiotics can enhance the proliferation of beneficial microbes or probiotics, to maximize sustainable changes in the human microbiome. Combinations of these approaches might provide synergistic and effective therapies for specific disorders. The human microbiome could be manipulated by such “smart” strategies to prevent and treat acute gastroenteritis, antibiotic-associated diarrhea and colitis, inflammatory bowel disease, irritable bowel syndrome, necrotizing enterocolitis, and a variety of other disorders.
The Metagenomics Era and Gastroenterology
The science of metagenomics and the international Human Microbiome Project1,2 has ushered in a new era for the field of gastroenterology. Microbes and their genetic content outnumber their mammalian counter-parts by 1–2 orders of magnitude,3 and the spatial topography of these populations within the gut shows a non-random distribution that ultimately benefits both microbe and host.4 New ribosomal RNA- and whole genome-based technologies have highlighted the potential importance of novel microbial species, with populations that differ between individuals, regions of the gut axis,5 and different mucosal layers at a single anatomical site.6 Culture-independent strategies such as high-throughput parallel sequencing and comparative genomics, metabolic profiling and functional genomics, fluorescence in situ hybridization, and phylogenetic microarrays will provide new insights into the composition, architecture, and functional roles of the human microbiota.7-9 These tools have provided key insights into many aspects of gastrointestinal disease, such as the discovery of reduced bacterial species diversity in patients with Crohn disease.10,11
As we begin to appreciate the functional significance of these dynamic communities, including their effect on human physiology and disease,12 new therapeutic approaches have emerged to provide a fresh perspective in the treatment of both acute and chronic disorders. Evidence suggests that perturbations of the gastrointestinal microbiota underlie many diseases and that therapeutic manipulation of microbial communities has the potential to ameliorate different gastrointestinal conditions.13 The composition of the intestinal microbiome may also affect mammalian physiology in extraintestinal compartments. For example, intestinal microbiome composition represented a key epigenetic factor modifying predisposition to type 1 diabetes in the nonobese diabetic mouse model.14 Changes in gastrointestinal microbial populations also correlated with specific patterns of metabolites excreted in the urine.15 These studies suggest that directed manipulation of the microbiome within the gastrointestinal tract may yield health benefits at remote sites. Human trials have shown effects of prebiotics and probiotics on systemic immune responses after oral intake, including overall health maintenance and reduction of duration of common infections. Pregnant women consuming oral probiotics conferred protection to their infants by reducing the risk of atopic eczema,16 and children given oral probiotics (Lactobacillus rhamnosus GG) showed a reduced risk of atopic eczema during a 4-year follow-up period.17 Probiotics may colonize mucosal surfaces outside of the gastrointestinal tract and confer health benefits at sites such as the oral cavity18 and the genitourinary tract19 after oral administration. Probiotic therapy also showed promise in the context of neurologic and psychiatric diseases,20 as well as in overall health maintenance, including reducing the duration of common illnesses such as upper respiratory infections and reducing absences from work or day care.21 Intentional manipulation of the human microbiome by prebiotics may also modulate systemic immunity.22 The plasticity of these microbial communities and their aggregate biological functions indicate similar compositional and functional plasticity in human physiology and the human metabonome (Figure 1).23
Figure 1.
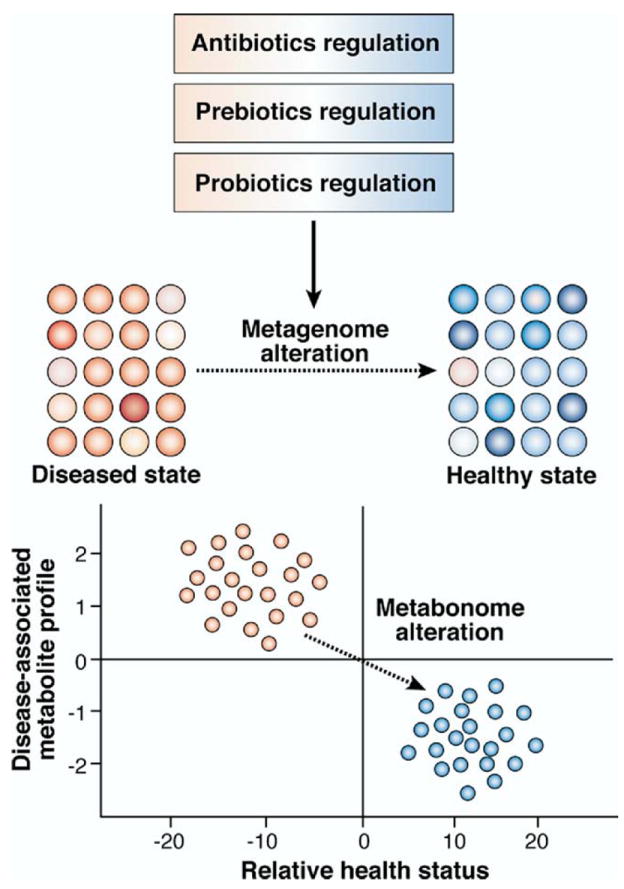
The gut microbiome as a therapeutic target: the “drug the extended” genome strategy. Perturbed metagenomic or metabonomic profiles associated with complex disease states can be restored to homeostasis with rationally selected antibiotic, probiotic, prebiotic, or combination treatment strategies. Adapted with permission from Macmillan Publishers Ltd: Nature Reviews Drug Discovery,23 copyright 2008. http://www.nature.com/nrd/.
The ideas of a core and variable human microbiome have been proposed and provide a conceptual framework for considering early development and relationships of the human microbiome with multiple physiologic functions (Figure 2).1 Microbial colonization of the human intestine within the first few days of life is an intricate process24 that results in a symbiotic relationship with the host that is likely to maintain health and homeostasis.25 Newborns are exposed to beneficial bacteria during passage through the birth canal and, subsequently, through breastfeeding. Commensal microbes can enter the breast milk from the skin of the breast (areola) or via the bloodstream after translocation from the maternal gut to the lactating mammary gland.26 Molecular techniques showed Escherichia coli, Clostridium, Bifidobacterium, Bacteroides, Streptococcus, Enterococcus, and Actinomyces species to be the dominant bacteria in healthy infants.27 Microarray-based bacterial population profiling was used to show that bacterial populations become established after 1 week of life, but remain in a relative state of flux until reaching equilibrium, an adult-like microbial population, by the end of infancy.28 A special window of opportunity for therapeutic intervention appears to exist during the first year of life, considering the drastically altered microbial profiles observed in both infants given antibiotics28 and those delivered by Cesarean section,29 compared with vaginally delivered neonates not receiving antibiotics. Studies in healthy infants indicated that formula supplementation with a single human breast milk– derived probiotic strain improved overall feeding tolerance by increasing gastric emptying rates and reducing fasting antral areas.30 Colonization of the gut by commensal bacteria is essential for normal development of gastrointestinal-associated lymphoid tissue (GALT); this effect has been shown by the profound architectural and immunologic defects observed in germ-free rats.31 Neonatology and pediatric gastroenterology are providing important information about the concurrent development of the gastrointestinal microbiota and the function of the mucosal immune system.
Figure 2.
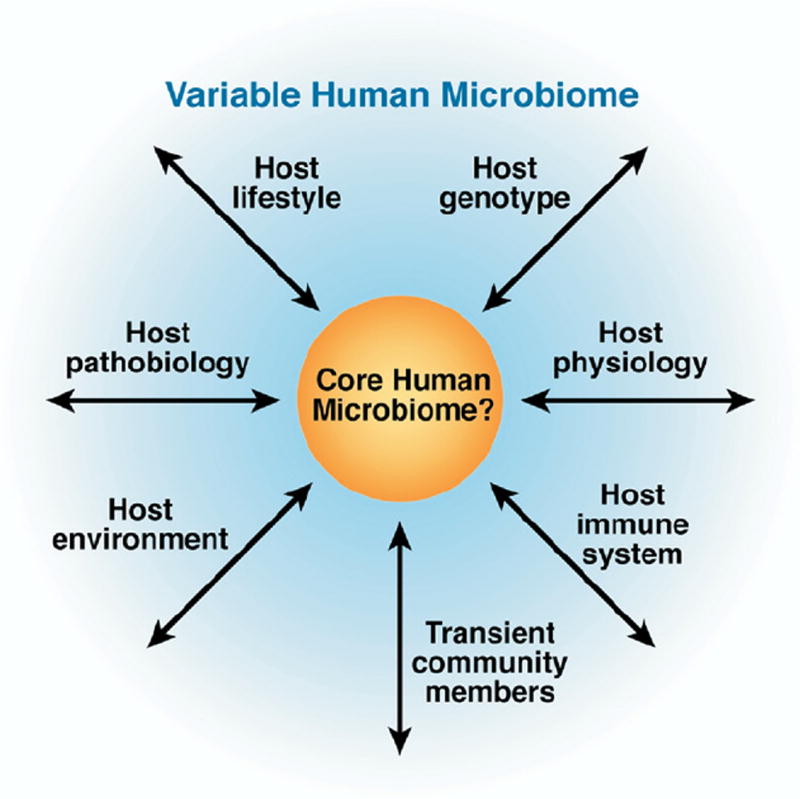
A super-organismal view of the human microbiome. Core and variable components of the human microbiome could have important implications for human health, including nutrient responsiveness, innate and adaptive immunity, and development. As the microbiome affects multiple aspects of human health and disease, host biology influences the composition and function of the commensal microbiota. A subset of microbial genes may be found in most healthy human beings (core microbiome), whereas variable components are present only in specific ethnic groups, age groups, geographic locations, or associated with specific dietary patterns or disease states. Manipulation of either the core or the variable parts of the human microbiome can affect human physiology, overall health status, and disease susceptibilities. Adapted with permission from Macmillan Publishers Ltd: Nature,1 copyright 2007. http://www.nature.com/nature/.
Although single probiotic strains may confer health benefits early in life, future studies are needed to determine whether combinations of diverse probiotic strains may yield more potent and sustainable benefits in children. Although it may seem intuitive that probiotic strategies would benefit from microbial diversity in oral formulations, individual strains may effectively serve as “starter” cultures to promote a sufficiently diverse, healthy microbial ecosystem. Similar to drugs, microbes may antagonize one another, so multimicrobe combinations should be considered carefully, especially in neonates and infants.
Lessons From Antibiotics
The suppression and elimination of microbial pathogens by antibiotics represents a time-tested approach in medical management. However, recent studies have highlighted the profound changes in microbial populations that result from applications of antimicrobial agents. Molecular profiling of infant fecal microbial communities after antibiotic treatment showed drastic reductions in total bacterial densities and alterations in population composition.28 These changes of the human-associated microbiota are usually temporary (rarely lasting >2 months), but long-term microbial population fluctuations have been reported in healthy adults.32 Next-generation DNA pyrosequencing technology was recently used to study fluoroquinolone-induced perturbations in the composition of the human microbiome.33 Although intestinal microbial populations have shown resilience as whole communities, several microbial taxa remained depleted 6 months after termination of oral ciprofloxacin therapy.34 Animal models are beginning to lend insights into the functional consequences of these perturbations in microbial populations. Microarray studies in rat pups, for example, showed amoxicillin-induced alterations in microbial populations that included depletion of the Lactobacillus genus; these alterations were associated with significant changes in the expression of nearly one-third of developmentally regulated genes.35 Furthermore, recent metabolomic studies have shown that depletion of gram-positive bacteria by vancomycin disrupts carbohydrate fermentation in mice; these changes resulted in increased quantities of unfermented oligosaccharides in the feces and reduced concentrations of short-chain fatty acids, including acetate, n-butyrate, propionate, and lactate.36 Thus, significant changes in the composition of gastrointestinal microbial communities can result in important functional differences in the host metabonome (Figure 1).
Antibiotic-induced alterations in the composition of the gastrointestinal microbiota increase disease risk by increasing susceptibility to gastrointestinal infections. Antibiotic-associated diarrhea and colitis can be caused by clostridial pathogens such as Clostridium difficile or Clostridium perfringens.37 Comparison of fecal molecular profiles by temporal temperature gradient gel electrophoresis has shown specific microbial patterns that represented permissive microbial communities and predicted C. difficile–associated disease development.32 Another recent study in mice showed increased susceptibility of animals to invasive salmonellosis after perturbations of the microbiota with streptomycin and vancomycin.38 Furthermore, antibiotic therapy for children infected with E. coli strain O157:H7 can increase the risk of hemolytic-uremic syndrome39 and prolong fecal shedding. Thus, unintentional changes of the human microbiome by antibiotics can influence susceptibility to enteric pathogens.
Not surprisingly, antimicrobial treatment for Helicobacter pylori induces marked disturbances in the intestinal microbiota. Stool cultures from patients with H. pylori taken before, during, and after triple therapy showed drastic alterations, including overall pruning of species diversity, with specific reductions in the level of Lactobacillus and Bifidobacterium from their pretreatment levels.40 A separate study showed overgrowth of potentially pathogenic organisms, including drug-resistant Enter-obacteriaceae and yeasts.41 These changes could account for reports of treatment-associated C. difficile colitis42 and other forms of diarrhea that can reduce patient compliance.43 Probiotics, when used in combination with standard antibiotic therapy for H. pylori infection, increased pathogen eradication and reduced the gastrointestinal side effects of conventional therapy, as concluded by a meta-analysis of 14 randomized trials.44 Evidence suggests that probiotic bacteria suppress gastrointestinal pathogens and potentiate antibiotic efficacy by production of antibacterial factors, including bacteriocins and small organic molecules.45,46 Small molecules secreted by probiotics, such as Lactobacillus acidophilus47 and some strains of Lactobacillus reuteri,48 reduced the level of expression of enterohemorrhagic E. coli virulence genes. Furthermore, a bacteriocin produced by Lactobacillus salivarius was responsible for preventing invasive infections in mice caused by foodborne Listeria monocytogenes.49 Clearly, “endogenous” antibiotics produced by indigenous bacteria can contribute to beneficial effects of probiotics.
Lessons from Probiotics
The concept of probiotics, a term that literally means “for life,” was introduced at the turn of the 20th century. Russian Nobel laureate Elie Metchnikoff, in his 1907 opus The Prolongation of Life: Optimistic Studies, proposed the heretical idea that ingesting microbes could have beneficial effects for human beings, especially to treat digestive diseases.50 Nearly a century later, British microbiologist Roy Fuller suggested that the beneficial effects are mediated by an improvement in intestinal microbial balance.51 The modern definition, drafted by joint expert consultation of the Food and Agricultural Organization of the United Nations and the World Health Organization in Argentina in 2001, identifies probiotics as “Live microorganisms, which, when consumed in adequate amounts, confer a health benefit on the host.”52 A wealth of new studies in both the clinical and basic sciences have sought to determine the safety and efficacy of probiotic therapies in the treatment of gastrointestinal disease.53 The most commonly studied organisms fall within the genera Bifidobacterium and Lactobacillus, but also include lactic acid bacteria such as Lactococcus and Streptococcus. Other probiotics and promising candidates include organisms of the genera Bacillus, Bacteroides, Enterococcus, Escherichia, Faecalibacterium, Propionibacterium, and the yeast Saccharomyces. Compared with the broader category of beneficial microbes, probiotics must be isolated as pure microorganisms, and single strains or combinations of strains must yield health benefit(s) in the target species in controlled human or animal trials.21
Probiotics are gaining widespread inclusion as new prevention strategies or therapies for multiple gastrointestinal diseases, as evidenced by multiple recent metaanalyses and systematic reviews (Table 1), although the practice of combining data from studies of different genera, species, strains, and doses of probiotics provides limited information about specific therapeutic interventions. Mechanisms of probiosis include remodeling of microbial communities and suppression of pathogens, immunomodulation by up-regulation of anti-inflammatory factors, immunomodulation by suppression of proinflammatory factors, enhancement of immunity, effects on epithelial cell differentiation, and proliferation and promotion of intestinal barrier function (Figure 3). Prophylactic manipulation of the microbiome may be important for disease prevention, including acute infections of the gastrointestinal tract. Several studies have examined the role of microbial supplementation to promote resilience of the host to infectious challenges. Molecular studies of microbial communities in a mouse model of antibiotic-associated diarrhea show that the probiotic yeast Saccharomyces cerevisiae var. boulardii, commonly referred to as S. boulardii, facilitated more rapid recovery of the normal microbiota to its pre-antibiotic state.54 Microbial supplementation could promote species diversity and increase resilience of microbial communities to challenges such as infection and concomitant antimicrobial therapy.
Table 1.
Large Systematic Reviews (>1000 patients) of Randomized Controlled Clinical Trials That Included Probiotics for the Treatment of Specific Gastrointestinal Diseases
| Indication | Patient population | N | Species | Result | Reference |
|---|---|---|---|---|---|
| Acute gastroenteritis | Adults and children with infectious diarrhea duration <14 days | 1917 | Lactobacillus spp. | Reduced risk of diarrhea at 3 days (RR, 0.66; 95% CI, 0.55– 0.77) | Allen et al, 200492 |
| Streptococcus spp. | Reduced duration by 30.48 (95% CI, 18.51– 42.46) hours | ||||
| E. faecium | |||||
| “S. boulardii” | |||||
| Children 1–60 months old with diarrhea duration <1 week | 1917 | Lactobacillus spp. | Reduced duration by 0.8 days (95% CI, 0.6–1.1 days) | Huang et al, 200293 | |
| Bifidobacterium spp. | |||||
| S. thermophilus | |||||
| B. subtilis | |||||
| E. faecium | |||||
| “S. boulardii” | |||||
| C. difficile-associated diarrhea or colitis | Children <18 years of age receiving antibiotic therapy for any reason | 1986 | Lactobacillus spp. | Per-protocol: reduced risk of AAD (RR, 0.49; 95% CI, 0.32–0.74) | Johnston et al, 200797 |
| Bifidobacterium spp. | Intention-to-treat: no significant benefit (RR, 0.90; 95% CI, 0.50–1.63) | ||||
| S. thermophilus | |||||
| “S. boulardii” | |||||
| Children and adults receiving antibiotics | 1214 | Lactobacillus spp. | Reduced risk of AAD (OR, 0.37; 95% CI, 0.26 – 0.53) | D’Souza et al, 2002100 | |
| B. longum | Bacteria reduced risk (OR, 0.34; 95% CI, 0.19 – 0.61) | ||||
| E. Faecium | Yeast reduced risk (OR, 0.39; 95% CI, 0.25–0.62) | ||||
| “S. boulardii” | |||||
| Irritable bowel syndrome | Children and adults with IBS established by Rome criteria | 1011 | Lactobacillus spp. | Improved clinical outcomes (RR, 1.22; 95% CI, 1.07–1.4) | Nikfar et al, 2008128 |
| Bifidobacterium spp. | |||||
| VSL#3 | |||||
| Propionibacterium freudenreichii | |||||
| Children and adults with IBS | 1404 | Lactobacillus spp. | Reduced global IBS symptoms (RR, 0.77; 95% CI, 0.62–0.94) | McFarland and Dublin 2008129 | |
| Bifidobacterium spp. | Reduced abdominal pain (RR, 0.78; 95% CI, 0.69–0.88) | ||||
| Streptococcus spp. | |||||
| VSL#3 | |||||
| B. subtilis | |||||
| L. lactis | |||||
| E. coli | |||||
| P. freudenreichii | |||||
| “S. boulardii” | |||||
| Necrotizing enterocolitis | Preterm infants <37 weeks and/or birth weight <2500 g | 1425 | Lactobacillus spp. | Reduced incidence of stage II–III | Alfaleh and Bassler 2008147 |
| Bifidobacterium spp. | NEC (RR, 0.32; 95% CI, 0.17–0.60) | ||||
| S. thermophilus | Reduced risk of mortality (RR, 0.43; 95% CI, 0.25–0.75) | ||||
| “S. boulardii” | No significant reduction in risk of nosocomial sepsis or number of days on total parenteral nutrition | ||||
| Preterm very low birthweight neonates <33 weeks gestation and birth weight <1500 g | 1393 | Lactobacillus spp. | Reduced risk of stage II–III NEC (RR, 0.36; 95% CI, 0.20–0.65) | Deshpande et al, 2007148 | |
| Bifidobacterium spp. | Reduced risk of death (RR, 0.47; 95% CI, 0.3– 0.73) | ||||
| S. thermophilus | Reduced number of days to full feeds (RR, −2.74; 05% CI, −4.98 to −0.51) | ||||
| “S. boulardii” | No significant reduction in risk of sepsis |
AAD, antibiotic-associated diarrhea; CI, confidence interval; OR, odds ratio; RR, relative risk.
Figure 3.
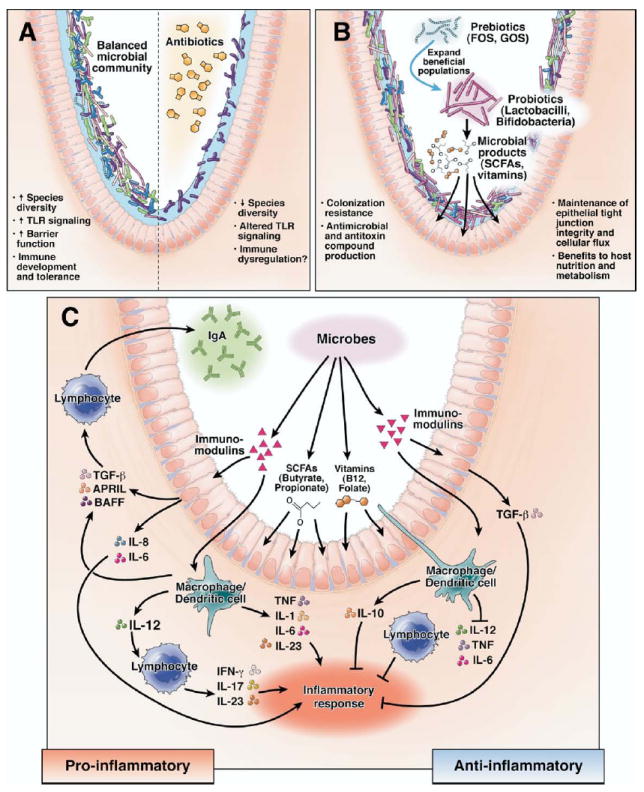
Microbial manipulation strategies and effects on intestinal biology. (A) The composition and aggregate functions of mixed microbial communities are dramatically altered by antibiotics. Various functions affected include gastrointestinal physiology and innate and adaptive immunity, resulting in increased host susceptibility to human disease. (B) Manipulation of the microbiota with rationally selected prebiotics or probiotics can inhibit pathogens, strengthen epithelial barrier function, and supply the host with key nutrients, including short-chain fatty acids (SCFAs) and vitamins. (C) Specific microbes modulate mucosal immunity by secreted factors and by direct interactions with immune cells and the intestinal epithelium. Anti-inflammatory responses are mediated by TGF-β production by epithelial cells and IL-10 from mononuclear cells. Immunostimulatory responses occur as a result of a wide variety of proinflammatory cytokines from stimulated epithelial cells, mononuclear cells, and lymphocytes, in addition to IgA production from B lymphocytes. FOS, fructo-oligosaccharides or oligo-fructose.
Several mechanisms of probiosis that suppress inflammation have been identified. Lactobacillus johnsonii induced transforming growth factor β (TGF-β) production in Caco-2 cells cocultured with leukocytes.55 High levels of TGF-β were also seen in the sera of infants given Bifidobacterium breve; increases in TGF-β levels are believed to result from increased Smad signaling within peripheral blood mononuclear cells.56 One study reported the ability of VSL#3 (the probiotic mixture containing 3 Bifidobacterium species, 4 Lactobacillus species, and Streptococcus thermophilus) or its 3 individual component Bifidobacterium species, but not the component Streptococcus or Lactobacillus species, to induce interleukin (IL)–10 production by human dendritic cells.57 In addition to increasing production of anti-inflammatory cytokines such as TGF-β or IL-10, probiotics stimulated proliferation of regulatory T-cell populations. Regulatory T cells that suppress T helper 1 and 2 effector responses were induced by Lactobacillus paracasei in mixed lymphocyte cultures.58 Recent studies have shown that regulatory T cells can be stimulated by bacterial-induced dendritic cell maturation.59
In addition to stimulation of immunoregulatory networks, probiotics can suppress proinflammatory signaling pathways in epithelial or immune cells. Avirulent Salmonella strains blocked ubiquitination and degradation of inhibitor of nuclear factor-κB (NF-κB α) (IκB-α), which increased levels of this inhibitory protein and blocked the NF-κB pathway, thus preventing synthesis of proinflammatory cytokines, including tumor necrosis factor (TNF) and IL-8 by human intestinal epithelial cells.60 In contrast to suppression of transcription factor activation, Bacteroides thetaiotaomicron induced nuclear clearance of the RelA subunit of NF-κB by nuclear per-oxisome proliferator activated receptor-γ.61 Thus, probiotics can impede entry or promote exit of key transcription factors. L. reuteri exhibited a strain-specific ability to suppress TNF production by lipopolysaccharide (LPS)–stimulated macrophages through the c-Jun-dependent activator protein-1 (AP-1) pathway.62 L. reuteri also suppressed NF-κB activation by preventing IκB-α degradation and inhibiting translocation of RelA into the nucleus.63 Perturbations of cell signaling pathways depend on the agonist used (eg, LPS or TNF). A 2-step model for probiotic-mediated immunomodulation can be proposed: one specific probiotic strain can suppress AP-1 signaling when cells are stimulated by microbial signals (LPS), and a different signaling pathway (NF-κB) can be inhibited when immune cells are stimulated by host cytokines (TNF).
In addition to suppressing inflammation, microbes, including probiotics, can stimulate immune responses by different mechanisms. One report cited the different abilities of various strains of Lactobacillus to induce IL-6, IL-12, and TNF production from murine dendritic cells.64 The E. coli Nissle strain induced proinflammatory IL-8 production by T84 and HT-29 colonocyte lines,65 whereas Lactobacillus casei induced phosphorylation and activation of NF-κB subunit RelA and induced IL-6 in primary intestinal epithelial cells from monoassociated rats.66 Certain subsets of activated T cells produce IL-23 and IL-17, cytokines linked to mucosal damage in Crohn disease67; the differentiation of T(H)17 lymphocytes68,69 and IL-17/IL-23 production by lymphocytes is regulated, in part, by commensal microbes70,71 and probiotics.72 Other immunomodulatory functions reported in the literature include increased phagocytosis capacity, stimulation of natural killer cell activity, enhanced cell-mediated immunity, and increased production of mucosal immunoglobulin (Ig)A.73 Commensal bacteria, including Lactobacillus plantarum and Bacillus subtilis, or bacterial components such as LPS or flagellin that serve as ligands for Toll-like receptors (TLRs), stimulated human colonocytes to produce the protein APRIL (a proliferation-inducing ligand). This protein mediates class-switch recombination of B cells to IgA2.74 Intriguingly, IgA seems to be essential for dampening the mucosal inflammatory response to commensal bacteria, given that Rag1−/− IgA-deficient mice generated a robust innate immune response to commensal Bacteroides fragilis.75
Recent studies have shown the efficacy of probiotics in improving intestinal barrier function. Proteins secreted by L. rhamnosus GG facilitated tight junction formation and reduced barrier permeability induced by hydrogen peroxide in Caco-2 cell monolayers via protein kinase C and mitogen-activated protein kinase pathways.76 Exposure of intestinal epithelial cells to VSL#3 prevented Salmonella-induced reduction in transepithelial resistance and increased mucin gene expression while stabilizing tight junctions.65 The single probiotic strain L. rhamnosus GG suppressed changes in tight junction proteins, barrier permeability, and transepithelial resistance induced by enterohemorrhagic E. coli O157:H7 in intestinal epithelial cell monolayers.77 L. acidophilus or its conditioned media increased Cl−/OH− exchange activity in Caco-2 cells by a phosphatidylinositol 3 kinase–mediated pathway78; this mechanism of action could account for attenuation of secretory diarrhea induced by enteric pathogens. A bacterial quorum-sensing molecule secreted by B. subtilis was internalized by human intestinal epithelial cells and stimulated production of a heat shock protein that can protect cell monolayers from oxidant-induced injury.79 Finally, commensal bacteria appear to maintain intestinal homeostasis through TLR stimulation. Mice deficient in MyD88, an adaptor molecule essential for TLR-mediated signaling, had impaired survival after administration of dextran sodium sulphate (DSS). This result was replicated in wild-type mice after depletion of their commensal microbiota by broad-spectrum antibiotics.80
Investigations of probiotics are yielding diverse mechanisms of action and multiple disease targets. Microbial mediators of biochemical function remain largely unknown, with recent studies beginning to unravel candidate signals linking microbe to host. More progress has been made recently with mammalian host signaling pathways and genes affected by commensals, beneficial microbes, and probiotics. Preliminary insights suggest that animal models are providing basic lessons in how beneficial microbes and probiotics may modulate signaling pathways and disease phenotypes. As more comprehensive human studies emerge, the detailed knowledge base linking specific probiotic strains with specific human diseases is being assembled.
Lessons From Prebiotics and Synbiotics
The term prebiotic was first introduced by Gibson and Roberfroid in 1995 and is defined as “a nondigestible food ingredient that beneficially affects the host by selectively stimulating the growth and/or activity of one or a limited number of bacteria in the colon, and thus improves host health.”81 Prebiotics are typically oligosaccharides or more complex saccharides that are selectively used by commensal bacteria, including species considered to be beneficial for the human host. Prebiotics must resist host digestion, absorption, and adsorption before fermentation by ≥1 species of the resident microbiota.82 Well-studied prebiotics, oligofructose, galacto-oligosaccharides (GOS), and lactulose, remodel intestinal microbial communities, but only specific classes of microbes that are already resident in the gastrointestinal tract can be stimulated by prebiotics. Importantly, prebiotics have begun to be tested as therapeutics for human diseases.83 If combined with compatible probiotics, prebiotics may theoretically enhance colonization, survival, and function of probiotics in addition to resident beneficial microbes. When fermented by gut bacteria, these carbohydrates lower luminal pH (creating an inhospitable environment for pathogens and stimulating mucin production) through their conversion to short-chain fatty acids, such as butyrate, which also serve as energy sources for intestinal epithelial cells.84 Fermented short-chain fatty acids also stimulate GALT immune cells, by direct interactions and by inducing epithelial cell production of stimulatory cytokines and chemokines.22
Synbiotics, a term also coined by Gibson and Roberfroid in 1995, refers to synergistic beneficial effects derived from “a mixture of probiotics and prebiotics that beneficially affects the host by improving the survival and implantation of live microbial dietary supplements in the gastrointestinal tract, by selectively stimulating the growth and/or by activating the metabolism of one or a limited number of health-promoting bacteria, and thus improving host welfare.”81 Synbiotics have the potential to promote the survival and increase the function of probiotics and resident beneficial microbes. Careful selection of probiotic strains and compatible prebiotics will maximize the potential effect of these strategies, in terms of sustainable changes of host-associated microbial communities. One study that examined biological activities of 8 different prebiotics, combined with 1 of 4 probiotics, found that several synbiotic combinations changed the composition of the fecal microbiota to a greater extent than their respective probiotic components alone.85 A double-blind randomized controlled trial (RCT) with 31 infants showed that L. plantarum, when administered in a synbiotic combination with oligofructose, successfully colonized 100% of infants at 2 months and 32% at 6 months after a 1-week administration.86 Another recent RCT examined 925 infants at risk for allergic disorders and found that daily administration of a 4-species probiotics mixture, combined with GOS, reduced antibiotic prescriptions and respiratory infections.87 These studies suggest that effective combinations of probiotics and prebiotics result in sustainable changes in microbial composition and benefits for the human host. However, studies have not clearly shown synergies of prebiotics/probiotics combinations in vivo, and future studies are needed to explore relative efficacies and mechanisms of action of such mixtures.
Acute Gastroenteritis
Although specific microbial pathogens have been identified as causative agents of acute gastroenteritis, antibiotic treatment strategies are limited in scope and can exacerbate toxin-mediated diseases in this context. One successful antibiotics-based strategy includes rifaximin for the prevention and treatment of traveler’s diarrhea.88 In contrast to antibiotics, microbial supplementation by probiotics has been studied as an approach for prevention or attenuation of acute gastroenteritis. Manipulation of the gastrointestinal microbiota by probiotics can reduce the duration of illness, presumably by suppression of pathogens or modulation of mucosal immunity. In a seminal RCT that enrolled 40 children hospitalized with acute watery (mostly rotavirus-associated) diarrhea, Shornikova et al89 showed that L. reuteri (strain SD2112; 1010–1011 colony forming units [cfu]/d for the duration of hospitalization or up to 5 days) reduced the duration of illness by an average of 1.2 days compared with the placebo group.89 L. paracasei (strain ST11; 1 × 1010 cfu/d for 5 days) reduced the duration of illness in a double-blind RCT of infants and children with moderate non–rotavirus diarrhea.90 In a RCT with 571 children, 5 days of either L. rhamnosus (strain GG; 1.2 × 1010 cfu/d) or a mixture of L. delbrueckii var bulgaricus (strain LMG-P17550; 2 × 109 cfu/d), L. acidophilus (strain LMG-P17549; 2 × 109 cfu/d), S. thermophilus (strain LMG-17503; 2 × 109 cfu/d), and Bifidobacterium bifidum (strain LMG-P17500; 1 × 109 cfu/dose) reduced average disease duration by 36.5 hours and 45.0 hours, respectively. However, a similar effect was not observed with either S. boulardii (strain It; 1 × 1010 yeasts/d), Bacillus clausii (strains O/C84, N/R84, T84, and SIN84; 2 × 109 cfu/d), or Enterococcus faecium (strain SF68; 1.5 × 108 cfu/d).91 A systematic review of 23 RCTs reported a mean reduction in the duration of acute gastroenteritis of 30.48 hours after treatment with probiotics, compared with placebo,92 whereas another large meta-analysis of 18 studies found an overall reduction in the duration of disease by 0.8 days.93
Clinical studies of synbiotic preparations in the treatment of specific gastrointestinal diseases are limited,94 but a combination of prebiotics and probiotics has been shown to effectively reduce the duration of acute gastroenteritis. One prospective RCT that enrolled 496 children in day care centers found that children consuming a synbiotic blend of Bifidobacterium lactis (strain CNCM I-3446), oligofructose, and acacia gum experienced a 20% reduction in the number of days with diarrhea.95 The potential role(s) of prebiotics without probiotics for prevention or attenuation of acute gastroenteritis remains unclear.
Antibiotic-Associated Diarrhea and Colitis
Antibiotic-induced alterations of the gastrointestinal microbiota clearly increase the risk of antibiotic-associated gastrointestinal disorders in susceptible individuals. Intuitively, the concomitant addition of beneficial microbes to treatment regimens for these disorders offers new opportunities to promote microbial diversity and resilience of the host. A series of systematic reviews showed promising but incomplete evidence to recommend probiotic preparations to treat C. difficile–associated diarrhea in children or adults96 or to prevent antibiotic-associated diarrhea in children.97 Among the probiotic strains showing the most promise in this context are L. rhamnosus GG and S. boulardii.98 In the elderly, L. casei (strain DN-114 001; 1.94 × 1010 cfu/d), L. bulgaricus (1.94 × 109 cfu/d), and S. thermophilus (1.94 × 1010 cfu/d), administered throughout the course of antibiotics and for 1 week after cessation, significantly reduced the risk of developing antibiotic-associated diarrhea in a double-blind RCT of 135 hospital patients.99 A large number of trials have examined the efficacy of probiotics given in conjunction with antibiotics to reduce the occurrence of either antibiotic-associated diarrhea or C. difficile–associated disease, with both yeast and bacterial probiotics showing a moderate but significant benefit.100
Combinations of antibiotics and prebiotics have also been tested for treatment of antibiotic-associated diarrhea and colitis. In a RCT enrolling 142 patients with C. difficile–associated diarrhea, individuals given the prebiotic oligofructose (12 g/d for 30 days) along with anti-clostridial therapy (metronidazole and vancomycin) had significantly fewer relapses and shorter hospital stays per relapse than those receiving antibiotics alone.101 One randomized, double-blind parallel group study examined the effects of a combination of antibiotics, probiotics, and prebiotics. Thirty healthy volunteers were given cefpodoxime along with L. acidophilus (strain NCFB 1748; 1 × 1011 cfu/d), B. longum (strain BB 536; 2.5 × 1010 cfu/d), and oligofructose (15 g/d) for 21 days; oligofructose alone; or placebo. C. difficile was found in the stool of only 1 individual given the synbiotic combination compared with 6 people given only the prebiotic and 6 individuals receiving placebo.102 Ultimately, the most effective strategies could use various combinations of antibiotics to suppress or eliminate pathogens, probiotics to supplement altered microbial communities, and prebiotics to replenish microorganisms diminished by changes in diet or therapy.
Inflammatory Bowel Disease
Microbial dysbiosis has been implicated in inflammatory bowel disease (IBD),103 and numerous studies have documented differences in gastrointestinal microbial populations between healthy individuals and patients with IBD. In the IL-10 – deficient mouse model of colitis, replacement of missing lactobacilli populations ameliorated cecal inflammation.104-107 A recent review of probiotic therapies for the induction and maintenance of remission in IBD concluded that beneficial effects were modest and strain specific.108
Promising preliminary results were observed in 34 patients with refractory ulcerative colitis that were given the probiotic preparation VSL#3 (3.6 × 1012 bacteria/d for 6 weeks); 77% responded to therapy, with no adverse effects reported.109 Treatment with the same probiotic mixture (9 × 1011 bacteria/d for 12 months) improved IBD questionnaire scores and prevented episodes of acute pouchitis in patients with ulcerative colitis and ileal pouch-anal anastomoses in a double-blind placebo-controlled trial.110 In a small double-blind RCT, treatment with VSL#3 (6 × 1011 bacteria/d for 12 months or until relapse) maintained remission in patients with chronic pouchitis.111 In rat pups with DSS-induced colitis, the VSL#3 mixture also attenuated disease, significantly lowering both disease activity and histopathologic indices.112 However, the probiotic L. salivarius (subsp. salivarius 433118) yielded no benefits in 2 different mouse models of colitis.113 Careful selection of correct probiotic strains, strain combinations, and doses might be crucial for sustainable effects, in terms of maintenance of remission or treatment of acute disease.
In addition to microbial supplementation, selective stimulation of microbial populations that reduce intestinal inflammation or maintain immune homeostasis could offer promising avenues to new therapies. One randomized double-blind crossover trial examined the efficacy of prebiotics in 24 patients with IBD. In patients with ileal pouch-anal anastomoses given the fructose polymer inulin (24 g/d for 3 weeks), mucosal inflammation was reduced at the pouch site, accompanied by increased butyrate, reduced pH and bile acids, and lower concentrations of B. fragilis.114 Published trials evaluating the efficacy of prebiotics in Crohn disease have also shown promising results, but are limited in size. For example, 10 patients with active Crohn disease given oligofructose (15 g/d for 3 weeks) had a significant aggregate reduction in the Harvey Bradshaw disease index, from 9.8 to 6.9, along with increased numbers of fecal bifidobacteria and a trend toward increased numbers of IL-10 –positive dendritic cells.115 Multiple studies reported a benefit of prebiotics in animal models of IBD, including oligofructose or lactulose in rats given trinitrobenzene sulphonic acid (TNBS), oligofructose/inulin in HLA-B27 transgenic rats, and inulin in rats given DSS.116 However, analogous to probiotics, careful prebiotic selection is essential, given that oligofructose, alone or in a synbiotic combination, failed to produce a benefit in DSS-challenged rats.117 Synbiotics were effective in one small double-blind RCT that included 18 patients with active ulcerative colitis; treatment for 1 month with a synbiotic mixture of a natural Bifidobacterium longum isolate (4 × 1011 bacteria/d) and inulin-oligofructose (12 g/d), a prebiotic selectively used by this probiotic agent in vitro, significantly reduced mucosal inflammatory markers and showed a trend toward histologic and sigmoidoscopic score improvements.118
Results from trials of synbiotics in patients with Crohn disease are less convincing than for patients with other forms of IBD, primarily because of the small numbers of patients enrolled in the Crohn disease studies. One double-blind RCT examined 30 patients with active Crohn disease who had just undergone resection. No differences were observed in endoscopic scores or clinical relapse rates when patients given a mixture of 4 lactic acid–producing bacteria (Pediococcus pentosaceus, L. paracasei subsp. paracasei, Lactococcus raffinolactis, and L. plantarum strain 2362; 1 × 1010 of each bacterium/d) with 4 fermentable fibers (β-glucans, inulin, pectin. and resistant starch; 2.5 g of each carbohydrate/d) for 24 months were compared with those given placebo.119 Crohn disease could be relatively refractory to microbial manipulation, or this disorder might require different approaches for therapeutic microbiology. Deficiencies of Firmicutes such as Faecalibacterium prausnitzii could determine susceptibility to Crohn disease11 and could indicate new directions for microbiome-targeted therapies. F. prausnitzii– based microbial supplementation of mice challenged with TNBS reduced intestinal inflammation and injury120; this organism was associated with the modulation of ≥8 metabolites in the urinary compartment alone.15 Probiotic strategies, based on metagenomics, metabonomics, and new classes of microbes, might be important for future management of Crohn disease and other intestinal disorders.
In patients with IBD, antimicrobial therapy is usually reserved for those with septic complications such as intraabdominal abscess, perianal fistula, small intestinal bowel overgrowth, toxic megacolon, and postoperative and other infections.121 Treatment with either metronidazole (1000 mg/d) or ciprofloxacin (20 mg/kg/d) for 2 weeks significantly reduced the pouchitis disease activity index, along with symptomatic, endoscopic, and histologic scores. Specific antimicrobial agents could be more effective; ciprofloxacin yielded greater benefits without the associated side effects seen with metronidazole therapy.122 A limited number of trials have tested the efficacy of broad-spectrum antibiotics in patients with primary active disease. More recent trials in patients with Crohn disease found that rifaximin (800–1600 mg/d for 12 weeks) reduced the number of treatment failures but did not facilitate induction of remission123 and that antimycobacterial therapy (clarithromycin 750 mg/d, rifabutin 450 mg/d, and clofazimine 50 mg/d for 2 years) significantly improved the short-term remission rate compared with controls.124 However, the role of Mycobacterium avium subsp. paratuberculosis as a causative agent of Crohn disease remains controversial125; these studies have highlighted the challenges of antibiotic strategies for treatment of IBD. Recent animal studies have provided encouraging results for the potential effect of antimicrobial strategies. In a mouse model of severe IBD caused by loss of TGF-β and IL-10 signaling, ciprofloxacin and metronidazole prevented intestinal inflammation and death.126 Another study reported that long-term exposure to the macrolide roxithromycin delayed the development of colitis and attenuated inflammatory damage in IL-10 – deficient mice. These findings were associated with reduced inflammatory cytokine production by mononuclear cells and minimal changes in the microbiota.127
Irritable Bowel Syndrome and Related Disorders
Irritable bowel syndrome (IBS) has been characterized as constipation-predominant, diarrhea-predominant, or as a disorder with alternating patterns of constipation or diarrhea predominance. IBS includes a constellation of symptoms such as abdominal pain, bloating, and altered defecation patterns. Microbial manipulation could provide tangible benefits for intestinal fermentation or gas production, intestinal motility patterns, and antinociceptive or anti-inflammatory effects that result from changes in the host-associated microbiome. One large systematic review evaluating > 1000 patients with IBS concluded that probiotic therapy significantly improved clinical outcomes,128 whereas another report examining 20 RCTs found that probiotics consumption was associated with an improvement in global IBS symptoms, especially reduced abdominal pain.129 In particular, 8 weeks of Bifidobacterium infantis (strain 35624; 1 × 1010 bacteria/d), but not L. salivarius (subsp. salivarius UCC4331; 1 × 1010 bacteria/d), improved cardinal IBS symptoms of abdominal pain or discomfort, bloating or distension, and bowel movement difficulty, while increasing the serum IL-10/IL-12 ratio, suggesting a generalized anti-inflammatory effect.130 In a more recent double-blind RCT, patients with IBS receiving a probiotic mixture (total 2.5 × 1010 cfu/d for 8 weeks) consisting of 2 strains of L. acidophilus (NCIMB 30157 and NCIMB 30156), B. bifidum (strain NCIMB 30172), and B. infantis (strain NCIMB 30153) reported a significant improvement in symptom severity scores and quality of life, compared with patients taking placebo.131
Relevant to IBS, probiotics can affect gastrointestinal motility and nociception by causing fundamental changes in the composition and function of the intestinal microbiota. In a randomized trial, the probiotic L. reuteri (strain ATCC 55730; 1 × 108 bacteria/d for 28 days) reduced crying times in breastfed colicky infants132; the same probiotic strain (1 × 108 cfu/d for 30 days) reduced regurgitation and mean crying times while increasing gastric emptying rates in preterm neonates.30 These studies highlight the potential effect of intentional manipulation of microbial communities on physiologic variables such as intestinal motility and pain perception. Animal studies have provided some evidence for mechanisms of action for the results from pediatric clinical trials. In mice, administration of L. paracasei (strain NCC2461) reversed the emergence of visceral hypersensitivity to colorectal distension after antibiotic-induced perturbations of the microbiota. This benefit of abdominal pain reduction was associated with decreased quantities of substance P in the visceral plexus.133 In addition to suppressing pain signaling by reducing quantities of key mediators, beneficial microbes can up-regulate analgesic receptors in the intestine. L. acidophilus (strain NCFM) increased expression of opioid receptors in human intestinal epithelial cells, providing a possible mechanism for its mediation of analgesic effects in the gut.134
Few studies have examined prebiotics as a treatment strategy for IBS, and concerns linger that the addition of fermentation substrates could worsen intestinal gas production. One double-blind crossover trial with 21 patients with IBS showed that 4 weeks of oligofructose therapy (6 g/d) had no effect on symptom scores, whole gut transit time, fecal weight or pH, or fasting breath hydrogen.135 However, a double-blind, controlled trial involving 20 preterm infants found that a prebiotic combination of oligofructose and GOS (1.8 g/d for 14 days) improved feeding tolerance by reducing stool viscosity and gastrointestinal transit time, compared with control formula.136 Promising results with prebiotics have provided encouragement for possible synbiotics-based approaches for the management of IBS. A parallel group study comparing L. paracasei (strain B21060; 1 × 1010 cfu/d for 12 weeks) plus a prebiotic mixture versus the prebiotic mixture alone in 267 patients with IBS found that the synbiotic treatment significantly increased the number of patients who rated their pain as absent or mild compared with baseline. Furthermore, subgroup analysis within the diarrhea-predominant IBS population showed a significant reduction in number of bowel movements and abdominal pain in the synbiotic treatment group.137
Microbial dysbiosis and the relative preponderance of specific classes of microbes could contribute to the pathogenesis of IBS. In 2000, Pimentel et al138 published results from a study of 47 patients who were codiagnosed with IBS and small intestinal bacterial overgrowth and treated with antibiotics. Individuals with evidence of successful treatment of bacterial overgrowth, as determined by the lactulose breath test, were more likely than those who failed eradication to have resolved IBS, based on the Rome criteria.138 This study linked bacterial overgrowth to IBS and inspired other small, prospective RCTs that reported positive effects of antibiotics, specifically of neomycin139 and rifaximin,140,141 for modest amelioration of IBS symptoms. Rifaximin is the only antibiotic that has consistently yielded a long-term benefit, although further studies are recommended.142 Careful selection of an appropriate antimicrobial agent is critical, especially because antimicrobial agents have been shown to trigger IBS. One prospective case– control study of 91 patients in a general medical practice found that those taking antibiotics for nongastrointestinal symptoms were >3 times as likely to report symptoms associated with IBS than were age-matched controls not prescribed antibiotics.143 The specific antibiotics tested and their antimicrobial spectrum might account for differences in clinical outcomes, because indigenous bacterial populations respond differently to different antibiotics.
Necrotizing Enterocolitis
Considering the wide fluctuations observed in intestinal microbial populations during infancy and the profound effects of antibiotic treatment in neonates, it is important to carefully consider the selection of microbiological therapeutics for patients with necrotizing enterocolitis (NEC). Despite the lack of any specific microbial cause for NEC and the significant questions that remain about the efficacy of antimicrobial strategies for this disorder, agents such as ampicillin plus cefotaxime or an aminoglycoside, depending on local resistance patterns, are currently recommended as standard therapy for the treatment of NEC.144 Relatively little research exists on the use of antibiotics to prevent the development of NEC in low-birthweight or premature infants. A systematic Cochrane review of 5 RCTs that evaluated gentamicin, kanamycin, and vancomycin found insufficient evidence to support enteral antibiotic use as prophylaxis for NEC.145 One retrospective study examined newborns admitted to a neonatal intensive care unit with central venous and arterial catheters and found that administration of amoxicillin-clavulanate and gentamicin was inversely correlated with NEC.146
Trials of intentional microbial manipulation with probiotics have generated promising agents for the prevention or treatment of NEC. Two recent systematic reviews evaluated RCTs that tested the ability of probiotic supplementation to prevent NEC in preterm neonates. Both meta-analyses found that probiotic therapy reduced the risk of NEC by approximately two-thirds and reduced the risk of all-cause mortality by 50%.147,148 One trial, involving 145 neonates that each weighed <1500 g, found that supplementation with a probiotic mixture containing B. infantis, B. bifidus, and S. thermophilus (3.5 × 108 cfu/d of each bacterium until 36 weeks postconceptual age) reduced incidence of NEC, severity of NEC, and infant mortality.149 Preterm piglets delivered by Cesarean section and fed parenterally with a mixture of 1 Bifidobacterium and 4 Lactobacillus species had reduced clinical NEC scores, enhanced brush border enzyme activity, and improved ratios of beneficial to pathogenic bacteria, compared with control (formula-fed) piglets.150 Probiotics might be useful for the medical management of this disease by one of several mechanisms. In a rat model of NEC, animals given IκB kinase inhibitors benefited through decreased mortality and intestinal injury, suggesting one potential mechanism of probiosis, namely, suppression of inflammation by the NF-κB signaling pathway.151 Studies with B. thetaiotaomicron (strain VPI 5482) reported the ability of commensal bacteria to stimulate angiogenesis in mouse intestinal villi.152 This finding suggests another potential role for probiotics in the treatment of NEC by alleviation of hypoxic-ischemic injury153 and indicates the variety of potential benefits conferred by microbes on host physiology.
Future Perspectives: What Lies Ahead?
As the global efforts in human microbiome-related research and therapeutic microbiology expand,154 the knowledge base of the diversity present in the gastrointestinal tract and the functional consequences of that diversity will be more deeply appreciated. Careful selection of drugs, nutrients, and bacterial strains is essential because antibiotics, probiotics, and prebiotics have a variety of biological effects, each with implications for the human microbiome. Careful preclinical screens, driven by improved understanding of the fundamental mechanisms that underlie perturbations of microbial communities, will be essential as gastroenterology enters the metagenomics era.
The current reality is that the menu of useful and established antibiotics, prebiotics, and probiotics remains rather limited in scope. To address regulatory constraints and ethical concerns,155 scientific investigations must continue to increase our knowledge of microbes and their role in health and disease states. In concert with improved understanding of mechanisms of action, the menu must expand in all 3 categories so that rational selection of effective microbial manipulation strategies may yield tangible benefits in human and veterinary medicine. As new commensal microbes are discovered and established commensals are explored in greater detail, next-generation antibiotics, including antimicrobial peptides and new drug classes, will be identified. Instead of mining soil or environmental microbes for antibiotics as in the past, the lion’s share of novel antimicrobial agents in the 21st century may be discovered by mining the human and mammalian microbiomes. These compounds may be expected to be evolutionarily tailored to preserve health-associated microbiomes, while fortifying defenses against unwelcome invaders or pathogens.
New prebiotics will be discovered as studies continue to explore the glycome of human breast milk, saliva, and the intestinal milieu. Novel plant-derived oligosaccharides and polysaccharides may contribute to the currently limited menu of prebiotics. Entirely new biochemical classes of prebiotics may include peptides and lipids as different types of compounds and associated changes are screened by metagenomics and metabonomics. For example, prebiotic substrates could alter endogenous biochemical pathways. Amino acids such as arginine and glutamine are produced by some commensal bacteria and have protective effects in multiple models of gastrointestinal injury156; they might also protect against NEC.157 Prebiotics could also stimulate the growth of beneficial microbes that produce lactate used by neighboring commensal bacteria, resulting in production of fuel sources such as short-chain fatty acids for intestinal epithelial cells.158,159 No fundamental reason exists for the stark limitations with respect to available prebiotics. Commensal microbes use a wide variety of organic substrates, and more detailed exploration of the metabolic pathways inside microbes may facilitate identification of novel pre-biotics that selectively stimulate entire classes of beneficial microbes.
Similar to prebiotics, the current probiotics “universe” remains limited in scope and largely confined to lactic acid bacteria. As with prebiotics, no fundamental barrier constrains consideration of probiotics to a few selected genera. Current classes of probiotics were mostly developed because of their historical importance in dairy products (eg, yogurt) or adaptability to food or supplement formulations. As human and mammalian microbiome research expands, many heretofore unappreciated or undiscovered microbes may give rise to new classes of probiotics. In addition to development of natural probiotics, recombinant DNA technology will facilitate the creation of “designer” or engineered probiotics. As mechanisms of probiosis are uncovered at the molecular level, bacteria could be manipulated to express certain genes of interest. One example is a nonpathogenic E. coli that expresses surface proteins that bind and neutralize cholera toxin.160 The engineered probiotic Lactobacillus gasseri overproduced the reactive oxygen species–neutralizing manganese superoxide dismutase and significantly reduced inflammation in IL-10 – deficient mice.161 An uncontrolled phase 1 trial of 10 patients has shown that a genetically modified strain of Lactococcus lactis, engineered to overproduce IL-10, was well-tolerated and showed promise in treating adults with Crohn disease.162 Recombinant probiotics and novel natural prebiotics could be important parts of future multiagent treatment regimens in gastroenterology, whereas natural probiotics and prebiotics might be important components of new functional food formulations for human nutrition (Figure 4).
Figure 4.
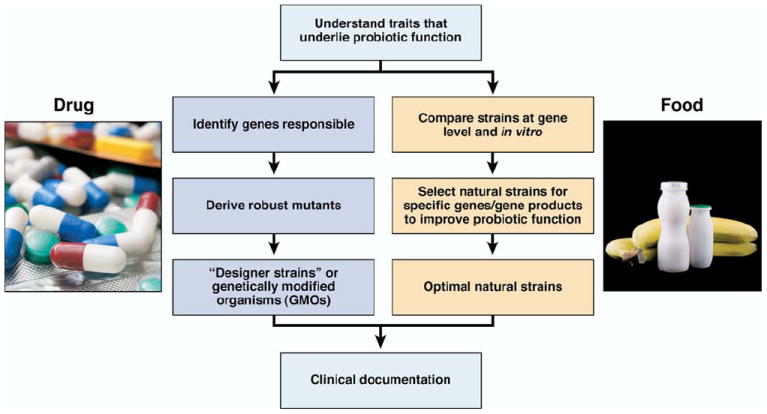
Selection of microbes for probiotics-based strategies. As emerging metagenomics approaches improve our understanding of the fundamental biology underlying probiosis, this knowledge will facilitate probiotic engineering and rational selection of natural strains to prevent and treat a variety of gastrointestinal diseases. Natural strains may be useful in functional foods, and recombinant probiotics might be used to target specific gastrointestinal disorders.
In addition to the effects on the gastrointestinal disorders discussed in this review, manipulation of the human intestinal microbiome could prevent or attenuate traveler’s diarrhea, reduce the long-term risk of gastrointestinal neoplasia, diminish the effect of radiation or chemical-induced injury in the intestine, and correct nutritional imbalances that result from deficiencies or excesses of specific microbial populations. A deeper understanding of the ability of certain microbial populations to harvest energy from the diet163 could lead to therapeutic interventions for obesity and malnutrition.164 Potential beneficiaries include the large global population of undernourished children. Especially promising is the mounting evidence indicating that intestinal bacterial fermentation products increase the effectiveness of oral rehydration therapy in severely dehydrated children and adults in developing countries.165 As relatively low-cost therapeutics, probiotics represent a promising avenue for health interventions targeting those living in severely impoverished and medically underserved regions of the world.
Appropriate use of the knowledge uncovered by the gut microbiome and metabonomics research will require an understanding of how commensal, probiotic, and pathogenic microorganisms interact with one another and the mammalian host. Identification of key substrates and signaling mediators will help explain microbe–microbe and microbe– host interactions at the molecular, cellular, and population levels. Therapeutic approaches will target specific microorganisms, as well as metabolites of entire microbial communities. Deliberate manipulation of the human microbiome with “smart” antibiotics, probiotics, and prebiotics enables scientists and physicians to consider predictive remodeling of human-associated microbial communities in more sophisticated ways. As we progress from the era of relatively simplistic antibiotics-only approaches, the knowledge of the human microbiome and its function will enable personalized medicine to become truly holistic with respect to microbes and man. In addition to considering human genotypes, gastroenterologists will be able to consider the human microbiome when creating customized disease prevention and treatment regimens for individual patients.
Acknowledgments
Funding
This work was supported by National Institutes of Health (NIH) National Institute of Digestive and Kidney Disease (R01 DK065075 and P30 DK56338), NIH National Center for Complementary and Alternative Medicine (R01 AT004326 and R21 AT003482), and the US Department of Defense (HR0011-08-1-0009).
Abbreviations used in this paper
- AP-1
activator protein-1
- APRIL
a proliferation-inducing ligand
- cfu
colony-forming units
- DSS
dextran sodium sulfate
- GALT
gastrointestinal-associated lymphoid tissue
- GOS
galacto-oligosaccharides
- IBD
inflammatory bowel disease
- IBS
irritable bowel syndrome
- Ig
immunoglobulin
- IκB-α
inhibitor of NF-κB α
- IL
interleukin
- LPS
lipopolysaccharide
- NEC
necrotizing enterocolitis
- NF-κB
nuclear factor-κB; peroxisome proliferator activated receptor-γ
- RCT
randomized controlled trial
- TGF-β
transforming growth factor beta
- TLR
Toll-like receptor
- TNBS
trinitrobenzene sulphonic acid
- TNF
tumor necrosis factor
Biographies


Footnotes
Conflicts of interest
The authors disclose the following: Dr. Versalovic has received unrestricted research support from Biogaia AB, Stockholm, Sweden, and has received honoraria from Group Danone, Paris, France.
References
- 1.Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pennisi E. Metagenomics. Massive microbial sequence project proposed. Science. 2007;315:1781. doi: 10.1126/science.315.5820.1781a. [DOI] [PubMed] [Google Scholar]
- 3.Backhed F, Ley RE, Sonnenburg JL, et al. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 4.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 5.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zoetendal EG, von Wright A, Vilpponen-Salmela T, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- 8.Medini D, Serruto D, Parkhill J, et al. Microbiology in the post-genomic era. Nat Rev Microbiol. 2008;6:419–430. doi: 10.1038/nrmicro1901. [DOI] [PubMed] [Google Scholar]
- 9.Zaneveld J, Turnbaugh PJ, Lozupone C, et al. Host-bacterial coevolution and the search for new drug targets. Curr Opin Chem Biol. 2008;12:109–114. doi: 10.1016/j.cbpa.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swidsinski A, Loening-Baucke V, Vaneechoutte M, et al. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis. 2008;14:147–161. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- 12.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarner F. Probiotics in gastrointestinal diseases. In: Versalovic J, Wilson M, editors. Therapeutic microbiology: probiotics and related strategies. Washington, DC: ASM Press; 2008. pp. 255–269. [Google Scholar]
- 14.Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M, Wang B, Zhang M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kukkonen K, Savilahti E, Haahtela T, et al. Probiotics and prebiotic galacto-oligosaccharides in the prevention of allergic diseases: a randomized, double-blind, placebo-controlled trial. J Allergy Clin Immunol. 2007;119:192–198. doi: 10.1016/j.jaci.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Kalliomaki M, Salminen S, Poussa T, et al. Probiotics and prevention of atopic disease: 4-year follow-up of a randomised placebo-controlled trial. Lancet. 2003;361:1869–1871. doi: 10.1016/S0140-6736(03)13490-3. [DOI] [PubMed] [Google Scholar]
- 18.Caglar E, Cildir SK, Ergeneli S, et al. Salivary mutans streptococci and lactobacilli levels after ingestion of the probiotic bacterium Lactobacillus reuteri ATCC 55730 by straws or tablets. Acta Odontol Scand. 2006;64:314–318. doi: 10.1080/00016350600801709. [DOI] [PubMed] [Google Scholar]
- 19.Anukam K, Osazuwa E, Ahonkhai I, et al. Augmentation of antimicrobial metronidazole therapy of bacterial vaginosis with oral probiotic Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14: randomized, double-blind, placebo controlled trial. Microbes Infect. 2006;8:1450–1454. doi: 10.1016/j.micinf.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Forsythe P, Bienenstock J. Probiotics in neurology and psychiatry. In: Versalovic J, Wilson M, editors. Therapeutic microbiology: probiotics and related strategies. Washington, DC: ASM Press; 2008. pp. 285–298. [Google Scholar]
- 21.Sanders ME. Use of probiotics and yogurts in maintenance of health. J Clin Gastroenterol. 2008;42(Suppl 2):S71–S74. doi: 10.1097/MCG.0b013e3181621e87. [DOI] [PubMed] [Google Scholar]
- 22.Lomax AR, Calder PC. Prebiotics, immune function, infection and inflammation: a review of the evidence. Br J Nutr. 2008:1–26. doi: 10.1017/S0007114508055608. [DOI] [PubMed] [Google Scholar]
- 23.Jia W, Li H, Zhao L, et al. Gut microbiota: a potential new territory for drug targeting. Nat Rev Drug Discov. 2008;7:123–129. doi: 10.1038/nrd2505. [DOI] [PubMed] [Google Scholar]
- 24.Macfarlane S, Woodmansey EJ, Macfarlane GT. Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl Environ Microbiol. 2005;71:7483–7492. doi: 10.1128/AEM.71.11.7483-7492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 26.Perez PF, Dore J, Leclerc M, et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics. 2007;119:e724–e732. doi: 10.1542/peds.2006-1649. [DOI] [PubMed] [Google Scholar]
- 27.Favier CF, de Vos WM, Akkermans AD. Development of bacterial and bifidobacterial communities in feces of newborn babies. Anaerobe. 2003;9:219–229. doi: 10.1016/j.anaerobe.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Palmer C, Bik EM, Digiulio DB, et al. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biasucci G, Benenati B, Morelli L, et al. Cesarean delivery may affect the early biodiversity of intestinal bacteria. J Nutr. 2008;138(suppl):1796S–1800S. doi: 10.1093/jn/138.9.1796S. [DOI] [PubMed] [Google Scholar]
- 30.Indrio F, Riezzo G, Raimondi F, et al. The effects of probiotics on feeding tolerance, bowel habits, and gastrointestinal motility in preterm newborns. J Pediatr. 2008;152:801–806. doi: 10.1016/j.jpeds.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Yamanaka T, Helgeland L, Farstad IN, et al. Microbial colonization drives lymphocyte accumulation and differentiation in the follicle-associated epithelium of Peyer’s patches. J Immunol. 2003;170:816–822. doi: 10.4049/jimmunol.170.2.816. [DOI] [PubMed] [Google Scholar]
- 32.De La Cochetiere MF, Durand T, Lalande V, et al. Effect of antibiotic therapy on human fecal microbiota and the relation to the development of Clostridium difficile. Microb Ecol. 2008;56:395–402. doi: 10.1007/s00248-007-9356-5. [DOI] [PubMed] [Google Scholar]
- 33.Huse SM, Dethlefsen L, Huber JA, et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dethlefsen L, Huse S, Sogin ML, et al. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumann A, Nutten S, Donnicola D, et al. Neonatal antibiotic treatment alters gastrointestinal tract developmental gene expression and intestinal barrier transcriptome. Physiol Genomics. 2005;23:235–245. doi: 10.1152/physiolgenomics.00057.2005. [DOI] [PubMed] [Google Scholar]
- 36.Yap IK, Li JV, Saric J, et al. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res. 2008;7:3718–3728. doi: 10.1021/pr700864x. [DOI] [PubMed] [Google Scholar]
- 37.Bartlett JG. Clinical practice. Antibiotic-associated diarrhea. N Engl J Med. 2002;346:334–339. doi: 10.1056/NEJMcp011603. [DOI] [PubMed] [Google Scholar]
- 38.Sekirov I, Tam NM, Jogova M, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong CS, Jelacic S, Habeeb RL, et al. The risk of the hemolyticuremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N Engl J Med. 2000;342:1930–1936. doi: 10.1056/NEJM200006293422601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buhling A, Radun D, Muller WA, et al. Influence of anti-Helicobacter triple-therapy with metronidazole, omeprazole and clarithromycin on intestinal microflora. Aliment Pharmacol Ther. 2001;15:1445–1452. doi: 10.1046/j.1365-2036.2001.01033.x. [DOI] [PubMed] [Google Scholar]
- 41.Adamsson I, Nord CE, Lundquist P, et al. Comparative effects of omeprazole, amoxycillin plus metronidazole versus omeprazole, clarithromycin plus metronidazole on the oral, gastric and intestinal microflora in Helicobacter pylori-infected patients. J Antimicrob Chemother. 1999;44:629–640. doi: 10.1093/jac/44.5.629. [DOI] [PubMed] [Google Scholar]
- 42.Nawaz A, Mohammed I, Ahsan K, et al. Clostridium difficile colitis associated with treatment of Helicobacter pylori infection. Am J Gastroenterol. 1998;93:1175–1176. doi: 10.1111/j.1572-0241.1998.00358.x. [DOI] [PubMed] [Google Scholar]
- 43.Bell GD, Powell K, Burridge SM, et al. Experience with “triple” anti-Helicobacter pylori eradication therapy: side effects and the importance of testing the pre-treatment bacterial isolate for metronidazole resistance. Aliment Pharmacol Ther. 1992;6:427–435. doi: 10.1111/j.1365-2036.1992.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 44.Tong JL, Ran ZH, Shen J, et al. Meta-analysis: the effect of supplementation with probiotics on eradication rates and adverse events during Helicobacter pylori eradication therapy. Aliment Pharmacol Ther. 2007;25:155–168. doi: 10.1111/j.1365-2036.2006.03179.x. [DOI] [PubMed] [Google Scholar]
- 45.Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 46.Spinler JK, Taweechotipatr M, Rognerud CL, et al. Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe. 2008;14:166–171. doi: 10.1016/j.anaerobe.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Medellin-Pena MJ, Wang H, Johnson R, et al. Probiotics affect virulence-related gene expression in Escherichia coli O157:H7. Appl Environ Microbiol. 2007;73:4259–4267. doi: 10.1128/AEM.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jelcic I, Hufner E, Schmidt H, et al. Repression of the locus of the enterocyte effacement-encoded regulator of gene transcription of Escherichia coli O157:H7 by Lactobacillus reuteri culture supernatants is LuxS and strain dependent. Appl Environ Microbiol. 2008;74:3310–3314. doi: 10.1128/AEM.00072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corr SC, Li Y, Riedel CU, et al. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci U S A. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metchnikoff E. The prolongation of life: optimistic studies. New York, NY: Springer Publishing Company, Inc; 2004. [Google Scholar]
- 51.Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989;66:365–378. [PubMed] [Google Scholar]
- 52.FAO/WHO. Report of a Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. Basel, Switzerland: World Health Organization; 2001. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. [Google Scholar]
- 53.Walker R, Buckley M. Probiotic microbes: the scientific basis. Washington, DC: American Academy of Microbiology; 2006. [PubMed] [Google Scholar]
- 54.Barc MC, Charrin-Sarnel C, Rochet V, et al. Molecular analysis of the digestive microbiota in a gnotobiotic mouse model during antibiotic treatment: influence of Saccharomyces boulardii. Anaerobe. 2008;14:229–233. doi: 10.1016/j.anaerobe.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 55.Haller D, Bode C, Hammes WP, et al. Non-pathogenic bacteria elicit a differential cytokine response by intestinal epithelial cell/leucocyte co-cultures. Gut. 2000;47:79–87. doi: 10.1136/gut.47.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujii T, Ohtsuka Y, Lee T, et al. Bifidobacterium breve enhances transforming growth factor beta1 signaling by regulating Smad7 expression in preterm infants. J Pediatr Gastroenterol Nutr. 2006;43:83–88. doi: 10.1097/01.mpg.0000228100.04702.f8. [DOI] [PubMed] [Google Scholar]
- 57.Hart AL, Lammers K, Brigidi P, et al. Modulation of human dendritic cell phenotype and function by probiotic bacteria. Gut. 2004;53:1602–1609. doi: 10.1136/gut.2003.037325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von der Weid T, Bulliard C, Schiffrin EJ. Induction by a lactic acid bacterium of a population of CD4(+) T cells with low proliferative capacity that produce transforming growth factor beta and interleukin-10. Clin Diagn Lab Immunol. 2001;8:695–701. doi: 10.1128/CDLI.8.4.695-701.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baba N, Samson S, Bourdet-Sicard R, et al. Commensal bacteria trigger a full dendritic cell maturation program that promotes the expansion of non-Tr1 suppressor T cells. J Leukoc Biol. 2008;84:468–476. doi: 10.1189/jlb.0108017. [DOI] [PubMed] [Google Scholar]
- 60.Neish AS, Gewirtz AT, Zeng H, et al. Prokaryotic regulation of epithelial responses by inhibition of IkappaB-alpha ubiquitination. Science. 2000;289:1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 61.Kelly D, Campbell JI, King TP, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 62.Lin YP, Thibodeaux CH, Pena JA, et al. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflamm Bowel Dis. 2008;14:1068–1083. doi: 10.1002/ibd.20448. [DOI] [PubMed] [Google Scholar]
- 63.Iyer C, Kosters A, Sethi G, et al. Probiotic Lactobacillus reuteri promotes TNF-induced apoptosis in human myeloid leukemia-derived cells by modulation of NF-kappaB and MAPK signalling. Cell Microbiol. 2008;10:1442–1452. doi: 10.1111/j.1462-5822.2008.01137.x. [DOI] [PubMed] [Google Scholar]
- 64.Christensen HR, Frokiaer H, Pestka JJ. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J Immunol. 2002;168:171–178. doi: 10.4049/jimmunol.168.1.171. [DOI] [PubMed] [Google Scholar]
- 65.Otte JM, Podolsky DK. Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am J Physiol Gastrointest Liver Physiol. 2004;286:G613–G626. doi: 10.1152/ajpgi.00341.2003. [DOI] [PubMed] [Google Scholar]
- 66.Ruiz PA, Hoffmann M, Szcesny S, et al. Innate mechanisms for Bifidobacterium lactis to activate transient pro-inflammatory host responses in intestinal epithelial cells after the colonization of germ-free rats. Immunology. 2005;115:441–450. doi: 10.1111/j.1365-2567.2005.02176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holtta V, Klemetti P, Sipponen T, et al. IL-23/IL-17 immunity as a hallmark of Crohn’s disease. Inflamm Bowel Dis. 2008;14:1175–1184. doi: 10.1002/ibd.20475. [DOI] [PubMed] [Google Scholar]
- 68.Atarashi K, Nishimura J, Shima T, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 69.Zaph C, Du Y, Saenz SA, et al. Commensal-dependent expression of IL-25 regulates the IL-23-IL-17 axis in the intestine. J Exp Med. 2008;205:2191–2198. doi: 10.1084/jem.20080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 71.Smits HH, van Beelen AJ, Hessle C, et al. Commensal Gramnegative bacteria prime human dendritic cells for enhanced IL-23 and IL-27 expression and enhanced Th1 development. Eur J Immunol. 2004;34:1371–1380. doi: 10.1002/eji.200324815. [DOI] [PubMed] [Google Scholar]
- 72.So JS, Lee CG, Kwon HK, et al. Lactobacillus casei potentiates induction of oral tolerance in experimental arthritis. Mol Immunol. 2008;46:172–180. doi: 10.1016/j.molimm.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 73.Delcenserie V, Martel D, Lamoureux M, et al. Immunomodulatory effects of probiotics in the intestinal tract. Curr Issues Mol Biol. 2008;10:37–54. [PubMed] [Google Scholar]
- 74.He B, Xu W, Santini PA, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 75.Peterson DA, McNulty NP, Guruge JL, et al. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 76.Seth A, Yan F, Polk DB, et al. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1060–G1069. doi: 10.1152/ajpgi.00202.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson-Henry KC, Donato KA, Shen-Tu G, et al. Lactobacillus rhamnosus strain GG prevents enterohemorrhagic Escherichia coli O157:H7-induced changes in epithelial barrier function. Infect Immun. 2008;76:1340–1348. doi: 10.1128/IAI.00778-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borthakur A, Gill RK, Tyagi S, et al. The probiotic Lactobacillus acidophilus stimulates chloride/hydroxyl exchange activity in human intestinal epithelial cells. J Nutr. 2008;138:1355–1359. doi: 10.1093/jn/138.7.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fujiya M, Musch MW, Nakagawa Y, et al. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe. 2007;1:299–308. doi: 10.1016/j.chom.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 80.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 81.Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 82.Gibson GR. Prebiotics as gut microflora management tools. J Clin Gastroenterol. 2008;42(Suppl 2):S75–S79. doi: 10.1097/MCG.0b013e31815ed097. [DOI] [PubMed] [Google Scholar]
- 83.Rowland I. Prebiotics in human medicine. In: Versalovic J, Wilson M, editors. Therapeutic microbiology: probiotics and related strategies. Washington DC: ASM Press; 2008. pp. 299–306. [Google Scholar]
- 84.Sakata T. Stimulatory effect of short-chain fatty acids on epithelial cell proliferation in the rat intestine: a possible explanation for trophic effects of fermentable fibre, gut microbes and luminal trophic factors. Br J Nutr. 1987;58:95–103. doi: 10.1079/bjn19870073. [DOI] [PubMed] [Google Scholar]
- 85.Saulnier DM, Gibson GR, Kolida S. In vitro effects of selected synbiotics on the human faecal microbiota composition. FEMS Microbiol Ecol. 2008;66:516–527. doi: 10.1111/j.1574-6941.2008.00561.x. [DOI] [PubMed] [Google Scholar]
- 86.Panigrahi P, Parida S, Pradhan L, et al. Long-term colonization of a Lactobacillus plantarum synbiotic preparation in the neonatal gut. J Pediatr Gastroenterol Nutr. 2008;47:45–53. doi: 10.1097/MPG.0b013e31815a5f2c. [DOI] [PubMed] [Google Scholar]
- 87.Kukkonen K, Savilahti E, Haahtela T, et al. Long-term safety and impact on infection rates of postnatal probiotic and prebiotic (synbiotic) treatment: randomized, double-blind, placebo-controlled trial. Pediatrics. 2008;122:8–12. doi: 10.1542/peds.2007-1192. [DOI] [PubMed] [Google Scholar]
- 88.Taylor DN, Bourgeois AL, Ericsson CD, et al. A randomized, double-blind, multicenter study of rifaximin compared with placebo and with ciprofloxacin in the treatment of travelers’ diarrhea. Am J Trop Med Hyg. 2006;74:1060–1066. [PubMed] [Google Scholar]
- 89.Shornikova AV, Casas IA, Isolauri E, et al. Lactobacillus reuteri as a therapeutic agent in acute diarrhea in young children. J Pediatr Gastroenterol Nutr. 1997;24:399–404. doi: 10.1097/00005176-199704000-00008. [DOI] [PubMed] [Google Scholar]
- 90.Sarker SA, Sultana S, Fuchs GJ, et al. Lactobacillus paracasei strain ST11 has no effect on rotavirus but ameliorates the outcome of nonrotavirus diarrhea in children from Bangladesh. Pediatrics. 2005;116:e221–e228. doi: 10.1542/peds.2004-2334. [DOI] [PubMed] [Google Scholar]
- 91.Canani RB, Cirillo P, Terrin G, et al. Probiotics for treatment of acute diarrhoea in children: randomised clinical trial of five different preparations. BMJ. 2007;335:340. doi: 10.1136/bmj.39272.581736.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Allen SJ, Okoko B, Martinez E, et al. Probiotics for treating infectious diarrhoea. Cochrane Database Syst Rev. 2004 doi: 10.1002/14651858.CD003048.pub2. CD003048. [DOI] [PubMed] [Google Scholar]
- 93.Huang JS, Bousvaros A, Lee JW, et al. Efficacy of probiotic use in acute diarrhea in children: a meta-analysis. Dig Dis Sci. 2002;47:2625–2634. doi: 10.1023/a:1020501202369. [DOI] [PubMed] [Google Scholar]
- 94.Bengmark S. Synbiotics in human medicine. In: Versalovic J, Wilson M, editors. Therapeutic microbiology: probiotics and related strategies. Washington, DC: ASM Press; 2008. pp. 307–321. [Google Scholar]
- 95.Binns CW, Lee AH, Harding H, et al. The CUPDAY Study: prebiotic-probiotic milk product in 1-3-year-old children attending childcare centres. Acta Paediatr. 2007;96:1646–1650. doi: 10.1111/j.1651-2227.2007.00508.x. [DOI] [PubMed] [Google Scholar]
- 96.Pillai A, Nelson R. Probiotics for treatment of Clostridium difficile-associated colitis in adults. Cochrane Database Syst Rev. 2008 doi: 10.1002/14651858.CD004611.pub2. CD004611. [DOI] [PubMed] [Google Scholar]
- 97.Johnston BC, Supina AL, Ospina M, et al. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev. 2007 doi: 10.1002/14651858.CD004827.pub2. CD004827. [DOI] [PubMed] [Google Scholar]
- 98.Segarra-Newnham M. Probiotics for Clostridium difficile-associated diarrhea: focus on Lactobacillus rhamnosus GG and Saccharomyces boulardii. Ann Pharmacother. 2007;41:1212–1221. doi: 10.1345/aph.1K110. [DOI] [PubMed] [Google Scholar]
- 99.Hickson M, D’Souza AL, Muthu N, et al. Use of probiotic Lactobacillus preparation to prevent diarrhoea associated with antibiotics: randomised double blind placebo controlled trial. BMJ. 2007;335:80. doi: 10.1136/bmj.39231.599815.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.D’Souza AL, Rajkumar C, Cooke J, et al. Probiotics in prevention of antibiotic associated diarrhoea: meta-analysis. BMJ. 2002;324:1361. doi: 10.1136/bmj.324.7350.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lewis S, Burmeister S, Brazier J. Effect of the prebiotic oligofructose on relapse of Clostridium difficile-associated diarrhea: a randomized, controlled study. Clin Gastroenterol Hepatol. 2005;3:442–448. doi: 10.1016/s1542-3565(04)00677-9. [DOI] [PubMed] [Google Scholar]
- 102.Orrhage K, Sjostedt S, Nord CE. Effect of supplements with lactic acid bacteria and oligofructose on the intestinal micro-flora during administration of cefpodoxime proxetil. J Antimicrob Chemother. 2000;46:603–612. doi: 10.1093/jac/46.4.603. [DOI] [PubMed] [Google Scholar]
- 103.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 104.Pena JA, Rogers AB, Ge Z, et al. Probiotic Lactobacillus spp. diminish Helicobacter hepaticus-induced inflammatory bowel disease in interleukin-10-deficient mice. Infect Immun. 2005;73:912–920. doi: 10.1128/IAI.73.2.912-920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McCarthy J, O’Mahony L, O’Callaghan L, et al. Double blind, placebo controlled trial of two probiotic strains in interleukin 10 knockout mice and mechanistic link with cytokine balance. Gut. 2003;52:975–980. doi: 10.1136/gut.52.7.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Madsen KL, Doyle JS, Jewell LD, et al. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology. 1999;116:1107–1114. doi: 10.1016/s0016-5085(99)70013-2. [DOI] [PubMed] [Google Scholar]
- 107.Schultz M, Veltkamp C, Dieleman LA, et al. Lactobacillus plantarum 299V in the treatment and prevention of spontaneous colitis in interleukin-10-deficient mice. Inflamm Bowel Dis. 2002;8:71–80. doi: 10.1097/00054725-200203000-00001. [DOI] [PubMed] [Google Scholar]
- 108.Isaacs K, Herfarth H. Role of probiotic therapy in IBD. Inflamm Bowel Dis. 2008;14:1597–1605. doi: 10.1002/ibd.20465. [DOI] [PubMed] [Google Scholar]
- 109.Bibiloni R, Fedorak RN, Tannock GW, et al. VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am J Gastroenterol. 2005;100:1539–1546. doi: 10.1111/j.1572-0241.2005.41794.x. [DOI] [PubMed] [Google Scholar]
- 110.Gionchetti P, Rizzello F, Helwig U, et al. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology. 2003;124:1202–1209. doi: 10.1016/s0016-5085(03)00171-9. [DOI] [PubMed] [Google Scholar]
- 111.Mimura T, Rizzello F, Helwig U, et al. Once daily high dose probiotic therapy (VSL#3) for maintaining remission in recurrent or refractory pouchitis. Gut. 2004;53:108–114. doi: 10.1136/gut.53.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fitzpatrick LR, Hertzog KL, Quatse AL, et al. Effects of the probiotic formulation VSL#3 on colitis in weanling rats. J Pediatr Gastroenterol Nutr. 2007;44:561–570. doi: 10.1097/MPG.0b013e31803bda51. [DOI] [PubMed] [Google Scholar]
- 113.Feighery LM, Smith P, O’Mahony L, et al. Effects of Lactobacillus salivarius 433118 on intestinal inflammation, immunity status and in vitro colon function in two mouse models of inflammatory bowel disease. Dig Dis Sci. 2008;53:2495–2506. doi: 10.1007/s10620-007-0157-y. [DOI] [PubMed] [Google Scholar]
- 114.Welters CF, Heineman E, Thunnissen FB, et al. Effect of dietary inulin supplementation on inflammation of pouch mucosa in patients with an ileal pouch-anal anastomosis. Dis Colon Rectum. 2002;45:621–627. doi: 10.1007/s10350-004-6257-2. [DOI] [PubMed] [Google Scholar]
- 115.Lindsay JO, Whelan K, Stagg AJ, et al. Clinical, microbiological, and immunological effects of fructo-oligosaccharide in patients with Crohn’s disease. Gut. 2006;55:348–355. doi: 10.1136/gut.2005.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Macfarlane S, Macfarlane GT, Cummings JH. Review article: prebiotics in the gastrointestinal tract. Aliment Pharmacol Ther. 2006;24:701–714. doi: 10.1111/j.1365-2036.2006.03042.x. [DOI] [PubMed] [Google Scholar]
- 117.Geier MS, Butler RN, Giffard PM, et al. Prebiotic and synbiotic fructooligosaccharide administration fails to reduce the severity of experimental colitis in rats. Dis Colon Rectum. 2007;50:1061–1069. doi: 10.1007/s10350-007-0213-x. [DOI] [PubMed] [Google Scholar]
- 118.Furrie E, Macfarlane S, Kennedy A, et al. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut. 2005;54:242–249. doi: 10.1136/gut.2004.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chermesh I, Tamir A, Reshef R, et al. Failure of Synbiotic 2000 to prevent postoperative recurrence of Crohn’s disease. Dig Dis Sci. 2007;52:385–389. doi: 10.1007/s10620-006-9549-7. [DOI] [PubMed] [Google Scholar]
- 120.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16730–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Isaacs KL, Sartor RB. Treatment of inflammatory bowel disease with antibiotics. Gastroenterol Clin North Am. 2004;33:335–345. X. doi: 10.1016/j.gtc.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 122.Shen B, Achkar JP, Lashner BA, et al. A randomized clinical trial of ciprofloxacin and metronidazole to treat acute pouchitis. Inflamm Bowel Dis. 2001;7:301–305. doi: 10.1097/00054725-200111000-00004. [DOI] [PubMed] [Google Scholar]
- 123.Prantera C, Lochs H, Campieri M, et al. Antibiotic treatment of Crohn’s disease: results of a multicentre, double blind, randomized, placebo-controlled trial with rifaximin. Aliment Pharmacol Ther. 2006;23:1117–1125. doi: 10.1111/j.1365-2036.2006.02879.x. [DOI] [PubMed] [Google Scholar]
- 124.Selby W, Pavli P, Crotty B, et al. Two-year combination antibiotic therapy with clarithromycin, rifabutin, and clofazimine for Crohn’s disease. Gastroenterology. 2007;132:2313–2319. doi: 10.1053/j.gastro.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 125.Kuenstner JT. The Australian antibiotic trial in Crohn’s disease: alternative conclusions from the same study[letter] Gastroenterology. 2007;133:1742–1743. doi: 10.1053/j.gastro.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 126.Kang SS, Bloom SM, Norian LA, et al. An antibiotic-responsive mouse model of fulminant ulcerative colitis. PLoS Med. 2008;5:e41. doi: 10.1371/journal.pmed.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tamagawa H, Hiroi T, Mizushima T, et al. Therapeutic effects of roxithromycin in interleukin-10-deficient colitis. Inflamm Bowel Dis. 2007;13:547–556. doi: 10.1002/ibd.20093. [DOI] [PubMed] [Google Scholar]
- 128.Nikfar S, Rahimi R, Rahimi F, et al. Efficacy of probiotics in irritable bowel syndrome: a meta-analysis of randomized, controlled trials. Dis Colon Rectum. 2008;51:1775–1780. doi: 10.1007/s10350-008-9335-z. [DOI] [PubMed] [Google Scholar]
- 129.McFarland LV, Dublin S. Meta-analysis of probiotics for the treatment of irritable bowel syndrome. World J Gastroenterol. 2008;14:2650–2661. doi: 10.3748/wjg.14.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.O’Mahony L, McCarthy J, Kelly P, et al. Lactobacillus and Bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128:541–551. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 131.Williams E, Stimpson J, Wang D, et al. Clinical trial: a multistrain probiotic preparation significantly reduces symptoms of irritable bowel syndrome in a double-blind placebo-controlled study. Aliment Pharmacol Ther. 2009;29:97–103. doi: 10.1111/j.1365-2036.2008.03848.x. published online ahead of print September 9, 2008. [DOI] [PubMed] [Google Scholar]
- 132.Savino F, Pelle E, Palumeri E, et al. Lactobacillus reuteri (American Type Culture Collection Strain 55730) versus simethicone in the treatment of infantile colic: a prospective randomized study. Pediatrics. 2007;119:e124–e1230. doi: 10.1542/peds.2006-1222. [DOI] [PubMed] [Google Scholar]
- 133.Verdu EF, Bercik P, Verma-Gandhu M, et al. Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut. 2006;55:182–190. doi: 10.1136/gut.2005.066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rousseaux C, Thuru X, Gelot A, et al. Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nat Med. 2007;13:35–37. doi: 10.1038/nm1521. [DOI] [PubMed] [Google Scholar]
- 135.Hunter JO, Tuffnell Q, Lee AJ. Controlled trial of oligofructose in the management of irritable bowel syndrome. J Nutr. 1999;12(suppl):1451S–1453S. doi: 10.1093/jn/129.7.1451S. [DOI] [PubMed] [Google Scholar]
- 136.Mihatsch WA, Hoegel J, Pohlandt F. Prebiotic oligosaccharides reduce stool viscosity and accelerate gastrointestinal transport in preterm infants. Acta Paediatr. 2006;95:843–848. doi: 10.1080/08035250500486652. [DOI] [PubMed] [Google Scholar]
- 137.Andriulli A, Neri M, Loguercio C, et al. Clinical trial on the efficacy of a new symbiotic formulation, Flortec, in patients with irritable bowel syndrome: a multicenter, randomized study. J Clin Gastroenterol. 2008;42(Suppl 3 Pt 2):S218–S223. doi: 10.1097/MCG.0b013e31817fadd6. [DOI] [PubMed] [Google Scholar]
- 138.Pimentel M, Chow EJ, Lin HC. Eradication of small intestinal bacterial overgrowth reduces symptoms of irritable bowel syndrome. Am J Gastroenterol. 2000;95:3503–3506. doi: 10.1111/j.1572-0241.2000.03368.x. [DOI] [PubMed] [Google Scholar]
- 139.Pimentel M, Chow EJ, Lin HC. Normalization of lactulose breath testing correlates with symptom improvement in irritable bowel syndrome. a double-blind, randomized, placebo-controlled study. Am J Gastroenterol. 2003;98:412–419. doi: 10.1111/j.1572-0241.2003.07234.x. [DOI] [PubMed] [Google Scholar]
- 140.Sharara AI, Aoun E, Abdul-Baki H, et al. A randomized double-blind placebo-controlled trial of rifaximin in patients with abdominal bloating and flatulence. Am J Gastroenterol. 2006;101:326–333. doi: 10.1111/j.1572-0241.2006.00458.x. [DOI] [PubMed] [Google Scholar]
- 141.Pimentel M, Park S, Mirocha J, et al. The effect of a nonabsorbed oral antibiotic (rifaximin) on the symptoms of the irritable bowel syndrome: a randomized trial. Ann Intern Med. 2006;145:557–563. doi: 10.7326/0003-4819-145-8-200610170-00004. [DOI] [PubMed] [Google Scholar]
- 142.Frissora CL, Cash BD. Review article: the role of antibiotics vs. conventional pharmacotherapy in treating symptoms of irritable bowel syndrome. Aliment Pharmacol Ther. 2007;25:1271–1281. doi: 10.1111/j.1365-2036.2007.03313.x. [DOI] [PubMed] [Google Scholar]
- 143.Maxwell PR, Rink E, Kumar D, et al. Antibiotics increase functional abdominal symptoms. Am J Gastroenterol. 2002;97:104–108. doi: 10.1111/j.1572-0241.2002.05428.x. [DOI] [PubMed] [Google Scholar]
- 144.Brook I. Microbiology and management of neonatal necrotizing enterocolitis. Am J Perinatol. 2008;25:111–118. doi: 10.1055/s-2008-1040346. [DOI] [PubMed] [Google Scholar]
- 145.Bury RG, Tudehope D. Enteral antibiotics for preventing necrotizing enterocolitis in low birthweight or preterm infants. Cochrane Database Syst Rev. 2001 doi: 10.1002/14651858.CD000405. CD000405. [DOI] [PubMed] [Google Scholar]
- 146.Krediet TG, van Lelyveld N, Vijlbrief DC, et al. Microbiological factors associated with neonatal necrotizing enterocolitis: protective effect of early antibiotic treatment. Acta Paediatr. 2003;92:1180–1182. [PubMed] [Google Scholar]
- 147.Alfaleh K, Bassler D. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev. 2008 doi: 10.1002/14651858.CD005496.pub2. CD005496. [DOI] [PubMed] [Google Scholar]
- 148.Deshpande G, Rao S, Patole S. Probiotics for prevention of necrotising enterocolitis in preterm neonates with very low birth-weight: a systematic review of randomised controlled trials. Lancet. 2007;369:1614–1620. doi: 10.1016/S0140-6736(07)60748-X. [DOI] [PubMed] [Google Scholar]
- 149.Bin-Nun A, Bromiker R, Wilschanski M, et al. Oral probiotics prevent necrotizing enterocolitis in very low birth weight neonates. J Pediatr. 2005;147:192–196. doi: 10.1016/j.jpeds.2005.03.054. [DOI] [PubMed] [Google Scholar]
- 150.Siggers RH, Siggers J, Boye M, et al. Early administration of probiotics alters bacterial colonization and limits diet-induced gut dysfunction and severity of necrotizing enterocolitis in preterm pigs. J Nutr. 2008;138:1437–1444. doi: 10.1093/jn/138.8.1437. [DOI] [PubMed] [Google Scholar]
- 151.De Plaen IG, Liu SX, Tian R, et al. Inhibition of nuclear factor-kappaB ameliorates bowel injury and prolongs survival in a neonatal rat model of necrotizing enterocolitis. Pediatr Res. 2007;61:716–721. doi: 10.1203/pdr.0b013e3180534219. [DOI] [PubMed] [Google Scholar]
- 152.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lin PW, Stoll BJ. Necrotising enterocolitis. Lancet. 2006;368:1271–1283. doi: 10.1016/S0140-6736(06)69525-1. [DOI] [PubMed] [Google Scholar]
- 154.Versalovic JWM. Therapeutic microbiology: probiotics and related strategies. Washington, DC: ASM Press; 2008. [Google Scholar]
- 155.McGuire AL, Colgrove J, Whitney SN, et al. Ethical, legal, and social considerations in conducting the Human Microbiome Project. Genome Res. 2008;18:1861–1864. doi: 10.1101/gr.081653.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Braga-Neto MB, Warren CA, Oria RB, et al. Alanyl-glutamine and glutamine supplementation improves 5-fluorouracil-induced intestinal epithelium damage in vitro. Dig Dis Sci. 2008;53:2687–2696. doi: 10.1007/s10620-008-0215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Becker RM, Wu G, Galanko JA, et al. Reduced serum amino acid concentrations in infants with necrotizing enterocolitis. J Pediatr. 2000;137:785–793. doi: 10.1067/mpd.2000.109145. [DOI] [PubMed] [Google Scholar]
- 158.Louis P, Scott KP, Duncan SH, et al. Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol. 2007;102:1197–1208. doi: 10.1111/j.1365-2672.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- 159.Duncan SH, Hold GL, Barcenilla A, et al. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int J Syst Evol Microbiol. 2002;52:1615–1620. doi: 10.1099/00207713-52-5-1615. [DOI] [PubMed] [Google Scholar]
- 160.Focareta A, Paton JC, Morona R, et al. A recombinant probiotic for treatment and prevention of cholera. Gastroenterology. 2006;130:1688–1695. doi: 10.1053/j.gastro.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 161.Carroll IM, Andrus JM, Bruno-Barcena JM, et al. Anti-inflammatory properties of Lactobacillus gasseri expressing manganese superoxide dismutase using the interleukin 10-deficient mouse model of colitis. Am J Physiol Gastrointest Liver Physiol. 2007;293:G729–G738. doi: 10.1152/ajpgi.00132.2007. [DOI] [PubMed] [Google Scholar]
- 162.Braat H, Rottiers P, Hommes DW, et al. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin Gastroenterol Hepatol. 2006;4:754–759. doi: 10.1016/j.cgh.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 163.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 164.Guerrant RL, Oria RB, Moore SR, et al. Malnutrition as an enteric infectious disease with long-term effects on child development. Nutr Rev. 2008;66:487–505. doi: 10.1111/j.1753-4887.2008.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Petri WA, Jr, Miller M, Binder HJ, et al. Enteric infections, diarrhea, and their impact on function and development. J Clin Invest. 2008;118:1277–1290. doi: 10.1172/JCI34005. [DOI] [PMC free article] [PubMed] [Google Scholar]


