Abstract
The lone star tick (Amblyomma americanum) is an abundant and aggressive biter of humans, domestic animals, and wildlife in the southeastern-central USA and an important vector of several known and suspected zoonotic bacterial pathogens. However, the biological drivers of bacterial community variation in this tick are still poorly defined. Knowing the community context in which tick-borne bacterial pathogens exist and evolve is required to fully understand the ecology and immunobiology of the ticks and to design effective public health and veterinary interventions. We performed a metagenomic survey of the bacterial communities of questing A. americanum and tested 131 individuals (66 nymphs, 24 males, and 41 females) from five sites in three states. Pyrosequencing was performed with barcoded eubacterial primers targeting variable 16S rRNA gene regions 5–3. The bacterial communities were dominated by Rickettsia (likely R. amblyommii) and an obligate Coxiella symbiont, together accounting for 6.7–100% of sequences per tick. DNAs from Midichloria, Borrelia, Wolbachia, Ehrlichia, Pseudomonas, or unidentified Bacillales, Enterobacteriaceae, or Rhizobiales groups were also detected frequently. Wolbachia and Midichloria significantly co-occurred in Georgia (p<0.00001), but not in other states. The significance of the Midichloria-Wolbachia co-occurrence is unknown. Among ticks collected in Georgia, nymphs differed from adults in both the composition (p = 0.002) and structure (p = 0.002) of their bacterial communities. Adults differed only in their community structure (p = 0.002) with males containing more Rickettsia and females containing more Coxiella. Comparisons among adult ticks collected in New York and North Carolina supported the findings from the Georgia collection despite differences in geography, collection date, and sample handling, implying that the differences detected are consistent attributes. The data also suggest that some members of the bacterial community change during the tick life cycle and that some sex-specific attributes may be detectable in nymphs.
Introduction
Ticks transmit a greater diversity of pathogens to humans and domestic animals than any other vector group [1], and the lone star tick (Amblyomma americanum) is the most common human-biting tick in the southeastern United States [2]. During the last century A. americanum has expanded its range into the northeastern and mid-western states [3]–[6], further increasing the number of people and domestic animals exposed to the tick and the pathogens it transmits.
Amblyomma americanum is a three-host tick that is nonspecific in its host use in the immature (larval and nymphal) life stages and has a preference for white-tailed deer (Odocoileus virginianus) as adults (reviewed in [6]). As many pathogens are acquired by ticks from their vertebrate hosts, the lack of host specificity throughout much of its life perfectly positions this tick as a vector of multiple zoonotic diseases, including ehrlichioses [7], rickettsioses, tularemia, and perhaps even Southern Tick-Associated Rash Illness (reviewed in [6]). Other bacteria of undefined pathogenicity (Candidatus “Midichloria mitochondrii” [8], [9], Borrelia lonestari [10]) and several viruses (Lone Star virus [11], Heartland virus [12]) are also associated with this species. Larvae, nymphs, and adult females feed once from a single host before becoming quiescent in the leaf litter and either molting into the next life stage (immatures) or ovipositing (adult females). Adult males take multiple smaller blood meals while seeking feeding females [13].
Tick-borne disease ecology motivates many tick microbiological studies, which frequently use specific assays to survey for the presence of known vertebrate pathogens. Yet many vertebrate pathogens are neither prevalent in the vector population (e.g. [14]) nor abundant within pathogen-infected ticks (e.g. [15]) and may represent only a minority population within the tick microbiome. This suggests that the interactions between vertebrate pathogens and other more common and abundant bacteria found in ticks may be important to the abundance and distribution of pathogen-infected ticks in the environment. Several previous surveys of the bacterial communities of A. americanum have provided useful inventories of community members. Sanger sequencing by Clay et al. [16] and Heise et al. [17] and pyrosequencing by Ponnusamy et al. [18] and Yuan [19] of bacterial 16S rRNA gene fragments from A. americanum confirmed the common presence of a Coxiella that is a likely obligate symbiont [20], [21]. They also detected the low pathogenicity spotted fever group Rickettsia Candidatus “Rickettsia amblyommii” (hereafter R. amblyommii) as an abundant and common bacterial community member. Additional Rickettsia species [17] and a novel Arsenophonus were also identified [16], [19], as well as a number of diverse taxa frequently found both environmentally and as animal associates, most commonly Pseudomonas [16], [17], [19], Enterobacteracea [16], [17], and Bacillaceae [17], [19].
While the A. americanum bacterial community has been well documented, the nature and basis for intraspecific differences are less well explored. Menchaca et al. [22] used semi-conductor sequencing to describe changes in the bacterial communities of laboratory colony derived nymphs and adults to test the effects of engorgement, molting, age, and environmental conditions on bacterial communities. They found an interesting trend towards reduction in community diversity with tick age and environmental stressors under extremely controlled conditions. When pyrosequencing bacterial 16S amplicons from a small sample of 12 A. americanum, Ponnusamy et al. found more unique bacterial taxa in males and nymphs than in females. In other ixodid tick species, tick life stage and sex have been used to explain variation in whole bacterial communities [23], [24] and focal bacterial taxa [25]–[27], although the mechanisms behind these differences remain unclear.
Here we characterize the life stage and sex specific bacterial communities of wild caught A. americanum collected at five sites in three states using pyrosequencing of variable regions 5–3 of the bacterial rrs (16S rRNA) gene. Samples of nymphs and adults from two sites in Georgia were compared to detect differences in community composition and structure between life stages and sexes from a single geographic area. Archived DNAs from adult A. americanum were also compared to see if sex-specific bacterial communities were a general phenomenon in this species or a region-specific occurrence. The geographical distribution of our sample sites in combination with our larger sample sizes enabled the identification of significant life stage and sex specific patterns of bacterial community composition and structure that have not been reported from previous studies.
Methods
Ethics statement
Tick collection in Georgia was performed in state parks with permission from the Georgia Department of Natural Resources (permits #29-WBH-10-135 and #29-WBH-11-49) and park management. Ticks from other states were collected as described in Mixson et al. [14].
Tick collection and DNA extraction
All A. americanum ticks were collected by running a 1 m2 flannel cloth over vegetation. The primary tick DNA sample set (hereafter referred to as the Georgia collection) was collected in 2010–2011 from two ecologically similar forested state parks very near to Atlanta, Georgia (Table 1) and stored in 70% ethanol at 4°C. DNA extraction was performed as described in Bermúdez et al. [28] with the Promega (Madison, WI, USA) Wizard SV 96 Genomic DNA Purification System. Ticks were treated with sequential pre-extraction surface washes of 10% bleach, 70% ethanol, and distilled water to reduce surface contamination. DNAs were stored at 4°C until used. Tick DNAs from our archives (hereafter referred to as the archival collection) were obtained from ticks from three sites on barrier islands in New York (1 site) and North Carolina (2 sites) in 2002–2003 (Table 1), stored whole at −20°C, and then individually processed for DNA extraction using the QIAamp Mini Kit (Qiagen, Hilden, Germany) as described in Mixson et al. [14]. These tick samples were not pre-treated prior to DNA extraction to reduce surface contaminants as they were collected and processed under older protocols for a different purpose than this experiment. DNA extraction was performed during 2002–2004, and the DNAs were stored at 4°C until used in this study. DNAs extracted following this manufacturer’s protocols are stable at least 16 years at 4°C [29].
Table 1. Life stage and geographic origin of Amblyomma americanum ticks used in bacterial community analyses.
| Tick DNA Set | Collection Site (Year) | Nymph | Adult Male | Adult Female | Total |
| Archival | Bodie Island, NC (July 2002) | NT | 5 | 5 | 10 |
| Archival | Buxton Woods, NC (July 2002) | NT | 4 | 5 | 9 |
| Archival | Shelter Island, NY (July 2003) | NT | 3 | 5 | 8 |
| Archival Total | NT | 12 | 15 | 27 | |
| Georgia | Panola Mountain, GA (2010–11)§ | 30 | 8 | 14 | 52 |
| Georgia | Sweetwater Creek, GA (2010–11)* | 36 | 4 | 12 | 52 |
| Georgia Total | 66 | 12 | 26 | 104 |
Nymphs collected in July 2010; adults in July 2010 and May 2011.
*Nymphs collected in August 2010; adults in May 2011.
NT = Not tested.
454 library preparation and sequencing
PCR primer and barcode designs were obtained from the Human Microbiome Project’s 16S 454 Sequencing Protocol. The primer sequences included eubacterial rrs primers 357F and 926R, which target variable regions 5–3 [30]. Five prime modifications of primers were made for compatibility with the Roche (Indianapolis, Indiana, USA) 454 sequencer’s Titanium chemistry. Modified primer sequences were F: 5′-CTA TGC GCC TTG CCA GCC CGC TCA GCC TAC GGG AGG CAG CAG-3′ and R: 5′-CGT ATC GCC TCC CTC GCG CCA TCA G (barcode) CCC GTC AAT TCM TTT RAG T-3′. Barcode sequences are provided in the supporting information (Table S1). The expected amplicon size was approximately 643 bp.
For PCR, each 20 µL reaction mix contained either 1x AccuPrime PCR Buffer II (Invitrogen, Carlsbad, California, USA), 0.5 U AccuPrime Taq High Fidelity, 13.5 µL water, 0.3 µM each forward and reverse primer, and 2 µL DNA or 15 µL Platinum PCR SuperMix High Fidelity (Invitrogen), and 1 µL water. We have found no significant differences in bacterial community composition when using these two enzymes for primary PCR (data comparing tick DNAs amplified with both polymerases are described further below and in the results). All reactions were performed in an Eppendorf Master Gradient thermocycler (Brinkmann Instruments, Westbury, New York, USA) with the following program: one step of 94°C (2 min), 35 cycles of 94°C (30 s), 50°C (30 s), and 68°C (1 min), and one step of 68°C (5 min), followed by holding at 4°C. Amplicons were analyzed by electrophoresis on 1% agarose gels stained with ethidium bromide to ensure reaction success and then quantitated using the Quant-iT PicoGreen dsDNA kit (Invitrogen) modified from the manufacturer’s protocol to 50% of the recommended final assay volume. Equal quantities of each PCR product were pooled, and the pool was purified using the Wizard SV Gel and PCR Clean-Up System (Promega). The pool was sequenced by the Centers for Disease Control and Prevention (CDC) Biotechnology Core Facility Branch on a Roche 454 GS-FLX sequencer.
To determine if the two polymerases that we used differentially biased the bacterial communities identified from these tick DNAs, we amplified 50 tick DNAs belonging to the archival collection once with each of the polymerases used in this experiment as described above. Tick DNA and barcode pairings were held constant between the libraries. A single library was produced from the AccuPrime amplicons, and two libraries were produced from the Platinum amplicons to control for variability introduced by random sampling, pipetting error, and bead loading on the 454 sequencer. Each library was sequenced on one quarter of a plate (Table S2). Twenty-six of 50 DNAs were sequenced to a depth of at least 1000 high quality reads across all three treatments and were used in the analysis of polymerase effects.
Life stage effects were analyzed using amplicons from six quarters from three sequencing plates (Table S2). Some DNAs were sequenced more than once, but no duplicates were used in the analyses. Samples not analyzed here were replicates of reported samples, belonged to variable groups that were not sufficiently sampled, or returned insufficient numbers of high quality sequences for further analyses.
Bioinformatic analysis
The bioinformatic pipeline was executed in the software package mothur (version 1.31.2) [31]. The quality control pipeline was modeled on the Schloss SOP pipeline available on the mothur website [32]. Briefly, flow data were denoised and the resulting sequences were discarded if the reverse primer sequence or barcode could not be identified, or if barcode or primer sequences had more than 1 or 2 errors, respectively. Sequences were also discarded that contained homopolymer runs >8 bp or ambiguous nucleotide calls. Sequence trimming was performed based on a sliding window of 50 bp with a threshold quality score of 35. The remaining sequences were aligned to the SILVA SEED bacterial 16S database [33] using 8mer searching to choose a template sequence and the Needleman-Wunsch algorithm to perform the alignment. The SILVA SEED database was modified to include several sequences of interest, eg. Arsenophonus and Candidatus ‘Midichloria mitochondrii’. Aligned sequences were then screened by starting positions to eliminate sequences that mapped outside the target region of the rrs. Finally, chimeric sequences were identified using the chimera.uchime function in mothur and then removed from further analyses.
All sequences passing the quality control pipeline were assigned to operational taxonomic units (OTUs) as described in the Schloss SOP. To summarize, all high quality sequence reads were ranked by abundance and rare reads differing from abundant reads by a single base were assumed to be sequencing errors and clustered with their similar sequence. An uncorrected pairwise distance matrix was calculated between the aligned sequence reads; both internal and terminal gaps were penalized once. Reads were then clustered into OTUs using the average neighbor method at a 3% distance. Taxonomic assignment of individual reads was determined by using mothur’s Bayesian classifier and requiring a bootstrap confidence of 80% on 100 iterations (kmer size = 8). A consensus taxonomy was determined for each OTU based on the individual sequence taxonomies within each. Genus level identification is provided whenever possible; otherwise the lowest taxonomic rank applying to all members of the OTU is given. Singleton OTUs were removed prior to statistical analyses [34]. Representative highest abundance sequences from OTUs are deposited in GenBank under accession numbers KJ130495–KJ130517; additional information can be found in Table S3. The full data set is available in the NCBI SRA under study accession number SRP042723.
Bacterial diversity analyses
Good’s community coverage estimate [35], rarefaction curves, and alpha diversity were calculated in the software mothur using a subsample size of 1000 sequences and 1000 iterations. The inverse Simpson diversity index (1/D) was used for alpha diversity because it has a clear biological interpretation and is less affected by sample size [36] and the presence of spurious OTUs [37] than similar measures. Analysis of variance (ANOVA) and Tukey’s honestly significant difference (HSD) post hoc tests were performed on the R statistical platform (version 3.0.1) [38].
Bacterial community analyses
We used community ecology methods to analyze both the OTU composition (presence/absence) and structure (abundance) of bacterial communities observed within each tick. As no one metric adequately compares all aspects of community assemblages, we measured the distance between bacterial communities using two metrics that each emphasize different community characteristics. The Jaccard dissimilarity index performs pairwise comparisons of communities based on the presence or absence of OTUs [39], while the Bray-Curtis dissimilarity index weights the abundance of OTUs in its calculations [40]. Each tick bacterial community was rarefied to 1000 sequences, and Jaccard and Bray-Curtis calculations were performed in R with the package “vegan” (version 2.0–7) [41] using the vegdist command.
To partition variation within each distance matrix, we used a non-parametric permutational multivariate analysis of variance (PERMANOVA) as implemented in the vegan function adonis (permutations = 1000) [41]. As PERMANOVA can sometimes be affected by differences in within group variation, each matrix was also tested for homogeneity of group dispersions via the vegan function betadisper [41]. Significance values were obtained by permutation (n = 1000). Corrections for multiple comparisons in the PERMANOVA and dispersion tests were performed using Holm’s method [42].
To visualize data sets and to corroborate the results of the PERMANOVAs, a non-metric multi-dimensional scaling (NMDS) was performed on each distance matrix. This ordination technique represents highly dimensional data by maximizing the correlation in rank-order between the original data set and a two dimensional representation [43], [44]. The relative location of each tick’s bacterial community within the ordination space can be interpreted with the addition of species (treating OTUs as species for this purpose) scores to the plot. Bacterial communities closer to a given species score in ordination space have greater values for that species than those communities located farther away [45]. The ordinations were produced using the function metaMDS in vegan (maximum permutations = 1000) with square root and Wisconsin double standardization transformations used according to default settings [41]. The goodness of fit for each variable’s group centroids were evaluated using the command envfit [41], and the species scores were calculated as weighted averages of the bacterial community scores [41]. Groups are considered significantly different for α = 0.05 if the 95% confidence intervals did not overlap.
To determine which OTUs differed in abundance between groups we used the Metastats method of White et al. [46] as implemented in mothur. In an effort to control type II error rates, we only interpreted those p-values that had an associated false discovery rate (i.e., the proportion of false positives expected in a set of reported significant results) of q≤0.05 [46], [47].
Polymerase effects analysis
For the archival tick DNAs successfully sequenced across the AccuPrime, Platinum 1, and Platinum 2 libraries, we processed the data as described above in bioinformatic analysis, rarefied each community to 1000 sequences, and compared the bacterial communities from each library originating from the same tick DNA. Jaccard and Bray-Curtis dissimilarity values were calculated pairwise for each tick’s three bacterial communities. For each dissimilarity metric an ANOVA was used to compare the means of each of the three comparison groups (Platinum 1 and Platinum 2, Platinum 1 and Accuprime, Platinum 2 and Accuprime) across ticks. The means would differ only if a polymerase produced a differentially biased bacterial community composition or structure as measured by the Jaccard or Bray-Curtis metrics, respectively. Bartlett’s test [48] was used to ensure homogeneity of group variances. All statistics were performed on the R platform.
Co-infection analysis
The co-occurrence of the most abundant OTUs (≥100 sequences in the rarefied data) was evaluated using a probabilistic model to detect pairs that occurred together significantly more or less often than if OTU assemblages were random [49]. The analysis software was kindly provided by the test’s author (v1.0). The Pearson correlation coefficient (r) was calculated using the cor.test function in the R statistical platform [38] with a two-sided alternative hypothesis.
Results
Effect of Polymerase on Bacterial Community Diversity
The Accuprime polymerase library produced 114,250 raw sequences compared to the 82,843 and 93,834 sequences produced by the Platinum polymerase libraries. The distribution of raw sequence lengths (Figure S1), the proportion of sequences removed as sequencing errors, alignment errors, chimeras, and contaminants (Figure S2), and the proportion of sequences retained as high quality sequences (Figure S2) were similar between the polymerases. Bartlett’s test for homogeneity of variances was non-significant for both the Jaccard and Bray-Curtis metrics. ANOVA detected no difference in the mean level of dissimilarity between bacterial communities originating from the same tick DNA but amplified with different polymerases (Platinum 1 and Accuprime, Platinum 2 and Accuprime) and bacterial communities that originated from the same amplification of the same tick DNA (Platinum 1 and Platinum 2) (Jaccard: F(2,75) = 2.74, p>0.07; Bray-Curtis: F(2,75) = 1.40, p>0.25). Because the AccuPrime and Platinum SuperMix polymerases did not differentially bias bacterial community detection, bacterial communities amplified using these polymerases were pooled in all life stage analyses below.
Life stage library characteristics
We obtained 350,501 high quality 16S rRNA gene sequence reads from 131 A. americanum tick samples from five sites (Tables 1 and S2). These sequences belonged to two tick DNA sample sets, referred to as the archival and Georgia tick DNA sets. Additional tick DNA samples sequenced on the same plates were not included in the analyses because they were inferior replicates of retained tick samples, produced low sequence yields (<1000), or belonged to variable groups that were insufficiently sampled for analysis. The data were dominated by sequences from high-abundance OTUs belonging to Rickettsia and Coxiella, which comprised 35.0% and 59.4% of sequences, respectively, and were found in 88.5% and 99.2% of the ticks, respectively. The sequence reads averaged 255.6 bp in length after trimming and comprised 408 operational taxonomic units (OTUs) when clustered at 97% identity. One hundred seventy OTUs were comprised of a single sequence (singletons) and were removed [34]. To facilitate comparison of bacterial communities between samples, each bacterial community was then rarefied to 1000 sequences. This reduced the total number of OTUs to 212, of which 42 contained only a single sequence. The 170 OTUs containing >1 sequence represented a minimum of 99 bacterial families and 99 genera.
Bacterial diversity
The Georgia DNA set’s rarefaction curve approached an asymptote, indicating that the available bacterial diversity was well sampled (Figure S3). This conclusion was supported by the Good’s coverage estimate for each tick sample, which had a mean of 0.998 (range 0.994–1.00). In contrast, the bacterial diversity of the archival DNAs, from ticks which were not surface decontaminated prior to DNA extraction, was less well sampled and the rarefaction curve had a steeper slope. However, while adding additional tick DNAs samples to this set would have detected additional OTUs, the communities of each tick in the data set were individually well sampled (Good’s coverage estimate, mean = 0.993, range = 0.976–1.00).
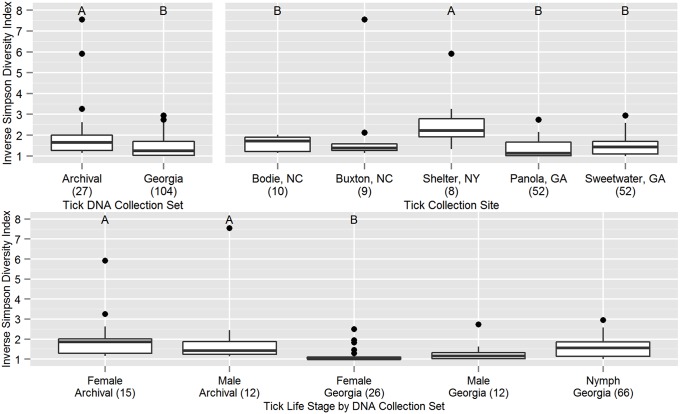
Alpha diversity was estimated using the inverse Simpson index and compared between DNA sample sets and across life stages and collection sites (Figure 1). Significant differences were found for all variables considered (ANOVA with Holm’s correction for multiple comparisons, DNA collections: F(1,129) = 15.9, p = 0.00023; life stages: F(4,126) = 5.21, p = 0.00064; sites: F(4,126) = 6.83, p = 0.00016). Among life stages from the different DNA collection sets, adult females from Georgia had lower alpha diversity than adult males (Tukey’s HSD, p = 0.013) and females (p = 0.0017) from the archival DNA collection. Comparisons across collection sites found a higher alpha diversity among ticks from Shelter Island compared to those from Bodie Island (Tukey’s HSD, p = 0.022), Sweetwater Creek (p = 0.00059) and Panola Mountain (p = 6.9×10−5).
Figure 1. Comparisons of bacterial community alpha diversity by tick DNA set, life stage, and collection site.
Box and whiskers plots represent the first and third quartiles (hinges), the median (bold line), and 1.5 times the interquartile range (error bars). Outlier points are plotted individually. Within each of three panels, n = 131 (see Table 1) and groups labeled A are significantly different from groups labeled B at α = 0.05 (Tukey’s HSD). Group sample sizes are given in parentheses after the group name. For the tick DNA sample set comparison, the archival tick DNAs were older and processed under different extraction protocols than the Georgia samples.
Bacterial community composition and structure
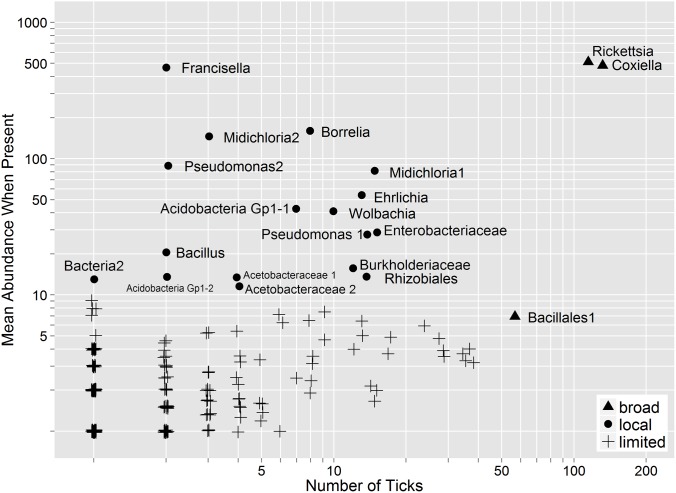
Of the 212 OTUs analyzed across both DNA collections, only Rickettsia, Coxiella, and a Bacillales group were observed in more than one third of all standardized samples (Figure 2). An additional 17 OTUs composed ≥1% of the community when detected, but were detected in one third or fewer of the tick DNA samples. No individual OTU was present in all rarefied samples, including Coxiella, a proposed obligate symbiont of A. americanum [21]. Of the two negative samples, the male from Panola Mountain was Coxiella positive prior to rarefaction, while the nymph from Sweetwater Creek was positive in a replicate sequencing run (data not shown).
Figure 2. Bacterial operational taxonomic unit (OTU) sequence abundance detected in tick samples.
OTUs present in greater than one third of all tick samples were considered broadly distributed. OTUs present at a mean abundance ≥1% of the community (i.e. 10 sequences) were considered locally abundant. Sequence abundance values are slightly offset to reveal overlapping OTUs.
DNAs from ticks collected in Georgia differed from those in the archival DNA collection in the age of the samples, the tick life stages represented, and the DNA extraction protocols used. Bacterial community structure was quite different between these two groups of ticks (PERMANOVA on Bray-Curtis distance matrix, F = 5.87, R2 = 0.044, p = 0.012) (Figure S4), but they did not differ in their dispersion (F = 1.09, p = 0.28). The bacterial communities of the Georgia tick DNAs contained 12 OTUs not found in the archival tick DNA bacterial communities, while the archival tick DNAs contained 61 OTUS not found among the Georgia tick bacterial communities. An additional 7 OTUs were present in both DNA collections but differentially abundant (Table S4). Greater than 93% (75/80) of the OTUs that were differentially abundant between the Georgia and archival tick DNAs have no confirmed association with the internal microbiota of ticks and therefore may have been derived from the external environment.
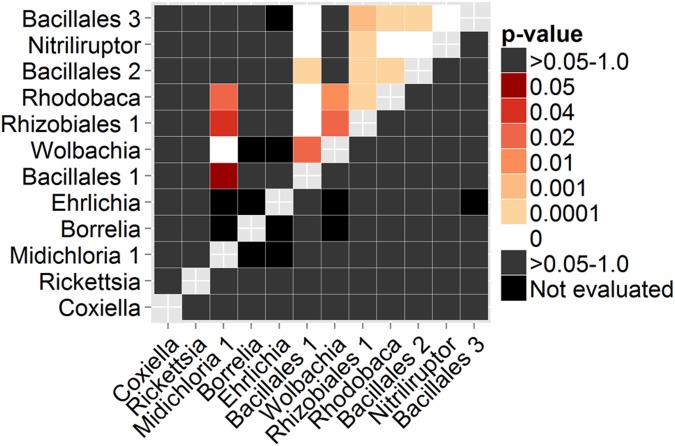
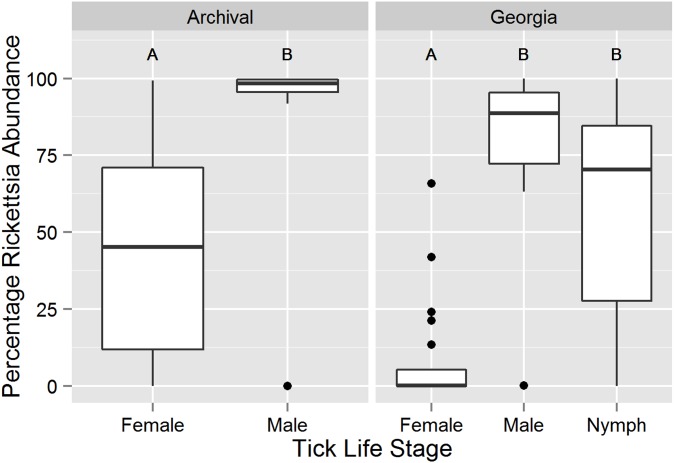
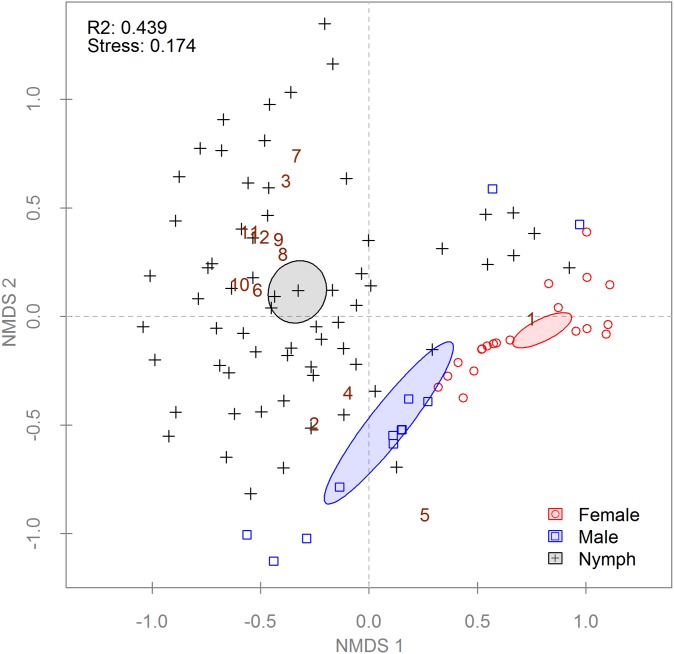
Twenty-two positive associations were detected among the 12 most abundant (>100 sequences) OTUs from the Georgia DNA collection (Figure 3). The Bacillales 1, Rhodobaca, and Rhizobiales 1 OTUs had the largest number of associations. The Wolbachia and Midichloria 1 OTUs were strongly positively associated. Two positive associations were found among the most abundant OTUs from the archival DNA collection (Rhizobiales and Acidobacteria Gp1, Burkholderiaceae and Methylobacterium, p<0.04). In general no significant associations were detected between the Rickettsia and Coxiella OTUs and the other OTUs due to the near ubiquity of Rickettsia and Coxiella in these samples. However, there was a significant inverse relationship between the abundance of Rickettsia and Coxiella across both sample sets (Georgia: n = 104, Pearson’s r = −0.976, p = 2.2×10−16; archival: n = 27, Pearson’s r = −0.689, p = 6.97×10−6). The life stages differed in the dominant bacteria (ANOVA on percentage Rickettsia out of total Rickettsia and Coxiella sequences, Georgia: F(2,101) = 26.8, p = 4.68×10−10; archival: F(1,25) = 14.0, p = 0.00097) (Figure 4). Within the Georgia collection, females differed from males and nymphs (Tukey’s HSD, males: p = 1.0×10−7, nymphs: p<1.0×10−7) (Figure 4).
Figure 3. Significance of co-infection patterns between selected operational taxonomic units (OTUs).
Only data from the Georgia tick DNA sample collection is shown. The upper triangle represents p-values of positive associations (codetection in the same sample) between OTUs and the lower triangle the p-values of negative associations. Black cells were not evaluated due to low expected co-occurrence (<1) between the OTUs [49].
Figure 4. Variation in percentage of Rickettsia among total Rickettsia and Coxiella sequences by tick life stage.
The number of unstandardized Rickettsia and Coxiella sequences from each tick sample (Georgia n = 104, archival n = 27) were summed and the percentage of Rickettsia sequences were calculated. Box and whiskers plots represent the first and third quartiles (hinges), the median (bold line), and 1.5 times the interquartile range (error bars). Outlier points are plotted individually. Within each DNA collection, life stages labeled with different letters are significantly different at α = 0.05.
Life stage and sex specific bacterial communities
Ticks from the Georgia collection were analyzed to detect differences between female, male, and nymphal bacterial communities (n = 104). Samples from Sweetwater Creek and Panola Mountain, Georgia were pooled because no differences were detected between the sites (data not shown). PERMANOVA found significant differences between life stages and sexes using both distance metrics (Table 2). Differences in within group variation (i.e. dispersion) were also detected with both distance matrices (Table 2) [50]. Results from the PERMANOVA were corroborated using ordination by NMDS. When only the presence or absence of OTUs was considered (Figure S5), nymphal communities were different from adults, but male and female communities did not differ. However, differences were detected between males and females when the ordination was weighted for OTU abundance (Figure 5). Comparison of the mean relative abundance of OTUs detected 8 of 13 which were differentially abundant between the tick life stages and sexes (Figure S6).
Table 2. Multivariate analysis of the effect of sex and life stage on the relatedness of Amblyomma americanum bacterial communities.
| PERMANOVA | Dispersion Test | ||||||
| Collection | Distance Metric | Metric Type | F | R2 | p * | F | p * |
| Georgia | Jaccard | Community composition | 17.3 | 0.255 | 0.0020 | 12.5 | 0.0020 |
| Georgia | Bray-Curtis | Community structure | 24.9 | 0.330 | 0.0020 | 8.77 | 0.0020 |
| Archival | Jaccard | Community composition | 1.63 | 0.0611 | 0.026 | 0.533 | 0.96 |
| Archival | Bray-Curtis | Community structure | 8.28 | 0.249 | 0.0020 | 0.345 | 0.96 |
Tick DNAs from the archival collection (n = 27) were older, collected in a different geographical region, and processed using different protocols than tick DNAs from the Georgia collection (n = 104).
*Corrected for multiple comparisons using Holm’s method; α = 0.05.
Figure 5. Non-metric multi-dimensional scaling (NMDS) of a Bray-Curtis distance matrix describing Georgian Amblyomma americanum bacterial communities.
Each point symbolizes a single tick’s community (n = 104); some points may overlap completely. Point and ellipse colors indicate life stage; ellipses represent 95% confidence intervals around life stage centroids. Non-overlapping centroids are considered significantly different at α = 0.05. R2 values in the upper left corner of plots describe the amount of variation in the data set explained by the groupings. The stress value given is a measure of the disagreement between the rank order in the original data set and that in the NMDS (lower numbers indicate better agreement). Brown numbers indicate the species scores for select OTUs as follows: (1) Coxiella, (2) Rickettsia, (3) Midichloria 1, (4) Borrelia, (5) Ehrlichia, (6) Bacillales 1, (7) Wolbachia, (8) Rhizobiales 1, (9) Rhodobaca, (10) Bacillales 2, (11) Nitriliruptor, (12) Bacillales 3.
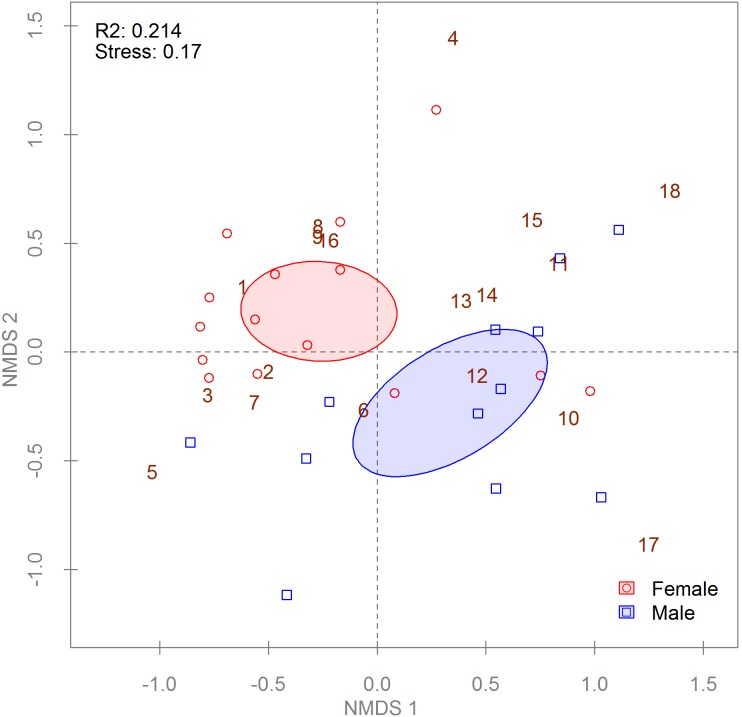
To determine if an effect of sex on tick bacterial communities could be detected in other parts of the A. americanum range, we applied PERMANOVA to Jaccard and Bray-Curtis distance matrices with data from the archival DNA samples from adult ticks collected outside of Georgia (n = 27) (Table 1). Ticks from all three sites were pooled for analysis because no site effect was detected (data not shown). PERMANOVA found significant differences for both distance matrices (Table 2), but NMDS found there was a difference between males and females only in the relative abundance of bacterial community members (Bray-Curtis metric, Figure 6) and not in community membership (Jaccard metric, Figure S7). Analysis of the OTUs containing ≥100 sequences each found 2 of 18 were differentially abundant between groups, with females possessing significantly higher Coxiella abundance and having higher Francisella abundance (p<0.001).
Figure 6. Non-metric multi-dimensional scaling (NMDS) of a Bray-Curtis distance matrix describing archival Amblyomma americanum bacterial communities.
Each point symbolizes a single tick’s community (n = 27); some points may overlap completely. Point and ellipse colors indicate life stage; ellipses represent 95% confidence intervals around life stage centroids. Non-overlapping centroids are considered significantly different at α = 0.05. R2 values in the upper left corner of plots describe the amount of variation in the data set explained by the groupings. The stress value is a measure of the disagreement between the rank order in the original data set and that in the NMDS (lower numbers indicate better agreement). Numbers indicate the species scores for the plot as follows: (1) Coxiella, (2) Rickettsia, (3) Midichloria 1, (4) Borrelia, (5) Francisella, (6) Midichloria 2, (7) Ehrlichia, (8) Enterobacteriaceae, (9) Pseudomonas 1, (10) Burkholderiaceae, (11) Acidobacteria Gp1, (12) Pseudomonas 2, (13) Rhizobiales 2, (14) Bacillus, (15) Acetobacteraceae 1, (16) Acetobacteraceae 2, (17) Bacteria 1, (18) Bacteria 2.
Discussion
454 pyrosequencing of barcoded rrs variable region 5–3 amplicons was used here to characterize the bacterial communities of 131 individual A. americanum ticks, including nymphs and adult males and females. The ticks were all questing, field-collected individuals obtained from five sites in three states; as such they are probably at least representative of the diversity of bacterial communities that occur in A. americanum found in the eastern states comprising its range.
The exact relationship of 454 sequence read abundance to the biological abundance of a given bacteria in the community is difficult to prove as amplification efficiency varies both between bacterial targets (e.g. template GC content [37], primer-template matching [51], 16S copy number variation per chromosome and per cell) and laboratory protocols (e.g. primer bar-code bias [52], PCR conditions [53]). In this system, while previous work has shown a good correlation between 454 read abundance and targeted qPCR quantitation [54], it is not currently possible to confirm such a relationship for all potential members of the A. americanum bacterial community. In general, while comparing communities across protocols is problematic due to the many potential sources of bias, 454 metagenome results are reproducible for a given protocol [37], [55], [56]; however, some authors have found that this is not always true [34].
We document here significant differences in the bacterial communities of the life stages and sexes of questing A. americanum. Males and females in both the Georgia and archival DNA collections differed in their bacterial community structure (Figures 5 and 6) but not composition (Figures S5 and S7). However, nymphs differed from adults in both bacterial community composition and structure (Figures S5 and 5, respectively), with many of the more abundant OTUs appearing primarily in nymphs (Figure S6). Many, but not all, of these OTUs belonged to taxa with both environmental and arthropod-associated members. While the nymphs from Georgia were surface decontaminated prior to DNA extraction, our method was unlikely to remove all exoskeleton-associated bacteria. The higher surface to volume ratio of nymphs compared to adults also provides a larger relative area for contamination. However, Menchaca et al. [22] found high levels of Bradyrhizobiaceae in both nymphs and adults derived from laboratory colonies and held indoors, suggesting a more persistent association for at least one of these taxa.
As we expected from previous investigations of the bacterial community of A. americanum [14], [16], [19], [22], [57], [58] (Table S3), Rickettsia and Coxiella were by far the dominant OTUs identified in our samples across all life stages, sexes, and DNA collections. The A. americanum-associated Coxiella is regarded as an obligate symbiont of the lone star tick due to its ubiquitous presence in previous surveys, limited evidence for a reduced genome size relative to the free-living relative (Coxiella burnetii), transovarial and transstadial maintenance, and the reduced viability and fecundity of antibiotic treated ticks [16], [20], [21], [59]. While one tick had no detectable Coxiella sequences in the data presented here, we later detected low levels of Coxiella in a replicate data set. This suggests that the abundance of Coxiella was at the limit of detection for 454 in this sample. Our data provide further support for the hypothesis that Coxiella is an obligate symbiont of A. americanum, although its population size in some individuals may be very low relative to other members of the bacterial community.
Fragments of the rrs are insensitive for the identification of different species of Rickettsia. However, abundant previous work has shown that the most common Rickettsia in A. americanum is R. amblyommii (eg. [14], [16], [57], [58], [60], [61]), a member of the spotted fever group with poorly defined human pathogenicity [62]–[64]. Despite efficient vertical [65] and probable horizontal (reviewed in [66]) transmission in A. americanum, infection rates in populations across the tick’s range vary widely (0–84%) [14], [16], [60], [61]. Our archival tick DNAs (Table 1) had been previously tested for R. amblyommii using direct PCR of the gene encoding the rOmpA protein followed by restriction fragment length polymorphism analysis; techniques and data were described in Mixson et al. [14]. Of these 27 adult ticks, two were negative by direct PCR but contained at least a few Rickettsia sequences when analyzed here (n = 1–160 sequences per tick, original unrarefied data). While R. amblyommii generally occurs as a high density, disseminated infection in A. americanum [67], taken together these data indicate that some R. amblyommii infections are below the detection limit of direct PCR and may not always be reliably detected by that insensitive technique (this has been affirmed by qPCR comparison of varying and often low R. amblyommii levels in other tick samples, Dasch unpublished results). Rickettsia amblyommii infection rates are therefore likely to be somewhat higher than has been previously reported, although whether or not the bacterium is actually ubiquitous or transcriptionally active in all samples remains to be determined. If naturally uninfected ticks exist, these low level infections may represent individuals that had acquired the bacterium during the preceding blood meal.
There was a strong inverse relationship between the abundance of Rickettsia and Coxiella in the bacterial communities of the adult A. americanum ticks from both the Georgia and archival DNA collections. Interestingly, the dominant bacterium varied with the sex of adults such that females produced more Coxiella sequences and males more Rickettsia sequences (Figure 4). The relatively higher Coxiella to Rickettsia ratio observed in females is supported by the qPCR data of Jasinskas et al. [20], but differs from the report of Ponnusamy et al. [18], who detected an increase in Rickettsia in females and low levels of Coxiella across all A. americanum samples using pyrosequenced V1-3 rrs fragments. Given the extensive evidence for Coxiella’s abundance in most A. americanum samples [16], [17], [19], [22] and the small sample analyzed by Ponnusamy et al., it is likely that either some bias against Coxiella existed in the Ponnusamy et al. protocol or that a non-representative sample was drawn from their population. If Coxiella is obligate for the survival of A. americanum [21], the number of bacteria may increase in females to ensure 100% transmission to eggs. An alternative, but not mutually exclusive, explanation is that males may contain higher numbers of R. amblyommii to facilitate increased locomotion [68], which would increase reproductive fitness if it enabled more mating opportunities for males. The distribution of nymphal bacterial communities across the gradient from Rickettsia dominated to Coxiella dominated suggests that sex-biased bacterial communities may arise in unfed questing nymphs (Figure 4). It will be difficult to prove this hypothesis until molecular sexing tools are available for A. americanum.
The only previously documented difference in A. americanum bacterial communities was decreased species richness in females relative to males and nymphs [18], but similar patterns to those reported here have been noted for other ixodid tick species. In Rhipicephalus turanicus, Coxiella sequences composed 90% of the total detected by bacterial 16S pyrosequencing in both males and females, but Rickettsia were relatively more abundant in infected males than in females [25]. Males also had greater bacterial richness than females, although there was no difference in diversity. In Ixodes scapularis, Moreno et al. used temporal temperature gradient gel electrophoresis of bacterial 16S amplicons to “finger print” the bacterial communities of ticks and found differences between nymphs, males, and females [23]. Francisella have been shown to increase in Dermacentor variabilis nymphs relative to larvae using 16S amplicon pyrosequencing [24]. Studies of individual bacteria have also revealed sex and life stage biased prevalence. For example, Candidatus ‘Midichloria mitochondrii’ (hereafter “M. mitochondrii”) is detected in only 40% of males but 100% of all other life stages of Ixodes ricinus [27]. Borrelia and Anaplasma infection rates also differ between male and female I. ricinus [26].
We also identified other potentially important intracellular bacteria in A. americanum. Two OTUs of M. mitochondrii were found, which contained all three of the genotypes detected from A. americanum in our previous work [8]. Operational taxonomic unit Midichloria 1 corresponds to genotypes A and B, and OTU Midichloria 2 is equivalent to genotype C. Wolbachia sequences were also identified in A. americanum from Georgia. Wolbachia have been previously reported in A. americanum by Zhang et al. [69], but a direct comparison is not possible because the two studies used different regions of the rrs gene. Wolbachia sequences were detected in our unrarefied data from 11 nymphs and 2 females collected from the two Georgia sites (Panola Mountain nymphs: 4/30, females: 2/14; Sweetwater Creek nymphs: 7/36, females: 0/12). These infection rates were at least two times greater than the highest minimum infection rates previously observed in nymph and female populations in Maryland [69], which could be a result of the increased sensitivity of Next-Gen sequencing versus direct PCR, the dilution effect of pooling of nymphs in earlier work, or simply a statistical artifact of our smaller sample sizes. As in previous work [69], we detected no Wolbachia-positive males, although our sample sizes for males were lower than those for females and nymphs. These Wolbachia have been previously typed as supergroup F [69], which also contains Wolbachia that infect scorpions [70], lice [71], filarial nematodes [72], and other hosts (reviewed in [73]). Interestingly, 11/13 of these Wolbachia positive ticks were also positive for genotype A of M. mitochondrii, which to date has only been found in Georgia [8]. This co-infection was highly significant, despite the co-clustering of M. mitochondrii genotype A in OTU Midichloria 1 with genotype B (p<0.0001) (Figure 3). There are at least three potential mechanisms that may lead to this degree of co-infection, including (i) stable vertical transmission of both bacteria within one or more tick lineages, (ii) repeated, simultaneous horizontal transmission of both bacteria from a single source, or (iii) differential survival of M. mitochondrii-infected ticks when attacked by Wolbachia-bearing insects or nematodes. Further work will be needed to differentiate between these hypotheses.
The bacterial OTUs that we detected across all tick samples were very similar to those identified by Yuan [19], who also used wild caught ticks and similar sequencing methods. Our results also did not differ substantively from those of Clay et al. [16] or Heise et al. [17], except for the absence of Arsenophonus in our samples [16]. However, our results were generally quite different from the previous work of Menchaca et al. [22], despite having used a similar fragment of the rrs. While both studies detected Coxiella, the ticks from the previous study contained a large proportion of taxa found rarely or not at all in our data, such as Clostridia, Caulobacteraceae, and Hyphomicrobiaceae. Whether these differences result from the different methods used by Menchaca et al. (Ion Torrent sequencing of nested PCR products), the use of laboratory raised ticks, or their exclusive feeding of ticks on a domestic chicken remains to be determined. However, some evidence for D. variabilis and I. scapularis ticks suggests that the vertebrate hosts have little effect on bacterial community structure [24], while some laboratory colonies of ticks have been observed to differ in some members of their bacterial community [17].
Most of the bacterial taxa identified in this study are broadly distributed in the environment with members found in soil, plants, and arthropods (Table S3). Many were previously identified in similar deep sequencing studies of other tick species (Table S3), but the low taxonomic resolution provided by short rrs fragments for these very diverse groups does not preclude similar surface contaminants on ticks collected on different continents. The enrichment of bacterial communities from the archival tick DNAs extracted without surface disinfection with potential environmental bacteria (Table S4) suggests that they are indeed contaminants, but the confounding variables of geographic location, DNA extraction methods, and tick life stage prevent definitive analysis here. Previous comparisons of surface decontaminated versus untreated A. americanum adults supports Sphingobacteria as a surface contaminant, but not other high abundance OTUs [22]. Several of these OTUs (Actinobacteria, Pseudomonas, Bacillales, Rhizobiales, and Burkholderiaceae) were also commonly found in A. americanum samples collected in the Midwestern USA [16], Texas and Missouri [19], and laboratory colonies [17]. Further work with bacterial species-specific methods should be undertaken to determine the topological relationship of these bacteria to ticks.
In summary, we have found a pattern of sex-specific bacterial community structure that was present irrespective of geographic origin of our A. americanum samples and significant differences in both the composition and structure of nymphal bacterial communities relative to adults. We also documented a novel association between M. mitochondrii and Wolbachia in ticks from Georgia. Given the innate limitations of the sequencing of rrs amplicon fragments, this metagenomic work has been effective as a discovery tool for future targeted studies. However, the extreme dominance of R. amblyommii and Coxiella in the A. americanum bacterial community makes it difficult to detect and analyze other community members in a statistically robust way. Future metagenomic studies would benefit from targeted depletion of the most prevalent and predominant bacteria to increase detection rates for other rarer but potentially important community members.
Supporting Information
Comparison of raw 454 read length distributions between replicate libraries created with different polymerases. Each library was produced with the same set of DNA templates and barcoded primers. The polymerase varied between panels A and B/C, while B and C were produced from the same amplification products but pooled and sequenced independently. Panel A represents the library created with AccuPrime Taq, and panels B and C represent Platinum SuperMix replicates 1 and 2, respectively. The horizontal dashed line marks 100,000 on the log scaled y-axis on each plot.
(TIF)
Comparison of quality control filtering between replicate 454 libraries created with different polymerases. Each library was produced with the same set of DNA templates and barcoded primers. Libraries Platinum 1 and Platinum 2 where produced from the same amplification products but pooled and sequenced independently. The total number of raw sequences per library is given at the top of each bar. Note that the categories given in the key are in the same order as they appear in the plot; contaminants were present in all three libraries but accounted for <0.04% of each library.
(TIF)
Bacterial operational taxonomic unit rarefaction curve for Amblyomma americanum tick DNA sample sets. The shaded regions represent the conditional 95% confidence interval obtained from 1000 randomizations of the data.
(TIF)
Non-metric multi-dimensional scaling (NMDS) of a Bray-Curtis distance matrix describing Amblyomma americanum bacterial communities. Each of the 131 points symbolizes a single tick’s community; some points may overlap completely. Point and ellipse colors (red = Georgia ticks, black = archival) indicate the tick DNA collection of origin; ellipses represent 95% confidence intervals around group centroids. Non-overlapping centroids are considered significantly different at α = 0.05. The R2 value in the upper left corner of the plot describes the amount of variation in the data set explained by the groupings. The stress value is a measure of the disagreement between the rank order in the original data set and that in the NMDS (lower numbers indicate better agreement). Operational taxonomic units that differ in mean relative abundance between the groups are given in Table S4.
(TIF)
Non-metric multi-dimensional scaling (NMDS) of a Jaccard distance matrix describing Georgian Amblyomma americanum bacterial communities. Each point symbolizes a single tick’s community (n = 104); some points may overlap completely. Point and ellipse colors indicate life stage; ellipses represent 95% confidence intervals around life stage centroids. Non-overlapping centroids are considered significantly different at α = 0.05. R2 values in the upper left corner of plots describe the amount of variation in the data set explained by the groupings. The stress value is a measure of the disagreement between the rank order in the original data set and that in the NMDS (lower numbers indicate better agreement). Brown numbers indicate the species scores for select OTUs as follows: (1) Coxiella, (2) Rickettsia, (3) Midichloria 1, (4) Borrelia, (5) Ehrlichia, (6) Bacillales 1, (7) Wolbachia, (8) Rhizobiales 1, (9) Rhodobaca, (10) Bacillales 2, (11) Nitriliruptor, (12) Bacillales 3.
(TIF)
Mean relative abundance of operational taxonomic units (OTUs) from Georgian ticks by life stage. Significant differences (p<0.05) are labeled within rows by the letters that appear in cells. For example, within a row cells labeled A are significantly different from those labeled B or C but not cells labeled A. Empty cells do not differ from any group. Note the discontinuity between the white-black and yellow-red scales.
(TIF)
Non-metric multi-dimensional scaling (NMDS) of a Jaccard distance matrix describing archival Amblyomma americanum bacterial communities. Each point symbolizes a single tick’s community (n = 27); some points may overlap completely. Point and ellipse colors indicate life stage; ellipses represent 95% confidence intervals around life stage centroids. Non-overlapping centroids are considered significantly different at α = 0.05. R2 values in the upper left corner of plots describe the amount of variation in the data set explained by the groupings. The stress value given is a measure of the disagreement between the rank order in the original data set and that in the NMDS (lower numbers indicate better agreement). Numbers indicate the species scores for the plot as follows: (1) Coxiella, (2) Rickettsia, (3) Midichloria 1, (4) Borrelia, (5) Francisella, (6) Midichloria 2, (7) Ehrlichia, (8) Enterobacteriaceae, (9) Pseudomonas 1, (10) Burkholderiaceae, (11) Acidobacteria Gp1, (12) Pseudomonas 2, (13) Rhizobiales 2, (14) Bacillus, (15) Acetobacteraceae 1, (16) Acetobacteraceae 2, (17) Bacteria 1, (18) Bacteria 2.
(TIF)
Barcode sequences used to label eubacterial rrs gene variable regions 5–3 primers for sample multiplexing during pyrosequencing.
(DOCX)
454 sequencing statistics from Titanium FLX plates.
(DOCX)
Summary of previous reports of most abundant operational taxonomic units (OTUs) from Amblyomma americanum .
(DOCX)
Metastats results comparing the differential abundance of bacterial operational taxonomic units between Amblyomma americanum DNA collections.
(XLSX)
Acknowledgments
We thank A. Michael Frace of the CDC Biotechnology Core Facility and Timothy Read of Emory University for their assistance with sequencing, Maria Zambrano of the Rickettsial Zoonoses Branch, CDC for her assistance in the lab, James Taylor of Emory University for his helpful discussions on analysis, and Robert Newkirk for his assistance with graphic preparation. We are grateful to Ellen Dugan, Deema Elchoufi, Ian Fried, Jasmine Hensley, R. Ryan Lash, Robert Newkirk, Gwen Parker, Audrey Zeis, and Gene Zhou for their aid with tick collections in Georgia and DNA extraction. We also thank Scott Campbell and Howie Ginsberg for providing some tick samples used here and described in previous publications. Primer synthesis was kindly provided by the CDC Biotechnology Core Facility. We also thank two anonymous reviewers for their valuable feedback. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Funding Statement
This work was funded in part by the appointment of A. J. Williams-Newkirk to the Research Participation Program at the CDC administered by the Oak Ridge Institute for Science and Education. The remainder of the funding was provided by United States Congressional funding to the Department of Health and Human Services and the Department of Homeland Security. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jongejan F, Uilenberg G (2004) The global importance of ticks. Parasitology 129: S3–S14 10.1017/S0031182004005967 [DOI] [PubMed] [Google Scholar]
- 2. Merten HA, Durden LA (2000) A state-by-state survey of ticks recorded from humans in the United States. J Vector Ecol 25: 102–113. [PubMed] [Google Scholar]
- 3. Ginsberg HS, Ewing CP, O’Connell AF, Bosler EM, Daley JG, et al. (1991) Increased population densities of Amblyomma americanum (Acari: Ixodidae) on Long Island, New York. J Parasitol 77: 493–495 10.2307/3283144 [DOI] [PubMed] [Google Scholar]
- 4. Keirans JE, Lacombe EH (1998) First records of Amblyomma americanum, Ixodes (Ixodes) dentatus, and Ixodes (Ceratixodes) uriae (Acari: Ixodidae) from Maine. J Parasitol 84: 629–631 10.2307/3284739 [DOI] [PubMed] [Google Scholar]
- 5. Means RG, White DJ (1997) New distribution records of Amblyomma americanum (L.) (Acari: Ixodidae) in New York State. J Vector Ecol 22: 133–145. [PubMed] [Google Scholar]
- 6. Childs JE, Paddock CD (2003) The ascendancy of Amblyomma americanum as a vector of pathogens affecting humans in the United States. Annu Rev Entomol 48: 307–337 10.1146/annurev.ento.48.091801.112728 [DOI] [PubMed] [Google Scholar]
- 7. Buller RS, Arens M, Hmiel SP, Paddock CD, Sumner JW, et al. (1999) Ehrlichia ewingii, a newly recognized agent of human ehrlichiosis. N Engl J Med 341: 148–155 10.1056/NEJM199907153410303 [DOI] [PubMed] [Google Scholar]
- 8. Williams-Newkirk AJ, Rowe LA, Mixson-Hayden TR, Dasch GA (2012) Presence, genetic variability, and potential significance of “Candidatus Midichloria mitochondrii” in the lone star tick Amblyomma americanum . Exp Appl Acarol 58: 291–300 10.1007/s10493-012-9582-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bazzocchi C, Mariconti M, Sassera D, Rinaldi L, Martin E, et al. (2013) Molecular and serological evidence for the circulation of the tick symbiont Midichloria (Rickettsiales: Midichloriaceae) in different mammalian species. Parasit Vectors 6: 1–7 10.1186/1756-3305-6-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. James AM, Liveris D, Wormser GP, Schwartz I, Montecalvo MA, et al. (2001) Borrelia lonestari infection after a bite by an Amblyomma americanum tick. J Infect Dis 183: 1810–1814 10.1086/320721 [DOI] [PubMed] [Google Scholar]
- 11. Kokernot RH, Calisher CH, Stannard LJ, Hayes J (1969) Arbovirus studies in the Ohio-Mississippi basin, 1964–1967 VII. Lone Star virus, a hitherto unknown agent isolated from the tick Amblyomma americanum (Linn.). Am J Trop Med Hyg 18: 789–795. [PubMed] [Google Scholar]
- 12. Savage HM, Godsey MS, Lambert A, Panella NA, Burkhalter KL, et al. (2013) First detection of Heartland virus (Bunyaviridae: Phlebovirus) from field collected arthropods. Am J Trop Med Hyg 89: 445–452 10.4269/ajtmh.13-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gladney WJ, Drummond RO (1970) Mating behavior and reproduction of the lone star tick, Amblyomma americanum . Ann Entomol Soc Am 63: 1036–1039. [DOI] [PubMed] [Google Scholar]
- 14. Mixson TR, Campbell SR, Gill JS, Ginsberg HS, Reichard MV, et al. (2006) Prevalence of Ehrlichia, Borrelia, and rickettsial agents in Amblyomma americanum (Acari: Ixodidae) collected from nine states. J Med Entomol 43: 1261–1268 10.1603/0022-2585(2006)431261:POEBAR2.0.CO2 [DOI] [PubMed] [Google Scholar]
- 15.Corrigan J (2012) Investigation of spotted fever group Rickettsia in dogs and ticks in northern California. MS Thesis, Humboldt State University. Available: http://humboldt-dspace.calstate.edu/handle/2148/980?show=full. Accessed 2013 December 31.
- 16. Clay K, Klyachko O, Grindle N, Civitello D, Oleske D, et al. (2008) Microbial communities and interactions in the lone star tick, Amblyomma americanum . Mol Ecol 17: 4371–4381 10.1111/j.1365-294X.2008.03914.x [DOI] [PubMed] [Google Scholar]
- 17. Heise SR, Elshahed MS, Little SE (2010) Bacterial diversity in Amblyomma americanum (Acari: Ixodidae) with a focus on members of the genus Rickettsia . J Med Entomol 47: 258–268 10.1603/me09197 [DOI] [PubMed] [Google Scholar]
- 18. Ponnusamy L, Gonzalez A, Treuren WV, Weiss S, Parobek CM, et al. (2014) Diversity of Rickettsiales in the microbiome of the lone star tick, Amblyomma americanum . Appl Environ Microbiol 80: 354–359 10.1128/AEM.02987-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan D (2010) A metagenomic study of the tick midgut. MS Thesis, The University of Texas. Available: http://digitalcommons.library.tmc.edu/utgsbs_dissertations/85. Accessed 2013 July 17.
- 20. Jasinskas A, Zhong J, Barbour AG (2007) Highly prevalent Coxiella sp. bacterium in the tick vector Amblyomma americanum . Appl Environ Microbiol 73: 334–336 10.1128/aem.02009-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhong JM, Jasinskas A, Barbour AG (2007) Antibiotic treatment of the tick vector Amblyomma americanum reduced reproductive fitness. PLoS One 2: e405 10.1371/journal.pone.0000405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Menchaca AC, Visi DK, Strey OF, Teel PD, Kalinowski K, et al. (2013) Preliminary assessment of microbiome changes following blood-feeding and survivorship in the Amblyomma americanum nymph-to-adult transition using semiconductor sequencing. PLoS ONE 8: e67129 10.1371/journal.pone.0067129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moreno CX, Moy F, Daniels TJ, Godfrey HP, Cabello FC (2006) Molecular analysis of microbial communities identified in different developmental stages of Ixodes scapularis ticks from Westchester and Dutchess Counties, New York. Environ Microbiol 8: 761–772. [DOI] [PubMed] [Google Scholar]
- 24. Hawlena H, Rynkiewicz E, Toh E, Alfred A, Durden LA, et al. (2013) The arthropod, but not the vertebrate host or its environment, dictates bacterial community composition of fleas and ticks. ISME J 7: 221–223 10.1038/ismej.2012.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lalzar I, Harrus S, Mumcuoglu KY, Gottlieb Y (2012) Composition and seasonal variation of Rhipicephalus turanicus and Rhipicephalus sanguineus bacterial communities. Appl Environ Microbiol 78: 4110–4116 10.1128/AEM.00323-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Halos L, Bord S, Cotté V, Gasqui P, Abrial D, et al. (2010) Ecological factors characterizing the prevalence of bacterial tick-borne pathogens in Ixodes ricinus ticks in pastures and woodlands. Appl Environ Microbiol 76: 4413–4420 10.1128/AEM.00610-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lo N, Beninati T, Sassera D, Bouman EAP, Santagati S, et al. (2006) Widespread distribution and high prevalence of an alpha-proteobacterial symbiont in the tick Ixodes ricinus . Environ Microbiol 8: 1280–1287 10.1111/j.1462-2920.2006.01024.x [DOI] [PubMed] [Google Scholar]
- 28. Bermúdez SE, Eremeeva ME, Karpathy SE, Samudio F, Zambrano ML, et al. (2009) Detection and identification of rickettsial agents in ticks from domestic mammals in eastern Panama. J Med Entomol 46: 856–861 10.1603/033.046.0417 [DOI] [PubMed] [Google Scholar]
- 29.Hartmann C, Lennartz K, Ibrahim H, Coz A, Kasper Y, et al. (8/20102) Application Note: Stable 16-year Storage of DNA Purified with the QIAamp DNA Blood Mini Kit. Available: http://www.qiagen.com/knowledge-and-support/resource-center/resource-download.aspx?id=705c6944-4633-4101-8500-a6642d253a0e&lang=en. Accessed 2014 April 6.
- 30.Jumpstart Consortium Human Microbiome Project Data Generation Working Group (2010) 16S 454 Sequencing Protocol HMP Consortium (version 4.2.2). Available: http://www.hmpdacc.org/doc/16S_Sequencing_SOP_4.2.2.pdf. Accessed 2014 April 6.
- 31. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schloss PD (2013) Schloss SOP. Mothur Wiki. Available: http://www.mothur.org/wiki/Schloss_SOP. Accessed 2013 September 30.
- 33. Schloss PD (2009) A high-throughput DNA sequence aligner for microbial ecology studies. PLoS ONE 4: e8230 10.1371/journal.pone.0008230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou J, Wu L, Deng Y, Zhi X, Jiang Y-H, et al. (2011) Reproducibility and quantitation of amplicon sequencing-based detection. ISME J 5: 1303–1313 10.1038/ismej.2011.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Esty WW (1986) The efficiency of Good’s nonparametric coverage estimator. Ann Stat 14: 1257–1260 10.1214/aos/1176350066 [DOI] [Google Scholar]
- 36. Hill TCJ, Walsh KA, Harris JA, Moffett BF (2003) Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol 43: 1–11 10.1111/j.1574-6941.2003.tb01040.x [DOI] [PubMed] [Google Scholar]
- 37. Pinto AJ, Raskin L (2012) PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS ONE 7: e43093 10.1371/journal.pone.0043093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R Core Team (2013) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available: http://www.R-project.org.
- 39. Jaccard P (1908) Nouvelles recherches sur la distribution florale. Société Vaudoise Sci Nat 44: 223–270. [Google Scholar]
- 40. Bray JR, Curtis JT (1957) An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27: 326–349 10.2307/1942268 [DOI] [Google Scholar]
- 41.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, et al. (2013) vegan: Community Ecology Package. Available: http://cran.r-project.org/web/packages/vegan/index.html.
- 42. Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6: 65–70. [Google Scholar]
- 43. Faith DP, Minchin PR, Belbin L (1987) Compositional dissimilarity as a robust measure of ecological distance. Vegetatio 69: 57–68 10.2307/20038103 [DOI] [Google Scholar]
- 44. Minchin PR (1987) An evaluation of the relative robustness of techniques for ecological ordination. Vegetatio 69: 89–107 10.2307/20038106 [DOI] [Google Scholar]
- 45.Kindt R, Coe R (2005) Chapter 10: analysis of ecological distance by ordination. Tree diversity analysis: a manual and software for common statistical methods for ecological and biodiversity studies. Nairobi, Kenya: World Agroforestry Centre. 153–196.
- 46. White JR, Nagarajan N, Pop M (2009) Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5: e1000352 10.1371/journal.pcbi.1000352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pawitan Y, Michiels S, Koscielny S, Gusnanto A, Ploner A (2005) False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics 21: 3017–3024 10.1093/bioinformatics/bti448 [DOI] [PubMed] [Google Scholar]
- 48. Bartlett MS (1937) Properties of sufficiency and statistical tests. Proc R Soc Lond Ser A 160: 268–282. [Google Scholar]
- 49. Veech JA (2013) A probabilistic model for analysing species co-occurrence. Glob Ecol Biogeogr 22: 252–260 10.1111/j.1466-8238.2012.00789.x [DOI] [Google Scholar]
- 50. Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46 10.1111/j.1442-9993.2001.01070.pp.x [DOI] [Google Scholar]
- 51. Hong S, Bunge J, Leslin C, Jeon S, Epstein SS (2009) Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J 3: 1365–1373 10.1038/ismej.2009.89 [DOI] [PubMed] [Google Scholar]
- 52. Berry D, Mahfoudh KB, Wagner M, Loy A (2011) Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl Environ Microbiol 77: 7846–7849 10.1128/AEM.05220-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF (2005) PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl Environ Microbiol 71: 8966–8969 10.1128/AEM.71.12.8966-8969.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clay K, Fuqua C (2010) The tick microbiome: diversity, distribution and influence of the internal microbial community for a blood-feeding disease vector. Critical Needs and Gaps in Understanding Prevention, Amelioration, and Resolution of Lyme and Other Tick-Borne Diseases: The Short-Term and Long-Term Outcomes. Washington, D. C.: Institute of Medicine Committee on Lyme Disease and Other Tick-Borne Diseases: The State of the Science. Available: iom.edu/∼/media/Files/Activity%20Files/Disease/TickBorne/08-The-Tick-Microbiome.pdf.
- 55. Kauserud H, Kumar S, Brysting AK, Nordén J, Carlsen T (2012) High consistency between replicate 454 pyrosequencing analyses of ectomycorrhizal plant root samples. Mycorrhiza 22: 309–315 10.1007/s00572-011-0403-1 [DOI] [PubMed] [Google Scholar]
- 56. Pilloni G, Granitsiotis MS, Engel M, Lueders T (2012) Testing the limits of 454 pyrotag sequencing: reproducibility, quantitative assessment and comparison to T-RFLP fingerprinting of aquifer microbes. PLoS ONE 7: e40467 10.1371/journal.pone.0040467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moncayo AC, Cohen SB, Fritzen CM, Huang E, Yabsley MJ, et al. (2010) Absence of Rickettsia rickettsii and occurrence of other spotted fever group rickettsiae in ticks from Tennessee. Am J Trop Med Hyg 83: 653–657 10.4269/ajtmh.2010.09-0197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fritzen CM, Huang J, Westby K, Freye JD, Dunlap B, et al. (2011) Infection prevalences of common tick-borne pathogens in adult lone star ticks (Amblyomma americanum) and American dog ticks (Dermacentor variabilis) in Kentucky. Am J Trop Med Hyg 85: 718–723 10.4269/ajtmh.2011.10-0583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Klyachko O, Stein BD, Grindle N, Clay K, Fuqua C (2007) Localization and visualization of a Coxiella-type symbiont within the lone star tick, Amblyomma americanum . Appl Environ Microbiol 73: 6584–6594 10.1128/AEM.00537-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jiang J, Yarina T, Miller MK, Stromdahl EY, Richards AL (2010) Molecular detection of Rickettsia amblyommii in Amblyomma americanum parasitizing humans. Vector-Borne Zoonotic Dis 10: 329–340 10.1089/vbz.2009.0061 [DOI] [PubMed] [Google Scholar]
- 61. Zhang X, Ren X, Norris DE, Rasgon JL (2012) Distribution and infection frequency of “Candidatus Rickettsia amblyommii” in Maryland populations of the lone star tick (Amblyomma americanum) and culture in an Anopheles gambiae mosquito cell line. Ticks Tick-Borne Dis 3: 38–42 10.1016/j.ttbdis.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dasch G, Kelly D, Richards A, Sanchez J, Rives C (1993) Western blotting analysis of sera from military personnel exhibiting serological reactivity to spotted fever group rickettsiae. Am J Trop Med Hyg 49: 220. [Google Scholar]
- 63. Apperson CS, Engber B, Nicholson WL, Mead DG, Engel J, et al. (2008) Tick-borne diseases in North Carolina: is “Rickettsia amblyommii” a possible cause of rickettsiosis reported as Rocky Mountain spotted fever? Vector-Borne Zoonotic Dis 8: 597–606 10.1089/vbz.2007.0271 [DOI] [PubMed] [Google Scholar]
- 64. Billeter SA, Blanton HL, Little SE, Levy MG, Breitschwerdt EB (2007) Detection of “Rickettsia amblyommii” in association with a tick bite rash. Vector-Borne Zoonotic Dis 7: 607–610 10.1089/vbz.2007.0121 [DOI] [PubMed] [Google Scholar]
- 65. Stromdahl EY, Vince MA, Billingsley PM, Dobbs NA, Williamson PC (2008) Rickettsia amblyommii infecting Amblyomma americanum larvae. Vector-Borne Zoonotic Dis 8: 15–24 10.1089/vbz.2007.0138 [DOI] [PubMed] [Google Scholar]
- 66. Smith MP, Ponnusamy L, Jiang J, Ayyash LA, Richards AL, et al. (2010) Bacterial pathogens in ixodid ticks from a Piedmont county in North Carolina: prevalence of rickettsial organisms. Vector-Borne Zoonotic Dis 10: 939–952 10.1089/vbz.2009.0178 [DOI] [PubMed] [Google Scholar]
- 67. Zanetti AS, Pornwiroon W, Kearney MT, Macaluso KR (2008) Characterization of rickettsial infection in Amblyomma americanum (Acari: Ixodidae) by quantitative real-time polymerase chain reaction. J Med Entomol 45: 267–275 10.1603/0022-2585(2008)45267:CORIIA2.0.CO2 [DOI] [PubMed] [Google Scholar]
- 68. Kagemann J, Clay K (2013) Effects of infection by Arsenophonus and Rickettsia bacteria on the locomotive ability of the ticks Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis . J Med Entomol 50: 155–162 10.1603/ME12086 [DOI] [PubMed] [Google Scholar]
- 69. Zhang X, Norris DE, Rasgon JL (2011) Distribution and molecular characterization of Wolbachia endosymbionts and filarial nematodes in Maryland populations of the lone star tick (Amblyomma americanum). FEMS Microbiol Ecol 77: 50–56 10.1111/j.1574-6941.2011.01089.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Baldo L, Prendini L, Corthals A, Werren JH (2007) Wolbachia are present in southern African scorpions and cluster with supergroup F. Curr Microbiol. 55: 367–373 10.1007/s00284-007-9009-4 [DOI] [PubMed] [Google Scholar]
- 71. Covacin C, Barker SC (2007) Supergroup F Wolbachia bacteria parasitise lice (Insecta: Phthiraptera). Parasitol Res 100: 479–485 10.1007/s00436-006-0309-6 [DOI] [PubMed] [Google Scholar]
- 72. Casiraghi M, Favia G, Cancrini G, Bartoloni A, Bandi C (2001) Molecular identification of Wolbachia from the filarial nematode Mansonella ozzardi . Parasitol Res 87: 417–420 10.1007/s004360000368 [DOI] [PubMed] [Google Scholar]
- 73. Lefoulon E, Gavotte L, Junker K, Barbuto M, Uni S, et al. (2012) A new type F Wolbachia from Splendidofilariinae (Onchocercidae) supports the recent emergence of this supergroup. Int J Parasitol 42: 1025–1036 10.1016/j.ijpara.2012.09.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of raw 454 read length distributions between replicate libraries created with different polymerases. Each library was produced with the same set of DNA templates and barcoded primers. The polymerase varied between panels A and B/C, while B and C were produced from the same amplification products but pooled and sequenced independently. Panel A represents the library created with AccuPrime Taq, and panels B and C represent Platinum SuperMix replicates 1 and 2, respectively. The horizontal dashed line marks 100,000 on the log scaled y-axis on each plot.
(TIF)
Comparison of quality control filtering between replicate 454 libraries created with different polymerases. Each library was produced with the same set of DNA templates and barcoded primers. Libraries Platinum 1 and Platinum 2 where produced from the same amplification products but pooled and sequenced independently. The total number of raw sequences per library is given at the top of each bar. Note that the categories given in the key are in the same order as they appear in the plot; contaminants were present in all three libraries but accounted for <0.04% of each library.
(TIF)
Bacterial operational taxonomic unit rarefaction curve for Amblyomma americanum tick DNA sample sets. The shaded regions represent the conditional 95% confidence interval obtained from 1000 randomizations of the data.
(TIF)
Non-metric multi-dimensional scaling (NMDS) of a Bray-Curtis distance matrix describing Amblyomma americanum bacterial communities. Each of the 131 points symbolizes a single tick’s community; some points may overlap completely. Point and ellipse colors (red = Georgia ticks, black = archival) indicate the tick DNA collection of origin; ellipses represent 95% confidence intervals around group centroids. Non-overlapping centroids are considered significantly different at α = 0.05. The R2 value in the upper left corner of the plot describes the amount of variation in the data set explained by the groupings. The stress value is a measure of the disagreement between the rank order in the original data set and that in the NMDS (lower numbers indicate better agreement). Operational taxonomic units that differ in mean relative abundance between the groups are given in Table S4.
(TIF)
Non-metric multi-dimensional scaling (NMDS) of a Jaccard distance matrix describing Georgian Amblyomma americanum bacterial communities. Each point symbolizes a single tick’s community (n = 104); some points may overlap completely. Point and ellipse colors indicate life stage; ellipses represent 95% confidence intervals around life stage centroids. Non-overlapping centroids are considered significantly different at α = 0.05. R2 values in the upper left corner of plots describe the amount of variation in the data set explained by the groupings. The stress value is a measure of the disagreement between the rank order in the original data set and that in the NMDS (lower numbers indicate better agreement). Brown numbers indicate the species scores for select OTUs as follows: (1) Coxiella, (2) Rickettsia, (3) Midichloria 1, (4) Borrelia, (5) Ehrlichia, (6) Bacillales 1, (7) Wolbachia, (8) Rhizobiales 1, (9) Rhodobaca, (10) Bacillales 2, (11) Nitriliruptor, (12) Bacillales 3.
(TIF)
Mean relative abundance of operational taxonomic units (OTUs) from Georgian ticks by life stage. Significant differences (p<0.05) are labeled within rows by the letters that appear in cells. For example, within a row cells labeled A are significantly different from those labeled B or C but not cells labeled A. Empty cells do not differ from any group. Note the discontinuity between the white-black and yellow-red scales.
(TIF)
Non-metric multi-dimensional scaling (NMDS) of a Jaccard distance matrix describing archival Amblyomma americanum bacterial communities. Each point symbolizes a single tick’s community (n = 27); some points may overlap completely. Point and ellipse colors indicate life stage; ellipses represent 95% confidence intervals around life stage centroids. Non-overlapping centroids are considered significantly different at α = 0.05. R2 values in the upper left corner of plots describe the amount of variation in the data set explained by the groupings. The stress value given is a measure of the disagreement between the rank order in the original data set and that in the NMDS (lower numbers indicate better agreement). Numbers indicate the species scores for the plot as follows: (1) Coxiella, (2) Rickettsia, (3) Midichloria 1, (4) Borrelia, (5) Francisella, (6) Midichloria 2, (7) Ehrlichia, (8) Enterobacteriaceae, (9) Pseudomonas 1, (10) Burkholderiaceae, (11) Acidobacteria Gp1, (12) Pseudomonas 2, (13) Rhizobiales 2, (14) Bacillus, (15) Acetobacteraceae 1, (16) Acetobacteraceae 2, (17) Bacteria 1, (18) Bacteria 2.
(TIF)
Barcode sequences used to label eubacterial rrs gene variable regions 5–3 primers for sample multiplexing during pyrosequencing.
(DOCX)
454 sequencing statistics from Titanium FLX plates.
(DOCX)
Summary of previous reports of most abundant operational taxonomic units (OTUs) from Amblyomma americanum .
(DOCX)
Metastats results comparing the differential abundance of bacterial operational taxonomic units between Amblyomma americanum DNA collections.
(XLSX)