Abstract
Comparisons of human genomes show that more base pairs are altered as a result of structural variation — including copy number variation — than as a result of point mutations. Here we review advances and challenges in the discovery and genotyping of structural variation. The recent application of massively parallel sequencing methods has complemented microarray-based methods and has led to an exponential increase in the discovery of smaller structural-variation events. Some global discovery biases remain, but the integration of experimental and computational approaches is proving fruitful for accurate characterization of the copy, content and structure of variable regions. We argue that the long-term goal should be routine, cost-effective and high quality de novo assembly of human genomes to comprehensively assess all classes of structural variation.
The spectrum of human genetic variation ranges from the single base pair to large chromosomal events, but it has become apparent that human genomes differ more as a consequence of structural variation than of single-base-pair differences1-6. Structural variation was originally defined as insertions, deletions and inversions greater than 1 kb in size7. With the sequencing of human genomes now becoming routine8, the operational spectrum of structural variants (SVs) and copy number variants (CNVs) has widened to include much smaller events (for example, those >50 bp in length). The challenge now is to discover the full extent of structural variation and to be able to genotype it routinely in order to understand its effects on human disease, complex traits and evolution.
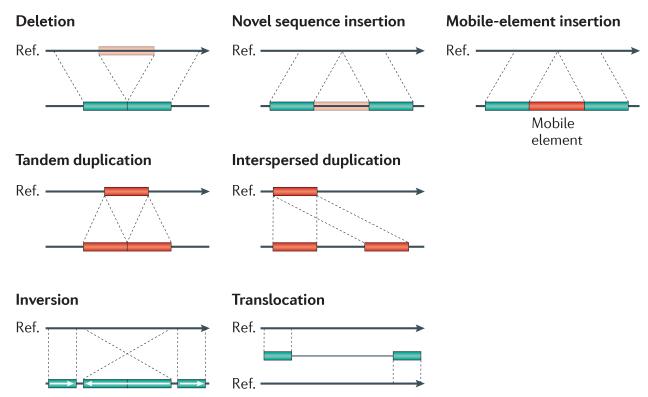
At least two distinct models have been proposed with respect to associations between disease and structural variation. The first involves large variants (typically gains and losses several hundred kilobase pairs in length) that are individually rare in the population (<1%) but collectively account for a significant fraction of disease, as seen for some neurological and neurocognitive disorders9-12. The second includes multicopy gene families that are commonly copy number variable and contribute to disease susceptibility, as seen for traits related to immune gene functions13,14. The discovery and genotyping of structural variation has been central to understanding these disease associations. Systematic and comprehensive assessment of structural variation has been problematic owing to the complexity and multifaceted features of SVs. Ideally, SV discovery and genotyping requires accurate prediction of three features: copy, content and structure. In practice, this goal has remained elusive because SVs tend to reside within repetitive DNA, which makes their characterization more difficult. SVs vary widely in size and there are numerous classes of structural variation: deletions, translocations, inversions, mobile elements, tandem duplications and novel insertions (FIG. 1). Within the past 5 years, a variety of computational and experimental methods has emerged; typically each focuses on a particular class of structural variation limited by frequency and size range of the events.
Figure 1. Classes of structural variation.
Traditionally, structural variation refers to genomic alterations that are larger than 1 kb in length, but advances in discovery techniques have led to the detection of smaller events. Currently, >50 bp is used as an operational demarcation between indels and copy number variants (CNVs). The schematic depicts deletions, novel sequence insertions, mobile-element insertions, tandem and interspersed segmental duplications, inversions and translocations in a test genome (lower line) when compared with the reference genome.
In this Review, we consider current methods for discovery and then for genotyping, including experimental approaches using microarrays, single-molecule analysis and sequencing-based computational approaches. The distinction between discovery and genotyping is important. Once a variant has been detected, validated and characterized at the sequence level (discovery), a different suite of methods may be applied to infer genotypes with relaxed thresholds. We discuss recent advances in the genetic characterization of germline structural variation — recognizing that the methods may be applied, in principle, to the study of somatic structural variation — and highlight current deficiencies, as well as areas for future development.
Hybridization-based microarray approaches
Microarrays have been the experimental workhorse of CNV discovery and genotyping1,6,15-18. These are represented primarily by array comparative genomic hybridization (array CGH) and SNP microarrays. Both hybridization-based technologies infer copy number gains or losses compared to a reference sample or population, but differ in the details and application of the molecular assays.
Array CGH
Array CGH platforms are based on the principle of comparative hybridization of two labelled samples (test and reference) to a set of hybridization targets (typically long oligonucleotides or, historically, bacterial artificial chromosome (BAC) clones). The signal ratio is then used as a proxy for copy number (see BOX 1 for details). An important consideration is the effect of the reference sample on the copy-number profile. For example, when only one sample is examined, a loss in the reference sample is indistinguishable from a gain in the test sample. For this reason, a well-characterized reference is key to interpretation of array CGH data19. Early studies of germline CNVs were based on BAC arrays or low-resolution oligonucleotide platforms and allowed detection of CNVs typically greater than 100 kb1,2,6 (BOX 2). These initial studies highlighted the incredible number of CNVs observed in healthy individuals; however, the breakpoints of these alterations were not sufficiently well-defined to allow accurate assessment of the proportion of the genome altered or its gene content. This led to a drastic overestimation of the extent of copy-number polymorphism using large-insert BAC clones2, which was subsequently refined by oligonucleotide microarrays or sequence-based studies of the same DNA samples4,5,20,21.
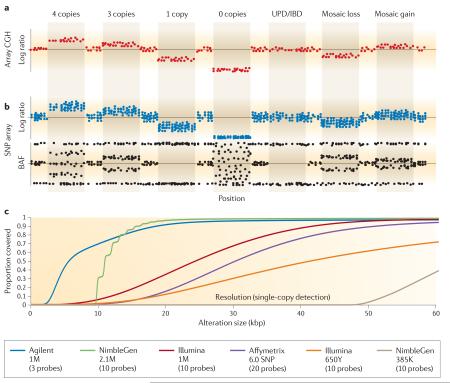
Box 1. Array CGH versus SNP microarray detection.
In array comparative genomic hybridization (array CGH), the signal ratio between a test and reference sample is normalized and converted to a log2 ratio, which acts as a proxy for copy number18,25,112. An increased log2 ratio represents a gain in copy number in the test compared with the reference; conversely, a decrease indicates a loss in copy number (see the figure, part a). SNP arrays generate a similar metric by comparing the signal intensities for the sample being analysed to a collection of reference hybridizations, or the rest of the population being analysed (part b, upper panel). The log ratio metric for SNP arrays demonstrates a lower per-probe signal-to-noise ratio (SNR) than array CGH (compare a and b in the figure); however, SNP arrays offer an additional metric that enables a more comprehensive assignment of copy number than does array CGH. This metric, termed B allele frequency (BAF) (part b, lower panel), can be calculated as the proportion of the total allele signal (A + B) explained by a single allele (A). The BAF has a significantly higher per-probe SNR than the log ratio data and can be interpreted as follows: a BAF of 0 represents the genotype (A/A or A/–), whereas 0.5 represents (A/B) and 1 represents (B/B or B/–). Different BAF values occur for AAB and ABB genotypes or more complex genotypes (for example, AAAB, AABB and BBBA). Homozygous deletions result in a failure of the BAF to cluster23,24. Thus, the BAF may be used to accurately assign copy numbers from 0 to 4 in diploid regions of the genome. The BAF also allows detection of copy-neutral events such as segmental uniparental disomy (segmental UPD) or whole-chromosome UPD and identity by descent (IBD), which results when a segment of one chromosome is replaced by the other allele without a change in copy number (this is therefore not detectable by array CGH)24. An additional advantage of the BAF is that it can be used to reliably detect and type low-level mosaic gains and losses24,113,114 (see the figure, part b).
Another important consideration in choosing an array platform is the ability to detect alterations in the size range being investigated. Array resolution is complicated by non-uniform probe distributions and differing SNRs between platforms, and as a result two platforms cannot be compared by simply counting the number of probes included. The number of probes required to detect a single-copy alteration varies between platforms, with Agilent Technologies offering the highest per-probe performance25,26,32. Part c of the figure shows the probe coverage of several major array platforms as determined by ResCalc25. This represents the theoretical ability to detect a copy number variant at any given location in the genome. In practice, however, thresholds of copy-number detection are typically greater owing to variable probe performance (BOX 2). Although alterations can, theoretically, be detected with a single probe using the Agilent platform, we set the detection limit to a more realistic (in a discovery context) three probes. The other major array platforms tend to require more probes, with Roche NimbleGen34 and Illumina16 platforms requiring ten probes, and Affymetrix39 requiring 20 probes to reliably detect a single-copy alteration.
Box 2. Copy number variant size distribution and discovery methods.
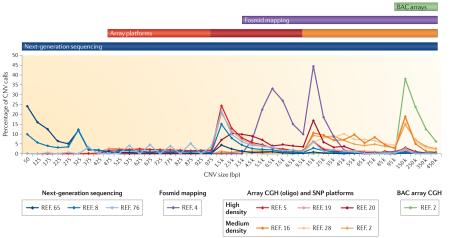
Different copy number variant (CNV) discovery methods are better able to identify CNVs of different sizes. The figure shows the proportion of CNV calls within a given size range for several recently published discovery efforts. Each of the different platforms tends to detect the largest proportion of events at the smallest size ranges to which they are sensitive, as expected owing to the increasing frequency of CNVs at smaller sizes. In terms of performance, next-generation sequencing (NGS)8,65,76 offers the widest possible range of detection, followed by ultra-high-resolution arrays (42 million (42M) probe, Roche NimbleGen5, and 24M probe, Agilent Technologies19), high-resolution SNP arrays20 and fosmid end-sequence pair mapping4, and finally medium-density oligonucleotide microarrays2,16,28 and bacterial artificial chromosome (BAC) platforms2. The size ranges detectable by each platform are illustrated by coloured bars above the plot indicating the lower limit of detection. Local peaks at 1 kb, 15 kb and 150 kb are due to changing bin sizes in the plot.
In addition to the size range of alterations detected, it is important to consider the differential abilities of assays to detect multiple subtypes of genomic alterations. The table shows the numbers of events detected for various categories of genomic alterations — deletions, novel insertions, inversions, duplications and mobile-element insertions (MEIs) — as reported in the database dbVar for the associated publications42. This highlights the significant bias of array platforms to deletion events, as well as the use of fosmid and sequencing-based platforms in detecting events missed by array technologies. Array CGH, array comparative genomic hybridization; CNP, copy-number polymorphism; ESP, end-sequence pair.
Currently, Roche NimbleGen and Agilent Technologies are the major suppliers of whole-genome array CGH platforms and routinely produce arrays with up to 2.1 million (2.1M) and 1M long oligonucleotides (50–75-mers), respectively, per microarray. Detection of a CNV typically requires a signal from at least 3 to 10 consecutive probes (BOX 1); as a result, SNP and CGH microarrays can routinely detect anywhere from dozens to several hundred events per genome depending on the platform applied (BOXES 1,2). Two studies have recently used ultra-high-resolution arrays (24M to 42M probes) for array CGH-based SV discovery in samples from HapMap individuals5,19. Although such high-density arrays are not practical for a large number of samples (30 and 40 samples were used in these studies), these approaches enabled the discovery of CNVs down to 500 bp, with breakpoints precise enough to allow the identification of sequence motifs at a subset of variants. One key advantage of array CGH platforms is the availability of custom, high-probe-density arrays from both major manufacturers. This has led to their widespread adoption in clinical diagnostics, essentially replacing karyotype analysis as the primary means of detecting copy-number alterations among children with developmental delay22.
SNP arrays
SNP microarray platforms are also based on hybridization, with a few key differences from CGH technologies. First, hybridization is performed on a single sample per microarray, and log-transformed ratios are generated by clustering the intensities measured at each probe across many samples20,23,24. Second, SNP platforms take advantage of probe designs that are specific to single-nucleotide differences between DNA sequences, either by single-base-extension methods (Illumina) or differential hybridization (Affymetrix)20,23,24. One key disadvantage is that, per probe, SNP microarrays tend to offer lower signal-to-noise ratio than do the best array CGH platforms. This is apparent in comparisons of array CGH and SNP platforms in terms of detection of CNVs by a purely ratio-based approach24-27. However, a key advantage of SNP microarrays is the use of SNP allele-specific probes to increase CNV sensitivity, distinguish alleles and identify regions of uniparental disomy through the calculation of a metric termed B allele frequency (BAF) (BOX 1).
SNP arrays have proved popular in CNV-detection studies, historically as complements to array CGH platforms for fine-mapping regions2 and currently in the large-scale discovery of CNVs in a broad variety of populations16,20,23,28,29. Early SNP arrays demonstrated poor coverage of CNV regions, but recent arrays (such as the Affymetrix 6.0 SNP and Illumina 1M platforms) incorporate better SNP selection criteria for complex regions of the genome and non-polymorphic copynumber probes (which are examined for log ratios but not BAF)20,23,30. Another important consideration is the choice of population because the average heterozygosity affects the proportion of SNPs that will generate a meaningful BAF signal (typically, heterozygosity is 30–40% in Illumina platforms). This is particularly relevant when dealing with populations that may have experienced a drastic bottleneck, as opposed to more outbred populations, and thus may affect the number of probes needed to identify an alteration23,24. Some studies combine array CGH and SNP platforms to offer higher confidence in CNV detection2,20,30.
Limitations
Microarrays are limited to detecting copy-number differences of sequences present in the reference assembly used to design the probes31, provide no information on the location of duplicated copies and are generally unable to resolve breakpoints at the single-base-pair level. Both array CGH and SNP platforms tend to suffer reduced sensitivity in the detection of single-copy gains (3 to 2 copy-number ratio) compared with deletions (1 to 2 copy-number ratio)16,23,25,32. This is particularly challenging when gains encompass only a few probes and SNP arrays may not contain sufficient probe density to use the BAF measurement. Thus, smaller events detected by array platforms are overwhelmingly deletions, partly owing to an ascertainment bias5,16,20. Homozygous deletions are the easiest class to detect regardless of platform and can be detected with fewer probes than single-copy gains and losses23,25.
An important practical consideration is that the various commercial array platforms offer different probe densities and per-probe performance in detecting alterations25,26,32-35 (BOX 1). Not surprisingly, numerous copynumber detection algorithms are available to call CNVs from microarrays, which, depending on parameter optimization, can lead to significant differences in detection and complications in downstream interpretation30,36-41. Current recommended solutions include using consensus calls from multiple algorithms30, using multiple samples to refine CNV calls20,39, selecting algorithms designed specifically for the platform being examined30,41 and optimizing parameters in conjunction with manual curation for a subset of events16,41. Although most algorithms and microarray platforms perform comparably for the detection of large events, smaller events are more challenging to routinely detect, with all single microarray array platforms (except custom arrays targeted to specific loci) losing sensitivity below 10 kb (BOX 2). As a result, small events are under-represented in databases such as dbVar42 (BOX 2) and the systematic discovery of pathogenic CNVs below 25 kb remains unexplored in most studies of disease16,43,44.
Perhaps the most important limitation of arrays is the use of hybridization-based assays in repeat-rich and duplicated regions. Array CGH and SNP platforms assume each location to be diploid in the reference genome, which is not valid in duplicated sequence. The signal for a 5 to 4 copy ratio, or other complex patterns, will not fit the expected results for a diploid reference sequence and may drop below the assay’s sensitivity to discriminate signals15. This is particularly challenging because CNVs have a strong positive correlation with segmental duplications and many breakpoints lie in duplicated regions5,15,16,45. Consequently, the accurate boundaries and copy numbers of these events will require additional technologies.
Advantages
Microarrays offer a distinct advantage in terms of throughput and cost. Large CNVs are individually very rare in the general population, yet 8% of individuals have a CNV of >500 kb in their genome16. Determining the pathogenic significance of any particular event in a rare-variant disease model requires screening of thousands of affected individuals and controls. Given the low cost of array CGH and SNP platforms and the large collection of public SNP data available from genome-wide association studies46, microarray data provide an opportunity to assay the CNV landscape of large data sets. For example, an analysis of 2,493 Illumina SNP profiles was used to generate a comprehensive picture of large CNVs in the 0.5–1% frequency range16. Expansion of sample sizes in future studies will aid the design of genotyping assays to examine even larger populations and increase our understanding of human disease.
Single-molecule analysis
Microarray approaches cannot identify balanced structural variants or, in the case of duplication, specify the location of a duplicated sequence (BOX 2); understanding the structure and location has traditionally required visualization at the single-molecule level. Approaches such as fluorescent in situ hybridization (FISH), fiber-FISH and spectral karyotyping provided our first glimpses of common and rare genome structural variation47. However, their low throughput and low resolution limit their application to a few individuals and to particularly large structural differences (~500 kb to 5 Mb). To improve resolution and scalability, many groups are developing methods for the direct visualization of structure in stretched DNA fragments at a large scale. For example, optical mapping, a technique originally developed to analyse yeast genomes48, was recently applied to human genome SV analysis49. This method is based on a modification of traditional restriction mapping, wherein restriction digestion is performed on immobilized DNA; this allows the identification of the fragment sizes and changes in their relative order on the basis of comparison to an in silico digested version of the reference genome sequence. This powerful technique allows fine-scale structural analysis of genomes, detecting inversions and translocations, as well as copynumber alterations, and their locations4,49,50. Although capable of detecting novel insertions, the technique is limited by its dependence on a reference genome, and currently has very limited throughput.
DNA barcoding methodologies are a promising alternative that may one day allow high-throughput detection of balanced structural differences; these methods include scanning fluorescently nick-labelled DNA molecules in nano-channel flow cells51 or nanoslits52 and the use of SNP-specific labelling of stretched DNA for haplotype resolution53. Similarly, absolute copy-number estimations made by amplifying single molecules using emulsion picolitre droplet PCR54 or single-molecule sequencing of human genomes55 offer tremendous potential to understand structural changes at the cellular level.
Sequencing-based computational approaches
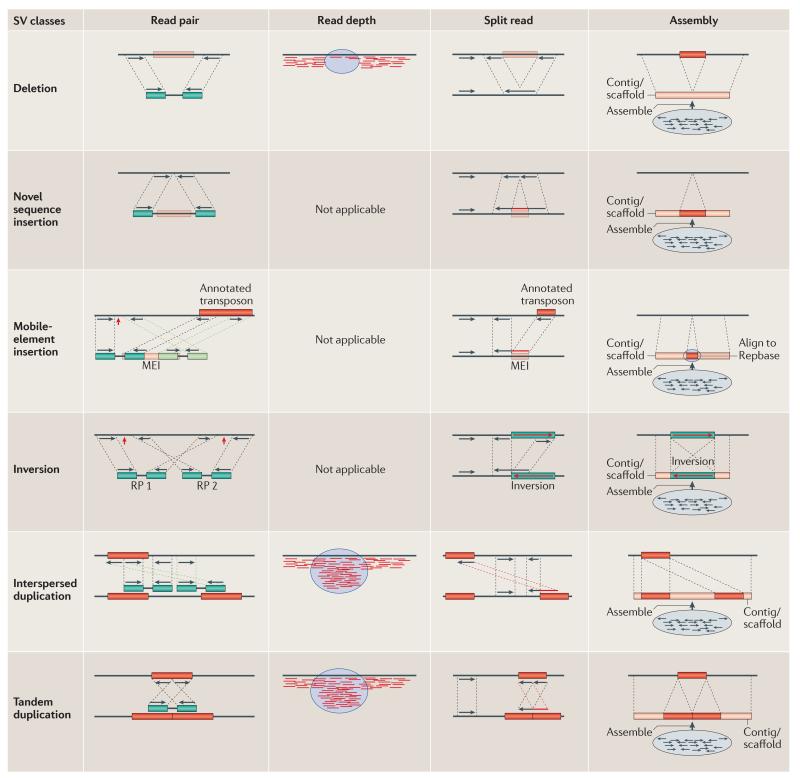
The advent of next-generation sequencing (NGS) technologies56-59 promises to revolutionize structural variation studies and, ultimately, replace microarrays as the platform for discovery and genotyping. However, NGS approaches present substantial computational and bioinformatics challenges. Most of the current algorithms for SV discovery are modelled on computational methods that were first developed to analyse capillary sequencing reads and fully sequenced large-insert clones3,60. There are four general types of strategy61,62, all of which focus on mapping sequence reads to the reference genome and subsequently identifying discordant signatures or patterns that are diagnostic of different classes of SV (described below and shown in FIG. 2).
Figure 2. Structural variation sequence signatures.
There are four general sequence-based analytical approaches used to detect structural variation. Theoretically, read-pair (RP), split-read and assembly methods can be used to discover variants from all classes of structural variant (SV), but each has different biases depending on the underlying sequence content of the variants and the data properties of the sequence reads. However, read-depth approaches can be used to detect only losses (deletions) and gains (duplications), and cannot discriminate between tandem and interspersed duplications. Briefly, read-pair methods analyse the mapping information of paired-end reads and their discordancy from the expected span size and mapped strand properties. Sensitivity, specificity and breakpoint accuracy are dependent on the read length, insert size and physical coverage3,4,59,62,65,66,68,69. Breakpoints are indicated by red arrows. Read-depth analysis examines the increase and decrease in sequence coverage to detect duplications and deletions, respectively, and predict absolute copy numbers of genomic intervals45,62,74-76. Split-read algorithms are capable of detecting exact breakpoints of all variant classes by analysing the sequence alignment of the reads and the reference genome; however, they usually require longer reads than the other methods and have less power in repeat- and duplication-rich loci62,78,79. Assembly algorithms83-86,115 have the most power to detect SVs of all classes at the breakpoint resolution, but assembling short sequences and inserts often result in contig/scaffold fragmentation in regions with high repeat and duplication content89. MEI, mobile-element insertion. Repbase is a database of repetitive elements.
Read-pair technologies
Read-pair methods assess the span and orientation of paired-end reads and cluster ‘discordant’ pairs in which the mapping span and/or orientation of the read pairs are inconsistent with the reference genome (FIG. 2). Most classes of variation can, in principle, be detected. Read pairs that map too far apart define deletions, those found too close together are indicative of insertions, and orientation inconsistencies can delineate inversions and a specific class of tandem duplications3,4,59,63. Read pairs in which only one end clusters and the others do not map to the reference have been used to flag variant sequences not included in the reference genome (novel insertions). The read-pair method is the most widely applied approach and was first demonstrated using BAC end sequences generated from the breast cancer cell line MCF-7 (REF. 60). It was subsequently applied to germline genetic variation using a fosmid end sequence library3. Later, it was applied to next-generation, paired-end data generated by the 454 FLX platform59. There are now many computational tools based on a read-pair approach, including PEMer64, VariationHunter65-67, BreakDancer68, MoDIL69, MoGUL70, HYDRA71, Corona58 and SPANNER (REFS 8,62 and C. Stewart and colleagues, personal communication).
Read-depth methods
Read-depth approaches assume a random (typically Poisson or modified Poisson) distribution in mapping depth and investigate the divergence from this distribution to discover duplications and deletions in the sequenced sample45. The basic idea is that duplicated regions will show significantly higher read depth and deletions will show reduced read depth when compared to diploid regions (FIG. 2). Read-depth approaches using NGS data were first applied to define rearrangements in cancer72,73, and segmental duplication and absolute copy-number maps in human genomes74,75. Methods that attempt to discover smaller deletions and duplications at better breakpoint resolution include the event-wise-testing (EWT) algorithm76 and CNVnator8,62,77.
Split-read approaches
Split-read methods are capable of detecting deletions and small insertions down to single-base-pair resolution and were first applied to longer Sanger sequencing reads78. The aim is to define the breakpoint of a structural variant on the basis of a ‘split’ sequence-read signature (that is, the alignment to the genome is broken; a continuous stretch of gaps in the read indicates a deletion or in the reference indicates an insertion; FIG. 2). Extensions of this approach may also detect mobile-element insertions (MEIs) if the reads are sufficiently long to span the mobile element (for example, >400 bp for Alu elements)62 to characterize the full sequence content. Alternatively, if the read length is shorter but the MEI breakpoint is in a unique sequence, a split-read approach can be used to anchor the insertion62. Application of this method to NGS data sets is currently limited owing to the difficulty in aligning shorter reads; however, the Pindel algorithm79 uses paired-end reads to reduce the search space for potential split reads, thus reducing the computational overhead of the local gapped alignment of short sequences to the reference genome.
Sequence assembly
In theory, all forms of structural variation could be accurately typed for copy, content and structure if the underlying sequence reads were long and accurate enough to allow de novo assembly. In practice, sequence-assembly approaches are still in their infancy and typically use a combination of de novo and local-assembly algorithms to generate sequence contigs that are then compared to a reference genome (FIG. 2). Local sequence assembly of fosmid clones with discordant read pairs has been used to systematically discover structural variation in 17 human genomes4,31,63 (BOX 2). Approaches that involve library construction, clone array and end sequencing are too laborious and prohibitively expensive to be widely adopted. Ideally, complete genome sequencing followed by de novo assembly and comparison to a high-quality reference could identify thousands of structural variants. For example, a genome assembly from capillary sequence reads from a human individual has been used to characterize 12,178 structural variants80-82. Well-known de novo assembly algorithms for next-generation whole-genome shotgun (NG-WGS) data include EULER-USR83, ABySS84, SOAPdenovo85 and ALLPATHS-LG86. Using the Cortex assembler8,62, variant assembly can be done entirely de novo or with different degrees of information from a reference, and Cortex has the ability to simultaneously assemble multiple genomes and call SVs between samples without the need for a reference. The NovelSeq framework87 merges de novo and local-assembly methods to characterize novel sequence insertions and, finally, TIGRA8,62 aims to improve breakpoint estimations in SV discovery.
Limitations
None of the four main approaches to discovering structural variation using sequence data is comprehensive. When many algorithms and experimental methods are applied to the same DNA samples, a significant fraction of the validated variants remains unique to a particular approach (FIG. 3). Each method has different strengths and weaknesses in detection, depending on the variant type or the properties of the underlying sequence at the SV locus. Although read depth is the only sequencing-based method to accurately predict absolute copy numbers74,75, the breakpoint resolution is often poor. Read-pair approaches are powerful, but resolving ambiguous mapping assignments in repetitive regions is challenging and accurate prediction of SV breakpoints depends on very tight fragment size distributions, which can make library construction difficult and costly61. On the basis of typical NGS fragment sizes, more than 90% of the discovered events are less than 1 kb and most of these are deletions rather than insertions8,62 (BOX 2). Similarly, split-read algorithms can be devised to detect a wide range of SV classes with exact breakpoint resolution; however, split read is currently reliable only in the unique regions of the genome. Sequence assembly promises to be the most versatile method by facilitating pair-wise genome comparisons; however, it has been shown to be heavily biased against repeats and duplications owing to assembly collapse over such regions88,89. Its application to SV detection is not routine and will require substantial development. The realization that the computational approaches outlined above can discover only a subset of structural variants and have various biases in detection has prompted the recent development of algorithms that incorporate multiple methodologies to improve sensitivity and specificity. Three algorithms, SPANNER8,62, CNVer90 and Genome STRiP91, combine read-pair and read-depth methods in different ways to more reliably detect CNVs.
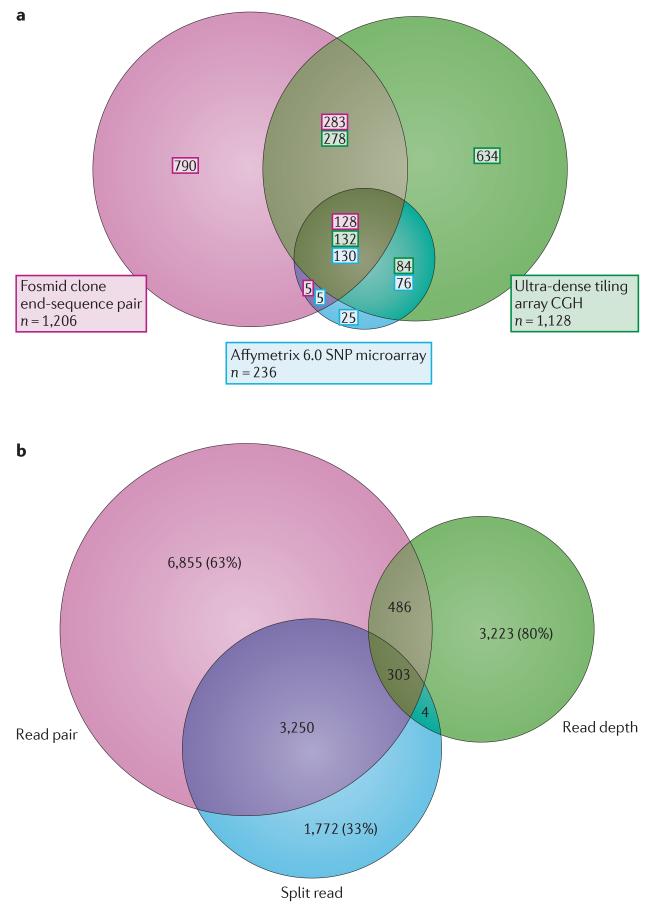
Figure 3. Copy number variant discovery biases.
a | Three different technologies have been applied to copy number variant (CNV) discovery for DNA obtained from the same five individual genomes (NA18517, NA19240, NA12878, NA19129 and NA12156). The experimental methods are: fosmid paired-end sequencing4,63, array comparative genomic hybridization (array CGH)5 and SNP microarray genotyping20. In this Venn diagram, only copy-number gains and losses of >5 kb are compared. SNP microarray CNVs in this study20 are biased towards common copy-number polymorphisms, which explains, in part, the fewer calls and the greater overlap with the other data sets. The fosmid end-sequence pair method also detects inversions, which are not considered in this analysis. b | This Venn diagram shows the numbers of unique and shared structural variants (SVs) found by different sequencing-based discovery approaches that have been used in the 1000 Genomes Project and shows that the approaches are complementary62. Read-pair, read-depth and split-read methods (involving 14 distinct algorithms) were applied to the same 185 genomic DNA samples. The proportion of the total number of SVs discovered by one approach that is unique to that approach may be as high as ~80%. Read-pair and split-read methods show the greatest extent of overlap. Read depth and split read are the most discordant approaches, with fewer than 20% of SVs detected by one approach detected by the other (assembly approaches are not compared as they are still in the development stage). The main differences in SV detection between these approaches are primarily found in duplication- and repeat-rich regions. Part a is modified, with permission, from REF. 63 © (2010) Elsevier. Part b is modified, with permission, from REF. 62 © (2011) Macmillan Publishing Ltd. All rights reserved.
Perhaps the greatest problem in using NGS to discover structural variation is the nature of the data. Sequence reads generated by the NGS platforms are considerably shorter than those produced by the capillary-based methods. Owing to the complex nature of human genomes (for example, widespread common repeats and segmental duplications), there is considerable read-mapping ambiguity. Longer reads and inserts are needed to ameliorate this bias by increasing the specificity in read mapping. It is estimated, however, that >1.5% of the human genome cannot be covered uniquely even with read lengths of 1 kb92. Another concern is sequence coverage, defined as the average number of times each base pair in the genome is represented in an aligned read. Sequence coverage is an important factor in achieving high sensitivity and specificity in SV detection (see below). Some projects may opt to sequence samples at low coverage for cost efficiency (for example, the 1000 Genomes Project uses two- to sixfold coverage8); however, this reduces the power to discover structural variation. To help ameliorate this effect, the read-pair-based MoGUL algorithm70 pools the mapping data of several individuals to detect common CNVs in a population (this will, however, still have reduced sensitivity to lower-frequency variants). A newer version of VariationHunter67 uses a similar pooling strategy, yet provides more sensitivity in detecting rare variation.
Finally, storage and analysis of NGS data requires a substantial investment in computational resources. Today, the raw sequencing data are stored in FASTQ files that contain a minimal amount of information: read name, sequence and the associated quality values. There is an urgent need for improvements in the efficiency of data processing as it is projected that the number of sequenced genomes generated worldwide will exceed 30,000 by the end of 2011 (REF. 93). Public access to such data would provide a rich resource for SV discovery.
Advantages
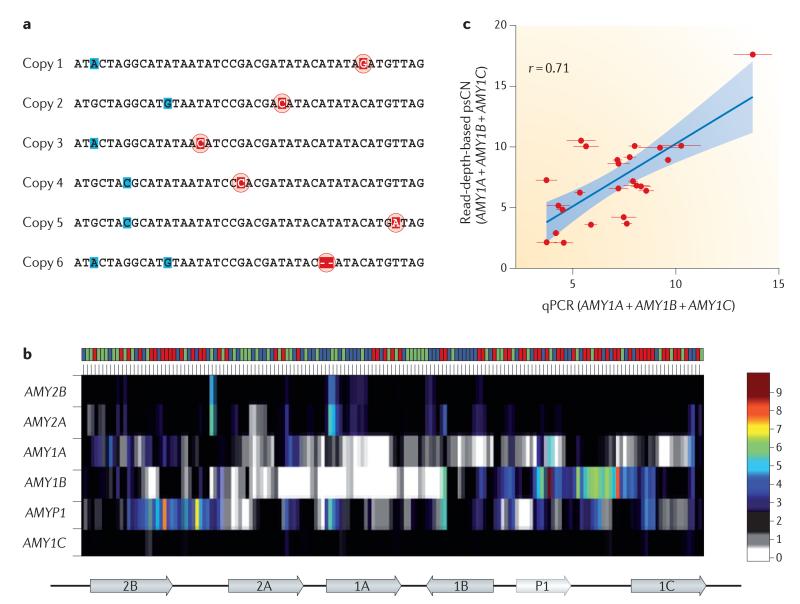
The most important benefit of NGS technologies is that it is possible to discover a multitude of variant classes (BOX 2) with a single sequencing experiment. In addition, the sequence data are largely unbiased and present a potential for understanding the complete spectrum of genetic variation. Genome-wide analysis without a priori information is possible, and the specificity and linear dynamic range response of NGS data offer many advantages for estimation of copy number. Through analysis of read depth, and uniquely identifying paralogous sequence variants (termed singly unique nucleotides (SUNs)), we can now begin to accurately estimate the absolute copy number of duplicated regions of the human genome74,75 (FIG. 4). Characterization of absolute copy numbers and the ability to distinguish between paralogous copies of duplicated gene families are necessary to better genotype these dynamic regions of the genome, which, in turn, is indispensible for understanding the phenotypic effect of duplications and their evolutionary importance.
Figure 4. Genotyping duplicated paralogues using next-generation sequencing.
a | Singly unique nucleotide (SUN) identifiers that distinguish paralogues from each other (red) are shown in the multiple sequence alignment of duplicated genes. These are distinguished from paralogous sequence variants that are not unique to a specific copy (blue). b | Read depth is measured at the SUN positions and used to estimate the copy number of each specific member of the amylase gene family. Across the top, each column represents a different individual from the 1000 Genomes Pilot Project. The colours represent the population identifiers: YRI (Yoruba in Ibadan, Nigeria) is shown in blue; CEU (Utah residents with northern and western European ancestry) is shown in green; and CHB/JPT (Chinese from Beijing, China, and Japanese from Tokyo, Japan) is shown in red. The corresponding copy-number prediction is depicted as a heat map. The pancreatic amylase genes (AMY2A and AMY2B) show little variation compared with the salivary amylase gene family (AMY1 genes). AMYP1 is a pseudogene. AMY1B shows the greatest copy-number variability, ranging from 0 to 9 copies. A schematic of the gene cluster is shown underneath the heat map; 2B represents AMY2B, and so forth. c | Aggregate paralogue-specific copy number (psCN) genotypes of AMY1 paralogues with estimates obtained by quantitative PCR (qPCR) directed at the three functional AMY1 copies compared across 25 JPT individuals. These data show that the qPCR and read-depth data correlate. Data for part b and the y axis of part c are taken from REF. 75; data for the x axis of part c are taken from REF. 116.
Genotyping
Discovery techniques are complicated by the need to analyse data blind to the possible location of CNVs; stringent thresholds must therefore be applied to control false positives. By contrast, genotyping techniques offer increased power to detect CNVs once the variant is known, and more relaxed thresholds may be applied than for discovery23. Genotyping platforms need to assay fewer probes per locus than discovery technologies and thus can be performed at a reduced cost for a larger number of samples. In addition to sensitivity and specificity, the main considerations for genotyping are the number of target loci, cost and throughput. Genotyping of CNVs requires more than the accurate determination of zygosity. For example, the multicopy nature of duplicated regions requires phase determination such that a genotype of 2/3 versus 4/1 can be deduced from an absolute copy number of 5. Genotyping will be further enhanced by imputation from SNP data, and there is a pressing need to integrate SNP and CNV alleles. However, multicopy regions of the genome are often resistant to SNP imputation owing to recurrent mutations and difficulty in assigning SNP genotypes.
PCR-based techniques
Conventional PCR across sequenced breakpoints59 and quantitative PCR provide the ability to rapidly and accurately screen a large number of samples at a very low cost per assay94-96. However, they are typically limited to a small number of loci. Multiplex ligation-dependent probe amplification (MLPA), multiplex amplifiable probe hybridization (MAPH) and multiplex amplicon quantification (MAQ) methodologies can simultaneously assay a larger number of loci (~40) on the basis of the quantification of PCR fragments in capillary electrophoresis experiments or microarray design97-99. These approaches require several optimization steps and are best applied when a large number of samples are to be analysed at a relatively low cost per sample. Alternatively, digital or single-molecule PCR allows screening of a large number of samples and sites in an emulsion or a microfluidic device. A significant benefit of this developing technology is that the enumeration of single-molecule PCR results, by real-time PCR or flow cytometry, allows the detection of events located on a single DNA fragment, which allows analysis of rare subpopulations or individual alleles100-103.
SNP-array-based techniques
Another approach for the validation of a moderate number of loci in a large number of samples is the use of customizable SNP-based assays such as the Illumina BeadXpress system. Using the GoldenGate assay, in which labelled probes are ligated in the presence of a target sequence, the readout is similar to Illumina SNP arrays and, as such, copy-number states can be determined using allelic ratios. Experiments can be performed in a 96-well plate format that can inter-rogate up to 384 probes in a single well104,105. Adaption of this system through the development of specific algorithms for copy-number analysis and probe selection has demonstrated utility in the detection of rare (<1%) and common (>1%) CNVs with a low false discovery rate using five probes per region of interest23,104,105. Although the initial set-up cost for an assay can be high, the system can process thousands of samples at a low cost per sample, generating combined SNP and copy-number genotypes in a matter of weeks. An alternative approach is to use off-the-shelf or custom SNP platforms that contain tag SNPs, which allow imputation of the presence of a CNV. Although not all sites may be imputable, this approach demonstrates the potential for using genome-wide association study data to characterize common CNVs in large populations32.
Array CGH-based techniques
Customized array CGH can be applied to genotype the largest number of CNV loci, as recently demonstrated by the Wellcome Trust Case Control Consortium (WTCCC)32. The WTCCC developed custom ~105,000 probe Agilent arrays covering 11,107 CNV loci discovered in various studies and applied this to a large population of ~19,000 samples; they successfully genotyped 4,000 CNVs. The same array was used by Conrad et al.5 to validate 8,599 CNVs (of which they genotyped 5,238) from their ultra-high-density array calls in the HapMap data set. Both studies found that some regions were more readily genotyped than others, with deletions typically being easier to genotype than duplications or multiallelic loci; they also found that multiple normalization algorithms may be required for different loci32. Hence, targeted arrays share the same limitations as discovery arrays in the context of genomic regions that can be profiled. However, the cost-effectiveness of multiplex, targeted arrays for genotyping enables analysis of much larger populations than in a discovery context.
Sequencing-based approaches
As genome- and exome-sequencing data sets become routine, there has been a shift to genotyping using computational analysis of sequencing data. Several of the SV-detection algorithms mentioned above can accurately distinguish the homozygous versus heterozygous states of the discovered SVs8,62,90. However, genotyping common SVs across many genomes using discovery algorithms takes a substantial amount of time and resources. When the structural variants are known a priori, genotyping of such variants in larger cohorts can be performed quickly using methods specifically developed for the purpose. To this end, BreakSeq106 was developed to build a library of SV breakpoints discovered in the literature, followed by validation using PCR. Raw reads from a newly sequenced genome were aligned to this breakpoint library to rapidly genotype the common structural variants. Kidd et al.31 introduced a similar concept, termed diagnostic k-mer analysis, to genotype sequenced novel insertions using the NG-WGS data that can also discriminate heterozygous from homozygous insertions. Read-depth-based algorithms such as CopySeq107 can genotype CNVs, and the SUN concept75 described above can be used to genotype duplicated genome segments. Combining methods can improve accuracy in genotyping, as it can in discovery. Genome-STRiP, for example, considers read-pair, read-depth and split-read approaches but, importantly, puts this in the context of a population-genetics framework to substantially improve the sensitivity and specificity of deletion polymorphism genotyping91. These genotyping approaches quickly identify the existence (or lack of) common SVs in the genome of a human individual; however, most require an extensive database of structural variants sequenced at the breakpoint level3-5,59,63,108.
An immediate benefit of NGS data sets is the potential to inform and improve existing experimental genotyping platforms, including SNP microarrays and array CGH19,75 (FIG. 5). SNP microarray genotyping platforms, for example, assume a diploid state as the baseline copy for the population average and predict copy-number differences on the basis of that assumption. In addition, the underlying mosaic structure of several segments of the genome make the task of CNV detection using SNP microarrays more difficult. A comparison between sequence read-depth and SNP copy-number predictions reveals a discrepancy for 30% of genotype calls when the same DNA samples are compared20,75. Most of these discrepancies map to duplicated regions and, if adjusted by a defined integer, a near-perfect concordance is achieved. Thus, sequence read depth can be used to more accurately genotype copy number from SNP microarrays. Similarly, analysing the same samples using both read depth and array CGH can be used to calibrate copy number for any specific region of the genome (FIG. 5b). Absolute copy-number estimates for duplicated genes, in conjunction with single-channel intensity data from array CGH experiments, allow for more accurate predictions of copy number on the basis of subtle changes in the dynamic range response. Thus, greater accuracy can be achieved in predicting copy number in duplicated regions using more affordable, high-throughput array CGH experiments. This reiterative complementarity of computational and experimental methodologies will be crucial to improving accuracy in detection and genotyping.
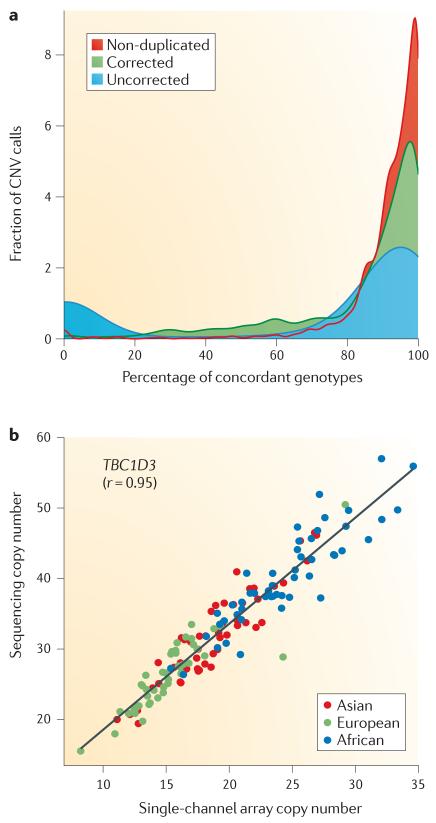
Figure 5. Improved copy number variant genotyping by the integration of computational and experimental approaches.
a | Absolute copy-number predictions made using sequence read depth75 are compared to copy-number genotype calls made using SNP microarrays (Affymetrix 6.0)20 on DNA from the same 114 individuals. The comparison shows good concordance in unique regions of the human genome (non-duplicated, red) when compared to all CNVs, including duplicated regions (uncorrected, blue). 94% of the discrepancies contain segmental duplications corresponding to 300 gene models. Analysis of the regions suggests population average copy numbers that differ from n = 2 (diploid). Readjusting the population average copy by an integer value using the read-depth estimations within the population ameliorates this bias (corrected, green) (change from 70% to 83% concordance). b | Single-channel array comparative genomic hybridization (array CGH) data (Agilent Technologies) is highly correlated with read-depth-based copy-number predictions for the highly duplicated TBC1D3 gene family. This calibration with absolute copy-number prediction allows for a more accurate prediction of the copy number of duplicated regions for future array CGH experiments. Part b is modified, with permission, from REF. 75 © (2010) American Association for the Advancement of Science.
Future directions
With respect to the structural variation of the human genome, this is an exciting time for the field of human genetics. Significant advances have been made in understanding variation in the copy, content and structure of the human genome. Genotyping of structural variation is now possible on an unprecedented scale, and the past 2 years have seen discovery increase by orders of magnitude (BOX 2). The widespread application of affordable microarray hybridization-based approaches to thousands of normal and disease samples has provided glimpses of the landscape of larger CNVs6,10,11,16,22; we now appreciate that rare CNVs are collectively quite common in the general population and that the pattern of such variation is significantly different in individuals with neurocognitive and neuropsychiatric disease109. Here, the main bottlenecks are proving the pathogenicity of individual loci (which requires tens of thousands of unaffected individuals) and exploring smaller events (<25 kb) in disease populations. The application of NGS through read-depth, read-pair and split-read methods has expanded the spectrum of human genome structural variation to include tens of thousands of smaller events (>50 bp)8. More importantly, the specificity and dynamic range response of NGS provides unparalleled accuracy in terms of the content and absolute copy-number prediction74,75. As a result, the veil has been lifted on a large class of previously inaccessible genetic variants.
The most serious challenges that remain are the absence of a ‘gold standard’ for assessment of disparate discovery and genotyping methods, and the remaining biases in global discovery. There is no human genome published so far for which the complete spectrum of structural variation has been resolved. There is also no commercial platform than can claim to be comprehensive in terms of either genotyping or discovery. Different experimental methods and computational analyses of NGS data sets applied to the same human DNA samples show disappointingly low levels of overlap — a general trend that has persisted over years of SV-discovery effort (FIG. 2). The limitations of computational bandwidth not-withstanding, there is currently no suite of algorithms that could be applied to systematically resolve all classes of structural variants. Biases remain in terms of content, size and class, with most discovery efforts focused on deletions in unique sequences, as evidenced by the 1000 Genomes Project8 and other published SV-discovery data sets (BOX 2).
How can these deficiencies be overcome? Improvements in NGS technology that increase accuracy and read length would facilitate the discovery and genotyping of structural variation in more complex regions of the genome. The long-term goal should be the de novo assembly of human genomes to a standard comparable to or better than that of the current human reference genome (GRCh37). On the basis of the experiences of sequencing and finishing the first human genome, this will ideally require the sequencing of large molecules >100 kb in length with accuracies in excess of 99.9% — a feat beyond the reach of current third-generation platforms. An interim solution is the integration of computational and experimental methods. For example, the read-depth approach can be used to correct reference effects in array data19,75. Several experimental approaches (such as FISH and optical maps) and sequence analyses (such as read-depth and clone sequencing) are typically required to resolve the architecture of complex regions of the genome50,110. Massively parallel sequencing of large molecules111 may provide an important step towards resolving the complete spectrum of structural variation, including balanced translocation and inversion events. As more sequence breakpoints become resolved for more difficult classes of structural variation, we will improve our ability to genotype using BreakSeq or diagnostic k-mer approaches31,106. Understanding copy, content and structure is an iterative process of evaluation that uses many orthogonal approaches and computational analyses. This effort requires committed investments from research laboratories, private industry and science funding agencies. Although getting to these answers will not be easy, the yield with respect to patterns of human genetic variation and insight into the architecture of human genetic disease will be worth the effort.
Box 2 Table.
| Method | Samples | Deletions |
Novel insertions |
Inversions |
Duplications |
MEIs |
Refs | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calls | Median length |
Calls | Median length |
Calls | Median length |
Calls | Median length |
Calls | Median length |
|||
| SNP microarray* | 270 | 1,122 | 6,216 | - | - | - | - | 442 | 14,122 | - | - | 20 |
|
| ||||||||||||
| SNP microarray‡ | 2,493 | 9,963 | 50,265 | - | - | - | - | 3,880 | 108,336 | - | - | 16 |
|
| ||||||||||||
| Fosmid ESP | 8 | 1,843 | 8,657 | 560 | 7,594 | 1,146 | 77,119 | 1,768 | 8,429 | - | - | 4 |
|
| ||||||||||||
| Array CGH§ | 40 | 7,909 | 2,284 | - | - | - | - | 4,740 | 5,265 | - | - | 5 |
|
| ||||||||||||
| Array CGH∥ | 30 | 14,597 | 2,439 | - | - | - | - | 5,502 | 3,835 | - | - | 19 |
|
| ||||||||||||
| NGS | 185 | 22,025 | 742 | 128 | 98 | - | - | 501¶ | 138 | 5,371 | 291 | 8 |
Affymetrix 6.0 SNP (CNP calls only).
Illumina 300K, 550K and 650K.
Custom 42M probe, NimbleGen (unique CNV loci).
Custom 24M probe, Agilent.
Tandem duplications only.
Acknowledgements
We thank J. Kidd, G. Cooper and S. Girirajan for valuable comments in the preparation of this review; P. Sudmant, F. Antonacci and J. Kitzman for their help in creating the figures; and T. Brown for proofreading the text. We also thank the authors of the algorithms that were unpublished during the preparation of this manuscript for sharing pre-prints and extended descriptions (S. McCarroll, K. Chen, A. Abyzov, Z. Iqbal and C. Stewart). B.P.C. is supported by a fellowship from the Canadian Institutes of Health Research. E. E.E. is an investigator of the Howard Hughes Medical Institute.
Glossary
- Structural variant
(SV). Genomic rearrangements that affect >50 bp of sequence, including deletions, novel insertions, inversions, mobile-element transpositions, duplications and translocations.
- Copy number variant
(CNV). Also defined as unbalanced structural variants; variants that change the number of base pairs in the genome.
- Mobile elements
DNA sequences that move location within the genome. Active mobile elements (transposons) in the human genome include Alu, L1 and SVA sequences.
- Array comparative genomic hybridization
(Array CGH). A technique based on competitively hybridizing fluorescently labelled test and reference samples to a known target DNA sequence immobilized on a solid glass substrate and then interrogating the hybridization ratio.
- SNP microarrays
Hybridization-based assays in which the target DNA sequences are discriminated on the basis of a single base difference. Assays are processed with a single sample per array and perform both SNP genotyping and copy-number interrogation.
- Single-base extension
Single-base-extension reactions use a primer that binds to a region of interest and follow this with an extension reaction that allows the incorporation of a single base after the primer.
- Segmental uniparental disomy
Uniparental disomy (often abbreviated UPD) is a cryptic alteration in which two copies of a chromosome or segment (segmental UPD) are present, but derive from a single parent.
- Nano-channel flow cells
Specialized flow cells narrow enough for a single DNA molecule to pass through in linear form without having sufficient room to fold over on itself.
- Nanoslits
Narrow channels (~1μm wide) on specialized silicon substrates. They are loaded with linear stretched DNA strands by applying a charge to microchannels on the substrates that contain electrodes.
- Emulsion picolitre droplet PCR
Emulsion PCR is based on the generation of independent PCR reaction by emulsifying the aqueous reagents in oil such that each droplet becomes a separate PCR reaction. Reagents are diluted such that each droplet contains a single target sequence.
- Paired-end reads
Two reads sequenced from the start and end of the same molecule (such as a fosmid, bacterial artificial chromosome or next-generation sequence fragment).
- Fosmid end sequence library
Paired-end sequences from a collection of bacterial cloning vectors that can carry an average of 40 kb of DNA.
- Tag SNP
A SNP in strong linkage disequilibrium with a set of SNPs or a copy number variant.
Footnotes
Competing interests statement
E.E.E. declares competing financial interests: see Web version for details.
References
- 1.Iafrate A J, et al. Detection of large-scale variation in the human genome. Nature Genet. 2004;36:949–951. doi: 10.1038/ng1416. The first report of CNVs in the human genome using array CGH.
- 2.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuzun E, et al. Fine-scale structural variation of the human genome. Nature Genet. 2005;37:727–732. doi: 10.1038/ng1562. The first study to implement a paired-end sequencing approach to study structural variation.
- 4.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conrad D F, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. This study represents the first application of an ultra-high-density CGH array.
- 6.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 7.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nature Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 8.The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. A milestone paper describing the pilot phase of the 1000 Genomes Project, the most extensive study on genomic variation in human genomes to date.
- 9.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. The first study to report CNVs in a common complex neuropsychiatric disease.
- 10.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nature Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 11.de Vries BB, et al. Diagnostic genome profiling in mental retardation. Am. J. Hum. Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stankiewicz P, Lupski JR. Genomic architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 13.Fellermann K, et al. A chromosome 8 gene-cluster polymorphism with low human b-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am. J. Hum. Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aitman TJ, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 15.Locke DP, et al. BAC microarray analysis of 15q11-q13 rearrangements and the impact of segmental duplications. J. Med. Genet. 2004;41:175–182. doi: 10.1136/jmg.2003.013813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itsara A, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snijders AM, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nature Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 18.Pinkel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 19.Park H, et al. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nature Genet. 2010;42:400–405. doi: 10.1038/ng.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nature Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 21.Perry GH, et al. The fine-scale and complex architecture of human copy-number variation. Am. J. Hum. Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller DT, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nature Genet. 2008;40:1199–1203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peiffer DA, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coe BP, et al. Resolving the resolution of array CGH. Genomics. 2007;89:647–653. doi: 10.1016/j.ygeno.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Greshock J, et al. A comparison of DNA copy number profiling platforms. Cancer Res. 2007;67:10173–10180. doi: 10.1158/0008-5472.CAN-07-2102. [DOI] [PubMed] [Google Scholar]
- 27.Curtis C, et al. The pitfalls of platform comparison: DNA copy number array technologies assessed. BMC Genomics. 2009;10:588. doi: 10.1186/1471-2164-10-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakobsson M, et al. Genotype, haplotype and copynumber variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 29.Gusev A, et al. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 2009;19:318–326. doi: 10.1101/gr.081398.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winchester L, Yau C, Ragoussis J. Comparing CNV detection methods for SNP arrays. Brief. Funct. Genomic. Proteomic. 2009;8:353–366. doi: 10.1093/bfgp/elp017. [DOI] [PubMed] [Google Scholar]
- 31.Kidd JM, et al. Characterization of missing human genome sequences and copy-number polymorphic insertions. Nature Methods. 2010;7:365–371. doi: 10.1038/nmeth.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wellcome Trust Case Control Consortium Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paris PL, et al. High resolution oligonucleotide CGH using DNA from archived prostate tissue. The Prostate. 2007;67:1447–1455. doi: 10.1002/pros.20632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hehir-Kwa JY, et al. Genome-wide copy number profiling on high-density bacterial artificial chromosomes, single-nucleotide polymorphisms, and oligonucleotide microarrays: a platform comparison based on statistical power analysis. DNA Res. 2007;14:1–11. doi: 10.1093/dnares/dsm002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wicker N, et al. A new look towards BAC-based array CGH through a comprehensive comparison with oligo-based array CGH. BMC Genomics. 2007;8:84. doi: 10.1186/1471-2164-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van de Wiel MA, et al. CGHcall: calling aberrations for array CGH tumor profiles. Bioinformatics (Oxford, England) 2007;23:892–894. doi: 10.1093/bioinformatics/btm030. [DOI] [PubMed] [Google Scholar]
- 37.van Wieringen WN, van de Wiel MA, Ylstra B. Normalized, segmented or called aCGH data? Cancer Inform. 2007;3:321–327. [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nature Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coe BP, Chari R, MacAulay C, Lam WL. FACADE: a fast and sensitive algorithm for the segmentation and calling of high resolution array CGH data. Nucleic Acids Res. 2010;38:e157. doi: 10.1093/nar/gkq548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dellinger AE, et al. Comparative analyses of seven algorithms for copy number variant identification from single nucleotide polymorphism arrays. Nucleic Acids Res. 2010;38:e105. doi: 10.1093/nar/gkq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Church DM, et al. Public data archives for genomic structural variation. Nature Genet. 2010;42:813–814. doi: 10.1038/ng1010-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 44.Heinzen EL, et al. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am. J. Hum. Genet. 2010;86:707–718. doi: 10.1016/j.ajhg.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 46.Mailman MD, et al. The NCBI dbGaP database of genotypes and phenotypes. Nature Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trask BJ, et al. Large multi-chromosomal duplications encompass many members of the olfactory receptor gene family in the human genome. Hum. Mol. Genet. 1998;7:2007–2020. doi: 10.1093/hmg/7.13.2007. [DOI] [PubMed] [Google Scholar]
- 48.Schwartz DC, et al. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science. 1993;262:110–114. doi: 10.1126/science.8211116. [DOI] [PubMed] [Google Scholar]
- 49.Teague B, et al. High-resolution human genome structure by single-molecule analysis. Proc. Natl Acad. Sci. USA. 2010;107:10848–10853. doi: 10.1073/pnas.0914638107. Application of the optical mapping technology to characterize human genome structure.
- 50.Antonacci F, et al. A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nature Genet. 2010;42:745–750. doi: 10.1038/ng.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das SK, et al. Single molecule linear analysis of DNA in nano-channel labeled with sequence specific fluorescent probes. Nucleic Acids Res. 2010;38:e177. doi: 10.1093/nar/gkq673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jo K, et al. A single-molecule barcoding system using nanoslits for DNA analysis. Proc. Natl Acad. Sci. USA. 2007;104:2673–2678. doi: 10.1073/pnas.0611151104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao M, et al. Direct determination of haplotypes from single DNA molecules. Nature Methods. 2009;6:199–201. doi: 10.1038/nmeth.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beer NR, et al. On-chip, real-time, single-copy polymerase chain reaction in picoliter droplets. Anal. Chem. 2007;79:8471–8475. doi: 10.1021/ac701809w. [DOI] [PubMed] [Google Scholar]
- 55.Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nature Biotech. 2009;27:847–852. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 57.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKernan KJ, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korbel J O, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. The first study in SV discovery using second-generation sequencing technologies.
- 60.Volik S, et al. End-sequence profiling: sequence-based analysis of aberrant genomes. Proc. Natl Acad. Sci. USA. 2003;100:7696–7701. doi: 10.1073/pnas.1232418100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nature Methods. 2009;6:S13–S20. doi: 10.1038/nmeth.1374. An extensive review on sequencing-based methods for discovering structural variation.
- 62.Mills R E, et al. Mapping copy number variation at fine scale by population scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. Describes the SV discovery and analysis efforts of the 1000 Genomes Project.
- 63.Kidd JM, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korbel JO, et al. PEMer: a computational framework with simulation-based error models for inferring genomic structural variants from massive paired-end sequencing data. Genome Biol. 2009;10:R23. doi: 10.1186/gb-2009-10-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hormozdiari F, Alkan C, Eichler EE, Sahinalp SC. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res. 2009;19:1270–1278. doi: 10.1101/gr.088633.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hormozdiari F, et al. Next-generation VariationHunter: combinatorial algorithms for transposon insertion discovery. Bioinformatics (Oxford, England) 2010;26:i350–i357. doi: 10.1093/bioinformatics/btq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hormozdiari F, Hajirasouliha I,A,M, Eichler EE, Sahinalp SC. Simultaneous structural variation discovery in multiple paired-end sequenced genomes. Proc. RECOMB. 2011 doi: 10.1101/gr.120501.111. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nature Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee S, Hormozdiari F, Alkan C, Brudno M. MoDIL: detecting small indels from clone-end sequencing with mixtures of distributions. Nature Methods. 2009;6:473–474. doi: 10.1038/nmeth.f.256. [DOI] [PubMed] [Google Scholar]
- 70.Lee S, Xing E, Brudno M. MoGUL: detecting common insertions and deletions in a population. Proc. RECOMB 2010. 2010;6044:357–368. [Google Scholar]
- 71.Quinlan AR, et al. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 2010;20:623–635. doi: 10.1101/gr.102970.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Campbell P J, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nature Genet. 2008;40:722–729. doi: 10.1038/ng.128. This manuscript describes the use of NGS technologies to characterize rearrangements in cancer.
- 73.Chiang DY, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nature Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alkan C, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nature Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. The first publication to describe methods to predict absolute copy numbers of duplicated segments.
- 75.Sudmant P H, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. Provides copy-number maps in 159 genomes and describes the SUN method to accurately genotype duplications and characterize paralogue-specific copy numbers.
- 76.Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. Feb 7 2011; doi: 10.1101/gr.114876.110. doi:10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mills RE, et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics (Oxford, England) 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xing J, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pang AW, et al. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 2010;11:R52. doi: 10.1186/gb-2010-11-5-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chaisson MJ, Brinza D, Pevzner PA. De novo fragment assembly with short mate-paired reads: does the read length matter? Genome Res. 2009;19:336–346. doi: 10.1101/gr.079053.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2009;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl Acad. Sci. USA. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hajirasouliha I, et al. Detection and characterization of novel sequence insertions using paired-end next-generation sequencing. Bioinformatics (Oxford, England) 2010;26:1277–1283. doi: 10.1093/bioinformatics/btq152. The first computational framework to merge local and de novo sequence assembly methods to characterize novel sequence insertions using NGS technology.
- 88.She X, et al. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- 89.Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nature Methods. 2011;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Medvedev P, Fiume M, Dzamba M, Smith T, Brudno M. Detecting copy number variation with mated short reads. Genome Res. 2010;20:1613–1622. doi: 10.1101/gr.106344.110. The first algorithm to incorporate both read-depth and read-pair methods for accurate CNV discovery.
- 91.Handsaker RE, Korn JM, Nemesh J, McCarroll SA. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nature Genet. 2011 Feb 13; doi: 10.1038/ng.768. doi:10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schatz MC, Delcher AL, Salzberg SL. Assembly of large genomes using second-generation sequencing. Genome Res. 2010;20:1165–1173. doi: 10.1101/gr.101360.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Human genome: genomes by the thousand. Nature. 2010;467:1026–1027. doi: 10.1038/4671026a. [DOI] [PubMed] [Google Scholar]
- 94.Weksberg R, et al. A method for accurate detection of genomic microdeletions using real-time quantitative PCR. BMC Genomics. 2005;6:180. doi: 10.1186/1471-2164-6-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schaeffeler E, Schwab M, Eichelbaum M, Zanger UM. CYP2D6 genotyping strategy based on gene copy number determination by TaqMan real-time PCR. Hum. Mutation. 2003;22:476–485. doi: 10.1002/humu.10280. [DOI] [PubMed] [Google Scholar]
- 96.Gomez-Curet I, et al. Robust quantification of the SMN gene copy number by real-time TaqMan PCR. Neurogenetics. 2007;8:271–278. doi: 10.1007/s10048-007-0093-1. [DOI] [PubMed] [Google Scholar]
- 97.Armour JA, Sismani C, Patsalis PC, Cross G. Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res. 2000;28:605–609. doi: 10.1093/nar/28.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kumps C, et al. Multiplex amplicon quantification (MAQ), a fast and efficient method for the simultaneous detection of copy number alterations in neuroblastoma. BMC Genomics. 2010;11:298. doi: 10.1186/1471-2164-11-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schouten JP, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fan HC, Blumenfeld YJ, El-Sayed YY, Chueh J, Quake SR. Microfluidic digital PCR enables rapid prenatal diagnosis of fetal aneuploidy. Am. J. Obstet. Gynecol. 2009;200:543.e1–543.e7. doi: 10.1016/j.ajog.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shen F, Du W, Kreutz JE, Fok A, Ismagilov RF. Digital PCR on a SlipChip. Lab Chip. 2010;10:2666–2672. doi: 10.1039/c004521g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Diehl F, et al. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nature Methods. 2006;3:551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- 103.Weaver S, et al. Taking qPCR to a higher level: analysis of CNV reveals the power of high throughput qPCR to enhance quantitative resolution. Methods (San Diego, California) 2010;50:271–276. doi: 10.1016/j.ymeth.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 104.Mefford HC, et al. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 2009;19:1579–1585. doi: 10.1101/gr.094987.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zerr T, Cooper G M, Eichler E E, Nickerson D A. Targeted interrogation of copy number variation using SCIMMkit. Bioinformatics (Oxford, England) 2010;26:120–122. doi: 10.1093/bioinformatics/btp606. References 104 and 105 describe an experimental method to rapidly and efficiently genotype thousands of cases for disease-associated candidate regions.
- 106.Lam HY, et al. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nature Biotech. 2010;28:47–55. doi: 10.1038/nbt.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Waszak SM, et al. Systematic inference of copynumber genotypes from personal genome sequencing data reveals extensive olfactory receptor gene content diversity. PLoS Comput. Biol. 2010;6:e1000988. doi: 10.1371/journal.pcbi.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Conrad DF, et al. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nature Genet. 2010;42:385–391. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Itsara A, et al. De novo rates and selection of large copy number variation. Genome Res. 2010;20:1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zody MC, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nature Genet. 2008;40:1076–1083. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kitzman JO, et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nature Biotech. 2011;29:59–63. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Oostlander AE, Meijer GA, Ylstra B. Microarray-based comparative genomic hybridization and its applications in human genetics. Clin. Genet. 2004;66:488–495. doi: 10.1111/j.1399-0004.2004.00322.x. [DOI] [PubMed] [Google Scholar]
- 113.Conlin LK, et al. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum. Mol. Genet. 2010;19:1263–1275. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rodriguez-Santiago B, et al. Mosaic uniparental disomies and aneuploidies as large structural variants of the human genome. Am. J Hum. Genet. 2010;87:129–138. doi: 10.1016/j.ajhg.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Perry GH, et al. Diet and the evolution of human amylase gene copy number variation. Nature Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]