Abstract
Turnover of integrin-based focal adhesions (FAs) with the extracellular matrix (ECM) is essential for coordinated cell movement. In collectively migrating human keratinocytes, FAs assemble near the leading edge, grow and mature as a result of contractile forces, and disassemble underneath the advancing cell body. We report that clustering of microtubule-associated CLASP1 and CLASP2 proteins around FAs temporally correlates with FA turnover. CLASPs and LL5β, which recruits CLASPs to FAs, facilitate FA disassembly. CLASPs are further required for FA-associated ECM degradation, and matrix metalloprotease inhibition slows FA disassembly similar to CLASP or LL5β depletion. Finally, CLASP-mediated microtubuletethering at FAs establishes a FA-directed transport pathway for delivery, docking and localized fusion of exocytic vesicles near FAs. We propose that CLASPs couple microtubule organization, vesicle transport and cell interactions with the ECM, establishing a local secretion pathway that facilitates FA turnover by severing cell-matrix connections.
INTRODUCTION
Cell migration is essential for development, tissue remodelling and wound healing, and requires coordination of intracellular signalling and cytoskeleton dynamics to generate traction forces. These forces are transmitted to the cell exterior by focal adhesions (FAs), stratified protein structures that connect the actin cytoskeleton to the extracellular matrix (ECM). FAs are initiated outside the cell by ECM-binding of integrin trans-membrane receptors, promoting integrin clustering and recruitment of intracellular adaptor proteins1. Nascent adhesions either rapidly turn over or connect to the actin cytoskeleton, and grow and mature in response to actomyosin contractile forces. Mature FAs thus mechanically couple the ECM to the actin cytoskeleton and provide anchor points required for cell migration2. However, FAs also have to release and disassemble underneath the cell body for productive forward movement, and an unresolved question is how FA disassembly is spatially and temporally controlled in a migrating cell.
Microtubules regulate FA disassembly3, and repeated targeting by dynamic microtubules promotes FA turnover4. Global FA disassembly is also induced by microtubule regrowth after nocodazole removal, which involves FAK, Rho GTPases and the endocytic machinery5-7. However, it is not known how FA turnover is locally controlled in migrating cells. Several +TIPs that bind to growing microtubule plus ends8, including the adenomatous polyposis coli protein, the spectraplakin MACF1/ACF7, and CLASP proteins, are closely associated with FAs9-11. CLASPs promote the stability of peripheral microtubules11,12, but it is not known why CLASP-mediated microtubule stabilization is important for cell migration.
By using quantitative live-cell imaging of migrating epithelial cells in which FAs display highly coordinated turnover, we find that CLASPs tether microtubules to FAs. CLASPs facilitate disassembly of mature FAs, are required for FA-associated ECM degradation, and we identify FAs as hotspots of exocytosis. Based on these data, we propose that FA disassembly can be facilitated through targeted, local exocytosis and ECM degradation releasing integrin-matrix connections. CLASPs are thus central to coupling the organization of intracellular vesicle transport to the remodelling of cell-matrix interactions, highlighting a previously unappreciated molecular pathway controlling FA turnover.
RESULTS
FA-associated CLASP-decorated microtubule clusters correlate with FA disassembly
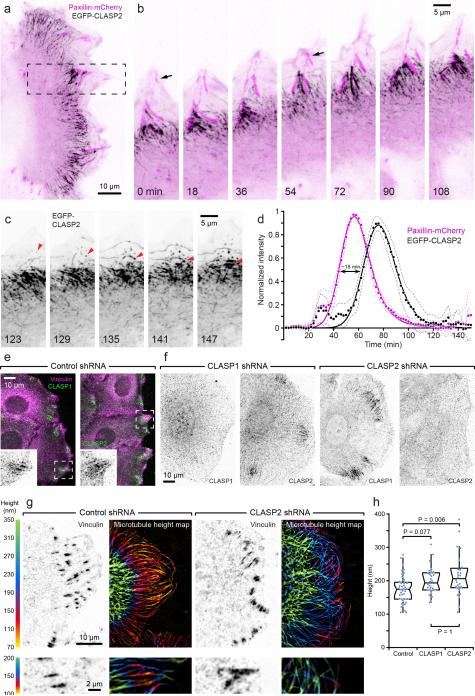
Wounding of a confluent HaCaT cell monolayer induces sheet migration in which cells retain cell-cell contacts11. FAs labelled by stable expression of paxillin-mCherry displayed highly coordinated turnover dynamics at the edge of these migrating cell sheets. In contrast to randomly migrating cells in which many nascent FAs turn over rapidly1, most FAs that appeared near the leading edge of migrating HaCaT cells, matured, and turned over in a highly coordinated manner as the cells moved forward. Thus, HaCaT sheet migration is a good model for investigating the dynamics of a homogenous FA population involved in cell migration. Transiently expressed EGFP-CLASP2 consistently accumulated along microtubules around >90% of mature FAs at the border between cell body and lamella/lamellipodia (Supplementary Video 1). These EGFP-CLASP2-decorated microtubules did not overlap with FAs, but instead surrounded and appeared to engulf FAs shortly before FA disassembly (Fig. 1a, b). In addition, EGFP-CLASP2 punctae appeared near the leading edge, flowed inward and accumulated around FAs, and appeared to capture microtubules at areas adjacent to FAs (Fig. 1c). Quantitative analysis of fluorescence intensity dynamics of paxillin-mCherry and EGFP-CLASP2 turnover confirmed that CLASP2-decorated microtubule clusters begin to assemble around mature FAs (Fig. 1d). Further CLASP2 accumulation correlated with FA disassembly, suggesting a role of CLASPs during the disassembly phase of FA turnover, and CLASP2-decorated MT clusters disappeared after complete FA disassembly. Both human CLASP isoforms, CLASP1 and CLASP2, similarly and independently localized around FAs, as indicated by immunofluorescence with isoform-specific antibodies (Fig. 1e, f). These data demonstrate a tight spatial and temporal correlation between FA turnover and the accumulation of CLASP-decorated microtubules, suggesting that CLASPs act to target and/or transiently capture microtubules at mature FAs.
Figure 1. Mature FAs recruit CLASP2-decorated microtubules.
(a) Spinning disk confocal microscopy of a migrating HaCaT epithelial cell expressing EGFP-CLASP2 (black) and paxillin-mCherry (magenta). In this and subsequent figures, images from live time-lapse sequences are contrast-inverted to better visualize intracellular protein dynamics. See methods for details. (b) EGFP-CLASP2 and paxillin-mCherry dynamics at the advancing cell edge in the region indicated in (a). Arrows indicate the birth of two FAs that are engulfed by CLASP2-decorated microtubules prior to disassembly. Elapsed time is in minutes. (c) Continuation of the same sequence showing only the EGFP-CLASP2 channel to highlight EGFP-CLASP2 particles that appear near the leading edge, increase in intensity while flowing retrograde, and often appear to capture growing microtubules (red arrowheads). (d) Turnover dynamics of paxillin-mCherry-labelled FAs and surrounding EGFP-CLASP2. Fluorescence intensity profiles measured as a function of time were normalized to the maximum paxillin-mCherry fluorescence intensity for each FA and aligned relative to each other (n = 20 FAs). The solid line is an exponentially-modified Gaussian curve fit, and dashed lines 95% confidence intervals. (e) Immunofluorescence of endogenous CLASP1 and CLASP2. Insets show only the CLASP channel. In this and subsequent figures, single channels are displayed with inverted contrast for improved display. (f) Immunofluorescence demonstrating isoform-specific shRNA-mediated CLASP depletion. (g) Scanning angle interference microscopy (SAIM) of microtubule height above the substrate in control and CLASP2-depleted cells. Bottom panels show areas from different cells at higher magnification and a shallower height map. Note the decrease of microtubule height specifically near FAs in control cells that is largely absent in CLASP2-depleted cells. (h) Quantification of microtubule height near FAs larger than 1 μm2 in control and CLASP-depleted cells. n = 40 (control shRNA); 28 (CLASP1 shRNA); 28 (CLASP2 shRNA) cells from two experiments. Box-and-whisker plot shows median, 1st and 3rd quartile (box), and 95% confidence intervals (notches) with whiskers extending to the furthest observations within ±1.5 times the interquartile range. Dots are individual data points, and source data are included in Supplementary Table 3. p values were calculated by non-parametric Kruskal-Wallis analysis of variance with Bonferroni error correction.
CLASPs anchor microtubules near FAs
To test whether CLASPs specifically mediate microtubule interactions with FAs, we generated HaCaT cell lines stably expressing either a control non-targeting shRNA or shRNAs targeting either CLASP1 or CLASP2. Out of four independent shRNA sequences tested for each CLASP isoform, two resulted in specific reduction of either CLASP1 or CLASP2 protein levels by >90% (Supplementary Fig. 1e; Fig. 1f). We were unable to generate CLASP1/2 double-depleted cell lines, likely due to mitotic defects13. Depletion of either CLASP isoform resulted in fragmentation of the Golgi apparatus (Supplementary Fig. 1a, f) indicating significant knockdown of CLASP function in either CLASP1 or CLASP2-depleted cells14.
Consistent with previous reports, the peripheral microtubule array was less dense and disorganized in CLASP depleted cells (Supplementary Fig. 1b)11,12. To determine whether CLASPs specifically mediate microtubule interactions with FAs, we used Scanning Angle Interference Microscopy (SAIM), a super-resolution technique that allows measurements of the distance of fluorescent-labelled structures from the ventral cell surface at ~5 nm axial resolution15. Consistent with previous TIRF studies, in control cells, microtubules descended toward the ventral cell surface as microtubules approached the cell periphery (Fig. 1g)16. However, axial resolution in TIRF is limited, and the improved resolution achieved by SAIM indicated that microtubules further approached the ventral cell surface in close proximity to FAs (Fig. 1g). The height of these FA-associated microtubules above the substrate-medium interface was often markedly less than 150 nm, indicating axial co-localization of these microtubules with the top of the FA plaque15,17. Strikingly, in CLASP-depleted cells, microtubules did not descend toward FAs and remained significantly above the height of FA-associated microtubules in control cells (Fig. 1g, h), indicating that CLASPs participate in tethering microtubules to FAs.
CLASPs facilitate FA turnover during epithelial sheet migration
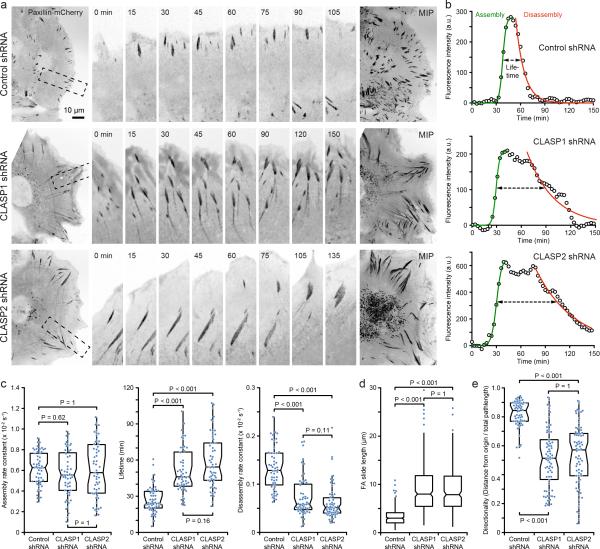
The striking spatiotemporal correlation between CLASP cluster formation and the disassembly phase of FA turnover indicated a role for CLASP-decorated MTs during FA disassembly. To test this hypothesis, we analysed FAs by immunofluorescence of endogenous paxillin. Depletion of either CLASP1 or CLASP2 resulted in a 3-5-fold increase in FA size (Supplementary Fig. 1d, g) associated with thick F-actin bundles with increased myosin phosphorylation (Supplementary Fig. 1b, c, d), and CLASP-depleted cells appeared larger and flatter. Because these findings are consistent with a FA turnover defect, we compared FA dynamics in control and CLASP1- or CLASP2-depleted paxillin-mCherry-expressing, migrating HaCaT keratinocytes by time-lapse spinning disk confocal microscopy (Fig. 2a; Supplementary Video 2). FAs that matured at the base of the lamella appeared both larger and longer-lived in CLASP-depleted cells. To quantify the dynamics of these FAs, paxillin-mCherry fluorescence intensity profiles as a function of time were fitted with a logistic function during the assembly phase, and a single exponential decay during the disassembly phase (Fig. 2b)18-20. FA lifetime was defined as the time during which the paxillin-mCherry fluorescence intensity remained above the half maximum. While the FA assembly rate was unchanged, the disassembly rate was significantly decreased and FA lifetime was increased by about two-fold in CLASP depleted cells (Fig. 2c). Consistent with a FA disassembly defect, other aspects of FA dynamics were aberrant in CLASP-depleted cells. Individual FAs often underwent cycles of failed disassembly and reassembly, and large, mature FAs often started to slide toward the cell centre (Fig. 2d) eventually undergoing disassembly once they appeared to reach a critical tension threshold. Such extensive FA sliding was not observed in control cells. Coordinated FA turnover is important for productive cell migration, and, the directionality of migrating CLASP-depleted cells was reduced compared with control cells that consistently moved in nearly straight paths perpendicular to the edge of the cell monolayer (Fig. 2e; Supplementary Fig. 2). Taken together, these data demonstrate that CLASPs are key players in controlling the disassembly phase of FA dynamics.
Figure 2. CLASPs facilitate FA disassembly in migrating epithelial cells.
(a) Time-lapse sequences of paxillin-mCherry dynamics in control and CLASP-depleted migrating HaCaT cells. The regions indicated are shown at higher magnification. The maximum intensity projections (MIP) over the entire three hour time-lapse sequences on the right further illustrate FA turnover defects and sliding. Elapsed time is in minutes. (b) Examples of turnover dynamics of representative FAs in control and CLASP-depleted cells used for calculation of FA dynamics parameters in (c). Data points are 3-frame running averages of FA fluorescence intensity. The green solid line is a logistic fit of the FA assembly phase, the red line a single exponential decay fit of the disassembly phase. The dashed arrow indicates FA lifetime as defined by fluorescence intensity above the half-maximum of the fit. (c) Analysis of FA assembly rates, lifetime and disassembly rates in control and CLASP-depleted migrating HaCaT cells. n = 54 (control shRNA); 53 (CLASP1 shRNA); 55 (CLASP2 shRNA) FAs from three experiments. (d) Length of FA sliding measured from MIPs. n = 405 (control shRNA); 217 (CLASP1 shRNA); 404 (CLASP2 shRNA) FAs. Representative data set of three experiments. Only outliers are shown as individual data points. (e) Quantification of directed migration of control and CLASP-depleted HaCaT cells at the edge of a cell monolayer. n = 62 (control shRNA); 71 (CLASP1 shRNA); 71 (CLASP2 shRNA) cells. Representative data set of three experiments. Box-and-whisker plots show median, 1st and 3rd quartile (box), and 95% confidence intervals (notches) with whiskers extending to the furthest observations within ±1.5 times the interquartile range. Dots are individual data points, and source data for (c), (d), and (e) are included in Supplementary Table 3. p values were calculated by non-parametric Kruskal-Wallis analysis of variance with Bonferroni error correction.
FAs direct CLASP cluster assembly
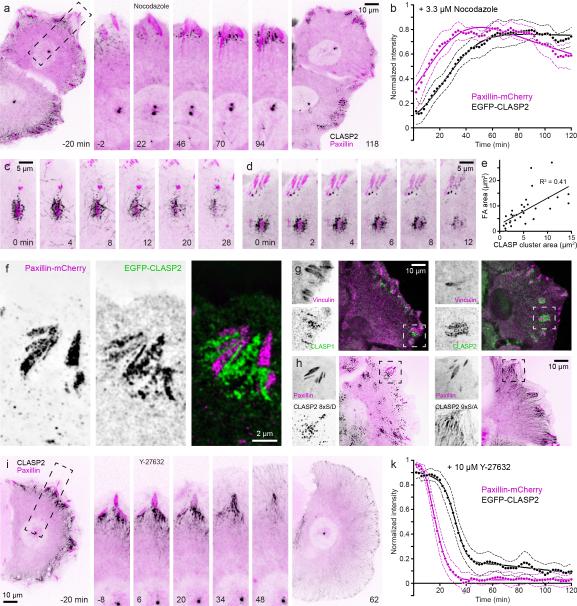
We next analysed what determined the formation and maintenance of cortical FA-associated CLASP accumulation. Notably, microtubules were not required. Upon treatment with 3.3 μM nocodazole, which depolymerized nearly all microtubules and increased contractility and FA assembly21, EGFP-CLASP2-labeled growing microtubule ends rapidly disappeared. In contrast, FA-associated EGFP-CLASP2 clusters continued to increase in fluorescence intensity as FAs increased in size (Fig. 3a, b; Supplementary Video 3). These microtubule-independent CLASP clusters were embedded in, but did not overlap with the FA plaque (Fig. 3f), and their size positively correlated with FA size (Fig. 3e). This indicates that a FA-generated, dose-dependent signal determines CLASP cluster assembly. After nocodazole washout, EGFP-CLASP2 at FAs rapidly re-associated with newly growing microtubules prior to FA disassembly (Fig. 3c; Supplementary Video 4), suggesting that FA-associated CLASP clusters participate in microtubule capture to promote FA disassembly. Endogenous CLASP1 and CLASP2 formed similar clusters around FAs in nocodazole-treated cells (Fig. 3g), demonstrating that these microtubule-independent CLASP clusters were not artefacts of EGFP-CLASP2 overexpression. In addition, EGFP-CLASP2 8xS/D that has reduced microtubule-binding activity 11, accumulated in similar punctate clusters around FAs (Fig. 3h). Nonphosphorylatable EGFP-CLASP2 9xS/A decorated microtubules also accumulated around FAs (Fig. 3h), indicating that CLASP2 accumulation around FAs is independent of GSK3β phosphorylation and microtubule association. Next, we tested whether CLASP clusters depended on FAs. Maintenance of mature FAs requires myosin-mediated contractility22. Contractility inhibition by the Rho-kinase inhibitor Y-27632 resulted in synchronized, rapid FA disassembly. EGFP-CLASP2-decorated microtubules immediately filled the area previously occupied by the FA (Fig. 3i, Supplementary Video 5), and EGFP-CLASP2-decorated microtubule clusters disappeared ~15 min after Y-27632 induced FA disassembly (Fig 3k). Microtubules were not required for CLASP cluster disassembly as Y-27632 also rapidly induced FA and associated EGFP-CLASP2 disassembly in cells in which microtubules had first been depolymerized with nocodazole (Fig. 3d). Together, these data show that FAs recruit and maintain associated CLASP clusters independent of CLASP interactions with microtubules.
Figure 3. CLASP clusters around FAs do not depend on microtubules.
(a) Time-lapse sequence of EGFP-CLASP2 (black) and paxillin-mCherry (magenta) dynamics in HaCaT epithelial cells treated with 3.3 μM nocodazole at t = 0 min. (b) Average dynamics of individual paxillin-mCherry-labeled FAs and surrounding EGFP-CLASP2 in nocodazole-treated cells (n = 16 FAs). The solid line is an exponentially-modified Gaussian curve fit, and dashed lines are 95% confidence intervals. (c) Time-lapse sequence of EGFP-CLASP2 (black) and paxillin-mCherry (magenta) dynamics after nocodazole washout, illustrating microtubule repolymerization and capture at FA-associated CLASP clusters, and subsequent FA disassembly. (d) Time-lapse sequence of EGFP-CLASP2 (black) and paxillin-mCherry (magenta) dynamics in HaCaT cells in which microtubules were depolymerized by 3.3 μM nocodazole for 90 minutes prior to the addition of 10 μM Rho-kinase inhibitor Y-27632, resulting in disassembly of both FAs and associated CLASP clusters. (e) Scatter plot of the correlation between FA size and microtubule-independent EGFP-CLASP2 cluster. (f) Structured illumination super-resolution microscopy (SIM) of EGFP-CLASP2 and paxillin-mCherry expressing nocodazole-treated HaCaT cells, illustrating close intercalation of CLASP clusters and FAs. (g) Localization of endogenous CLASP1 (left) and CLASP2 (right) around FAs in 3.3 μM nocodazole-treated HaCaT cells. (h) HaCaT cells expressing paxillin-mCherry and either phosphomimetic EGFP-CLASP2 8xS/D (left) or non-phosphorylatable EGFP-CLASP2 9xS/A (right). (i) Time-lapse sequence of EGFP-CLASP2 (black) and paxillin-mCherry (magenta) dynamics in HaCaT epithelial cells treated with 10 μM Y-27632 at t = 0 min. (k) Average dynamics of individual paxillin-mCherry-labeled FAs and surrounding EGFP-CLASP2 in 10 μM Y-27632-treated cells (n = 20 FAs). The solid line is an exponentially-modified Gaussian curve fit, and dashed lines are 95% confidence intervals. All elapsed time is in minutes.
LL5β is required for CLASP-mediated FA turnover
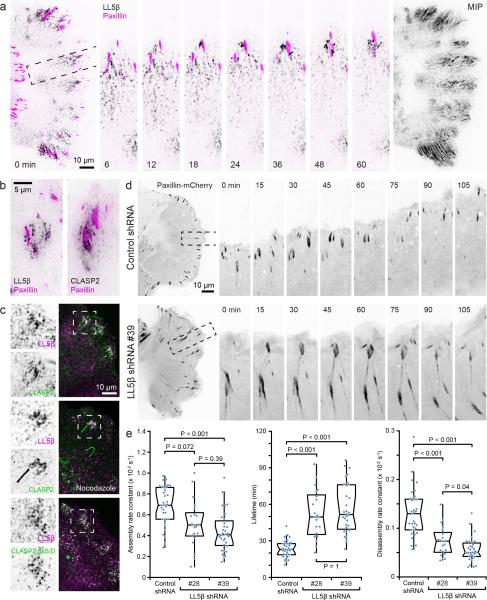
We next determined whether LL5β, a peripheral membrane protein that binds CLASPs and is required for CLASP recruitment to the cell cortex23, displayed FA-associated dynamics similar to CLASPs. In contrast to non-motile HeLa cells in which cortical LL5β is static23, LL5β clusters in migrating HaCaT keratinocytes were highly dynamic, and appeared and grew in the vicinity of maturing FAs (Fig. 4a; Supplementary Video 6). Similar to CLASPs, FA-associated LL5β fluorescence intensity reached a maximum when mCherrypaxillin fluorescence peaked and the FA had started to disassemble. In addition, small, punctate LL5β particles appeared to be released from these FA-associated sites and underwent retrograde flow toward the cell body during which they slowly disappeared. Similar to CLASPs, cortical FA-associated LL5β clusters depended on FAs as they quickly disappeared in Y-27632 treated cells (Supplementary Fig. 3a). In contrast, LL5β clusters did not require microtubules or phosphoinositide 3-kinase (PI3K) activity. EGFP-CLASP2 and endogenous LL5β staining partially overlapped around FAs (Fig. 4c), which was further enhanced when CLASP2 binding to microtubules was eliminated either by microtubule depolymerization or in cells expressing EGFP-CLASP2 8xS/D (Fig 4c). In addition, both EGFP-LL5β and EGFP-CLASP2 localized to similar punctate FA-associated structures by TIRF microscopy (Fig. 4b). Overall, the localization and dynamics of these LL5β clusters were similar to the spatiotemporal correlation of CLASPs with FAs.
Figure 4. LL5β is required for CLASP-mediated focal adhesion turnover.
(a) Time-lapse sequence of EGFP-LL5β (black) and paxillin-mCherry (magenta) dynamics in a migrating HaCaT epithelial cell at the edge of a cell monolayer. The image on the right shows a maximum intensity projection (MIP) of only the LL5β channel over the entire 3 hour sequence, illustrating the complex dynamics of LL5β particles near FAs. Elapsed time is in minutes. (b) Total internal reflection (TIRF) microscopy of EGFP-LL5β and EGFP-CLASP2 near paxillin-mCherry labeled FAs, demonstrating similar punctate pattern at the ventral cell surface. (c) LL5β immunofluorescence in HaCaT cells expressing the indicated EGFP-CLASP2 constructs indicating increased overlap of LL5β and CLASPs in the absence of CLASP-microtubule binding. (d) Time-lapse sequences of paxillin-mCherry dynamics in control and LL5β-depleted migrating HaCaT cells. The regions indicated are shown at higher magnification. Elapsed time is in minutes. (e) Analysis of FA assembly rates, lifetime and disassembly rates in control and LL5β-depleted migrating HaCaT cells. n = 33 (control shRNA); 21 (LL5β shRNA #28); 32 (LL5β shRNA #39) FAs from three experiments. Box-and-whisker plots show median, 1st and 3rd quartile (box), and 95% confidence intervals (notches) with whiskers extending to the furthest observations within ±1.5 times the interquartile range. Dots are individual data points, and source data are included in Supplementary Table 3. p values were calculated by non-parametric Kruskal-Wallis analysis of variance with Bonferroni error correction.
If LL5β recruits CLASPs to FAs, we expected LL5β to also be required for normal FA disassembly dynamics. To test this hypothesis, we generated LL5β-depleted HaCaT cell lines using two independent shRNA sequences that reduced LL5β expression levels by ~90% (Supplementary Fig. 3b, c). As expected, LL5β depletion resulted in loss of FA-associated CLASPs (Supplementary Fig. 3d). In contrast, LL5β accumulation around FAs was not affected in CLASP-depleted cells (Supplementary Fig. 3e), confirming that LL5β recruits CLASPs to the cell cortex23. Cytoskeleton phenotypes (Supplementary Fig. 3f, g) and FA turnover defects in LL5β-depleted cells were similar to what we observed in CLASP-depleted cells. Compared with control, FAs were larger in LL5β-depleted cells, and displayed 2-3-fold increased lifetimes and decreased disassembly rates (Fig. 4d, e). In contrast to CLASP-depleted cells, LL5β-depleted cells also exhibited a moderate FA assembly defect, suggesting additional CLASP-independent functions. Together, these data demonstrate that LL5β-mediated CLASP recruitment around FAs facilitates FA turnover.
CLASPs are required for FA-associated extracellular matrix degradation
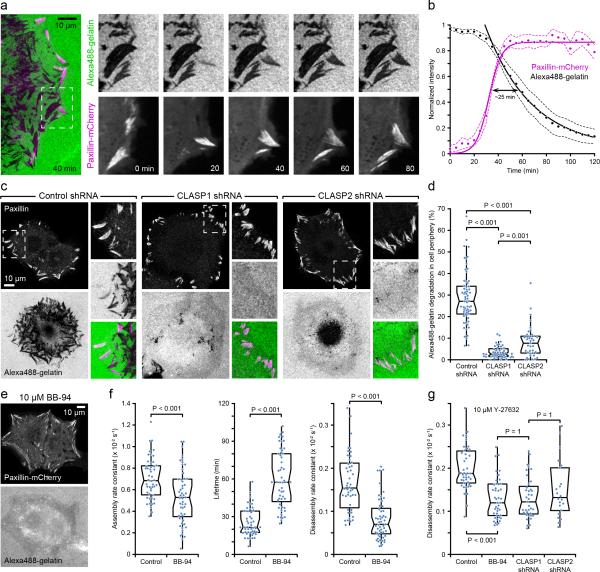
ECM degradation mediated by the membrane-associated matrix metalloprotease MT1-MMP occurs in the vicinity of FA sites in invasive cancer cells24. To determine if FA-associated ECM degradation is a normal cell process and a mechanism that could control FA turnover dynamics, we examined whether FA-associated ECM degradation occurred in non-transformed migrating epithelial cells plated on fluorescently labelled gelatine. HaCaT cells spread slowly on gelatine-coated coverslips and exhibited little ECM degradation at early time points, consistent with low invasiveness of precancerous HaCaT cells25. After 24 hours however, in addition to punctate ECM degradation underneath the cell centre, peripheral areas of ECM degradation sharply delineated FAs creating a pattern of footprints of many generations of FAs during cell spreading. Time-lapse microscopy and analysis of fluorescence intensities further revealed a strong spatial and temporal correlation of paxillin-mCherry FA turnover and Alexa-488 gelatine degradation (Fig. 5a, b; Supplementary Video 7) similar to CLASP accumulation around FAs. In CLASP-depleted cells, residual gelatine degradation localized to the cell centre and did not correlate with FAs, demonstrating that CLASPs are required for local FA-associated ECM degradation (Fig. 5c, d).
Figure 5. CLASPS are required for FA-associated ECM degradation.
(a) Time-lapse sequence of Alexa488-gelatine degradation in a paxillin-mCherry-expressing cell illustrating spatiotemporal correlation with FA turnover. The region indicated is shown at higher magnification. Areas of matrix degradation appear as dark areas in the Alexa488-gelatine images. Elapsed time is indicated in minutes. (b) Average turnover dynamics of Alexa488-gelatine at individual paxillin-mCherry-labeled FAs. Fluorescence intensity profiles measured as a function of time were normalized to the maximum Alexa488-gelatine and paxillin-mCherry fluorescence intensities, and aligned relative to the half maximum of FA assembly (n = 20 FAs). Because FA lifetime is highly variable in spreading HaCaT cells, the FA disassembly phase was not included in this quantification. Solid lines are a logistic fit for FA assembly and a single exponential decay for ECM degradation. Dashed lines are 95% confidence intervals. (c) Matrix degradation by control and CLASP-depleted HaCaT cells after 24 hours of spreading on Alexa488-gelatine-coated coverslips (green). Cells were also immunostained for paxillin (magenta). (d) Quantification of the area of gelatin degradation in the periphery of control and CLASP-depleted cells. n = 64 (control shRNA); 50 (CLASP1 shRNA); 39 (CLASP2 shRNA) cells from three experiments. (e) Alexa488-gelatine degradation is inhibited in BB-94 treated cells. (f) Analysis of FA assembly rates, lifetime and disassembly rates in control and BB-94-treated migrating HaCaT cells. n = 48 (control); 53 (BB-94) FAs from three experiments. (g) Analysis of Y-27632-induced (10 μM) FA disassembly in control, BB-94-treated, and CLASP-depleted HaCaT cells. n = 43 (control); 43 (BB-94); 37 (CLASP1 shRNA); 22 (CLASP2 shRNA) FAs. Box-and-whisker plots show median, 1st and 3rd quartile (box), and 95% confidence intervals (notches) with whiskers extending to the furthest observations within ±1.5 times the interquartile range. Dots are individual data points, and source data for (d), (f), and (g) are included in Supplementary Table 3. p values were calculated by non-parametric Kruskal-Wallis analysis of variance with Bonferroni error correction.
To determine if proteolytic ECM degradation near FAs is a mechanism by which CLASPs facilitate FA disassembly, we treated cells with BB-94, a broad-spectrum MMP inhibitor26. As expected, BB-94 completely blocked FA-associated Alexa-488 gelatin degradation24 (Fig. 5e). In addition, BB-94 inhibited FA disassembly (Fig. 5f) similar to what we observed in CLASP- or LL5β-depleted cells (Fig. 2c, 4e). This demonstrates that, like CLASPs, extracellular MMP activity facilitates FA turnover. Because inhibition of actomyosin contractility induces rapid and simultaneous disassembly of mature FAs, we asked to what extent this depended on CLASPs or MMP activity. Y-27632 still induced FA disassembly in both BB-94-treated or CLASP-depleted cells at a slightly reduced rate comparable with normal FA disassembly in migrating cells (Fig. 5g). This indicates that although FA-associated ECM degradation facilitates FA disassembly, inhibition of FA-associated contractile forces can still trigger dissociation of the intracellular FA plaque in the absence of MMP activity and ECM degradation.
CLASPs direct FA-associated exocytosis
Our results implicate FA-associated ECM degradation as a potential mechanism by which CLASPs facilitate FA turnover. We thus asked how CLASPs could guide local MMP activity. Because CLASP clusters connect microtubules to mature FAs, we hypothesized that intracellular vesicle transport to and/or from FAs along FA-associated microtubule tracks may participate in CLASP-mediated FA disassembly. Recent reports implicated endocytosis in FA disassembly in fibroblasts5,6,27. However, we could not detect obvious overlap of EGFP-CLASP2 localization with clathrin heavy chain or dynamin II immunofluorescence in either control migrating HaCaT cells, nocodazole-treated cells in which CLASPs maximally accumulated around FAs, or cells in which FA disassembly was acutely induced by nocodazole washout (Supplementary Fig. 4), indicating that CLASP clusters are likely not involved in an endocytosis-mediated mechanism of FA turnover.
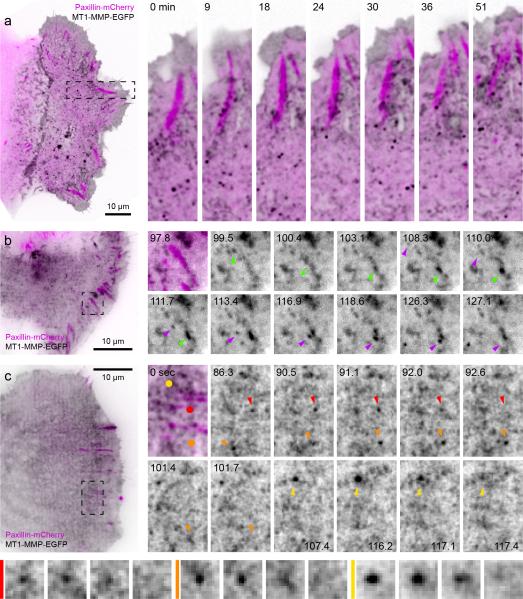
To test whether CLASP-stabilized microtubule tracks toward FAs could instead serve as anterograde transport pathways mediating local ECM degradation through FA-associated MMP exocytosis, we analyzed MT1-MMP-EGFP dynamics in relation to mCherry-paxillin FA turnover24. A large fraction of MT1-MMP-EGFP localized to the plasma membrane indicating that the C-terminal cytoplasmic EGFP does not disrupt MT1-MMP exocytosis. In addition, membrane-bound as well as MT1-MMP-EGFP in smaller intracellular vesicles was enriched along mature FAs reminiscent of FA-associated CLASP clusters (Fig. 6a). We next used TIRF microscopy to better visualize MT1-MMP-EGFP exocytosis. Although membrane-bound MT1-MMP-EGFP largely obfuscated vesicle trafficking, MT1-MMP-EGFP vesicles became more visible after extensive photobleaching of the plasma membrane pool. This allowed us to observe examples of transport, docking and sudden disappearance of MT1-MMP-EGFP vesicles near FAs characteristic of exocytosis (Fig. 6b, c; Supplementary Video 8). In an attempt to improve visualization of the vesicular pool of MT1-MMP, we tested a different MT1-MMP reporter construct28. MT1-MMP-mCherry-TM brightly localized to the lumen of large intracellular compartments that evolved through macropinocytosis, during which mCherry fluorescence was released from the membrane (Supplementary Fig. 5a, b), thus likely representing endosomes or lysosomes. Comparatively little MT1-MMP-mCherry-TM localized to the plasma membrane and we were unable to directly observe MT1-MMP-mCherry-TM exocytosis.
Figure 6. MT1-MMP dynamics in migrating epithelial cells.
(a) Spinning disk confocal microscopy of a migrating HaCaT epithelial cell expressing MT1-MMP-EGFP and paxillin-mCherry. The regions indicated are shown at higher magnification. Elapsed time is in minutes. (b, c) High speed TIRF microscopy of MT1-MMP-EGFP dynamics. Coloured arrowheads indicate individual MT1-MMP-EGFP vesicles moving toward and rapidly disappearing near FAs. MT1-MMP-EGFP vesicle fusion events identified by coloured dots and arrowheads in (c) are shown magnified in the bottom panel. Elapsed time is in seconds.
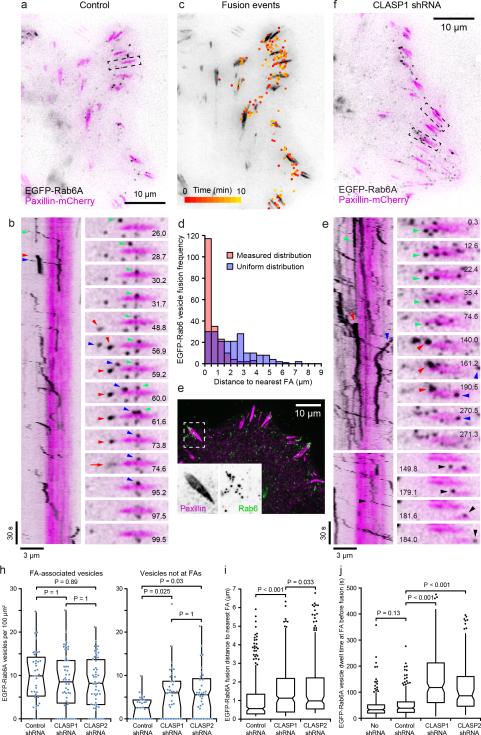
In order to quantify CLASP-mediated FA-associated exocytosis, we next analysed EGFP-Rab6A dynamics based on the rationale that CLASP and LL5β bind factors required for Rab6/Rab8-mediated exocytosis23,29,30. Although Rab6 was originally implicated in retrograde Golgi-to-ER transport31 and ER tubules are continuously pulled toward the leading edge of migrating HaCaT cells, we did not observe a specific enrichment of ER around FAs (Supplementary Fig. 5c). To test if Rab6-positive vesicles are delivered to FAs, we next analysed EGFP-Rab6A dynamics in HaCaT cells expressing paxillin-mCherry by TIRF microscopy. This allowed us to clearly visualize EGFP-Rab6A vesicle dynamics and exocytosis near the ventral cell surface (Fig. 7a, b; Supplementary Video 9). In control cells, EGFP-Rab6A vesicles moved along linear tracks, often stopped at membrane domains adjacent to FAs, and after a short period of pause rapidly disappeared (Fig. 7b). EGFP-Rab6A vesicle disappearance in most cases was not accompanied by a track leaving the FA, and was often associated with a brief burst of plasma membrane-associated fluorescence (Supplementary Fig. 6a). Docked EGFP-Rab6A vesicles disappeared within ~500 milliseconds during which the EGFP-Rab6A signal spread laterally, but the total amount of fluorescence did not decrease (Supplementary Fig. 6b), strongly indicating two-dimensional diffusion of EGFP-Rab6A characteristic for vesicle fusion with the plasma membrane30,32.
Figure 7. Targeting of Rab6A-mediated exocytosis to FAs depends on CLASPs.
(a) TIRF image of control HaCaT cell expressing EGFP-Rab6A (black) and paxillin-mCherry (magenta). (b) Kymograph and time-lapse sequence of the region in (a). Pausing vesicles generate vertical and moving vesicles diagonal tracks. Coloured arrowheads correlate specific vesicles in the kymograph and time-lapse sequence. Red arrow: Burst of EGFP-Rab6A diffusion characteristic for a plasma membrane fusion event. (c) Map of all observed (~200) EGFP-Rab6A fusion events in a 10 min time-lapse sequence overlaid onto the paxillinmCherry channel, illustrating clustering near FAs. (d) Histogram of EGFP-Rab6A fusion distance to the nearest FA compared with a uniform distribution (20x20 pixel grid) for the cell in (c). (e) Immunofluorescence showing endogenous Rab6 vesicles around FAs. (f) TIRF image of CLASP1-depleted cell expressing EGFP-Rab6A and paxillin-mCherry. (g) Kymographs and time-lapse sequences of the regions in (f), illustrating different types of aberrant EGFP-Rab6A vesicle dynamics. Black arrowhead: Paused FA-associated vesicle that fails to fuse and continues to move on. (h) Analysis of EGFP-Rab6A vesicle number in regions surrounding FAs (left; n = 40 (control shRNA); 48 (CLASP1 shRNA); 52 (CLASP2 shRNA)) or identically sized regions between FAs (right; n = 27 (control shRNA); 43 (CLASP1 shRNA); 38 (CLASP2 shRNA)) from three experiments. (i) Analysis of the distance of fusion events from the nearest FA. n = 399 (control shRNA); 347 (CLASP1 shRNA); 300 (CLASP2 shRNA) fusion events from three cells per condition. Six extreme outliers (> 5x the interquartile range from median) were removed from this dataset. (j) Quantification of vesicle dwell time, the duration between a EGFP-Rab6A vesicle stopping near a FA and vesicle disappearance, determined by measuring the length of vertical kymograph track segments. n = 254 (no shRNA); n = 298 (control shRNA); 234 (CLASP1 shRNA); 149 (CLASP2 shRNA) vesicles from two experiments. Box-and-whisker plots show median, 1st and 3rd quartile (box), and 95% confidence intervals (notches) with whiskers extending to the furthest observations within ±1.5 times the interquartile range. Dots are individual data points, outliers only in (i) and (j). Source data for (h), (i), and (j) are included in Supplementary Table 3. p values were calculated by non-parametric Kruskal-Wallis analysis of variance with Bonferroni error correction.
To analyze the spatial distribution of Rab6A exocytosis, we mapped all visible fusion events in TIRF time-lapse series (Fig. 7c) and measured the distance to the nearest FA. Compared with a uniform distribution, Rab6A exocytosis was highly enriched near FAs (Fig. 7d), and in control cells over two thirds of Rab6A fusion events occurred within one micrometre of a FA. This is consistent with our kymography analysis (Fig. 7b), as well as an enrichment of EGFP-Rab6A (Fig. 7h) or endogenous Rab6 vesicles near FAs (Fig. 7e). In contrast, in CLASP-depleted cells, EGFP-Rab6A vesicle dynamics was aberrant, and the overall number of EGFP-Rab6A vesicles appeared increased (Fig. 7f ,g; Supplementary Video 10). EGFPRab6A vesicles as well as fusion events were more randomly distributed and did not localize as specifically near FAs (Fig. 7h, i), and the dwell time between vesicles stopping near a FA and fusion was increased ~3-fold in CLASP-depleted cells (Fig. 7j). Of note, this increased dwell time made it very difficult to observe fusion events in CLASP-depleted cells because EGFP-Rab6A vesicles often bleached before fusion occurred, and it is thus not possible to accurately determine the absolute number of fusion events. Together these data demonstrate the existence of a FA-associated exocytosis pathway in migrating epithelial cells, and that CLASPs are required for the spatial fidelity of exocytic vesicle targeting to allow for efficient fusion at specific membrane sites near FAs.
DISCUSSION
In this study, we identify CLASPs as links connecting microtubules to regulated FA turnover. Mature FAs recruit CLASPs independent of microtubules through direct interactions with LL5β23. FAs are thus likely the source of a biochemical or mechanical signal mediating LL5β and CLASP recruitment. Although the PH-domain of LL5β binds PIP3 33 , PI3K inhibition does not abolish FA-associated LL5β clusters in HaCaT or in HeLa cells23. In migrating HaCaT cells, LL5β and CLASP particles appear near the leading edge and move toward the cell interior at a rate similar to F-actin retrograde flow consistent with LL5β binding to filamins33−35. Similar to LL5β, the laminin-receptor α3β1 integrin localizes to a zone adjacent to FAs in primary keratinocytes36, and cortical LL5β accumulation depends on α3β1 integrin engagement37. Thus, the formation of FA-associated LL5β and CLASP clusters likely requires a complex interplay of signalling, adhesion and F-actin dynamics. In any case, the time delay between FA assembly and LL5β/CLASP recruitment establishes a feedback mechanism facilitating disassembly of large, mature FAs required for controlled cell movement.
Although FA turnover may involve proteolytic cleavage of intracellular FA components38,39 and endocytosis5,6, mechanisms that disassemble mature FAs underneath the advancing cell body are incompletely understood. For example, it is unlikely that endocytosis initiates FA turnover as this would require internalization of active, ECM-engaged integrin molecules. In contrast, FAs rapidly disassemble when contractile forces are released, and we hypothesize that integrin detachment from the ECM can trigger FA disassembly. Recent findings show that MT1-MMP mediates ECM degradation near FAs in invasive cancer cells24,40, and is required for directional cell migration41. Our data demonstrate that sharply delineated FA-associated ECM degradation also occurs during non-invasive HaCaT cell spreading, consistent with the activity of a membrane-bound protease. This indicates that FA-associated ECM degradation is a universal process during normal and pathological tissue remodelling. In support of the hypothesis that FA-associated ECM degradation facilitates FA turnover, we find that FA-associated ECM degradation depends on CLASPs, and that either CLASP depletion or inhibition of MMP activity elicits strikingly similar inhibition of FA turnover. In addition, without MT1-MMP, cells cannot internalize integrins42, indicating that this requires cleavage of extracellular ECM-integrin interactions. As expected, FA disassembly is still triggered by of actomyosin contractility inhibition that bypasses detachment from the ECM. However, because MMPs have other targets in addition to ECM components, alternative mechanisms are possible. For example, MT1-MMP activates other MMPs43, and alters the extracellular signalling environment by releasing latent ECM-bound growth factors44. MT1-MMP also inhibits ADAM-dependent cleavage of FGFR2, preventing ectodomain shedding, which reduces downstream signalling45. To what extent such MMP modulation of signalling networks contributes to FA dynamics remains to be determined.
Our results raise the question of how CLASP-tethered microtubules promote FA-associated ECM degradation. Although MMPs are crucially important for normal and pathological ECM remodelling46,47, surprisingly little is known about how MMP activity is spatially and temporally controlled. MT1-MMP may be retained or activated by direct interactions with FA components24, but it is not known how MT1-MMP is transported to FAs. Although we were able to observe examples of EGFP-MT1-MMP exocytosis near FAs, complex intracellular dynamics and plasma membrane localization precluded quantitative analysis. We used EGFP-Rab6A as a different marker to test whether a FA-directed exocytosis pathway exists because the Rab6-docking protein ELKS, that is required for Rab6A-mediated exocytosis23,29,30, is biochemically linked to CLASPs and LL5β. Remarkably, we find that Rab6A vesicle docking and fusion with the plasma membrane is highly enriched near FAs demonstrating FA-directed exocytosis in migrating cells. Interestingly, accumulation of Rab6 vesicles around FAs is evident in the original paper implicating Rab6 in retrograde Golgi-to-ER transport31. Although we cannot exclude that Rab6-mediated retrograde transport occurs near FAs, we did not observe specific ER-FA interactions. Rab6A and Rab8 localize to the same exocytic vesicles29, and MT1-MMP secretion requires Rab848. Rab6A is thus an appealing candidate mediating MT1-MMP exocytosis, although a recent screen of MT1-MMP-mCherry-TM vesicle associated Rab GTPases did not identify Rab6A49. This may be due to MT1-MMP-mCherry-TM prominently labelling endosomal and lysosomal compartments making it difficult to distinguish smaller and dimmer exocytic vesicles. This highlights the necessity for improved reporters of MT1-MMP dynamics and the importance of future work examining the spatial distribution of other Rab GTPases potentially involved in MT1-MMP exocytosis, and how secretory pathways to FAs and invadopodia are related28,50.
In any case, we demonstrate that membrane domains surrounding FAs are exocytosis hotspots and it is likely that FA-associated exocytosis underlies CLASP-mediated FA turnover and ECM degradation. Interestingly, MACF1/ACF7, a +TIP that guides microtubules along F-actin fibres also facilitates FA turnover10,51. Thus, CLASPs and MACF1/ACF7 may participate in different aspects of the same process, but their functional relationship is not understood, and little is known how MACF1/ACF7 is involved in secretory vesicle trafficking52. Collectively our data support a mechanism in which targeted MT1-MMP delivery to FAs requires CLASP-stabilized FA-associated microtubule tracks, and promotes FA turnover through local release of cell-matrix interactions. Although our main readout in the current study is a FA turnover defect, FA-associated exocytosis may be more broadly important for ECM remodelling and play central roles in normal and pathological tissue dynamics. In addition, other exocytic cargo is likely transported along these FA-associated microtubule tracks, and it will be important to determine to what extent this pathway is involved in more physiological 3D cell migration systems in which directed vesicle trafficking to the protruding cell tip depends on +TIP functions53.
METHODS
Cell Culture
Immortalized HaCaT cells11,54, HEK-293FT (Invitrogen, R700-07) and, HEK-293A cells were cultured in DMEM, 10% FBS (Invitrogen, 26140), 100 i.u./ml penicillin, 100 μg/ml streptomycin (Invitrogen, 15070063), 2mM L-Glutamine (Invitrogen #25030), and 0.1 mM MEM non-essential amino acids (Invitrogen,11140-050). 293FT cells were maintained in 0.5 mg/ml Geneticin/G418 (Invitrogen, 10131-035). According to NIH recommendations, the identity of HaCaT cells was established by short tandem repeat (STR) profiling (DDC Medical, Fairfield OH). The profile below is identical to a recently published profile of the HaCaT cell line55 indicated in bold except for the two underlined loci: D3S1358 (16); TH01 (9.3); D21S11 (28, 30.2); D18S51 (12); Penta E (7,12); D5S818 (11,12); D13S317 (10,12); D7S820 (9,11); D16S539 (9,12); CSF1PO (9,11); Penta D (11,13); AMEL (x); vWA (16,17); D8S1179 (14); TPOX (11,14); FGA (24); D19S433 (13,14); D2S1338 (17,25).
HaCaT cell migration assays were performed as previously described56. Briefly, #1.5 coverslips (64-0713, Warner Instruments) were cleaned by heating and sonication for 20 minutes in Versa-Clean dishwashing detergent (04-342, Fischer) diluted in ddH2O, followed by 20 minutes sonication in ddH2O. Coverslips were rinsed multiple times in ddH2O then stored in 100% ethanol and flame-sterilized prior to coating with fibronectin (11051407001, Roche, 10 μg/ml). HaCaT cells were seeded on fibronectin-coated coverslips at a density to ensure confluency within 24-48 hours. Cells were rinsed and incubated in Ca2+/Mg2+-free PBS for 5 minutes to loosen cell-cell contacts. Coverslips were placed, cell side up onto Parafilm. A stainless-steel, flat edged razor blade was used to swipe half the coverslip, removing cells from one side of the monolayer to create a monolayer edge. Wounded monolayers were then incubated with 0.25% Trypsin-EDTA for 10 seconds, and rinsed 5 times with media to remove cell debris. For live-cell microscopy, coverslips were mounted into sealed, custom-made imaging chambers with media containing 20 mM HEPES, pH 7.5 and allowed to recover overnight at 37°C, 5% CO2 prior to imaging.
DNA Constructs, Adeno- and Lentiviral Vectors
Adenovirus particles for transient expression of wild-type EGFP-CLASP2(340-1362) and the nonphosphorylatable EGFP-CLASP2(340-1362) 9xS/A mutant were as described11. The phosphomimetic EGFP-CLASP2(340-1362) 8xS/D construct was subcloned into pENTR/D-TOPO and recombined into pAd/CMV/V5-DEST following the manufacturer's instructions (Invitrogen #K2400-20, 42-0200). High-titre adenovirus stocks were produced and purified by CsCl2 density gradient ultracentrifugation57, and cells were infected with adenovirus as described previously11. In brief, the adenovirus titre was adjusted for optimal transduction levels in HaCaT cells and cells were infected for 5 hours, after which the tissue culture medium was replaced.
To generate stable fluorescent protein expressing HaCaT cell lines, paxillin-mCherry58, EGFPRab6A59 and EGFP-LL5β33 were subcloned into pENTR/D-TOPO and recombined into pLenti6/V5-DEST (Invitrogen, 35-1271). Lentivirus particles were packaged using Virapower (Invitrogen, 44-2050), and HaCaT cells were transduced with pLenti6 lentivirus particles following the manufacturer's instructions as previously described56. Briefly, HaCaT cells were seeded to be 70% confluent on the day of infection in a 35 or 60 mm dish. Cells were incubated in Ca2+/Mg2+-free PBS for 5 minutes at 37°C, 5% CO2, to loosen cell-cell contacts prior to the addition of virus particles. Infected HaCaT cells were selected with 10 μg/ml blasticidin (Invitrogen, R210–01) for 7 days and FACS sorted to select for low expression levels. Dual EGFP-Rab6A/paxillin-mCherry and EGFP-LL5β/paxillin-mCherry cell lines were made by transducing HaCaT cells stably expressing paxillin-mCherry with the respective EGFP-protein pLenti6 virus and FACS sorted for cells expressing both EGFP and mCherry simultaneously. Cells were maintained under blasticidin selection, which was removed 24 hours prior to experimental use.
Lentivirus-mediated shRNA
Validated shRNA pLKO.1 target gene sets were purchased from Open Biosystems (CLASP1 RHS4533-NM_001142273, CLASP2 RHS4533-NM_015097, LL5β RHS4533-NM_001134437). A non-targeting shRNA sequence was used as control that has no known target in the mammalian genome60. All shRNA sequences used are listed in Supplementary Table 1. For shRNA lentivirus production, 293-FT cells were co-transfected with pLKO.1 plasmid and ViraPower packaging mix according to the manufacturer's protocol and as described previously53,61. Lentivirus-containing supernatants were harvested after 48 hours, centrifuged, aliquoted, snap frozen in liquid nitrogen, and stored at -80°C. HaCaT cells plated in 35 or 60 mm dishes were infected with viral supernatant supplemented with 4 μg/ml Sequabrene (S-2667, Sigma) 4 hours after plating. After 18 hours, cells were transferred to 10 cm dishes in fresh medium, and 4 μg/ml puromycin (A11138-02, Invitrogen) was added 24 hours later. Cells were maintained in puromycin selection media for 7 days prior to experimental use. Four shRNA sequences were tested for each protein and the two independent shRNAs listed in supplementary Table 1 that yielded the best depletion were used for experiments.
Antibodies, Immunofluorescence and Immunoblotting
Primary antibodies used for immunofluorescence (and immunoblotting) including specific dilutions used are listed in supplementary Table 2. All secondary fluorescently labelled antibodies were from Invitrogen except highly cross absorbed secondary antibodies form Jackson Immuno Research (Supplementary Table 2). For immunofluorescence of phalloidin-labeled F-actin, EGFP-CLASP2, paxillin-mCherry, clathrin HC, dynamin-II, LL5β, paxillin, E-cadherin, GM130, Rab6A, and phospho-myosin, samples were fixed with 4% paraformaldehyde (EM grade, 15710, Electron Microscopy Sciences) in BRB80 buffer (80 mM K-PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA) for 20 minutes at room temperature, washed 3 times with Tris-buffered saline (TBS), and permeabilized with 0.25% Triton X-100 in TBS for 5 minutes. Samples were blocked overnight at 4°C in blocking buffer (2% BSA, 0.1% Triton X-100, 0.1% NaN3 in PBS). Primary antibodies were diluted in blocking buffer. Samples were rinsed 3-5 times in blocking buffer 5-10 minutes each before incubating in secondary antibodies (1:500) for 45 minutes at room temperature. Secondary antibodies and phalloidin were diluted in blocking buffer. Samples were rinsed 3-5 times in TBS, 5-10 minutes each, then mounted in either Mowiol mounting medium (0.1 M Tris-HCl pH 8.5, 25% glycerol, 10% Mowiol 4-88; 475904, Calbiochem) or stored in 0.1% n-propyl gallate (P-3130, Sigma) in PBS for TIRF imaging and stored at 4°C.
For immunofluorescence of CLASP1 and CLASP2, samples were fixed in −20°C methanol for 5 minutes, before three rehydration washes with PBS for 10 minutes each. Samples were blocked overnight at 4°C in blocking buffer before processing for immunofluorescence as described above.
Samples processed for microtubules and SAIM were fixed in 0.25% glutaraldehyde (EM grade, 16220, Electron Microscopy Sciences) for 2 minutes, and an additional 20 minutes in the same fixative with 0.1% Triton X-100. Following three 20 minute incubations in PBS with 0.2% NaBH4 (452882, Sigma-Aldrich) to quench glutaraldehyde-induced autofluorescence, samples were blocked overnight at 4°C in blocking buffer before processing for immunofluorescence as described above.
For immunoblotting, cells were lysed in ice-cold RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and protease inhibitors) containing phosphatase inhibitors (50 mM NaF, 1 mM NaVO4, 1 mM ethylene glycol tetraacetic acid, 10 mM sodium pyrophosphate, 1 mM β-glycerol phosphate, and 10 nM calyculin A) for 5 minutes on ice, scraped off the tissue culture dish, transferred to an Eppendorf tube, and incubated on ice for an additional 5 minutes. Cell lysates were centrifuged at 13000 rpm for 5 minutes and supernatants transferred to new tubes for either a BCA assay or to one containing SDS-page sample buffer. Samples were heated to 100°C for 5 minutes, and centrifuged prior to loading. 20 γ of protein was loaded per lane onto a 7.5% Tris-glycine gel. Proteins were separated by SDS-page. Proteins were transferred by wet-transfer for 2 hours at 75 Volts onto nitrocellulose (Biorad Mini Trans Blot Apparatus). Membranes were incubated overnight in blocking buffer (5% non-fat dry milk, TBS, 0.1% Tween-20) at 4°C. Primary antibodies were diluted in blocking buffer as indicated in the table above. Membranes were incubated for 2 hours at room temperature in a 50 ml conical tube on a blood-roller, then washed 3-5 times in TBS, 0.1% Tween-20. Secondary horse radish peroxidase (HRP) conjugated antibodies were diluted 1:10000 in blocking buffer and incubated with membranes at room temperature for 45 minutes, washed 3-5 times with TBS, 0.1% Tween-20, and rinsed once in TBS prior to incubation with ECL (NEL102001, PerkinElmer). Membranes were imaged using a FluorChemQ gel documentation system (92-14116-00, Alpha Innotech), cropped in Adobe Photoshop CS5 and assembled in Adobe Illustrator CS5. LUTs were adjusted linearly to enable comparison of protein of interest to loading controls.
Gelatine Degradation Assay
Gelatine-coated coverslips were prepared essentially as described24,62. In brief, 0.2% porcine gelatin (G-1890, Sigma) was labelled with Alexa488 succinimidyl ester (A20000, Invitrogen). Cleaned coverslips were coated with a solution of 0.1% w/v poly-l-lysine (P4707, Sigma) for 20 minutes at 22°C, and subsequently washed 3 times with PBS before incubating with 0.5% glutaraldehyde (16220, Electron Microscopy Services) for 15 minutes at 22°C. Coverslips were rinsed 3 times with PBS then inverted onto drops of Alexa488-gelatin pre-heated to 37°C (diluted 1:4 with unlabelled 0.2% gelatin) for 10 minutes at 22°C. Gelatine-coated coverslips were then placed in tissue culture dishes and cross-linked with 0.5% glutaraldehyde for 15 minutes to prevent tearing of the gelatine resulting from contractile forces generated by adherent cells. Glutaraldehyde was quenched in 5 mg/ml NaBH4 (452882, Sigma-Aldrich), for 15 minutes at 22°C. Coverslips were rinsed 3 times in PBS before dipping in 70% ethanol for sterilization. Coated coverslips were rinsed 3 times in DMEM media and soaked overnight at 4°C to ensure all traces of ethanol were removed before experimental use. Coverslips were stored at 4°C in PBS for up to a week.
HaCaT cells were seeded 24 hours before the assay to attain ~50% confluency the following day. On the day of the assay cells were rinsed in PBS and incubated with 5 mM EDTA in PBS for 5 minutes to loosen cell-cell contacts. Cells were then incubated in 0.1% w/v crystalline trypsin (T0303, Sigma) in Hanks Balanced Salt Solution (HBSS, 14025-076, Invitrogen) for 5 minutes at 37°C. The detached cells were transferred into 5 ml HBSS and centrifuged at 2000 rpm at 22°C for 5 minutes. The supernatant was aspirated and the cell pellet resuspended in 1 ml complete DMEM. 200 μl of the resuspended cell solution was added to gelatine-coated coverslips in a 12-well dish containing 1 ml of DMEM per well and allowed to adhere at 37°C in a 5% CO2 incubator. Cells were fixed with 4% PFA in BRB80 at indicated time points and processed for immunofluorescence.
Images were acquired by spinning disk confocal microscopy as described below, and regions of interest (ROIs) were selected using the “Simple ROI editor” and the “Bezier” tool in NIS elements around the cell periphery within 10 μm of the cell edge. Regions were thresholded to include areas of Alexa488-gelatine degradation. Degradation area was normalized to ROI total area to give percentage degradation.
Other Reagents
Y-27632 (Y0501, Sigma) 10 mM stock in H2O used at 10 μM. Nocodazole (M1404, Sigma) 10 mM stock in DMSO (D2650, Sigma), used at 3.3 μM. LY-294002 (L9908, Sigma) 10 mM stock in DMSO, used at 10 μM. BB94 (Batimastat 196440, Millipore) 10 mM stock in DMSO used at 10 μM.
Microscopy, Image Processing, and Data Analysis
Spinning disk confocal live cell imaging was done on an environmentally controlled Nikon TI inverted microscope stand equipped with a Borealis-modified Yokogawa CSU-X1 confocal head (Spectral Applied Research) and a Clara cooled scientific grade interline CCD camera (Andor). Details of this microscope system are published elsewhere56. Intracellular fluorescent protein-tagged protein dynamics were imaged at 37°C using a 60× 1.49 NA CFI Apochromat TIRF objective (Nikon).
TIRF images were acquired on the same TI inverted microscope stand (Nikon) equipped with a motorized TIRF illuminator (Nikon), an iXon EMCCD camera (Andor), and a 100× 1.49 NA CFI Apochromat TIRF objective (Nikon) using 1.5× intermediate magnification. TIRF imaging of EGFP-Rab6a vesicle dynamics was done at ~1 frame per second, but the paxillin-mCherry channel was imaged only every 10th frame to limit photobleaching, increase acquisition speed, and because FAs are relatively static over these short time scales. Emission wavelengths were separated using a filterwheel (Sutter) mounted between microscope and EMCCD camera.
Phase contrast time-lapse sequences were acquired on a TE 2000 inverted microscope stand (Nikon) with a CoolSNAP HQ2 scientific grade interline CCD camera (Photometrics) and a 40× 0.6 NA CFI Plan Fluor ELWD DM objective (Nikon).
All microscope hardware was controlled by NIS Elements software (Nikon), and image processing and analysis was performed in NIS Elements. Generally, for display purposes image contrast was linearly adjusted on the 14- or 16-bit raw data, images were low pass filtered in NIS Elements (Detail level: 2), and processed with an unsharp mask filter (Power: 0.5; Area: 7). Figures were assembled using Adobe Photoshop and Illustrator CS5 (Adobe). Movies were assembled in Quicktime Pro (Apple).
All statistical analysis was performed in Excel (Microsoft) and the Analyse-it plug in for Excel (Analyse-it Software, Ltd). Least square curve fitting was done using the Solver function in Excel (Microsoft). All data and statistical analysis are included in Supplementary Table 3. Details on specific image and data analysis protocols are outlined below:
Black/magenta Image Overlays
To better visualize the dynamics of fine CLASP (or LL5β) structures around FAs, all live cell imaging data are displayed with inverted contrast in a black/magenta overlay. These overlays were generated in NIS Elements 4.0 as follows:
The multi-wavelength data set was separated into two individual time-lapse sequences, e.g. CLASP and paxillin.
Both channels were corrected for photobleaching over time using “Equalize Intensity in Time” with the following settings: Histogram Stretching; Bottom: Minimal Intensity in whole frame; Top: Mean Intensity in Selected Area; selecting the entire image as significant image area.
Both channels were filtered with a “Low-Pass Filter” (Detail: 2) to reduce pixel noise, and with an “Unsharp Mask” (Power: 0.5; Area: 7) to slightly enhance contrast of dim structures.
Background was subtracted using “Subtract Background using ROI” with a background ROI in an image area outside the cell, and the following settings: Calculate background ROI on Current Image; Offset being approximately equal to the standard deviation of the background (25 for Clara CCD camera) to avoid signal clipping; Apply to All Frames.
Maximum image intensities were made comparable by dividing the brighter channel by a number using “Image Operations”. For example, if the CLASP channel was approximately twice as bright as the paxillin channel, divide CLASP channel by 2: CLASP/2.
The two channels were added using “ND Image Arithmetics”: CLASP/2 + paxillin.
Channels were combined into a color image with “Merge Channels” (Red: CLASP/2; Green: CLASP/2 + paxillin; Blue: CLASP/2)
The merged time-lapse sequence was contrast-inverted “Complement Colors”, and the low end of the LUT set to the same grey value for all three channels. This results in the CLASP channel appearing in black and the paxillin channel in magenta.
Focal Adhesion Size
Prior to fixation, cell monolayers were wounded as described above. Cells were stained for paxillin and imaged by spinning disk confocal (60×; 0.107 μm/pixel). Images were cropped to include only the front of migrating cells at the edge of the cell monolayer. Images were thresholded for bright objects, and focal adhesion areas were measures using the “Object Count” function in NIS Elements.
Focal Adhesion Turnover
Paxilin-mCherry time lapse sequences of migrating HaCaT cells were acquired for 3 hours at 3 min intervals. Regions of interest were defined using the Bezier ROI tool to outline individual adhesions. If the FA significantly changed size and location and moved out of the ROI over time, ROIs were redrawn to include all fluorescence using the “redraw” function in the NIS Elements ROI tool. Fluorescence intensity as a function of time was measured from the unprocessed image data using the “Time measurement” tool in NIS Elements. Cytoplasmic background was subtracted by using duplicated ROIs adjacent to FAs. Fluorescence intensity as a function of time was similarly measured in dual wavelengths experiments demonstrating spatiotemporal correlation of paxilin-mCherry and EGFP-CLASP2 turnover.
To quantify FA turnover dynamics, time lapse fluorescence intensity data were smoothed with a 3-frame moving average to reduce noise. The assembly phase was fitted with a logistic function, in which fmax is the maximum fluorescence intensity, klog is the rate constant and t1/2 the time at half maximum:
The disassembly phase was fitted with a single exponential decay with the rate constant kexp:
Least square curve fitting was done using the Solver function in Excel (Microsoft). FA lifetime was defined as the time during which the fluorescence intensity remained above the half maximum and was calculated from the assembly and disassembly curve fits:
Of note, we likely underestimate FA disassembly defects because FAs that did not assemble and disassemble within the 3-hour time-lapse sequence were not included in our analysis.
Rab6A Vesicle Number
Random frames were selected from EGFP-Rab6A TIRF time-lapse sequences. ROIs were drawn using the “Bezier” tool in NIS elements of approximately 15-35 μm2, around the largest FA in each cell, then duplicated and placed over FAs and adjacent cortical areas which did not contain a FA. Areas were selected in the paxillin-mCherry channel independent of the EGFP-Rab6A channel in order not to bias region selection. Vesicles were then counted in each ROI. The number was normalized to the ROI area and multiplied by a factor to calculate vesicles per 100 μm2.
Rab6A Kymographs and Analysis of Vesicle Dwell Time
EGFP-Rab6A TIRF time-lapse sequences were rotated such that FAs to be analysed were aligned with the horizontal image axis. Images were filtered as described above to enhance contrast of Rab6A vesicles. The image sequence was then cropped closely around FAs as indicated by the dashed box below in Supplementary Figure 6a. The vertical width of the crop (y-axis) was 20 pixels. A sequence of x-t kymographs was generated in the “Show Slices View” in NIS Elements by right-clicking the x-t-view and using the “Switch Axes and Create New Document” command. The resulting image sequence is shown in Supplementary Figure 6b and corresponds to 20 kymographs for each of the horizontal (x-axis) lines of pixels in the cropped time-lapse sequence. To better visualize the tracks of vesicles that moved at paths that were not parallel to the horizontal axis, maximum intensity projections were generated of either the entire x-t kymograph sequence (Supplementary Figure 6c) or only of selected planes. EGFP-Rab6A vesicle dwell time was defined as the time between a vesicle stopped moving until it disappeared, and measured as the length of vertical EGFP-Rab6A tracks in the kymographs using the NIS elements “Annotations and Measurements” vertical measurement tool.
Repeatability of Experiments
Figures 1e, 1f, 3c, 3d, 3f-h, 4a-c, 6, and 7e contain representative images of live cell or immunofluorescence experiments that are not otherwise quantified. Each of these experiments was performed at least three times.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health grant R01 GM079139 to T.W., and American Heart Association postdoctoral fellowship 10POST3870021 to S.S. Research was conducted in a facility constructed with support from the Research Facilities Improvement Program grant C06 RR16490, and on a microscope system funded by Shared Equipment Grant S10 RR26758 from the National Center for Research Resources of the National Institutes of Health. We also thank Christopher O'Connell at Nikon, Melville, NY, for imaging on the Nikon SIM system, Bradley Webb for help with the gelatine degradation assay, Anna Akhmanova, David Bryant, Mark McNiven, and Jim Norman for reagents, and Diane Barber, David Bryant, Alpha Yap and members of the Wittmann and Barber labs for discussions and comments on the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS:
S.J.S. and T.W. conceived the project. S.J.S., M.P, H.P., A.E., S.G., and T.W generated reagents, conducted experiments, and analysed data. M.P. contributed SAIM data and analysis. S.J.S. and T.W. wrote the manuscript and assembled figures.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol. 2010;26:315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stehbens S, Wittmann T. Targeting and transport: How microtubules control focal adhesion dynamics. J Cell Biol. 2012;198:481–489. doi: 10.1083/jcb.201206050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaverina I, Krylyshkina O, Small JV. Microtubule targeting of substrate contacts promotes their relaxation and dissociation. J Cell Biol. 1999;146:1033–1044. doi: 10.1083/jcb.146.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol. 2005;7:581–590. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- 6.Ezratty EJ, Bertaux C, Marcantonio EE, Gundersen GG. Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells. J Cell Biol. 2009;187:733–747. doi: 10.1083/jcb.200904054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rooney C, White G, Nazgiewicz A, Woodcock SA, Anderson KI, Ballestrem C, Malliri A. The Rac activator STEF (Tiam2) regulates cell migration by microtubule-mediated focal adhesion disassembly. EMBO Rep. 2010;11:292–298. doi: 10.1038/embor.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar P, Wittmann T. +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 2012;22:418–428. doi: 10.1016/j.tcb.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto S, Fumoto K, Okamoto T, Kaibuchi K, Kikuchi A. Binding of APC and dishevelled mediates Wnt5a-regulated focal adhesion dynamics in migrating cells. EMBO J. 2010;29:1192–1204. doi: 10.1038/emboj.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X, Kodama A, Fuchs E. ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity. Cell. 2008;135:137–148. doi: 10.1016/j.cell.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar P, Lyle KS, Gierke S, Matov A, Danuser G, Wittmann T. GSK3beta phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J Cell Biol. 2009;184:895–908. doi: 10.1083/jcb.200901042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, Akhmanova A. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mimori-Kiyosue Y, Grigoriev I, Sasaki H, Matsui C, Akhmanova A, Tsukita S, Vorobjev I. Mammalian CLASPs are required for mitotic spindle organization and kinetochore alignment. Genes Cells. 2006;11:845–857. doi: 10.1111/j.1365-2443.2006.00990.x. [DOI] [PubMed] [Google Scholar]
- 14.Miller PM, Folkmann AW, Maia AR, Efimova N, Efimov A, Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paszek MJ, Dufort CC, Rubashkin MG, Davidson MW, Thorn KS, Liphardt JT, Weaver VM. Scanning angle interference microscopy reveals cell dynamics at the nanoscale. Nat Methods. 2012 doi: 10.1038/nmeth.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krylyshkina O, Anderson KI, Kaverina I, Upmann I, Manstein DJ, Small JV, Toomre DK. Nanometer targeting of microtubules to focal adhesions. J Cell Biol. 2003;161:853–859. doi: 10.1083/jcb.200301102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meenderink LM, Ryzhova LM, Donato DM, Gochberg DF, Kaverina I, Hanks SK. P130Cas Src-binding and substrate domains have distinct roles in sustaining focal adhesion disassembly and promoting cell migration. PLoS One. 2010;5:e13412. doi: 10.1371/journal.pone.0013412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 20.Stehbens S, Wittmann T. Analysis of focal adhesion turnover: A quantitative live cell imaging example. Methods Cell Biol in press. 2014 doi: 10.1016/B978-0-12-420138-5.00018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang YC, Nalbant P, Birkenfeld J, Chang ZF, Bokoch GM. GEF-H1 couples nocodazole-induced microtubule disassembly to cell contractility via RhoA. Mol Biol Cell. 2008;19:2147–2153. doi: 10.1091/mbc.E07-12-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K, Kano Y, Amano M, Kaibuchi K, Fujiwara K. Stress fiber organization regulated by MLCK and Rho-kinase in cultured human fibroblasts. Am J Physiol Cell Physiol. 2001;280:C1669–C1679. doi: 10.1152/ajpcell.2001.280.6.C1669. [DOI] [PubMed] [Google Scholar]
- 23.Lansbergen G, Grigoriev I, Mimori-Kiyosue Y, Ohtsuka T, Higa S, Kitajima I, Demmers J, Galjart N, Houtsmuller AB, Grosveld F, Akhmanova A. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev Cell. 2006;11:21–32. doi: 10.1016/j.devcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, McNiven MA. Invasive matrix degradation at focal adhesions occurs via protease recruitment by a FAK-p130Cas complex. J Cell Biol. 2012;196:375–385. doi: 10.1083/jcb.201105153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massimi P, Zori P, Roberts S, Banks L. Differential regulation of cell-cell contact, invasion and anoikis by hScrib and hDlg in keratinocytes. PLoS One. 2012;7:e40279. doi: 10.1371/journal.pone.0040279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies B, Brown PD, East N, Crimmin MJ, Balkwill FR. A synthetic matrix metalloproteinase inhibitor decreases tumor burden and prolongs survival of mice bearing human ovarian carcinoma xenografts. Cancer Res. 1993;53:2087–2091. [PubMed] [Google Scholar]
- 27.Chao WT, Kunz J. Focal adhesion disassembly requires clathrin-dependent endocytosis of integrins. FEBS Lett. 2009;583:1337–1343. doi: 10.1016/j.febslet.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X, Zech T, McDonald L, Gonzalez EG, Li A, Macpherson I, Schwarz JP, Spence H, Futo K, Timpson P, Nixon C, Ma Y, Anton IM, Visegrady B, Insall RH, Oien K, Blyth K, Norman JC, Machesky LM. N-WASP coordinates the delivery and F-actin-mediated capture of MT1-MMP at invasive pseudopods. J Cell Biol. 2012;199:527–544. doi: 10.1083/jcb.201203025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grigoriev I, Yu KL, Martinez-Sanchez E, Serra-Marques A, Smal I, Meijering E, Demmers J, Peranen J, Pasterkamp RJ, van der Sluijs P, Hoogenraad CC, Akhmanova A. Rab6, Rab8, and MICAL3 cooperate in controlling docking and fusion of exocytotic carriers. Curr Biol. 2011;21:967–974. doi: 10.1016/j.cub.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 30.Grigoriev I, Splinter D, Keijzer N, Wulf PS, Demmers J, Ohtsuka T, Modesti M, Maly IV, Grosveld F, Hoogenraad CC, Akhmanova A. Rab6 regulates transport and targeting of exocytotic carriers. Dev Cell. 2007;13:305–314. doi: 10.1016/j.devcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 31.White J, Johannes L, Mallard F, Girod A, Grill S, Reinsch S, Keller P, Tzschaschel B, Echard A, Goud B, Stelzer EH. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J Cell Biol. 1999;147:743–760. doi: 10.1083/jcb.147.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toomre D, Steyer JA, Keller P, Almers W, Simons K. Fusion of constitutive membrane traffic with the cell surface observed by evanescent wave microscopy. J Cell Biol. 2000;149:33–40. doi: 10.1083/jcb.149.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paranavitane V, Coadwell WJ, Eguinoa A, Hawkins PT, Stephens L. LL5beta is a phosphatidylinositol (3,4,5)-trisphosphate sensor that can bind the cytoskeletal adaptor, gamma-filamin. J Biol Chem. 2003;278:1328–1335. doi: 10.1074/jbc.M208352200. [DOI] [PubMed] [Google Scholar]
- 34.Takabayashi T, Xie MJ, Takeuchi S, Kawasaki M, Yagi H, Okamoto M, Tariqur RM, Malik F, Kuroda K, Kubota C, Fujieda S, Nagano T, Sato M. LL5beta directs the translocation of filamin A and SHIP2 to sites of phosphatidylinositol 3,4,5-triphosphate (PtdIns(3,4,5)P3) accumulation, and PtdIns(3,4,5)P3 localization is mutually modified by co-recruited SHIP2. J Biol Chem. 2010;285:16155–16165. doi: 10.1074/jbc.M109.081901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adh Migr. 2011;5:160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter WG, Wayner EA, Bouchard TS, Kaur P. The role of integrins alpha 2 beta 1 and alpha 3 beta 1 in cell-cell and cell-substrate adhesion of human epidermal cells. J Cell Biol. 1990;110:1387–1404. doi: 10.1083/jcb.110.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hotta A, Kawakatsu T, Nakatani T, Sato T, Matsui C, Sukezane T, Akagi T, Hamaji T, Grigoriev I, Akhmanova A, Takai Y, Mimori-Kiyosue Y. Laminin-based cell adhesion anchors microtubule plus ends to the epithelial cell basal cortex through LL5alpha/beta. J Cell Biol. 2010;189:901–917. doi: 10.1083/jcb.200910095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franco SJ, Huttenlocher A. Regulating cell migration: calpains make the cut. J Cell Sci. 2005;118:3829–3838. doi: 10.1242/jcs.02562. [DOI] [PubMed] [Google Scholar]
- 39.Storr SJ, Carragher NO, Frame MC, Parr T, Martin SG. The calpain system and cancer. Nat Rev Cancer. 2011;11:364–374. doi: 10.1038/nrc3050. [DOI] [PubMed] [Google Scholar]
- 40.Takino T, Saeki H, Miyamori H, Kudo T, Sato H. Inhibition of membrane-type 1 matrix metalloproteinase at cell-matrix adhesions. Cancer Res. 2007;67:11621–11629. doi: 10.1158/0008-5472.CAN-07-5251. [DOI] [PubMed] [Google Scholar]
- 41.Gonzalo P, Guadamillas MC, Hernandez-Riquer MV, Pollan A, Grande-Garcia A, Bartolome RA, Vasanji A, Ambrogio C, Chiarle R, Teixido J, Risteli J, Apte SS, del Pozo MA, Arroyo AG. MT1-MMP is required for myeloid cell fusion via regulation of Rac1 signaling. Dev Cell. 2010;18:77–89. doi: 10.1016/j.devcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi F, Sottile J. MT1-MMP regulates the turnover and endocytosis of extracellular matrix fibronectin. J Cell Sci. 2011;124:4039–4050. doi: 10.1242/jcs.087858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Overall CM, Dean RA. Degradomics: systems biology of the protease web. Pleiotropic roles of MMPs in cancer. Cancer Metastasis Rev. 2006;25:69–75. doi: 10.1007/s10555-006-7890-0. [DOI] [PubMed] [Google Scholar]
- 44.Mu D, Cambier S, Fjellbirkeland L, Baron JL, Munger JS, Kawakatsu H, Sheppard D, Broaddus VC, Nishimura SL. The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. J Cell Biol. 2002;157:493–507. doi: 10.1083/jcb.200109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan KM, Wong HL, Jin G, Liu B, Cao R, Cao Y, Lehti K, Tryggvason K, Zhou Z. MT1-MMP inactivates ADAM9 to regulate FGFR2 signaling and calvarial osteogenesis. Dev Cell. 2012;22:1176–1190. doi: 10.1016/j.devcel.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 46.Rowe RG, Weiss SJ. Breaching the basement membrane: who, when and how? Trends Cell Biol. 2008;18:560–574. doi: 10.1016/j.tcb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 47.Friedl P, Wolf K. Tube travel: the role of proteases in individual and collective cancer cell invasion. Cancer Res. 2008;68:7247–7249. doi: 10.1158/0008-5472.CAN-08-0784. [DOI] [PubMed] [Google Scholar]
- 48.Bravo-Cordero JJ, Marrero-Diaz R, Megias D, Genis L, Garcia-Grande A, Garcia MA, Arroyo AG, Montoya MC. MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J. 2007;26:1499–1510. doi: 10.1038/sj.emboj.7601606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiesner C, Azzouzi KE, Linder S. A specific subset of RabGTPases controls cell surface exposure of MT1-MMP, extracellular matrix degradation and 3D invasion of macrophages. J Cell Sci. 2013 doi: 10.1242/jcs.122358. [DOI] [PubMed] [Google Scholar]
- 50.Monteiro P, Rosse C, Castro-Castro A, Irondelle M, Lagoutte E, Paul-Gilloteaux P, Desnos C, Formstecher E, Darchen F, Perrais D, Gautreau A, Hertzog M, Chavrier P. Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. J Cell Biol. 2013 doi: 10.1083/jcb.201306162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suozzi KC, Wu X, Fuchs E. Spectraplakins: Master orchestrators of cytoskeletal dynamics. J Cell Biol. 2012;197:465–475. doi: 10.1083/jcb.201112034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burgo A, Proux-Gillardeaux V, Sotirakis E, Bun P, Casano A, Verraes A, Liem RK, Formstecher E, Coppey-Moisan M, Galli T. A molecular network for the transport of the TI-VAMP/VAMP7 vesicles from cell center to periphery. Dev Cell. 2012;23:166–180. doi: 10.1016/j.devcel.2012.04.019. [DOI] [PubMed] [Google Scholar]
- 53.Gierke S, Wittmann T. EB1-recruited microtubule +TIP complexes coordinate protrusion dynamics during 3D epithelial remodeling. Curr Biol. 2012;22:753–762. doi: 10.1016/j.cub.2012.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao M, Sano D, Pickering CR, Jasser SA, Henderson YC, Clayman GL, Sturgis EM, Ow TJ, Lotan R, Carey TE, Sacks PG, Grandis JR, Sidransky D, Heldin NE, Myers JN. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin Cancer Res. 2011;17:7248–7264. doi: 10.1158/1078-0432.CCR-11-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stehbens S, Pemble H, Murrow L, Wittmann T. Imaging intracellular protein dynamics by spinning disk confocal microscopy. Methods Enzymol. 2012;504:293–313. doi: 10.1016/B978-0-12-391857-4.00015-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gierke S, Kumar P, Wittmann T. Analysis of microtubule polymerization dynamics in live cells. Methods Cell Biol. 2010;97:15–33. doi: 10.1016/S0091-679X(10)97002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- 59.Matanis T, Akhmanova A, Wulf P, Del NE, Weide T, Stepanova T, Galjart N, Grosveld F, Goud B, De Zeeuw CI, Barnekow A, Hoogenraad CC. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat Cell Biol. 2002;4:986–992. doi: 10.1038/ncb891. [DOI] [PubMed] [Google Scholar]
- 60.Bryant DM, Datta A, Rodriguez-Fraticelli AE, Peranen J, Martin-Belmonte F, Mostov KE. A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol. 2010;12:1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 62.Thompson O, Kleino I, Crimaldi L, Gimona M, Saksela K, Winder SJ. Dystroglycan, Tks5 and Src mediated assembly of podosomes in myoblasts. PLoS One. 2008;3:e3638. doi: 10.1371/journal.pone.0003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.