SUMMARY
Fe-S clusters are essential across the biological world, yet how cells regulate expression of Fe-S cluster biogenesis pathways to cope with changes in Fe-S cluster demand is not well understood. Here, we describe the mechanism by which IscR, a [2Fe-2S] cluster-containing regulator of Escherichia coli, adjusts the synthesis of the Isc Fe-S biogenesis pathway to maintain Fe-S homeostasis. Our data indicate that a negative feedback loop operates to repress transcription of the iscRSUA-hscBA-fdx operon, encoding IscR and the Isc machinery, through binding of [2Fe-2S]-IscR to two upstream binding sites. IscR was shown to require primarily the Isc pathway for synthesis of its Fe-S cluster, providing a link between IscR activity and demands for Fe-S clusters through the levels of the Isc system. Surprisingly, the isc operon was more repressed under anaerobic conditions, indicating increased Fe-S cluster occupancy of IscR and decreased Fe-S cluster biogenesis demand relative to aerobic conditions. Consistent with this notion, overexpression of a Fe-S protein under aerobic conditions, but not under anaerobic conditions, led to derepression of PiscR. Together, these data show how transcriptional control of iscRSUA-hscBA-fdx by [2Fe-2S]-IscR allows E. coli to respond efficiently to varying Fe-S demands.
Keywords: IscR, Fe-S biogenesis, Isc, Suf, autoregulation
INTRODUCTION
Iron-sulfur (Fe-S) proteins are nearly ubiquitous across all domains of life and carry out many important cellular functions such as respiration, photosynthesis, nitrogen fixation, and gene regulation (Beinert, 2000, Fontecave, 2006, Kiley and Beinert, 2003). In Escherichia coli, the majority of Fe-S cluster biogenesis under non-stress conditions is proposed to be catalyzed by the highly conserved, housekeeping Isc (iron-sulfur cluster) pathway, encoded by the iscRSUA-hscBA-fdx (isc) operon (Johnson et al., 2005, Py and Barras, 2010, Py et al., 2011). The current model for Isc-mediated Fe-S biogenesis in E. coli is that IscS, a cysteine desulfurase, provides the sulfur to build a transient Fe-S cluster on the scaffold protein IscU. HscB and HscA form a complex with IscU to apparently facilitate transfer of a Fe-S cluster from IscU to a subset of apo-proteins. Alternatively, IscA, an A type carrier, promotes cluster transfer by delivering the Fe-S cluster from the scaffold to target apo-proteins. Other genes in the operon encode a [2Fe-2S] ferredoxin (Fdx) and the transcription factor IscR. Although transcription of the isc operon is repressed by IscR (Schwartz et al., 2001), a [2Fe-2S] cluster-containing protein encoded by the first gene of this operon, a detailed understanding of how this autoregulatory mechanism occurs and ultimately how Fe-S biogenesis is regulated in vivo remains to be elucidated.
While IscR was first discovered for its role in regulating transcription of the Isc system (Schwartz et al., 2001), subsequent genome-wide transcription profiling data revealed that IscR controls the expression of more than 40 genes in E. coli (Giel et al., 2006).Among these genes are those encoding for anaerobic respiratory Fe-S enzymes (periplasmic nitrate reductase, hydrogenases-1 and −2) and additional proteins with roles in Fe-S formation, including ErpA, NfuA, and the Suf system. IscR also directly activates the expression of the alternate Fe-S biogenesis pathway encoded by the sufABCDSE (suf) operon (Nesbit et al., 2009, Yeo et al., 2006), which includes proteins that exhibit functional similarity to those of the Isc pathway (Py and Barras, 2010, Py et al., 2011, Johnson et al., 2005). However, Suf is proposed to function primarily under oxidative and nitrosative stress and iron-limiting conditions (Justino et al., 2005, Mukhopadhyay et al., 2004, Outten et al., 2004, Zheng et al., 2001). Nevertheless, the transcription profiling results implicated a broader role for IscR in sensing the cellular Fe-S status and thus controlling the expression of the Isc and Suf systems accordingly.
Further analysis of promoters directly regulated by IscR revealed two classes of DNA target sites for IscR site-specific binding, denoted as Type 1 and Type 2 sites (Giel et al., 2006). The nucleotide sequences vary between Type 1 (Giel et al., 2006) and Type 2 (Nesbit et al., 2009) sites, and it is not yet clear how a dimer of IscR distinguishes between them (Nesbit et al., 2009).Of the IscR-dependent promoters identified thus far, most contain a single Type 1 or Type 2 site (Giel et al., 2006). In contrast, DNase I footprinting has revealed that IscR binds to three individual sites within the iscR promoter region (PiscR) (Giel et al., 2006). One region of protection, which encompasses nucleotides −67 to −14 relative to the +1 transcription start site, includes two Type 1 IscR binding sites: site A (−65 to −41) and site B (−40 to −16) (Giel et al., 2006). The remaining IscR site, located from +9 to +26, does not share sequence similarity with other known IscR binding sites and is referred to as PiscR site C (Giel et al., 2006). The contribution of each site to repression of the isc operon in vivo is not known.
A previous study provided important insight that repression of the isc operon by IscR may allow cells to homeostatically regulate the synthesis of the Isc Fe-S biogenesis pathway (Schwartz et al., 2001). In this model, IscR [2Fe-2S] cluster occupancy would be a reporter of the cellular Fe-S status, tuning synthesis of the Isc pathway in response to changing demands for Fe-S biogenesis. In support of the model, the presence of [2Fe-2S]-IscR was demonstrated in vivo by Mössbauer spectroscopy and repression of PiscR was shown to require [2Fe-2S]-IscR, since clusterless IscR mutants failed to repress PiscR (Fleischhacker et al., 2012, Yeo et al., 2006). In addition, strains lacking IscS and HscA showed decreased IscR repression, indicating a role of the Isc pathway in maintaining the [2Fe-2S] form of IscR (Schwartz et al., 2001). Nevertheless, the effect of other Isc proteins or the Suf pathway on [2Fe-2S]-IscR activity has not been examined. In addition, whether the decrease in in vivo repression observed with the clusterless mutants can be explained by a lack of DNA binding is of particular interest because IscR target promoters containing Type 2 sites (such as those in the sufABCDSE, napFDAGHBC, hyaABCDEF, and hybOABCDEFG operons) do not require the [2Fe-2S] cluster form of IscR either for binding DNA in vitro (Nesbit et al., 2009) or for transcriptional regulation in vivo (Nesbit et al., 2009, Yeo et al., 2006). Together, the unique features of the [2Fe-2S] cluster-dependent and -independent modes of IscR regulation have led to the proposal that conditions that increase the Fe-S demand will lead to decreased [2Fe-2S]-IscR levels, de-repression of the isc operon, elevated apo-IscR levels, and accordingly, increased regulation of promoters with Type 2 sites by apo-IscR. Such differences in [2Fe-2S] occupancy of IscR may explain why the isc operon is expressed at higher levels under aerobic conditions (Giel et al., 2006) since the demand for Fe-S biogenesis is expected to be greater because of the O2 sensitivity of some Fe-S clusters (Imlay, 2006, Imlay, 2008). Thus, determining whether the [2Fe-2S] cluster occupancy of IscR differs between aerobic and anaerobic conditions is an important question to address.
In this study, we dissected the mechanism by which IscR negatively regulates PiscR to better understand how cells maintain Fe-S homeostasis. We first established roles for the three predicted IscR binding sites in repressing PiscR transcription in vivo. The requirement of the [2Fe-2S]-containing form of IscR for site-specific binding in vitro and regulation of PiscR under aerobic and anaerobic conditions in vivo was analyzed using IscR mutants that cannot ligate an Fe-S cluster but are otherwise functional. We report the role of other Isc pathway components and the Suf system in providing Fe-S clusters to IscR under aerobic and anaerobic growth conditions. To address whether differences in [2Fe-2S] cluster occupancy for IscR exist between aerobic and anaerobic growth, an in vivo titration experiment was performed to determine the level of IscR protein required for PiscR repression for both conditions. Finally, to examine whether IscR responds to cellular demands for Fe-S clusters, we tested whether overexpression of an Fe-S protein affected transcription from PiscR. Together, our data indicate that [2Fe-2S]-IscR regulates expression of the Isc pathway via a negative feedback mechanism based on the cellular Fe-S demand.
RESULTS
The two Type 1 sites play a role in negative autoregulation of the iscR promoter
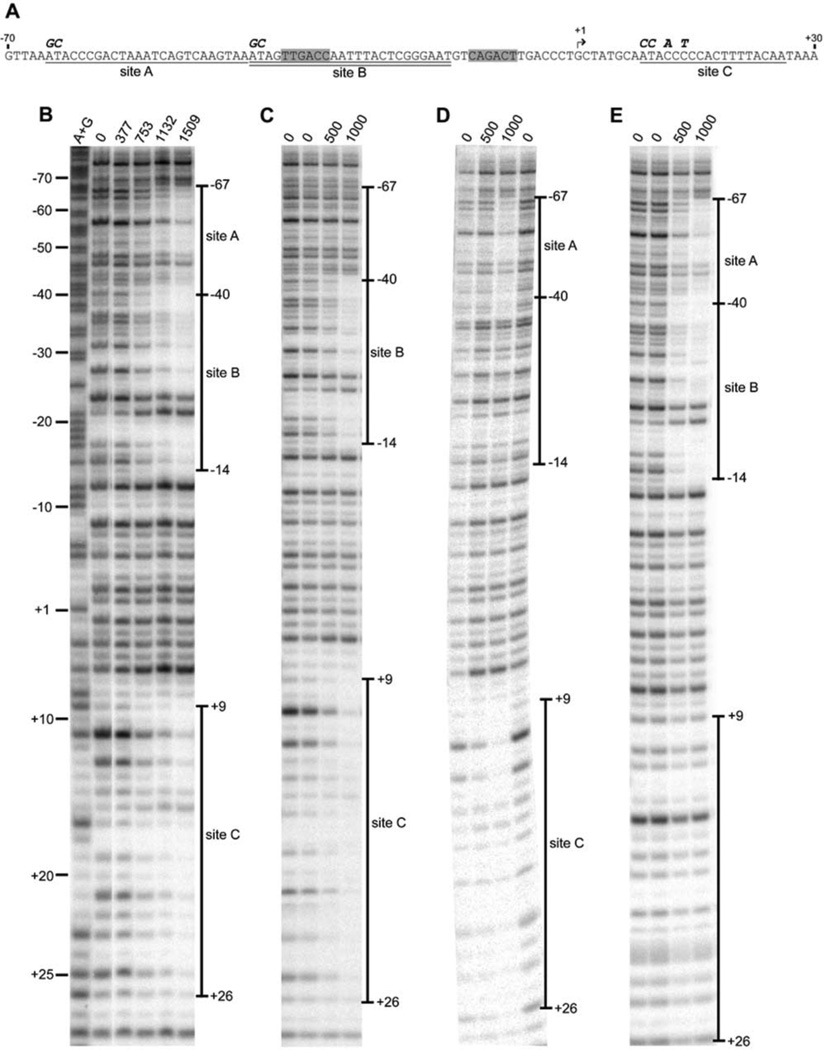
To examine the contribution of each of the three IscR binding sites (A, B, and C; Fig. 1A) within the iscR promoter region to negative autoregulation, mutations in each individual site were constructed and their effect on IscR binding was determined. Since sites A and B are representative of the Type 1 IscR binding site, the first AT of each site, which are among the most highly conserved bases within the Type I site (Giel et al., 2006), were substituted with GC (Fig. 1A). Using a wild-type DNA fragment for comparison (Fig. 1B), DNase I footprinting showed that when site A was mutated, anaerobically isolated [2Fe-2S]-IscR no longer protected the region (−67 to −41 bp relative to the +1 iscR transcription start site) encompassing site A, but still protected the regions (−40 to −14 bp and +9 to +26 bp) encompassing sites B and C in a protein concentration-dependent manner (Fig. 1C). In an analogous fashion, the DNA fragment containing the mutated site B showed IscR-mediated protection in the regions encompassing sites A and C, but not site B (Fig. 1D). Site C is not as well-conserved among enterobacteria and does not resemble other known IscR binding sites (Giel et al., 2006). Nevertheless, upon mutating the base pairs located at positions +9, +10, +12, and +14 within this site (Fig. 1A), protection of site C from DNase I cleavage was eliminated, while there was no effect on IscR binding to sites A and B (Fig. 1E). Thus, our data demonstrate that mutation of sites A, B, and C within PiscR decreases in vitro binding of IscR to each individual site but does not affect IscR binding to the remaining intact sites.
Fig. 1.
In vitro binding of anaerobically purified [2Fe-2S]-IscR to wild-type and mutated IscR binding sites within PiscR. A) The IscR binding sites A (underlined), B (double underlined), C (underlined), the transcriptional start site (bent arrow), the −35 and −10 promoter elements (shaded),and the bases substituted in this study (bold italics) are indicated. DNase I footprints of IscR bound to DNA fragments containing the wild-type PiscR region (panel B) or promoter regions with mutations in sites A, B, or C (panels C, D, and E, respectively). The amount of IscR protein (nM) present in each reaction and the extent of the footprint relative to the +1 transcription start site are denoted. Samples were electrophoresed with Maxam-Gilbert (A + G) ladders. The lane order in panel B was digitally rearranged for presentation purposes.
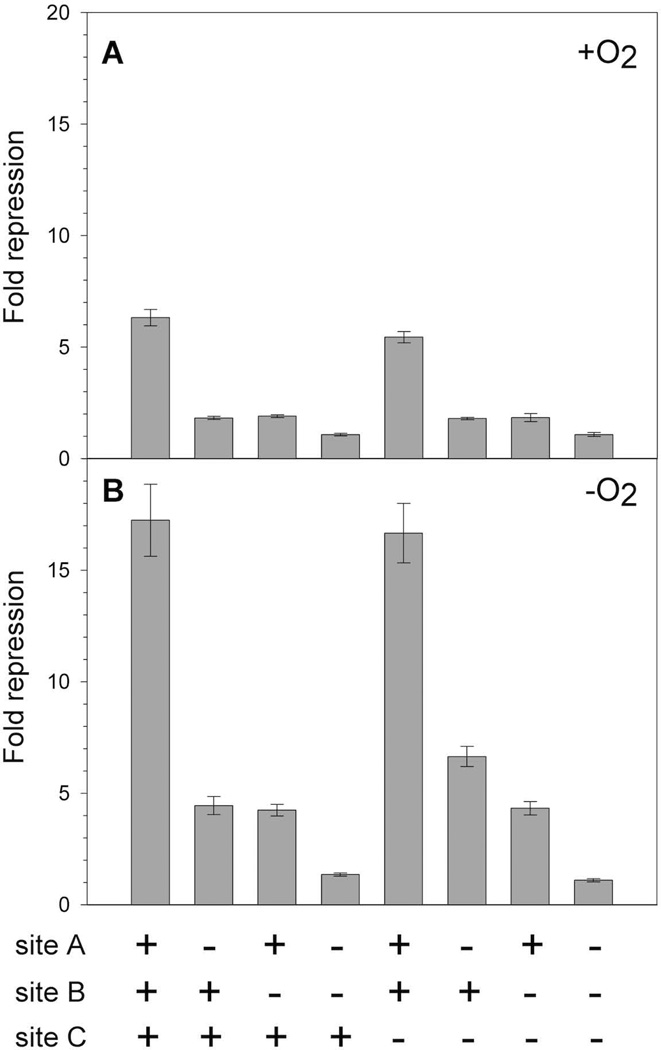
We determined the effects of the mutated IscR sites on in vivo PiscR repression via β-galactosidase assays using aerobically or anaerobically grown strains with chromosomal PiscR -lacZ fusions containing the mutations in sites A, B, or C (Fig. 2). As expected from previous results (Giel et al., 2006), IscR repressed PiscR expression more under anaerobic (17-fold) than aerobic (6-fold) growth conditions. Nevertheless, under both growth conditions, mutation of either site A or B caused a 3- to 4-fold loss in repression compared to the intact promoter, indicating a defect in PiscR repression. When sites A and B were both mutated, repression was eliminated, similar to that observed in a strain lacking IscR. Surprisingly, mutation of site C had no effect on PiscR expression. Furthermore, mutation of site C did not result in any further significant defect in PiscR repression when sites A or B were also mutated. Thus, our data suggest that both Type 1 sites A and B are necessary for full IscR-mediated repression of PiscR in vivo. Since sites A and B overlap sequences (−65 to −16 bp) of the −35 and extended −10 promoter elements required for σ70 binding (Record et al., 1996) and a region potentially encompassing an UP element important for recognition by αCTD (Gourse et al., 2000), it appears that IscR binding of both Type 1 sites is required to fully prevent RNA polymerase from binding PiscR.
Fig. 2.
In vivo effects of mutations in IscR binding sites A, B, and C within PiscR. β-galactosidase activity from wild-type (PK7571) or mutated (PK8521, PK8528, PK8582, PK8551, PK8801, PK8808, PK8820) PiscR-lacZ fusions (recombined at the chromosomal lac region) was measured in cells grown under A) aerobic or B) anaerobic conditions in MOPS minimal media with glucose (0.2%). The amount of IscR-dependent repression (Fold repression) was determined by dividing the β-galactosidase activity present in the strain lacking IscR (PK7572) by the β-galactosidase activity measured for each strain. The presence (+) or absence (−) of wild-type IscR binding sites A, B, and C in the PiscR-lacZ fusion is indicated below the figure. Error bars represent the propagation of standard errors for three biological replicates.
IscR requires its Fe-S cluster to repress expression from the iscR promoter in vivo under aerobic and anaerobic conditions
To establish the extent of the IscR [2Fe-2S] cluster requirement for negative autoregulation under both aerobic and anaerobic growth conditions, we measured PiscR repression in strains expressing IscR mutants that are defective in cluster binding. These mutants contain one or more alanine substitutions of three of the four IscR [2Fe-2S] cluster ligands (C92, C98, C104) (Fleischhacker et al., 2012) and were previously shown to contain no detectable Fe-S cluster upon isolation (Nesbit et al., 2009). Under either aerobic or anaerobic conditions, strains containing chromosomally encoded IscR-C92A, IscR-C98A, IscR-C104A, or IscR-C92A/C98A/C104A exhibited a nearly complete defect in PiscR-lacZ repression (Fig. 3), indicating that under both growth conditions, IscR requires its [2Fe-2S] cluster to repress PiscR. Measurements of IscR protein in these strains showed that each of the IscR variants was present at levels ∼2-fold and ∼8-fold greater than wild-type IscR under aerobic and anaerobic conditions, respectively, (Table 2) (Nesbit et al., 2009), indicating that the loss in PiscR repression did not result from a defect in protein accumulation. Furthermore, the same mutants were previously shown to be otherwise functional at a subset of IscR-regulated promoters that contain the Type 2 IscR binding motif and do not require the Fe-S form of IscR for binding (Nesbit et al., 2009). Taken together, our data support the notion that IscR requires its [2Fe-2S] cluster to negatively regulate PiscR under both aerobic and anaerobic conditions and this occurs by [2Fe-S]-IscR binding to the two Type 1 sites within the iscR promoter region.
Fig. 3.
β-galactosidase activity from PiscR fused to lacZ integrated at the λatt site was determined in strains containing wild-type IscR (PK6364) or chromosomal mutants defective in [2Fe-2S] cluster binding [IscR-C92A (PK7854), IscR-C98A (PK7855), IscR-C104A (PK7856), and IscR-C92A/C98A/C104A (PK7898)]. Strains were grown under A) aerobic or B) anaerobic conditions in MOPS minimal media containing glucose (0.2%). Fold repression was determined by dividing the β-galactosidase activity present in the strain lacking IscR (PK6512) by the β-galactosidase activity measured for each strain and error bars represent the propagation of standard errors for three biological replicates.
Table 2.
In vivo levels of IscR protein measured via quantitative Western blotting.
| IscR monomers, µM | IscR monomers/cell | |||
|---|---|---|---|---|
| Chromosome | Aerobic | Anaerobic | Aerobic | Anaerobic |
| wild-type IscR | 17.1 ± 1.2 | 1.8 ± 0.1 | 10,300 ± 750 | 1060 ± 80 |
| IscR- C92A/C98A/C104A |
29.9 ± 2.0 | 13.1 ± 1.2 | 18,000 ± 1200 | 7900 ± 750 |
[2Fe-2S]-IscR binds with greater affinity to the PiscR sites A and B in vitro than does apo-IscR
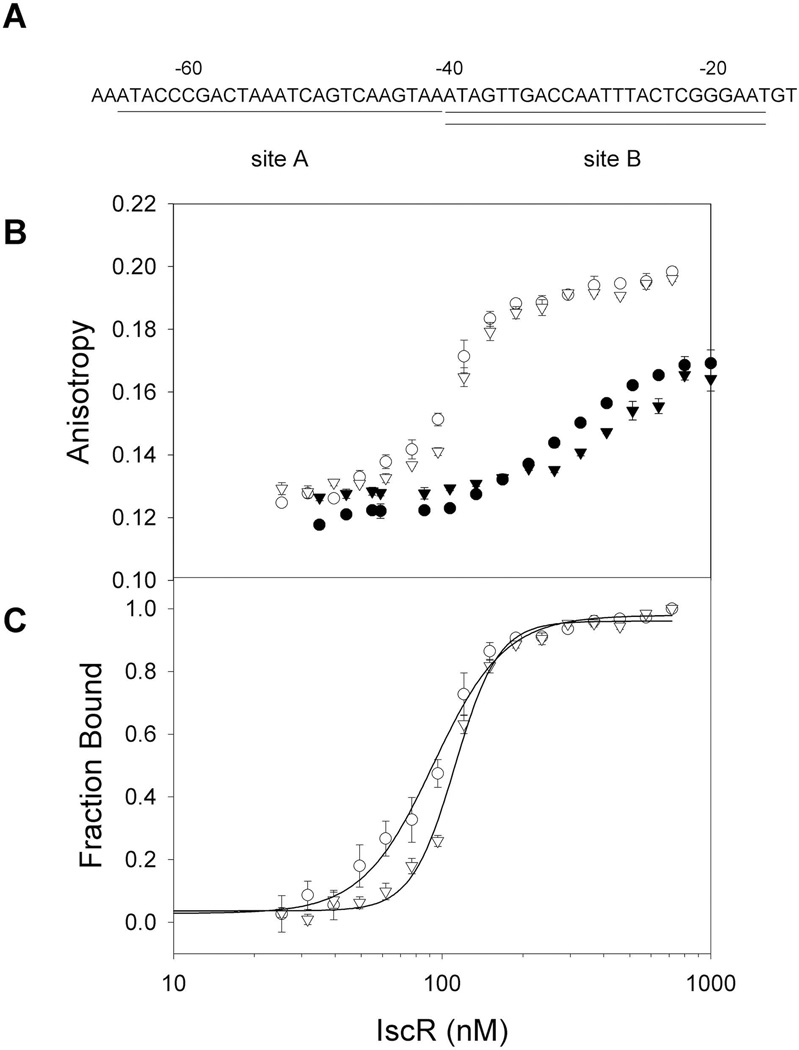
Previous in vitro studies demonstrated that [2Fe-2S]-IscR and IscR-C92A/C98A/C104A bind with equal affinity to PhyaA (Nesbit et al., 2009), which contains a Type 2 IscR binding site. However, the above in vivo data raise the question as to whether [2Fe-2S]-IscR binds with higher affinity than apo-IscR to the Type 1 sites within PiscR. To address this question, fluorescence anisotropy assays were performed under anaerobic conditions with DNA fragments containing either site A or B from PiscR (Fig. 4). Curve fitting the fraction bound with the Hill equation indicates that the apparent Kd of monomeric [2Fe-2S]-IscR is 93 ± 5 nM for PiscR site A and 112 ± 2 nM for PiscR site B (Fig. 4C). Since the purified wild-type IscR used in this experiment was ≥50% occupied with [2Fe-2S] clusters, it is likely that the actual Kd values for PiscR sites A and B may be lower than the apparent Kd values determined. In contrast, Kd values could not be determined for IscR-C92A/C98A/C104A because saturation was not obtained even at the highest protein concentration tested (1.0 µM)(Fig. 4B). Thus, our data suggest that [2Fe-2S]-IscR has a much higher binding affinity for the PiscR A and B sites than does clusterless IscR. Furthermore, the observed difference in [2Fe-2S]-IscR and clusterless IscR binding affinity for the two PiscR Type 1 sites provides an explanation as to why the [2Fe-2S] cluster is required for IscR to repress PiscR in vivo.
Fig. 4.
Binding isotherms of [2Fe-2S]-IscR and apo-IscR for the two Type 1 sites within the PiscR region. A) Sequences of IscR binding sites A (underlined) and B (double underlined) within PiscR. Numbers indicate the distance relative to the +1 transcription start site. B) DNA binding isotherms of wild-type [2Fe-2S]-IscR (open symbols) and the clusterless mutant protein IscR-C92A/C98A/C104A (closed symbols) measured as a change in anisotropy under anaerobic conditions. Both forms of IscR protein were incubated with 5 nM fluorescently labeled DNA containing either site A (circles) or site B (triangles) in 40 mM Tris-Cl (pH 7.9) and 150 mM KCl. C) Fraction bound corrected for fluorescence quenching of labeled PiscR sites A or B bound by [2Fe-2S]-IscR as a function of protein concentration. Error bars represent the standard errors of triplicate experiments.
IscR requires a functional Isc system to fully repress PiscR in vivo
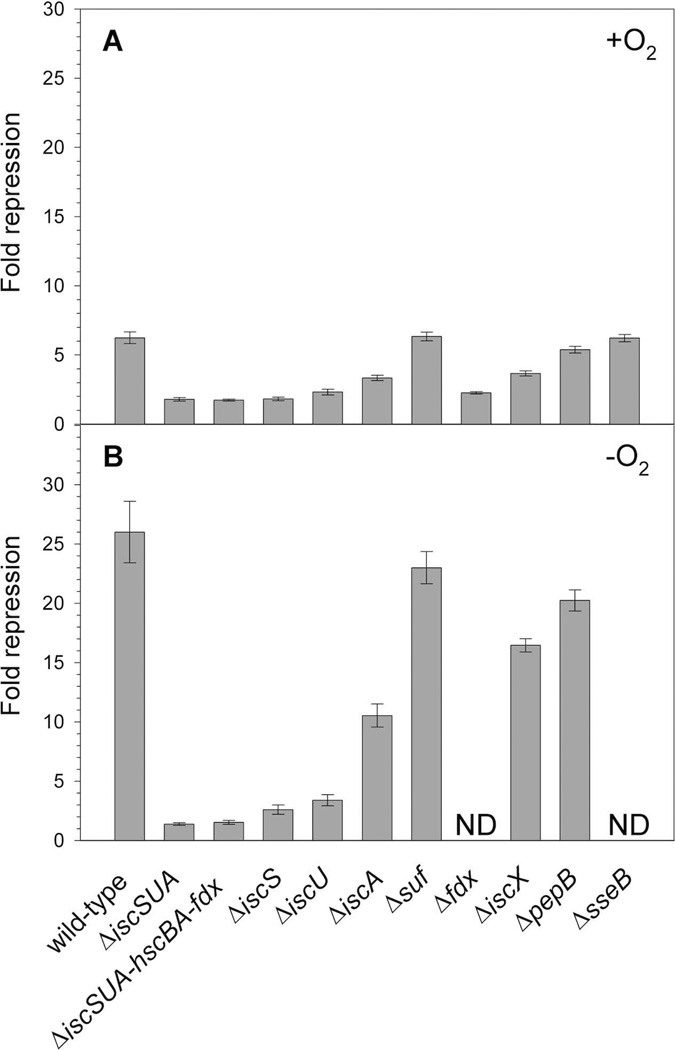
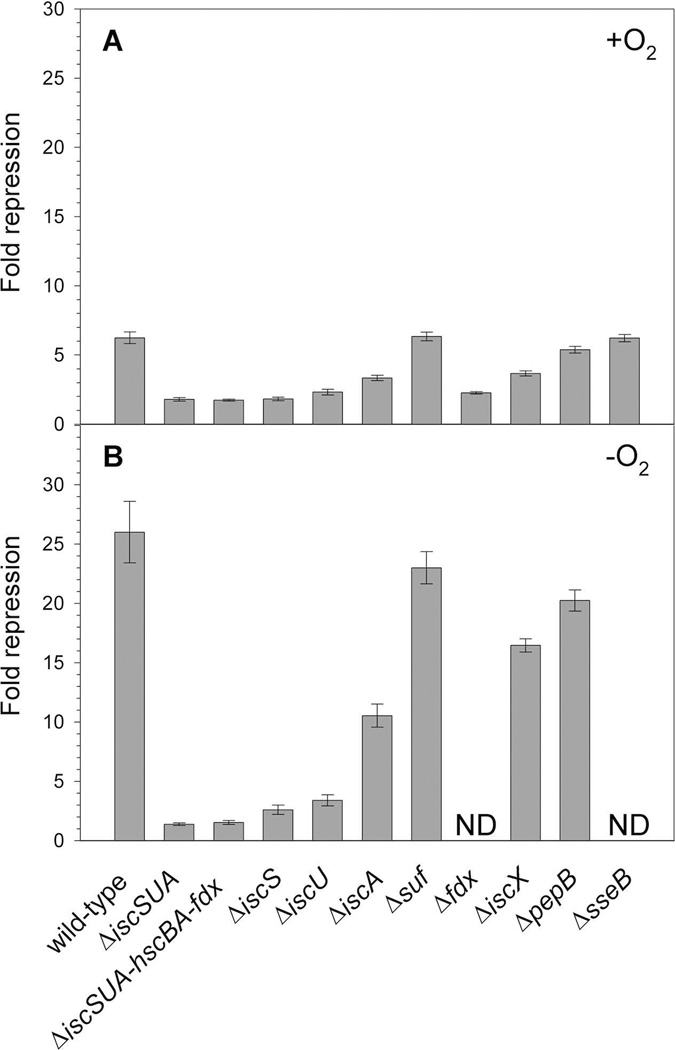
PiscR repression requires [2Fe-2S]-IscR and is decreased in strains lacking iscS or hscA (Schwartz et al., 2001), supporting the model that the Isc machinery synthesizes the [2Fe-2S] cluster for IscR. However, since iscS and hscA are reported to have roles beyond Fe-S cluster assembly (Py and Barras, 2010, Johnson et al., 2005), we characterized the role of other members of the Isc pathway in supplying IscR its [2Fe-2S] cluster. Strains lacking individual components such as IscS, IscU, or Fdx yielded a similar defect in repression as the strain lacking the entire pathway (ΔiscSUA-hscBA-fdx), suggesting that these proteins are necessary for the biogenesis of [2Fe-2S]-IscR (Fig. 5). Strains lacking IscA had a smaller effect (∼2-fold) on IscR activity (Fig. 5).Quantitative Western blots revealed that under aerobic growth conditions, levels of IscR are increased about 4- and 2-fold in ΔiscU and ΔiscA strains, respectively (data not shown), indicating that IscR protein was present but unable to fully repress PiscR. Finally, we found that elimination of the Isc pathway or individual components showed a similar defect in PiscR repression under both aerobic and anaerobic growth conditions (Fig. 5). Interestingly, these results contrast those of a previous study that revealed that the [4Fe-4S] cluster-containing transcription factor FNR had a more stringent requirement for the Isc pathway under aerobic conditions than anaerobic conditions (Mettert et al., 2008).
Fig. 5.
β-galactosidase activity was measured in wild-type (PK6364) or mutant (PK8120, PK8122, PK7783, PK6828, PK7759, PK6564, PK6826, PK6848, PK7540, PK7751) strains containing PiscR fused to lacZ integrated at the λatt site. Cultures were grown under A) aerobic or B) anaerobic conditions in MOPS minimal media supplemented with glucose (0.2%); for the wild-type, ΔiscSUA, ΔiscSUA-hscBA-fdx, ΔiscS, ΔiscU, ΔiscA strains, media were also supplemented with nicotinic acid (12.5 µg ml−1), and thiamine (2 µg ml−1). Fold repression was determined by dividing the β-galactosidase activity present in the strain lacking IscR (PK6512), grown in the presence or absence of nicotinic acid and thiamine, by the β-galactosidase activity measured for each strain and error bars represent the propagation of standard errors for three biological replicates. ND, not determined.
Other Fe-S biogenesis proteins are not necessary for [2Fe-2S]-IscR activity when the Isc pathway is functional
We also examined whether other Fe-S biogenesis proteins contribute to IscR cluster assembly since a small amount of [2Fe-2S]-IscR activity remained in the strain lacking the Isc pathway (Fig. 5). Under either aerobic or anaerobic conditions, deletion of the suf operon had no effect on PiscR repression (Fig. 5), indicating that the Suf pathway plays no apparent role in IscR function under nonstress conditions when the Isc system is present. In a strain lacking NfuA, an IscA/NifU-like protein that facilitates Fe-S cluster transfer (Bandyopadhyay et al., 2008, Angelini et al., 2008)and whose expression is repressed by IscR (Giel et al., 2006), there was a small defect (∼2-fold) in IscR repression, but only under anaerobic conditions (data not shown). In addition, PiscR repression was not affected in strains lacking CsdA, a cysteine desulfurase (Trotter et al., 2009), or YtfE, a protein implicated in Fe-S cluster assembly or repair (Justino et al., 2009)(data not shown). Finally, deletion of the genes encoding IscX, PepB, and SseB, which are located downstream of iscRSUA-hscBA-fdx and are repressed by IscR (Giel et al., 2006), were also tested for their effects on IscR function, although only IscX has been suggested to have a possible function in Fe-S biogenesis (Pastore et al., 2006, Shimomura et al., 2005). Only the strain lacking IscX showed any defect, slightly less than a 2-fold effect under either aerobic or anaerobic conditions (Fig. 5). Thus, in the presence of the Isc pathway, other Fe-S assembly proteins do not have a major effect on IscR-dependent repression of PiscR, thus supporting our model that the Isc pathway provides the [2Fe-2S] cluster required for IscR to fully repress PiscR expression under aerobic and anaerobic conditions.
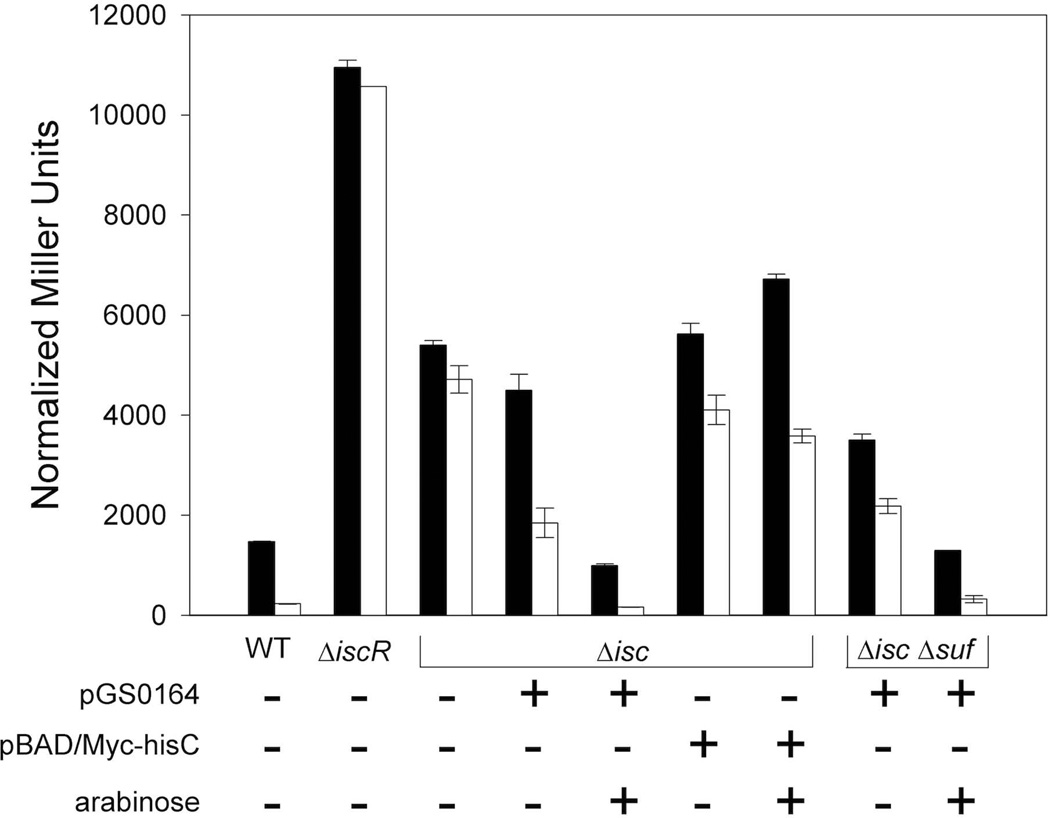
In the absence of the Isc machinery, elevated expression of the Suf pathway can fully restore [2Fe-2S]-IscR activity
Unlike regulation of PiscR, regulation of the suf operon by IscR does not require IscR to ligate a [2Fe-2S] cluster (Nesbit et al., 2009, Yeo et al., 2006). Thus, we considered the possibility that upregulation of the Suf machinery by apo-IscR and subsequent transfer of a Suf-generated [2Fe-2S] cluster to IscR may explain the residual [2Fe-2S]-IscR-dependent activity measured in the ΔiscSUA-hscBA-fdx strain. To address whether IscR could serve as a substrate for the Suf machinery, PiscR expression was measured in strains that contained the ΔiscSUA-hscBA-fdx or ΔiscSUA-hscBA-fdx ΔsufABCDSE alleles and a plasmid with the suf operon under control of PBAD. As expected, the strain lacking both the Isc and the Suf pathways was viable only when the suf plasmid, not the vector, was present since deletion of both pathways is lethal (Py et al., 2011). Furthermore, when 10 mM arabinose was present in the growth medium to induce suf operon expression, strains containing the suf plasmid had PiscR activity similar to that of the wild-type strain under aerobic and anaerobic growth conditions (Fig 6). This activity correlated with at least 30-fold higher levels of SufD protein compared to that of the wild-type strain as determined by Western blot analyses (data not shown). Thus, the Suf pathway, when expressed at elevated levels, appears to be able to substitute for the Isc pathway by inserting a [2Fe-2S] cluster into IscR, and this may account for the residual [2Fe-2S]-IscR activity measured in the strain lacking the Isc pathway.
Fig. 6.
β-galactosidase activity from λ-PiscR-lacZ was determined in aerobically (black) or anaerobically (white) grown wild-type (PK6364), ΔiscSUA-hscBA-fdx [Δisc (PK8122, PK8614, PK8615)], and ΔiscSUA-hscBA-fdxΔsufABCDSE [ΔiscΔsuf (PK8618)] strains. The ΔiscSUA-hscBA-fdx strain derivatives contained either the plasmid expressing the sufABCDSE operon from PBAD (pGS0164), the vector alone (pBAD/Myc-hisC), or no vector. All cultures were grown in M9 minimal media containing glucose (0.2%), nicotinic acid (12.5 µg ml−1), and thiamine (2 µg ml−1). The plasmid-containing strains were also grown in the presence of Ap (50 µg ml−1) and where indicated, arabinose (0.2%). Error bars represent the standard errors of three biological replicates.
IscR [2Fe-2S] cluster occupancy is greater under anaerobic growth conditions
In the wild-type strain, repression of PiscR was ∼3–5-fold greater under anaerobic conditions than aerobic conditions despite the fact that there is ∼10-fold less IscR protein under anaerobic conditions (Figs. 2, 3, and 5, Table 2). Since in vitro DNA binding by [2Fe-2S]-IscR is not sensitive to the O2-dependent change in the [2Fe-2S] cluster oxidation state (Fleischhacker et al., 2012), a simple interpretation of our in vivo results is that the fraction of IscR containing a [2Fe-2S] cluster may be higher under anaerobic conditions. This, in turn, would lead to more PiscR repression and less IscR protein. This notion was tested by measuring the levels of IscR protein required for PiscR repression under both growth conditions. A strain containing chromosomally-encoded iscR and iscSUA-hscBA-fdx under control of Ptac and PBAD, respectively, was grown in medium containing 20 mM arabinose and various concentrations of IPTG (0–640 µM). We subsequently measured PiscR-lacZ repression which, according to our in vivo data, requires the [2Fe-2S] form of IscR, and the amount of IscR present at each IPTG concentration was quantified by Western blots. Under both growth conditions, the amount of PiscR repression increased with increasing levels of IscR; however, more IscR protein was needed to fully repress PiscR under aerobic conditions compared to anaerobic conditions (Fig S1). In fact, half-maximal PiscR repression required ∼9-fold more IscR protein aerobically than anaerobically (955 ± 93 nM vs. 107 ± 14 nM monomeric IscR). Assuming that the concentration of [2Fe-2S]-IscR is equivalent to the IscR protein concentration required for 50% repression, our data indicate that under aerobic conditions IscR is at least 9-fold less occupied with [2Fe-2S] clusters than under anaerobic conditions. Thus, we conclude that the difference in the amount of PiscR repression under aerobic and anaerobic conditions can be explained by O2-dependent changes in the cluster occupancy of IscR.
The iscR promoter is derepressed in response to increased Fe-S demand
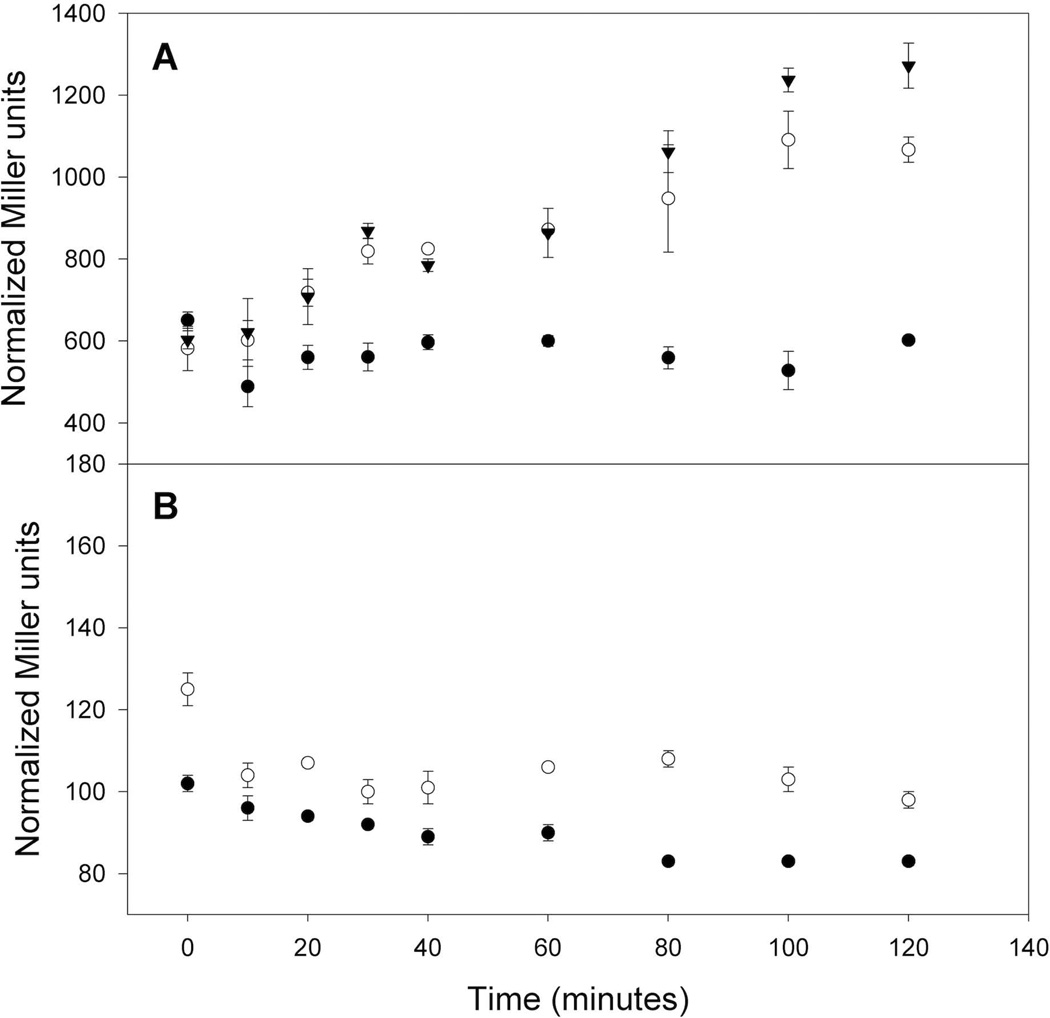
Because the Isc requirement for biogenesis of the [4Fe-4S] cluster of FNR is greater under aerobic than anaerobic conditions (Mettert et al., 2008), we considered the possibility that acquisition of the [2Fe-2S] cluster by IscR is reduced under aerobic conditions because of the competition with other substrates for the Isc machinery. Such a competition mechanism could explain how IscR senses the cellular Fe-S demands and correspondingly adjusts expression of the Isc pathway. If IscR senses the cellular Fe-S status through its [2Fe-2S] cluster, then conditions that increase the demand for Fe-S clusters are predicted to affect [2Fe-2S]-IscR activity. This notion was tested by measuring PiscR-lacZ activity upon overexpression of the [4Fe-4S] protein FNR from Ptac. Since FNR does not regulate PiscR (Kang et al., 2005, Giel, 2007), we expected that changes in iscR transcription may result from the competition between FNR and IscR for the Isc machinery. Furthermore, because the [4Fe-4S] cluster of FNR is not stable under aerobic conditions (Khoroshilova et al., 1997), we also overexpressed the mutant protein FNR-L28H, whose [4Fe-4S] cluster is more resistant to destruction by oxygen (Bates et al., 2000). Under aerobic conditions, PiscR was derepressed within 20 minutes of addition of IPTG to a final concentration of 100 µM to the strain containing either the Ptac-fnr or the Ptac-fnr-L28H expression plasmid but not the vector control (Fig. 7A). Furthermore, no derepression was observed for the Ptac-fnr strain when no IPTG was added (data not shown). Thus, under aerobic conditions, an increase in the Fe-S cluster demand via overexpression of FNR relieves repression of the isc operon, presumably due to the competition between FNR and IscR for acquiring Fe-S clusters from the Isc machinery.
Fig. 7.
Activity from PiscR-lacZ integrated at the λatt site was measured in strains (PK7329, PK7333, PK7328) containing plasmids with IPTG-inducible wild-type fnr (open circles), IPTG-inducible fnr-L28H (triangles) or the vector control (closed circles). Strains were grown under A) aerobic or B) anaerobic conditions in M9 minimal media containing glucose (0.2%), Ap (50 µg ml−1), and Cm (20 µg ml−1). When cells reached an OD600 of 0.2 (0 minutes), IPTG was added to a final concentration of 100 µM, samples were taken at various time points, and β-galactosidase activity (Miller units) was assayed and normalized for cell number as explained in Experimental Procedures. Error bars represent the standard errors of three biological replicates.
In contrast, derepression of PiscR was not observed when FNR was overexpressed under anaerobic conditions (Fig. 7B), despite the accumulation of FNR protein to similar levels as under aerobic conditions (data not shown). This suggests that the Isc machinery can meet an increase in Fe-S demand under anaerobic conditions such that no competition between FNR and IscR was detected. Thus, the Isc pathway seems to be more buffered against changes in Fe-S demand under anaerobic conditions, likely due to less general cluster turnover under these conditions. In summary, this apparent difference in the cellular Fe-S demand between aerobic and anaerobic conditions highlights the significance of negative feedback regulation of the isc operon.
DISCUSSION
The findings of this study have revealed new mechanistic insight as to how IscR regulates transcription of PiscR to maintain Fe-S cluster homeostasis in E. coli. We have defined the requirement for the two Type 1 IscR binding sites within PiscR for full repression of the isc operon and have shown that these sites are preferentially bound by the [2Fe-2S] form of IscR. Examination of PiscR repression under aerobic and anaerobic growth has revealed that the increased repression of isc under anaerobic conditions is due to increased IscR [2Fe-2S] cluster occupancy under these conditions. Furthermore, decreased isc repression under aerobic conditions suggests that there may be more competition between IscR and substrate proteins for the Isc machinery when O2 is present. Consistent with this notion, overexpression of an Fe-S protein under aerobic conditions, but not under anaerobic conditions, led to derepression of PiscR. In particular, our results shed new insight on how IscR responds to the presence of O2 to satisfy the cellular need for Fe-S cluster biogenesis.
IscR binding sites A and B are critical for IscR-mediated repression of PiscR
Of the three IscR binding sites present within PiscR (Giel et al., 2006), we found that only the two adjacent Type 1 IscR binding sites, A and B, are responsible for PiscR repression in vivo under standard growth conditions. The requirement for the [2Fe-2S] form of IscR for repression also correlated well with the increased affinity of [2Fe-2S]-IscR for either site A or B. Nevertheless, the in vivo concentration of the clusterless mutant IscR-C92A/C98A/C104A is sufficiently high (≥17 µM; Table 2) such that some binding of the mutant might be expected even though its binding characteristics in vitro were poor. Perhaps an additional mechanism exists in vivo to prevent repression of PiscR by IscR apo-protein. Because apo-IscR binds to many locations on the E. coli chromosome (Myers and Kiley, unpublished), one possibility is that apo-IscR is bound at other higher affinity sites across the genome. Thus, like many transcription factors, regulation of promoters by IscR is likely dependent on a hierarchy of binding affinities.
Since both IscR sites A and B span the region that RNA polymerase binds, the mechanism of IscR repression can be simply explained by promoter occlusion. Indeed, DNase I footprinting experiments at wild-type PiscR showed that addition of IscR at the same time as RNAP resulted in a protection pattern virtually identical to the one for IscR alone (Giel, 2007), indicating that binding of IscR to PiscR occludes RNAP. Since most IscR-regulated promoters contain just a single IscR binding site (Giel et al., 2006), perhaps the presence of multiple IscR binding sites within the PiscR region may be important to extend the dynamic range to regulate Fe-S biogenesis in vivo. While our footprinting experiments indicate that [2Fe-2S]-IscR could bind independently to these two sites, the use of linear DNA in these experiments does not rule out the possibility that some protein-protein interactions occur when present on supercoiled chromosomal DNA.
The Isc pathway is the major system for IscR [2Fe-2S] cluster biogenesis
Deletion of the iscSUA-hscBA-fdx operon resulted in decreased PiscR repression, suggesting that IscR primarily receives its [2Fe-2S] cluster from the Isc pathway. When the Isc pathway was absent, however, a small amount of [2Fe-2S]-IscR activity remained, suggesting that IscR could acquire [2Fe-2S] clusters from another source. Similar results were also observed for the transcription factor FNR and the enzymes isopropylmalate isomerase, NADH dehydrogenase I, and IspG/H, which all require Fe-S clusters for function (Mettert et al., 2008, Jang and Imlay, 2010, Vinella et al., 2009). Since apo-IscR is sufficient to activate PsufA transcription (Nesbit et al., 2009, Yeo et al., 2006), we propose that in the ΔiscSUA-hscBA-fdx strain, upregulation of the Suf pathway by apo-IscR may explain the residual [2Fe-2S]-IscR activity. Consistent with this notion, sufA promoter expression and SufD protein levels were elevated 2- to 3-fold in a mutant strain lacking the Isc pathway compared to the wild-type strain (Mettert et al., 2008). Furthermore, our studies indicate that IscR can in fact serve as a substrate for the Suf machinery in the ΔiscSUA-hscBA-fdx mutant when the suf operon is overexpressed. Since the Isc system has been shown to be present but non-functional during H2O2 stress (Jang and Imlay, 2010), the ability of an Isc substrate protein to also receive Fe-S clusters from the Suf pathway would be particularly advantageous for E. coli under conditions of oxidative stress.
IscR senses and responds to the different Fe-S demands that exist between aerobic and anaerobic growth conditions
Our finding that IscR is more sensitive to changes in Fe-S demand under aerobic conditions makes physiological sense. Considering that greater than 150 Fe-S proteins exist in E. coli (Py et al., 2011) and that some Fe-S clusters are sensitive to O2 and/or reactive oxygen species (Imlay, 2006, Imlay, 2008), it is likely that Fe-S clusters are continually being damaged or destroyed during aerobic growth, thereby increasing the levels of substrate proteins that need Fe-S biogenesis or repair. As a result, there would be more competition between IscR and substrate proteins for the Isc machinery under aerobic conditions than anaerobic conditions. In support of this notion, we showed that aerobic but not anaerobic overexpression of either wild-type [4Fe-4S]-FNR or the [4Fe-4S]-FNR-L28H mutant derivative that contains an O2-resistant cluster (Bates et al., 2000) relieved PiscR repression. Why IscR may not compete as well with other substrate proteins is not known but may be related to the fact that IscR binds its [2Fe-2S] cluster with an atypical ligation scheme of three cysteines and one histidine (Fleischhacker et al., 2012) perhaps making it a poor substrate for the Isc machinery.
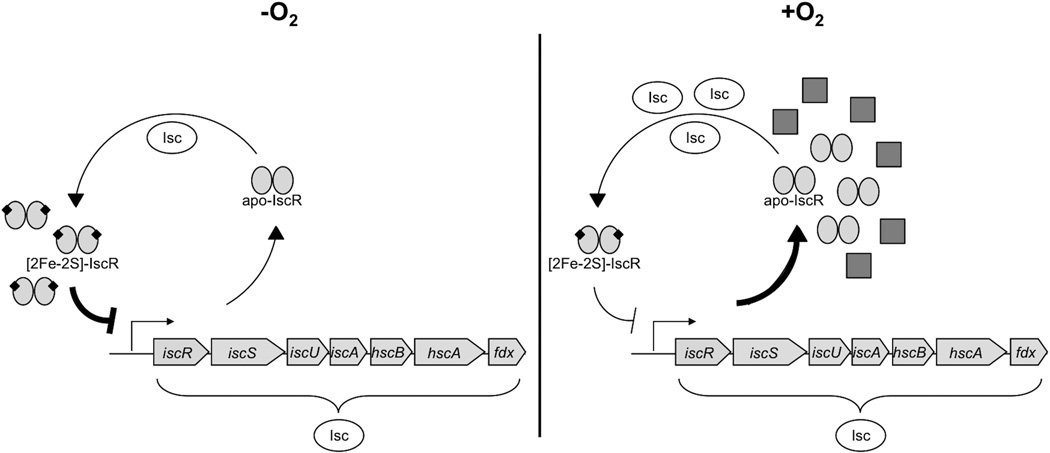
In sum, we propose that under aerobic conditions, the sensitivity of some Fe-S clusters to O2 and/or reactive oxygen species results in a higher rate of cluster turnover, which in turn leads to increased synthesis of the isc operon to meet the demands for Fe-S biogenesis. In contrast, under anaerobic conditions, the Isc machinery appears to satisfy the Fe-S demand more efficiently due to decreased general cluster turnover, and thus less competition among substrate proteins, under these conditions (Fig. 8). However, we cannot rule out the possibility that additional mechanisms may also contribute to regulating Isc Fe-S cluster biogenesis under anaerobic growth. Taken together, the different needs for Isc Fe-S biogenesis under aerobic and anaerobic conditions emphasize the important role of IscR as an Fe-S sensor, enabling E. coli to respond appropriately to environments of varying O2 tension.
Fig. 8.
A model for the differential demand in Isc-mediated Fe-S cluster biogenesis between aerobic and anaerobic growth conditions is shown. In the presence of O2, the need for Fe-S clusters is predicted to be high due to increased rates of general Fe-S cluster turnover under aerobic conditions, and thus elevated levels of apoprotein substrates (gray squares). As a result, there is competition between these substrates and IscR in acquiring Fe-S clusters from the Isc pathway, resulting in low IscR [2Fe-2S] cluster occupancy and thus less repression of iscR-SUA-hscBA-fdx. Under anaerobic conditions, the general rate of cluster turnover is decreased compared to aerobic conditions. The low demand for Fe-S biogenesis in the absence of O2 leads to increased IscR [2Fe-2S] cluster occupancy, and in turn, more repression of the Isc pathway.
EXPERIMENTAL PROCEDURES
Bacterial strain and plasmid construction
Strains and plasmids described in this work are listed in Table 1 and sequences of primers used are available upon request. The λ-PiscR-lacZ reporter fusion (−476 to +74 bp relative to the IscR start codon) (Schwartz et al., 2001) is located within the λatt site, whereas the PiscR-lacZ reporter fusion (−428 to +144 bp relative to the IscR start codon) replaces the wildtype lac promoter and was constructed by a method previously described (Giel et al., 2006, Kang et al., 2005). In the latter case, a lacI-kan-PiscR-lacZ fragment from pPK8508 was PCR-amplified and recombined onto the chromosome in the lac promoter region of BW25993/pKD46. PiscR-lacZ fusions containing mutations within PiscR were recombined onto the chromosome in the same manner after introducing mutations into pPK8508 using QuikChange (Stratagene). The resulting kan-promoter-lacZ constructs were transduced into the appropriate strain backgrounds using P1vir and confirmed by colony PCR and DNA sequencing.
Table 1.
Strains and plasmids used in this work.
| Strain | Relevant Genotype or Phenotype | Source |
|---|---|---|
| MG1655 | λ− F− rph-1 | Laboratory stock |
| PK8039 | MG1655 yhgI-lacZ | (Giel et al., 2006) |
| PK9110 | PK8039 nadA∷Tn10λcI857 Δ(cro-bioA) | This study |
| PK9116 | PK9110 but nadA+, TetS | This study |
| PK9120 | PK9116 mutS104∷mini-Tn10 | This study |
| PK9133 | PK9120 cat-FRT-araC-PBAD | This study |
| PK9520 | PK9133 bla-Ptac replacing −100 to +28 bp of PiscR relative to the +1 transcription start site |
This study |
| PK6364 | MG1655 λ-iscR’-lacZ | (Schwartz et al., 2001) |
| PK6512 | PK6364 ΔiscR | This study |
| PK8120 | PK6364 ΔiscSUA∷cat | This study |
| PK8122 | PK6364 ΔiscSUA-hscBA-fdx∷cat | This study |
| PK8614 | PK8122 + pGS0164 | This study |
| PK8618 | PK8614 ΔsufABCDSE∷kan | This study |
| PK8615 | PK8122 + pBAD/Myc-hisC | This study |
| PK7783 | PK6364 ΔiscS | This study |
| PK6828 | PK6364 ΔiscU | This study |
| PK7759 | PK6364 ΔiscA | This study |
| PK6564 | PK6364 ΔsufABCDSE∷kan | This study |
| PK6826 | PK6364 fdx∷kan | This study |
| PK6848 | PK6364 ΔiscX | This study |
| PK7540 | PK6364 ΔpepB | This study |
| PK7751 | PK6364 sseB∷Tn5KAN2 | This study |
| PK7854 | PK6364 iscR-C92A zfh-3601∷FRT | This study |
| PK7855 | PK6364 iscR-C98A zfh-3601∷FRT | This study |
| PK7856 | PK6364 iscR-C104 zfh-3601∷FRT | This study |
| PK7898 | PK6364 iscR-C92A/C98A/C104A zfh-3601∷FRT | This study |
| PK8616 | PK6364 ΔcsdA∷cat | This study |
| PK8617 | PK6364 ΔytfE∷kan | This study |
| PK7325 | PK6364 + pACYClacIQ-CAM | This study |
| PK7329 | PK6364 + pACYClacIQ-CAM + pDHB60 | This study |
| PK7333 | PK6364 + pACYClacIQ-CAM + pPK7332 | This study |
| PK7328 | PK6364 + pACYClacIQ-CAM + pPK7307 | This study |
| PK7571 | MG1655 PiscR-lacZ (in lac region) | This study |
| PK8151 | PK7571 ΔlacY | This study |
| PK9523 | PK8151 bla-Ptac-iscR-FRT-cat-FRT-araC-PBAD-iscS + pACYClacIQ |
This study |
| PK8677 | PK8151 ΔiscR + pACYClacIQ | This study |
| PK8521 | MG1655 PiscR (site A mutated)-lacZ | This study |
| PK8528 | MG1655 PiscR (site B mutated)-lacZ | This study |
| PK8582 | MG1655 PiscR (sites A & B mutated)-lacZ | This study |
| PK8551 | MG1655 PiscR (site C mutated)-lacZ | This study |
| PK8801 | MG1655 PiscR (sites A & C mutated)-lacZ | This study |
| PK8808 | MG1655 PiscR (sites B & C mutated)-lacZ | This study |
| PK8820 | MG1655 PiscR (sites A, B, & C mutated)-lacZ | This study |
| PK4854 | MG1655 ΔiscR | (Schwartz et al., 2001) |
| PK7572 | PK4854 PiscR-lacZ | This study |
| PK8523 | PK4854 PiscR (site A mutated)-lacZ | This study |
| PK8530 | PK4854 PiscR (site B mutated)-lacZ | This study |
| PK8584 | PK4854 PiscR (sites A & B mutated)-lacZ | This study |
| PK8553 | PK4854 PiscR (site C mutated)-lacZ | This study |
| PK8803 | PK4854 PiscR (sites A & C mutated)-lacZ | This study |
| PK8810 | PK4854 PiscR (sites B & C mutated)-lacZ | This study |
| PK8822 | PK4854 PiscR (sites A, B, & C mutated)-lacZ | This study |
| PK7599 | BL21 himA∷tet + pPK6161 | (Nesbit et al., 2009) |
| PK7881 | BL21 himA∷tetΔiscR∷kan + pPK7862 | (Nesbit et al., 2009) |
| DY329 | W3110 ΔlacU169nadA∷Tn10gal490 λcI857 Δ(cro- bioA) |
(Yu et al., 2000) |
| CSH115 | F-, ara-600, Δ(gpt-lac)5 λ−, mutS104∷mini-Tn10 relA1?, spoT1?, thi-1 |
The Coli Genetic Stock Center (Yale) |
| BW25993 | lacIqhsdR514ΔaraBADAH33ΔrhaBADLD78 | (Datsenko and Wanner, 2000) |
| Plasmid | ||
| pPK6161 | iscR cloned into pET-11a | (Schwartz et al., 2001) |
| pPK7862 | iscR-C92A/C98A/C104A cloned into pET-11a | (Nesbit et al., 2009) |
| pPK823 | fnr cloned into pET-11a | (Lazazzera et al., 1993) |
| pPK1868 | fnr-L28H cloned into pET-11a | (Bates et al., 2000) |
| pDHB60 | ApR, Ptac (pBR322-derived) | (Boyd et al., 2000) |
| pPK7332 | XbaI-HindIII fnr fragment from pPK823 cloned into XbaI and HindIII sites of pDHB60 |
This study |
| pPK7307 | XbaI-HindIII fnr-L28H fragment from pPK1868 cloned into XbaI and HindIII sites of pDHB60 |
This study |
| pKD13 | FRT-kan-FRT | (Datsenko and Wanner, 2000) |
| pKD32 | Same as pKD13 but with FRT-cat-FRT | B. L. Wanner |
| pCP20 | ApR | (Datsenko and Wanner, 2000) |
| pKD46 | Phage λgam-bet-exo genes under ParaB control | B. L. Wanner |
| pBR322 | ApR | (Bolivar et al., 1977) |
| pPK7035 | pBR322 with kan from pHP45Ω and BamHI-NdeI fragment from pRS1553 |
(Kang et al., 2005) |
| pPK8508 | pPK7035 with PiscR (−428 to +144 bp relative to the IscR start codon) cloned into BamHI and XhoI sites |
This study |
| pBAD/Myc- hisC |
ApR | Invitrogen |
| pGS0164 | sufABCDSE cloned into pBAD/Myc-hisC | (Outten et al., 2003) |
| pPK9125 | FRT-cat-FRT from pKD32 cloned into SphI site of pBAD/Myc-hisC |
This study |
| pACYC184 | TetR, CmR cloning vector | (Chang and Cohen, 1978) |
| pPK5960 | pACYC184 with StuI-EcoRV 985 bp fragment containing iscR and part of iscS |
(Schwartz et al., 2001) |
| pPK9112 |
iscRS fragment from pPK5960 cloned into EcoRV site of pBR322 |
This study |
| pPK9127 | FRT-cat-FRT-araC-PBAD fragment from pPK9125 cloned into EcoRI site of pPK9112 |
This study |
| pRZ7411 | HindIII-BamHI fragment of fnr in pACYC184 | (Lazazzera et al., 1993) |
| pPK9003 | pRZ7411 based vector with bla from pBR322 at AflII site and Ptac from pPK7332 at AflII and NdeI sites |
This study |
| pACYClacIQ | lacIQ in pACYC184, TetR | (Boyd et al., 1987) |
| pACYClacIQ CAM |
lacIQ in pACYC184, CmR | (Derman et al., 1993) |
| pPK6511 | PiscR cloned into pUC19-spf’ | (Schwartz et al., 2001) |
| pPK8515 | pPK6511 site A mutated | This study |
| pPK6806 | pPK6511 site B mutated | This study |
| pPK8547 | pPK6511 site C mutated | This study |
In frame deletions of iscR, iscSUA, iscSUA-hscBA-fdx, iscS, iscU, iscA, iscX, pepB, csdA, and lacY were constructed by replacing the coding region(s) with a CmR or KanR cassette flanked by FLP recognition target (FRT) sites from plasmid pKD32 or pKD13, respectively, as described previously (Datsenko and Wanner, 2000). Transduction with P1 vir was used to move the cat or kan allele to the appropriate strain backgrounds. In some cases, the CmR or KanR cassette was removed by transforming strains with pCP20, encoding FLP recombinase (Datsenko and Wanner, 2000) and by screening for Cm or Kan sensitivity. All gene deletions were confirmed by colony PCR. Previously constructed alleles encoding IscR mutants in which cysteine residues were substituted with alanine (Nesbit et al., 2009)were transduced into PK6364 using P1 vir and selecting for KanR. The KanR cassette was subsequently removed by transforming strains with pCP20. The following existing alleles were also transduced into appropriate strain backgrounds using P1vir∷sseB with a Tn5KAN2 insertion at position 150, fdx∷kan, ΔsufABCDSE∷kan, and ΔytfE∷kan.
Construction of bla-Ptac-iscR-cat-araC-PBAD-iscSUA on the chromosome of strain PK9520 was performed in several steps. First, a MG1655 recombineering strain (PK9120) was constructed by transducing nadA∷Tn10 [λcI857Δ(cro-bioA)] from DY329 into PK8039 using P1 vir and selecting for TetR at 30°C to form PK9110. The nadA∷Tn10 was removed by transducing to nicotinic acid prototrophy using P1 vir MG1655 and screening for TetS at 30°C to make PK9116. Colonies retaining the biotin requirement were then transduced with P1 vir mutS104∷mini-Tn10 from CSH115 to form PK9120. Next, FRT-cat-FRT was PCR-amplified from pKD32 and cloned into the SphI site of pBAD/Myc-hisC to form pPK9125. FRT-cat-FRT-araC-PBAD was amplified and inserted between iscR and iscS in pPK9112, using the EcoRI site 6 nt after the iscR stop codon to form pPK9127. To obtain FRT-cat-FRT-araC-PBAD upstream of iscS on the chromosome, this construct was amplified with primers containing ends with homology to this region of the chromosome, electroporated into PK9120, and selected for CmR. The bla-Ptac region from pPK9003 was amplified and electroporated into the above PK9120 derivative (PK9133) to replace −100 to +28 bp of PiscR (relative to the +1 transcription start site) with bla-Ptac, forming PK9520. After verification by DNA sequencing, bla-Ptac-iscR-FRT-cat-FRT-araC-PBAD-iscS was transduced into PK8151 using P1 vir and selected for CmR and ApR with 10 mM arabinose to derepress PBAD-iscSUA. Finally, the strain was transformed with pACYClacIQ to form PK9523.
Wild-type fnr and fnr-L28H were cloned into pDHB60 by digesting pPK823 or pPK1868, respectively, with HindIII and XbaI, followed by ligation of these fragments into the HindIII and XbaI sites of pDHB60. PK6364 was transformed with pACYClacIQ-CAM, and the resulting strain was transformed with pDHB60, pPK7332 (fnr), or pPK7307 (fnr-L28H).
β-galactosidase assays
Strains were grown aerobically by shaking at 250 rpm to an OD600 ∼0.2 or anaerobically in screw-capped tubes to an OD600 ∼0.1 at 37° C in MOPS minimal medium (Neidhardt et al., 1974) supplemented with 0.2% glucose (w/v) or M9 minimal medium (Miller, 1972) containing 0.2% glucose (w/v), 1mM MgSO4, 2.5 µg ml−1 ferric ammonium citrate, 2 µg ml−1 thiamine, and 0.02% ammonium molybdate (w/v). As indicated in the figure legends, media were also supplemented with 12.5 µg ml−1 nicotinic acid and 2 µg ml−1 thiamine for some experiments. For the FNR overexpression experiment, strains were grown under aerobic or anaerobic conditions by sparging (Sutton and Kiley, 2003). For all β-galactosidase assays, Cm or Tet were added to culture samples at final concentrations of 20 or 10 µg ml−1, respectively, to terminate cell growth and any further protein synthesis; cells were placed on ice until assayed for β-galactosidase activity as previously described (Miller, 1972). Because of aerobic and anaerobic cell count differences, the Miller units from aerobically grown strains were multiplied by 1.55, similar to previous studies (Mettert and Kiley, 2007). Assays were repeated at least three independent times and the standard errors for data plotted as “Fold Repression” were calculated using a propagation of standard error formula (Ku, 1966).
For the FNR overexpression experiment, strains were grown in M9 minimal medium (described in the above paragragh) containing 50 µg ml−1 Ap, and 20 µg ml−1 Cm under aerobic or anaerobic conditions by sparging (Sutton and Kiley, 2003). FNR expression was induced by adding a final concentration of 100 µM IPTG at OD600 of 0.2. Samples were removed at various time points and processed as above, and β-galactosidase assays were performed in triplicate for each time point. The experiment was repeated on at least two separate occasions.
Western blot analysis
Strains were grown as for β-galactosidase assays, and IscR, SufD, or FNR levels were measured by Western blots as described previously (Nesbit et al., 2009, Mettert et al., 2008, Sutton et al., 2004). IscR was quantified using isolated proteins as standards, followed by imaging as described (Nesbit et al., 2009, Mettert et al., 2008). The cytoplasmic concentration of IscR (µM) was calculated using the molecular weight determined for monomeric IscR (17,336 Da) (Schwartz et al., 2001), the number of cells per ml of culture sampled as previously determined via viable cell counts (Mettert et al., 2008), and the estimated cell volume of ∼1 × 10−15 L (Kubitschek, 1990).
Protein purification
Purifications of [2Fe-2S] cluster-containing wild-type IscR (Giel et al., 2006) and IscR-C92A/C98A/C104A (Nesbit et al., 2009) were performed and the protein concentration and iron and sulfide content were determined as previously described (Beinert, 1983, Kennedy et al., 1984, Khoroshilova et al., 1995). For all in vitro experiments, wild-type IscR was ≥50% occupied with [2Fe-2S] clusters. For simplicity, all protein concentrations herein are reported as monomers, although IscR is mainly a dimer in solution (Nesbit et al., 2009).
DNase I footprinting assays
Mutations within PiscR were introduced into pPK6511 using QuikChange (Stratagene), generating pPK8515, pPK6806, and pPK8547, and DNA fragments containing the wild-type or mutated iscR promoter region were isolated from their respective plasmids after digestion with XbaI and HindIII. Klenow fragment (NEB) and [α-32P]dCTP were used to label the XbaI end of the fragment (comprised of bases −161 to +38 relative to the PiscR transcription start site). Assays were performed as described (Giel et al., 2006) except the incubation time of protein with DNA prior to the addition of DNase I was 10 minutes.
Fluorescence anisotropy
DNA binding isotherms were generated under anaerobic conditions by measuring changes in fluorescence polarization when IscR bound dsDNA as described (Nesbit et al., 2009), and assays were repeated on three independent occasions. Briefly, 30-mer dsDNA contained the following Type 1 sites (underlined): PiscR site A (5'-AAATACCCGACTAAATCAGTCAAGTAAATA-3') or PiscR site B (5'-AAATAGTTGACCAATTTACTCGGGAATGTC-3'). [2Fe-2S]-IscR (20 to 718 nM) or IscR-C92A/C98A/C104A (35 to 1000 nM) was incubated with 5 nM Texas Red labeled-DNA, 40 mM Tris-Cl (pH 7.9), and 150 mM KCl for 10 min at room temperature under anaerobic conditions. For wild-type IscR, the fraction bound was determined as previously described (Nesbit et al., 2009).
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge the contributions of Helmut Beinert (deceased) in the early stages of this work and Sarah Teter for helping with strain construction. We thank Jon Beckwith, Fred Blattner, Larry Vickery, F. Wayne Outten, Stephen Spiro, and Gisela Storz for providing plasmids or strains and James Keck for the use of his Beacon 2000 fluorescence polarization fluorometer. This work was supported by NIH grant GM45844 to P.J.K. J.L.G., A.D.N., and A.S.F. were trainees of the NIH Biotechnology Predoctoral Training Grant GM08349, the NIH Molecular Biosciences Predoctoral Training Grant GM07215, and the NIH Postdoctoral Training Grant F32GM085987, respectively, all to the University of Wisconsin-Madison.
REFERENCES
- Angelini S, Gerez C, Ollagnier-de Choudens S, Sanakis Y, Fontecave M, Barras F, Py B. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J Biol Chem. 2008;283:14084–14091. doi: 10.1074/jbc.M709405200. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Naik SG, O'Carroll IP, Huynh BH, Dean DR, Johnson MK, Dos Santos PC. A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J Biol Chem. 2008;283:14092–14099. doi: 10.1074/jbc.M709161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates DM, Popescu CV, Khoroshilova N, Vogt K, Beinert H, Münck E, Kiley PJ. Substitution of leucine 28 with histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S]2+ cluster to oxygen. J Biol Chem. 2000;275:6234–6240. doi: 10.1074/jbc.275.9.6234. [DOI] [PubMed] [Google Scholar]
- Beinert H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal Biochem. 1983;131:373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- Beinert H. Iron-sulfur proteins: ancient structures, still full of surprises. J Biol Inorg Chem. 2000;5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, et al. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- Boyd D, Manoil C, Beckwith J. Determinants of membrane protein topology. Proc Natl Acad Sci USA. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D, Weiss DS, Chen JC, Beckwith J. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J Bacteriol. 2000;182:842–847. doi: 10.1128/jb.182.3.842-847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derman AI, Puziss JW, Bassford PJ, Jr, Beckwith J. A signal sequence is not required for protein export in prlA mutants of Escherichia coli . EMBO J. 1993;12:879–888. doi: 10.1002/j.1460-2075.1993.tb05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischhacker AS, Stubna A, Hsueh KL, Guo Y, Teter SJ, Rose JC, et al. Characterization of the [2Fe-2S] cluster of Escherichia coli transcription factor IscR. Biochemistry. 2012;51:4453–4462. doi: 10.1021/bi3003204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecave M. Iron-sulfur clusters: ever-expanding roles. Nat Chem Biol. 2006;2:171–174. doi: 10.1038/nchembio0406-171. [DOI] [PubMed] [Google Scholar]
- Giel JL. Role of IscR in regulation of iron-sulfur biogenesis in Escherichia coli: Identification of the IscR regulon and mechanisms of autoregulation. University of Wisconsin-Madison; 2007. Ph.D. dissertation. [Google Scholar]
- Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli . Mol Microbiol. 2006;60:1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- Gourse RL, Ross W, Gaal T. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol. 2000;37:687–695. doi: 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol. 2006;59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Imlay JA. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol Microbiol. 2010;78:1448–1467. doi: 10.1111/j.1365-2958.2010.07418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological Fe-S clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Justino MC, Baptista JM, Saraiva LM. Di-iron proteins of the Ric family are involved in iron-sulfur cluster repair. Biometals. 2009;22:99–108. doi: 10.1007/s10534-008-9191-2. [DOI] [PubMed] [Google Scholar]
- Justino MC, Vicente JB, Teixeira M, Saraiva LM. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J Biol Chem. 2005;280:2636–2643. doi: 10.1074/jbc.M411070200. [DOI] [PubMed] [Google Scholar]
- Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol. 2005;187:1135–1160. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MC, Kent TA, Emptage M, Merkle H, Beinert H, Münck E. Evidence for the formation of a linear [3Fe-4S] cluster in partially unfolded aconitase. J Biol Chem. 1984;259:14463–14471. [PubMed] [Google Scholar]
- Khoroshilova N, Beinert H, Kiley PJ. Association of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA binding. Proc Natl Acad Sci USA. 1995;92:2499–2503. doi: 10.1073/pnas.92.7.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoroshilova N, Popescu C, Münck E, Beinert H, Kiley PJ. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc Natl Acad Sci USA. 1997;94:6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiley PJ, Beinert H. The role of Fe-S proteins in sensing and regulation in bacteria. Curr Opin Microbiol. 2003;6:181–185. doi: 10.1016/s1369-5274(03)00039-0. [DOI] [PubMed] [Google Scholar]
- Ku HH. Notes on the use of propagation of error formulas. J Res. 1966;70C:263–273. [Google Scholar]
- Kubitschek HE. Cell volume increase in Escherichia coli after shifts to richer media. J Bacteriol. 1990;172:94–101. doi: 10.1128/jb.172.1.94-101.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazazzera BA, Bates DM, Kiley PJ. The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev. 1993;7:1993–2005. doi: 10.1101/gad.7.10.1993. [DOI] [PubMed] [Google Scholar]
- Mettert EL, Kiley PJ. Contributions of [4Fe-4S]-FNR and integration host factor to fnr transcriptional regulation. J Bacteriol. 2007;189:3036–3043. doi: 10.1128/JB.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettert EL, Outten FW, Wanta B, Kiley PJ. The impact of O2 on the Fe-S cluster biogenesis requirements of Escherichia coli FNR. J Mol Biol. 2008;384:798–811. doi: 10.1016/j.jmb.2008.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Mukhopadhyay P, Zheng M, Bedzyk LA, LaRossa RA, Storz G. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc Natl Acad Sci USA. 2004;101:745–750. doi: 10.1073/pnas.0307741100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbit AD, Giel JL, Rose JC, Kiley PJ. Sequence-specific binding to a subset of IscR-regulated promoters does not require IscR Fe-S cluster ligation. J Mol Biol. 2009;387:28–41. doi: 10.1016/j.jmb.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli . Mol Microbiol. 2004;52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- Outten FW, Wood MJ, Munoz FM, Storz G. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli . J BiolChem. 2003;278:45713–45719. doi: 10.1074/jbc.M308004200. [DOI] [PubMed] [Google Scholar]
- Pastore C, Adinolfi S, Huynen MA, Rybin V, Martin S, Mayer M, et al. YfhJ, a molecular adaptor in iron-sulfur cluster formation or a frataxin-like protein? Structure. 2006;14:857–867. doi: 10.1016/j.str.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Py B, Barras F. Building Fe-S proteins: bacterial strategies. Nat Rev Microbiol. 2010;8:436–446. doi: 10.1038/nrmicro2356. [DOI] [PubMed] [Google Scholar]
- Py B, Moreau PL, Barras F. Fe-S clusters, fragile sentinels of the cell. Curr Opinion Microbiol. 2011;14:218–223. doi: 10.1016/j.mib.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Record MTJ, Reznikoff WS, Craig ML, McQuade KL, Schlax PJ. Escherichia coli RNA polymerase (Eσ70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, et al., editors. Escherichia Coli and Salmonella: Cellular and Molecular Biology. 2nd edn. Washington, DC: American Society for Microbiology; 1996. pp. 792–820. [Google Scholar]
- Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, Kiley PJ. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci USA. 2001;98:14895–14900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura Y, Takahashi Y, Kakuta Y, Fukuyama K. Crystal structure of Escherichia coli YfhJ protein, a member of the ISC machinery involved in assembly of iron-sulfur clusters. Proteins. 2005;60:566–569. doi: 10.1002/prot.20481. [DOI] [PubMed] [Google Scholar]
- Sutton VR, Kiley PJ. Techniques for studying the oxygen-sensitive transcription factor FNR from Escherichia coli . Methods Enzymol. 2003;370:300–312. doi: 10.1016/S0076-6879(03)70027-5. [DOI] [PubMed] [Google Scholar]
- Sutton VR, Mettert EL, Beinert H, Kiley PJ. Kinetic analysis of the oxidative conversion of the [4Fe-4S]2+ cluster of FNR to a [2Fe-2S]2+ cluster. J Bacteriol. 2004;186:8018–8025. doi: 10.1128/JB.186.23.8018-8025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter V, Vinella D, Loiseau L, Ollagnier de Choudens S, Fontecave M, Barras F. The CsdA cysteine desulphurase promotes Fe/S biogenesis by recruiting Suf components and participates to a new sulphur transfer pathway by recruiting CsdL (ex-YgdL), a ubiquitin-modifying-like protein. Mol Microbiol. 2009;74:1527–1542. doi: 10.1111/j.1365-2958.2009.06954.x. [DOI] [PubMed] [Google Scholar]
- Vinella D, Brochier-Armanet C, Loiseau L, Talla E, Barras F. Iron-sulfur (Fe/S) protein biogenesis: phylogenomic and genetic studies of A-type carriers. PLoS Genet. 2009;5:e1000497. doi: 10.1371/journal.pgen.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo WS, Lee JH, Lee KC, Roe JH. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol Microbiol. 2006;61:206–218. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli . Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.