Abstract
Salmonella and other pathogenic bacteria use the type III secretion system (T3SS) to inject virulence proteins into human cells to initiate infections. The structural component of the T3SS contains a needle and a needle tip. The needle is assembled from PrgI needle protomers and the needle tip is capped with several copies of the SipD tip protein. How a tip protein docks on the needle is unclear. A crystal structure of a PrgI-SipD fusion protein docked on the PrgI needle results in steric clash of SipD at the needle tip when modeled on the recent atomic structure of the needle. Thus, there is currently no good model of how SipD is docked on the PrgI needle tip. Previously, we showed by NMR paramagnetic relaxation enhancement (PRE) methods that a specific region in the SipD coiled-coil is the binding site for PrgI. Others have hypothesized that a domain of the tip protein – the N-terminal α-helical hairpin, has to swing away during the assembly of the needle apparatus. Here, we show by PRE methods that a truncated form of SipD lacking the α-helical hairpin domain binds more tightly to PrgI. Further, PRE-based structure calculations revealed multiple PrgI binding sites on the SipD coiled-coil. Our PRE results together with the recent NMR-derived atomic structure of the Salmonella needle suggest a possible model of how SipD might dock at the PrgI needle tip. SipD and PrgI are conserved in other bacterial T3SSs, thus our results have wider implication in understanding other needle-tip complexes.
Keywords: Salmonella, type III secretion, SipD, PrgI, NMR
Introduction
Many Gram-negative bacterial pathogens utilize the T3SS to inject virulence proteins into eukaryotic cells to cause disease [1,2]. The main structural component of the T3SS is the needle complex, which is a macromolecular structure assembled in the bacterial envelope from over 20 different proteins and consists of a membrane bound base, an external needle, a tip, and a translocon (Fig. 1A). In Salmonella, the needle is assembled from the polymerization of approximately 120 copies of the needle protein PrgI [3-5]. PrgI (80 residues) [6,7] and other T3SS needle protomers [8,9] have α-helical hairpin structures flanked by flexible tails. At the distal end of the T3SS needle docks a tip complex [10,11], which in Salmonella is formed by the tip protein SipD (343 residues) [12]. The crystal structure of SipD [13] and homologs [14,15] show three common structural features: an N-terminal α-helical hairpin domain, a long central coiled-coil, and a distal domain (Fig. 1B). Prior to contact with host cell membrane, the T3SS needle apparatus is pre-assembled where the SipD tip complex is docked at the needle tip but the translocon comprising the membrane proteins SipB and SipC is still missing [12]. When Salmonella detect eukaryotic cells, a conformational change occurs at the needle tip in a process that is not yet well understood that allows the translocon proteins SipB and SipC to transit through the needle and dock at the SipD tip complex. SipB and SipC form a translocon pore on the host cell membrane to complete the assembly of the needle apparatus and allow the passage of effector proteins directly into the host cell [12]. The effectors then modulate host cell biology for the pathogen’s benefit.
Fig. 1.
Cartoon of T3SS and structure of SipD. (A) Cartoon of the Salmonella T3SS. (B) Crystal structure of an intact form of SipD (residues 39-343, PDB 3NZZ) [13], and (B) a model of a truncated form, SipDt (residues 101-343). SipD and SipDt are colored by structural feature as red (N-terminal α-helical hairpin), green (helix α3), gray (coiled-coil); and cyan (distal domain).
How the tip protein binds to the needle protein is not well understood. Using an intact form of SipD, we recently showed by PRE that PrgI interacts with a specific region of the SipD coiled-coil [16]. Others have hypothesized that the α-helical hairpin domain in the tip proteins functions as a self-chaperone [15] that prevents the tip protein from self-oligomerizing within the bacterial cytosol. And upon assembly at the needle tip, the α-helical hairpin domain is not required for interaction with the needle and thereby swings away [17]. Thus, a truncated form of SipD (here referred to as SipDt, Fig. 1C) lacking the α-helical hairpin domain should interact with PrgI. To determine how a truncated SipD interact with PrgI and dock at the needle tip, Lunelli et al. [18] determined the crystal structure of a PrgI-SipDt fusion protein comprising of PrgI at the N-terminus, a short 5-residue (GGSGG) linker, and a truncated SipD (residues 127-343) lacking the α-helical hairpin domain (Fig. 2A). The crystal structure of the PrgI-SipDt fusion protein could be docked at the needle tip of the model of the needle derived by electron microscopy (EM) [18]. Since then, however, a seminal contribution from solid-state NMR (ssNMR) by Loquet et al. [3] showed a completely different orientation of PrgI compared to the EM-derived atomic models of T3SS needles [4,9,19]. The ssNMR atomic model of the needle places the N-terminus of needle protomers outside the needle and the C-terminus inside the needle channel (Fig. 2B, C) [3,20], which is completely opposite to the orientation of needle protomers suggested by EM [4,9,19]. When the crystal structure of the PrgI-SipDt fusion protein [18] is docked on the current model of the PrgI needle, a massive steric clash of SipD occurs at the needle tip (Fig. 2D). The crystal structure of the PrgI-SipDt fusion protein is thus incompatible with the current atomic model of the needle and could not be used in modeling SipD at the needle tip.
Fig. 2.
Docking of PrgI-SipDt fusion protein at the needle tip. (A) Structure of PrgI-SipDt fusion protein (PDB 3ZQB, [18]) colored as green (SipDt) and red (PrgI). (B,C) Atomic model of the Salmonella needle derived by solid-state NMR (PDB 2LPZ, [3]) shown as orthogonal views. (D) Docking two PrgI-SipDt molecules at the needle tip by superposition of PrgI in the fusion protein (red) with two adjacent PrgI protomers at the needle tip results in steric clash of SipDt (green and cyan).
Here, we extend our previous PRE studies characterizing the PrgI-SipD interaction by using a truncated form of SipD lacking the α-helical hairpin. In contrast to the crystallography results of PrgI-SipDt fusion protein wherein PrgI packs on one contiguous surface of the SipD coiled-coil [18], our PRE results suggest there are multiple PrgI binding surfaces on the SipD coiled-coil. Based on our PRE results, we propose a model of SipD docked on the needle tip without resulting in steric clash of SipD.
Results
PRE of truncated SipD-PrgI interaction
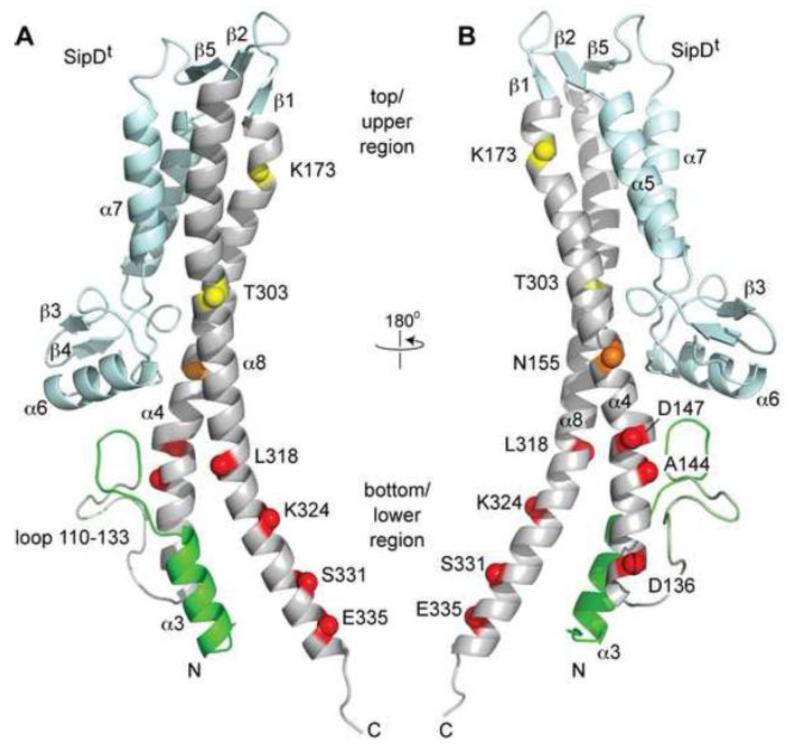
To identify how SipDt (a truncated form of SipD lacking the α-helical hairpin) binds to PrgI, we used our previous PRE approach [16]. Single time-point PREs were determined for ten SipDt cysteine mutants spread across the length of the coiled-coil in residues D136, A144, D147, N155, K173, T303, L318, K324, S331 and E335 (Fig. 3). The structure of SipDt shown in Fig. 3 is oriented with an arbitrary “top/bottom” or “upper/lower” perspective used here to describe the various parts of the coiled-coil. Spin labels attached to residues on the lower half of the coiled-coil on helix α4 (D136, A144 and D147, Fig. 3B) and helix α8 (L318, K324, S331 and E335, Fig. 3), induced strong PRE effect upon titration into PrgI and peak intensity ratios (Ipar / Idia) were in the range of 0.30 and 0.67 (Fig. 4). D136 at the extreme bottom of helix α4 exhibited the strongest PRE effect with (Ipar / Idia) ratio of 0.30 (Fig. 4E). On the other hand, when the spin label was attached to K173 (Fig. 4A) or T303 (Fig. 4F), which were located on the upper half of the coiled-coil (Fig. 3), SipDt showed relatively weaker PRE effect on PrgI with Ipar / Idia ratios at 0.91 and 0.83, respectively. In line with these, N155 (Fig. 4B) in the middle of helix α4 showed relatively moderate PRE effect (Ipar / Idia = 0.73). Thus, single time point PREs showed that PrgI mostly interacted with the lower half of the SipDt coiled-coil spanned by residues D147 to D136 on helix α4 (Fig. 4C-E) and residues L318 to S331 on helix α8 (Fig. 4G-J).
Fig. 3.
Location of PRE spin labels on SipDt. (A,B) The PRE spin labels on the SipDt coiled-coil are depicted as spheres and colored based on weak (yellow), medium (orange), and strong PRE effect with PrgI. SipDt as depicted here is oriented with an arbitrary “top/bottom” or “upper/lower” used herein to describe the various parts of the coiled-coil.
Fig. 4.
Single time-point PREs of spin labeled SipDt titrated into PrgI. The ten SipDt spin labels are at position (A) K173, (B) N155, (C) D147, (D) A144, (E) D136, (F) T303, (G) L318, (H) K324, (I) S331, and (J) E335. The locations of the spin labels on helix α4 and α8 are indicated by black spheres and the average Ipar/Idia values are indicated on each plot.
To quantify intermolecular PREs and obtain restraints for PRE-based structure calculations of the PrgI-SipDt complex, we determined the PRE 1H relaxation rates (Λ2) using the two time-point method [21]. As the total acquisition time for the two time-point PRE method was much longer compared to the single time-point method, the two time-point PREs were only acquired for the four cysteine mutants in positions A144 and D147 on helix α4 and K324 and E335 on helix α8 that showed the strongest single time-point PREs. As a negative control, Λ2 were also determined for K173, which showed the weakest single time-point PRE. Results of two time-point PREs showed that the four spin labels at A144, D147, K324 and E335 showed strong PRE effect, with average Λ2 values of 27.51 s−1, 26 s−1, 32.9 s−1 and 22.5 s−1, respectively (Fig. 5), whereas K173 showed the lowest Λ2 average value of 7.0 s−1.
Fig. 5.
Two time-point PREs. Two time-point Λ2 values of backbone 1HN were acquired for the SipDt spin labels at positions: (A) A144, (B) D147, (C) K324, (D) E335 and (E) K173.
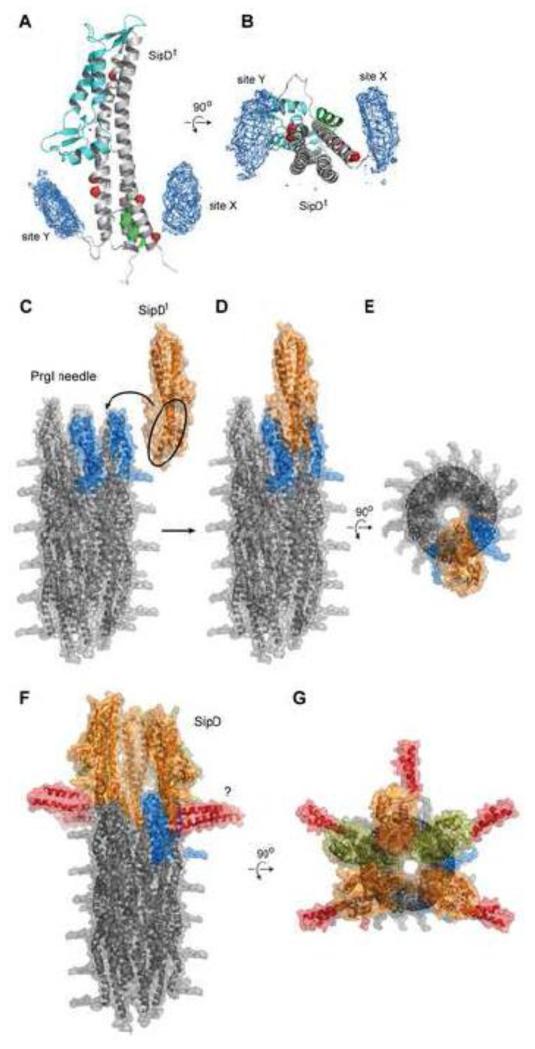
Results of PRE structure calculations using Λ2 values from the spin labels at A144, D147, K324 and E335 as distance restraints revealed that two populations of PrgI were localized around the lower half of the SipDt coiled-coil region (Fig. 6). One PrgI population was partly localized at the position previously occupied by the α-helical hairpin facing the lower half of the SipD coiled-coil (labeled site X in Fig. 6A,B). PRE results suggested that PrgI binds to SipD at the position previously occupied by the α-helical hairpin. The binding site X on SipD was not identified as a PrgI binding surface by the crystal structure of the PrgI-SipDt fusion protein and is a unique contribution from PRE. Our PRE results are the first direct experimental demonstration that a truncated T3SS tip protein interacts with its cognate needle protomer at the site previously occupied by the α-helical hairpin, and this information has a direct bearing on determining how a tip protein docks at the needle tip. A second PrgI population was localized on another face of the coiled-coil opposite from binding site X (labeled site Y in Fig. 6A,B). Results of PRE structure calculations therefore suggested that a truncated SipD could bind simultaneously to two PrgI molecules at two sites, and that both PrgI binding sites X and Y were located at the lower half of the SipD coiled-coil and on opposite faces of the coiled-coil (Fig. 6A,B). Another unique contribution revealed by PRE is that there are two PrgI binding sites on SipD, and this information is also important in modeling SipD at the needle tip. Results from crystallography [18], ITC [18], as well as fluorescence polarization [this work] thus far has suggested only one binding site. Binding site Y (Fig. 6A) overlaps with the PrgI binding site on SipD in the crystal structure of the PrgI-SipDt fusion protein (Fig. 2A) [18].
Fig. 6.
Results of PRE structure calculations and model of SipD at the needle tip. (A,B) PRE structure calculations revealed two binding sites of PrgI around the SipDt coiled-coil. The PrgI binding sites are labeled site X and site Y and shown as atomic probability maps around SipDt with the Cα positions of the spin labels shown as red spheres. (C,D, E) Model of how the SipD coiled-coil will insert in-between the spaces at the needle tip of the assembled Salmonella needle (PDB 2LPZ). (F,G) Model of full-length SipD at the needle tip, which could accommodate 5 SipD molecules (colored orange and olive) with the N-terminal α-helical hairpin (red) splayed outside the PrgI needle. (F) The question mark denotes that the conformation of the N-terminal α-helical hairpin is unknown in the assembled needle-tip complex.
PRE-based model of SipD docked at the needle tip
The PRE results regarding the PrgI-SipDt interaction together with the ssNMR-derived atomic model of the Salmonella needle [3] constrain the possible orientations of SipDt at the needle tip and thus suggest a possible model of how SipD might dock at the needle tip (Fig. 6C,D,E). In the atomic structure of the Salmonella needle [3], PrgI molecules are arranged in a spiral symmetry with the PrgI α-helical hairpin slightly titled away from the wall of the needle. Further, at the needle tip, the PrgI α-helical hairpins are separated from each other by 25 Å [3], which is the distance between the center of mass of two adjacent PrgI α-helical hairpins. Because our PRE data suggested that SipDt could interact with two PrgI molecules, we propose that the SipD central coiled-coil could fit into the gaps between the PrgI α-helical hairpins at the needle tip. The average distance between the Cα atoms that spans the coiled-coil region at the bottom of SipD is ~17 Å, which could be easily accommodated between the two PrgI α-helical hairpins at the needle tip. In this model, the region corresponding to the SipD N-terminal α-helical hairpin will be splayed outside the needle (Fig. 6F,G). Consistent with the pentameric state proposed by others [15,17,18], a pentameric SipD complex could be accommodated at the needle tip (Fig. 6F,G). Further, our PRE model of how SipD docks at the needle tip reveals the regions of SipD that are exposed at the needle tip and therefore could be potential sites for interaction with the translocon. This could be experimentally tested using recombinant SipD and translocon proteins. Finally, we expect SipD to follow the spiral symmetry of the needle protomers, however, how this symmetry translates into the translocon remains unknown.
Biological tests of the model
To test the biological relevance of our PRE-derived PrgI-SipD model, we assayed the effect of mutations on the PrgI binding site at the SipD coiled-coil region on the ability of Salmonella to invade cultured human epithelial cells. The ability of Salmonella to invade eukaryotic cells has been shown to be strictly dependent on SipD function [22]. Point mutations were introduced in five residues (S148, V157, S313, D320 and V323) located at the lower half of the SipD coiled-coil at the binding interface with PrgI. The mutations were designed to alter the bulkiness of the side-chain (S148E, V157K, S313E and V323K) or the surface electrostatics (S148E, V157K, S313E, D320R and V323K). Results showed that the S148E mutation did not alter invasiveness (Fig. 7A), suggesting that S148 may not be crucial for PrgI-SipD interaction. Other mutations, such as point mutations on the lower half of helix α4 (V157K) and helix α8 (D320R and V323K), significantly decreased invasiveness. Among these, D320R and V323K located at the bottom of helix α8 showed only 25% invasiveness, while V157K (on helix α4) and S313E (on helix α8), showed invasiveness between 50-60% of wild type. We reasoned that point mutations may not cause large alterations in protein-protein interaction surfaces to drastically affect invasiveness; hence, double and triple mutations that alter contiguous surfaces were designed. The double and triple mutations on SipD rendered Salmonella completely non-invasive (Fig. 7A). Thus, altering the PrgI binding surface on SipD as predicted by the PRE can drastically affect SipD function and Salmonella invasiveness.
Fig. 7.
Decreased Salmonella invasiveness is correlated with decreased PrgI-SipD interaction. (A) Mutations of SipD residues at the PrgI-SipD binding interface reduced Salmonella invasiveness. (B,C) The noninvasive SipD mutations abrogated the SipDt-PrgI interaction as determined by fluorescence polarization.
To correlate the observed loss of function of SipD to its binding interaction with PrgI, we used fluorescence polarization (FP) to quantify the binding affinities of PrgI with the various SipD mutants. PrgI bound to SipDt with an apparent KD of 108 ± 10 μM (Fig. 7C), however the dissociation constant of PrgI with SipD could not be determined by FP because it was too weak as indicated by the FP plot failing to reach saturation (Fig. 7B). Because the FP assay could be only carried out with the SipDt construct, mutations were introduced in SipDt and tested for in vitro binding affinity with PrgI. The S148E point mutant, which showed a wild type phenotype in the invasion assay, displayed a binding affinity (apparent KD of 189± 20 μM) to PrgI similar to SipDt. The other SipD point mutants that showed decreased invasiveness (S313E, V157K, D320R, and V323K) also showed decreased binding affinities to PrgI compared to SipDt (Fig. 7B). In contrast, the double and triple mutations that rendered Salmonella totally non-invasive resulted in Kd that were too weak to be determined by FP (Fig. 7C). Thus, the decreased in vitro binding affinities between PrgI and SipD correlated with decreased Salmonella invasion assay.
To further explore the binding of SipD to PrgI in vivo, we used a previously described surface display immunofluorescence-based assay that specifically detects SipD only when anchored at the needle tip [12]. In contrast to wild type, the SipD triple mutant (D320R / D323K / S327R) was not detected at the bacterial surface (Fig. 8 A-C) despite the fact that this mutant was expressed and secreted by the type III secretion system at wild type levels (Fig. 8D). It has been previously shown that loss-of-function mutations in the SipD family of proteins leads to enhance type III secretion. It is thought that the properly positioned SipD complex at the tip acts as a “lid” that prevents the premature secretion of effector proteins before contact with cells. We found that a Salmonella strain expressing the SipD triple mutant (D320R / D323K / S327R) exhibit enhanced type III secretion equivalent to the absence of SipD.
Fig. 8.
A non-invasive SipD triple mutant (D320R/V323K/S327R) failed to be displayed on the bacterial surface. (A) Salmonella sipD− knockout strain probed with antibodies to lipopolysaccharide (red) and Flag-tag (green) transformed with plasmid that supplied no SipD (left panel); Flag-tagged SipD (middle panel), Flag-tagged mutant SipD (right panel). (B) Protein expression and secretion of SipD and other T3SS proteins were not affected as probed by immunoblotting.
Electron microscopy examination of needle complexes obtained from a Salmonella strain expressing the SipD triple mutant showed that they were properly assembled and exhibited a normal appearance (Fig. 9). These results confirmed that the phenotypes associated with mutations in SipD were not the result of gross morphological changes in the needle complex. Taken together, these results indicate that the SipD triple mutant, which showed a marked decrease in PrgI binding affinity, was unable to assemble at the tip of the needle complex in vivo. In other words, the lack of PrgI-SipD interaction observed in the SipD triple mutant is not due to overt structural defects of the needle complex itself but rather due to the specific disruption of the PrgI binding surface.
Fig. 9.
Needle complex assembly and stability is not affected by mutations in SipD. (A) Electron micrographs of purified needle complex from a Salmonella sipD− knockout strain transformed with plasmid that supplied no SipD (left panel); Flag-tagged SipD (middle panel), Flag-tagged mutant SipD (right panel). (B) Purified needle complexes were probed with antibodies against proteins that form the needle complex (InvG, PrgH, PrgK, PrgI, and PrgJ).
Discussion
An essential step in the assembly of the T3SS needle complex is the anchoring of several copies of SipD at the tip of the needle. Recently, we showed by using PRE that the Salmonella PrgI protein binds at the lower half of the SipD coiled-coil [16]. Here, we extended our results by showing that PrgI binds more tightly to a truncated form of SipD (termed SipDt here) lacking the N-terminal α-helical hairpin (Fig. 1). This work was motivated by observations that the N-terminal α-helical hairpin functions mainly as self-chaperone to the tip protein [15] and that it is partially dispensable for bacterial invasiveness [23]. Based on structural analysis, others have proposed that the N-terminal α-helical hairpin may be splayed out to allow the anchoring of SipD at the tip [15,17]. Furthermore, using isothermal calorimetry, Lunelli et al. [18] showed that the PrgI-SipD interaction could only be detected when the N-terminal α-helical hairpin was removed from SipD. Understanding how SipDt interacts with PrgI is important because this information is needed to model how SipD is docked onto the assembled PrgI needle tip. To this end, Lunelli et al. [18] determined the crystal structure of a PrgI-SipDt fusion protein and used that structure to dock SipD on the needle tip. When the PrgI-SipDt fusion protein was docked on the current model of the PrgI needle [3], however, there was a massive steric clash between SipD molecules at the needle tip (Fig. 2). Thus the PrgI-SipDt crystal structure could not be used to dock SipD at the needle tip. We reasoned that the 5-residue linker (with the sequence GGSGG) between PrgI and SipDt was perhaps too short and artificially constrained PrgI to SipDt in an orientation that could not be used to dock SipD at the needle tip without resulting in steric clash of SipD molecules. Here we used PRE methods (Fig. 3-6) to identify specific regions in the SipDt coiled-coil as the binding sites for PrgI (Fig. 6) and propose an alternate model (Fig. 6) that did not result in the steric clash of SipD at the needle tip.
Our PRE results identified two binding sites for PrgI on the SipD coiled-coil and suggested that the SipD coiled-coil region could insert between PrgI protomers at the needle tip (Fig. 6). Our model predicts that upon assembly at the needle tip, the N-terminal α-helical hairpin of SipD will be splayed outside the needle [15,17]. This scenario is consistent with the observation that the major epitopes of the Shigella tip protein IpaD that are exposed on the bacterial surface are in residues 13-121, which correspond to the region encompassing the N-terminal α-helical hairpin [24]. It should be noted that during the assembly of the needle apparatus, the tip protein must pass through a narrow, 25 Å diameter needle channel [4,9], which at best could only fit two tightly packed α-helices. Hence, the tip protein likely passes through the channel in a partially unfolded state. Others have also suggested that the tip protein N-terminal region could swing away to allow the passage of the rest of the tip protein through the needle channel [14]. Subsequently, upon reaching the needle tip, the central coiled-coil could be exposed for interaction with the needle protomer.
We have thus shown using PRE methods to characterize the binding interaction of SipD with PrgI, based on which site-directed mutagenesis could render Salmonella non-invasive (Fig. 7). The Salmonella invasion assay by itself, however, could not directly link the non-invasiveness of a particular SipD mutation to defective SipD-PrgI interaction because of the multifunctional nature of SipD because it binds to other proteins and bile salts and defects in those interactions could render SipD nonfunctional. Additionally, there are many things that could go wrong during the assembly of the needle apparatus, which after all is assembled from over 20 different proteins in a hierarchical and multi-step manner. Thus, we used a series of experimental approaches to link the non-invasiveness of SipD mutations specifically to the disruption of the PrgI-SipD binding interface that was identifed by PRE methods.
We used fluorescence polarization to correlate the relative binding affinities of the PrgI-SipD interaction with respect to SipD loss of function mutants (Fig. 7). Determination of the binding affinities between PrgI and SipD is challenging because of the poor solution behavior of the native forms and weak binding affinities. Currently, there are only two methods that have been successfully used to measure the binding affinities of PrgI with SipD – isothermal calorimetry [18], and shown herein, fluorescence polarization. In both ITC and FP methods, the truncated version of SipD lacking the N-terminal α-helical hairpin was used because it yielded a measurable binding affinity with PrgI, whereas the intact SipD construct was beyond the detection limit of fluorescence polarization. The results of fluorescence polarization (Fig. 7) suggested that the non-invasiveness of the SipD triple mutants, which were designed to disrupt the PrgI-SipD interaction on the basis of the PRE results, were due to the decreased binding affinities to PrgI at a molecular level. Further, a cell-based assay using Flag-tagged SipD demonstrated that the non-invasive triple mutation (D320R V323K S327R) prevented SipD from localizing on the bacterial surface (Fig. 8). Electron microscopy showed that Salmonella was able to properly assemble the base and the PrgI needle itself (Fig. 9), therefore the non-invasive phenotype of the SipD mutants were due solely to the inability of SipD to bind to PrgI and anchor on the needle tip. Without SipD anchored at the needle tip, the translocon cannot be assembled rendering Salmonella incapable of injecting effectors directly into eukaryotic cells and thereby incapable of invading eukaryotic cells.
The model of SipD at the needle tip is also useful in developing a working model of the interface beween SipD and the translocon. PrgI and SipD are conserved in other T3SSs, thus our results will advance understanding of the needle-tip interaction in other bacterial pathogens.
Materials and Methods
Protein expression and purification
The expression and purification of a monomeric form of PrgI lacking the C-terminal five residues and replaced with a his-tag has been described previously [6]. SipDt (residues 101-343) was subcloned in the NdeI/SalI sites of the expression plasmid pDZ1 [13] and expressed and purified following similar protocol used for the intact SipD [13]. To eliminate complications arising from multiple cysteines during the spin labeling, all the SipDt constructs used in the PRE experiments contained the background C244S mutation. A total of 10 cysteine point mutations were introduced by PCR in the following residues of SipDt: D136, A144, D147, N155, K173, T303, L318, K324, S331 and E335 (Fig. 3). These residues are located along helix α4 and α8 of the central coiled-coil (Fig. 3). All mutagenesis were confirmed by DNA sequencing. 15N-labeled PrgI and SipDt were obtained by transforming E. coli BL21(DE) with the appropriate expression plasmid and growing cells in M9 minimal media with 1 g/L [15N] ammonium chloride as the sole nitrogen source whereas unlabeled proteins were obtained by growing E. coli in LB medium.
Fluorescence polarization
For fluorescence polarization (FP), a PrgI double mutant, V65A/V67A, which rendered full-length PrgI to be soluble and functional [7] was used. A site-directed mutant L31C was used to attach the fluorophore as follows: PrgI was dialyzed into 1× PBS pH 7.4 with a 10 fold excess of tris(2-carboxyethyl)phosphine (TCEP) to reduce disulfide bonds for 30 minutes at 25°C. A 20 fold excess of Alexa Fluor 488-C5-Maleimide (Invitrogen) was resuspended in 1× PBS pH 7.4 and added dropwise to the protein solution with mixing and allowed to react for 3 hours at 25°C in the dark. Excess dye was removed by dialysis in 1× PBS pH 7.4 followed by passage through a centrifugal filter unit with molecular weight cut off of 3 kDa (Millipore). Increasing concentrations of SipD were titrated into 25 nM PrgI-fluorophore and FP data were collected at 25°C using a Varian Cary Eclipse Fluorescence Spectrophotometer. The average of 5 data points was subtracted from a base polarization value of 25 nM PrgI-fluorophore in 1× PBS pH 7.4 alone. The difference in millipolarization (ΔmP) was plotted against increasing concentrations of SipD. The FP binding curve was fitted using the Hill Equation and Origin (OriginLab, Northampton, MA).
Bioassays
Site directed mutations in SipD within the plasmid pRK2 [13] were introduced by PCR and used to determine the effect of SipD mutations on the ability of Salmonella to invade a cultured human epithelial cell line Henle 407 (ATCC #CCL6) following protocol described previously [13]. The S. typhimurium strains (wild type and sipD− knockout) used in the Salmonella invasion assay have been described previously [13].
For electron microscopy, needle complexes were purified as described elsewhere [5,25]. Purified needle complexes were applied to glow discharged carbon coated Cu-grids and stained with 2% phosphotungstic acid (PTA), pH 7.0. Images were acquired using a Tecnai Biotwin TEM (FEI Company, Hillsboro) at 80 kV using the Morada Soft Imaging system and a 6 M pixel CCD camera (Olympus, Munster, Germany).
Immunoblotting was used to determine the level of protein expression and secretion of T3SS proteins and performed as described [12,26] using antibodies specific to the SptP effector protein and the structural proteins SipB, SipC, InvJ, PrgH, PrgK, InvG, PrgJ, and PrgI.
Immunofluorescence microscopy was used to assay the localization of SipD on the bacterial surface using mouse monoclonal anti-Flag M2 (Sigma) and a rabbit polyclonal anti-S. typhimurium lipopolysaccharide (LPS; Difco Laboratories) antibodies as described elsewhere[12]. S. typhimurium strain with a nonpolar sipD knockout, ΔsipD [22] was transformed with a plasmid pWSK129 [27] that supplied exogenous internally 3×Flag tagged sipD (p3×FsipD). The triple mutation (D320R V323K S327R) was introduced in p3xFsipD.
NMR spectroscopy
NMR data was acquired at 25°C on a Bruker Avance 800 MHz spectrometer equipped with a cryoprobe, processed with NMRPipe [28] and analyzed using NMRView [29]. The spin label MTSL (S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methane-sulfonothioate, Toronto Research Chemicals) was attached to the SipDt cysteine mutants as described previously [16]. All the ten SipDt cysteine mutants were 100% spin labeled as confirmed by electrospray ionization mass spectrometry.
For PRE data acquisition, two 2D proton-nitrogen correlation spectra were acquired for two samples, one for 15N-labeled PrgI complexed with diamagnetic SipDt, and another for 15N-labeled PrgI complexed with paramagnetic SipDt. The protein concentrations, buffer conditions, NMR data acquisition and processing parameters were kept identical for both samples. Initially, single time-point PREs were determined for the 10 SipDt cysteine mutants (D136C, A144C, D147C, N155C, K173C, T303C, L318C, K324C, S331C and E335C) in complex with 15N-labeled PrgI following the PRE method described previously [16]. In the single time-point PRE method, two identical 2D 1H-15N HSQC [30] spectra were acquired for 0.6 mM 15N-labeled PrgI complexed with 0.3 mM diamagnetic or paramagnetic SipDt. Typical acquisition parameters were 8 scans, 18 ppm 15N sweep width centered at 118 ppm, 216 15N complex points and a recycle delay of 5 sec to ensure that the magnetization returned to equilibrium before the next scan. Each 2D 1H-15N HSQC spectrum took about 2.30 hrs to complete. The single time-point PRE for each non-overlapped PrgI backbone amide peaks was calculated from the peak intensity ratio (Ipar / Idia).
For the SipDt mutants (D136C, A144C, K324C and E335C) that showed strong single time-point PREs with PrgI, two time-point PREs [21] were acquired following the method described previously [16]. As a control, two time-point PREs were also acquired for K173C, which showed weak single time-point PRE. For acquiring two time-point PREs, the optimal delay between the two time points [21] was found to be 10 ms. Typically, for a PrgI-SipDt complex at 1:0.5 molar ratio, the two-time point PRE data was acquired with 108 scans and 18 ppm 15N sweep width centered at 118 ppm. Each 2D 1H-15N HSQC spectra had 2048 1H complex points and 300 15N complex points. The total acquisition time was ~23 hrs to complete the two HSQC spectra for each cysteine mutant. For each non-overlapped peak, the PRE 1H transverse relaxation rate (Λ2) was determined as described [16] and used as restraints in structure calculations of the PrgI-SipDt complex using Xplor-NIH [31]. The protocol for PRE structure calculation has been described earlier [16]. Briefly, each paramagnetic probe was represented by three conformers with the structure of SipDt fixed. An ensemble of three conformers was used to represent PrgI and these were allowed to freely rotate and translate. Jointly refining against intermolecular PREs of A144C, D147C, K173C, K324C and E335C, 128 structures were calculated and 60 structures were selected for their lowest PRE Q-factors below 0.35 [21]. All structure figures were rendered using Pymol [32].
Research Highlights.
SipD interacts with PrgI to form the tip of the type III secretion needle complex.
PRE methods were used to determine how SipD interacts with PrgI.
PRE revealed multiple PrgI binding sites on the SipD coiled-coil.
PRE results allow modeling of SipD at the needle tip.
Acknowledgements
Supported by the Ministry of Science and Technology of China grant 2013CB910200 (C.T.) and NIH grants T32-GM008359 (A.C.M.), AI030492 (J.G.), and AI074856 (R.N.D.). C.T. is an International Early Career Scientist of Howard Hughes Medical Institute. We are grateful to Carey Johnson and Mark Richter (University of Kansas) for helpful discussion on the fluorescence spectroscopy part.
Abbreviations
- NMR
nuclear magnetic resonance
- PRE
paramagnetic relaxation enhancement
- SipDt
truncated form of SipD lacking the N-terminal α-helical hairpin
- T3SS
type III secretion system
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cornelis GR. The type III secretion injectisome. Nat. Rev. Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 2.Chatterjee S, Chaudhury S, McShan AC, Kaur K, De Guzman RN. Structure and Biophysics of Type III Secretion in Bacteria. Biochemistry. 2013;52:2508–2517. doi: 10.1021/bi400160a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loquet A, Sgourakis NG, Gupta R, Giller K, Riedel D, Goosmann C, Griesinger C, Kolbe M, Baker D, Becker S, Lange A. Atomic model of the type III secretion system needle. Nature. 2012;486:276–279. doi: 10.1038/nature11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galkin VE, Schmied WH, Schraidt O, Marlovits TC, Egelman EH. The structure of the Salmonella typhimurium type III secretion system needle shows divergence from the flagellar system. J. Mol. Biol. 2010;396:1392–1397. doi: 10.1016/j.jmb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galan JE, Unger VM. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Ouellette AN, Egan CE, Rathinavelan T, Im W, De Guzman RN. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J. Mol. Biol. 2007;371:1304–1314. doi: 10.1016/j.jmb.2007.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poyraz O, Schmidt H, Seidel K, Delissen F, Ader C, Tenenboim H, Goosmann C, Laube B, Thunemann AF, Zychlinsky A, Baldus M, Lange A, Griesinger C, Kolbe M. Protein refolding is required for assembly of the type three secretion needle. Nat. Struct. Mol. Biol. 2010;17:788–792. doi: 10.1038/nsmb.1822. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Wang Y, Picking WL, Picking WD, De Guzman RN. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J. Mol. Biol. 2006;359:322–330. doi: 10.1016/j.jmb.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 9.Deane JE, Roversi P, Cordes FS, Johnson S, Kenjale R, Daniell S, Booy F, Picking WD, Picking WL, Blocker AJ, Lea SM. Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12529–12533. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller CA, Broz P, Muller SA, Ringler P, Erne-Brand F, Sorg I, Kuhn M, Engel A, Cornelis GR. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science. 2005;310:674–676. doi: 10.1126/science.1118476. [DOI] [PubMed] [Google Scholar]
- 11.Espina M, Olive AJ, Kenjale R, Moore DS, Ausar SF, Kaminski RW, Oaks EV, Middaugh CR, Picking WD, Picking WL. IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri. Infect. Immun. 2006;74:4391–4400. doi: 10.1128/IAI.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lara-Tejero M, Galan JE. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect. Immun. 2009;77:2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee S, Zhong D, Nordhues BA, Battaile KP, Lovell SW, De Guzman RN. The Crystal Structure of the Salmonella Type III Secretion System Tip Protein SipD in Complex with Deoxycholate and Chenodeoxycholate. Protein Sci. 2011;20:75–86. doi: 10.1002/pro.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erskine PT, Knight MJ, Ruaux A, Mikolajek H, Wong Fat Sang N, Withers J, Gill R, Wood SP, Wood M, Fox GC, Cooper JB. High Resolution Structure of BipD: An Invasion Protein Associated with the Type III Secretion System of Burkholderia pseudomallei. J. Mol. Biol. 2006;363:125–136. doi: 10.1016/j.jmb.2006.07.069. [DOI] [PubMed] [Google Scholar]
- 15.Johnson S, Roversi P, Espina M, Olive A, Deane JE, Birket S, Field T, Picking WD, Blocker AJ, Galyov EE, Picking WL, Lea SM. Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J. Biol. Chem. 2007;282:4035–4044. doi: 10.1074/jbc.M607945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rathinavelan T, Tang C, De Guzman RN. Characterization of the Interaction between the Salmonella Type III Secretion System Tip Protein SipD and the Needle Protein PrgI by Paramagnetic Relaxation Enhancement. J. Biol. Chem. 2011;286:4922–4930. doi: 10.1074/jbc.M110.159434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blocker AJ, Deane JE, Veenendaal AK, Roversi P, Hodgkinson JL, Johnson S, Lea SM. What’s the point of the type III secretion system needle? Proc. Natl. Acad. Sci. U.S.A. 2008;105:6507–6513. doi: 10.1073/pnas.0708344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lunelli M, Hurwitz R, Lambers J, Kolbe M. Crystal structure of PrgI-SipD: insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands. PLoS Pathog. 2011;7:e1002163. doi: 10.1371/journal.ppat.1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujii T, Cheung M, Blanco A, Kato T, Blocker AJ, Namba K. Structure of a type III secretion needle at 7-A resolution provides insights into its assembly and signaling mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4461–4466. doi: 10.1073/pnas.1116126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demers JP, Sgourakis NG, Gupta R, Loquet A, Giller K, Riedel D, Laube B, Kolbe M, Baker D, Becker S, Lange A. The common structural architecture of Shigella flexneri and Salmonella typhimurium type three secretion needles. PLoS Pathog. 2013;9:e1003245. doi: 10.1371/journal.ppat.1003245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwahara J, Tang C, Marius Clore G. Practical aspects of 1H transverse paramagnetic relaxation enhancement measurements on macromolecules. J. Magn. Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaniga K, Trollinger D, Galan JE. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Picking WL, Nishioka H, Hearn PD, Baxter MA, Harrington AT, Blocker A, Picking WD. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect. Immun. 2005;73:1432–1440. doi: 10.1128/IAI.73.3.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turbyfill KR, Mertz JA, Mallett CP, Oaks EV. Identification of epitope and surface-exposed domains of Shigella flexneri invasion plasmid antigen D (IpaD) Infect. Immun. 1998;66:1999–2006. doi: 10.1128/iai.66.5.1999-2006.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 26.Zhong D, Lefebre M, Kaur K, McDowell MA, Gdowski C, Jo S, Wang Y, Benedict SH, Lea SM, Galan JE, De Guzman RN. The Salmonella Type III Secretion System Inner Rod Protein PrgJ Is Partially Folded. J. Biol. Chem. 2012;287:25303–25311. doi: 10.1074/jbc.M112.381574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 28.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 29.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 30.Grzesiek S, Bax A. The importance of not saturating H2O in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 1993;115:12593–12594. [Google Scholar]
- 31.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor–NIH for NMR molecular structure determination. Prog. NMR Spectroscopy. 2006;48:47–62. [Google Scholar]
- 32.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, California, U.S.A.: 2002. [Google Scholar]