Abstract
Many members of the DEXD/H-box helicase family play important roles in the innate immune system against viral infection. We, therefore, isolated double-stranded RNA (dsRNA) complex in myeloid dendritic cells (mDCs). We found that DHX15, a DEXDc helicase family member, is one of the components of this complex. Knockdown of DHX15 expression by short hairpin RNA efficiently reduced the ability of mDCs to produce interferon beta (IFN-β), IL-6 and TNF-α in response to dsRNA and RNA virus. DHX15 specifically bound poly I:C via its HELICc domain. DHX15 interacted with MAVS and formed a complex following stimulation with poly I:C. The N-terminal domain containing a DEXDc motif in DHX15 bound the C-terminus of MAVS. DHX15 is required to activate IRF3 phosphorylation, as well as NF-κB and MAPK signaling during RNA virus infection. We, therefore, identified DHX15 as a new RNA virus sensor mediated by MAVS to activate the immune responses to RNA.
Keywords: Dendritic cells, Cytokine Receptors, Signal transduction, Innate immunity, RNA virus
Introduction
The innate immune system is the first line of defense against pathogenic microbial infection. During virus infection, viral nucleic acid can be recognized by immune cells, including dendritic cells, macrophages and fibroblasts, to rapidly initiate the inflammation. There are at least five types of pattern recognition receptors (PRRs) in immune cells that sense different forms of viral nucleic acids and induce IFN and proinflammatory cytokines responses. 1) Endosomal Toll-like receptors. TLR3 senses double-stranded RNA (1); TLR7 and TLR8 sense single-stranded RNA (ssRNA) (2); TLR9 senses CpG DNA (3, 4). 2) Retinoid acid-inducible gene I-like helicases (RLH) receptors. RLH receptors sense RNA in the cytosol. Melanoma differentiation factor 5 (MDA5) senses dsRNA (5, 6); Retinoic acid-inducible gene (RIG-I) senses ssRNA that has a 5′ triphosphate (6–8). 3) NOD-like receptors (NLRs). Several NLR family members form inflammasome complexes in response to various danger signals. However, nucleotide-binding oligomerization domain 2 (Nod2) can also sense viral ssRNA to activate IRF3 and induce IFN-β (9). 4) Pyrin and HIN domain-containing (PYHIN) family members. Absent in melanoma 2 (AIM2) and interferon gamma-inducible protein 16 (IFI16) are very important members in the PYHIN family that play roles in the innate immune system. Although AIM2 assembles a complex with dsDNA and the adaptor molecule of apoptosis-associated speck-like protein containing a CARD (ASC) to activate inflammasome (10–16), IFI16 senses dsDNA to induce a type I IFN response mediated by STING (17). 5) Other cytosolic nucleic acid sensors including Pol III, Ku70, LRRFIP1 and DEXD/H-box family helicases. Pol III can function as a sensor of B-form DNA in a RIG-I-dependent manner (18, 19). Ku70 can recognize viral nucleic acids to induce type III IFN (20, 21). LRRFIP1 senses both dsRNA and dsDNA to induce IFN-β (22). Most interestingly, besides RLH members, other DEXD/H-box family helicases, including DDX1, DDX3, DHX9, DDX21, DHX33, DHX36, DDX41 and DDX60, are reported to sense viral RNA or viral DNA (23–32).
Helicases are enzymes that catalyze the unwinding of double-stranded nucleic acids. Many studies have shown that helicases also play an essential and broad role in recognizing viral RNA and viral DNA in immune cells. RLH members, as well as DDX3, DHX9 and DHX33, use MAVS as the adaptor to sense viral RNA (24, 27, 29, 33). DDX1, DDX21 and DHX36 form a complex with TRIF to sense dsRNA (23). Mediated by MyD88, DHX9 and DHX36 sense CpG DNA (26). DHX60 can function as a sensor of dsRNA to promote RLH receptor-dependent signaling (32). However, it is unclear whether other helicases are involved in sensing viral RNA or viral DNA.
By isolating dsRNA/protein complexes from splenic murine dendritic cells, we found that DHX15, a member of DEXD/H-box family helicase, forms a complex with poly I:C, a synthetic analog of double-stranded RNA. The yeast homolog of DHX15 may recruit Prp43 to remove the lariat-intron in late-stage RNA splicing activity and play a critical role in modulating pre-mRNA splicing (34). However, the function of DHX15 in the innate immune system is unknown. In this study, we show that DHX15 acts as an important RNA sensor in myeloid dendritic cells. DHX15 could specifically bind poly I:C but not poly dG:dC. DHX15 induces the production of type I IFN and proinflammatory cytokines in response to dsRNA and RNA virus in a MAVS-dependent manner. DHX15 is also required to activate IRF3 phosphorylation, as well as NF-κB and MAPK signaling during RNA reovirus infection.
Materials and Methods
Reagents
Sendai virus was purchased from The American Type Culture Collection (ATCC, Manassas, VA). Reovirus was purchased from Advanced Biotechnologies, Inc. (Columbia, MD). Poly I:C, poly dG:dC and biotin-labeled poly I:C were purchased from InvivoGen (San Diego, CA). Lipofectamine™ 2000 was purchased from Invitrogen (Carlsbad, CA). DHX15 antibody was purchased from Abcam (Cambridge, MA) and used for immunoprecipitation and immunoblotting. The following antibodies were used for immunoblotting: anti-IRF3 (Santa Cruz, Dallas, TX); anti-DDX21 (Novus Biologicals, Littleton, CO); anti-MAVS, anti-STING, anti-Erk1/2, anti-p38, anti-p65, anti-phospho-Erk1/2, anti-phospho-p38, anti-phospho-p65 and anti-phospho-IRF3 (Cell Signaling, Danvers, MA); anti-DHX41, anti-GAPDH-HRP, anti-Flag-HRP, anti-HA-HRP and anti-Myc-HRP (Sigma, St. Louis, MO). Anti-HA and anti-Myc beads were purchased from Sigma. Protein A/G beads and NeutAvidin beads were purchased from Thermo Scientific (Rockford, IL).
D2SC mDC cell culture
DC cell line D2SC, a line derived from murine splenocytes, was maintained in IMDM containing 5% heat-inactivated fetal calf serum (Sigma) and 1% penicillin-streptomycin (Invitrogen). D2SC cells were infected with the lentiviral vector carrying a DHX15-, MAVS- or STING-targeting shRNA or a scrambled shRNA. After 24 hours of growth, cells were selected by adding puromycin (2 ng/ml) in the culture medium. Knockdown efficiency and specificity was confirmed by immunoblot. shRNA-treated cells were stimulated with one of the following nucleic acids plus Lipofectamine™ 2000: poly I:C (5.0 μg/ml), poly dG:dC (2.5 μg/ml), RNA virus of Sendai virus or reovirus, and DNA virus of HSV-1. Each virus was at a multiplicity of infection of 10.
MEF Cell Culture
The MEF cells were maintained in Dulbecco’s Modified Eagle Medium containing 10% fetal calf serum and 1% penicillin-streptomycin (Invitrogen-GIBCO). MEF cells were infected with the lentiviral vector carrying a DHX15-targeting shRNA sequence or a scrambled shRNA. After 2 days of growth, cells were selected by adding puromycin (1 ng/ml) in the medium. The knockdown efficiency of DHX15 was confirmed by immunoblot. shRNA-treated cells were stimulated with poly I:C (5 μg/ml) plus Lipofectamine™ 2000.
Preparation of GM-CSF-derived DC
Single-cell suspensions of bone marrow cells were cultured in RPMI 1640 medium containing 10% heat-inactivated fetal calf serum and 1% penicillin-streptomycin supplemented with murine GM-CSF at a final concentration of 25 ng/ml (R&D Systems, Minneapolis, MN). Fresh GM-CSF was provided on day 4 of culture. The day-6 cells were harvested to knock down DHX15, MAVS and STING by shRNA. The knockdown efficiency was detected with western blotting 36 hours post shRNA treatment.
RNA interference
shRNA lentiviral vectors were purchased from Thermo Scientific. Targets for DHX15: clone TRCN0000113044 (DHX15-a) and clone TRCN0000113043 (DHX15-b). Target for MAVS: clone TRCN0000124770. The shRNA lentiviral transduction particles targeting STING (clone TRCN0000346266) were purchase from Sigma. siRNAs were purchased from GE Dharmacon-Thermo Scientific as following: ONTARGETplus SMARTpool, catalogue number E-011250-00-0005 for DHX15, catalogue number D-001810-10-20 for Non-targeting Pool control. siRNAs were transfected into HEK293T cells with Lipofectamine™ 2000.
Plasmid construction
The cDNA of murine DHX15 and MAVS were purchased from OriGene (Rockville, MD). The full coding region or truncated coding region of DHX15 and MAVS were amplified by PCR, cloned into pCMV-HA or pCMV-Myc vectors (Clontech, Mountain View, CA) and then transformed into Escherichia coli-competent cells (One Shot® TOP10, Invitrogen). The resultant plasmids were transfected into HEK293T cells, and the expressed recombinant proteins were purified with Anti-HA or anti-Myc beads.
Identification of DHX15-binding protein complex from D2SC Cells
One hundred million D2SC cells were lysed in NP40 lysis buffer (50 mM Tris-Cl [pH7.5], 1 mM EDTA, 150 mM NaCl, 1.0% NP40, and 10% Glycerol), and subjected to ultracentrifugation. Cleared lysate was incubated with 20 μg DHX15 antibody overnight, and then incubated with protein A/G beads for one more hour. Beads were washed extensively with lysis buffer. Binding proteins were separated on a 4%–20% gradient polyacrylamide gel, and stained with EZBlue (Sigma). All protein bands were analyzed by liquid chromatography–mass spectrometry.
Immunoprecipitation and immunoblotting
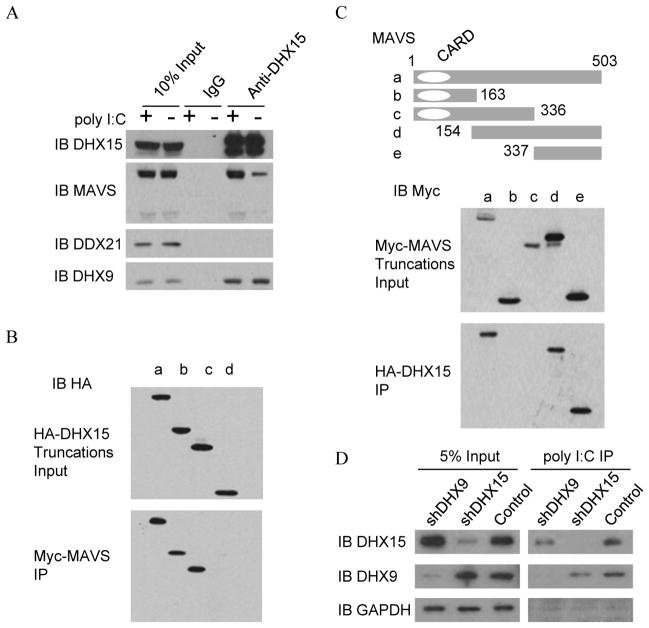
For poly I:C and poly dG:dC in vivo binding assays, D2SC cells or shRNA treated D2SC cells were unstimulated (−) or stimulated (+) with biotin-labeled poly I:C or poly dG:dC for 2 hours. Whole-cell lysates were prepared from these cells, and then incubated with NeutAvidin beads. Bound complexes were pelleted by centrifugation and analyzed by immunoblot with anti-DHX15, anti-DDX41 and anti-DDX21. Lysates from HEK293T cells transfected with HA-tagged DHX15 plasmids were incubated with anti-HA beads. Proteins were eluted from beads after washing 6 times with PBS. To ascertain DHX15 binding specificity, Flag tagged DHX15 protein (OriGene) was incubated for 2 hr with biotin-labeled poly I:C in the absence or presence of increasing amounts (0.5, 5, 50 μg/ml) of un-labeled poly I:C or poly dG:dC. Following incubation with NeutAvidin beads, bound complexes were pelleted by centrifugation and analyzed by immunoblot with anti-Flag-HRP antibody. For DHX15:MAVS interaction analysis, D2SC cells were unstimulated (−) or stimulated (+) with poly I:C for 2 hours; whole-cell lysates were prepared from the cells and then immunoprecipitated with anti-DHX15 antibody. Bound proteins were analyzed by immunoblotting with anti-DHX15, anti-MAVS, anti-DDX21 or anti-DHX9 antibodies. To map region(s) in DHX15 required for interaction with MAVS, Myc-MAVS was incubated for 1 hour with HA-DHX15 or DHX15 truncations proteins, individually, followed by incubation for 1 more hour after adding anti-Myc beads. Bound proteins were pelleted by centrifugation and analyzed by immunoblotting with anti-HA-HRP antibody. To map region(s) in MAVS required for interaction with DHX15, HA-DHX15 protein was incubated with purified Myc-MAVS or MAVS truncation proteins for 1 hour; anti-HA beads were added and incubated for 1 more hour. Bound complexes were pelleted by centrifugation and analyzed by immunoblotting with anti-Myc-HRP antibodies.
ELISA
The concentrations of IL-6 and TNF-α in culture supernatants were measured using the kits from R&D Systems, Inc. The IFN-β in culture supernatants was analyzed using a commercially available IFN-β ELISA kit (PBL Interferon Source, Piscataway, NJ), according to the manufacturer’s instructions.
Luciferase reporter gene assay
D2SC cells were seeded on 48-well plates (0.5 × 106 cells/well) and then transfected with 100 ng NF-κB luciferase and 2 ng Renilla luciferase reporter vectors plus 50, 100, or 200 ng DHX15 or DHX15 mutant expression vector. Empty control vector was added so that a total of 500 ng vector DNA was transfected into each well of cells. At 24 h posttransfection, cells were stimulated with 5.0 μg/ml poly I:C delivered by Lipofectamine™ 2000. Cells were harvested after 6 h of stimulation. The luciferase activity in the total cell lysate was detected with the Dual-Luciferase Reporter Assay (Promega, Madison, WI).
MAVS aggregation
DHX15-targeting shRNA or a scrambled shRNA treated D2SC were prepared. These shRNA-treated cells were stimulated with poly I:C (5.0 μg/ml) plus Lipofectamine™ 2000 for 6 hr or without stimulation. Crude mitochondrial extracts were then prepared and analyzed by semidenaturing detergent agarose gel electrophoresis (SDD-AGE) or SDS-PAGE using a MAVS antibody.
Signal transduction
D2SC cells were lysed in RIPA buffer after a 0, 60, 90 or 120 minute infection with reovirus at a multiplicity of infection of 10. Lysates were resolved by 4–20% SDS-PAGE and blotted with antibodies recognizing unphosphorylated or phosphorylated indicated proteins.
Results
DHX15 senses poly I:C, but not poly dG:dC in D2SC mDCs
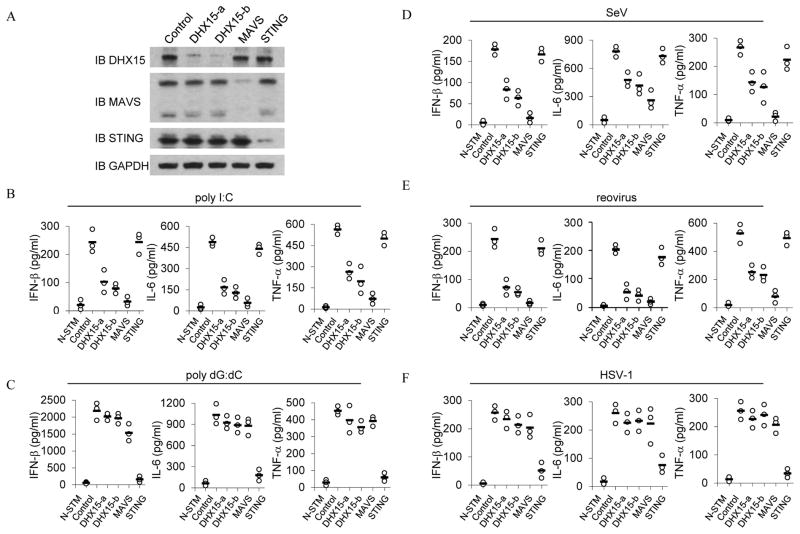
Poly I:C is a synthetic analog of double stranded RNA, a molecular pattern associated with RNA viral infection. Poly I:C with the length of 1.5–8 kb has been frequently used to induce type I IFN responses. We therefore used this poly I:C complex to identify proteins in splenic murine dendritic cells that bound to it by immunoprecipitation (IP) and liquid chromatography–mass spectrometry. D2SC mDCs were initially incubated with culture medium or biotin-labeled poly I:C for 4 hours. Whole cell lysates from the treated D2SC cells were prepared and subjected to purification with avidin-conjugated beads. The proteins bound to biotin-labeled poly I:C were separated by gradient polyacrylamide gel electrophoresis and analyzed by LC-MS. We found that a novel member of the DEXD/H-box helicase family DHX15 was in this complex. Because many members of this family, including RIG-I, LGP2, MDA5, DDX1, DDX3, DDX21, DHX9, DDX21 DHX36 and DHX60 have been shown to play important roles in sensing dsRNA and viral infection, we decided to investigate the function of DHX15 in sensing dsRNA and RNA virus. We established stable D2SC cell lines that expressed small heteroduplex RNA (shRNA) to knockdown expression of DHX15, MAVS or STING. Two distinct DHX15-targeting shRNA constructs (DHX15-a and DHX15-b) were selected to knock down expression of DHX15. A scrambled shRNA targeting sequence served as the control. Knockdown efficiency of each cell line was assessed by immunoblot (Figure 1A). IFN-β, TNF-α and IL-6 production was measured after control and knockdown shRNA in D2SC cells. The cells were then stimulated with poly I:C. As shown in Figure 1B, control D2SC mDCs produced high levels of IFN-β, IL-6 and TNF-α following this stimulation. Production of these cytokines was strongly attenuated in DHX15- and MAVS-knockdown D2SC mDCs in response to poly I:C. The production of these cytokines was not affected or was only slightly affected in STING-knockdown D2SC mDCs, which confirmed a previous report showing that STING plays a critical role in DNA sensing but no role in poly I:C sensing (35).
Figure 1.
DHX15 senses poly I:C and RNA viruses, but not DNA or DNA virus in mDCs. (A) Immunoblot (IB) showing the knockdown efficiency of shRNAs targeting the indicated genes in D2SC cells, a mouse mDC cell line. Non-targeting shRNA served as a control (first left lane). GAPDH blots are shown as loading controls (lower panel). ELISA of IFN-β, IL-6 and TNF-α production from D2SC cells with the indicated shRNA after a 20 hour stimulation with (B) 5.0 μg/ml of poly I:C, (C) 2.5 μg/ml of poly dG:dC, (D) Sendai virus, (E) reovirus, or (F) HSV-1. Poly I:C and poly dG:dC were delivered to the cells by Lipofectamine™ 2000. Viruses were added to the cells at a multiplicity of infection (MOI) = 10. N-STM, scrambled shRNA treated D2SC cells without stimulation.
We next determined whether DHX15 senses DNA in D2SC cells. The cytokine production was measured after culturing control and knockdown shRNA in D2SC cells. The cells were then stimulated with poly dG:dC. As shown in Figure 1C, DHX15-knockdown as well as MAVS-knockdown in D2SC mDCs had little effect on IFN-β, IL-6 and TNF-α production in response to poly dG:dC. This observation confirmed a previous report showing that the RNA sensing adaptor molecule of MAVS is not required for cytokine production in response to cytosolic DNA (36). However, STING-knockdown led to over 90% reduction in cytokine production in response to poly dG:dC.
DHX15 senses RNA viruses, but not DNA virus in D2SC mDCs
To determine the function of DHX15 in sensing viral infection, cytokine production was measured after culturing control and knockdown shRNA in D2SC cells with two RNA viruses (Sendai virus or reovirus). DHX15-knockdown D2SC had 50% to 70% reduction in IFN-β, IL-6 and TNF-α production in response to Sendai virus or reovirus (Figure 1D and 1E). While MAVS-knockdown D2SC mDCs had over 90% reduction in IFN-β levels, STING-knockdown D2SC cells had little effect on cytokine production in response to these RNA viruses. To further determine whether DHX15 senses DNA virus, cytokine levels were measured after culturing control and knockdown D2SC cells with HSV-1 virus. As shown in Figure 1F, DHX15- and MAVS-knockdown in D2SC mDCs had little effect on IFN-β, IL-6 and TNF-α production by D2SC cells in response to HSV-1 infection. In contrast, STING-knockdown D2SC mDCs had 80% reduction in cytokine production. These data indicate that DHX15 plays an important role in sensing RNA virus infection, but not DNA virus infection.
DHX15 plays an important role in bone marrow-derived DCs in responses to poly I:C
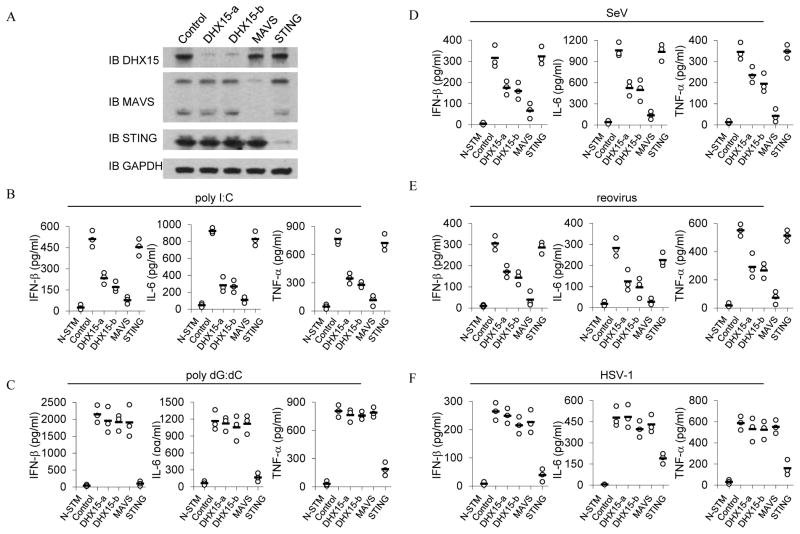
To determine whether DHX15 senses poly I:C in primary cells, GM-CSF-derived DCs were prepared and treated with shRNA to knockdown expression of DHX15, MAVS or STING. Figure 2A shows the specificity and efficiency of shRNA knockdown at the protein level without affecting the expression of non-targeted proteins in bone marrow-derived DCs (BMDCs). Control and knockdown BMDCs were examined for their IFN-β, IL-6 and TNF-α responses following stimulation with poly I:C or poly dG:dC. As shown in Figure 2B, DHX15- and MAVS-knockdown BMDCs had much lower levels of IFN-β, IL-6 and TNF-α in response to poly I:C when compared with levels produced by the control cells. In contrast, STING-knockdown BMDCs produced normal levels of cytokines in response to poly I:C. STING-knockdown in BMDCs abolishes their cytokine response to poly dG:dC; while DHX15- and MAVS-knockdown in BMDCs had little effect on IFN-β, IL-6 and TNF-α production in response to poly dG:dC DNA. These data indicate that DHX15 and MAVS play critical roles in sensing poly I:C but no role in sensing poly dG:dC in BM-derived DCs.
Figure 2.
DHX15 plays an important role in sensing RNA and RNA viruses in bone marrow-derived DCs. (A) Immunoblot (IB) showing the knockdown efficiency of shRNA targeting the indicated genes in BMDCs. Non-targeting shRNA served as a control (first left lane). GAPDH blots are shown as loading controls (lower panel). ELISA of IFN-β, IL-6 and TNF-α production from BMDCs with the indicated shRNA after a 20 hour stimulation with (B) 5.0 μg/ml of poly I:C, (C) 2.5 μg/ml of poly dG:dC, (D) Sendai virus, (E) reovirus, or (F) HSV-1. Poly I:C and poly dG:dC were delivered to the cells by Lipofectamine™ 2000. Viruses were added to the cells at a multiplicity of infection (MOI) = 10. N-STM, scrambled shRNA treated BMDCs without stimulation.
DHX15 plays an important role in bone marrow-derived DCs in responses to RNA viruses
To determine whether DHX15 senses RNA virus in primary cells, DHX15- MAVS- or STING-knockdown BMDCs were infected with Sendai virus, reovirus or HSV-1. DHX15-knockdown BMDCs had about 50% reduction in IFN-β, IL-6 and TNF-α production in response to Sendai virus or reovirus (Figure 2D and 2E). MAVS-knockdown in BMDCs abolishes their cytokine response to RNA viruses. STING-knockdown BMDCs have normal cytokine production in response to RNA viruses and strongly attenuated cytokine production in response to DNA virus. On the contrary, knockdown of DHX15 or MAVS in BMDCs had little effect on IFN-β, IL-6 and TNF-α production in response to HSV-1 infection. These data indicate that DHX15 plays an important role in sensing RNA virus infection but not DNA virus infection in BMDCs.
DHX15 is independent of MDA5 to sense poly I:C with the length of 1.5–8 kb
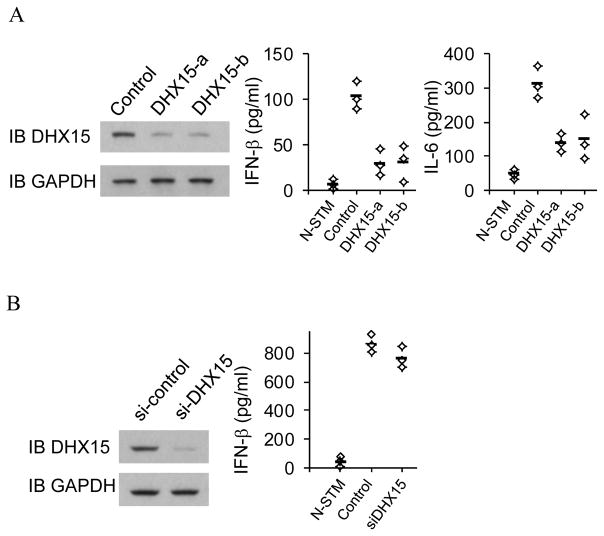
To determine whether DHX15 senses poly I:C independent of RLH receptors, MEF cells were prepared from MDA5 deficient mice. Expression of DHX15 was knocked down by shRNA (left panel, Figure 3A). Residual responses to poly I:C with the length of 1.5–8 kb in MDA5 deficient MEF are detectable, which agrees with a previous report (6). These residual responses were decreased substantially after knockdown of DHX15, suggesting that DHX15 senses long poly I:C independent of MDA5 (Right panel, Figure 3A). In addition, RIG-I plays no role in sensing poly I:C with the length of 1.5–8 kb (23). These data indicate that DHX15 is independent of RLH receptors to sense long poly I:C.
Figure 3.
DHX15 is independent of MDA5 to sense poly I:C. (A) Left panel, Immunoblot (IB) showing the knockdown efficiency of shRNA targeting DHX15 in MDA5 deficient MEF. Non-targeting shRNA (Control) served as a control. GAPDH blots are shown as loading controls. Right panel, ELISA of IFN-β and IL-6 production from MDA5-deficient MEF with the indicated shRNA after a 20 hour stimulation with 5.0 μg/ml of poly I:C delivered to the cells by Lipofectamine™ 2000. N-STM, scrambled shRNA treated MDA5-deficient MEF without stimulation. (B) Left panel, Immunoblot (IB) showing the knockdown efficiency of siRNA targeting DHX15 in HEK293T cells. Non-targeting siRNA (si-control) served as a control. GAPDH blots are shown as loading controls. Right panel, ELISA of IFN-β production from HEK293T cells with the indicated siRNA after a 20 hour stimulation with 5.0 μg/ml of poly I:C delivered to the cells by Lipofectamine™ 2000. N-STM, scrambled siRNA treated HEK293T cells without stimulation.
To further determine whether DHX15 senses poly I:C in other cell type, Expression of DHX15 in HEK293T cells was knocked down by siRNA (left panel, Figure 3B). Control and knockdown HEK293T cells were examined for their IFN-β response following stimulation with poly I:C. As shown in right panel, Figure 3B, DHX15-knockdown HEK293T cells have normal IFN-β production in response to poly I:C, indicating DHX15 plays no role in sensing poly I:C in HEK293T cells.
DHX15 binds poly I:C directly
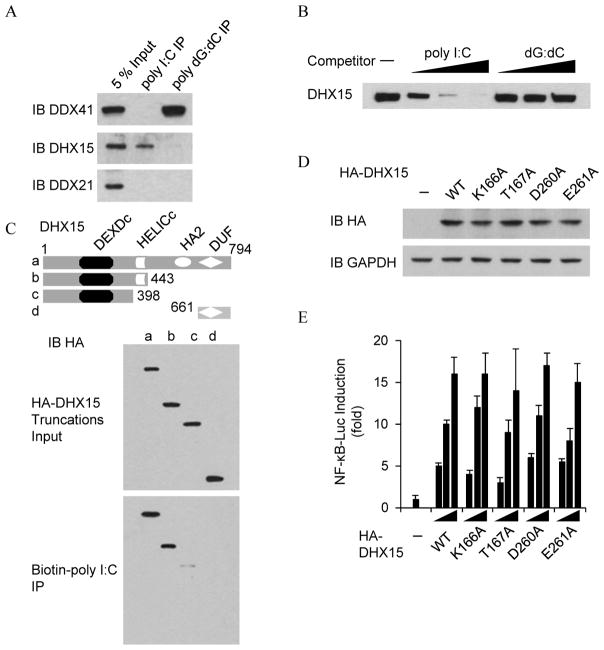
To determine whether DHX15 specifically binds dsRNA, we firstly conducted pulldown assays in which biotin-labeled poly I:C or biotin-labeled poly dG:dC was transfected into D2SC cells. Whole cell lysates from the treated D2SC cells were prepared and subjected to purification with avidin-conjugated beads. The proteins bound to biotin-labeled poly I:C or biotin-labeled poly dG:dC were analyzed by immunoblotting with anti-DHX41, anti-DHX15 or anti-DDX21 antibody. As shown in Figure 4A, poly I:C can pull down DHX15, but not DDX41. In contrast, poly dG:dC can pull down DDX41, but not DHX15. DDX21, the bridge of DDX1-DDX21-DHX36 RNA sensing complex (23), cannot be pulled down either poly I:C or poly dG:dC. We then conducted immunoprecipitation and immunoblotting assays in which a Flag-tagged DHX15 recombinant protein was incubated with biotin-labeled poly I:C in the absence or presence of increasing amounts (0.5, 5, 50 μg/ml) of un-labeled poly I:C or poly dG:dC. Following incubation with NeutAvidin beads, bound complexes were pelleted by centrifugation and analyzed by immunoblot with anti-Flag-HRP antibody. Only unlabeled poly I:C could block the binding of biotin-labeled poly I:C to DHX15 (Figure 4B). We lastly conducted mapping assays in which we repeated pulldown assays using biotin-labeled poly I:C and truncated versions of DHX15. As shown in Figure 4C, the HELICc domain of DHX15 was required for poly I:C binding. These data indicate that DHX15 directly and specifically binds poly I:C.
Figure 4.
DHX15 binds poly I:C via HELICc domain. (A) Immunoblot (IB) of indicated proteins precipitated with NeutAvidin beads from whole-cell lysates of biotin-labeled poly I:C or biotin-labeled poly dG:dC-stimulated D2SC cells. Whole-cell lysate of unstimulated D2SC cells (Input) served as a control. (B) Immunoblot using anti-Flag antibody of pulldown competition assays in which 0.5, 5, or 50 μg/ml of poly I:C or poly dG:dC was added to a mixture of Flag-DHX15 protein plus biotinylated poly I:C, followed by addition of NA-beads. (C) Immunoblot using anti-HA antibody of pulldown assays in which HA-tagged serial truncations of DHX15 were individually incubated with biotinylated poly I:C, followed by addition of NA-beads. Top, schematic representations of full-length and serial truncations of DHX15. a, DHX15 full size; b, DHX15 deleted C-terminal HA2 and DUF domains; c, N-terminus of DHX15 with DEXDc domain; d, C-terminus of DHX15 with DUF domain. DEXDc: DEAD-like helicases superfamily domain; HELICc: helicase C-terminal domain. HA2: Helicase associated domain; DUF: domain of unknown function. Numbers denote amino acid residues. (D) Anti-HA antibody blotting (IB HA) showing the expression of HA fusions in D2SC cells transfected with empty vector (−) or expression vectors for wild type DHX15 or its mutants individually. GAPDH blots are shown as loading controls. (E) Activation of the NF-κB promoter in D2SC cells transfected with an NF-κB luciferase reporter (NF-κB-Luc; 100 ng) plus increasing concentrations (50, 100 or 200 ng) of expression vectors for wild type DHX15 or its mutants individually. At 24 h posttransfection, cells were stimulated for 6 h with 5.0 μg/ml poly I:C delivered by Lipofectamine™ 2000 and then harvested. Results are presented relative to those of cells transfected with empty vector alone (−). Data are representative of three independent experiments (mean and s.d. in e).
To determine whether the ATPase activity of DHX15 is required to sense poly I:C, the Walk A motif and the Walk B motif in DHX15 were mutated by replacing lysine 166, threonine 167, aspartic acid 260 or glutamic acid 261 with alanine individually. These mutants were ATPase deficient (37). Wild type and mutants of DHX15 were overexpressed in D2SC mDCs individually (Figure 4D). The transactivation assays showed that overexpression of wild type DHX15 or a DHX15 mutant led to an equal increased NF-κB promoter activation following stimulation with poly I:C (Figure 4E), indicating that DHX15 is independent of ATPase activity to sense poly I:C.
DHX9 and MAVS are in DHX15-binding protein complex
To identify DHX15 binding proteins, we performed immunoprecipitation with antibody to DHX15 in the mouse mDC line D2SC, followed by protein sequencing by liquid chromatography–mass spectrometry. We obtained approximately 25 unique sequences with five or more ‘Peptide hits’ (Table 1). We identified helicase DHX9 is also in the group of DHX15-interacting proteins.
Table 1.
The identification of DHX15-binding protein complex from D2SC Cells. Sequencing of anti-DHX15 immunoprecipitation complex by liquid chromatography–mass spectrometry. Peptides hits: the number of peptides ions matched that associated protein.
| Gene ID | Symbol | Peptide hits | Gene description |
|---|---|---|---|
| 13204 | DHX15 | 198 | DEAH (Asp-Glu-Ala-His) box polypeptide 15 |
| 54723 | TFIP11 | 44 | tuftelin interacting protein 11 |
| 67429 | NUDCD1 | 30 | NudC domain containing 1 |
| 217207 | DHX8 | 20 | DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| 19704 | UPF1 | 13 | UPF1 regulator of nonsense transcripts homolog (yeast) |
| 13211 | DHX9 | 11 | DEAH (Asp-Glu-Ala-His) box polypeptide 9 |
| 27967 | CHERP | 10 | calcium homeostasis endoplasmic reticulum protein |
| 15568 | ELAVL1 | 10 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 |
| 13665 | EIF2S1 | 9 | eukaryotic translation initiation factor 2, subunit 1 alpha |
| 105445 | DOCK9 | 8 | dedicator of cytokinesis 9 |
| 329755284 | MAVS | 8 | mitochondrial antiviral-signaling protein isoform 2 |
| 216238 | EEA1 | 7 | early endosome antigen 1 |
| 26900 | DDX3Y | 7 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked |
MAVS is recruited to DHX15 complex upon poly I:C stimulation
Because MAVS is the critical adaptor molecule in the RNA-sensing pathway and because our data have also shown that dendritic cells with knockdown of DHX15 or MAVS displayed similar cytokine responses to poly I:C and RNA virus, we next determined whether DHX15 senses poly I:C and RNA virus in a MAVS-dependent manner. We performed anti-DHX15 immunoprecipitation experiments in the whole cell lysate from the unstimulated (−) or stimulated (+) D2SC cells with poly I:C. As shown in Figure 5A, endogenous DHX15 associates with MAVS and DHX9 in both unstimulated and stimulated D2SC cells. Notably, much more MAVS was precipitated by anti-DHX15 in mDCs after stimulation with poly I:C. In contrast, anti-DHX15 did not precipitate DDX21. To map the domains in DHX15 and MAVS that mediate their interaction, we incubated a recombinant Myc-MAVS protein with either a full-length or truncated HA-DHX15 protein, followed by pulldown with anti-Myc beads. As indicated in Figure 5B, the N terminal domain, which contains the DEXDc motif, of DHX15 was required for DHX15 to interact with MAVS. Likewise, we conducted reciprocal experiments using truncated versions of MAVS and full-length DHX15 to identify the MAVS binding site to DHX15. We found that it is the C terminal domain instead of the CARD domain of MAVS that is required for MAVS interaction with DHX15 (Figure 5C).
Figure 5.
N-terminal domain including the DEXDc motif of DHX15 and C-terminal region of MAVS are required for DHX15:MAVS interaction. (A) Immunoblot (IB) of indicated proteins precipitated with anti-DHX15 antibody from whole-cell lysates of unstimulated (−) or poly I:C stimulated (+) D2SC cells. IgG antibody served as a control; Input, D2SC lysates used in immunoprecipitation assays; (B) Immunoblot (IB) using anti-HA antibody of pulldown assays in which purified Myc-MAVS was incubated with full-length or truncated HA-DHX15 protein, followed by addition of anti-Myc beads; (C) Immunoblot (IB) using anti-Myc antibody of pulldown assays in which purified HA-DHX15 was incubated with full-length or truncated Myc-MAVS protein, followed by addition of anti-HA beads. Top, schematic representations of full-length and serial truncations of MAVS. CARD: N-terminal caspase recruitment domain. Numbers denote amino acid residues. (D) Immunoblot (IB) of indicated proteins precipitated with NeutAvidin beads from whole-cell lysates of biotin-labeled poly I:C-stimulated DHX15- or DHX9-knockdown D2SC cells. Scrambled shRNA treated D2SC cells served as a control.
Knockdown of DHX9 reduces DHX15 binding RNA
Since DHX9 is in DHX15-binding protein complex, and interacts with DHX15 at endogenous protein level, we next determine whether DHX9 affects DHX15 binding RNA. As shown in Figure 5D, DHX9-knockdown or DHX15-knockdown reduces DHX9-DHX15 complex binding poly I:C.
DHX15 activates IRF3, NF-κB and MAPKs that are downstream of MAVS signaling upon reovirus infection
Since knockdown of DHX15 in mDCs resulted in the reduction of cytokine production in response to poly I:C and RNA viruses, we next determine whether DHX15 is required to activate MAVS signaling. Scrambled shRNA treated (control) and DHX15-knockdown D2SC cells were stimulated for 6 hr with 5.0 μg/ml of poly I:C delivered by Lipofectamine™ 2000 or without stimulation. Crude mitochondrial extracts were prepared and analyzed by SDD-AGE and SDS-PAGE followed by western blotting (Figure 6A). In control D2SC cells, the aggregation of MAVS was detected after poly I:C stimulation. The aggregation of MAVS was barely detected in DHX15-knockdown D2SC cells, indicating DHX15 is required for MAVS aggregation in response to poly I:C. We next investigated whether DHX15 is required to activate IRF3, NF-κB and MAPKs that are downstream of MAVS signaling that is induced by reovirus. Scrambled shRNA (control) and DHX15- and MAVS-knockdown D2SC cells were infected with reovirus, and total cell extracts were prepared and analyzed by SDS-PAGE followed by western blotting (Figure 6B). In control D2SC cells, the phosphorylation of Erk1/2, p38, p65 and IRF3 was detected at 90 to 120 minutes after viral infection. The phosphorylation of IRF3, NF-κB and MAPKs was barely detectable in DHX15- or MAVS-knockdown D2SC cells at these time points. These data suggest that DHX15 is important for activating IRF3, NF-κB and MAPKs signaling upon reovirus infection.
Figure 6.
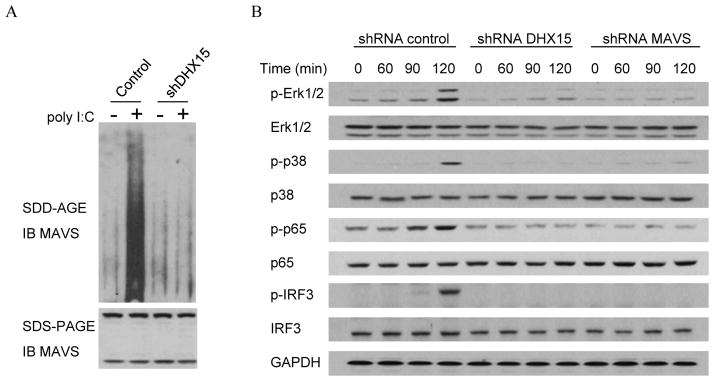
(A) DHX15 is required to induce MAVS aggregation. Immunoblot (IB) of MAVS by SDD-AGE and SDS-PAGE in crude mitochondrial extracts from DHX15-knockdown D2SC after 6 hr stimulation with 5.0 μg/ml of poly I:C delivered by Lipofectamine™ 2000 or without stimulation. Scrambled shRNA treated D2SC cells served as a control. (B) DHX15 and MAVS are required for activating the NF-κB, IRF3 and MAPK signal transduction pathways upon reovirus infection. Immunoblot of indicated proteins from lysates of D2SC with indicated shRNA after infection with reovirus at a multiplicity of infection of 10. GAPDH served as loading control.
Discussion
The innate immune system detects invading pathogenic microorganisms, mainly by recognizing the pathogenic nucleic acids through PRRs. After sensing viral RNA or viral DNA, antiviral responses are triggered in the immune cells to produce IFNs and inflammatory cytokines. IFNs are perhaps the most important line of innate antiviral defense. The induced inflammatory cytokines of IL-6 and TNF-α are likely to profoundly modulate both innate and adaptive immune responses. In this study, we found that DHX15 plays an important role in sensing viral RNA to produce both IFN and inflammatory cytokines mediated by MAVS in mDCs. The HELICc domain of DHX15 was required to bind poly I:C specifically. DHX15 interacts with MAVS through the N-terminal DEXDc domain and C-terminal domain. Critically, we have demonstrated that the observed association between DHX15 and MAVS is enhanced with the stimulation of poly I:C. Furthermore, targeting DHX15 with shRNA, and thus inhibiting reovirus-induced IRF3 activation and NF-κB and MAPK signaling, resulted in reduced IFN-β and cytokine responses to RNA virus infection. These results suggest that DHX15 is an important RNA sensor that is dependent on MAVS to sense pathogenic RNA.
Recent studies have led to the identification of a large number of viral nucleic acids sensors. An interesting question is how these sensors cooperate in viral nucleic acid sensing. The first possibility is that different sensors operate in different cell types. Pol III can sense B-form DNA in HEK293T cells through RIG-I (18, 19). However, the B-form DNA sensing pathway is independent of RIG-I in murine cells (38). DDX41 plays a critical role in mDCs and THP1 cells in sensing dsDNA and DNA viruses (30). Knockdown of DDX41 also resulted in diminished levels of IRF3 phosphorylation following DNA virus (adenovirus) infection in macrophage cell line RAW 264.7 (31). However, neither Pol III nor p204, the homolog of IFI16 in mouse, is required to activate IRF3 upon adenovirus infection in RAW 264.7 cells (31). DNA-Pkcs appears to be essential for DNA-induced IFN response in fibroblasts (39), but it is dispensable in bone marrow derived macrophages (40). The dsDNA and DNA virus sensing pathway is dependent on STING in the endothelial MS1 cell line (41). However, DDX41, AIM2 and p204 are not required to activate IRF3 in response to viral dsDNA in MS1 cells, indicating that there should be an unknown receptor playing a critical role in sensing DNA virus in MS1 cells. Though MAVS is the critical adaptor in some cell types to sense RNA virus, the RIG-I/MAVS pathway is not required in plasmacytoid dendritic cells in response to RNA virus infection (36). Indeed, TLR7 plays a critical role in sensing RNA viruses such as influenza A virus (2, 42). The second possibility is that different sensors recognize different subcellular localizations of nucleic acids. TLR3 senses endosomal poly I:C (1). RLH receptors preferentially sense cytosolic dsRNA. Interestingly, many helicases, including DDX1, DDX3, DHX9, DHX15, DDX21, DHX36, RIG-I and DDX60, have also been identified to play important roles in sensing cytosolic RNA or poly I:C. These helicases may forms different complexes to sense different size of RNA: MDA5 was found to sense the long form of poly I:C with a size of over 1000 bp (43); while the DDX1, DDX21 and DHX36 complex was found to sense both long and short poly I:C (23). RIG-I was found to preferentially sense short poly I:C or ssRNA with a 5′ triphosphate (7, 8). We have found that DHX15 is independent of MDA5 to play an important role to sense long poly I:C. DHX15 can pull down DHX9 at endogenous protein level, and knockdown of DHX9 reduces RNA binding DHX15, indicating that DHX15 and DHX9 forms a complex to sense poly I:C. Though DDX3 can bind viral RNA, DDX3 forms a complex with MAVS, and is an enhancer of MAVS against RNA virus (24). DDX60 can bind both viral RNA and viral DNA. DDX60 forms complex with RLR to promote RLR-mediated antivirus activity (32). Different helicases may play role in sensing RNA at different stages: DDX1-DDX21-DHX36 forms complex with TRIF (23) and DHX33 forms complex with MAVS (29) in the steady stage. DHX15 forms complex with MAVS in 2hr stimulation with poly I:C. By contrast, the expression of RIG-I or MDA5 is very low in the steady stage. The protein level of RIG-I is still un-detectable even after 7 hr of VSV infection (24), though IFN-β is induced before 4 to 6 hr VSV infection. Taken together, all these data suggest that DDX1-DDX21-DHX36-TRIF and DHX33-MAVS has a more important role in the initial sensing of poly I:C and in triggering the initial type I IFN response. DHX15-DHX9-MAVS complex induces second wave IRF3 activation and IFN production. These induced type I IFN production upregulated MDA5 or RIG-I, which then plays a more important role in the amplification phase of the type I IFN response to cytosolic dsRNA. Previous studies have shown that reovirus can activate multiple RNA sensors, including PKR (44) and DHX9 (27). We found that DHX15 can sense reovirus infection in mDCs. These studies suggest that evolutionary pressure may have forced the innate immune system to develop redundant sensors for detecting reovirus infection. In the sensing pathway of RLH receptors, both RIG-I and MDA5 interact with MAVS via a CARD domain (33, 45). DHX15 does not contain a CARD domain. Interestingly, we found that DHX15 binds MAVS via the C-terminal region of MAVS instead of an N-terminal CARD domain, indicating that the C-terminus is as important as the CARD domain of MAVS in sensing viral RNA.
While this paper was under revision, the antiviral activity of DHX15 in HeLa cells was reported (37). Similar with the role playing in mDCs, DHX15 also plays an important role to sense poly I:C and RNA virus to activate the NF-κB and MAPKs pathways and produce type I IFN and proinflammatory cytokines in HeLa cells. DHX15 forms a complex with MAVS after poly I:C stimulation. Interestingly, DHX15 plays no role to activate IRF3 in HeLa cells, though knockdown of DHX15 results an 80 to 90% of decrease of IFN-β mRNA in response to transfected poly I:C or encephalomyocarditis virus infection. It needs to further clarify how DHX15 deficiency blocks the production of type I IFN while IRF3 is activated. The NF-κB luciferase reporter assay indicates that DHX15 may also mediates the signaling of downstream of MAVS in HEK293 cells, although the relationship between DHX15 and MAVS in different cell types requires further investigation.
Acknowledgments
We thank all of our colleagues in our laboratory. We thank Dr. Carson Harrod for editing the manuscript.
This work was supported by NIH grant R37 AI091947 to YJL.
The abbreviations used are
- DHX15
DEAH (Asp-Glu-Ala-His) box helicase 15
- PRRs
pattern recognition receptors
- RLH
retinoid acid-inducible gene I-like helicases
- IFN-β
interferon beta
- BMDC
bone marrow-derived dendritic cell
- DC
dendritic cell
- MDA5
Melanoma differentiation factor 5
- RIG-I
Retinoic acid-inducible gene
- NLRs
NOD-like receptors
- Nod2
nucleotide-binding oligomerization domain 2
- PYHIN
Pyrin and HIN domain-containing
- AIM2
absent in melanoma 2
- IFI16
interferon gamma-inducible protein 16
- ASC
apoptosis-associated speck-like protein containing a CARD
- HA
hemagglutinin
- HA2
Helicase associated domain
- HelicC
helicase C terminal domain
- IRF3
IFN regulatory factor 3
- mDC
myeloid dendritic cell
- poly dG:dC
poly deoxyguanylic-deoxycytidylic acid
- poly I:C
polyinosine-polycytidylic acid
- shRNA
small heteroduplex RNA
- CARD
N-terminal caspase recruitment domain
- HSV-1
Herpes simplex virus 1
- PKR
protein kinase receptor
- Pol III
polymerase III
- SDD-AGE
semidenaturing detergent agarose gel electrophoresis
Footnotes
Disclosures
The authors have no financial conflict of interest.
References
- 1.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 2.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 3.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 4.Bauer S, Kirschning CJ, Häcker H, Redecke V, Hausmann S, Akira S, Wagner H, Lipford GB. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci USA. 2001;98:9237–9242. doi: 10.1073/pnas.161293498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 6.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 7.Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 8.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 11.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 14.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O’Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci USA. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters NE, Ferguson BJ, Mazzon M, Fahy AS, Krysztofinska E, Arribas-Bosacoma R, Pearl RLH, Ren H, Smith GL. A mechanism for the inhibition of DNA-PK-mediated DNA sensing by a virus. PLoS Pathog. 2013;9:e1003649. doi: 10.1371/journal.ppat.1003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, Imamichi H, Huang DW, Lempicki RA, Baseler MW, et al. Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol. 2011;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 23.Zhiqiang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, Liu YJ. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshiumi H, Sakai K, Matsumoto M, Seya T. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur J Immunol. 2010;40:940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]
- 25.Ariumi Y, Kuroki M, Abe K, Dansako H, Ikeda M, Wakita T, Kato N. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol. 2007;81:13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J, et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Yuan B, Lu N, Facchinetti V, Liu YJ. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J Immunol. 2011;187:4501–4508. doi: 10.4049/jimmunol.1101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitoma H, Hanabuchi S, Kim T, Bao M, Zhang Z, Sugimoto N, Liu YJ. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39:123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Lu N, Yuan B, Weng L, Wang F, Liu YJ, Zhang Z. The interaction between the helicase DHX33 and IPS-1 as a novel pathway to sense double-stranded RNA and RNA viruses in myeloid dendritic cells. Cell Mol Immunol. 2014;11:49–57. doi: 10.1038/cmi.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stein SC, Falck-Pedersen E. Sensing adenovirus infection: activation of interferon regulatory factor 3 in RAW 264.7 cells. J Virol. 2012;86:4527–4537. doi: 10.1128/JVI.07071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol Cell Biol. 2011;31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 34.Wen X, Tannukit S, Paine ML. TFIP11 interacts with mDEAH9, an RNA helicase involved in spliceosome disassembly. Int J Mol Sci. 2008;9:2105–2113. doi: 10.3390/ijms9112105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Mosallanejad K, Sekine Y, Ishikura-Kinoshita S, Kumagai K, Nagano T, Matsuzawa A, Takeda K, Naguro I, Ichijo H. The DEAH-Box RNA Helicase DHX15 Activates NF-κB and MAPK Signaling Downstream of MAVS During Antiviral Responses. Sci Signal. 2014;7:ra40. doi: 10.1126/scisignal.2004841. [DOI] [PubMed] [Google Scholar]
- 38.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 39.Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife. 2012;1:e00047. doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Stein SC, Lam E, Falck-Pedersen E. Cell-specific regulation of nucleic acid sensor cascades: a controlling interest in the antiviral response. J Virol. 2012;86:13303–13312. doi: 10.1128/JVI.02296-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iparraguirre A, Tobias JW, Hensley SE, Masek KS, Cavanagh LL, Rendl M, Hunter CA, Ertl HC, von Andrian UH, Weninger W. Two distinct activation states of plasmacytoid dendritic cells induced by influenza virus and CpG 1826 oligonucleotide. J Leukoc Biol. 2008;83:610–620. doi: 10.1189/jlb.0807511. [DOI] [PubMed] [Google Scholar]
- 43.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Errington F, Steele L, Prestwich R, Harrington KJ, Pandha HS, Vidal L, de Bono J, Selby P, Coffey M, Vile R, Melcher A. Reovirus Activates Human Dendritic Cells to Promote Innate Antitumor Immunity. J Immunol. 2008;180:6018–6026. doi: 10.4049/jimmunol.180.9.6018. [DOI] [PubMed] [Google Scholar]
- 45.Potter JA, Randall RE, Taylor GL. Crystal structure of human IPS-1/MAVS/VISA/Cardif caspase activation recruitment domain. BMC Struct Biol. 2008;8:11. doi: 10.1186/1472-6807-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]