Abstract
Sulfation is a common post-translational modification of tyrosine residues in eukaryotes; however, detection using traditional liquid chromatography-mass spectrometry (LC-MS) methods is challenging based on poor ionization efficiency in the positive ion mode and facile neutral loss upon collisional activation. In the present study, 193 nm ultraviolet photodissociation (UVPD) is applied to sulfopeptide anions to generate diagnostic sequence ions which do not undergo appreciable neutral loss of sulfate even using higher energy photoirradiation parameters. At the same time, neutral loss of sulfate is observed from the precursor and charge reduced precursor ions, a spectral feature that is useful for differentiating tyrosine sulfation from the nominally isobaric tyrosine phosphorylation. LC-MS detection limits for UVPD analysis in the negative mode were determined to be around 100 fmol for three sulfated peptides, caerulein, cionin, and leu-enkephalin. The LC-UVPD-MS method was applied for analysis of bovine fibrinogen, and its key sulfated peptide was confidently identified.
Introduction
The comprehensive identification of protein post-translational modifications (PTMs) continues to be an important goal of proteomics research in order to gain a better understanding of biological systems, especially in the context of how PTMs influence protein structure and function [1]. Despite advancements in analytical technology, particularly in mass spectrometry (MS), PTM mapping remains a challenging task based on the diverse array of PTMs, their low abundance and lability, and their unique chemical properties, thus driving the development of new techniques to aid in characterization. O-sulfation, first discovered in 1954 on bovine fibrinogen, is a primary modification of tyrosine with the potential for sulfate addition on up to an estimated 1% of all tyrosine residues of the total protein in an organism [2–4]. Modification is limited to secretory and transmembrane proteins that have traversed the trans-Golgi network where two membrane-bound tyrosylprotein sulfotransferase enzymes (TPST1 and TPST2) catalyze the transfer of sulfate from adenosine 3’-phosphate 5’-phosphosulfate (PAPS) to the tyrosine phenol [5–11]. The primary function of tyrosine sulfation is the modulation of proteinprotein interactions in the extracellular region [12–15]. More specifically, sulfation has been shown to play a profound role in numerous physiological and pathological processes, including hormonal regulation, hemostasis, inflammation and viral entry into host cells [16,17]. However, other role(s) for tyrosine sulfation in protein function may exist.
Despite the biological significance of tyrosine sulfation, the sulfoproteome remains largely unexplored due to the analytical challenges associated with characterization using mass spectrometry. Several properties of sulfated peptides, including an often very acidic amino acid sequence and the labile sulfo-ester bond, present major handicaps for conventional positive mode MS analysis [18,19]. Traditionally soft ionization techniques such as electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI) result in partial or complete loss of the modification in the positive mode. Sulfopeptides that remain intact during ionization and the first stage of mass analysis undergo the predominant neutral loss of sulfate upon collisional induced dissociation (CID) and any product ions observed likewise exhibit loss of modification [20,21]. Electron-based activation (ETD and ECD) also promote sulfate loss from product ions [22]; however, modification retention has been observed for highly basic sulfopeptides likely due to formation of a salt bridge between the acidic sulfo-moiety and arginine side chains [23]. For more acidic peptides, gas-phase adduction using metal cations or guanidinium groups has been used to generate stabilizing salt bridges, making sulfation site localization possible upon ECD [24–26]. An alternative strategy for site localization in the positive mode takes advantage of the lability of sulfate in a subtractive-based identification method. In this method free tyrosine residues are acetylated prior to MS analysis so that any unmodified tyrosine residues detected must necessarily originate from sulfate loss in the mass spectrometer [27,28]. While effective, these techniques rely on quantitative reaction of unmodified tyrosine and require more front-end sample processing.
Mass spectrometry analysis in the negative mode can provide a more direct approach for the detection of tyrosine sulfation based on the greater stability of sulfopetides as gas-phase anions. The consistent detection of intact deprotonated sulfopeptides upon ESI is a significant advantage compared to the prevalent decomposition of protonated sulfopeptides during ESI; however, there remains a need for improved MS/MS characterization. The primary fragmentation pathway for CID of peptide anions is neutral loss of sulfate and while this information is useful for confirming the presence of sulfation, the lack of peptide backbone fragments is an impediment [29–31]. Alternative activations methods including metastable atom-activated dissociation (MAD) [32], which uses a beam of high kinetic energy helium atoms for ion activation, and negative ion electron capture dissociation (niECD) [33], have shown promise for tyrosine sulfation mapping. Both techniques provide a high level of peptide sequence coverage without significant losses of the sulfate modifications. Specifically, MAD results in the formation of a diverse array of fragment ions, including a, b, c, x, y and z, and niECD favors formation of c and z fragment anions. Long activation times and/or extensive spectral averaging are required for optimal results for these two methods, limiting the compatibility of these methods with online LC-MS methods.
Ultraviolet photodissociation (UVPD) at 193 nm is a fast activation method that has shown merits for peptide sequencing in both the positive and negative modes [34–40], including analysis of peptides decorated with acidic PTMs [38,41,42]. Upon UVPD activation, peptide anions dissociate into predominantly a and x type ions with other ion series including c, z, and y observed less frequently. The unique UVPD fragmentation behavior of peptide anions has been incorporated into a database search engine (MassMatrix) to effectively streamline data interpretation and make possible the analysis of more complex proteomic samples [39]. Like electron based activation methods, UVPD is a fragmentation technique that does not promote neutral loss of post-translational modifications, one of the considerable disadvantages of CID. For example, in the UVPD analysis of deprotonated phosphorylated peptides, backbone cleavage remains the primary fragmentation pathway, thus allowing both the peptide sequence and the site of modification to be determined [38,40]. Similar promising results have recently been obtained for a sulfated peptide derived from the Ax21 protein in the gram negative bacterium Xanthomonas oryzae pv. oryzae [42]. This first successful identification of tyrosine sulfation using UVPD has prompted a more in-depth investigation of UVPD analysis of deprotonated sulfopeptides. In the present study, several figures of merit which are relevant for sulfate mapping using UVPD are evaluated. These include: the overall peptide sequence coverage, changes in modification retention as a function of the laser settings, MS1 and MS2 sensitivity, and the compatibility with online LCMS analysis.
Experimental
Materials
LCMS grade solvents and mobile phase additives were obtained from Fisher Scientific (Fairlawn, NJ). Other reagents were obtained from Sigma (St. Louis, Mo) Peptides GlpQDsYTGWMDF-NH2 (caerulein), RDsYTGWNleDF-NH2 (Thr28,Nle31-cholecystokinin-33 sulfated), Ac-DpYVPML-NH2, RRLIEDAEpYAARG-NH2, Ac-IpYGEF-NH2 (P60c-src Substrate II, phosphorylated), and TSTEPQpYQPGENL (Pp60c-src 521–553) were purchased from American Peptide Company (Sunnyvale, Ca); GDFEEIPEEsYLQ (hirudin fragment 54–65) and NsYsYGWMDF-NH2 (cionin) were purchased from Sigma; and sYGGFL (leucine-enkephalin sulfated) was purchased from Phoenix Pharmaceuticals (Burlingame, Ca). Bovine fibrinogen was obtained from Calbiochem. The protein was reduced with 5 mM dithiothreitol (DTT) at 55 °C for 45 minute. Iodoacetamide was then added to 15 mM, and alkylation proceeded for 45 minutes in the dark. Additional DTT was added to quench the alkylation. Trypsin (Promega, Madison, WI) was added in a 1:20 enzyme: protein ratio, and digestion occurred overnight at 37 °C.
Mass spectrometry, liquid chromatography, ultraviolet photodissociation
All experiments were conducted on a Thermo Fisher Scientific Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a Coherent ExciStar XS excimer laser (Santa Clara, Ca) operated at 193 nm and 500 Hz as previously described [43,44]. For direct infusion at 4 µL/min, 10 µM peptide solutions were prepared in water containing 25% methanol and either 0.1% formic acid, 0.1% ammonium hydroxide, or 5 mM ammonium acetate for analysis in the positive, negative, or both ion modes, respectively. For positive mode analysis, the ESI source was operated at 3.5 kV with a sheath gas flow of 4 units and auxiliary and sweep gases both zero. Orbitrap MS1 and MS2 automatic gain control (AGC) targets were 1,000,000 and 50,000, respectively. For negative ion mode experiments, the heated ESI source (HESI) was used and the source parameters were tuned before each analysis in order to optimize the spray stability. Optimum HESI temperatures ranged from 40–60°C with source voltage 2.3–3 kV. A high sheath gas flow between 25–40 units improved desolvation, while lower amounts of auxiliary gas and sweep gas were needed (both were operated between 0–5 units). Orbitrap AGC targets were 1,000,000 for MS1 and 100,000 for MS2 in negative mode. In both polarities CID and higher energy collision induced dissociation (HCD) were performed using 35% normalized collision energy with activation times of 10 and 0.1 ms, respectively. To optimize the UVPD conditions, a variety of energies including 1, 2, 3, 4, and 5 mJ were used with the number of laser pulses ranging from 1 to 6. Ultimately, 3 pulses at 2 mJ were selected for LCMS experiments.
Liquid chromatography was performed on a Dionex Ultimate 3000 capillary LC operated at a flow rate of 4 µL/min. Mobile phase A was 5 mM ammonium acetate in water, and mobile phase B was 5 mM ammonium acetate in 90% methanol, 10% water. Peptide solutions were prepared in 100% mobile phase A for separation on a 3 × 150 mm Agilent Zorbax Extend-C18 column with 3.5 µm particle size (Santa Clara, CA). Separations of an equimolar mixture of caerulein, cionin, and leu-enkephalin were accomplished using a linear gradient that increased from 25% B to 60% B over 15 minutes. Doubly deprotonated caerulein (m/z 674.7162) and cionin (m/z 625.6651) and singly deprotonated leuenkephalin (m/z 634.2178) were targeted for UVPD using a precursor mass list with associated 10 ppm m/z tolerance. Dynamic exclusion was disabled and MS1 (400–2000 m/z) and MS2 scans, both collected in the Orbitrap at resolution 15000, were alternated over the course of the LCMS run.
For UVPD analysis of tryptic bovine fibrinogen in the negative mode, about 5 µg (≈14.7 pmol) of protein digest was injected, and the percent B was increased linearly from 2% to 35% over 45 minutes. A top ten data dependent scan program was used in which the first scan was a negative full FTMS survey scan over m/z 400–2000 at resolution 120,000 followed by 10 UVPD events on the ten most abundant ions from scan event 1. Dynamic exclusion was enabled for 25 seconds with a single repeat count. For UVPD at 15000 resolution, the isolation width was set to 3 m/z, the HCD normalized collision energy was 1% and the activation time was 6 ms in order to generate 3 laser pulses. After the initial analysis of fibrinogen, an additional segment from time 16.85–18.40 min was included to continually target a sulfated peptide of interest, GlpFPTDsYDEGQDDRPK (fibrinopeptide B), for UVPD. During this segment the m/z of interest, 935.35160, was isolated using an increased width of 6 Da and dissociated using 3 laser pulses each at 2 mJ.
Database search
Results from LCMS analysis of fibrinogen were interpreted using the MassMatrix database search algorithm [45–48]. The experimental data was searched against the fibrinogen bovine FASTA and a reversed decoy database. Trypsin (no P rule) was specified for digestion while a maximum of 3 missed cleavages. The minimum peptide length was 5 amino acids while the maximum length was 75 amino acids. The peptide mass tolerance was 20 ppm and the fragment mass tolerance was ±0.02 Da. MassMatrix uses three independent statistical scores including, pp, pp2, and pptag, to evaluate quality of peptide-spectrum matches, and the minimum output for each score was defined as 5.0, 5.0, and 1.3 respectively. Iodoacetamide derivatization of cysteine was a fixed modification, while sulfated tyrosine and pyroglutamate from glutamate were variable modifications.
Results and Discussion
UVPD, CID, and HCD Analysis of Sulfopeptide Cations and Anions
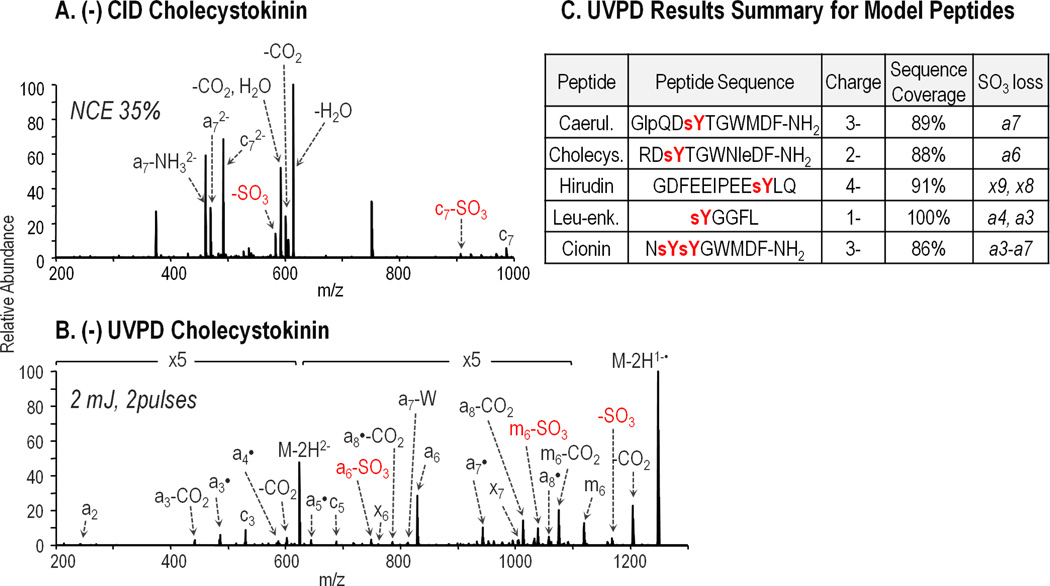
Four singly sulfated peptides including caerulein (GlpQDsYTGWMDF-NH2), cholecystokinin (RDsYTGWLDF-NH2), hirudin (GDFEEIPEEsYLQ), and leu-enkephalin (sYGGFL), and one doubly sulfated peptide, cionin (NsYsYGWMDF-NH2) were analyzed in both positive and negative modes and characterized using CID, HCD and UVPD. The spectral quality of MS survey scans in the positive mode was generally poor, even for cholecystokinin, the most basic of the five peptides analyzed and thus the most amenable to positive mode ionization. Supplemental Figure 1A and 1B show ESI mass spectra for cholecystokinin sprayed in both acidic (0.1% formic acid containing) and neutral (5 mM ammonium acetate containing) solutions. For both conditions, singly protonated cholecystokinin was the most abundant charge state. A doubly protonated species was also observed but in low abundance relative to an extensive array of sodium and potassium adducts. Further, spontaneous loss of SO3 was observed during ESI for all charge states, with neutral loss ions appearing about 40% abundance relative to 2+ sulfated precursor ions and about 3% abundance relative to singly charged sulfated precursor ions. Upon CID of singly protonated cholecystokinin, loss of SO3 was the predominant product ion observed with b6 through b8 ions (all with SO3 loss) observed only after magnifying the spectrum 50 times (Supplemental Figure 2A). The higher energy deposition of HCD facilitated extensive backbone fragmentation of 1+ cholecystokinin (Supplemental Figure 2B); however, all product ions lacked the sulfate modification. UVPD is an even higher energy process, with a single 193 nm photon having an energy of 6.4 eV. As a result, fragmentation can occur at each bond along the peptide backbone to generate a, b, c, x, y, and z ions. Supplemental Figure 3 displays the bond cleavages of the peptide backbone to produce these types of fragment ions. UVPD performed similarly (Supplemental Figure 2C), producing an array of backbone fragments including a, b, and c ions, none of which retained the SO3 modification, allowing the amino acid sequence to be determined while the actual site of sulfation could not be pinpointed.
Although doubly protonated cholecystokinin was also present in the ESI mass spectrum, this charge state was more difficult to analyze based on instability during the ion isolation stage prior to MS/MS activation. Supplemental Figure 4 shows a drastic decrease in ion abundance as well as a shift from the normal isotope distribution of 2+ cholecystokinin using an isolation width of 2 m/z without applying normalized collision energy. Subsequent isolation using widths of 4, 6, and 8 m/z were performed while maintaining 0% normalized collision energy to determine the optimal isolation width. At isolation widths of 6 and 8 m/z, the isotope distribution appeared normal and the ion abundance was on scale with that observed in the ESI mass spectrum prior to isolation. Despite these improvements at increased isolation widths, SO3 loss is observed following ion isolation further demonstrating the lability of the sulfoester bond. Activation using CID, HCD, and UVPD (Supplemental Figure 5A–C) resulted in results similar to those obtained for 1+ cholecystokinin. Upon ETD (Supplemental Figure 5D), three sulfated c ions, c6–c8, were generated, accounting for 38% sequence coverage. Using negative electrospray conditions, sulfopeptides ionized more readily and exhibited no loss of SO3. Supplemental Figure 6 shows the ESI mass spectra for cholecystokinin sprayed from pH neutral solution containing 5 mM ammonium acetate and basic solution containing 0.1% ammonium hydroxide. Electrospray using both solutions yielded an abundant 2- sulfated precursor ion with minimal salt adducts. The CID spectrum for doubly deprotonated cholecystokinin (Figure 1A) was dominated by the neutral losses of H2O, CO2, and SO3, with water loss being the preferred fragmentation pathway. A few product ions were derived from cleavage of the peptide backbone, including a7 and c7. While these fragment ions retained the SO3 modification, alone they did not provide sufficient information to sequence the peptide. HCD results for deprotonated cholecystokinin (Supplemental Figure 7) mirrored those obtained using CID although more energy was required to obtain the same MS/MS results. In contrast, UVPD of deprotonated cholecystokinin (Figure 1B) provided nearly complete sequence coverage afforded by the broad series of a and x ions with only a single missed cleavage between the first and second amino acid residues (no a1 or x8 ions). Also, significant SO3 loss was only observed for one product ion, a6, for which the analogous intact sulfated product ion was also observed and in greater abundance relative to the corresponding sulfate loss ion. Nearly full sequence coverage was likewise obtained upon UVPD of deprotonated caerulein, hirudin, and leu-enkephalin analyzed in 3-, 4-, and 1- charge states, respectively. These results highlight the lack of charge state dependence on UVPD performance, thus making ion abundance the most important factor in choosing a particular charge state for dissociation. Some loss of SO3 from product ions was also observed for each peptide (listed in Figure 1C), but again the modified form of each ion was always detected. Interestingly, the UVPD results for di-sulfated cionin were very different from the UVPD results for the singly sulfated peptides. While an extensive series of a ions was generated, each ion with the exception of a2 also underwent neutral loss of one out of the two SO3 modifications (Supplemental Figure 8). This result may be rationalized based on the side-by-side positioning of the sulfotyrosine residues in the peptide sequence and the ensuing instability caused by simultaneous deprotonation of both of the sulfate moieties, thus causing proton driven loss of one sulfate [33,49].
Figure 1.
MS/MS spectra for 2- charged cholecystokinin (RDsYTGWNleDF-NH2), m/z 624.24, using (A) CID at normalized collision energy (NCE) 35% and (B) UVPD using 3 pulses at 2 mJ. “-W” denotes sidechain loss from tryptophan (C9H7N, 129 Da). Products ion that have lost SO3 are annotated in red font. UVPD data for the other model sulfopeptides is summarized in part (C) with sequence coverage listed for the most abundant charge state of each peptide. Product ions from which SO3 loss is observed are also listed.
In addition to formation of diagnostic sequence ions, charge reduction of the deprotonated precursor ion via electron photodetachment [50] and concomitant loss of CO2 and SO3 from these ions were also dominant fragmentation pathways upon UVPD. Several amino acid side chains also proved to be labile upon UVPD including the tryptophan side chains (C9H7N, 129 Da) and glutamic acid side chains (C3H4O2, 72 Da), which were observed as abundant neutral losses from the precursor and charge reduced radical ions. These side chain losses have also been reported previously following negative electron transfer dissociation (NETD) and 266 nm UVPD of proton deficient radical cations [51,52]. Combinations of CO2, SO3, and peptide side chain neutral losses were also commonly observed upon UVPD.
Differentiating sY from pY
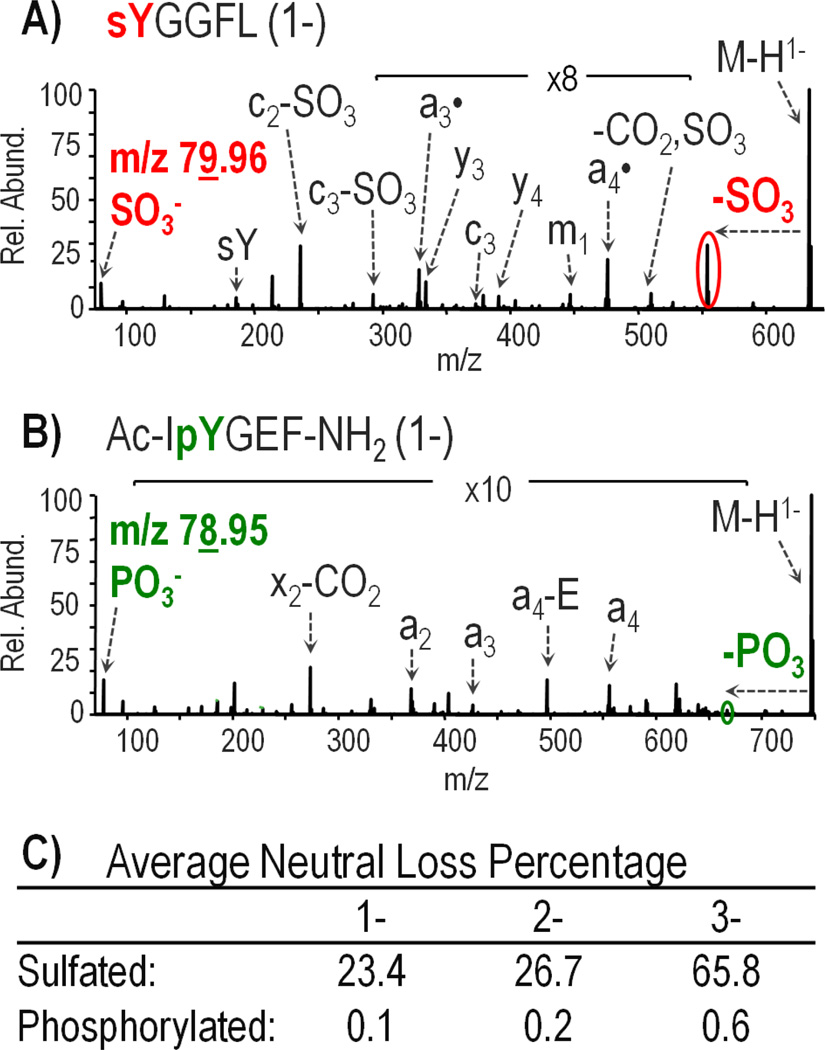
After thorough analysis of UVPD fragmentation of deprotonated sulfated peptides, UVPD of deprotonated phospho-tyrosine containing peptides was examined to determine if the two modifications could be distinguished. Differentiating sulfotyrosine from phosphotyrosine is challenging based on the nearly isobaric nature of the modifications, with sulfation adding 79.956 Da and phosphorylation adding 79.966 Da. Other studies have approached the problem by exploiting differences in the neutral loss characteristics of phosphotyrosine- and sulfotyrosine-containing peptide anions upon CID [30]. Based on the UVPD fragmentation behavior of four deprotonated phosphotyrosine-containing peptides including TSTEPQpYQPGENL, Ac-DpYVPML-NH2, RRLIEDAEpYAARG, and Ac-IpYGEF-NH2, it appears that a similar method monitoring sulfo and phospho neutral losses can be used for UVPD. An example of the comparative UVPD spectra obtained for a deprotonated sulfopeptide and phosphopeptide is shown in Figure 2 (for sulfated leu-enkephalin and phosphorylated P60-src substrate IIAc-IpYGEF-NH2). Figure 2C displays the average relative percentage of loss of SO3 and PO3 from all the singly sulfated and phosphorylated peptides in the 1-, 2-, and 3- charge states (doubly sulfated cionin is excluded). For all phosphopeptides in all charge states, the percentage neutral loss was below 1%. The sulfopeptides, in contrast, showed a much greater extent of neutral loss with 23%, 27%, and 66% loss from 1-, 2-, and 3- precursor and charge-reduced ions. For doubly and triply deprotonated precursor ions, the greatest neutral loss of the modification was observed from the 1- charge reduced ion derived from one electron photodetachment from 2- precursors and from two electron photodetachment products from 3- precursors. Finally, distinct reporter ions of m/z 79.96 and m/z 78.95 were detected in the UVPD mass spectra for deprotonated sulfotyrosine and phosphotyrosine peptides, corresponding to SO3− and PO3− ions, respectively. A one dalton difference is easily distinguished using current mass spectrometry instrumentation; however, for detection in an Orbitrap mass analyzer (which UVPD necessitates since it is undertaken in the HCD cell of the instrument) the m/z of the precursor ion must be low enough such that the reporter ions fall within 1/20 of the precursor m/z. The UVPD spectra of deprotonated peptides sYGGFL (leu-enkephalin) and Ac-IpYGEF-NH2 in Figure 2 showcase both the detection of low mass reporter ions and the significant difference in the degree of SO3 and PO3 neutral loss from the precursor ion, thus allowing phosphorylation and sulfation to be readily differentiated.
Figure 2.
UVPD mass spectra of singly deprotonated peptides: (A) sulfated leu-enkephalin and (B) phosphorylated P60c-src substrate II Ac-IpYGEF-NH2 using 3 laser pulses at 2 mJ. (C) Average percentage of the neutral loss product observed from the selected precursor and charge reduced precursor ions. Cionin is excluded from the sulfated peptide averages.
Laser Parameter Optimization
Photodissociation offers a high degree of tunability in terms of the laser energy and the number of pulses applied for MS/MS activation, enabling the selection of different UVPD parameters to suit particular applications. For sulfopeptide analysis, the optimal UVPD settings should maximize the abundance of sequence ions while minimizing the confounding neutral loss of SO3 from these ions. Also, the neutral loss of SO3 from the precursor ion and the charge-reduced precursor ion would ideally be prominent, as these ions provide further evidence for the presence of sulfation on the peptide and help to differentiate sulfotyrosine from phosphotyrosine. To establish the laser conditions that best meet these criteria, sulfopeptide analysis was undertaken using a matrix of 30 different laser parameters derived from combinations of 1, 2, 3, 4, 5, or 6 pulses at energies of 1, 2, 3, 4, or 5 mJ. Various precursor charge states were accessed using ammonium hydroxide containing solutions (Supplemental Figure 9A) including 1-, 2-, 3- for caerulein and hirudin and 1-, 2- for cholecystokinin and leu-enkephalin were analyzed in order to assess the impact of charge state on UVPD fragmentation. Hirudin, the most acidic peptide, was also observed in 4- and 5- charge states; however, UVPD data from these precursors was not included for the laser parameter optimization because of the lack of supporting data from other peptides that produce ions in charge states beyond 3-. Additionally, when peptides were analyzed in 5 mM ammonium acetate buffers which are more analogous to those used for LC-MS, charge states did not exceed 3- (Supplemental Figure 9B).
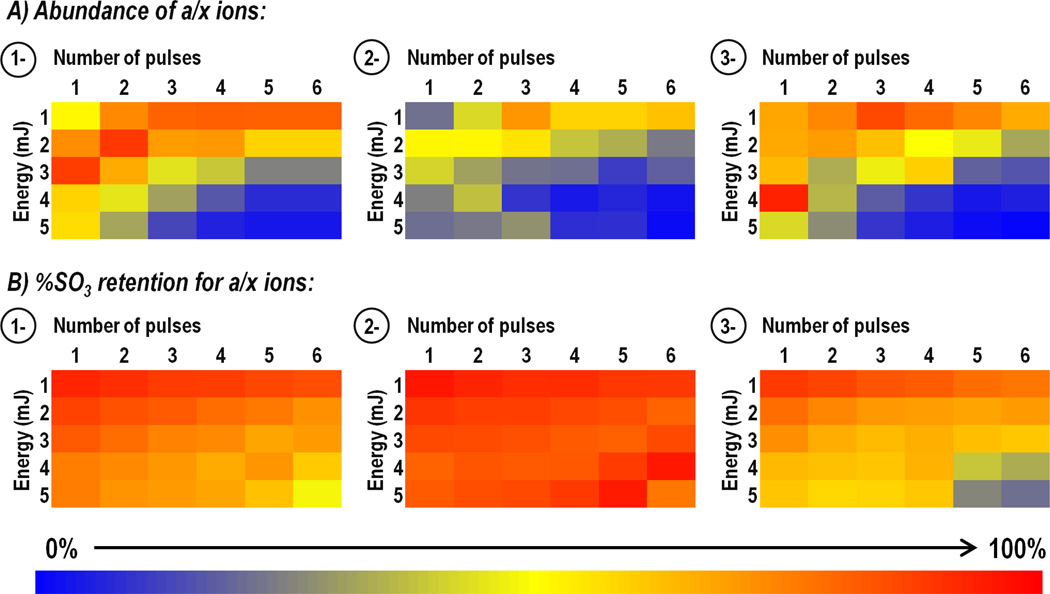
For each peptide, laser settings were evaluated based on the abundance of the neutral loss of sulfate from the precursor and/or charge-reduced precursor as well as the abundance of three singly deprotonated a or x products, all containing tyrosine to allow both sulfated and nonsulfated forms of each ion to be monitored. Specifically these were a4, a7, a8 (m/z 568.13, 912.28, 1043.32) from caerulein; a3, a6, a7 (m/z 485.15, 829.29, 942.38) from cholecystokinin; a8, a9, a10 (m/z 1124.41, 1253.45, 1259.44) from hirudin; and a2, a3, a4 (m/z 271.04, 328.06, 475.13) from leu-enkephalin. To determine which laser conditions promoted the most efficient generation of informative sequence ions, the abundance of sulfated product ions were summed and charted as a function of laser conditions. Absolute abundances were then normalized to 100% for each peptide prior to averaging the values for all peptides of the same precursor charge state in order to avoid biasing the results towards the most abundant product ions arising from a single peptide. Figure 3A shows the results for 1-, 2-, and 3- peptides as heat maps in which the color red represents the greatest abundance of product ions and blue represents the lowest abundance of product ions. While there is no single combination of laser energy and pulse number that outperformed all others, there is a clear trend of greater a/x ion abundance at lower energies and fewer laser pulses. This trend was consistent for 1-, 2-, and 3- precursor ions. These results align well with previous UVPD studies that favored the use of minimal laser pulses and energy for fragmentation of deprotonated peptides [38–40,42]. When the abundances of SO3 neutral loss ions from precursor and charge-reduced precursor ions were tabulated, normalized, and averaged across peptides (Supplemental Figure 10), the results showed the same pattern that was observed for a/x sequence ions, with the SO3 neutral loss ions increasing in abundance as fewer laser pulses at lower energy were used for activation.
Figure 3.
Impact of laser parameters: Optimization of abundances of a/x sequence ions and minimization of loss of sulfate. For each peptide, the abundances of three singly charged Y-containing a/x product ions were monitored in both their sulfated and non-sulfated forms. Specifically these were a4, a7, a8 from caerulein; a3, a6, a7 from cholecystokinin; x8, x9, a10 from hirudin; and a2, a3, a4 from leu-enkephalin. A) The summed abundances of sulfated product ions (normalized to 100%) is charted as a function of both laser energy (1–5 mJ) and number of pulses (1–6). B) Percent sulfate retention is determined for each set of laser conditions based on the total abundance of the sulfated products ions divided by the total abundance of both sulfated and non-sulfated (neutral loss) product ions. Heat maps for 1- and 2- precursor ions (top and middle, respectively) contain data from all four peptides, whereas heat maps for 3- precursor ions (bottom) contain data only from hirudin and caerulein. The deepest red shade indicates the most optimum outcome.
While the neutral loss of SO3 from precursor and charge-reduced precursor ions is a useful marker for peptide sulfation, loss of SO3 from product ions is an undesirable outcome of UVPD that can prevent localization of the sulfation sites. Thus, in addition to monitoring the sulfated product ion abundance, it is also critical to note the abundances of non-sulfated product ions so that laser conditions which promote this neutral loss can be identified and avoided. Changes in sulfate retention are presented as percentages based on the abundance of the three sulfated a/x ions chosen for each peptide (listed above) and the abundance of the same a/x ions found 79 Da lower in mass after sulfate neutral loss using the following equation:
The percentage SO3 retention for peptides of the same charge state is averaged and displayed as heat maps in Figure 3B, with red representing 100% sulfate retention and blue 0%. Interestingly, the decrease in the abundance of sulfated products observed at high energy using many pulses does not correspond with a significant increase in the abundance of non-sulfated, neutral loss ions. Instead the percentage of SO3 retention remains relatively constant across all laser conditions and only under the most energetic dissociation conditions (4–5 mJ, 5–6 pulse) does the extent of SO3 loss greatly increase, showing that UVPD is a robust method for sulfation mapping. Based on these analyses, the optimum UVPD parameters were 2–3 laser pulses at 2–3 mJ.
MS1 and MS2 Limit of Detection
Another important performance metric to consider for negative UVPD analysis of sulfopeptides is the absolute limit of detection (LOD) because of the generally low abundance of modified peptides in biological samples. Detection limits are especially pertinent for negative mode LC-MS workflows because of the lower flux of precursor ions generated compared to using the positive ESI mode [53–56]. Several LC mobile phase additives have been reported to improve ESI efficiency in the negative mode including acetic acid (pH 4) and ammonium hydroxide (pH 11) [57,58]; however, prolonged exposure to acidic solutions may result in hydrolysis of sulfate and thus should be avoided in favor of higher pH alternatives [59]. At the same time, high pH solutions were damaging to the LC system hardware and column and ultimately were abandoned in favor of more robust separations at neutral pH using 5 mM ammonium acetate-containing mobile phases. Additionally methanol was used instead of acetonitrile as the organic mobile phase constituent to further boost the ESI efficiency [57].
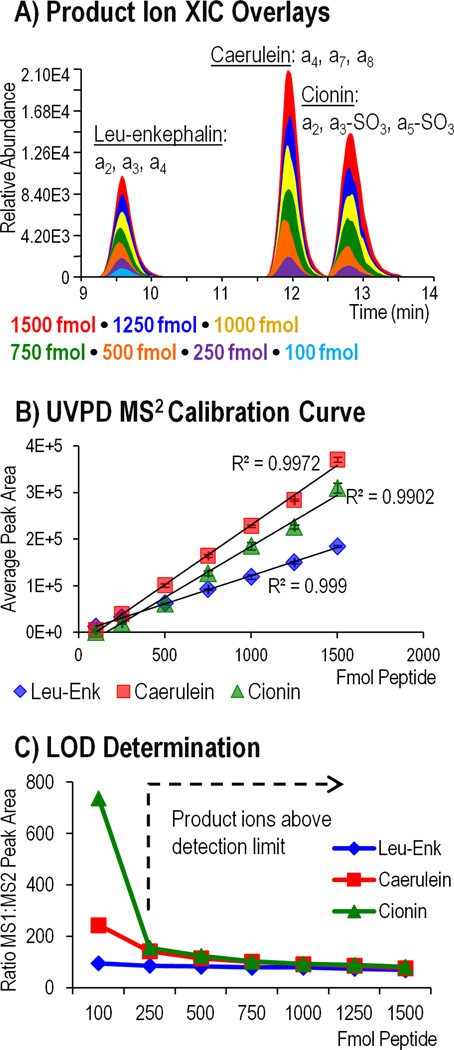
For the LOD determination, seven solutions containing 100, 250, 500, 750, 1000, 1250, and 1500 femtomoles each of caerulein, cionin, and leu-enkephalin were analyzed by LCMS. A precursor mass list was used to target each peptide in its most abundant charge state (1- for leu-enkephalin and 2- for caerulein and cionin), and MS1 and MS2 scans were alternated to ensure an even distribution of MS survey spectra and UVPD mass spectra across each eluting peak. MS extracted ion chromatograms (XIC) were generated for each peptide, and peak areas were plotted against the femtomole amount injected to create a linear calibration curve (Supplemental Figure 11A and 11B). Each data point on the calibration curve has an associated error bar showing the standard deviation in the peak area for four replicate runs, with high reproducibility observed for measurements made at 500 femtomoles and greater (≤3%RSD) and slightly lower reproducibility (≤20%RSD) at the most dilute concentrations. Assuming the limit of detection was equal to three times the standard deviation of the peak area, divided by the slope of the calibration curve, then the resulting MS1 LODs for caerulein, cionin, and leuenkephalin were determined to be 113 fmol, 182 fmol, and 96 fmol, respectively.
To evaluate the MS/MS LODs, XICs were generated based on the sum of the three most abundant a-type product ions for each peptide including: a4, a7, a8 for caerulein; a2, a3, a4 for leuenkephalin; and a2, a3-SO3, a5-SO3 for cionin. XICs and calibration curves for each peptide are displayed in Figure 4 A–B. When the calibration curve was used to calculate MS/MS LOD, the detection limits for each peptide were lower than those obtained for MS1, a result which is logically unsound because an ion cannot be selected for MS/MS activation if it is not first detected by MS1. Following this result, a more empirical measure of MS/MS LOD was employed based on the characteristic decrease in product ion abundance relative to precursor ion abundance upon UVPD. This was systematically accomplished using the MS1 peak areas from the XICs in Supplemental Figure 11A and dividing each by the corresponding MS2 peak areas from Figure 4A. Figure 4C shows the plot of MS1:MS2 ratio (factor decrease in abundance from MS1 to MS2) as a function of the number of femtomoles injected. In the range of 1500-250 fmol, the MS1:MS2 peak area ratio for each peptide is about 100 which corresponds to a two order of magnitude difference in precursor and product ion abundance. For caerulein and cionin, an increase in the MS1:MS2 ratio is observed at 100 femtomoles, indicating a larger than characteristic drop in the product ion abundance following UVPD and suggesting that the detection limit has been exceeded. Leu-enkephalin did not exhibit the same increase and thus the limit of detection is expected to be lower than 100 fmol. Inspection of the MS/MS spectra at 250 and 100 femtomoles for each peptide (Supplemental Figure 12) confirmed the LOD results of 250 fmol for caerulein and cionin and near 100 fmol for leu-enkephalin.
Figure 4.
MS2 LOD determined for leu-enkephalin (1-), caerulein (2-), and cionin (2-) using three 2 mJ pulses for photodissociation. A) Extracted ion chromatograms (XIC) representing the summed abundance of three product ions from each peptide were generated for each dilution and overlaid. B) The area under the curve was plotted as a function of femtomoles of peptide injected to generate a calibration curve for each peptide. C) Precursor ion peak area (MS1) was divided by product ion abundance (MS2) and plotted against the number of femtomoles injected. Above the MS2 LOD, the MS1:MS2 ratio should remain consistent as represented by the flat region of the graph.
Because molar detection limits will be specific for different peptides, a more universal measure for the MS2 detection limit could be defined as the lowest precursor ion signal that can undergo UVPD and produce product ions which can be distinguished from background noise. Since, UVPD product ions are generally about two orders of magnitude lower in abundance compared to their precursor ion abundances, precursor ion abundances should be at least on the order of 10,000 units in order to detect the most abundant UVPD sequence ions and on the order of at least 100,000 units for the best quality UVPD data. Understanding the lower detection limits of UVPD can be useful for data dependent LCMS runs in which a threshold signal for MS2 can be defined, therefore making it possible to avoid activating ions that will not provide meaningful data. Also noteworthy is the abundance of the neutral loss of SO3 from precursor and charge reduced ions which is typically greater compared to the abundance of sequence ions. For 100 fmol injections of caerulein and cionin (Supplemental Figure 12 B–C), the SO3 neutral loss ion was observed despite a lack of corresponding sequence ions. Detection of these sulfate loss ions can provide evidence of peptide sulfation and enable specific ions to be targeted in subsequent analyses to obtain improved MS/MS results.
The MS1 and MS2 detection limits reported for the LC-UVPD-MS strategy are suitable for the analysis of isolated sulfopeptides in which several micrograms of total digest are routinely injected for LC-MS analysis. For a complex biological sample, enrichment of sulfated peptides would be necessary based on their low stoichiometric abundances relative to unmodified proteins in the sample. Various enrichment methods have been reported including ones that utilize immobilized metal affinity chromatography, weak anion exchange, and anti-sulfotyrosine monoclonal antibodies and we are currently integrating a robust enrichment method with the sensitive UVPD-MS approach[60–62]. Additionally, the UVPD method is not designed for direct peptide quantification per se, although this could be achieved using peptide standards to generate a calibration curve or through the use of various label or label-free methods for relative quantification between samples.
Analysis of Bovine Fibrinogen
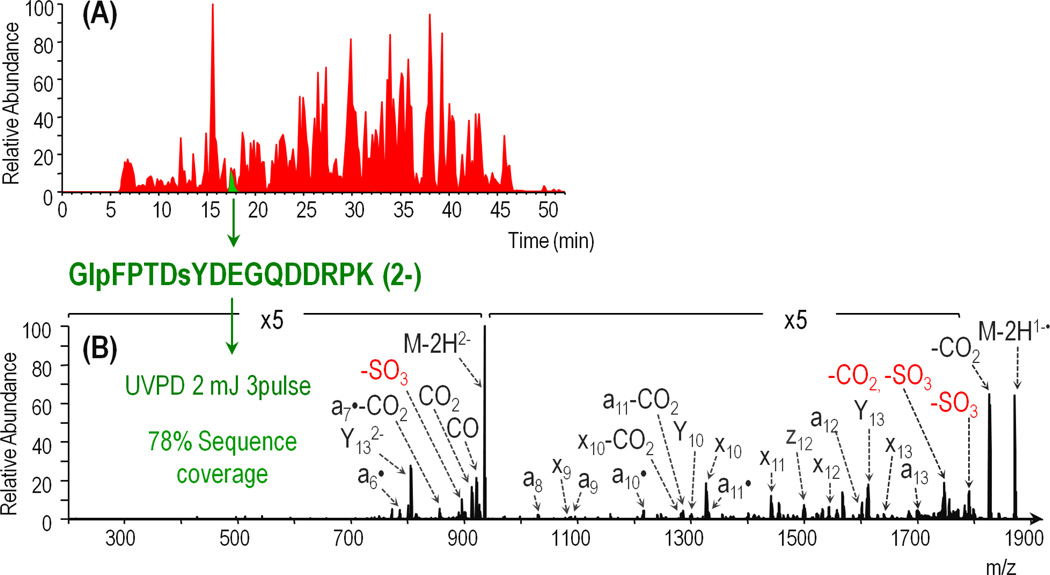
Bovine fibrinogen is a heterohexameric protein containing two sets of three non-identical alpha (615 residues, 67 kDa), beta (468 residues, 53 kDa), and gamma (444 residues, 50 kDa) chains. A single sulfo-modification is expected at tyrosine 6 on the beta chain. Trypsin-digested fibrinogen was analyzed by LCMS-UVPD in the negative mode using 3 pulses at 2 mJ based on the parameter optimization undertaken for the sulfopeptides. The base peak chromatogram is shown in red in Figure 5A with an XIC for the sulfopeptide of interest overlaid in green. The sequence coverage of the alpha, beta, and gamma chains obtained using UVPD was about 38%, 40%, and 19% respectively. The sulfated peptide, GlpFPTDsYDEGQDDRPK, was identified based on accurate precursor mass and the diagnostic 80 Da neutral loss ion; however, the quality of the MS/MS spectrum was poor due to a low precursor ion signal (E4 range). To increase the abundance of the sulfated peptide, a greater amount of sample was injected and the isolation width was increased from 3 m/z to 6 m/z. Additionally, the data dependent LCMS program was revised to include a targeted segment in which UVPD was performed on GlpFPTDsYDEGQDDRPK over the course of the peptide elution to allow MS/MS spectral averaging. The targeted run yielded an extensive series of a/x ions, including a6– a14 and x9 – x14, facilitated identification of the peptide. Because all of these product ions (with the exception of x9) contain the sulfation and there are cleavages at both the N-terminal and C-terminal sides of the sulfo-tyrosine, the site of sulfation was unambiguously assigned.
Figure 5.
A) Base peak chromatogram of tryptic bovine fibrinogen (red) with extracted ion chromatogram for sulfopeptide GlpFPTDsYDEGQDDRPK (2-) from the fibrinogen beta chain (green) overlaid. B) Negative UVPD mass spectrum (three pulses at 2 mJ) of GlpFPTDsYDEGQDDRPK (2-) from the average of 18 MS/MS scans acquired over 12 seconds. SO3 neutral loss products are annotated in red.
Conclusion
The present report demonstrates the suitability of 193 nm UVPD-MS for the analysis of sulfopeptide anions. For mono-sulfated peptides caerulein, cholecystokinin, leu-enkephalin, and hirudin, UVPD resulted in almost full sequence coverage across all charge states analyzed. Di-sulfated cionin also generated an extensive array of product ions; however, each ion only retained one of the two sulfate moieties. Systematic evaluation of laser parameters confirmed the stability of SO3 across various energies and numbers of pulses. This allowed the selection of laser parameters that maximized the abundance of sequence ions and SO3 loss ions which are useful for distinguishing between the nominally isobaric tyrosine sulfation and phosphorylation modifications. LC-UVPD-MS detection limits near 100 fmol were obtained for sulfated peptides, caerulein, cionin, and leu-enkephalin. The LC-UVPD-MS method was applied for analysis of bovine fibrinogen and the expected sulfated peptide, GlpFPTDsYDEGQDDRPK, was confidently identified. We anticipate that UVPD-LC-MS could be used for global sulfation analysis following enrichment of sulfated peptides from a biological sample.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Institute of Health Grants R21GM099028 (to J.S.B.) and RO1HD056022 (to K.L.M.), the Robert A. Welch Foundation (F1155 to J.S.B.) and institutional funds from the Oklahoma Medical Research Foundation (to K.L.M).
References
- 1.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotech. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 2.Bettelheim FR. TYROSINE-O-SULFATE IN A PEPTIDE FROM FIBRINOGEN. J. Am. Chem. Soc. 1954;76:2838–2839. [Google Scholar]
- 3.Huttner WB. Sulphation of tyrosine residues-a widespread modification of proteins. Nature. 1982;299:273–276. doi: 10.1038/299273a0. [DOI] [PubMed] [Google Scholar]
- 4.Baeuerle PA, Huttner WB. Tyrosine sulfation of yolk proteins 1, 2, and 3 in Drosophila melanogaster. Journal of Biological Chemistry. 1985;260:6434–6439. [PubMed] [Google Scholar]
- 5.Beisswanger Roland, Corbeil D, Vannier C, Thiele C, Dohrmann U, Kellner Roland, Ashman K, Niehrs Christof, Huttner Wieland B. Existence of Distinct Tyrosylprotein Sulfotransferase Genes: Molecular Characterization of Tyrosylprotein Sulfotransferase-2. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:11134–11139. doi: 10.1073/pnas.95.19.11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ouyang Y-B, Lane WS, Moore KL. Tyrosylprotein Sulfotransferase: Purification and Molecular Cloning of an Enzyme that Catalyzes Tyrosine O-Sulfation, a Common Posttranslational Modification of Eukaryotic Proteins. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2896–2901. doi: 10.1073/pnas.95.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouyang Y-B, Moore KL. Molecular Cloning and Expression of Human and Mouse Tyrosylprotein Sulfotransferase-2 and a Tyrosylprotein Sulfotransferase Homologue in Caenorhabditis elegans. Journal of Biological Chemistry. 1998;273:24770–24774. doi: 10.1074/jbc.273.38.24770. [DOI] [PubMed] [Google Scholar]
- 8.Seibert C, Cadene M, Sanfiz A, Chait BT, Sakmar TP. Tyrosine Sulfation of CCR5 N-Terminal Peptide by Tyrosylprotein Sulfotransferases 1 and 2 Follows a Discrete Pattern and Temporal Sequence. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11031–11036. doi: 10.1073/pnas.172380899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danan LM, Yu Z, Hoffhines AJ, Moore KL, Leary JA. Mass Spectrometric Kinetic Analysis of Human Tyrosylprotein Sulfotransferase-1 and −2. Journal of the American Society for Mass Spectrometry. 2008;19:1459–1466. doi: 10.1016/j.jasms.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danan LM, Yu Z, Ludden PJ, Jia W, Moore KL, Leary JA. Catalytic Mechanism of Golgi-Resident Human Tyrosylprotein Sulfotransferase-2: A Mass Spectrometry Approach. Journal of the American Society for Mass Spectrometry. 2010;21:1633–1642. doi: 10.1016/j.jasms.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore KL. The Biology and Enzymology of Protein Tyrosine O-Sulfation. The Journal of Biological Chemistry. 2003;278:24243–24246. doi: 10.1074/jbc.R300008200. [DOI] [PubMed] [Google Scholar]
- 12.Huttner WB. Tyrosine Sulfation and the Secretory Pathway. Annual Review of Physiology. 1988;50:363–376. doi: 10.1146/annurev.ph.50.030188.002051. [DOI] [PubMed] [Google Scholar]
- 13.Stone MJ, Chuang S, Hou X, Shoham M, Zhu JZ. Tyrosine sulfation: an increasingly recognised post-translational modification of secreted proteins. New Biotechnology. 2009;25:299–317. doi: 10.1016/j.nbt.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Hille A, Huttner WB. Occurrence of tyrosine sulfate in proteins -- a balance sheet 2. Membrane proteins. European Journal of Biochemistry. 1990;188:587–596. doi: 10.1111/j.1432-1033.1990.tb15439.x. [DOI] [PubMed] [Google Scholar]
- 15.Kehoe JW, Bertozzi CR. Tyrosine sulfation: a modulator of extracellular protein–protein interactions. Chemistry & Biology. 2000;7:R57–R61. doi: 10.1016/s1074-5521(00)00093-4. [DOI] [PubMed] [Google Scholar]
- 16.Hortin G. Sulfation of tyrosine residues in coagulation factor V. Blood. 1990;76:946–952. [PubMed] [Google Scholar]
- 17.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. Tyrosine Sulfation of the Amino Terminus of CCR5 Facilitates HIV-1 Entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 18.Monigatti F, Hekking B, Steen H. Protein sulfation analysis—A primer. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2006;1764:1904–1913. doi: 10.1016/j.bbapap.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Seibert C, Sakmar TP. Toward a framework for sulfoproteomics: Synthesis and characterization of sulfotyrosine-containing peptides. Biopolymers. 2008;90:459–477. doi: 10.1002/bip.20821. [DOI] [PubMed] [Google Scholar]
- 20.Nemeth-Cawley JF, Karnik S, Rouse JC. Analysis of sulfated peptides using positive electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 2001;36:1301–1311. doi: 10.1002/jms.235. [DOI] [PubMed] [Google Scholar]
- 21.Wolfender J-L, Chu F, Ball H, Wolfender F, Fainzilber M, Baldwin MA, Burlingame AL. Identification of tyrosine sulfation in Conus pennaceus conotoxins α-PnIA and α-PnIB: further investigation of labile sulfo- and phosphopeptides by electrospray, matrix-assisted laser desorption/ionization (MALDI) and atmospheric pressure MALDI mass spectrometry. J. Mass Spectrom. 1999;34:447–454. doi: 10.1002/(SICI)1096-9888(199904)34:4<447::AID-JMS801>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 22.Medzihradszky KF, Guan S, Maltby DA, Burlingame AL. Sulfopeptide Fragmentation in Electron-Capture and Electron-Transfer Dissociation. Journal of the American Society for Mass Spectrometry. 2007;18:1617–1624. doi: 10.1016/j.jasms.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JEP, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yagami T, Kitagawa K, Aida C, Fujiwara H, Futaki S. Stabilization of a tyrosine O-sulfate residue by a cationic functional group: formation of a conjugate acid–base pair. The Journal of Peptide Research. 2000;56:239–249. doi: 10.1034/j.1399-3011.2000.00746.x. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Håkansson K. Electron Capture Dissociation of Tyrosine O-Sulfated Peptides Complexed with Divalent Metal Cations. Anal. Chem. 2006;78:7570–7576. doi: 10.1021/ac061352c. [DOI] [PubMed] [Google Scholar]
- 26.Cantel S, Brunel L, Ohara K, Enjalbal C, Martinez J, Vasseur J-J, Smietana M. An innovative strategy for sulfopeptides analysis using MALDI-TOF MS reflectron positive ion mode. Proteomics. 2012;12:2247–2257. doi: 10.1002/pmic.201100525. [DOI] [PubMed] [Google Scholar]
- 27.Yu Y, Hoffhines AJ, Moore KL, Leary JA. Determination of the sites of tyrosine O-sulfation in peptides and proteins. Nat Meth. 2007;4:583–588. doi: 10.1038/nmeth1056. [DOI] [PubMed] [Google Scholar]
- 28.Kim J-S, Song S-U, Kim H-J. Simultaneous Identification of Tyrosine Phosphorylation and Sulfation Sites Utilizing Tyrosine-Specific Bromination. J. Am. Soc. Mass Spectrom. 2011;22:1916–1925. doi: 10.1007/s13361-011-0214-9. [DOI] [PubMed] [Google Scholar]
- 29.Drake SK, Hortin GL. Improved detection of intact tyrosine sulfate-containing peptides by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in linear negative ion mode. The International Journal of Biochemistry & Cell Biology. 2010;42:174–179. doi: 10.1016/j.biocel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Edelson-Averbukh M, Shevchenko A, Pipkorn R, Lehmann W. Discrimination Between Peptide O-Sulfo- and O-Phosphotyrosine Residues by Negative Ion Mode Electrospray Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2011;22:2256–2268. doi: 10.1007/s13361-011-0248-z. [DOI] [PubMed] [Google Scholar]
- 31.Gibson BW, Cohen P. [26] Liquid secondary ion mass spectrometry of phosphorylated and sulfated peptides and proteins. In: McCloskey James A., editor. Methods in Enzymology. Academic Press; 1990. pp. 480–501. [DOI] [PubMed] [Google Scholar]
- 32.Cook S, Jackson G. Metastable Atom-Activated Dissociation Mass Spectrometry of Phosphorylated and Sulfonated Peptides in Negative Ion Mode. J. Am. Soc. Mass Spectrom. 2011;22:1088–1099. doi: 10.1007/s13361-011-0123-y. [DOI] [PubMed] [Google Scholar]
- 33.Hersberger KE, Håkansson K. Characterization of O-Sulfopeptides by Negative Ion Mode Tandem Mass Spectrometry: Superior Performance of Negative Ion Electron Capture Dissociation. Anal. Chem. 2012;84:6370–6377. doi: 10.1021/ac301536r. [DOI] [PubMed] [Google Scholar]
- 34.Brodbelt JS. Shedding Light on the Frontier of Photodissociation. J. Am. Soc. Mass Spectrom. 2011;22:197–206. doi: 10.1007/s13361-010-0023-6. [DOI] [PubMed] [Google Scholar]
- 35.Ly T, Julian RR. Ultraviolet Photodissociation: Developments towards Applications for Mass-Spectrometry-Based Proteomics. Angewandte Chemie International Edition. 2009;48:7130–7137. doi: 10.1002/anie.200900613. [DOI] [PubMed] [Google Scholar]
- 36.Reilly JP. Ultraviolet photofragmentation of biomolecular ions. Mass Spectrom. Rev. 2009;28:425–447. doi: 10.1002/mas.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madsen JA, Boutz DR, Brodbelt JS. Ultrafast Ultraviolet Photodissociation at 193 nm and its Applicability to Proteomic Workflows. Journal of Proteome Research. 2010;9:4205–4214. doi: 10.1021/pr100515x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madsen JA, Kaoud TS, Dalby KN, Brodbelt JS. 193-nm photodissociation of singly and multiply charged peptide anions for acidic proteome characterization. Proteomics. 2011;11:1329–1334. doi: 10.1002/pmic.201000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madsen JA, Xu H, Robinson MR, Horton AP, Shaw JB, Giles DK, Kaoud TS, Dalby KN, Trent MS, Brodbelt JS. High-Throughput Database Search and Large-Scale Negative Polarity LC-MS/MS with Ultraviolet Photodissociation for Complex Proteomic Samples. Molecular & Cellular Proteomics. 2013;12:2604–2614. doi: 10.1074/mcp.O113.028258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaw J, Madsen J, Xu H, Brodbelt J. Systematic Comparison of Ultraviolet Photodissociation and Electron Transfer Dissociation for Peptide Anion Characterization. J. Am. Soc. Mass Spectrom. 2012;23:1707–1715. doi: 10.1007/s13361-012-0424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madsen JA, Ko BJ, Robotham SA, Xu H, Horton AP, Iwashkiw JA, Shaw JB, Feldman MF, Brodbelt JS. Concurrent Automated Sequencing of the Glycan and Peptide Portions of O-Linked Glycopeptide Anions. Anal. Chem. 2013;85:9253–9261. doi: 10.1021/ac4021177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han S-W, Lee S-W, Bahar O, Schwessinger B, Robinson MR, Shaw JB, Madsen JA, Brodbelt JS, Ronald PC. Tyrosine sulfation in a Gram-negative bacterium. Nat Commun. 2012;3:1153. doi: 10.1038/ncomms2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasicek L, Ledvina A, Shaw J, Griep-Raming J, Westphall M, Coon J, Brodbelt J. Implementing Photodissociation in an Orbitrap Mass Spectrometer. Journal of The American Society for Mass Spectrometry. 2011;22:1105–1108. doi: 10.1007/s13361-011-0119-7. 1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaw JB, Li W, Holden DD, Zhang Y, Griep-Raming J, Fellers RT, Early BP, Thomas PM, Kelleher NL, Brodbelt JS. Complete Protein Characterization Using Top-Down Mass Spectrometry and Ultraviolet Photodissociation. J. Am. Chem. Soc. 2013;135:12646–12651. doi: 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu H, Freitas M. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu H, Yang L, Freitas M. A robust linear regression based algorithm for automated evaluation of peptide identifications from shotgun proteomics by use of reversed-phase liquid chromatography retention time. BMC Bioinformatics. 2008;9:347. doi: 10.1186/1471-2105-9-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu H, Freitas MA. MassMatrix: A database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu H, Freitas MA. Monte Carlo Simulation-Based Algorithms for Analysis of Shotgun Proteomic Data. J. Proteome Res. 2008;7:2605–2615. doi: 10.1021/pr800002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yagami T, Kitagawa K, Aida C, Fujiwara H, Futaki S. Stabilization of a tyrosine O-sulfate residue by a cationic functional group: formation of a conjugate acid–base pair. The Journal of Peptide Research. 2000;56:239–249. doi: 10.1034/j.1399-3011.2000.00746.x. [DOI] [PubMed] [Google Scholar]
- 50.Antoine R, Joly L, Tabarin T, Broyer M, Dugourd P, Lemoine J. Photo-induced formation of radical anion peptides. Electron photodetachment dissociation experiments. Rapid Commun. Mass Spectrom. 2007;21:265–268. doi: 10.1002/rcm.2810. [DOI] [PubMed] [Google Scholar]
- 51.Rumachik N, McAlister G, Russell J, Bailey D, Wenger C, Coon J. Characterizing Peptide Neutral Losses Induced by Negative Electron-Transfer Dissociation (NETD) J. Am. Soc. Mass Spectrom. 2012;23:718–727. doi: 10.1007/s13361-011-0331-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Q, Nelson H, Ly T, Stoltz BM, Julian RR. Side Chain Chemistry Mediates Backbone Fragmentation in Hydrogen Deficient Peptide Radicals. J. Proteome Res. 2008;8:958–966. doi: 10.1021/pr800592t. [DOI] [PubMed] [Google Scholar]
- 53.Straub RF, Voyksner RD. Negative ion formation in electrospray mass spectrometry. Journal of the American Society for Mass Spectrometry. 1993;4:578–587. doi: 10.1016/1044-0305(93)85019-T. [DOI] [PubMed] [Google Scholar]
- 54.Yamashita M, Fenn JB. Negative ion production with the electrospray ion source. J. Phys. Chem. 1984;88:4671–4675. [Google Scholar]
- 55.Hiraoka K, Kudaka I. Negative-mode electrospray-mass spectrometry using nonaqueous solvents. Rapid Commun. Mass Spectrom. 1992;6:265–268. [Google Scholar]
- 56.Cech NB, Enke CG. Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom. Rev. 2001;20:362–387. doi: 10.1002/mas.10008. [DOI] [PubMed] [Google Scholar]
- 57.Zhang X, Clausen MR, Zhao X, Zheng H, Bertram HC. Enhancing the Power of Liquid Chromatography-Mass Spectrometry-Based Urine Metabolomics in Negative Ion Mode by Optimization of the Additive. Anal. Chem. 2012;84:7785–7792. doi: 10.1021/ac3013835. [DOI] [PubMed] [Google Scholar]
- 58.McAlister GC, Russell JD, Rumachik NG, Hebert AS, Syka JEP, Geer LY, Westphall MS, Pagliarini DJ, Coon JJ. Analysis of the Acidic Proteome with Negative Electron-Transfer Dissociation Mass Spectrometry. Anal. Chem. 2012;84:2875–2882. doi: 10.1021/ac203430u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balsved D, Bundgaard JR, Sen JW. Stability of tyrosine sulfate in acidic solutions. Analytical Biochemistry. 2007;363:70–76. doi: 10.1016/j.ab.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 60.Balderrama GD, Meneses EP, Orihuela LH, Hernández OV, Franco RC, Robles VP, Batista CVF. Analysis of sulfated peptides from the skin secretion of the Pachymedusa dacnicolor frog using IMAC-Ga enrichment and high-resolution mass spectrometry. Rapid Commun. Mass Spectrom. 2011;25:1017–1027. doi: 10.1002/rcm.4950. [DOI] [PubMed] [Google Scholar]
- 61.Amano Y, Shinohara H, Sakagami Y, Matsubayashi Y. Ion-selective enrichment of tyrosine-sulfated peptides from complex protein digests. Analytical Biochemistry. 2005;346:124–131. doi: 10.1016/j.ab.2005.06.047. [DOI] [PubMed] [Google Scholar]
- 62.Hoffhines AJ, Damoc E, Bridges KG, Leary JA, Moore KL. Detection and Purification of Tyrosine-sulfated Proteins Using a Novel Anti-sulfotyrosine Monoclonal Antibody. Journal of Biological Chemistry. 2006;281:37877–37887. doi: 10.1074/jbc.M609398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.