Abstract
Purpose
To prepare mesoporous silica-based delivery systems capable of simultaneous delivery of drugs and nucleic acids.
Methods
The surface of mesoporous silica nanoparticles (MSN) was modified with poly(ethylene glycol) (PEG) and poly(2-(dimethylamino)ethylmethacrylate) (PDMAEMA) or poly (2-(diethylamino)ethylmethacrylate) (PDEAEMA). The particles were then loaded with a lysosomotropic agent chloroquine (CQ) and complexed with plasmid DNA or siRNA. The ability of the synthesized particles to deliver combinations of CQ and nucleic acids was evaluated using luciferase plasmid DNA and siRNA targeting luciferase and GAPDH.
Results
The results show a slow partial MSN dissolution to form hollow silica nanoparticles in aqueous solution. The biological studies show that polycation-modified MSN are able to simultaneously deliver CQ with DNA and siRNA. The co-delivery of CQ and the nucleic acids leads to a significantly increased transfection and silencing activity of the complexes compared with MSN not loaded with CQ.
Conclusion
PEGylated MSN modified with polycations are promising delivery vectors for combination drug/nucleic acid therapies.
Keywords: gene delivery, mesoporous, nanoparticles, polycation, silica, siRNA
Introduction
Combinations of two or more chemotherapeutic agents with pharmacodynamically synergistic or additive effects are effectively used in a number of cancer therapy protocols (1). Combining traditional small-molecule chemotherapeutic agents with nucleic acid-based drugs, such as genes and siRNA, provides additional flexibility in achieving synergistic or additive targets for combination therapies (2,3). Although drug/gene combinations have been investigated and used in the treatment of cancer, no proven method of systemic delivery is available to achieve simultaneous targeted drug/gene delivery to tumors. Typical drug/gene therapy approaches rely on intratumoral injection of a gene carrying virus combined with systemic chemotherapy and are thus ill-suited for the treatment of metastatic disease (4). In most cases, a successful drug/gene or drug/siRNA combination requires delivery of both agents to the same population of tumor cells in a coordinated manner. Because pharmacokinetics and disposition profiles of small-molecule drugs and nucleic acid drugs differ greatly, current combination therapies suffer from reduced efficacy. Thus, systems capable of targeted delivery of drug/gene and drug/siRNA combinations are urgently needed.
Silica materials have been found to be biocompatible and bioresorbable in many biomedical applications (5). Mesoporous silica nanoparticles (MSN) are particularly promising delivery vectors for drug/gene and drug/siRNA combinations because of high drug-loading capacity and compatibility with a large number of conventional chemotherapeutic agents. MSN have large surface areas and ordered porous channels that can be used to encapsulate a variety of small molecule chemotherapeutics, including doxorubicin, paclitaxel, and camptothecin (6–9). The well-established surface chemistry of MSN allows easy surface modification with polycations, thus enabling preparation of hybrid organic-inorganic particles that can also serve as carriers of nucleic acids. Thus, a major advantage of MSN over other gene and siRNA delivery systems is that the porous structure of MSN allows both the binding of nucleic acids on the surface as well as the encapsulation of large amounts of small molecules within the particles.
The ability of polycation-modified MSN to deliver genes and siRNA has been demonstrated by several labs in cell culture (10,11). Xia et al. prepared cationic MSN vehicles by surface adsorption of polyethyleneimine (PEI) and showed that such particles can deliver both siRNA and plasmid DNA (10). The presence of PEI did not hinder the possibility to load poorly soluble drugs like paclitaxel into the MSN. Recently, simultaneous delivery of doxorubicin and Bcl-2-targeted siRNA by MSN has been shown to suppress multidrug resistance and substantially enhance the anticancer action of doxorubicin (12). The MSN were modified with amine-terminated polyamidoamine dendrimers to allow siRNA binding using a method reported by Lin's group (11). The dendrimer-modified MSN loaded with doxorubicin could efficiently deliver siRNA targeted against mRNA encoding an antiapoptotic protein Bcl-2. The combined drug/siRNA delivery resulted in a substantially enhanced activity against human ovarian cancer cells in vitro.
The present study describes the preparation of polycation-modified MSN that are further stabilized by surface modification with poly(ethylene glycol) (PEG). The ability of the prepared systems to improve delivery of siRNA and plasmid DNA by loading the particles with chloroquine, a lysosomotropic agent, is reported.
Materials and Methods
Materials
Tetraethylorthosilicate (TEOS), 3-mercaptopropyltri-methoxysilane (MPTMS), 3-aminopropyltriethoxysilane (APTES), N-cetyltrimethylammonium bromide (CTAB), chloroquine (CQ) diphosphate salt, ammonium molybdate tetrahydrate, oxalic acid, sodium sulfite, 4-methylaminophenol sulfate, conc. hydrochloric acid, conc. sulfuric acid, 2,2′-dipyridyl disulfide (Aldrithiol-2), 2,2′-azobisisobutyronitrile (AIBN), 3-mercaptopropionic acid (MPA), 2-(N,N-diethylamino)ethyl methacrylate (DEAEMA) and 2-(N,N-dimethylamino)ethyl methacrylate (DMAEMA) were purchased from Sigma-Aldrich. PEG(2 kDa)-maleimide (PEG-Mal) was obtained from Laysan Bio, Inc. (Arab, AL). Dulbecco's modified Eagle's medium (DMEM), phosphate-buffered saline (PBS), fetal bovine serum (FBS), and Lipofectamine™ 2000 reagent were purchased from Invitrogen. High-expression luciferase DNA plasmid (gWiz-Luc) containing luciferase reporter gene was from Aldevron (Fargo, ND). Luciferase-targeted siRNA was from Insight Genomics (Santa Barbara, CA), and siRNA targeting GAPDH (cat # AM4624) and control nontargeted siRNA (cat #AM4611) were from Ambion.
Polycation Synthesis and Characterization
Carboxyl-terminated polymers of DEAEMA (PDEAEMA-COOH) and DMAEMA (PDMAEMA-COOH) were synthesized by AIBN-initiated free radical polymerization in the presence of MPA as a chain transfer agent. Briefly, DEAEMA (0.0264 mol), MPA (8.33×10−4 mol), and AIBN (9.83×10−4 mol) were dissolved in anhydrous DMF, deoxygenated by a stream of nitrogen, and the polymerization was carried out at 70°C for 15 h. Polymerization of DMAEMA was carried out by a similar route but in toluene. The polymers were isolated by precipitation with petroleum ether, and the content of the terminal carboxyl groups was determined by 1H-NMR in CDCl3. Average molecular weight and polydispersity of the polymers were determined by size exclusion chromatography in 0.2 M acetate buffer, pH 4.5.
Synthesis of Mesoporous Silica Nanoparticles (MSN)
Thiol- and amino-group functionalized MSN were synthesized by the co-condensation of TEOS, MPTMS and APTES, using modifications from previously reported procedures for singly-functionalized materials (13,14). One g of CTAB was dissolved in 480 mL of de-ionized water made basic by the addition of 3.5 mL of 2.0 M NaOH, and the temperature raised to 80°C. To this solution, 5.0 mL TEOS was injected at a rate of ∼1.0 mL/min using a syringe pump while stirring. The injection of TEOS was immediately followed by the drop-wise addition of MPTMS (2.6 mmol, 0.49 mL) and APTES (2.6 mmol, 0.61 mL), respectively, into the system, to achieve a ratio of TEOS: MPTMS: APTES of 8.7: 1: 1. The system was maintained at 80°C for about 2 h, and the final product was isolated by centrifugation. The isolated product was washed with excess de-ionized water, followed by methanol, and dried. Removal of the template (CTAB) was carried out by refluxing the dried product in acidic methanol solution (18 mL conc. HCl, 320 mL CH3OH) at 80°C overnight. The product was again isolated by centrifugation and then washed first with methanol, subsequently with nanopure water, and dried overnight under active vacuum in a dessicator to yield free-flowing powder. The accessible thiol (15) and amino (16) groups on the surface of the nanoparticles are estimated using literature procedures.
For dissolution studies, a simpler monofunctionalized (thiol only) MSN system was prepared. In order to increase the yield of the final product, the reported procedure (15) was slightly modified. One g of CTAB was first dissolved in 600 mL de-ionized water followed by addition of 3.5 mL 2.0 M NaOH. The system was then raised to 80°C followed by the drop-wise addition of 10.0 mL TEOS at the rate of 1.0 mL/min. This was then followed by the drop-wise addition of 1.98 mL MPTMS. The system was then maintained at 80°C for about 2 h and isolated as described above for the doubly-functionalized MSN.
Surface Modification of MSN with PEG and Polycations
PEG-Mal (4.0 g) solution in DMF was added to MSN (0.1 g) suspension in DMF, and the mixture was stirred for 24 h at room temperature. MSN coated with PEG (PMSN) were isolated by centrifugation and washing with DMF. Then, PMSN were reacted with N-hydroxysuccinimidyl (NHS) ester of either PDEAMA-COOH or PDMAEMA-COOH in DMF for 24 h at room temperature. The respective polycation NHS esters were prepared by a standard NHS/dicyclohexylcarbodiimide activation and used immediately after isolation. PDMAEMA-PMSN and PDEAMA-PMSN were isolated by centrifugation and extensive washing with DMF before vacuum drying.
Dissolution Studies of Thiol-Functionalized MSN by Silicomolybdic Acid Spectrophotometric Method
Thiol-functionalized MSN were dispersed in (1) pure de-ionized water (2), an aqueous solution of 50 mM TRIS (pH 7.4) and (3) DMEM buffered with 10 mM HEPES (pH 7.4). Two different concentrations of MSN, 0.5 mg/mL and 2 mg/mL, were used for this study. The nanoparticles were stirred at RT for 24 h. The systems were then centrifuged, and the supernatant was collected and stored separately. The nanoparticles deposited at the bottom of the centrifuge tube were subjected to TEM analyses to evaluate the extent of dissolution. The silicomolybdic assay was carried out with the collected supernatant according to a previously reported procedure (17). One mL of the supernatant was diluted to 16.0 mL by the addition of de-ionized water. To this was added 1.5 mL of an aqueous solution prepared from 0.2 g ammonium molybdate tetrahydrate and 0.6 ml concentrated hydrochloric acid in a 10 mL volume-tricflask. The yellow silicomolybdic acid formed over 10 min. Because the molar extinction coefficient of this yellow compound was small, the compound was further reduced to the molybdenum blue complex by the addition of 7.5 mL of a an aqueous solution prepared from oxalic acid (2 g), 4-methylaminophenol sulphate (0.667 g), anhydrous sodium sulfate (0.4 g), and concentrated sulfuric acid (10 mL) diluted with de-ionized water (50 mL) in a 100 mL volumetric flask. The blue color was allowed to develop for about 2 h while stirring at RT, and the concentration of the absorbing species was estimated by application of Beer's Law to the optical absorbance spectrum. The molybdenum blue complex exhibits an absorbance maxima at λ=810 nm with a molar absorptivity of 44,700±150 lmol−1cm−1.
Characterization of Particles
Particle sizes and the morphology of the materials were investigated by transmission electron microscopy (TEM) on a JEOL 2010F Analytical Electron Microscope at 200 kV. TEM samples were prepared by placing a drop of a sonicated methanol dispersion of MSN or an acetone dispersion of PMSN-PDEAEMA on a carbon-coated copper grid. The surface area, average pore size, cumulative pore volume, and pore size distributions were determined from nitrogen adsorption/desorption isotherms (Supplemental Material, Figure S1) acquired at 77 K using a 30 s equilibrium interval on an ASAP 2010 Micromeritics porosimeter. MSN samples were degassed at 100°C for 24 h and PMSN-PDEAEMA samples at 50°C for 12 h, in order to remove any adsorbed molecules prior to the analysis. The surface areas were computed using the Brunauer-Emmett-Teller (BET) model. The cumulative pore volumes were obtained from the BJH (Barret-Joyner-Halenda) model, and the pore size distribution was obtained from density functional theory (DFT) modeling using the DFT package of the Micromeritics V2.00 software over the entire range of the adsorption isotherm. Thermal gravimetric analysis (TGA) in air (Perkin-Elmer Pyris 1, 5°C/minute) was used to quantify the amount of PDEAEMA-PEG grafted onto the MSN, and FT-IR Spectroscopy (FT-IR, FT/IR-4200, JASCO, Japan) was used to verify the chemical identity of the conjugated polymers on the nanoparticles.
Chloroquine Loading and Release
CQ diphosphate salt was converted to free base by treatment with NaOH before loading in the silica nano-particles (PMSN, PMSN-PDEAMA, PMSN-PDMAEMA). The loading was performed by incubating 0.12 g CQ with 0.04 g particles in 4 mL of ethanol. The mixture was stirred for 24 h at room temperature, and the particles loaded with the drug were centrifuged at 12,000 rpm, briefly washed with ethanol, and dried in vacuo at 50°C. The amount of encapsulated CQ was calculated from the difference in the concentration of the initial CQ solution relative to the content of CQ in the supernatant and the washings after loading from absorbance at 330 nm. In a typical CQ release experiment, vials containing the CQ-loaded nano-particles with or without DNA were incubated in 0.2 M buffers of different pH (acetate buffer, pH 4.5 and 5.5; phosphate buffer, pH 6.5 and 7.4) and kept in a revolver rotator at 37°C. The release medium was periodically removed and replaced with the same volume of fresh medium to approximate infinite sink conditions. CQ content in the release medium was determined from the absorbance at 330 nm based on a standard calibration curve.
Ethidium Bromide Exclusion
DNA condensation by the silica particles was determined by ethidium bromide exclusion assay. DNA (20 μg/mL) was mixed with ethidium bromide (1 μg/mL) in 30 mM acetate buffer (pH 5.5) and fluorescence measured using 366 nm excitation and 590 nm emission and set to 100%. Background fluorescence was set to 0% using ethidium bromide (1 μg/mL) solution alone. Fluorescence readings were then taken following a stepwise addition of the silica particles, and condensation curves for each MSN were constructed.
Size and Zeta-Potential Measurement
Hydrodynamic diameter and zeta potential of the nano-particles and their DNA or siRNA complexes were measured by dynamic light scattering (DLS) using a ZetaPlus Particle Size and Zeta Potential analyzer (Brookhaven Instruments) equipped with a 35 mW solid state laser (658 nm). Plasmid DNA and siRNA complexes were prepared at indicated MSN-to-nucleic acid ratio in 0.02 M sodium acetate buffer, pH 5.5. The final concentration of DNA and siRNA was 20 μg/ml and 30 pmol/ml, respectively. The DNA and siRNA complexes were diluted with triply distilled water and 0.1% diethylpyrocarbonate in water that was passed through a 0.2 μm membrane filter prior to use, respectively. All samples were measured in triplicate.
Cytotoxicity
Cytotoxicity of the silica nanoparticles and their DNA and siRNA complexes in B16F10 murine melanoma cells was determined by MTS assay using a commercially available kit (CellTiter 96® Aqueous Cell Proliferation Assay, Promega). Ten-thousand cells seeded in a 96-well plate 24 h before the experiment were incubated with the particles in 100 μL of DMEM/FBS. The medium was removed after 24 h and replaced with a mixture of 200 μL fresh DMEM and 20 μL MTS reagent. The cells were incubated for 1 h at 37°C, and the absorbance of each sample was then measured at 505 nm to determine cell viability. The results were expressed as mean percentage cell viability relative to untreated cells ± S.D.
DNA and siRNA Transfection
Complexes of the silica nanoparticles with gWIZ-Luc plasmid DNA, anti-luciferase siRNA, anti-GAPDH siRNA, or scrambled siRNA were formulated at various MSN-to-nucleic acid ratios (0.5–20) in 0.02 M sodium acetate buffer, pH 5.5. The mixture was gently pipetted and vortexed for 3–5 s. All transfection studies were performed in 24-well plates with cells plated 24 h before transfection at seeding densities of 60,000 B16F10 cells per well. In experiments with different w/w ratio, the dose of nucleic acid was kept constant. The cells were incubated for 4 h with the complexes (2.4 μg DNA, 30 nm siRNA) in 500 μL of complete growth medium. After 4 h of incubation, the transfection mixture was completely removed, and the cells were cultured for an additional 24 h prior to analysis. The total protein content of the samples was measured using a BCA protein assay kit (Pierce, USA) and used to normalize the results. Luciferase DNA transfections are expressed as relative light units (RLU)/mg cellular protein±S.D. of triplicate samples. Silencing of the GAPDH housekeeping gene was determined using a commercial kit (KDalert, Catalog # AM1639) using a Synergy 2 microplate luminometer (BioTek, USA). The total protein content of the samples was measured using a GAPDH assay protocol and was used to normalize the GAPDH activity. Silencing activity of siRNA complexes is expressed as % GAPDH knockdown, relative to scrambled control. To evaluate siRNA silencing of transiently expressed luciferase, B16F10 cells were first transfected with luciferase plasmid DNA using Lipofectamine.
Results and Discussion
Synthesis and Stability Studies of MSN
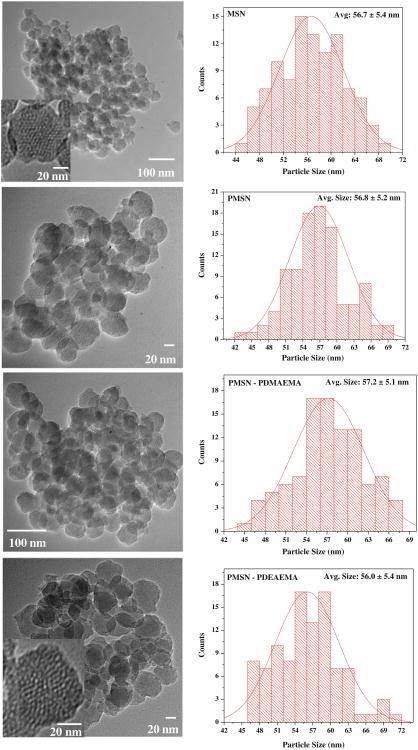
MSN co-functionalized with thiol and amino groups were prepared by co-condensation of TEOS, MPTMS and APTES in the presence of a mesostructured surfactant template of CTAB in water. Removal of the template by heating in acidic methanol yields silica nanoparticles with TEM-determined diameter of 56.7 nm and with 2–3 nm pore sizes displaying local hexagonal order (Fig. 1a). The presence of accessible pores was confirmed by porosimetry (Fig. S1). Modeling of nitrogen physisorption data revealed a BET surface area of over 400 m2/g and a narrow DFT pore size distribution with a peak at 2.0 nm (Fig. S2), consistent with the pore size estimated from TEM images. The quantity of accessible thiol and amine was assessed by colorimetric assays and found to be 9.12×10−5 mol -SH/g MSN and 1.42×10−4 moles of -NH2/g MSN, respectively (Fig. S3, S4).
Fig. 1.
TEM images and size distribution determined from TEM of the studied particles. From top to bottom: MSN, PMSN, PDMAEMA-PMSN, PDEAEMA-PMSN (insets: HRTEM showing porous structure of the particles).
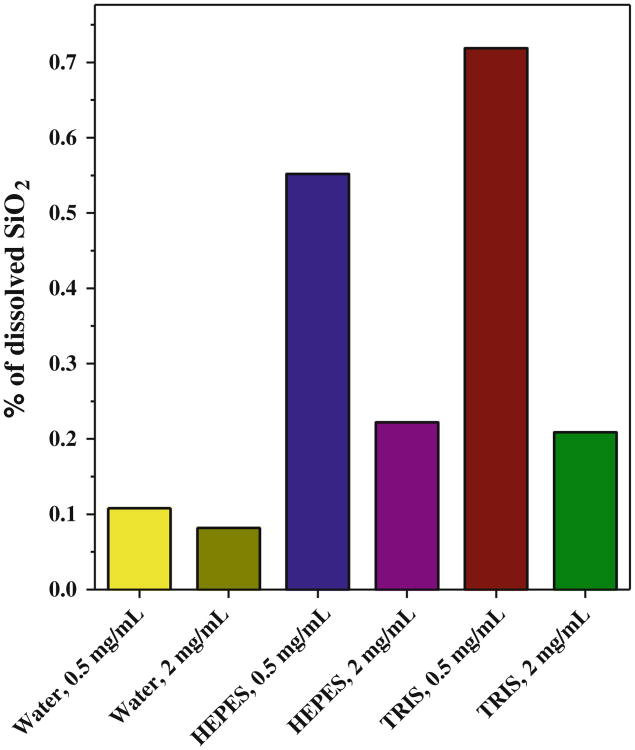
Easy dissolution of porous silica particles in simulated physiologic conditions has been described in numerous studies (18–20). In order to assess the stability of the synthesized MSN in an aqueous environment, a crucial factor for application in drug delivery, static dissolution studies of MSN at two different concentrations and incubations in three different buffers were conducted over 24 h. The extent of dissolution was probed qualitatively by TEM and quantitatively by a soluble silica assay. The TEM results in Fig. 2 show that during dissolution studies of 2 mg/mL of MSN, the mesoporous structure of the particles is preserved over the 24-h incubation period in pure water, suggesting little or no dissolution. Incubating the particles in DMEM and TRIS yielded samples wherein the linear pore channels are only evident in portions of the imaged particles, suggesting enhanced dissolution relative to pure water. This effect is exacerbated when the MSN concentration is low, 0.5 mg/mL, as evidenced by the nearly complete disappearance of mesopore channels and formation of hollow nanoparticles with similar diameter as the original particles. The formation of hollow nano-particles is consistent with the mechanism of MSN dissolution consisting of redeposition of silica at the entrance of the channels as described in a recent study (18). No such changes were observed in pure water, highlighting the importance of pH on silica dissolution (the pH of the 2 mg/mL MSN solution in deionized water was 5.1).
Fig. 2.
MSN dissolution. TEM images of MSN acquired after 24 h incubation of 0.5 and 2 mg/mL MSN in DMEM, TRIS, and deionized water.
Silica nanoparticles are hydrolyzed to form silicic acid, Si(OH)4, which, in the case of a systemic silica application, can then readily be excreted in the urine (21). The hydrolysis of silica proceeds from the surface and is catalyzed by OH−; hence, its rate increases rapidly with increasing pH and in particles with high surface area (18,19,22). To quantify the extent of dissolution of the mesoporous silica nanoparticles, the supernatant collected after incubation was subjected to the silicomolybdic acid spectrophotometric assay to measure the amount of soluble silicic acid (17). As shown in Fig. 3, the analysis indicated a higher percentage of dissolution for lower MSN concentrations (0.5 mg/mL), consistent with the TEM data. The most relevant results for the in vitro studies presented below were those in DMEM. The content of dissolved silica (silicic acid) after 24 h was 2.1 and 3.5 μg/mL for 0.5 and 2 mg/mL MSN, respectively. The observed silicic acid concentrations are below its equilibrium concentration (120– 140 μg/mL, pH 5-8), indicating slow kinetics of silica dissolution under the applied conditions (19). Our results confirm that MSN have sufficient stability and can largely maintain their mesoporous structure within the experimental timeframe (4 h incubation with cells, 24 h incubation after internalization into cells) of the described cell culture studies. While the observed extent of silica dissolution is acceptable for in vitro studies under static conditions, systemic application of MSN may require their additional stabilization. In such a case, the rate of hydrolysis can be reduced by a suitable modification of free silanol groups on the surface of silica (23).
Fig. 3.
MSN dissolution. The content of soluble silicic acid was determined by silicomolybdic acid assay after 24 incubation of MSN in deionized water, DMEM, and TRIS.
Polymer Modification of MSN
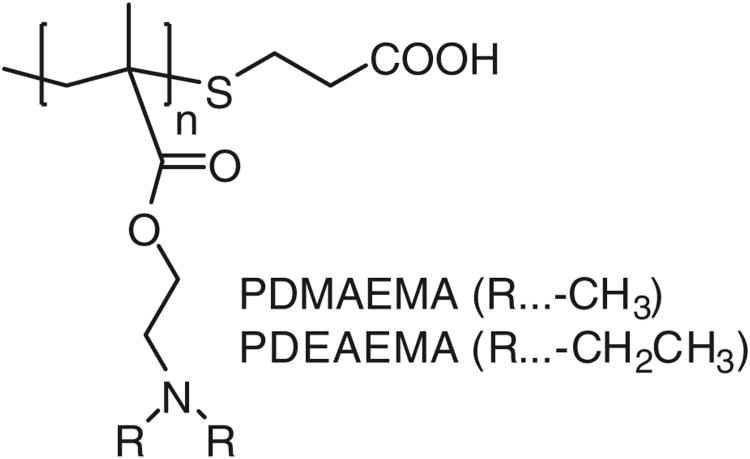
The use of MSN in drug/gene delivery requires additional means of steric stabilization due to their strong tendency to aggregate. Surface modification with polycations is known to prevent MSN aggregation, but complexing such particles with DNA or siRNA leads to an increased hydrophobicity and subsequent decrease in colloidal stability. Here, we modified the surface of MSN with polycations to enable DNA and siRNA binding, as well as with PEG to stabilize against aggregation. PEG2kDa-maleimide was reacted with the thiol groups of the MSN to covalently link PEG via a stable thioether bond, producing PEG-coated MSN (PMSN). Two types of carboxyl-terminated tertiary polyamines (Fig. 4) were synthesized by free radical polymerization in the presence of a chain transfer agent. The polycations were attached to the surface amines of PMSN via amide bond using standard carbodiimide chemistry. The resulting particles modified with both PEG and PDMAEMA (Mw 19.9 kDa) or PDEAEMA (Mw 22.9 kDa) were extensively washed to remove any free polycations that could interfere with the subsequent DNA and siRNA delivery experiments. We emphasize the fact that although the thiol and amine groups are present both in the pores of the particles and on the surface, the ∼2 nm pore size limits access of high molecular-weight polymers to the interior functional groups, thus restricting PEG and polycations to the surface of the particles (24). Accordingly, the synthesized particles contain both PEG and the polycations attached randomly to the surface. TEM analysis of the polymer-MSN composites revealed that the mesoporous structure and MSN size are essentially unchanged after polymer addition (Fig. 1).
Fig. 4.
Chemical structure of the polycations used for surface modification of MSN.
In order to verify the success of the attachment protocol, PDMAEMA-PMSN was characterized by IR spectroscopy, TGA and porosimetry. The presence of chemical functionalities in the composites inherent to the polymer was first confirmed by IR spectroscopy. Quantification of polymer content (PEG and polycations) was achieved by TGA. First, PEG content in PMSN was found to be 10.8 wt% from the difference of SiO2 content in the original MSN and PMSN (Fig. S5, Table S1). Polycation content was determined in a similar way and was found to be 8.2 wt% for PDEAEMA-PMSN and 36 wt% for PDMAEMA-PMSN. Finally, porosimetry revealed a significant decrease in surface area, consistent with pore blockage in the solid state. For example, the surface area of PDMAEMA-grafted MSN decreased to 70 m2/g, and DFT modeling revealed an absence of mesopores (Fig. S6). While porosimetry is not a quantitative assay for loading or modification, the absence of accessible pores in the solid state is consistent with what we would expect from successful surface modification of MSN by polymer (24).
Evaluation of DNA Complexation with MSN
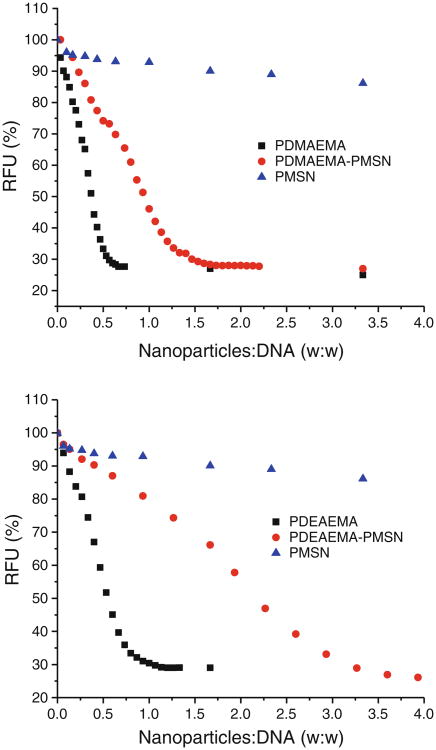
The ability of the prepared MSN to form complexes with plasmid DNA was evaluated by ethidium bromide exclusion assay (Fig. 5). As expected, both free polycations condensed DNA efficiently, while PEG-coated MSN (PMSN) did not condense DNA. Although PMSN contain free amines on the surface of the particles, their content is low, and they are shielded by PEG, as evidenced by their nearly neutral zeta potential (Fig. 6), thus preventing any significant DNA binding. Not surprisingly, of the two polycation-containing PMSN, the particles with the higher polycation content (PDMAEMA-PMSN) condensed DNA more efficiently (i.e., at a lower w:w ratio). However, the actual DNA binding ability of the two polycations attached to the particles was similar, as confirmed when re-plotting the ethidium bromide condensation data as RFU vs. ratio of polycation:DNA (not shown). The DNA condensation results show that the presence of PEG on the surface of the particles did not prevent DNA from binding to the polycations, most likely due to a relatively high molecular weight of both polycations relative to PEG.
Fig. 5.
DNA condensation by PEG-coated MSN (PMSN), PEG- and polycation-coated MSN (PDMAEMA-PMSN, PDEAEMA-PMSN) and free polycations (PDMAEMA, PDEAEMA). DNA condensation was determined by ethidium bromide exclusion assay using luciferase plasmid DNA and PDMAEMA-based (top) or PDEAEMA-based (bottom) particles (RFU: relative fluorescence units normalized to ethidium bromide/DNA fluorescence).
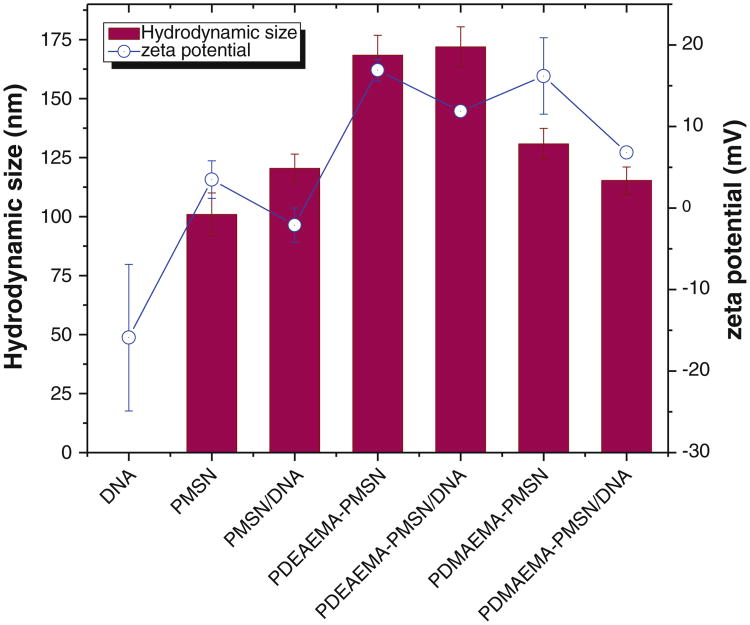
Fig. 6.
Hydrodynamic size and zeta potential of polymer-modified MSN and their DNA complexes. (particle:DNA w:w ratio 20).
Although the average size of the unmodified MSN was 56.7 nm by TEM (Fig. 1a), the unmodified particles exhibited colloidal instability (aggregation) in solution that prevented analysis of their size by DLS. Coating of the particles with PEG led to a substantially improved colloidal stability as demonstrated by the lack of aggregation and the observed hydrodynamic diameter of ∼100 nm (Fig. 6). Coating PMSN with the polycations further increased their hydrodynamic size to ∼130 nm in the case of PDMAEMA and to ∼170 nm in the case of PDEAEMA. The larger size of PDEAEMA-coated particles is most likely a reflection of some aggregation that is consistent with a more hydrophobic nature of PDEAEMA at neutral pH compared with PDMAEMA. The zeta potential of the particles also increased after polycation coating, from ∼3 mV for PMSN to ∼17 mV for both types of polycations. DNA binding resulted in decreased zeta potential of the particles but only small changes in their size. Increased scattering intensity, however, indicated increased molecular weight of the particles due to DNA binding. The absence of a large change in particle size suggests that DNA binding does not result in any significant aggregation of MSN and that the formed complexes most likely consist of a single MSN particle complexed with DNA. Because the complexes were analyzed at w:w 20 when there is a substantial excess of free MSN, caution has to be exercised when interpreting the DLS results due to distorting effect of the free particles on the measured hydrodynamic size.
CQ Release Studies
Various therapeutically active small molecules, including doxorubicin, paclitaxel, and ibuprofen have been successfully loaded into MSN. Here, we loaded chloroquine in MSN as a proof of principle that the particles can successfully deliver small molecule agents and nucleic acids. CQ is often used to enhance transfection of non-viral gene delivery vectors in vitro. CQ enhances transfection activity of polyplexes by several mechanisms, including (i) pH buffering of endocytic vesicles, (ii) protection of DNA against enzymatic degradation, and (iii) displacement of polycations from DNA in the polyplexes (25–28). Although CQ is safe to use in cell culture, to achieve the necessary concentrations for enhancing transfection in vivo requires toxic doses. We hypothesized that co-delivery of CQ with plasmid DNA and siRNA in a single particle may overcome the need for systemic exposure and eventually allow in vivo use of CQ. Internalization of CQ-loaded MSN carrying plasmid DNA into acidic endosomes or lysosomes was expected to result in rapid CQ release in endosomes and lysosomes due to its enhanced solubility in acidic pH.
CQ loading was conducted in ethanol to ensure that the polymers at the MSN surface are solvated and do not block the pore entrances. The presence of the PEG, nevertheless, reduced the total CQ loading from 0.73 mg/mg MSN in the case of unmodified MSN to 0.43 mg/mg MSN in the case of PMSN. When expressed per mg of SiO2, CQ loading decreased from 0.94 to 0.64 mg CQ/mg SiO2. Further decrease in CQ loading was observed in polycation-coated PMSN (0.15 and 0.32 mg/mg MSN in the case of PMSN-PDEAEMA and PMSN-PDMAEMA, respectively). Due to high polymer content in PDMAEMA- PMSN, the actual loading capacity of silica (SiO2) was in fact similar to that of the original MSN (1.04 vs. 0.94 mg CQ/mg SiO2) (Table S1).
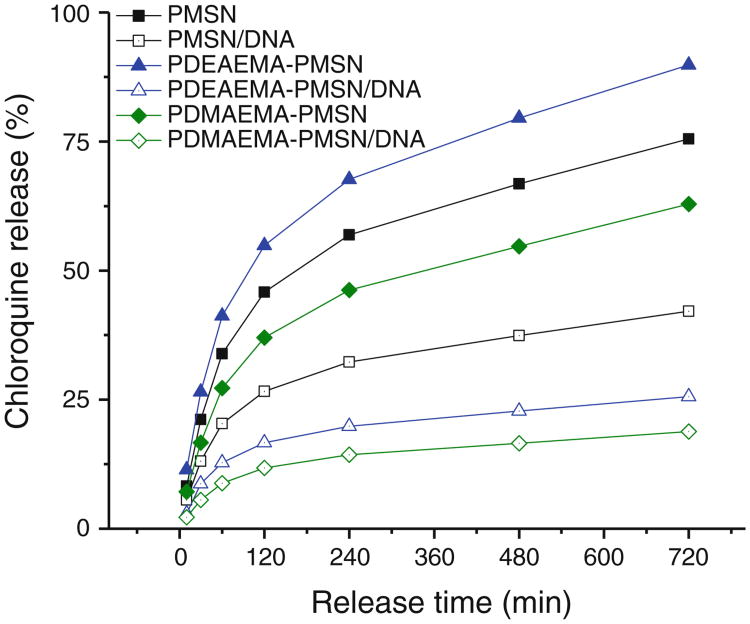
CQ release from PMSN unfortunately shows only a limited pH dependence which reduces its potential of selective release in acidic endo/lysosomes (not shown). CQ release data show fast release from PMSN (Fig. 7). The presence of polycations has a different effect on the rate of CQ release depending on the polycation type. However, because of the differences in polycation content in the particles, we are unable to determine if the differences are due to differences in chemical properties of the polycations or simply due to differences in the amount of polymer on the surface of the particles. The presence of the polycation layer containing complexed DNA creates an additional diffusional barrier for CQ release and may alter the release profile. We have therefore investigated the effect of DNA presence on the kinetics of CQ release. As shown in Fig. 7, the presence of DNA had a profound effect on CQ release in all cases, including PMSN. It is clear that the presence of DNA in the system reduces the rates of CQ release into the solution. The observation that even a mixture of PMSN with DNA, where no significant complexation between the particles and DNA was observed, causes similar, although not as profound, changes as in the case of polycation-coated particles, suggests that at least part of the observed changes are caused by CQ binding to the DNA. The additional decrease in the rate of release for the polycation-coated particles can be ascribed to the diffusional barrier created by DNA/polycation surface layer.
Fig. 7.
Kinetics of chloroquine release from polymer-modified MSN and their DNA complexes. (particle:DNA w:w ratio 20).
Cytotoxicity Studies
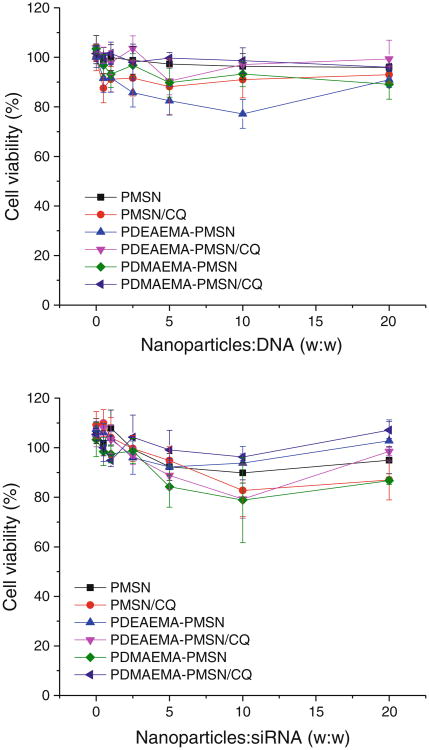
Existing reports suggest that MSN exhibit very low toxicity in vitro. In contrast, PDMAEMA and similar non-biodegradable polycations typically show significant toxicity both in vitro and in vivo (29,30). In addition to intracellular action, polycations exert their cytotoxicity also by perturbing plasma membranes. Surface modification of MSN with polycations can thus increase the toxicity of the particles. To evaluate cytotoxicity of the modified MSN, we have performed cell viability studies of DNA and siRNA complexes of the different types of MSN prepared in this study (Fig. 8). Our data suggest that all the tested formulations up to maximum particle:nucleic acid w/w ratio (20) used in subsequent transfections exhibit only limited signs of toxicity documented by the observed cell viability levels mostly above 80%. In addition, no statistically significant differences in cell viability were observed between DNA and siRNA complexes. The lack of observed toxicity of polycations immobilized on the surface of the nanoparticles is not surprising. Our recent study confirmed that toxicity of surface-adsorbed polycations is substantially decreased when compared with polycations in solution (31). In addition, the toxicity of polycations is also known to be reduced when complexed with DNA. These changes are related to the known effects of molecular weight, chain flexibility, and charge density on toxicity of polycations (32– 36). In general, flexible polycations with high molecular weight and charge density exhibit high cytotoxicity. Surface immobilization reduces polycation flexibility, while DNA complexation reduces both chain flexibility and charge density. All of these factors are then responsible for the observed low toxicity of polycation-coated MSN.
Fig. 8.
Toxicity of DNA (top) and siRNA (bottom) complexes of polymer-modified MSN with and without CQ in B16F10 cells.
Transfection Activity
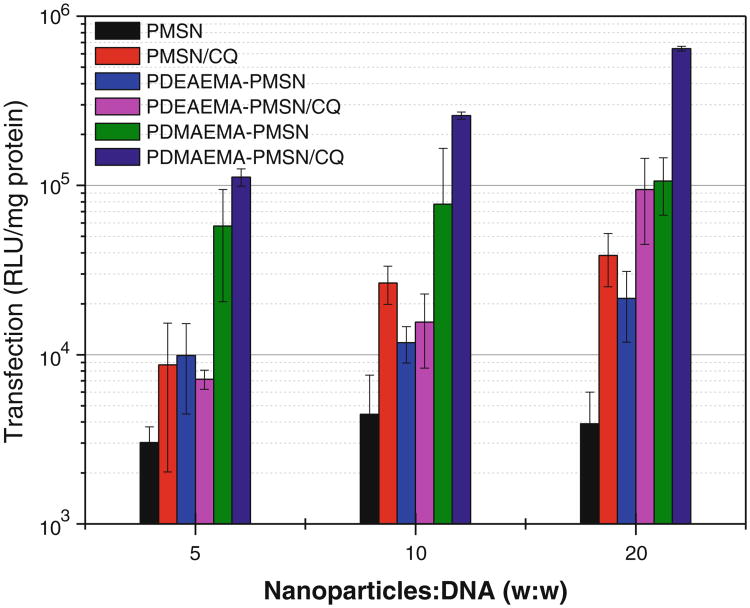
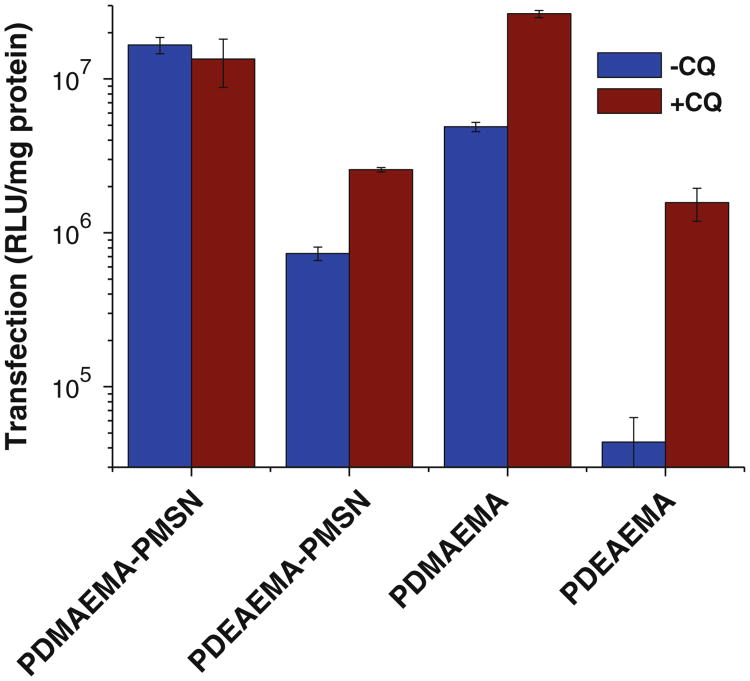
Transfection activity was first determined using luciferase plasmid DNA at three different w:w ratios of the particles to DNA using either empty or CQ-loaded particles (Fig. 9). In all cases, DNA complexes of PDMAEMA-PMSN exhibited the highest transfection activity. Although PDEAEMA-PMSN complexes showed higher transfection than PMSN in the absence of CQ, their overall activity was significantly lower than that of PDMAEMA-PMSN. Even comparison of the two formulations with similar polycation content (PDEAEMA-PMSN at w:w 20 and PDMAEMA-PMSN at w:w 5) shows higher activity of the PDMAEMA-containing particles. When CQ-loaded particles were used, PDEAEMA-PMSN showed higher transfection than PMSN only at the highest particle-to-DNA w:w ratio. The low observed transfection activity of PDEAEMA-PMSN is at least in part due to the lowest CQ loading in these particles. The enhancing effect of CQ on transfection activity increased with increasing w:w ratio, possibly suggesting the effect of the slow kinetics of CQ release from particles containing DNA and suboptimal loading in the particles at lower w:w ratios. To further evaluate the contribution of the mode of CQ delivery, we have conducted transfections with externally added CQ (Fig. 10). Our results show insensitivity of PDMAEMA-PMSN particles to CQ, while PDEAEMA-PMSN exhibited low sensitivity to externally added CQ. Both polycation-modified PSMN mediated higher transfection than the free polycations at corresponding w:w ratios (Fig. 10). Addition of external CQ increased transfection mediated by free polycations to levels comparable with those of polycation-modified PMSN.
Fig. 9.
Transfection activity of luciferase plasmid DNA complexes of polymer-modified MSN with and without CQ in B16F10 cells.
Fig. 10.
Effect of externally added CQ on the transfection activity of polymer-modified MSN and free polycations in B16F10 cells. (complexes with polycation-PMSN were made at w:w 20 and complexes with free polycations at the corresponding total polycation concentration).
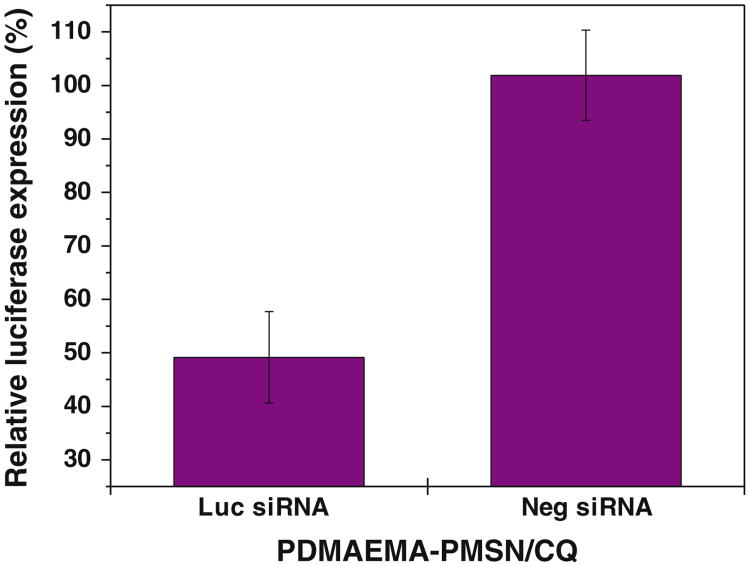
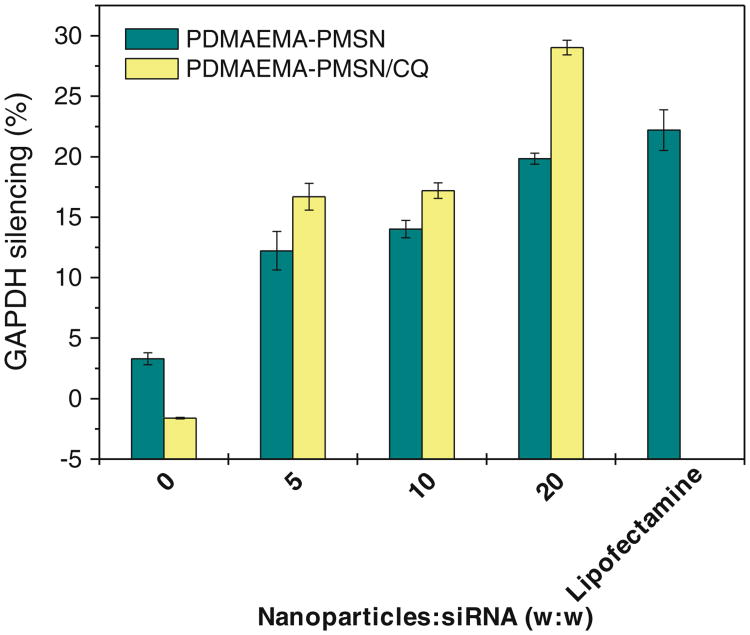
The ability of the polycation-coated PMSN to deliver siRNA was evaluated using only PDMAEMA-PMSN because of their superior performance in delivering DNA and because PDEAEMA-PMSN in our initial studies with siRNA were found to be very poor siRNA transfection agents. First, we have tested siRNA silencing of transiently expressed luciferase gene using CQ-loaded PDMAEMA-PMSN (Fig. 11). Luciferase expression decreased to ∼50% of the control non-transfected cells when anti-Luc siRNA was used, while no significant silencing was observed with control non-targeted siRNA. The ability of siRNA complexes of PDMAEMA-PMSN to silence endogenous gene expression was then determined using siRNA targeting GAPDH (Fig. 12). Naked siRNA mediated only low levels of GAPDH silencing (0–3%) regardless of the presence or absence of CQ. In contrast, significant silencing was observed for PDMAEMA-PMSN siRNA constructs, and the silencing efficiency increased with increasing particle: siRNA ratio, similar to our observations with plasmid DNA. Moreover, the use of CQ-loaded particles resulted in enhanced silencing activity at all tested ratios with the best improvement observed at the highest w:w ratio (from 20% to 29% GAPDH silencing).
Fig. 11.
Gene silencing by siRNA complexes of polymer-modified MSN. Luciferase silencing in transiently transfected cells using CQ-loaded PDMAEMA-PMSN particles (particle:siRNA w:w ratio 20) in B16F10 cells.
Fig. 12.
Silencing of endogenous GAPDH expression by siRNA complexes of PDMAEMA-PMSN.
Conclusion
In conclusion, this study described the synthesis of polycation- and PEG-coated mesoporous silica nanoparticles. The prepared particles were able to successfully deliver plasmid DNA and siRNA in cell culture. Loading the particles with chloroquine further enhanced transfection activity of both investigated nucleic acid types, confirming the suitability of the particles for combination drug/gene and drug/siRNA delivery. The observed loss of the mesoporous structure after incubation in cell culture media, however, raises concerns about the suitability of these particles for systemic delivery. A thorough investigation of the in vivo stability of the particles as well as determination of the effect of drug loading and polymer modification on particle dissolution are under investigation to further assess the usefulness of MSN in drug delivery.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant EB0043588 from the National Institute of Biomedical Imaging and Bioengineering.
Abbreviations
- APTES
3-aminopropyltriethoxysilane
- CQ
chloroquine
- CTAB
N-cetyltrimethylammonium bromide
- DMEM
Dulbecco's modified Eagle's medium
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- MPA
3-mercaptopropionic acid
- MPTMS
3-mercaptopropyltrimethoxysilane
- MSN
mesoporous silica nanoparticles
- PDEAEMA
poly(2-(diethylamino)ethylmethacrylate)
- PEG
poly(ethylene glycol)
- PEI
lyethyleneimine
- PDMAEMA
poly(2-(dimethylamino)ethylmethacrylate)
- PMSN
PEG-coated MSN
- TEOS
tetraethylorthosilicate
Footnotes
Electronic Supplementary Material: The online version of this article (doi:10.1007/s11095-010-0245-0) contains supplementary material, which is available to authorized users.
Contributor Information
Shanta Raj Bhattarai, Department of Pharmaceutical Sciences, Wayne State University, Detroit, Michigan 48202, USA.
Elayaraja Muthuswamy, Department of Chemistry, Wayne State University, Detroit, Michigan 48202, USA.
Amit Wani, Department of Pharmaceutical Sciences, Wayne State University, Detroit, Michigan 48202, USA.
Michal Brichacek, Department of Chemistry, Wayne State University, Detroit, Michigan 48202, USA.
Antonio L. Castañeda, Department of Chemistry, Wayne State University, Detroit, Michigan 48202, USA
Stephanie L. Brock, Department of Chemistry, Wayne State University, Detroit, Michigan 48202, USA
David Oupicky, Email: oupicky@wayne.edu, Department of Pharmaceutical Sciences, Wayne State University, Detroit, Michigan 48202, USA.
References
- 1.Jia J, Zhu F, Ma X, Cao ZW, Li YX, Chen YZ. Mechanisms of drug combinations: interaction and network perspectives. Nat Rev Drug Discov. 2009;8:111–28. doi: 10.1038/nrd2683. [DOI] [PubMed] [Google Scholar]
- 2.Yadav S, van Vlerken LE, Little SR, Amiji MM. Evaluations of combination MDR-1 gene silencing and paclitaxel administration in biodegradable polymeric nanoparticle formulations to overcome multidrug resistance in cancer cells. Cancer Chemother Pharmacol. 2009;63:711–22. doi: 10.1007/s00280-008-0790-y. [DOI] [PubMed] [Google Scholar]
- 3.Quist SR, Wang-Gohrke S, Kohler T, Kreienberg R, Runnebaum IB. Cooperative effect of adenoviral p53 gene therapy and standard chemotherapy in ovarian cancer cells independent of the endogenous p53 status. Cancer Gene Ther. 2004;11:547–54. doi: 10.1038/sj.cgt.7700727. [DOI] [PubMed] [Google Scholar]
- 4.Griffith TS, Stokes B, Kucaba TA, Earel JK, VanOosten RL, Brincks EL, et al. TRAIL gene therapy: from preclinical development to clinical application. Curr Gene Ther. 2009;9:9–19. doi: 10.2174/156652309787354612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viitala R, Jokinen M, Tuusa S, Rosenholm JB, Jalonen H. Adjustably bioresorbable sol-gel derived SiO2 matrices for release of large biologically active molecules. J Sol Gel Sci Technol. 2005;36:147–56. [Google Scholar]
- 6.Lebold T, Jung C, Michaelis J, Brauchle C. Nanostructured silica materials as drug-delivery systems for doxorubicin: single molecule and cellular studies. Nano Lett. 2009;9:2877–83. doi: 10.1021/nl9011112. [DOI] [PubMed] [Google Scholar]
- 7.Liong M, Lu J, Kovochich M, Xia T, Ruehm SG, Nel AE, et al. Multifunctional inorganic nanoparticles for imaging, targeting, and drug delivery. ACS Nano. 2008;2:889–96. doi: 10.1021/nn800072t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vivero-Escoto JL, Slowing II, Wu CW, Lin VS. Photoinduced intracellular controlled release drug delivery in human cells by gold-capped mesoporous silica nanosphere. J Am Chem Soc. 2009;131:3462–3. doi: 10.1021/ja900025f. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Liong M, Zink JI, Tamanoi F. Mesoporous silica nano-particles as a delivery system for hydrophobic anticancer drugs. Small. 2007;3:1341–6. doi: 10.1002/smll.200700005. [DOI] [PubMed] [Google Scholar]
- 10.Xia T, Kovochich M, Liong M, Meng H, Kabehie S, George S, et al. Polyethyleneimine coating enhances the cellular uptake of mesoporous silica nanoparticles and allows safe delivery of siRNA and DNA constructs. ACS Nano. 2009;3:3273–86. doi: 10.1021/nn900918w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radu DR, Lai CY, Jeftinija K, Rowe EW, Jeftinija S, Lin VSY. A polyamidoamine dendrimer-capped mesoporous silica nanosphere-based gene trasfection reagent. J Am Chem Soc. 2004;126:13216–7. doi: 10.1021/ja046275m. [DOI] [PubMed] [Google Scholar]
- 12.Chen AM, Zhang M, Wei D, Stueber D, Taratula O, Minko T, et al. Co-delivery of doxorubicin and Bcl-2 siRNA by mesoporous silica nanoparticles enhances the efficacy of chemotherapy in multidrug-resistant cancer cells. Small. 2009;5:2673–7. doi: 10.1002/smll.200900621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai CY, Trewyn BG, Jeftinija DM, Jeftinija K, Xu S, Jeftinija S, et al. A mesoporous silica nanosphere-based carrier system with chemically removable cds nanoparticle caps for stimuli-responsive controlled release of neurotransmitters and drug molecules. J Am Chem Soc. 2003;125:4451–9. doi: 10.1021/ja028650l. [DOI] [PubMed] [Google Scholar]
- 14.You YZ, Kalebaila KK, Brock SL, Oupický D. Temperature-controlled uptake and release in PNIPAM-modified porous silica nanoparticles. Chem Mater. 2008;20:3354–9. [Google Scholar]
- 15.Radu DR, Lai CY, Huang J, Shu X, Lin VSY. Fine-tuning the degree of organic functionalization of mesoporous silica nano-sphere materials via an interfacially designed co-condensation method. Chemical Communications. 2005:1264–1266. doi: 10.1039/b412618a. [DOI] [PubMed] [Google Scholar]
- 16.Rosenholm JM, Linden M. Wet-chemical analysis of surface concentration of accessible groups on different amino-functionalized mesoporous SBA-15 silicas. Chem Mater. 2007;19:5023–34. [Google Scholar]
- 17.Coradin T, Eglin D, Livage J. The silicomolybdic acid spectrophotometric method and its application to silicate/biopolymer interaction studies. Spectrosc Int J. 2004;18:567–76. [Google Scholar]
- 18.Galarneau A, Nader M, Guenneau F, Di Renzo F, Gedeon A. Understanding the stability in water of mesoporous SBA-15 and MCM-41. J Phys Chem C. 2007;111:8268–77. [Google Scholar]
- 19.Finnie KS, Waller DJ, Perret FL, Krause-Heuer AM, Lin HQ, Hanna JV, et al. Biodegradability of sol-gel silica microparticles for drug delivery. J Sol-Gel Sci Technol. 2009;49:12–8. [Google Scholar]
- 20.Lin YS, Haynes CL. Impacts of mesoporous silica nanoparticle size, pore ordering, and pore integrity on hemolytic activity. J Am Chem Soc. 2010;132:4834–42. doi: 10.1021/ja910846q. [DOI] [PubMed] [Google Scholar]
- 21.Jugdaohsingh R, Reffitt DM, Oldham C, Day JP, Fifield LK, Thompson RPH, et al. Oligomeric but not monomeric silica prevents aluminum absorption in humans. Am J Clin Nutr. 2000;71:944–9. doi: 10.1093/ajcn/71.4.944. [DOI] [PubMed] [Google Scholar]
- 22.Klause M, Rothhaar U, Bicker M, Ohling W. Dissolution of thin SiO2-coatings - characterization and evaluation. J Non-Cryst Solids. 2010;356:141–6. [Google Scholar]
- 23.Abou El Sherbini KS, Pape C, Rienetz O, Schiel D, Stosch R, Weidler PG, et al. Stabilization of n-aminopropyl silica gel against hydrolysis by blocking silanol groups with TiO2 or ZrO2. J Sol-Gel Sci Technol. 2010;53:587–97. [Google Scholar]
- 24.You YZ, Kalebaila KK, Brock SL, Oupicky D. Temperature-controlled uptake and release in PNIPAM-modified porous silica nanoparticles. Chem Mater. 2008;20:3354–9. [Google Scholar]
- 25.Yang S, Coles DJ, Esposito A, Mitchell DJ, Toth I, Minchin RF. Cellular uptake of self-assembled cationic peptide-DNA complexes: multifunctional role of the enhancer chloroquine. J Control Rel. 2009;135:159–65. doi: 10.1016/j.jconrel.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 26.Gabrielson NP, Pack DW. Efficient polyethylenimine-mediated gene delivery proceeds via a caveolar pathway in HeLa cells. J Control Release. 2009;136:54–61. doi: 10.1016/j.jconrel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Cheng J, Zeidan R, Mishra S, Liu A, Pun SH, Kulkarni RP, et al. Structure-function correlation of chloroquine and analogues as transgene expression enhancers in nonviral gene delivery. J Med Chem. 2006;49:6522–31. doi: 10.1021/jm060736s. [DOI] [PubMed] [Google Scholar]
- 28.Oupicky D, Carlisle RC, Seymour LW. Triggered intracellular activation of disulfide crosslinked polyelectrolyte gene delivery complexes with extended systemic circulation in vivo. Gene Ther. 2001;8:713–24. doi: 10.1038/sj.gt.3301446. [DOI] [PubMed] [Google Scholar]
- 29.Luten J, van Nostrum CF, De Smedt SC, Hennink WE. Biodegradable polymers as non-viral carriers for plasmid DNA delivery. J Control Rel. 2008;126:97–110. doi: 10.1016/j.jconrel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 30.de Wolf HK, de Raad M, Snel C, van Steenbergen MJ, Fens MH, Storm G, et al. Biodegradable poly(2-dimethylamino ethylamino) phosphazene for in vivo gene delivery to tumor cells. Effect of polymer molecular weight. Pharm Res. 2007;24:1572–80. doi: 10.1007/s11095-007-9299-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blacklock J, You YZ, Zhou QH, Mao G, Oupicky D. Gene delivery in vitro and in vivo from bioreducible multilayered polyelectrolyte films of plasmid DNA. Biomaterials. 2009;30:939–50. doi: 10.1016/j.biomaterials.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunter AC. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Adv Drug Deliv Rev. 2006;58:1523–31. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24:1121–31. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 34.Malik N, Wiwattanapatapee R, Klopsch R, Lorenz K, Frey H, Weener JW, et al. Dendrimers: relationship between structure and biocompatibility in vitro, and preliminary studies on the biodistribution of 125I-labelled polyamidoamine dendrimers in vivo. J Control Release. 2000;65:133–48. doi: 10.1016/s0168-3659(99)00246-1. [DOI] [PubMed] [Google Scholar]
- 35.Fischer D, Bieber T, Li Y, Elsasser HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999;16:1273–9. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- 36.Slita AV, Kasyanenko NA, Nazarova OV, Gavrilova II, Eropkina EM, Sirotkin AK, et al. DNA-polycation complexes: effect of polycation structure on physico-chemical and biological properties. J Biotechnol. 2007;127:679–93. doi: 10.1016/j.jbiotec.2006.07.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.