Summary
Background
Proteasome inhibitors are widely used in treatment of multiple myeloma and as research tools. Diminished proteasome function also may contribute to neuronal dysfunction. In response to these inhibitors, cells enhance the expression of proteasome subunits by the transcription factor Nrf1. Here we investigate the mechanisms by which decreased proteasome function triggers production of new proteasomes via Nrf1.
Results
Exposure of myeloma or neuronal cells to proteasome inhibitors (bortezomib, epoxomicin, MG132), but not to proteotoxic or ER stress, caused a 2–4-fold increase within 4h in mRNAs for all 26S subunits. In addition, p97 and its cofactors (Npl4, Ufd1, p47), PA200 and Usp14 were induced, but expression of immunoproteasome-specific subunits was suppressed. Nrf1 mediates this induction of proteasomes and p97, but only upon exposure to low concentrations of inhibitors that inhibit proteolysis partially. Surprisingly, high concentrations of these inhibitors prevent this compensatory response. Nrf1 is normally ER-bound, and its release requires its deglycosylation and ubiquitination. Normally ubiquitinated Nrf1 is rapidly degraded, but when partially inhibited, proteasomes carry out limited proteolysis and release the processed Nrf1 (lacking its N-terminal region) from the ER, which allows it to enter the nucleus and promote gene expression.
Conclusions
When fully active, proteasomes degrade Nrf1, but when partially inhibited, they perform limited proteolysis which generates the active form of Nrf1. This elegant mechanism allows cells to compensate for reduced proteasome function by enhancing production of 26S subunits and p97.
Introduction
The ubiquitin-proteasome system (UPS) catalyzes the degradation of most proteins in eukaryotic cells. In the UPS, substrates are targeted for degradation by 26S proteasomes by attachment of a chain of ubiquitins (Ub). Most ubiquitinated proteins are then rapidly degraded by the 26S proteasome. This ATP-dependent proteolytic complex consists of the 20S proteolytic particle capped by one or two 19S regulatory particles, which bind poly-ubiquitinated proteins and catalyze their unfolding and translocation into the 20S particle[1]. Proteasome function is also regulated by the association of the 20S with additional regulatory complexes[2], whose precise physiological importance is still unclear (e.g. PA28αβ, the γ-interferon-induced complex that functions in antigen presentation, PA28γ, and PA200/Blm10). Also associated with the 26S proteasome are the de-ubiquitinating enzymes (DUBs) Usp14 and Uch37, which help recycle Ub, but also regulate the particle’s (proteolytic and ATPase) activities[3]. The degradation of many Ub conjugates additionally requires the p97/VCP/Cdc48 ATPase complex, which functions with cofactors to extract ubiquitinated proteins from larger structures to facilitate their degradation by proteasomes[4]. The best characterized role of p97 is in the Endoplasmic Reticulum(ER)-associated degradation (ERAD)[5], by which misfolded proteins in the ER are ubiquitinated and extracted by p97 in complex with Npl4, Ufd1, and p47[6].
Proteasomes are essential for cell viability, but are especially important in multiple myeloma cells[7]. Consequently, proteasome inhibitors such as bortezomib (BTZ) or carfilzomib have become the preferred therapy for this cancer. One challenge in their use is the occurrence of drug resistance, but the responsible mechanisms are largely unclear[8]. Therefore, information on how cells compensate for proteasome inhibition is of appreciable interest. Upon proteasome inhibition, mammalian cells show increased expression of multiple 26S subunits, which elevate proteasome content and promotes survival[9–11]. This response limits the ability of proteasome inhibitors to kill myeloma cells. Thus, blocking this compensatory response may enhance the efficacy of this treatment. Conversely, decreased proteasome function due to the accumulation of aggregation-prone proteins seems to be important in the pathogenesis of neurodegenerative diseases[12], and pharmacological induction of proteasomes may enhance the cells’ degradative capacity and prevent the accumulation of toxic proteins.
In mammals, upon proteasome inhibition, the transcription factor Nrf1 mediates the induction of genes encoding many 26S subunits[9, 10]. Loss of Nrf1 makes cells more sensitive to killing by proteasome inhibitors. Nrf1, like its homolog, Nrf2, recognizes antioxidant response elements (ARE) in the promoters of many proteasome genes[9, 10]. However, Nrf2 induces 26S subunits only during oxidative stress, but not upon proteasome inhibition. Nrf1 is degraded by the UPS with a half-life of only ~12min)[10]. Upon proteasome inhibition, ubiquitinated Nrf1 is readily detected, and several Ub ligases (Hrd1, Fbw7, β-TRCP) have been implicated in Nrf1 ubiquitination [10, 13, 14]. It is currently believed that proteasome inhibitors activate Nrf1 by blocking its rapid degradation[10, 14]. One basic problem with this simple mechanism is that it does not explain why Nrf2 does not play a similar role, since it is also degraded by the UPS with a half-life of ~13 min[15]. An unusual characteristic of Nrf1 is that it is ER-associated. Therefore, Nrf1 activation requires its release from the ER via proteolytic processing by an unidentified protease[10], which presumably is activated upon proteasome inhibition.
It is also unclear whether proteasome inhibition also induces coordinately the expression of other components of the UPS, such as PA28, immunoproteasome subunits, proteasome assembly chaperones (POMP, p27, S5b, gankyrin)[16, 17], 26S-associated DUBs (Usp14 and Uch37), and p97 plus its major cofactors. Here we investigated whether exposure to proteasome inhibitors causes coordinate induction of all 26S subunits and these related factors. These studies have focused on neuroblastoma and myeloma cells, due to the importance of proteasome inhibitors in myeloma therapy and the apparent decreased proteasome function in neurodegenerative disease. These two types of cells are also interesting to study because myeloma cells are especially sensitive to killing by proteasome inhibitors but neurons are relatively insensitive. We also attempted to determine how quickly these genes are induced after proteasome inhibition, whether all proteasome inhibitors have similar effects, and how the degree of inhibition of proteolysis influences this response. These studies uncovered the surprising finding that high concentrations of these agents were less effective than low ones in inducing this Nrf1-dependent response. Studies to understand this unusual concentration dependence indicated that some proteasome function is necessary to catalyze the proteolytic processing of Nrf1 on the ER, which allows the processed Nrf1 (lacking its N-terminal region) to enter the nucleus and enhance expression of 26S subunits and p97.
Results
Proteasome inhibition rapidly induces 26S subunits independently of the unfolded protein response
In the neuroblastoma line SH-SY5Y, treatment with a low concentration (10nM) of BTZ for 16h caused a maximal (2–4-fold) increase in mRNAs for all 33 proteasome subunits (Fig. 1A,S1A), and about a 50% increase in their protein levels (Fig. 1B). The magnitude of this induction was similar for both 19S and 20S subunits, including Rpn6[18] or Rpn11[19] that were reported to be regulated independently, and the loosely associated subunit, Ecm29 (Fig. S1B). At higher concentrations (1µM), BTZ, caused a clear induction of these mRNAs in 4h (Fig. S1C), although the magnitude was much smaller than at 16h. A similar induction was detected in another neuroblastoma line, M17, the myeloma line, MM1.S, and HEK293A cells (Fig. S1D), although these lines differ in the degree of induction. SH-SY5Y cells induced 26S subunits much more strongly than others. Therefore, we mainly used SH-SY5Y cells in subsequent experiments. Several other proteasome inhibitors, including MG262 (Z-LLL-boronate) and epoxomicin (Epox), caused a similar induction of 26S subunits (Fig. S1E). Thus, their induction appears to be a general cellular mechanism to compensate for reduced proteasome function.
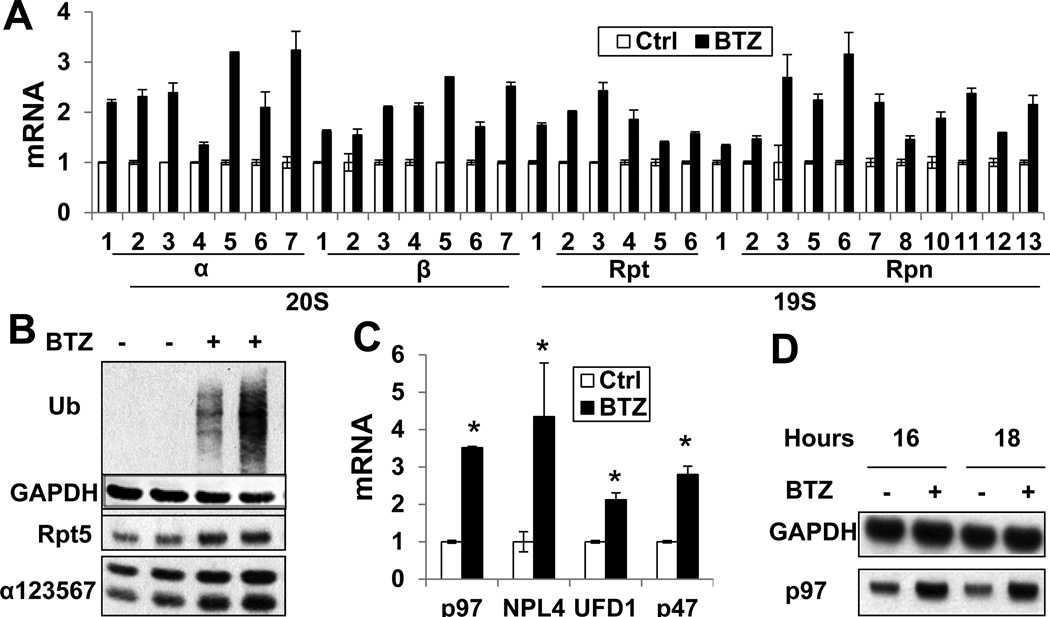
Figure 1. Proteasome inhibitor cause induction of all 26S proteasome subunits, p97, and its cofactors in ERAD (See also Fig. S1).
(A) Treatment of neuroblastoma SH-SY5Y cells with BTZ (10nM, 16h) induced mRNAs for all 26S subunits, measured by RT-PCR. (p<0.05 for all changes) (B) and increased the levels of Rpt5 and 20S subunits measured by Western blot (WB). Biological duplicates were assayed, and levels of Ub conjugates were measured to prove proteasome inhibition. (C) Treatment of SH-SY5Y cells with BTZ (10nM, 16h) also induced mRNAs for p97 and its cofactors, Npl4, Ufd1, and p47, and (D) increased content of p97 protein. (*: p<0.05). Error bars represent standard deviation (Error bar=SD).
The unfolded protein response (UPR) is triggered by the accumulation of misfolded proteins in the ER and is strongly induced by proteasome inhibitors[20]. We therefore tested whether the UPR per se induces 26S expression. Unlike BTZ, the UPR inducers, tunicamycin or thapsigargin, did not stimulate 26S induction (Fig. S1E–H). Also, low concentrations of BTZ induced 26S subunits without causing the UPR (i.e. eIF2α phosphorylation, Fig. S1F) Thus, proteasome induction is not mediated by the UPR.
Components of the UPS that are induced with 26S subunits
We next determined whether these inhibitors also stimulated expression of several proteasome-associated proteins. In response to γ-interferon, cells induce three distinct catalytic subunits (immunoproteasome subunits), β1i, β2i, and p5i, which are more efficient in generating peptides suitable for MHC-I antigen presentation[21]. Unlike the standard subunits, expression of immunoproteasome subunits decreased by 50–70% (Fig. S1I). PA28αβ are also induced by γ-interferon and can also enhance antigen presentation[22]. BTZ treatment caused a small decrease in PA28αβ mRNA expression. BTZ did not alter the expression of their nuclear homolog, PA28γ (Fig. S1I). However, BTZ treatment did induce the other nuclear activator, PA200/Blm10 (Fig. S1I).
The assembly of newly-synthesized subunits into mature proteasomes requires assembly chaperones. BTZ treatment induced the 20S-chaperone POMP[17], but not the chaperones involved in assembly of the 19S base, S5b, p27, or Gankyrin[16] (Fig. S1J). The 26S-associated DUBs, Usp14 and Uch37, catalyze disassembly of Ub chains on substrates and can promote protein deubiquitination without degradation[3], but they also allosterically regulate 20S gate opening and ATP hydrolysis[23]. Treatment with BTZ or MG132 for 16h induced the mRNA for Usp14 2-fold, but surprisingly did not affect Uch37 expression (Fig. S1K). P97 and its co-factors Npl4, Ufd1, and p47, are essential for degradation of many proteins, especially by ERAD. These genes were induced 2–5-fold by BTZ treatment coordinately with 26S subunits (Fig. 1C–D,S1C). However, BTZ did not induce Rad23A/B, which in ERAD shuttle Ub conjugates to proteasomes (Fig. S1L).
Thus, upon proteasome inhibition, mammalian cells induce all 26S components, Usp14, PA200, and the p97 complex, which together should enhance the cell’s capacity for proteolysis. Furthermore, BTZ treatment increased expression of the polyubiquitin gene UBB (2–3-fold, Fig. S1M). Since Ub conjugates accumulate upon proteasome inhibition and free Ub can be depleted[24], its increased production presumably compensates for decreased Ub recycling and enhances the cell’s capacity for proteolysis.
Proteasome inhibitors at high concentrations inhibit the induction of 26S subunits
All the agents used in this study inhibit preferentially the proteasome’s chymotrypsin-like activity but vary in their affinities for this site. MG132 is commonly used at much higher concentrations than BTZ, Epox, and MG262. Treatment of SH-SY5Y cells with 10µM MG132 for 16h blocked the chymotrypsin-like site without causing substantial cell death (Fig. S2A–C), but surprisingly, did not induce any α-subunits, 6 of the β-subunits, and many 19S subunits (Rpt4/5, Rpn3/8/10/12) (Fig. 2A). In addition, MG132 caused a smaller induction of Rpt3, and Rpn3/6/14) than did BTZ, though it caused a similar induction of Rpt2 and Rpn1/2/5/7/13. Strikingly, when cells were exposed to both MG132 (10µM) and BTZ (10nM) (Fig. S2DE), the inhibitory effect of MG132 was dominant over the induction of 26S and p97 by low concentrations of BTZ or other inhibitors. Furthermore, in contrast to 10µM, lower MG132 concentrations (1–2.5µM) did induce the expression of 26S subunits (Fig. S2F), as reported previously[9, 10].
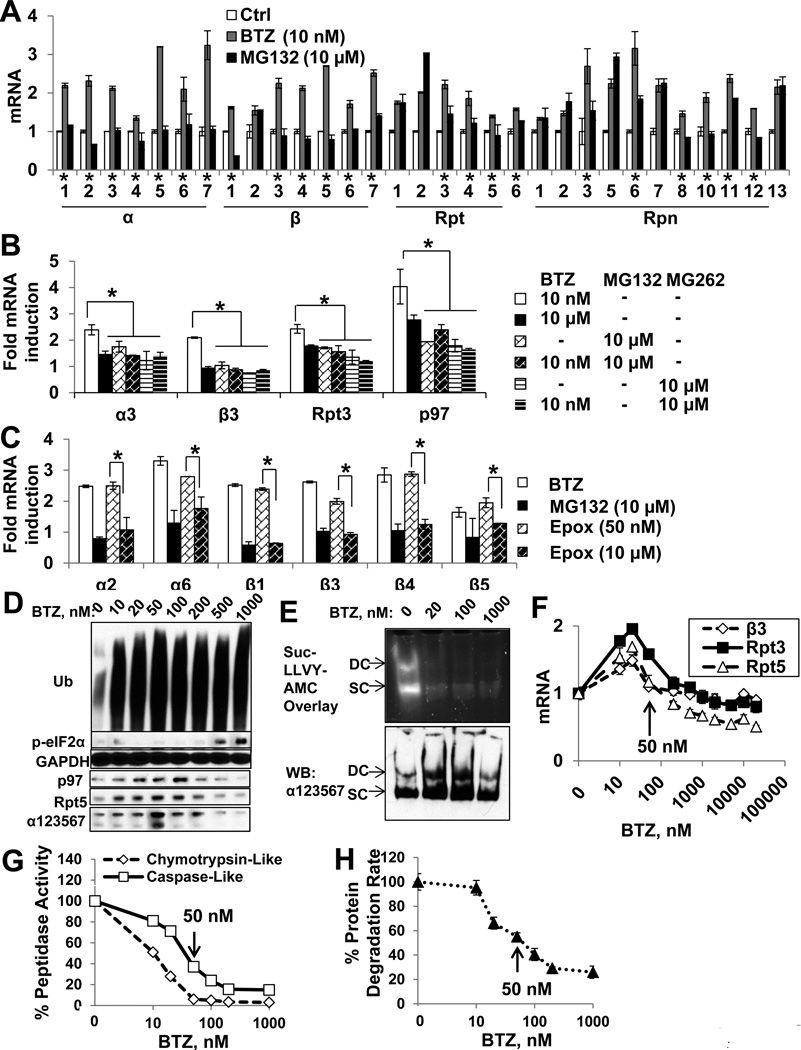
Figure 2. High concentrations of proteasome inhibitors block the induction of 26S subunits (See also Fig. S2).
(A) Unlike treatment of SH-SY5Y cells with low concentrations of BTZ (10nM), 10µM MG132 for 16h did not induce the mRNA for 20S subunits and many 19S subunits. (*:genes whose mRNA levels are lower, (p<0.05) in MG132-treated than BTZ-treated cells.) (B) High concentrations (10µM) of BTZ or MG262, alone or in combination with 10nM BTZ, suppressed the induction of a3, β3, Rpt3, and p97. (C) Treatment with Epox at 10µM, but not 50nM, suppressed the induction of 20S subunit mRNAs. *: p<0.05. (D–E) SH-SY5Y cells were treated with BTZ for 16h. (D) The level of 26S subunits and p97 were determined by WB. (E) Then singly or doubly-capped (SC and DC) 26S proteasomes were separated by native PAGE and their activity and amount were determined by Suc-LLVY-AMC overlay assay or WB with the anti-a123567 antibody. (F-H) BTZ inhibits 26S subunit expression in HEK293A cells at 50nM or higher. (G) Peptidase activities were measured in cell lysates with fluorogenic peptide substrates. (H) Protein degradation rate was measured by assaying hydrolysis of radiolabeled long-lived cell proteins to acid-soluble products. (F) β3, Rpt3, and Rpt5 mRNAs were measured. Arrows indicate 50nM, above which BTZ blocks protein degradation and 26S induction. Error bar=SD.
Thus, MG132 inhibits proteasome induction only at high concentrations, where this tripeptide aldehyde can also inhibit several cellular serine and cysteine proteases. Therefore, at such concentrations, MG132’s ability to block induction of 26S subunits and p97 might be due to an effect on these other proteases (see below) or to its causing a greater inhibition of proteasome function by inhibiting both the caspase-like and the chymotrypsin-like activities[25]. To test these possibilities, we investigated the effects of higher concentrations of the much more specific peptide boronate proteasome inhibitors, BTZ and MG262[26], and epoxomicin, an epoxyketone that specifically inactivates the 20S threonine proteases[27]. At high concentration (10µM), BTZ, MG262, and Epox, all suppressed the induction of 26S subunits and p97 (Fig. 2BC) without causing detectable cytotoxicity in 16h (Fig. S2BC), in sharp contrast to the increased expression of these genes with low concentrations (10nM BTZ, 50nM Epox, 1µM MG262 (Fig. 2BC,Fig. S3L)). Similarly, in HEK293A cells, high concentrations of BTZ inhibited the expression of 26S subunits (Fig. 2F). In addition, the levels of 26S and p97 protein, as well as the amount of assembled 26S particles, were increased by low, but not high concentrations of BTZ (Fig. 2DE).
At these high concentrations, the inhibitors all block multiple peptidase activities and thus markedly reduce cellular proteolysis[25]. Most likely these high concentrations prevent induction of 26S subunits by causing a greater inhibition of the proteasome. Therefore, we compared in HEK293A cells the effects of increasing BTZ concentrations on 26S expression, the extent of inhibition of degradation of long-lived cell proteins labelled with [3H]-phenylalanine, and the degree of inhibition of the 26S’s chymotrypsin-like and caspase-like activities in cell lysates (tested with specific fluorescent peptide substrates[28]). At 20nM, BTZ inhibits the chymotrypsin-like site > 50%, but 50nM is required to inhibit it by 100% and to inhibit the caspase-like activity and protein degradation rate by > 50% (Fig. 2GH). Intriguingly, in these cells, maximal induction of 26S subunits, β3, Rpt3, Rpt5, occurred with 20nM BTZ, but at 50nM and higher, BTZ inhibited the expression of these genes progressively (Fig. 2F). Therefore, greater inhibition of protein degradation (e.g. with high concentrations that block the chymotrypsin-like activity completely and also inhibit the caspase-like activity), suppressed the expression of 26S subunits and p97.
The induction of 26S subunits requires cleavage of the ER-bound transcription factor Nrf1 and some proteasome function
We confirmed the prior finding[9] that Nrf1 is the critical factor inducing 26S subunits after BTZ treatment by showing that stable Nrf1 knock-down in SH-SY5Y cells suppressed the induction of many 26S subunits mRNAs and decreased their basal mRNA levels (Fig. S3A,B). Thus, it appears important in determining proteasome content under normal conditions. By contrast, knock-down or overexpression of Nrf2 did not affect 26S induction upon BTZ treatment (Fig. S3C–F). Nrf1 is normally associated with the ER through its N-terminus, and deletion of the N-terminal 30 residues allows its translocation into the nucleus and the stimulation of gene expression[14, 29, 30]. Therefore, it was hypothesized that a proteolytic cleavage of Nrf1 near its N-terminus releases it from the ER[10], and during preparation of this manuscript, Nrf1 was reported to be cleaved before Leu104[31]. However, no specific protease has been implicated in this step. As discussed above, the most likely explanation for the ability of high concentrations of proteasome inhibitors to suppress induction is that the 26S itself processes Nrf1, and high concentrations of inhibitors block its capacity for limited proteolysis.
We therefore tested whether Nrf1 was proteolytically processed upon exposure to low but not high concentrations of proteasome inhibitors. Using an antibody that recognizes the C-terminus of Nrf1 (C-19), we could detect the full-length (FL) Nrf1 (~85kD) plus a smaller form (~75kD) in cells treated with 10nM BTZ (Fig. S3BGH). These two bands also reacted with another Nrf1 antibody (H-285, which recognizes residues 191–475), and levels of both were diminished by the Nrf1 shRNA (Fig. S3B). The appearance of this smaller form upon BTZ treatment strongly suggests that it is a processed (P) form of Nrf1. Inhibition of protein synthesis by CHX after 4h of BTZ (20nM) treatment caused a rapid decline in the levels of both FL and P forms(Fig. S3J). Thus, both forms of Nrf1 have short half-lives even in the presence of 10nM BTZ. Upon treatment with 20nM BTZ, the P form gradually accumulated, and its level was much higher at 8–12h than at 4h (Fig. S3J). However, ATP depletion during proteasome inhibitor treatment completely abolished Nrf1 processing (Fig. S3K), implying that generation of the P form involves an ATP-dependent process, such as the UPS (see below).
We next tested whether Nrf1 processing is blocked by high concentrations of proteasome inhibitors. Nrf1 was processed in both SH-SY5Y cells (Fig. 3A) and MM1.S cells (Fig. S3I) treated with 10nM BTZ, which caused 26S subunit induction (Fig. S1D), but not with 10µM MG132, which failed to stimulate their expression. Similarly, high concentrations (10µM) of BTZ, Epox, or MG262 also suppressed Nrf1 processing (Fig. 3AB), just as they prevent proteasome induction, while at low concentrations that permitted induction, these inhibitors caused Nrf1 processing (Fig. 3AB, S3L–N). Furthermore, treatment with 10µM MG132 or MG262 together with 10nM BTZ suppressed the processing of Nrf1 (Fig. 3A), just as these combinations repressed 26S expression (Fig. 2B). We confirmed these observations using both C-19 and H-285 antibodies to monitor Nrf1 processing. Because the H-285 antibody recognizes the processed Nrf1 much more strongly than C-19, H-285 was used in most subsequent experiments.
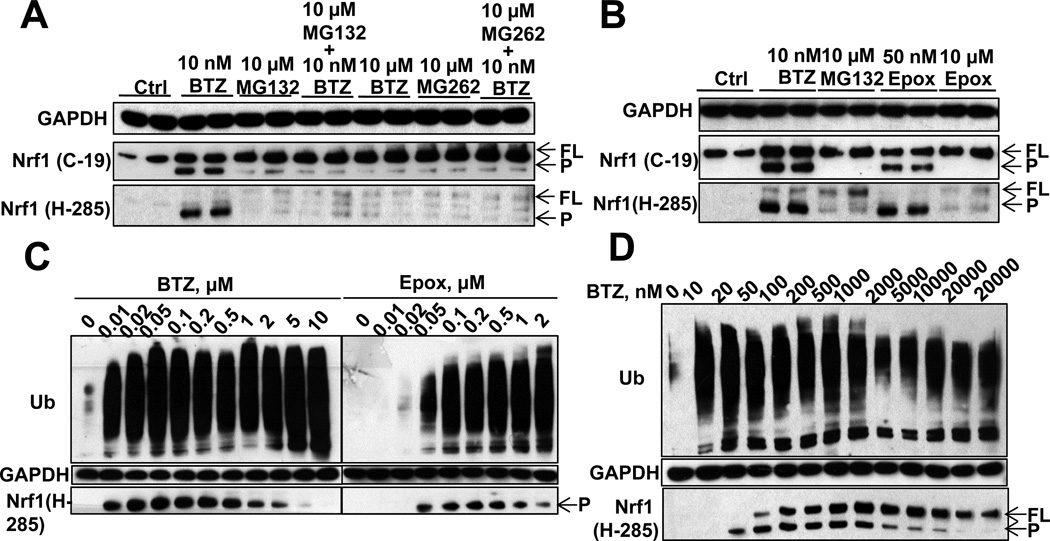
Figure 3. High concentrations of proteasome inhibitors block the proteolytic processing of Nrf1 (See also Fig. S3).
(A) Treatment of SH-SY5Y cells with high concentrations (10µM) of MG132, BTZ, or MG262 for 16h, alone or with 10nM BTZ, suppressed the processing of Nrf1. FL: full-length Nrf1; P: processed Nrf1 (B) 16h treatment with Epox at 10µM, but not 50nM, suppressed Nrf1 processing. (C) BTZ or Epox treatment (SH-SY5Y cells) for 16h at concentrations higher than 0.5µM decreases the processing Nrf1. (D) Treatment of HEK293A cells with BTZ for 16h at above 20nM causes the accumulation of FL Nrf1 and above 1µM decreases the level of processed Nrf1.
To determine the minimal concentrations at which proteasome inhibitors begin to block Nrf1 processing, we compared the effects of increasing concentrations of BTZ, Epox, and MG132 in SH-SY5Y cells. MG132 was able to enhance Nrf1 processing (Fig. S3N) and to induce subunit expression (Fig. S2F) at 1 or 2.5µM, but at higher concentrations, 5 or 10µM, both Nrf1 processing and proteasome gene expression were suppressed. When the concentrations of the more potent and specific inhibitors, BTZ and Epox, were raised above 0.5µM, the level of the P form started to decrease in SH-SY5Y (Fig. 3C) and HEK293A cells (Fig. 3D). A more sensitive indicator of the incomplete processing of Nrf1 was the accumulation of FL Nrf1. In SH-SY5Y cells, FL Nrf1 is present at very low concentrations (probably due to its very rapid degradation) and was barely detectable by the H-285 antibody. However, in HEK293A cells, BTZ (≥50nM) was able to cause FL-Nrf1 accumulation (Fig. 3D), and thus, at these concentrations, BTZ must already inhibit Nrf1 processing. Remarkably, 50nM is exactly the concentration at which BTZ suppressed the expression of 26S subunits (Fig. 2F). Also, 50nM is the concentration at which BTZ causes complete inhibition of the chymotrypsin-like activity, and also inhibits the caspase-like activity and cellular protein degradation > 50% (Fig. 2GH). These extensive correlations make it very likely that the proteasome is responsible for Nrf1 processing to the active form. Therefore, only when proteasomes are partially compromised by low inhibitor concentrations that block only partially the chymotrypsin-like site will there be sufficient 26S function for limited proteolysis of Nrf1. However, when multiple active sites are inhibited and protein degradation is reduced > 50%, Nrf1 processing cannot take place.
Although the proteasome is the only known enzyme in mammalian cells sensitive to epoxyketones[27], we carried out extensive tests of the unlikely possibility that Nrf1 is processed by an additional protease that is inhibited by peptide aldehydes and/or peptide boronates or an ER-associated intramembrane protease. By inhibiting these proteases using chemical inhibitors or siRNAs, we obtained strong data (Fig. S3O–W) indicating that the various potential candidates, including MG132-sensitive proteases (calpain, lysosomal cathepsins) or ER-associated intramembrane proteases (γ-secretase, signal peptidase, and ER-associated rhomboid family intramembrane proteases) do not play a role in processing Nrf1 upon proteasome inhibition and expression of 26S subunits (See Fig. S3O–W legend for detailed discussion). Together, these various negative observations and the extensive correlations shown above make it very likely that Nrf1 processing is mediated by the proteasome itself.
Proteolytic removal of Nrf1’s N-terminus is essential for nuclear entry
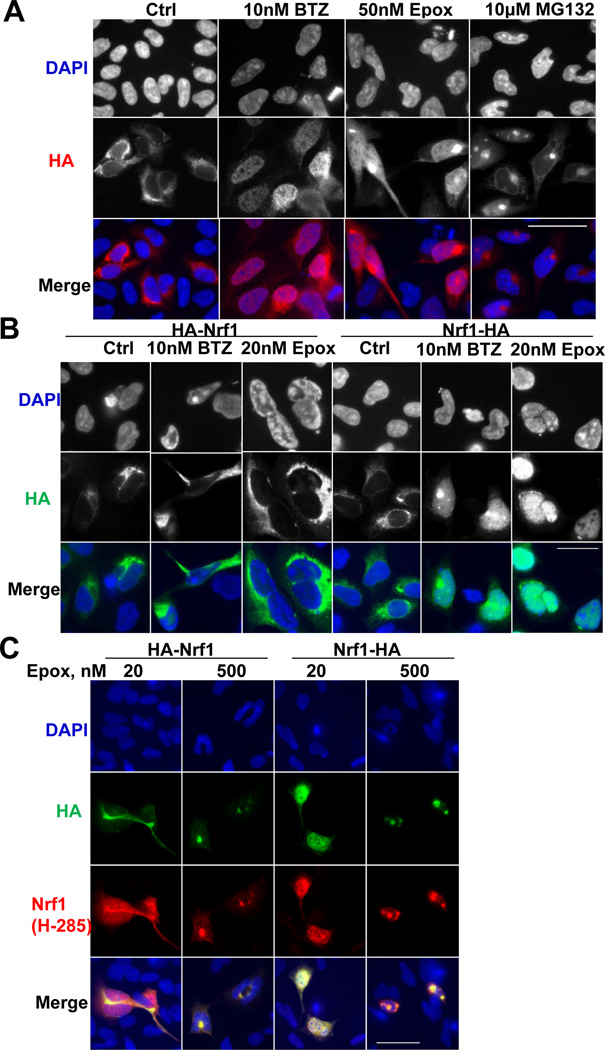
Nrf1 contains several ER transmembrane domains, but the topology of full-length Nrf1 has not been resolved. However, the N-terminal transmembrane domain (residues 2–30) is required for its ER association, and deletion of this region allows nuclear translocation[29]. To test if Nrf1 processing involves proteolytic removal of this N-terminal region, we expressed full-length Nrf1 tagged with HA at its N- or C-terminus. Full-length Nrf1 initially exists as a glycosylated form (G)[10, 30]. After Epox treatment, the G-species was first converted to the FL form, whose molecular weight decreased due to deglycosylation (Fig. 4A), as reported recently[10]. The identity of the G and FL species was clear because both retained the HA tag, whether it was on the N- or C-terminus (Fig. 4A), and the full-length sequence of the G-Form was confirmed by LC/MS/MS (Fig. S4). After deglycosylation, Nrf1 was proteolytically processed, as shown by LC/MS/MS analysis of the shorter form (Fig. S4). When Nrf1 was tagged at its N-terminus (HA-Nrf1), the HA tag was lost during Nrf1 processing, but not when it was tagged on its C-terminus (Nrf1-HA). The HA antibody therefore no longer reacted with processed HA-Nrf1 (Fig. 4A) even though Epox at low concentrations still induced processing of HA-Nrf1, which still reacted with the Nrf1 antibody. As expected, the processed Nrf1-HA was larger than the processed HA-Nrf1, which no longer contained the HA tag. However, we could not detect the N-terminal HA tag after it was cleaved off HA-Nrf1. Like the processing of endogenous Nrf1, Nrf1-HA processing was very sensitive to proteasome inhibition. An 80% inhibition of the chymotrypsin-like activity by 20nM Epox promoted Nrf1 processing, but a greater inhibition of the proteasome with 100nM Epox or with 20nM BTZ, which inhibits the chymotrypsin-like site completely and the caspase-like site, resulted in a decline in the P form and an accumulation of the FL form (Fig. 4B).
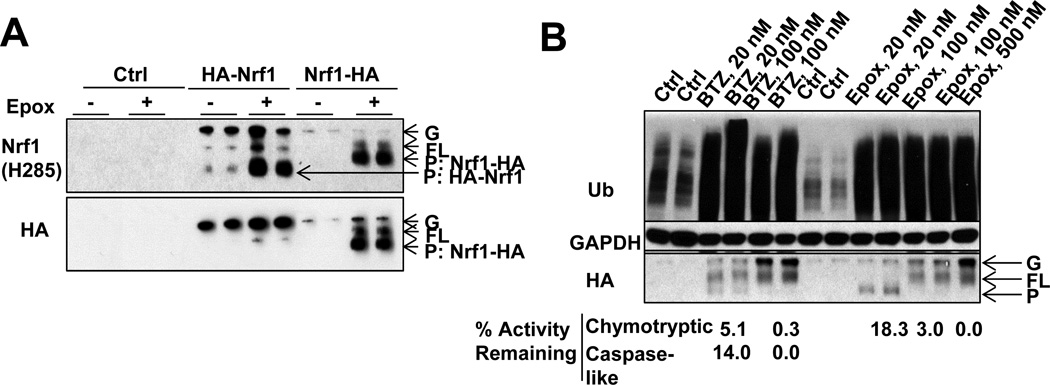
Figure 4. Low, but not high, concentrations of proteasome inhibitors, causes the processing of Nrf1 near its N-terminus (See also Fig. S4).
(A) Nrf1 tagged with HA at its N-terminus (HA-Nrf1) or C-terminus (Nrf1-HA) was expressed in HEK293A cells. Nrf1-HA and HA-Nrf1 exist in both a glycosylated form (G, which is hardly detectable for the endogenous Nrf1 under most conditions), and deglycosylated form (FL, because both full-length forms retain the HA epitope no matter if Nrf1 was tagged at its N- or C-termini). After treatment with Epox (20nM, 16h), the HA tag still remained on the processed form of Nrf1-HA, but not on the product of HA-Nrf1. (Biological duplicates assayed.) (B) The processing of Nrf1-HA is most efficient with 20nM Epox treatment that retains 18.3% Chymotrypsin-like (ChT-L) activity, while 20nM BTZ treatment (that retains only 5.1% chymotrypsin-like activity and 14.0% caspase-like activity) partially suppressed Nrf1-HA processing. Higher concentrations of BTZ or Epox blocked Nrf1-HA processing.
To test if the loss of the N-terminus actually allows Nrf1 entry into the nucleus[14, 29], HEK293A cells overexpressing Nrf1-HA were treated with proteasome inhibitors and Nrf1-HA was localized with an HA antibody. Nrf1-HA was initially cytoplasmic, as expected from its reported ER-association, but entered the nucleus after treatment with BTZ or Epox (Fig 5B). Although the H-285 antibody detected this nuclear translocation of HA-Nrf1 (Fig. 5C), the HA antibody could not detect any HA-Nrf1 in the nucleus (Fig. 5BC). Thus, Nrf1 could not enter the nucleus unless its N-terminus was removed. However, as expected, high inhibitor concentrations (10µM MG132 (Fig. 5A) or 500nM Epox (Fig. 5C)) blocked Nrf1 accumulation in the nucleus. Thus, some proteasome activity is essential for Nrf1 release from the ER and nuclear entry.
Figure 5. Nrf1 nuclear translocation with low, but not high concentrations, of proteasome inhibitors requires its N-terminal processing.
(A) HEK293A cells overexpressing Nrf1-HA were treated with BTZ (10nM), Epox (50nM), or MG132 (10µM) for 16h, and Nrf1-HA localization was detected by HA immunostaining. Nuclear translocation of Nrf1-HA occurred after treatment with BTZ and Epox, but not MG132. (B) After HEK293A cells overexpressing Nrf1-HA or HA-Nrf1 were treated with BTZ (10nM) or Epox (20nM) for 16h, HA immunostaining detected Nrf1-HA, but not HA-Nrf1, in the nuclei of the BTZ or Epox-treated cells. (C) HEK293A cells overexpressing Nrf1-HA or HA-Nrf1 were treated with Epox (20 or 500nM) for 16h. Nrf1 antibody, but not the HA antibody, could detect nuclear HA-Nrf1 upon 20nM Epox treatment. 500nM Epox, like 10µM MG132, did not induce Nrf1 nuclear translocation. All scale bars are 50 µm.
Nrf1 processing requires ubiquitination, but is not activated by heat shock
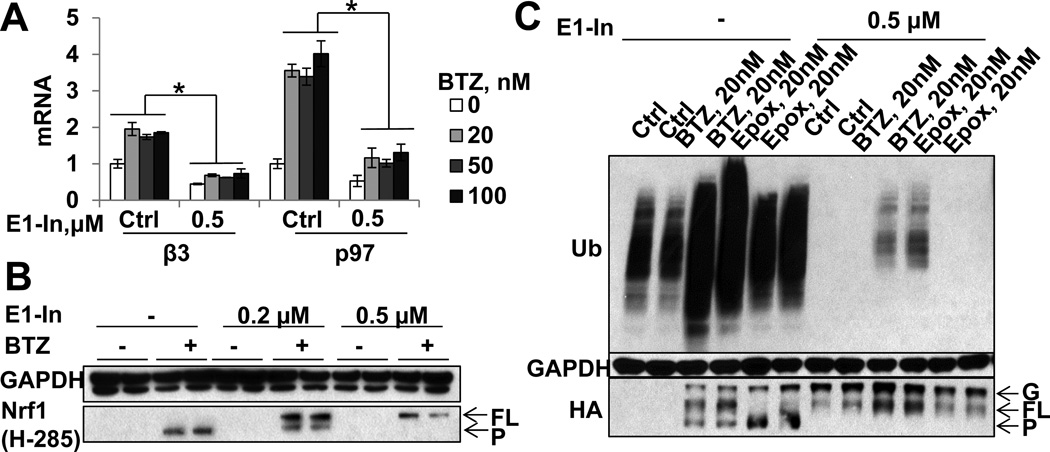
Nrf1 is normally poly-ubiquitinated and rapidly degraded by proteasomes[10, 13, 14]. It was therefore assumed that proteasome inhibitors cause gene induction simply by blocking Nrf1 degradation[10]. However, this model is inconsistent with our finding that high concentrations of these inhibitors prevent Nrf1 degradation but do not induce 26S subunit expression. Instead, Nrf1 activation and induction of 26S subunits and p97 require some proteasome activity to process Nrf1 and allow nuclear entry. To determine whether the processing of Nrf1 by the 26S also requires the ubiquitination of Nrf1, we treated cells for 16h with BTZ (10/100nM) and the specific inhibitor of ubiquitination, ML00603997[32]. This treatment almost completely depleted various cell lines of ubiquitinated proteins within 1h (Fig. S5AC), and was not toxic to the cells in 24h. Blocking ubiquitination dramatically suppressed BTZ-induced expression of 26S subunits and p97 below their basal mRNA level (Fig. 6A,S5B), and also completely suppressed the processing of endogenous Nrf1 induced by BTZ (Fig. 6B,S5AC). Similarly, inhibiting ubiquitination prevented the processing of overexpressed Nrf1-HA in HEK293A cells and caused it to accumulate in its FL form (Fig. 6C). Furthermore, this inhibitor alone caused the accumulation of FL Nrf1. Thus, ubiquitination is required for BTZ-induced Nrf1 processing and the induction of 26S subunits and p97.
Figure 6. Nrf1 processing and the expression of 26S subunits and p97 require Nrf1 ubiquitination (See also Fig. S5).
(A–B) SH-SY5Y cells were treated with the E1 inhibitor ML00603997 (E1-In, 0.5µM) for 1h, then treated with different concentrations of BTZ together with 0.5µM ML00603997 for 16h. E1-In suppressed both (A) expression of mRNA for p97 and the 26S subunit β3 and (B) the processing of Nrf1 (Biological duplicates assayed). (C) HEK293A cells expressing Nrf1-HA were treated with E1-In as in (A–B) and different concentrations of proteasome inhibitors. E1-In caused the accumulation of full-length Nrf1-HA by itself or in the presence of proteasome inhibitors. Error bar=SD.
A possible explanation why blocking ubiquitination reduces Nrf1 processing and proteasome production could be that reduced proteasome function activates Nrf1-mediated gene induction by causing an accumulation of misfolded or ubiquitinated proteins. However, merely increasing protein misfolding and the level of Ub conjugates in the cytosol by treating SH-SY5Y cells with arsenite or heat shock not only failed to promote Nrf1 processing, but surprisingly, even blocked this process and the induction of 26S subunits and p97 by BTZ (Fig. S5D–G). Therefore, Nrf1 processing and production of new proteasomes is not signaled by an accumulation of misfolded or ubiquitinated proteins, which actually reduces these processes. Alternatively, Nrf1 may need to be ubiquitinated in order to be processed by the proteasome. We tested this possibility using RA190[33], an inhibitor of Rpn13, one of two proteasome subunits that bind Ub chains. RA190 binds covalently to Rpn13’s Ub-binding domain and thus seems to block the association of conjugates with the 26S[33]. Rpn13 inhibition blocked the accumulation of processed Nrf1 in HEK293A and SH-SY5Y cells and reduced the induction of 26S expression by 20nM BTZ (Fig. S5H–L). Therefore, Nrf1 processing requires recognition of ubiquitinated species by the 26S. Most likely, E1 inhibition stops Nrf1 processing by blocking its ubiquitination and binding to the 26S and not by reducing cell’s content of Ub conjugates.
Nrf1 processing requires p97 function for its deglycosylation
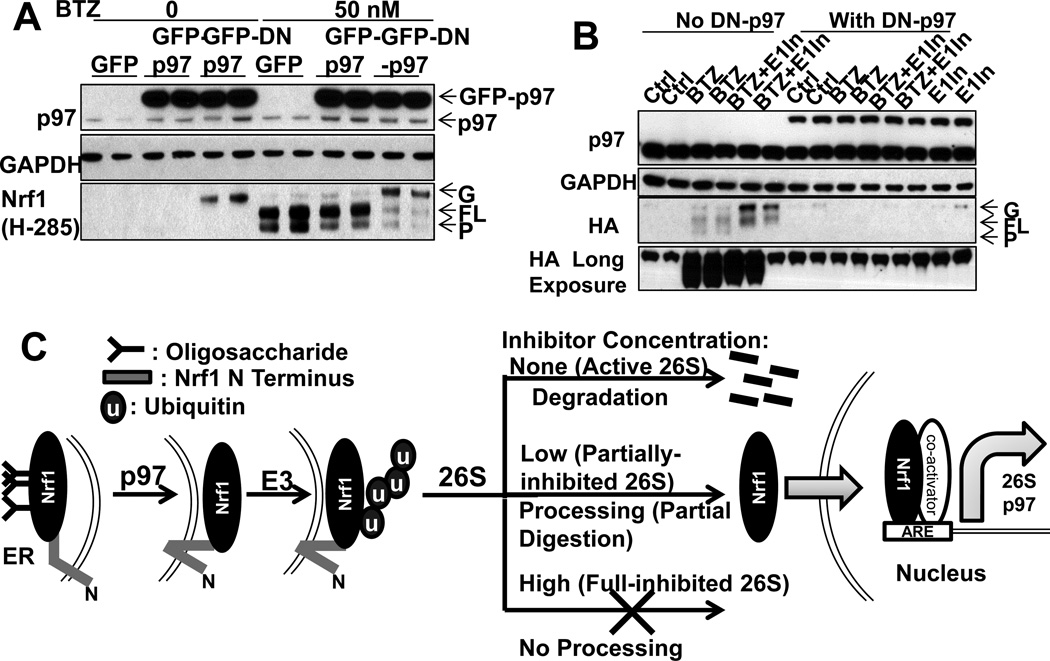
This ubiquitination and proteasome-mediated limited proteolysis of Nrf1 resembles the processing of three other transcription factors, NF-κB[34], Gli[35] and Spt23[36]). The processing of Spt23, which is also ER-associated, requires p97 to extract the processed Spt23 from the ER[36]. To test if p97 is also essential for Nrf1 processing, we expressed a dominant-negative mouse p97K524A (DN-p97) that cannot bind ATP[37], or treated cells with the p97 inhibitor NMS859, which also blocks ATP-binding[38]. As expected, DN-p97 and NMS859 both raised cellular levels of Ub conjugates (Fig. S6). Upon BTZ treatment (50nM, 4h), DN-p97 also caused an accumulation of the glycosylated form of Nrf1, but decreased the level of both FL-Nrf1 and the P form (Fig. 7A,S6B). The same effects were observed in HEK293A cells or SH-SY5Y cells treated with the p97 inhibitor (Fig. S6EF) at concentrations that do not cause cell death (Fig. S6D). The DN-p97 also blocked the formation of both the FL and P forms of ectopically-expressed Nrf1-HA (Fig. 7B). Therefore, p97 function is necessary for the production of the deglycosylated FL form. As these findings predict, the DN-p97 also prevented Nrf1 entry into the nucleus upon treatment with 20nM BTZ and reduced the expression of 26S subunits (Fig. S6C,H). Similarly, p97 inhibitor also abolished induction of 26S subunits and p97 in HEK293A and SH-SY5Y cells (Fig. S6G,I)
Figure 7. Nrf1 processing and the expression of 26S subunits and p97 require Nrf1 deglycosylation and p97 activity (See also Fig. S6).
(A) To test whether p97 function is essential for Nrf1 processing, HEK293A cells overexpressing GFP-p97, GFP-DN-p97, or GFP (control) were incubated with or without 50nM BTZ for 4h. GFP-DN-p97 (but not GFP-p97) reduced the level of full-length and processed Nrf1, but caused the accumulation of a 100kDa glycosylated form of Nrf1. (B) DN-p97 and Nrf1-HA were co-expressed in HEK293A cells. DN-p97 caused the sequestration of Nrf1-HA in its glycosylated form and blocked the formation of full-length and processed Nrf1 upon treatment (12h) with BTZ (20nM) or E1-In (0.5µM). (C) Model: Upon partial inhibition of proteasomes with low concentrations of inhibitors, cells induce 26S subunits and p97 via Nrf1. Although Nrf1 is normally degraded completely, partial inhibition of proteasomes by low concentrations of inhibitors favors limited degradation of the N-terminal part of Nrf1. Consequently, the processed C-terminal portion of Nrf1 is released from the ER and enters the nucleus to transcribe 26S subunits and p97. Complete inhibition of proteasomes by high concentrations of inhibitors blocks Nrf1 processing and transcription. Both Nrf1 degradation and its proteolytic processing require first deglycosylation and extraction of Nrf1 from the ER via p97 activity and then ubiquitination of Nrf1.
Because FL-Nrf1 accumulated upon E1 inhibition, while G-Nrf1 accumulated when p97 was inhibited, Nrf1 must be ubiquitinated after its deglycosylation. To test this conclusion, we treated HEK293A cells co-expressing Nrf1-HA and DN-p97 with the E1 inhibitor. Although blocking ubiquitination alone caused a build-up of Nrf1 in the FL form, inhibition of both E1 and p97 led to an accumulation of Nrf1 in the G-form (Fig. 7B). Therefore, Nrf1 must be ubiquitinated again after its deglycosylation.
Discussion
The increased production of new proteasomes after treatment with proteasome inhibitors is an important compensatory response that promotes cell survival and is likely to reduce therapeutic efficacy. As shown here, this response involves coordinate induction of all the standard proteasome subunits via Nrf1 as well as several other key UPS components. The 20S assembly chaperone POMP, but not chaperones involved in 19S base assembly (S5b, p27, Gankyrin) were induced, presumably because POMP is destroyed during 20S assembly. The simultaneous induction of PA200/Blm10 is interesting since PA200 can promote Ub-independent proteasomal degradation of certain proteins[39]. On the other hand, the lack of induction of immunoproteasome subunits and PA28αβ correlates with their functioning predominantly in antigen presentation and regulation by γ-interferon, rather than in protein degradation. In fact, expression of immunoproteasomes was suppressed by BTZ, as was also noted previously[10, 11], and none of these genes contains an ARE sequence in their promoters. Clearly, their production is regulated differently from standard proteasomes. Proteasome inhibition induced Usp14, but surprisingly, not the other 26S-associated DUB, Uch37, even though it, like Usp14, activates gate opening and ATP hydrolysis upon binding of Ub conjugates[23]. Nevertheless, the finding that Uch37 expression is not coordinately regulated suggests that these two 26S-associated DUBs serve distinct roles. Lastly, our finding that p97 and its cofactors (Ufd1, Npl4, and p47) were strongly induced together with proteasome subunits is consistent with their important general role in the UPS.
This simultaneous induction of UBB, PA200, Usp14, and p97 plus its ERAD cofactors suggests strongly that these proteins are all important in enhancing the cell’s proteolytic capacity. However, simultaneous induction does not prove that the mechanisms are the same. In fact, high concentrations of proteasome inhibitors inhibited the induction of p97 together with 26S subunits, but not the induction of Usp14, Ecm29, and UBB, which thus must not require Nrf1. Nrf1-dependent induction of all proteasome subunits and p97 differ sharply from the selective induction of Rpn6 by FoxO4[40]. This response to proteasome inhibition also clearly differs from the heat shock response and the UPR[20, 41], both of which are caused by the build-up of misfolded proteins and therefore can also be induced by proteasome inhibitors. Neither the UPR nor heat shock leads to Nrf1 processing or induction of 26S subunits. In fact, surprisingly, heat shock suppresses this response to proteasome inhibition (Fig. S5D–G). Among four cell lines tested, the induction of 26S subunits was strongest in SH-SY5Y cells, but this property is not general for all neuronal cells because another neuroblastoma line, M17, induced the expression of 26S subunits much more weakly (Fig. S1D) despite being more resistant to killing by BTZ than SH-SY5Y cells (data not shown). Therefore, although both neuroblastoma lines were much more resistant to killing by BTZ than the myeloma line MM1.S, this greater resistance cannot be attributed to a greater ability to induce the expression of 26S subunits.
Because Nrf1 is a short-lived protein, it was believed that upon proteasome inhibition, Nrf1 is activated simply because its degradation is prevented[10, 13, 14]. Such a mechanism is unable to explain our findings that 26S subunits and p97 are induced upon treatment with low, but not high concentrations of proteasome inhibitors. In fact, this mechanism would predict the exact opposite result, i.e. greater Nrf1 activation at higher concentrations. Our results instead support the type of mechanism illustrated in Fig. 7C, in which 26S proteasomes catalyze not only the complete degradation of Nrf1[14], but also limited proteolysis to release Nrf1’s active region from its N-terminal transmembrane domain. Since Nrf1 also regulate basal expression of 26S subunits (Fig. S3A), both Nrf1 degradation and processing probably also occur under normal conditions, but with reduced proteasome function, processing is favored.
A variety of observations support this model, especially our demonstration that some proteasome function is essential for Nrf1 processing and its translocation into the nucleus. Specifically, BTZ most efficiently induced Nrf1 processing and 26S subunit expression at low concentrations that only partially block the chymotrypsin-like site and cause only a minor (<40%) decrease in proteolysis (Fig. 2FGH,3D). By contrast, higher concentrations that completely block this site and also cause a large (>50%) inhibition of the caspase-like site and protein breakdown, inhibit Nrf1 processing, nuclear entry, and 26S induction. Thus, although active proteasomes tend to degrade Nrf1 completely, partially-inhibited 26S no longer digest ubiquitinated Nrf1 processively and tend to release partially-digested Nrf1, which activates transcription. When 26S function is blocked to a greater extent, the inactive Nrf1 precursor was stabilized, and the expression of 26S subunits and p97 blocked.
The precise mode of processing of ubiquitinated Nrf1 by the partially-inhibited 26S remains uncertain. Although the N-terminal HA tag on Nrf1 and the transmembrane segment were not detected after they were cleaved from HA-Nrf1, these observations do not distinguish whether Nrf1 is partially digested from its N-terminus, or as seems more likely, proteasomal cleavage is initiated from an internal loop near the transmembrane segment, followed by degradation of the N-terminal part, perhaps by an ER-associated peptidase (e.g. signal peptidases). While 26Sgenerally degrade proteins in a processive manner, incomplete degradation beginning from one end or an internal loop, and release of large fragments, are often observed with isolated proteasomes[42]. Perhaps simply the slowing of proteasomal function, as occurs with low concentrations of inhibitors, enhances the probability of release of Nrf1’s C-terminal part. The lack of processivity leading to its release may depend on the tightness of folding of its different domains, the direction of translocation into 20S[42], the site(s) of ubiquitination, and the length or number of Ub chains on the substrate. Three E3s have been reported to act on Nrf1: Hrd1, an ER-associated E3, β-TRCP, a nuclear E3 that probably catalyzes degradation of the mature Nrf1, and Fbw7, which acts on multiple growth-related proteins[13, 14]. Exactly how these E3s (or others) influence the complete degradation or processing of Nrf1 is an important issue for future research.
This mechanism (Fig. 7C) for Nrf1 maturation resembles the 26S-catalyzed processing of inactive precursors to three other transcription factors: NF-κB[34], Gli[35], and the ER-bound Spt23 in yeast by proteasomes and p97[36]. These other examples, like Nrf1 processing, require precursor ubiquitination and ATP. In these respects, Nrf1 resembles a typical ERAD substrate. Not surprisingly, p97 is important for Nrf1 degradation by fully-active proteasomes[10] as well as its processing by partially-inhibited 26S. Nrf1 may translocate across the ER membrane several times[29]. This complex topology should block the release of Nrf1 from the ER. Since p97 activity allows removal of these glycosyl chains (Fig. 7A,Fig. S6B), the p97 complex probably extracts most of Nrf1’s C-terminal part from the ER, so that the subsequent N-terminal proteolytic processing could release Nrf1. After the present studies were completed, Deshaies and colleagues reported studies of Nrf1’s ER topology based on susceptibility to protease K[31], and also concluded that Nrf1’s C-terminal part is initially localized in the ER lumen and was relocated into the cytosol by p97. We further demonstrated that after Nrf1 is deglycosylated and its C-terminal region relocated to the cytosol by p97, Nrf1 needs to be ubiquitinated again in order to be processed. Thus, p97 functions here differently than in Spt23 processing, where p97 seems to extract Spt23 from the ER after it has already been processed by the proteasome[36]. Since p97 is required for Nrf1 processing, p97 inhibition, like high concentrations of proteasome inhibitors, suppressed rather than activated the transcription of 26S genes. Because the p97 inhibitors have potential as chemotherapeutic agents[38], it is noteworthy that they do not elicit the compensatory production of more 26S.
The finding that partial proteasome inhibition triggers the induction of new 26S has clear implications for multiple myeloma therapy. Because this compensatory response promotes cancer survival[9–11] and increases the chances of drug resistance, the optimal treatment regimen should be one that inhibits multiple active sites and achieves more complete inhibition of proteasomes for longer periods. For the optimization of drug administration, monitoring Nrf1 processing may be a useful biomarker to evaluate the efficacy of proteasome inhibition and rates of new particle production. Further understanding of the mechanisms for Nrf1 processing may also indicate ways to block new proteasome production to improve the treatment of cancer or conversely, ways to enhance Nrf1 processing to promote clearance of misfolded proteins in neurodegenerative diseases.
Experimental Procedures
Detailed experimental procedures are presented in Supplemental Information
Gene overexpression or knock-down
Lipofectamine-2000 (Life Technologies) was used to transfect plasmids into HEK293A cells. Stable knock-downs in SH-SY5Y cells were performed by infection with lentiviral particles expressing shRNA and puromycin-resistant marker.
Immunostaining
Transfected cells were seeded on coverslips in 6-well plates. Treated cells were fixed with −20°C methanol. Images were taken at a Nikon Ti Inverted Fluorescence Microscope.
Real-Time RT-PCR
mRNA was extracted via TRIzol Reagent (Life Technologies), and cDNA was synthesized using Multiscribe Reverse Transcriptase (Applied Biosystems). Real-time RT-PCR was performed using ABsolute Blue QPCR ROX Mix (Thermo Scientific ) on a Bio-Rad C-1000 thermocycler.
Assay of proteasome function and protein degradation
Proteasomal peptidase activities were measured as described previously[28]. Native gels and overlay assay with Suc-LLVY-AMC were conducted as described[43]. The degradation rate of long-lived cell proteins was measured after labelling with L-Phe-[3,4,5-3H] (American Radiolabeled Chemicals) as described[44].
Supplementary Material
Highlights.
Nrf1 induces all 26S subunits and p97 upon treatment with proteasome inhibitors
Partially-inhibited proteasomes process Nrf1 to allow its nuclear translocation
Complete 26S inhibition blocks Nrf1 processing and transcriptional activity
Nrf1 processing requires its deglycosylation (involving p97), then ubiquitination.
Acknowledgement
Microscopy was performed at the Nikon Imaging Center at HMS. Z.S. is a Novartis Fellow of the Life Sciences Research Foundation and also held a fellowship from the Leukemia and Lymphoma Society. These studies were supported by grants from the NIGMS(GM051923-18). Drs. M. Wolfe, M. Hannink, and L. Dick, kindly provided valuable reagents.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annual review of biochemistry. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends in cell biology. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Molecular & cellular proteomics : MCP. 2011;10:R110 003871. doi: 10.1074/mcp.R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dantuma NP, Hoppe T. Growing sphere of influence: Cdc48/p97 orchestrates ubiquitin-dependent extraction from chromatin. Trends in cell biology. 2012;22:483–491. doi: 10.1016/j.tcb.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Ye Y. Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. Journal of structural biology. 2006;156:29–40. doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Bar-Nun S. The role of p97/Cdc48p in endoplasmic reticulum-associated degradation: from the immune system to yeast. Current topics in microbiology and immunology. 2005;300:95–125. doi: 10.1007/3-540-28007-3_5. [DOI] [PubMed] [Google Scholar]
- 7.Goldberg AL. Development of proteasome inhibitors as research tools and cancer drugs. The Journal of cell biology. 2012;199:583–588. doi: 10.1083/jcb.201210077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laubach JP, Mitsiades CS, Roccaro AM, Ghobrial IM, Anderson KC, Richardson PG. Clinical challenges associated with bortezomib therapy in multiple myeloma and Waldenstroms Macroglobulinemia. Leukemia & lymphoma. 2009;50:694–702. doi: 10.1080/10428190902866732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Molecular cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steffen J, Seeger M, Koch A, Kruger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Molecular cell. 2010;40:147–158. doi: 10.1016/j.molcel.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Meiners S, Heyken D, Weller A, Ludwig A, Stangl K, Kloetzel PM, Kruger E. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of Mammalian proteasomes. The Journal of biological chemistry. 2003;278:21517–21525. doi: 10.1074/jbc.M301032200. [DOI] [PubMed] [Google Scholar]
- 12.Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 13.Biswas M, Phan D, Watanabe M, Chan JY. The Fbw7 tumor suppressor regulates nuclear factor E2-related factor 1 transcription factor turnover through proteasome-mediated proteolysis. The Journal of biological chemistry. 2011;286:39282–39289. doi: 10.1074/jbc.M111.253807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsuchiya Y, Morita T, Kim M, Iemura S, Natsume T, Yamamoto M, Kobayashi A. Dual regulation of the transcriptional activity of Nrf1 by beta-TrCP- and Hrd1-dependent degradation mechanisms. Molecular and cellular biology. 2011;31:4500–4512. doi: 10.1128/MCB.05663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart D, Killeen E, Naquin R, Alam S, Alam J. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. The Journal of biological chemistry. 2003;278:2396–2402. doi: 10.1074/jbc.M209195200. [DOI] [PubMed] [Google Scholar]
- 16.Besche HC, Peth A, Goldberg AL. Getting to first base in proteasome assembly. Cell. 2009;138:25–28. doi: 10.1016/j.cell.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fricke B, Heink S, Steffen J, Kloetzel PM, Kruger E. The proteasome maturation protein POMP facilitates major steps of 20S proteasome formation at the endoplasmic reticulum. EMBO reports. 2007;8:1170–1175. doi: 10.1038/sj.embor.7401091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues AP, Manning G, Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 19.Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Molecular and cellular biology. 2009;29:1095–1106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nature reviews. Molecular cell biology. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka K, Kasahara M. The MHC class I ligand-generating system: roles of immunoproteasomes and the interferon-gamma-inducible proteasome activator PA28. Immunological reviews. 1998;163:161–176. doi: 10.1111/j.1600-065x.1998.tb01195.x. [DOI] [PubMed] [Google Scholar]
- 22.Cascio P, Hilton C, Kisselev AF, Rock KL, Goldberg AL. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. The EMBO journal. 2001;20:2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peth A, Kukushkin N, Bosse M, Goldberg AL. Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. The Journal of biological chemistry. 2013;288:7781–7790. doi: 10.1074/jbc.M112.441907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanna J, Meides A, Zhang DP, Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 25.Kisselev AF, Callard A, Goldberg AL. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. The Journal of biological chemistry. 2006;281:8582–8590. doi: 10.1074/jbc.M509043200. [DOI] [PubMed] [Google Scholar]
- 26.Adams J, Behnke M, Chen S, Cruickshank AA, Dick LR, Grenier L, Klunder JM, Ma YT, Plamondon L, Stein RL. Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorganic & medicinal chemistry letters. 1998;8:333–338. doi: 10.1016/s0960-894x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- 27.Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods in enzymology. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Hayes JD. Identification of topological determinants in the N-terminal domain of transcription factor Nrf1 that control its orientation in the endoplasmic reticulum membrane. The Biochemical journal. 2010;430:497–510. doi: 10.1042/BJ20100471. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Lucocq JM, Yamamoto M, Hayes JD. The NHB1 (N-terminal homology box 1) sequence in transcription factor Nrf1 is required to anchor it to the endoplasmic reticulum and also to enable its asparagineglycosylation. The Biochemical journal. 2007;408:161–172. doi: 10.1042/BJ20070761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radhakrishnan SK, den Besten W, Deshaies RJ. p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. eLife. 2014;3:e01856. doi: 10.7554/eLife.01856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen JJ, Tsu CA, Gavin JM, Milhollen MA, Bruzzese FJ, Mallender WD, Sintchak MD, Bump NJ, Yang X, Ma J, et al. Mechanistic studies of substrate-assisted inhibition of ubiquitin-activating enzyme by adenosine sulfamate analogues. The Journal of biological chemistry. 2011;286:40867–40877. doi: 10.1074/jbc.M111.279984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anchoori RK, Karanam B, Peng S, Wang JW, Jiang R, Tanno T, Orlowski RZ, Matsui W, Zhao M, Rudek MA, et al. A bisbenzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer. Cancer cell. 2013;24:791–805. doi: 10.1016/j.ccr.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 35.Schrader EK, Harstad KG, Holmgren RA, Matouschek A. A three-part signal governs differential processing of Gli1 and Gli3 proteins by the proteasome. The Journal of biological chemistry. 2011;286:39051–39058. doi: 10.1074/jbc.M111.274993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- 37.Kothe M, Ye Y, Wagner JS, De Luca HE, Kern E, Rapoport TA, Lencer WI. Role of p97 AAA-ATPase in the retrotranslocation of the cholera toxin A1 chain, a non-ubiquitinated substrate. The Journal of biological chemistry. 2005;280:28127–28132. doi: 10.1074/jbc.M503138200. [DOI] [PubMed] [Google Scholar]
- 38.Magnaghi P, D’Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D, Gasparri F, Cozzi L, Cucchi U, Orrenius C, et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nature chemical biology. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- 39.Qian MX, Pang Y, Liu CH, Haratake K, Du BY, Ji DY, Wang GF, Zhu QQ, Song W, Yu Y, et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell. 2013;153:1012–1024. doi: 10.1016/j.cell.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. The Journal of biological chemistry. 1997;272:9086–9092. doi: 10.1074/jbc.272.14.9086. [DOI] [PubMed] [Google Scholar]
- 42.Berko D, Tabachnick-Cherny S, Shental-Bechor D, Cascio P, Mioletti S, Levy Y, Admon A, Ziv T, Tirosh B, Goldberg AL, et al. The direction of protein entry into the proteasome determines the variety of products and depends on the force needed to unfold its two termini. Molecular cell. 2012;48:601–611. doi: 10.1016/j.molcel.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elsasser S, Schmidt M, Finley D. Characterization of the proteasome using native gel electrophoresis. Methods in enzymology. 2005;398:353–363. doi: 10.1016/S0076-6879(05)98029-4. [DOI] [PubMed] [Google Scholar]
- 44.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell metabolism. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.