Abstract
Background
Agarwood is derived from Aquilaria trees, the trade of which has come under strict control with a listing in Appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora. Many secondary metabolites of agarwood are known to have medicinal value to humans, including compounds that have been shown to elicit sedative effects and exhibit anti-cancer properties. However, little is known about the genome, transcriptome, and the biosynthetic pathways responsible for producing such secondary metabolites in agarwood.
Results
In this study, we present a draft genome and a putative pathway for cucurbitacin E and I, compounds with known medicinal value, from in vitro Aquilaria agallocha agarwood. DNA and RNA data are utilized to annotate many genes and protein functions in the draft genome. The expression changes for cucurbitacin E and I are shown to be consistent with known responses of A. agallocha to biotic stress and a set of homologous genes in Arabidopsis thaliana related to cucurbitacin bio-synthesis is presented and validated through qRT-PCR.
Conclusions
This study is the first attempt to identify cucurbitacin E and I from in vitro agarwood and the first draft genome for any species of Aquilaria. The results of this study will aid in future investigations of secondary metabolite pathways in Aquilaria and other non-model medicinal plants.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-578) contains supplementary material, which is available to authorized users.
Keywords: Agarwood, Cucurbitacin, Aquilaria, Genome
Background
Aquilaria agallocha is one of the largest producers of agarwood, a valuable product derived from Aquilaria and Gyrinops trees. Agarwood-producing tree species have become endangered due to the deforestation of tropical forests and the international trade of agarwood has come under strict control with a listing in Appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora. The use of agarwood is prevalent in many cultures, particularly in the Middle East and Asia where it has been used for over a thousand years. In particular, the use of agarwood is prevalent in religious ceremonies, herbal medicine, and as fragrances for perfumes and aromatherapy.
The main compounds related to the medicinal properties of agarwood are terpenes and phenylethyl chromone derivatives [1–3] which can be highly variable in content and composition among different agarwood-producing tree species. Previous studies have focused on sesquiterpenes, the most abundant terpenes compound in agarwood [4, 5]. However, little is known about triterpenoids in agarwood. Terpenoid content is induced under biotic stress as an immune response to resist various pathogens and its derivatives have been shown to exhibit anti-microorganism and anti-tumour functions [6, 7]. A specific compound of interest, cucurbitacin, is produced to combat fungal and bacterial pathogens [8]. Cucurbitacins have previously been isolated in Chinese medicinal herbs and have been shown to have pharmacological effects [6]. In particular, cucurbitacin I is known to repress cancer cell motility by perturbing actin dynamics and has also been known to exhibit cytotoxicity against MDA-MB-468 human breast cancer cells from animal models and indirectly interrupt actin dynamics [9]. The study of anti-tumour compounds and related pathways is thus an important field in agarwood research.
In this study, we identified cucurbitacin E and I from in vitro A. agallocha agarwood and present a draft genome for A. agallocha. In vitro materials were used to perform this study due to the lengthy growth period of resinous material in this species as well as to avoid contamination from microorganisms. Importantly, this process is applicable to plant factories for large-scale production in the future. The DNA and RNA sequence data were obtained using Illumina HiSeq 2000 sequencing technology, from which we performed de novo genome assembly and gene annotation. We inferred a putative pathway for cucurbitacin E and I from the genomic and transcriptomic data in order to better understand these important medicinal compounds in agarwood. The transcripts discovered to be related to the cucurbitacin pathway, were validated through qRT-PCR. To the best of our knowledge, this is the first draft genome for any species of Aquilaria as well as the first study to identify cucurbitacin E and I in agarwood from in vitro materials.
Results and discussion
Agarwood contains high cucurbitacin content
The strain of A. agallocha used in this study was originally derived from Myanmar and domesticated in Taiwan, after import. In vitro materials from this strain were analyzed using LC-ESI-MS where the presence of cucurbitacin I and E were detected (Additional file 1: Figures S1 and S2 respectively). After identification, in vitro callus, shoot and plant materials were analyzed for cucurbitacin E and I content (Figure 1a) where it was seen to be most abundant in in vitro plant. To the best of our knowledge, the cucurbitacin I content from in vitro A. agallocha produced agarwood is significantly higher than in any other Chinese medicinal herb studied previously, with an average concentration of 334.62 μg/g observed in this study as compared to previously reported concentrations of 0.55 μg/g and 25 μg/g from studies by Afifi et al. and Wu et al. respectively [10, 11].
Figure 1.
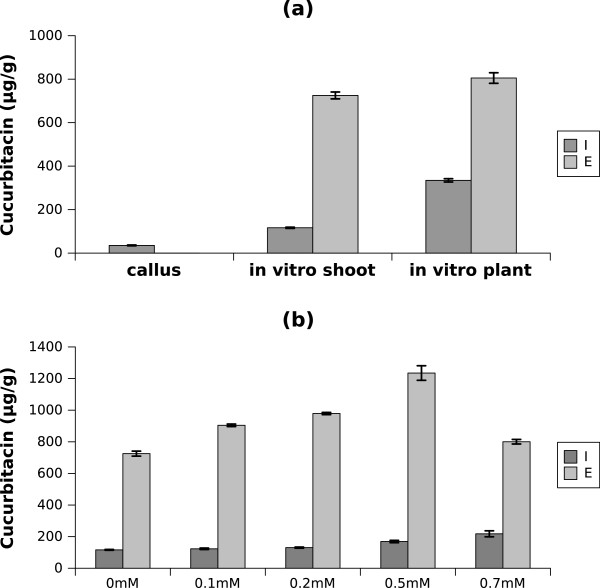
Agarwood endogenous cucurbitacin E and I content. (a) The endogenous cucurbitacin content was measured from different stages of agarwood plant. Data is represented as mean ± standard deviation (n = 5). (b) The endogenous cucurbitacin content of 2 month old agarwood in vitro shoot material was measured after treatment with various concentrations of methyl jasmonate (MJ). Data is represented as mean ± standard deviation (n = 5).
As mentioned, agarwood produces terpenoids under biotic stress [8]. To investigate whether pathogens induce cucurbitacin content, the in vitro material was treated with methyl jasmonate (MJ), an important signal in pathogen related pathways. Various concentrations of MJ were applied to mimic different levels of biotic stress and the change in cucurbitacin content in response to different levels of biotic stress was measured. It was seen that cucurbitacin content increased as MJ concentration increased, up to 218 μg/g of cucurbitacin I at 0.7 mM MJ and 1,235 μg/g of cucurbitacin E at 0.5 mM MJ (Figure 1b). These results reveal that cucurbitacin pathway related genes were induced by MJ, indicating that agarwood may produce cucurbitacins and triterpenoids under biotic stress. With consideration for both cucurbitacin E and I peak values, the 0 mM and 0.5 mM MJ treatment samples from in vitro shoot were chosen for RNA-seq analysis.
Assembly of a draft genome for A. agallocha
Although transcriptome assembly has become commonplace, the majority of transcripts obtained from transcriptome assembly are partial transcripts. Genome assembly may potentially allow for more complete transcript information to be obtained when compared to a de novo transcriptome procedure, thus easing primer design. As well, the sequence information of 5′ UTR regions is more easily obtainable from a genome assembly than from transcriptome studies. These factors allow for deeper studies of gene expression mechanisms.
The size of the haploid A. agallocha genome was estimated to be approximately 736 Mb by flow cytometry (Additional file 1: Figure S3). Five DNA libraries were constructed for a total of 144.3 Gb, which represents approximately 196X sequencing coverage (Table 1). The DNA libraries contained one pair-end library with a fragment length of 300 bp and four mate-pair libraries of various fragment lengths. For the A. agallocha transcriptome, two RNA libraries were constructed to represent MJ treatment conditions (0 mM and 0.5 mM MJ) where polyA + RNA from in vitro shoot was sequenced for a total of 12.5 Gb.
Table 1.
Sequencing read libraries for agarwood ( A. agallocha )
| Library | Type | Read length | No. read pairs | |
|---|---|---|---|---|
| DNA | 300 bp | paired-end | 101 bp | 384,629,812 |
| 2 k bp | mate-pair | 101 bp | 55,940,209 | |
| 4 k bp | mate-pair | 101 bp | 47,800,413 | |
| 5 k bp | mate-pair | 101 bp | 109,363,782 | |
| 10 k bp | mate-pair | 101 bp | 116,549,154 | |
| RNA | 0 mM MJ | paired-end | 91 bp | 40,919,476 |
| 0.5 mM MJ | paired-end | 91 bp | 27,676,735 |
The DNA libraries were utilized in a de novo assembly procedure where the resulting draft genome (NCBI BioProject: PRJNA240626) contained 28,482 scaffolds with an N50 of 126.4 kb, a 1.3 Mb longest sequence, and a total size of 728.5 Mb, approximately 98% genome coverage (Table 2).
Table 2.
Summary of DNA pair-end libraries and de novo genome assembly
| Stage | N50 (kb) | Ave. (kb) | Total length (Mb) | Longest (kb) | No. sequences |
|---|---|---|---|---|---|
| Contigs | 14.6 | 3.1 | 715.3 | 183.7 | 230,048 |
| +2 k | 43.8 | 7.7 | 716.1 | 517.2 | 92,539 |
| +4 k | 72.3 | 15.4 | 727.3 | 872.5 | 47,190 |
| +5 k | 94.0 | 20.2 | 727.5 | 995.7 | 36,097 |
| +10 k | 125.8 | 25.1 | 728.3 | 1289.8 | 29,057 |
| Gapfilled | 126.4 | 25.6 | 728.5 | 1291.6 | 28,482 |
Gene annotation
The TIGR plant repeat database [12] and Repbase (2012/04/18) were combined with ab initio repeat prediction to perform repeat masking. This resulted in 59.18% of the draft genome classified as repeat sequences, with 27.57% classified as Long Tandem Repeat (LTR) elements. Transposable elements are generally non-coding DNA sequences that can change its location within a genome and can play an important role in development and evolution [13]. Thus, annotation of transposable elements was performed in this study using the TIGR plant repeat database (Table 3).
Table 3.
Repeated sequences annotation of transposable elements via TIGR database
| Class | No. | Size (bp) |
|---|---|---|
| Retrotransposon | 747 | 142,807 |
| Transposon | 109 | 15,237 |
| Miniature Inverted-repeat Transposable Elements (MITE) | 1 | 62 |
| Centromere-specific retrotransposon | 1 | 57 |
| Centromere satellite | 10 | 778 |
| Unclassified centromere sequence | 8 | 1,770 |
| Telomere sequence | 11 | 2,215 |
| Telomere associated | 18 | 3,457 |
| rDNA 45S | 48 | 11,686 |
| rDNA 5S | 77 | 9,655 |
| Unclassified (total) | 361 | 38,797 |
RNA-seq data was aligned to the repeat-masked genome (achieving a mapping rate of 79.6%) to provide extrinsic support for gene prediction. Ab initio gene prediction combined with protein alignment resulted in annotations for 40,507 protein-coding genes, among which 3,257 genes encoded for multiple isoforms, representing 44,448 transcripts in total. On average, the predicted gene-models consisted of transcript lengths of 3,465.72 bp, coding lengths of 1,228.27 bp, and 5.48 exons per gene. A total of 66.7% of the predicted gene-models had matches in the NCBI non-redundant protein database, UniProt enzyme database, or matched a protein functional domain in Pfam. The transcriptome sequence data was able to be mapped to the draft genome at a mapping rate of 81.32%. As well, a total of 41.0% of gene-models were supported by 18,837 of 24,205 transcript sequences constructed from RNA-seq using the draft genome as a reference. A de novo assembled transcriptome was also aligned to the draft genome, where 33.0% of predicted exons were supported by 99,125 of 122,323 exons able to be mapped to the draft genome. Functional classification for the set of annotated transcripts was performed using Gene Ontology (GO) (Figure 2).
Figure 2.
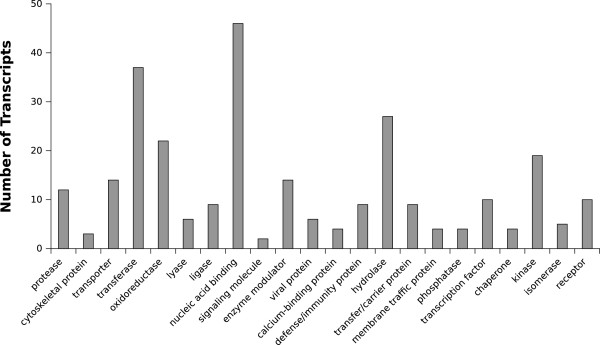
Functional classification of annotated proteins. The gene count for various protein classes as determined by Gene Ontology for the set of annotated proteins in A. agallocha agarwood.
We compared our gene set to those from a recent study by Xu et al. [5], where a de novo transcriptome assembly for Aquilaria sinensis was performed. In their study, 38,159 transcripts were annotated with an average assembled sequence length of 678.65 bp as compared to our results of 44,448 annotated transcripts with an average length coding length of 1,228.27 bp. As well, 35,479 transcripts in our gene set contained both start and stop codons, which is another indicator for completeness. For gene discovery, assembling a draft genome clearly aids in the number of genes discovered as well as in the completeness of the assembled sequence.
Differential expression
Under biotic stress and wounded conditions, MJ is an important signal in a plant’s defence system and will induce secondary metabolites. Thus, the set of annotated transcripts were analyzed for differential expression between the two treatment conditions, 0 mM and 0.5 mM MJ, in order to observe the effect of MJ on gene expression. The short reads from RNA-seq data were aligned to the set of annotated transcripts and the gene expression for each treatment condition was quantified, resulting in 4,827 differentially expressed genes (Additional file 2: Table S1) with at least a two-fold change in expression, of which 2,084 genes were up-regulated and 2,743 genes were down-regulated. Functional classification was performed for the set of differentially expressed genes, using GO (Figure 3). After treatment with 0.5 mM MJ, the activity of transcripts in the categories of metabolic processes and catalytic activity was observed to have increased, which is consistent with observations in previous studies [5].
Figure 3.
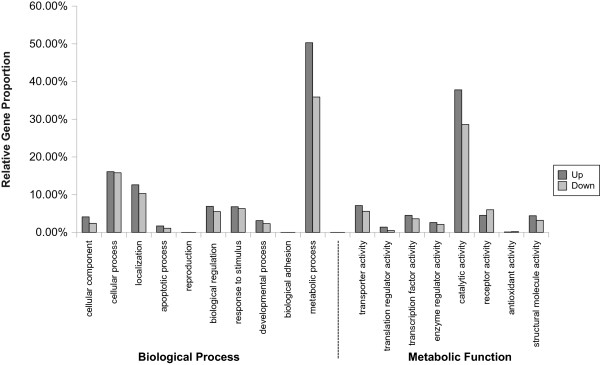
Functional classification of differentially expressed transcripts. Functional classifications of the set of differentially expressed transcripts through Gene Ontology, separated into up and down modes of regulation. It can be seen that metabolic processes and catalytic activity were enriched with up-regulated genes in the presence of biotic stress.
Putative pathway for cucurbitacin E and I
Cucurbitacins belong to the class of cytotoxic triterpenoids and are synthesized from MEP/DOXP and mevalonate pathways [8, 14]. Although the importance of medicinal compounds in A. agallocha agarwood is known, knowledge of its molecular mechanism is lacking and a transgenic line for this species is not able to be created. Thus, an assay of the cucurbitacin bio-synthetic pathway is currently not available.
To investigate the cucurbitacin pathway in A. agallocha, transcripts were annotated using Arabidopsis thaliana proteins as well as UniProt enzymes. The annotated transcripts were then used to infer a putative cucurbitacin pathway in A. agallocha by referring to the mevalonate pathway in A. thaliana from KEGG [15] as well as many differentially expressed cytochrome P450s (CYP450s) and S-adenosyl-L-methionine-dependent methyltransferases (SAM-Mtases) as putative downstream genes (Figure 4).
Figure 4.
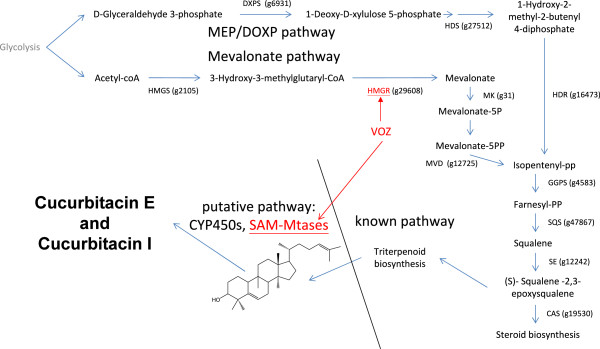
Cucurbitacin E and I pathways in A. agallocha. A schematic illustration showing cucurbitacin E and I bio-synthesis, including isoprenoid precursors via MEP and Mevalonate pathways. The VOZ transcription factor’s cis-element was observed to exist in HMGR.
We identified genes which encode for enzymes in the cucurbitacin E and I pathway (Table 4), including three important gene categories: HMGR [16], DXPS [17], and SQS [17]; as well as genes which encode for synthases in cucurbitacin metabolism. These gene expression results are consistent with the knowledge that cucurbitacins are synthesized to resist pathogens [8]. We also investigated the vascular plant one-zinc-finger transcription factor (VOZ), described as follows.
Table 4.
Enzymes identified in the cucurbitacin E and I pathway and their change in gene expression
| Enzyme | Enzyme | 0.5 mM/0 mM |
|---|---|---|
| Name | Commission | Ratio |
| 1-deoxy-D-xylulose-5-phosphate synthase (DXPS) | 2.2.1.7 | 7.1334 |
| 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) | 1.1.1.267 | 1.2901 |
| 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (MCT) | 2.7.7.60 | 0.4569 |
| 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK) | 2.7.1.148 | 1.8742 |
| 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MCS) | 4.6.1.12 | 0.8282 |
| 4-hydroxy-3-methylbut-2-enyl diphosphate synthase (HDS) | 1.17.7.1 | 5.4502 |
| 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR/IDS) | 1.17.1.2 | 5.9418 |
| acetyl-CoA acetyltransferase (AACT) | 2.3.1.9 | 1.5835 |
| hydroxymethylglutaryl-CoA synthase (HMGS) | 2.3.3.10 | 3.0714 |
| 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) | 1.1.1.34 | 20.4453 |
| mevalonate kinase (MK) | 2.7.1.36 | 2.2961 |
| phosphomevalonate kinase (PMK) | 2.7.4.2 | 1.6881 |
| diphosphomevalonate decarboxylase (MVD) | 4.1.1.33 | 2.3948 |
| isopentenyl-diphosphate delta-isomerase (IPI) | 5.3.3.2 | 1.9775 |
| geranylgeranyl pyrophosphate synthetase (GGPS) | 2.5.1.29 | 3.1042 |
| squalene synthetase (SQS) | 2.5.1.21 | 2.0913 |
| cycloartenol synthase (CAS) | 5.4.99.8 | 1.5239 |
| squalene monooxygenase (SE) | 1.14.13.132 | 1.5357 |
| vascular plant one-zinc-finger transcription factor (VOZ) | 1.3396 |
VOZs are transcription factors that are highly conserved in land plant evolution [18, 19]. It has been shown to bind to the cis-element GCGTNx7ACGC, which belongs to the NAC subgroup VIII-2 [20]. VOZs have been observed to be both positive and negative transcription factors of biotic and abiotic stress-response pathways, respectively, in A. thaliana. Although the voz1voz2-2 mutant did not notably change in endogenous ABA content, the voz1voz2 double mutant was inable to combat pathogens (Pseudomonas syringae and Colletotrichum higginsianum) due to low gene expression from defense-response genes [18]. This indicates that VOZs are positive regulators in the SA and MJ signaling pathways in land plants. Therefore, we speculated that VOZ plays a postive role in the cucurbitacin pathway. A transcript from our gene set homologous to VOZ in A. thaliana and Zea mays was able to be identified with identities of 71.04% and 69.40% respectively.
We identified HMGR as containing the VOZ cis-element in its promoter region. In can be seen from Table 4 that VOZ expression was slightly up-regulated and HMGR was significantly up-regulated after MJ treatment. This provides evidence that VOZ transcription factors are a positive regulator that play a role, directly or indirectly, in the cucurbitacin pathway and biotic stress-response related genes.
The putative pathway also includes CYP450s and SAM-Mtases (a subset of the differentially expressed genes from Additional file 2: Table S1). CYP450s are one of the largest gene families in plants and catalyzes most oxidation steps in secondary metabolism such as in the biosynthesis of defense compounds, pigment, and antioxidants [21, 22]. Putatively, CYP450s may catalyze the conversion of cucurbitadienol. SAM-Mtases may also act on cucurbitadienol by catalyzing methylation, as it is known that many compounds with anti-microorganism functions have cucurbitadienol backbones activated by methylation [23]. We annotated 161 cytochrome P450s and 66 S-adenosyl-L-methionine-dependent methyltransferases (SAM-Mtases) in the A. agallocha genome, of which, 66 CYP450s and 27 SAM-Mtases showed significant up-regulation. These genes can be considered candidate genes that are possibly involved in the cucurbitacin pathway. As well, we identified a small number of SAM-Mtases that contained the VOZ cis-element, though their expression was not observed to be significantly up-regulated.
The transcripts related to the cucurbitacin pathway and the VOZ transcription factor were validated using qRT-PCR (Figure 5). Our results are consistent with the expectation of cucurbitacin pathway genes being up-regulated in response to biotic stress. There is some disparity between qRT-PCR and RNA-seq values. However, this is likely due to differences in platform. The qRT-PCR primer sequences were designed for the 3′ ends of transcripts, which is highly stable, as compared to quantifying expression using the whole transcript with RNA-seq, which is typically not uniformly sequenced.
Figure 5.
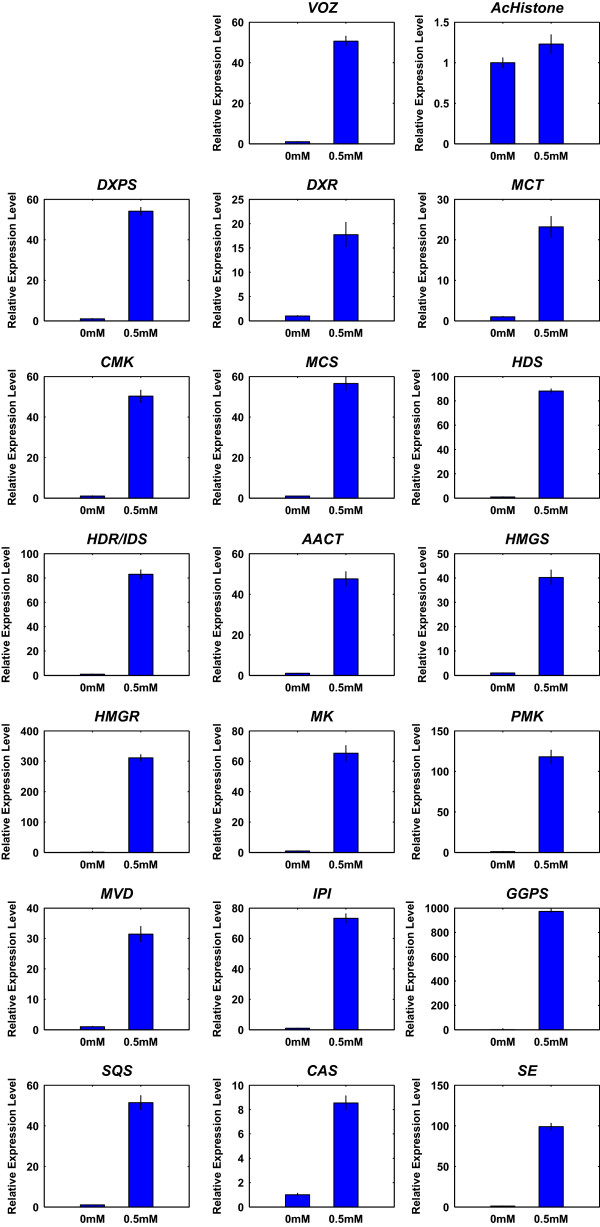
qRT-PCR validation. The qRT-PCR analysis results showing relative change in expression for each gene involved in the cucurbitacin pathway, as well as the VOZ transcription factor and the internal control AcHistone, after treatment with MJ.
Conclusion
Cucurbitacin, a compound of interest with medicinal value which exhibits anti-microorganism and anti-tumour properties was identified and observed to be abundant in in vitro A. agallocha agarwood. To better understand the genes and pathways related to cucurbitacin, a draft genome was assembled, from which, many genes and protein functions were able to be annotated and analyzed. The expression change for cucurbitacins was shown to be consistent with known responses of A. agallocha to biotic stress. As well, the DNA and RNA data was utilized to infer a putative pathway for cucurbitacin E and I in A. agallocha and a set of homologous genes in A. thaliana related to cucurbitacin bio-synthesis was presented. The VOZ transcription factor, a positive regulator of biotic stress-response pathways was identified as well as a putative regulation target among the genes related to the cucurbitacin pathway, HMGR, in which the VOZ cis-element was present in its promoter. The draft genome from this study can provide a resource for the investigation of secondary metabolite pathways not only in Aquilaria trees, but also in other non-model medicinal plants. The confirmation that cucurbitacins can be obtained from in vitro materials with a relatively high yield has positive implications with regards to plant factories to save on costs and time, as well as limiting the effects of research on this endangered species in the wild.
Methods
Plant materials for DNA and RNA extraction
All in vitro plant materials were grown under long-day conditions (16 h of light, 8 h of darkness) at 25°C. A plant regeneration system from shoot tips into in vitro plants was created using a tissue culture process similar to the processes described by He et al. [14]. Bio-assays were performed on the callus, in vitro shoot (2 months), and in vitro plant (4 months). DNA was extracted from 1 g of in vitro materials using the Plant Genomic DNA MiniKit (Maestrogen) following the manufacturer’s instructions. The DNA samples were sent to BGI for sequencing using HiSeq 2000. MJ treatment was performed according to the process described by Kumeta and Ito [4] where MJ was dissolved in DMSO to a concentration of 300 mM and then added to the culture at final concentrations of 0 mM, 0.1 mM, 0.2 mM, 0.5 mM, and 0.7 mM. RNA was extracted from 1 g of in vitro 0 mM and 0.5 mM MJ treated samples using RNeasy Plant MiniKit following the protocol prescribed by the manufacturer.
LC-ESI-MS
In vitro materials were ground with liquid nitrogen and mixed with 1 mL of methanol. Supernatant was collected by centrifugation (12000 rpm, 1 min). The LC-ESI-MS system consisted of an ultra-performance liquid chromatography system (Ultimate 3000 RSLC, Dionex) and an electrospray ionization source of quadrupole time-of-flight mass spectrometer (maXis HUR-QToF system, Bruker Daltonics). The autosampler was set at 4°C. Separation was performed with reversed-phase liquid chromatography on a BEH C8 column (2.1 × 100 mm, Walters). The elution started from 99% mobile phase A (0.1% formic acid in ultrapure water) and 1% mobile phase B (0.1% formic acid in ACN), held at 1% B for 1.5 min, raised to 60% B in 6 min, further raised to 90% in 0.5 min, and then lowered to 1% B in 0.5 min. The column was equilibrated by pumping 1% B for 4 min. The flow rate was set to 0.4 mL/min with an injection volume of 5 μL. LC-ESI-MS chromatogram were acquired under the following conditions: capillary voltage of 4500 V in positive ion mode, dry temperature of 190°C, dry gas flow maintained at 8 L/min, nebulizer gas at 1.4 bar, and acquisition range of m/z 100–1000.
Genome assembly
Five DNA libraries were constructed for a total of 144.3 Gb, which represents approximately 196X sequencing coverage (see Table 1) consisting of one paired-end library and four mate-pair libraries with various fragment lengths. First, the DNA paired-end read library was filtered according to base-call quality (25 of the first 35 bases from the 5′ end must be better than a quality score of 30 for read retention), removing sequence reads which contained ambiguous base-calls, and low complexity (sequences where 85% or more of the bases consist of the same nucleotide). The A. agallocha genome was assembled using String Graph Assembler (SGA) [24]. First, SGA was used to assemble the paired-end reads (kmer length of 41 for error correction, 65 and 79 for the minimum overlap and overlap lengths respectively) into contigs. Then, SSPACE [25] was used to construct scaffolds from the four mate-pair libraries (trimmed to 35 bp remaining at the 5′ end), which were utilized sequentially, from smallest to largest fragment size. Following scaffolding, gap closing was performed using GapFiller [26]. Sequences under 800 bp in length were excluded from the final assembly and subsequent analysis.
Gene annotation
Repeat masking was performed on the assembled genome using RepeatMasker [27] and RepeatModeler [28] along with the TIGR plant repeat database [12] and Repbase (2012/04/18). RNA-seq reads were aligned to the repeat masked genome and potential transcripts were assembled using TopHat followed by Cufflinks [29]. The RNA-seq alignment results and assembled transcript sequences were used to generate extrinsic data for the gene prediction tool Augustus [30], which was used to predict gene models and transcripts from the draft genome assembly. The quality of the gene prediction was checked by performing both reference based transcript assembly with the draft genome using TopHat and Cufflinks as well as a de novo assembly using Velvet [31] followed by Oases [32]. For the reference based assembly, RNA-seq reads were aligned to the draft genome using TopHat, after which Cufflinks was used to assemble transcripts. The Cufflinks assembled transcripts were checked against the predicted transcripts from Augustus using blastn. The Velvet assembled transcripts were aligned to the draft genome using TopHat and blat. The regions in the draft genome which were able to be mapped by Velvet assembled transcripts were overlapped with Augustus predicted exon regions in order to determine how many exon locations were successfully predicted by Augustus. Transcripts from predicted gene-models were aligned against the NCBI non-redundant set of proteins using blastp (E-value 1E-5) to find homologues. The best alignment for each transcript was retained as annotation. Functional classification for the set of annotated transcripts was performed using the webserver Panther and its GO gene analysis tool [33].
Differential expression
RNA-seq reads for the 0 mM and 0.5 mM MJ treatment conditions were individually aligned to the set of annotated transcripts using BWA [34]. For each condition, quantification of transcript expression was performed by using eXpress [35] to calculate the fragments per kilobase per million (FPKM) for each transcript. The fold change (0.5 mM/0 mM) was calculated for each transcript from the FPKM values. A transcript was denoted up-regulated or down-regulated if the log2 fold change was greater than 1 or less than −1, respectively, otherwise, a transcript was denoted non-differentially expressed. The fold change at the gene level was obtained by averaging the fold change ratios of all transcripts belonging to the same gene, as determined by annotation using blastp [36]. In other words, the fold change for transcripts assigned to the same A. thaliana gene ID was averaged to obtained a gene level fold change. Functional classification for the set of differentially expressed transcripts was performed using the webserver Panther and its GO gene analysis tool [33].
Putative cucurbitacin E and I pathway
From the differential expression analysis, transcripts which exhibited expression (FPKM > 0) in both 0 mM and 0.5 mM MJ treatment conditions were annotated with A. thaliana proteins from TAIR as well as UniProt enzymes for EC classifications using blastp (E-value 1E−5). The resulting set of annotated transcripts was used to infer a putative cucurbitacin E and I pathway by referring to the mevalonate pathway of A. thaliana from KEGG [15].
The promoter regions of genes related to the cucurbitacin pathway was assayed for the VOZ cis-element, GCGTNx7ACGC. A promoter region was denoted as 5,000 bp upstream from the gene TSS. A sequence motif search for the VOZ cis-element was performed in the promoter regions using Perl.
qRT-PCR validation
Validation of the curcubitacin I pathway transcripts as well as the transcription factor VOZ found in A. agallocha was performed using qRT–PCR analysis. The 0 mM and 0.5 mM MJ treated RNA samples were extracted from 1 g of four month old in vitro A. agallocha shoots using RNeasy Plant MiniKit following the protocol prescribed by the manufacturer. Primers pairs were designed for each transcript (Additional file 3: Table S2) with the ABI Prism 7500 sequence detection system (Applied Biosystems). Each primer pair was used to amplify the respective cDNA fragments using a cycling profile consisting of 58°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. The relative gene expression was determined by the comparative CT method, 2−ΔCT (ΔCT = CT, gene of interest – CT, control gene), using AcHistone as the internal control [5]. Four independent biological repeats were performed for each assay where the final expression value is the mean expression of the repeats.
Availability of supporting data
All supporting data used in this study is publicly available at NCBI under BioProject: PRJNA240626 (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA240626). Specifically, the genome assembly can be obtained under GenBank Assembly ID: GCA_000696445.1 (http://www.ncbi.nlm.nih.gov/assembly/GCA_000696445.1). The transcriptome sequences are under SRA accessions: SRX550129 (http://www.ncbi.nlm.nih.gov/sra/SRX550129), SRX540116 (http://www.ncbi.nlm.nih.gov/sra/SRX540116). As well, our A. agallocha specimen has been submitted to a herbarium at Herbarium, Research Center for Biodiversity, Academia Sinica, Taipei (HAST) under the accession number 137059 (http://www.hast.biodiv.tw/specimens/SpecimenDetailE.aspx?specimenOrderNum=137059).
Electronic supplementary material
Additional file 1: Figure S1: Identification of Cucurbitacin I (formula weight: 514.65 g) with LC-ESI-MS. Red represents the shoot tip sample mixed with cucurbitacin I standard. Green represents the shoot tip sample. Figure S2. Identification of Cucurbitacin E (formula weight: 556.69 g) with LC-ESI-MS. Red represents the shoot tip sample mixed with cucurbitacin E standard. Green represents the shoot tip sample. Figure S3. Genome size of A. agallocha by flow cytometry. The haploid genome size of A. agallocha was approximately 0.604-fold of that of the reference standard (CEN Singlet; 2C = 2.5 pg DNA, 1 pg = 978 Mb). (DOC 5 MB)
Additional file 2: Table S1: The set of 4,827 differentially expressed genes. (CSV 218 KB)
Additional file 3: Table S2: Gene specific primers for real-time PCR analysis of gene expression. (DOC 34 KB)
Acknowledgements
The authors would like to thank Academia Sinica and National Science Council of Republic of China, Taiwan, for the financial support under the grants: NSC 102-2313-B-001-001-MY3, 101-2313-B-001-002. The staff of TCX-D800 Metablomics Core, Technology Commons, College of Life Science, National Taiwan University for their help with the LC-ESI-MS analysis. The technical support was provided by Su-Hsin Huang at the Scientific Instrument Center’s Flow Cytometry Core Facility in Institute of Plant and Microbial Biology, Academia Sinica, Taipei, Taiwan. Dr. Ching-I Peng, the curator for Herbarium, Academia Sinica, Taipei for his help in sample preparations.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Author contributions
The initiation and financial responsibility of this study were from LFOC and HFL. Experiments were designed by CHC, CYC, and LFOC. Biological experiments were performed by TCYK, CHC, TYC, MJC, MHY. Sequencing data were analyzed by TCYK, CHC, TYC, MHY, CYC. The in vitro plant manipulation, sampling and quality were controlled by MJC and LCH. Supervision performed by LCH, STJ, CYC, LCH and LFOC. Manuscript was prepared by TCYK and CHC with input from the other coauthors. All authors read and approved the final manuscript.
Contributor Information
Chuan-Hung Chen, Email: Chentim19850301@gmail.com.
Tony Chien-Yen Kuo, Email: tonykuo403@gate.sinica.edu.tw.
Meng-Han Yang, Email: d95922001@ntu.edu.tw.
Ting-Ying Chien, Email: tinin@mars.csie.ntu.edu.tw.
Mei-Ju Chu, Email: meiju@gate.sinica.edu.tw.
Li-Chun Huang, Email: bolch@gate.sinica.edu.tw.
Chien-Yu Chen, Email: cychen@mars.csie.ntu.edu.tw.
Hsiao-Feng Lo, Email: hflo@ntu.edu.tw.
Shih-Tong Jeng, Email: stjeng@ntu.edu.tw.
Long-Fang O Chen, Email: ochenlf@gate.sinica.edu.tw.
References
- 1.Chen HQ, Wei JH, Yang JS, Zhang Z, Yang Y, Gao ZH, Sui C, Gong B. Chemical constituents of agarwood originating from the endemic genus Aquilaria plants. Chem Biodivers. 2012;9:236–250. doi: 10.1002/cbdv.201100077. [DOI] [PubMed] [Google Scholar]
- 2.Liu YY, Chen HQ, Yang Y, Zhang Z, Wei JH, Meng H, Chen WP, Feng JD, Gan BC, Chen XY, Gao ZH, Huang JQ, Chen B, Chen HJ. Whole-tree Agarwood-Inducing Technique: An Efficient Novel Technique for Producing High-Quality Agarwood in Cultivated Aquilaria sinensis Trees. Molecules. 2013;18:3086–3106. doi: 10.3390/molecules18033086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueda J, Imamura L, Tezuka Y, Tran QL, Tsuda M, Kadota S. New sesquiterpene from Vietnamese agarwood and its induction effect on brain-derived neurotrophic factor mRNA expression in vitro. Bioorg Med Chem. 2006;14:3571–3574. doi: 10.1016/j.bmc.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 4.Kumeta Y, Ito M. Characterization of delta-guaiene synthases from cultured cells of Aquilaria, responsible for the formation of the sesquiterpenes in agarwood. Plant Physiol. 2010;154:1998–2007. doi: 10.1104/pp.110.161828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y, Zhang Z, Wang M, Wei J, Chen H, Gao Z, Sui C, Luo H, Zhang X, Yang Y, Meng H, Li W. Identification of genes related to agarwood formation: transcriptome analysis of healthy and wounded tissues of Aquilaria sinensis. BMC Genomics. 2013;14:227. doi: 10.1186/1471-2164-14-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Momma K, Masuzawa Y, Nakai N, Chujo M, Murakami A, Kioka N, Kiyama Y, Akita T, Nagao M. Direct interaction of Cucurbitacin E isolated from Alsomitra macrocarpa to actin filament. Cytotechnology. 2008;56:33–39. doi: 10.1007/s10616-007-9100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Yang Y, Xue J, Wei J, Zhang Z. 2011. Molecules. 2011;16:4884–4896. doi: 10.3390/molecules16064884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JC, Chiu MH, Nie RL, Cordell GA, Qiu SX. Cucurbitacins and cucurbitane glycosides: structures and biological activities. Nat Prod Rep. 2005;22:386–399. doi: 10.1039/b418841c. [DOI] [PubMed] [Google Scholar]
- 9.Knecht DA, LaFleur RA, Kahsai AW, Argueta CE, Beshir AB, Fenteany G. Cucurbitacin I inhibits cell motility by indirectly interfering with actin dynamics. PLoS One. 2010;5:e14039. doi: 10.1371/journal.pone.0014039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu PL, Lin FW, Wu TS, Kuoh CS, Lee KH, Lee SJ. Cytotoxic and anti-HIV principles from the rhizomes of Begonia nantoensis. Chem Pharm Bull. 2004;52:345–349. doi: 10.1248/cpb.52.345. [DOI] [PubMed] [Google Scholar]
- 11.Ayyad SE, Abdel-Lateff A, Basaif SA, Shier T. Cucurbitacins-type triterpene with potent activity on mouse embryonic fibroblast from Cucumis prophetarum, cucurbitaceae. Pharmacognosy Res. 2011;3:189–193. doi: 10.4103/0974-8490.85006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouyang S, Buell CR. The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004;32:D360–D363. doi: 10.1093/nar/gkh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/S0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 14.He ML, Qi SY, Hu LJ. Rapid in vitro propagation of medicinally important Aquilaria agallocha. J Zhejiang Univ Sci B. 2005;6:849–852. doi: 10.1631/jzus.2005.B0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song AA, Abdullah JO, Abdullah MP, Shafee N, Othman R, Tan EF, Noor NM, Raha AR. Overexpressing 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) in the lactococcal mevalonate pathway for heterologous plant sesquiterpene production. PLoS One. 2012;7:e52444. doi: 10.1371/journal.pone.0052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benveniste P. Sterol metabolism. Arabidopsis Book. 2002;1:e0004. doi: 10.1199/tab.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakai Y, Nakahira Y, Sumida H, Takebayashi K, Nagasawa Y, Yamasaki K, Akiyama M, Ohme-Takagi M, Fujiwara S, Shiina T, Mitsuda N, Fukusaki E, Kubo Y, Sato MH. Vascular plant one-zinc-finger protein 1/2 transcription factors regulate abiotic and biotic stress responses in Arabidopsis. Plant J. 2013;73:761–775. doi: 10.1111/tpj.12069. [DOI] [PubMed] [Google Scholar]
- 19.Mitsuda N, Hisabori T, Takeyasu K, Sato MH. VOZ; isolation and characterization of novel vascular plant transcription factors with a one-zinc finger from Arabidopsis thaliana. Plant Cell Physiol. 2004;45:845–854. doi: 10.1093/pcp/pch101. [DOI] [PubMed] [Google Scholar]
- 20.Jensen MK, Hagedorn PH, de Torres-Zabala M, Grant MR, Rung JH, Collinge DB, Lyngkjaer MF. Transcriptional regulation by an NAC (NAM-ATAF1,2–CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 2008;56:867–880. doi: 10.1111/j.1365-313X.2008.03646.x. [DOI] [PubMed] [Google Scholar]
- 21.Bak S, Beisson F, Bishop G, Hamberger B, Hofer R, Paquette S, Werck-Reichhart D. Cytochromes p450. Arabidopsis Book. 2011;9:e0144. doi: 10.1199/tab.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coon MJ. Cytochrome P450: nature’s most versatile biological catalyst. Annu Rev Pharmacol Toxicol. 2005;45:1–25. doi: 10.1146/annurev.pharmtox.45.120403.100030. [DOI] [PubMed] [Google Scholar]
- 23.Struck AW, Thompson ML, Wong LS, Micklefield J. S-adenosyl-methionine-dependent methyltransferases: highly versatile enzymes in biocatalysis, biosynthesis and other biotechnological applications. Chembiochem. 2012;13:2642–2655. doi: 10.1002/cbic.201200556. [DOI] [PubMed] [Google Scholar]
- 24.Simpson JT, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2012;22:549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 26.Boetzer M, Pirovano W. Toward almost closed genomes with GapFiller. Genome Biol. 2012;13:R56. doi: 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smit AFA, Hubley R, Green P: Repeatmasker open-3.0.http://www.repeatmasker.org 1996–2010
- 28.Smit AFA, Hubley R: Repeatmodeler open-1.0.http://www.repeatmasker.org 2008–2010
- 29.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 31.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, Kitano H, Thomas PD. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1: Identification of Cucurbitacin I (formula weight: 514.65 g) with LC-ESI-MS. Red represents the shoot tip sample mixed with cucurbitacin I standard. Green represents the shoot tip sample. Figure S2. Identification of Cucurbitacin E (formula weight: 556.69 g) with LC-ESI-MS. Red represents the shoot tip sample mixed with cucurbitacin E standard. Green represents the shoot tip sample. Figure S3. Genome size of A. agallocha by flow cytometry. The haploid genome size of A. agallocha was approximately 0.604-fold of that of the reference standard (CEN Singlet; 2C = 2.5 pg DNA, 1 pg = 978 Mb). (DOC 5 MB)
Additional file 2: Table S1: The set of 4,827 differentially expressed genes. (CSV 218 KB)
Additional file 3: Table S2: Gene specific primers for real-time PCR analysis of gene expression. (DOC 34 KB)


