Abstract
Global health, whether in developed or developing countries, is in need of robust systems diagnostics for major diseases, such as HIV/AIDS, impacting the world populations. Fourier transform Infrared (FTIR) spectroscopy of serum is a quick and reagent-free methodology with which to analyze metabolic alterations such as those caused by disease or treatment. In this study, Attenuated Total Reflectance Fourier-Transform (ATR-FTIR) Spectroscopy was investigated as a means of distinguishing HIV-infected treatment-experienced (HIVpos ARTpos, n=39) and HIV-infected-treatment-naïve (HIVpos ARTneg, n=16) subjects from uninfected control subjects (n=30). Multivariate pattern recognition techniques, including partial least squares discriminant analysis (PLS-DA) and orthogonal partial least squares discriminant analysis (OPLS-DA), successfully distinguished sample classes, while univariate approaches identified significant differences (p<0.05) after Benjamini-Hochberg corrections. OPLS-DA discriminated between all groups with sensitivity, specificity, and accuracy of >90%. Compared to uninfected controls, HIVpos ARTpos and HIVpos ARTneg subjects displayed significant differences in spectral regions linked to lipids/fatty acids (3010 cm−1), carbohydrates (1299 cm−1; 1498 cm−1), glucose (1035 cm−1), and proteins (1600 cm−1; 1652 cm−1). These are all molecules shown by conventional biochemical analysis to be affected by HIV/ART interference. The biofluid metabolomics approach applied here successfully differentiated global metabolic profiles of HIV-infected patients and uninfected controls and detected potential biomarkers for development into indicators of host response to treatment and/or disease progression. Our findings therefore contribute to ongoing efforts for capacity-building in global health for robust omics science and systems diagnostics towards major diseases impacting population health.
Introduction
In addition to immune system disruption, HIV infection is also known to cause metabolic abnormalities ranging from dyslipidemia, hyperglycemia, insulin resistance, and diabetes (Butt et al., 2004; Dube et al., 2003; Friis-Moller et al., 2003; Koutkia and Grinspoon, 2003; Omech et al., 2012). Antiretroviral therapy (ART) can normalize CD4+ cell count leading to reduced morbidity and death in HIV-infected individuals (Vrisekoop et al., 2008), but the continued use of ART can lead to metabolic disorders (John et al., 2001; Koutkia and Grinspoon, 2003). HIV-1-infected individuals, and especially those on treatment, must be monitored constantly for the onset of metabolic complications in order to initiate interventions to delay and/or prevent severe metabolic diseases such as type II diabetes and heart disease (Butt et al., 2004; Friis-Moller et al., 2003).
The diagnosis and monitoring of metabolic abnormalities resulting from HIV infection and/or ART is based mainly on radioimmunoassays, dual-energy X-ray absorptiometry (DEXA), and numerous other colorimetric assays (Chang et al., 2004; Negredo et al., 2002; Taylor et al., 2007) where one metabolite or a group of related metabolites are detected at a time. These methods are not only laborious to perform but also expensive and can be subjective (Wohl et al., 2006). A possible solution to some of these issues is offered by metabolomics methodologies where multiple metabolites can be measured in a single analysis.
Several spectroscopic techniques have been employed to obtain metabolic profiles that distinguish diseased from healthy biofluids (Derenne et al., 2012; Edelstein, 2011; Graça et al., 2013; Johnston et al., 2010; Serkova and Niemann, 2006; Sitole et al., 2013). Previous studies from our group described the potential of both nuclear magnetic resonance (NMR) spectroscopy (Hewer et al., 2006; Philippeos et al., 2009) and mass spectrometry (MS) (Williams et al., 2012) in HIV/AIDS metabolomics. Here we explore the use of ATR-FTIR, a methodology incorporating a much simpler data collection approach, for the detection of metabolites in HIV-infected sera compared to negative controls with the secondary goal of determining whether metabolites detected in this manner would be comparable to those identified by NMR and MS-metabolomics and thus leading to the same conclusions.
Vibrational spectroscopic methods such as FTIR- and Raman spectroscopy are multipurpose techniques that offer advantages in simplicity, rapidity, low-cost, and minimal sample preparation. FTIR spectroscopy, in combination with multivariate statistical analysis of data, was shown to be a useful method of metabolic profiling of disease, since it accurately reflected the phenotype of a sample (Ellis et al., 2006). Very few studies have evaluated the potential of FTIR technology for distinguishing spectra from HIV-infected compared to uninfected individuals. Bahmani and co-workers (2009) demonstrated that visible–near-infrared (Vis-NIR) spectra, coupled with partial least square (PLS) regression could detect HIV infection in plasma samples. In their study, Bahmani et al. (2009) showed that plasma Vis-NIR spectroscopy combined with principal component analysis (PCA) and soft independent modelling of class analogy (SIMCA) allowed for a discrimination between samples from HIVpos untreated patients and uninfected controls. This makes sense because the effect of HIV on metabolism is dependent on viral load, with higher levels being more detrimental to the patient and the most extreme cases of metabolic interference being seen in the presence of ART. Data presented by Bahmani et al. (2009) suggested that vibrational changes of the functional groups in ArCH (857–890 nm), ROH (940–970 nm), and RNH2 (1030 nm) were sufficient to discriminate cases in the context of their HIV status. Since these functional groups (ArCH, ROH, and RNH2) are found in proteins, lipids, and saccharides, cautious inferences regarding metabolic irregularities associated with these molecules can be made.
Given the preliminary successes with Vis-NIR spectra able to discriminate HIV-infected compared to uninfected spectra, this current investigation attempted to show an improved discrimination between HIV infected (treated and untreated) and uninfected sera using FTIR spectroscopy in the mid-IR region (4000–400 cm−1). Since the mid-IR region detects vibrations of essential biomolecules, it is widely used in studies of spectral differences of biological samples (as reviewed by Ellis et al., 2006).
The metabolic differences detected by ATR-FTIR spectroscopy between the experimental groups is presented here for the first time. ATR-FTIR spectroscopy coupled with chemometrics successfully distinguished sera from HIV-infected patients and uninfected controls with distinctions visible in the presence of treatment. This data allowed for conclusions similar to those reached using data collected with more sensitive metabolomics methodologies such as NMR and MS.
Materials and Methods
Ethics approval
The ethics committee of the University of Pretoria (Faculties of Natural and Agricultural Sciences and Health Sciences) approved this study with protocol numbers E080-506-019 and 163/2008. All patient volunteers gave their consent for involvement in the study, and patient anonymity was maintained throughout. HIV-infected patients were randomly selected from the King's Hope Clinic in Diepsloot, west of Johannesburg, and from Eersterust Community Health Centre in Pretoria, South Africa. Clinical information on each participant was obtained from both the medical records, and from a questionnaire completed upon sample collection.
Sample collection
Fifty-five HIV-infected serum samples were collected, 39 of which were on anti-retroviral treatment regimens, predominantly non-nucleoside reverse transcriptase inhibitors (NNRTI)-based regimens. The remaining 16 HIV-infected samples were not on any retroviral treatment. Similarly, 30 uninfected controls were also collected. Sample collection, preparation, and storage occurred as described by Hewer et al. (2006) and Williams et al. (2013). Table 1 presents the most relevant clinical information for the patients.
Table 1.
Clinical Information for All Three Experimental Groups
| HIV status | |||
|---|---|---|---|
| HIV− | HIV+ ART+ | HIV+ ART− | |
| % Female | 90% | 84.6% | 87.5% |
| % Male | 10% | 15.3% | 12.5% |
| Mean age (years)±SD | 28.41±8.50 | 40.29±8.12 | 41.6±10.05 |
| Mean CD4 count (cells/mm3 blood) | ND | 357.227 | 188.950 |
| Viral load (copies/mL) | N/A | 17981.16 | 119933.95 |
ND=not done; N/A=not applicable.
Mid-ATR-FTIR spectroscopy of serum
Serum samples were liquefied slowly at room temperature. Following 30 min incubation in a 56°C water bath, 5–10 μL serum was transferred onto separate glass slides (Kevley Technologies, Ohio, USA) and air-dried overnight to form homogeneous dried films. Serum samples were then scraped off onto the diamond crystal surface area. IR spectra were acquired in absorbance mode (mid-IR source 4000–400 cm−1) using a Bruker FTIR V70x spectrometer (Bruker Optik GmbH, Karlsruhe, Germany) containing an ATR attachment and operated by OPUS (7) software. A diamond single reflection ATR accessory was used to procure the mid-IR spectra of each serum sample. A background absorption spectrum (for atmospheric correction) was taken after every 10 samples. The ATR diamond crystal was washed with ethanol and distilled water (three washes) and dried with tissue before use and between each analysis. Spectra were obtained at 4 cm−1 resolution and an average of 32 scans per sample was collected. Using OPUS software, rubberband baseline corrections and vector normalizations were performed for all the spectra. Differentiation was carried out using the Savitzky–Golay (SG) algorithm (Savitzky et al., 1964) with the following settings: number of coefficients, 17 (window size); polynomial fit order, 2; differentiation orders, 2.
Spectral processing
Processing of the serum spectra
Standardization of the data was done as described by Hewer et al. (2006). All spectra were normalized prior to multivariate analysis.
Linear discriminant analysis
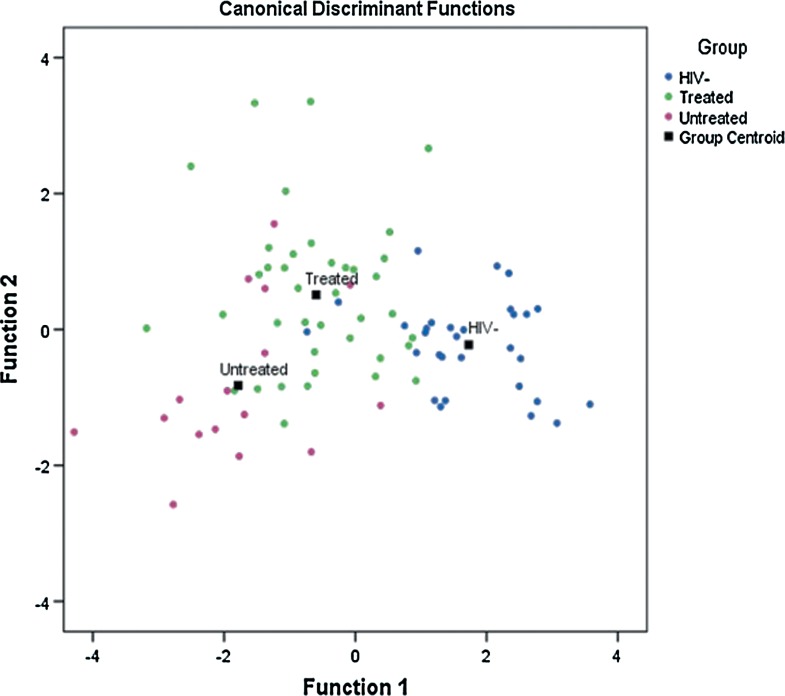
In order to classify cases into their respective groups (HIVpos ARTpos, HIVpos ARTneg and uninfected control), linear discriminant analysis (LDA) was used as described by Philippeos et al. (2009) where stepwise analysis was applied to the 1867 frequencies to obtain the best data set for the classification. The resulting classification was based on 8 out of the 1867 spectral variables, these 8 variables being the ones that presented the highest difference between the groups. A leave-one-out-at a time cross-validation (LOOCV) was done similar to that described by both Hewer et al. (2006) and Philippeos et al. (2009), where each sample was left out of the analysis (validation data) and then classified based on the other samples (training data). This was repeated such that each sample is left out at least once. The discriminant functions resulting from this analysis are depicted in the scatter plot (Fig. 1).
FIG. 1.
Representative scatter plot of ATR-FTIR spectra obtained from HIV infected (treated and untreated) and uninfected controls. The three experimental groups' discriminant functions are: 30 uninfected controls, 39 HIVpos ARTpos, and 16 HIVpos ARTneg.
Multivariate statistical analysis
Prior to multivariate analysis, an inclusion criterion was applied to the HIV-infected groups that was based on CD4 count. Only samples with CD4 count higher than 200 μ/L were included for the HIVpos ARTpos group, and only samples with CD4 count lower than 200 μ/L were included for the HIVpos ARTneg group. Also, the raw data of six control spectra produced values that were inconsistent with instrument controls and were excluded from further analysis. From the remaining samples, the average CD4+ count for the HIVpos ARTpos patients (n=30) was 357.22 cells/μL, HIVpos ARTneg patients (n=16) was 188.95 cells/μL, and the total number of individuals in the control group was 24.
Further analysis was performed using the SIMCA-P software package, version 13.0 (Umetrics, Umeå, Sweden). PCA was performed to observe inherent group clustering. Supervised PLS-DA was applied to obtain improved class discrimination. The leave-one-out cross-validation method was used to validate the quality of the PLS-DA models, and the R2 and Q2 parameters were used to define the quality of the models. R2 represents the total explained difference for the X matrix and indicates goodness of fit, and Q2 indicates the predictability of a model (Boccard et al., 2013; Szymanska et al., 2012). In biological samples a good predictive capability is indicated by Q2>0.5 and an excellent capability is indicated by Q2>0.9 (Sadeghi-Bazargani et al., 2011). The reliability of the models was further validated by the permutation test where n=200 (Lindgren et al., 1996). In addition, OPLS-DA was used for class discrimination and biomarker identification. OPLS-DA is a supervised method that uses an orthogonal signal filter to find the highest covariance between a given dataset and the sample class. The method was constructed using the FT-IR data as the X matrix, and the class information identifier for the different groups as the Y vector. The OPLS-DA model was performed to give a clearer separation since it removes variability that is not relevant to class separation within a given model. A more detailed explanation of the OPLS-DA methodology can be found in Worley and Powers (2013). Finally, in order to access the reliability of the OPLS-DA model, analysis of variance testing of cross-validated predictive residuals (CV-ANOVA) was performed. CV-ANOVA is generally calculated to highlight the significance of the two group separation in an OPLS-DA model (Ericksson et al., 2008; Wheelock and Wheelock, 2013). In order to identify the variables responsible for the separation of the groups, Variable Importance in Projection (VIP) values was used. VIP scores are used for variable selection as they give the discriminatory power of each variable (Wheelock and Wheelock, 2013). VIP values >1.0 indicate maximum discriminatory power, whereas those with values <1 indicate minimal discriminatory power (Chong et al., 2005). Significance tests using t-test was performed using SPSS version 19.0. Significance was determined after “BH” correction and considered significant when the p value <0.05.
Addressing potentially confounding factors
Since the samples were from a random selection of individuals, there were a number of differences within the groups, the biggest differences being age and gender. To gauge the effects of each of these factors on the metabolomics profiles, an OPLS-DA model was developed that included the effects of age and gender. This was done by comparing the original model with class information of Y=HIVpos ARTpos /HIVpos HIVneg patients and uninfected controls to: i) Age model with class information of Y=A (20–30 years) and B=(40–60years); and ii) Gender model with class information of Y=male and female. This comparison was done to identify whether age and gender were major casual factors. All these models were generated from the same dataset with the only difference being the class information. The age and gender models for the HIVpos ARTpos group are shown in the Supplementary Material section. (Supplementary material is available online at www.liebertpub.com/omi.)
Results
Population characterization
A total of 85 blood samples were collected, of which 30 were from uninfected healthy volunteers (uninfected controls), 39 HIV-infected individuals receiving antiretroviral treatment (HIVpos ARTpos patients), and 16 HIV-infected not on treatment (HIVpos ARTneg patients). The ages, viral load, and CD4 counts for the different patient groups are provided in Table 1.
FT-IR spectra of serum samples
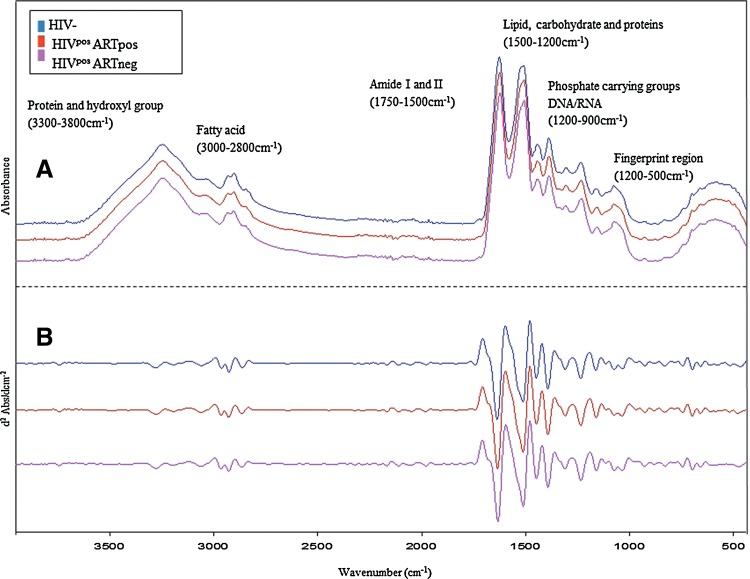
Representative averages of the serum FT-IR spectra of (a) uninfected volunteers, (b) HIVpos ARTpos patients, and (c) HIVpos ARTneg patients are shown in Figure 2A raw data and Figure 2B second derivative in the wavelength range of 400–4000 cm−1. The second derivative spectra highlight small differences that may not be visible in the raw spectra. The standard deviation and distance between the three groups is represented in Supplementary Figures S1 and S2. A few absorption features were identified from all three spectral types. These features include; the C=O stretching vibration modes of the amide I absorption range (1652 cm−1), coupled N-H functional group modes of amide II (1480–1580 cm−1), as well as the functional group modes existing in proteins and amino acids (C-N stretching vibration 1200–1500 cm−1). Also visible are the carbohydrates (1300–925 cm−1) and lipid/fatty acid (3300–2800 cm−1) absorption features. Visually, there were slight differences between the spectra of all three groups which became more pronounced following statistical evaluation of the data.
FIG. 2.
Representative averaged ATR-FTIR spectra of serum obtained from: (A) HIV uninfected controls (blue), HIVpos ARTpos (red), and HIVpos ARTneg (pink). (B) Second derivative spectra of HIV negative controls, HIVpos ARTpos and HIVpos ARTneg. Differences are visible amongst the three groups.
Linear discriminant analysis
LDA easily handles cases where the ‘within-class’ frequencies are unequal and maximizes the ratio of ‘between class’ variance to the ‘within-class’ variance in any particular data set, thereby guaranteeing maximal separation (Fukunaga, 1990). Stepwise LDA analysis was performed to confirm that an FT-IR metabolomics approach would be capable of distinguishing three different experimental groups as seen in previous HIV-NMR and MS-metabolomics studies. Table 2 shows the LDA classification of samples as HIVpos ARTpos, HIVpos ARTneg and uninfected controls, with original grouped cases and cross-validated cases. The original classification accuracy was 78.8% and 72.9% after cross-validation (Table 2). Both original and cross-validation analysis verified that classification of the samples into the three separate groups was possible. From the scatter plot (Fig. 1), the three groups show some clustering with a degree of overlap. This overlap is probably due to related metabolic processes occurring in all three groups. Nevertheless, LDA was able to confirm that ATR-FTIR serum spectra of HIVpos ARTpos, HIVpos ARTneg, and uninfected controls could be separated into separate groups.
Table 2.
Classification of Samples as HIVpos ARTpos and HIVpos ARTneg and Uninfected Controls
| Predicted Group Membership | ||||
|---|---|---|---|---|
| Group | HIV- | Treated | Untreated | Total |
| Original | ||||
| Count | ||||
| HIV- | 28 | 2 | 0 | 30 |
| treated | 4 | 28 | 7 | 39 |
| untreated | 1 | 4 | 11 | 16 |
| % | ||||
| HIV- | 93.3 | 6.7 | 0.0 | 100.0 |
| treated | 10.3 | 71.8 | 17.9 | 100.0 |
| untreated | 6.3 | 25.0 | 68.8 | 100.0 |
78.8% of original grouped cases correctly classified; 72.9% of cross-validated grouped cases correctly classified.
Discrimination between HIV-infected patients and uninfected controls
Unsupervised PCA was performed to obtain an indication of the variation between the mentioned experimental groups and to identify outliers. The PCA scores plots identified a few outliers. There are a number of reasons why an observation may be identified as an outlier, including differences in biochemical composition of samples, and presence of other disease and/or infections (tuberculosis, pneumonia, or diabetes). Given that the removal of outliers may at times be biased, the PCA scores plot, scores contribution plots, Hotellings T2, and DModX were used to compare and verify outliers prior to their removal. Nine outliers were identified and excluded before further analysis. Based on the contribution plot, comparisons between the outliers and the rest of the samples within the dataset, these nine outliers exhibited atypical intensities in the hydroxyl and protein spectral regions (3300–3800 cm−1). These observations are currently under further investigation.
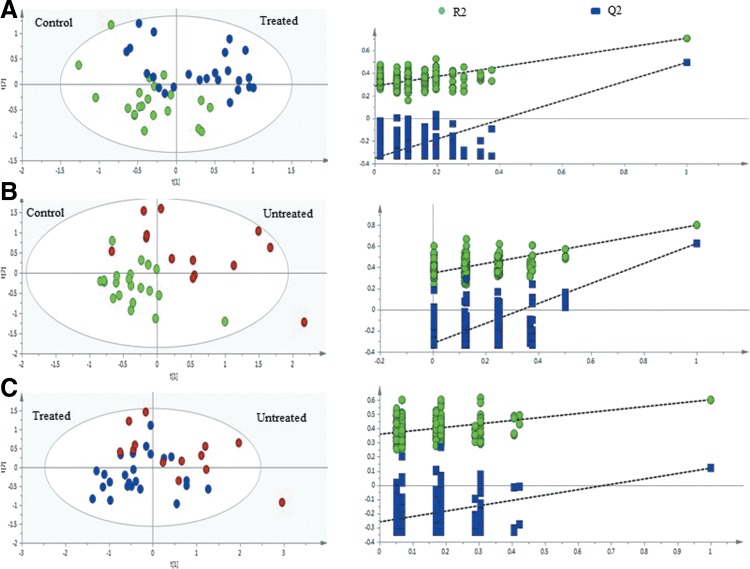
Since PCA is an unsupervised method, score plots could not reveal an obvious separation between groups. In order to get a clearer separation between the experimental groups, supervised PLS-DA was applied. The PLS-DA scores plot for HIVpos ARTpos patients versus uninfected controls (Fig. 3A) showed a minor separation (R2X=0.615, R2Y=0.709, Q2=0.495), indicating a difference between the metabolic profiles of HIVpos ARTpos patients and uninfected controls. Similarly, Figure 3B shows a minor difference between the HIVpos ARTneg patients and uninfected controls (R2X=0.695, R2Y=0.8, Q2=0.626). In Figure 3C, there was a degree of overlap between the HIVpos ARTpos and HIVpos ARTneg patients, which suggests that the metabolic profiles of those two groups are similar (R2X=0.671, R2Y=0.602, Q2=0.12). Sensitivity, specificity, and accuracy of the PLS-DA models were also calculated (Table 3). To assess the robustness of the PLS-DA models, permutation tests of 200 permutations were done. The results from the permutations show that the original PLS-DA models were valid, since the Q2 regression lines had a negative intercept and the permutated data on the left was lower than the original data on the right (Fig. 3, left panel). The PLS-DA analysis therefore demonstrated that there were metabolic differences between HIV-infected and uninfected controls.
FIG. 3.
Cross-validation of PLS-DA models obtained from 200 permutation tests. PLS-DA plots indicating the separation between (A) uninfected controls and HIVpos ARTpos patients, (B) uninfected controls and HIVposARTneg, and (C) HIVpos ARTpos and HIVpos ARTneg patients.
Table 3.
Prediction Results of OPLS-DA Model Based on FT-IR Spectra Obtained from HIV-Infected Serum and Uninfected Controls
| Sensitivitya (%) | Specificityb (%) | Accuracyc (%) Fischer's probabilityd | |
|---|---|---|---|
| HIVposARTpos vs. control | 100% | 95.24% | 95.2% 6.6e-12 |
| HIVposARTneg vs. control | 100% | 100% | 100% 1.1e-009 |
| HIVposARTpos vs. HIVposARTneg | 83.33% | 100% | 89.19% 6.7e-007 |
Sensitivity calculation was determined from the ratio of true positives (HIVpos ARTpos correctly predicted) to total number of modeled HIVpos ARTpos spectra. bSpecificity was calculated from the ratio of true negatives (control samples correctly predicted) to total number of modeled control spectra. cAccuracy was based on the ratio of total number of samples correctly classified to total number of samples predicted. dProbability of a model occurring by chance, satisfied when p<0.05. All calculations are done as widely described in the literature.
Identifying discriminatory metabolites
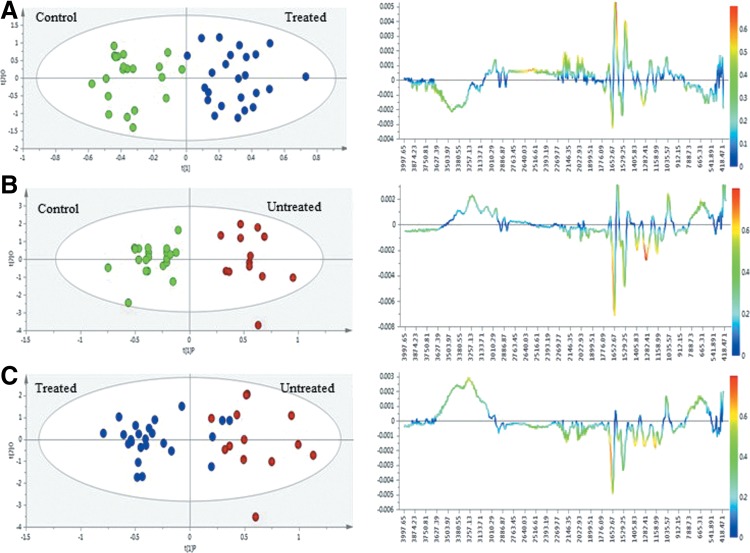
In order to identify the spectral regions/metabolites responsible for the discrimination of the three groups, OPLS-DA was carried out with a Pareto scaling approach to further maximize the group separation, as well as to establish a global overview of the characteristics of the HIV-infected groups. In the OPLS-DA score plots, the separation between the HIVpos ARTpos patients and uninfected controls was further improved with R2X=0.756 and Q2 X=0.58 (Fig. 4A, left panel). Also, a pronounced difference between the HIVpos ARTneg group and the uninfected control group (Fig. 4B, left panel) is apparent, with R2X=0.823 and Q2=0.674, respectively. The OPLS-DA score plot for the HIVpos ARTpos and HIVpos ARTneg groups showed some degree of separation, with four HIVpos ARTpos samples appearing in the HIVpos ARTneg cluster (Fig. 4C, left panel). The variables responsible for the discrimination in the score plots are shown in the corresponding coefficient loading plots (Fig. 4, right panel). In the loadings plots, the positive direction represents variables that are more prominent in the groups located in the positive direction of the score plot (principal component), while those in the negative directions denote variables that are prominent in the groups located in the negative direction of the score plot. The color scaling maps located on the right of the coefficient plot designates the contribution of variables in discriminating the groups in each score plot. In the color scaling, the red color indicates a more significant contribution than blue.
FIG. 4.
Multivariate statistical analysis of ATR-FTIR-based serum spectra obtained from HIV uninfected patients and uninfected controls. Scores plots (right panel) and loadings coefficient plots (left panel) generated from: (A) uninfected control vs. HIVpos ARTpos patients, (B) HIV uninfected control vs. HIVpos ARTneg, and (C) HIVpos ARTneg vs. HIVpos ARTneg patients. Assignments for spectral regions are shown in Figure 2.
Based on the loadings plot comparison between the HIVpos ARTpos group and the uninfected group (Fig. 4A, right panel), the HIVpos ARTpos group contained prominent spectral absorbance in the proteins (1600 cm−1), lipids (3010 cm−1), and glucose (1035 cm−1) spectral regions, while less prominent spectral absorbance in the fingerprint region (665 cm−1), some carbohydrate regions (1299 cm−1, 1158 cm−1; 1498 cm−1) as well as the hydroxyl/protein (3257-3627 cm−1) region could be observed. Compared with the uninfected control group, the HIVpos ARTneg group (Fig. 4B, right panel) was similar to the HIVpos ARTpos, except for the more prominent absorbance observed in the fingerprint (665 cm−1) hydroxyl/protein (3257–3627 cm−1) regions. When comparing the HIVpos ARTpos and HIVpos ARTneg groups (Fig. 4C, right panel), the spectral absorbance in proteins (amide I) and lipid ester (1700 cm−1) were similar between the two groups, while the absorbance in some carbohydrate regions were more prominent in the treated group. Additionally, the spectral absorbance of the hydroxyl (3257–3627 cm−1), fingerprint (665 cm−1), and glucose region (1035 cm−1) were prominent in the HIVpos ARTneg group. The variables responsible for the outcome of each OPLS-DA model are listed in Table 4 along with the possible band assignments. Using these specific variables, we built classification models in order to check if the discriminatory power of the variables increases, decreases, or remains the same for all three models. Since the results obtained showed no major difference between the two models, we continued with the original models.
Table 4.
Possible IR Band Assignments of Variables with Highest Discriminatory Power for: HIVposARTpos, HIVpos ARTneg and Uninfected Control Models
| Aavs. Bb | Aavs. Cc | Bbvs. Cc | |||||
|---|---|---|---|---|---|---|---|
| Variables (cm−1) | Literature Assignmentsd | P valuee | VIPf | P valuee | VIPf | P valuese | VIPf |
| 3000–2800 cm−1 | C-H str (asym) of –CH2-and –CH3- lipids | ||||||
| 3303 | 0.0728 | 2.1 | 0.1382 | 1.0 | 0.0127 | 1.8 | |
| 3266 | 0.2280 | 0.7 | 0.0296 | 1.7 | 0.0043 | 2.1 | |
| 3270 | 0.1793 | 0.9 | 0.0316 | 1.7 | 0.0042 | 2.1 | |
| 1700–1600 cm−1 | Amide I (proteins) | ||||||
| 1652 | 0.0098 | 2.6 | <0.0001 | 4.3 | 0.0001 | 3.1 | |
| 1654 | 0.0088 | 2.7 | <0.0001 | 4.3 | 0.0001 | 3.2 | |
| 1656 | 0.0088 | 2.5 | <0.0001 | 4.2 | <0.0001 | 3.1 | |
| 1650 | 0.0174 | 2.5 | <0.0001 | 4.2 | 0.0002 | 3.1 | |
| 1648 | 0.0507 | 2.1 | <0.0001 | 4.0 | 0.0004 | 3.0 | |
| 1617 | 0.0242 | 3.74 | 0.0533 | 2.4 | 0.0379 | 0.0 | |
| 1619 | 0.0088 | 3.72 | 0.0827 | 2.3 | 0.7441 | 0.1 | |
| 1616 | 0.0088 | 3.71 | 0.0378 | 2.4 | 0.8673 | 0.0 | |
| 1621 | 0.0088 | 3.64 | 0.1294 | 2.1 | 0.6948 | 0.1 | |
| 1614 | 0.0088 | 3.63 | 0.0316 | 2.4 | 0.8839 | 0.0 | |
| 1623 | 0.0098 | 3.51 | 0.1983 | 1.8 | 0.6424 | 0.3 | |
| 1600–1500 cm−1 | Amide II (proteins) | ||||||
| 1500 | 0.0088 | 2.83 | 0.1255 | 2.1 | 0.6517 | 2.1 | |
| 1504 | 0.0088 | 2.9 | 0.1983 | 1.0 | 0.3924 | 0.8 | |
| 1527 | 0.7846 | 0.3 | 0.0223 | 0.9 | 0.0127 | 2.0 | |
| 1498 | 0.0104 | 2.6 | 0.1134 | 1.0 | 0.7441 | ||
| 1350–1240 cm−1 | Amide III (proteins) | ||||||
| 1303 | 0.0088 | 2.1 | <0.0001 | 2.6 | 0.0147 | 1.3 | |
| 1299-1290 | 0.0083 | 2.1 | <0.0001 | 2.8 | 0.0005 | 1.5 | |
| 1200–900 cm−1 | C-O and C-C str, deformation of carbohydrates and glucose region | ||||||
| 1035 | 0.7876 | 0.8 | 0.0285 | 1.7 | 0.0345 | 1.0 | |
| 665 | 0.2730 | 01 | 0.0109 | 1.7 | 0.0391 | 1.6 | |
Assignments made based on literature guidelines (Gunasekaran et al., 2010; Jackson et al., 1996; Petrich et al., 2001).
The symbol A represents the uninfected control group, bRepresents the HIVposARTpos and cRepresents the HIVposARTneg.
All literature assignments were made based on literature findings.
P values calculated using the t-test and values that are significant after “BH” correction are indicated.
VIP values generated from the OPLS-DA model (values>1)
CV-ANOVA tests performed on the OPLS models resulted in p=0.00032 for HIVpos ARTpos versus uninfected controls, p=0.000963 for HIVpos ARTneg versus uninfected controls, and p=0.230 for HIVpos ARTpos versus HIVpos ARTneg.
Age and gender effects
This was a small exploratory volunteer-based study that was not designed to adjust for covariates. However, since age and gender are important factors in human metabolomics studies, we compared the original model to age and gender-specific models. The resulting age model (Supplementary Fig. S3B) showed that age had no influence on the original model's separation. Since both the original and age-specific models were not identical to each other, we can conclude that age was not the major variable contributing to the original separation. The gender-specific model (Supplementary Fig. S3C) was much more distinct in comparison to the age model. However, given the low predictability (Q2=0.142) and explained variation (R2=0.6), it is also evident that gender was not the primary discriminatory variable in the original OPLS-DA model. Given that gender was a greater confounding factor than age, we removed the seven male subjects and constructed a new OPLS-DA model (Supplementary Fig. S4) to gauge if the effects of gender were of significance. The removal of the male subjects seem to have had an effect on the OPLS-DA model, however this effect was not significant since the variables with discriminatory power remained similar for both models and model predictability (Q2=0.562; R2X=0.686) did not change dramatically.
Discussion
Numerous spectrometric metabolomics studies have identified spectral regions with which to discriminate HIV positive (treatment experienced and treatment-naive) from negative sera/plasma. The spectroscopic methodologies for which most data in this regard have been collected are NMR and MS (Sitole et al., 2013). The purpose of the current study was to explore the use of ATR-FTIR spectroscopy in the detection of potential biomarkers of HIV infection and/or antiretroviral treatment and the possibility of this methodology serving as a complementary tool for NMR and MS. The first application of mid- ATR-FTIR to HIV infected (treated) and uninfected sera, is presented here.
Data presented show that ATR-FTIR-based metabolic profiling can discriminate and identify functional groups that exist in metabolites that have been identified using other HIV-metabolomics spectroscopic techniques. These metabolites include lipids, carbohydrates, and proteins. Irregularities in the metabolic pathways involving these molecules have been implicated in the development of adverse metabolic effects in HIV-infected individuals (Butt et al., 2004; Dube et al., 2003; Friis-Moller et al., 2003; John et al., 2001; Koutkia and Grinspoon, 2003; Omech et al., 2012; Vrieskoop et al., 2008).
Carbohydrate, lipid, and protein metabolism
It has been well documented that HIV and antiretroviral treatment results in disruptions of lipid, carbohydrate, amino acid, and protein metabolism in HIVpos ARTpos individuals (Carr et al., 1998; Dube et al., 1997; Duong Van Huyen, 2003; Karamchand et al., 2008; Mulligan et al., 2000). Here, the most significant vibrations were detected in the proteins (1600 cm−1, 1652 cm−1), carbohydrate (1035 cm−1, 1158 cm−1, 1498 cm−1), and lipid/fatty acid (3010 cm−1) spectral regions. These results are in agreement with our previous observations where NMR-based metabolomics identified significant differences in lipids and glucose between HIVpos ARTpos, HIVpos ARTneg, and uninfected controls (Hewer et al., 2006; Philippeos et al., 2009).
In their Multicenter AIDS Cohort study, Brown et al. (2005), using both prevalence and incidence analysis, demonstrated that HIV-positive men on any ART regimen had a four times greater chance of developing diabetes than did their uninfected counterparts, suggesting that metabolites associated with lipid and glucose metabolism are likely biomarkers for discriminating among HIV-infected individuals on various ART regimens.
Elevations of cholesterol increased the risk of heart disease in HIV-positive persons (Friis-Moller et al., 2003). Significant differences in spectral regions (of C-H, CH2, and CH3 vibrations) of both cholesterol and phospholipids were observed here when comparing uninfected controls and HIV-positive individuals. This is suggestive of a relationship between antiretrovirals and changes in cholesterol levels. Because HIV infection and its treatment have been linked to elevated levels of low-density lipoprotein (LDL), data presented here is reason to assume that ATR-FTIR may be developed into a screening mechanism for virus/treatment-induced cardiovascular complications.
When comparing HIVpos ARTneg subjects to HIVpos ARTpos and uninfected controls, distinct differences were observed in the fingerprint region specifically between 665 cm−1–900 cm−1. The HIVpos ARTneg displayed larger differences in this region compared to samples from treated individuals; this is in agreement with Bahmani et al. (2009) who reported significant differences between HIV-infected (untreated) samples and healthy controls in this spectral region. Changes in this region are assigned to vibrations of ArCH, ROH, and RNH2 whose functional groups form part of the structure of the mentioned metabolites.
Possibly confounding factors
In studies involving metabolic changes due to any stimuli, age and gender can potentially be confounding factors, given the effect of these aspects on metabolism. In this study there were differences between age and gender in the three experimental groups, which is why these two factors were considered as class identifiers (Y-vector) in the statistical model (Supplementary Figs. S3 and S4). The age and gender statistical models were different from the original models where HIVpos ARTpos was compared to uninfected control subjects. The age and gender models show that neither age nor gender were confounding factors in the current study.
ATR-FTIR as a metabolomics tool
NMR and MS are the most widely used metabolomics tools, due to advanced levels of sensitivity and specificity. These methods are however very expensive, difficult to access in resource limited settings, and above all, require expert knowledge for instrument operation and data interpretation. The benefits of FTIR spectroscopy over NMR and MS is its lower cost and easier operation. FTIR cannot replace NMR and MS, but given the proven ability of this instrument in detecting and identifying differential metabolites (Ellis et al., 2006), this type of vibrational spectroscopy is capable of providing information in line with that of more sensitive techniques. Results obtained from this study are comparable to our previous findings (Hewer et al., 2006; Philippeos et al., 2009), confirming the ability of FTIR spectroscopy to produce data relatable to that of NMR and MS metabolomics. Also, given its proven ability as a diagnostic tool in other diseases (as highlighted by Dorling and Baker, 2013), ATR-FTIR may find useful application in HIV-metabolomics-based monitoring of disease progression or treatment success following further development and validation of the approach.
In conclusion, differences in the sera of HIV-infected (treated and untreated) individuals and uninfected controls were identified with a mid-ATR-FTIR metabolomics approach. The current study was limited in sample size but provided original insights for novel systems diagnostics for HIV/AIDS. This work has demonstrated the reliability, simplicity, and predictive ability of FTIR-based metabolomics in discriminating between the experimental groups studied in our sample. Future studies with larger subject numbers are warranted to expand upon the present findings.
Supplementary Material
Abbreviations Used
- AIDS
acquired immunodeficiency syndrome
- ANOVA
analysis of variance
- ART
antiretroviral therapy
- ATR
attenuated total reflectance
- BH
Benjamini-Hochberg
- CV-ANOVA
cross-validated predictive residuals
- DEXA
dual-energy X-ray absorptiometry
- DModX
distance to model
- FTIR
Fourier transform infrared spectroscopy
- HIV
human immunodeficiencyv
- IR
infrared
- LDA
linear discriminant analysis
- LDL
low-density lipoprotein
- LOOCV
leave-one-out-at a cross-validation
- MS
mass spectrometr
- Neg
negative
- NMR
nuclear magnetic resonance
- NNRTI
non-nucleoside reverse transcriptase inhibitor
- OPLS-DA
orthogonal partial least squares discriminant analysis
- PCA
principal component analysis
- PLS
partial least squares discriminant analysis
- Pos
positive
- SIMCA
soft independent modelling of class analog
- VIP
variable importance in projection
Acknowledgments
Financial support for this study was received from the Technology Innovation Agency (TIA) and the University of Pretoria.
Author Disclosure Statement
No competing financial interests exists.
References
- Bahmani MK, Khosravi A, Miri R, Yukieiwabu R, Shiikuta K, and Sakudo A. (2009). A spectroscopic characterization of human immunodeficiency virus type-1-infected plasma by principal component analysis and soft independent modeling of class analogy of visible and near-infrared spectra. Mol Med Rep 2, 805–809 [DOI] [PubMed] [Google Scholar]
- Bell SE, Fido LA, Speers SJ, Armstrong WJ, and Spratt S. (2005). Forensic analysis of architectural finishes using Fourier transform infrared and Raman spectroscopy. Part II: White paint. Appl Spectrosc 59, 1340–1646 [DOI] [PubMed] [Google Scholar]
- Boccard J, Douglas N, and Rutledge DN. (2013). A consensus orthogonal partial least squares discriminant analysis (OPLS-DA) strategy for multiblock Omics data fusion. Anal Chim Acta 769, 30–39 [DOI] [PubMed] [Google Scholar]
- Brown TT, Cole SR, Li X, et al. (2005). Antiretroviral therapy and the prevalence and incidence of diabetes mellitus in the multicenter AIDS Cohort Study. Arch Intern Med 165, 1179–1184 [DOI] [PubMed] [Google Scholar]
- Butt AA, Fultz SL, Kwoh CK, Kelley D, Skanderson M, and Justice AC. (2004). Risk of diabetes in HIV infected veterans pre- and post-HAART and the role of HCV coinfection. Hepatology 40, 115–119 [DOI] [PubMed] [Google Scholar]
- Carr A, Samaras K, Burton S, et al. (1998). A syndrome of peripheral lipodystrophy, hyperlipidaemia and insulin resistance due to HIV protease inhibitors. AIDS 12, F51–58 [DOI] [PubMed] [Google Scholar]
- Cassol E, Misra V, Holman A, Kamat A, Morgello S, and Gabuzda D. (2013). Plasma metabolomics identifies lipid abnormalities linked to markers of inflammation, microbial translocation, and hepatic function in HIV patients receiving protease inhibitors. BMC Infect Dis 13, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Lee PL, Yiannoutsos CT, et al. (2004). A multicenter in vivo proton-MRS study of HIV-associated dementia and its relationship to age. Neuroimage 23, 1336–1347 [DOI] [PubMed] [Google Scholar]
- Chong IG, and Jun CH. (2005). Performance of some variable selection methods when multicollinearity is present. Chemometr Intell Lab Syst 78, 103–112 [Google Scholar]
- Derenne A, Verdonck M, and Goormaghtigh E. (2012). The effect of anticancer drugs on seven cell lines monitored by FTIR spectroscopy. Analyst 137, 3255–3264 [DOI] [PubMed] [Google Scholar]
- Dorling KM, and Baker MJ. (2013). Highlighting attenuated total reflection Fourier transform infrared spectroscopy for rapid serum analysis. Trends Biotechnol 31, 327–328 [DOI] [PubMed] [Google Scholar]
- Dube MP, Johnson DL, Currier JS, and Leedom JM. (1997). Protease inhibitor-associated hyperglycemia. Lancet 350, 713–714 [DOI] [PubMed] [Google Scholar]
- Dubé MP, Stein JH, Aberg JA, et al. (2003). Guidelines for the evaluation and management of dyslipidemia in human immunodeficiency virus (HIV)-infected adults receiving antiretroviral therapy: Recommendations of the HIV Medicine Association of the Infectious Disease Society of America and the Adult AIDS Clinical Trials Group. Clin Infect Dis 37, 613–627 [DOI] [PubMed] [Google Scholar]
- Duong Van Huyen JP, Landau A, Piketty C, et al. (2003). Toxic effects of nucleoside reverse transcriptase inhibitors on the liver. Value of electron microscopy analysis for the diagnosis of mitochondrial cytopathy. Am J Clin Pathol 119, 546–555 [DOI] [PubMed] [Google Scholar]
- Edelstein CL. (2011). Biomarkers in Kidney Disease, 1st ed. (Academic Press as an imprint of Elsevier, London, UK: ) [Google Scholar]
- Ellis DI, and Goodacre R. (2006). Metabolic fingerprinting in disease diagnosis: Biomedical applications of infrared and Raman spectroscopy. Analyst 131, 875–885 [DOI] [PubMed] [Google Scholar]
- Ellis DI, Harrigan GG, and Goodacre R. (2003). Metabolic fingerprinting with Fourier transform infrared spectroscopy. In: Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis. Kluwer Academic Publishers, Boston, MA [Google Scholar]
- Eriksson L, Trygg J, and Wold S. (2008). CV-ANOVA for significance testing of PLS and OPLS models. J. Chemometr 22, 594–600 [Google Scholar]
- Fakunaga K. (1990). Introduction to Statistical Pattern Recognition, 2nd ed. Academic Press, San Diego, CA [Google Scholar]
- Falutz J. (2011). HIV infection, body composition changes and related metabolic complications: Contributing factors and evolving management strategies. Curr Opin Clin Nutr Metab Care 14, 255–260 [DOI] [PubMed] [Google Scholar]
- Friis-Moller N, Neber R, Reiss P, et al. (2003). Cardiovascular disease risk factors in HIV patients association with antiretroviral therapy. Results from the DAD study. AIDS 17, 1179–1193 [DOI] [PubMed] [Google Scholar]
- Gidman EA, Laurence M, Jones M, et al. (2007). A methodology for screening haemolymph of intertidal mussels, Mytilus edulis, using FT-IR spectroscopy as a tool for environmental assessment. Metabolomics 3, 465–473 [Google Scholar]
- Gkrania-Klotsas E, and Klotsas AE. (2007). HIV and HIV treatment: Effects on fats, glucose and lipids. Br Med Bull 84, 49–68 [DOI] [PubMed] [Google Scholar]
- Graça G, Moreira AS, Correia AJ, et al. (2013). Mid-infrared (MIR) metabolic fingerprinting of amniotic fluid: A possible avenue for early diagnosis of prenatal disorders? Anal Chim Acta 764, 24–31 [DOI] [PubMed] [Google Scholar]
- Gunasekaran S, Uthra D, Sailatha E, and Anita B. (2010). FTIR spectral study of jaundice blood samples before and after treatment. Asian J Chem 1, 51–56 [Google Scholar]
- Hewer R, Vorster J, Steffens FE, and Meyer D. (2006). Applying biofluid 1H NMR-based metabonomic techniques to distinguish between HIV-1 positive/AIDS patients on antiretroviral treatment and HIV-1 negative individuals. J Pharm Biomed Anal 41, 1442–1446 [DOI] [PubMed] [Google Scholar]
- Jain RG, Furfine ES, Pedneault L, White AL, and Lenhard JM. (2001). Metabolic complications associated with antiretroviral therapy. Antiviral Res 51, 151–177 [DOI] [PubMed] [Google Scholar]
- John M, Moore CB, James IR, et al. (2001). Chronic hyperlactatemia in HIV-infected patients taking antiretroviral therapy. AIDS 15, 717–723 [DOI] [PubMed] [Google Scholar]
- John M, Nolan D, and Mallal S. (2001). Antiretroviral therapy and the lipodystrophy syndrome. Antivir Ther 6, 9–20 [DOI] [PubMed] [Google Scholar]
- Johnston C, Dunn W, Broadhurst D, et al. (2010). Serum metabolite profiles differentiate Crohn's disease from ulcerative colitis and from healthy controls. Gut 59, A61–A61 [Google Scholar]
- Karamchand L, Dawood H, and Chuturgoon AA. (2008). Lymphocyte mitochondrial depolarization and apoptosis in HIV-1-infected HAART patients. J Acquir Immune Defic Syndr 48, 381–388 [DOI] [PubMed] [Google Scholar]
- Koutkia P, and Grinspoon S. (2003). HIV-associated lipodystrophy: Pathogenesis, prognosis, treatment, and controversies. Annu Rev Med 55, 303–317 [DOI] [PubMed] [Google Scholar]
- Lindgren F, Hansen B, Karcher W, Sjöström M, and Eriksson L. (1996). Model validation by permutation tests: Applications to variable selection. J Chemometr 10, 521–532 [Google Scholar]
- Mantsch HH, and Chapman D. (1996). Infrared Spectroscopy of Biological Molecules, 1st ed. Wiley-Liss, New York [Google Scholar]
- McComsey G. (2002). Update on mitochondrial toxicity of antiretrovirals and its links to lipodystrophy. AIDS Rev 4, 140–147 [PubMed] [Google Scholar]
- Mulligan K, Grunfeld C, Tai VW, et al. (2000). Hyperlipidemia and insulin resistance are induced by protease inhibitors independent of changes in body composition in patients with HIV infection. J Aquir Immune Defic Syndr 23, 35–43 [DOI] [PubMed] [Google Scholar]
- Munshi SU, Rewari BB, Bhavesh NS, and Jameel S. (2013). Nuclear magnetic resonance based profiling of biofluids reveals metabolic dysregulation in HIV-infected persons and those on anti-retroviral therapy. PLoS One 8, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negredo E, Ribalta J, Paredes R, et al. (2002). Reversal of atherogenic lipoprotein profile in HIV-1 infected patients with lipodystrophy after replacing protease inhibitors by nevirapine. AIDS 16, 1383–1389 [DOI] [PubMed] [Google Scholar]
- Nicolaou N, Xu Y, and Goodacre R. (2012). Fourier transform infrared spectroscopy and multivariate analysis for the detection and quantification of different milk species. J Dairy Sci 93, 5651–5660 [DOI] [PubMed] [Google Scholar]
- Omech B, Sempa J, Castelnuovo B, et al. (2012). Prevalence of HIV-associated metabolic abnormalities among patients taking first-line antiretroviral therapy in Uganda. ISRN AIDS 2012, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrich W. (2001). Mid-infrared and Raman spectroscopy for medical diagnostics. Appl Spectrosc Rev 36, 181–237 [Google Scholar]
- Philippeos C, Steffens FE, and Meyer D. (2009). Comparative 1H NMR-based metabonomic analysis of HIV-1 sera. J Biomol NMR 44, 127–137 [DOI] [PubMed] [Google Scholar]
- Powderly W. (2004). Metabolic complications associated with HIV infection and antiretroviral therapy. AIDS Patient Care STDS 18, 431–435 [DOI] [PubMed] [Google Scholar]
- Sadeghi-Bazargani H, Bangdiwala SI, Mohamma K, Maghsoudi H, and Mohammadi R. (2011). Compared application of the new OPLS-DA statistical model versus partial least squares regression to manage large numbers of variables in an injury case-control study. Sci Res Essays 6, 4369–4377 [Google Scholar]
- Sakudo A, Tsenkova R, Onozuka T, et al. (2005). A novel diagnostic method for human immunodeficiency virus type-1 in plasma by near-infrared spectroscopy. Microbiol Immunol 49, 695–701 [DOI] [PubMed] [Google Scholar]
- Salas-Salvado J, and Garcia-Lorda P. (2001). The metabolic puzzle during the evolution of HIV infection. Clin Nutr 20, 379–391 [DOI] [PubMed] [Google Scholar]
- Savitzky A, and Golay MJE. (1964). Smoothing and differentiation of data by simplified least square procedures Anal Chem 36, 1627–1639 [DOI] [PubMed]
- Serkova NJ, and Niemann CU. (2006). Pattern recognition and biomarker validation using quantitative 1H-NMR-based metabolomics. Expert Rev Mol Diagn 6, 717–731 [DOI] [PubMed] [Google Scholar]
- Sitole LJ, Williams AA, and Meyer D. (2013). Metabonomic analysis of HIV-infected biofluids. Mol BioSyst 9, 18–28 [DOI] [PubMed] [Google Scholar]
- Szymaçska E, Saccenti E, Smilde AK, and Westerhuis JA. (2012). Double-check validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 8, 3–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Schweinsburg BC, Alhassoon OM, et al. (2007). Effects of human immunodeficiency virus and methamphetamine on cerebral metabolites measured with magnetic resonance spectroscopy. J Neurovirol 13, 150–159 [DOI] [PubMed] [Google Scholar]
- Trygg J, and Wold S. (2002). Orthogonal projections to latent structures (O-PLS). J Chemometr 16, 119–128 [Google Scholar]
- UNAIDS (2009). AIDS epidemic update, Geneva, Switzerland: http://data.unaids.org/pub/report/2009/jc1700_epi_update_2009_en.pdf Accessed 5July2011 [Google Scholar]
- Vrisekoop N, van Gent R, de Boer AB, et al. (2008). Restoration of the CD4 T cell compartment after long-term highly active antiretroviral therapy without phenotypical signs of accelerated immunological aging. J Immunol 181, 1573–1581 [DOI] [PubMed] [Google Scholar]
- Wheelock AM, and Wheelock CE. (2013). Trials and tribulations of omics data analysis: Assessing quality of SIMCA-based multivariate models using examples from pulmonary medicine. Mol Biosyst 9, 2589–2596 [DOI] [PubMed] [Google Scholar]
- Williams A, Koekemoer G, Lindeque Z, Reinecke C, and Meyer D. (2012). Qualitative serum organic acid profiles of HIV-infected individuals not on antiretroviral treatment. Metabolomics 8, 804–818 [Google Scholar]
- Wohl DA, McComsey G, Tebas P, et al. (2006). Current concepts in the diagnosis and management of metabolic complications of HIV infection and its therapy. Clin Infect Dis 43, 645–653 [DOI] [PubMed] [Google Scholar]
- Worley B, and Powers R. (2013). Multivariate analysis in metabolomics. Curr Metabolomics 1, 92–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
References
Supplementary Data: Supplementary information data associated with this manuscript can be found in the Supplementary Data.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.