Abstract
The goal of structural biology is to reveal details of the molecular structure of proteins in order to understand their function and mechanism. X-ray crystallography and NMR are the two best methods for atomic level structure determination. However, these methods require milligram quantities of proteins. In this chapter a reproducible methodology for large-scale protein production applicable to a diverse set of proteins is described. The approach is based on protein expression in E. coli as a fusion with a cleavable affinity tag that was tested on over 20,000 proteins. Specifically, a protocol for fermentation of large quantities of native proteins in disposable culture vessels is presented. A modified protocol that allows for the production of selenium-labeled proteins in defined media is also offered. Finally, a method for the purification of His6-tagged proteins on immobilized metal affinity chromatography columns that generates high-purity material is described in detail.
Keywords: Protein expression, Protein purification, Disposable vessel fermentation, Selenomethionine-labeling, IMAC, His-tag, High-throughput
1 Introduction
Structural genomics uses X-ray crystallography and NMR spectroscopy for structure determination and requires milligram quantities of high-quality proteins. Protein samples must be soluble, highly homogeneous, and free of critical contaminants (especially proteases, metals, lipids, and other impurities). In structural genomics it is also important that numerous proteins or their constructs are produced in parallel and in a highly reproducible manner. To achieve these objectives, every step in the production of a protein must be carefully evaluated, optimized, and accelerated, including gene cloning, protein expression, and purification. Over the years, much effort has been put into optimizing Escherichia coli as an expression host for proteins from other bacteria and higher organisms [1–8]. A variety of classes of proteins, from prokaryotic to human and from single domain to protein complexes, can be produced in E. coli [1, 2, 9–12]. Because these proteins vary greatly in their properties, to facilitate purification each protein is typically fused with an N- or a C-terminal affinity tag. A hexahistidine (His6)-tag is most commonly used as it allows the use of immobilized metal affinity chromatography (IMAC) for all soluble proteins [4, 7, 13–15].
Rapid technological advances have been made during the last decade in structural genomics pipelines that use robotic hardware, software, process parallelization, improved fermentation, and semiautomated purification methods [16, 17]. Developments have also been made to reduce the time and labor of protein production methods [13, 18–21]. An example of an improvement in the fermentation step of protein production is the growth of high cell density bacterial cultures in polyethylene terephthalate (PET) bottles [19]. Expression of proteins using optimized minimal media in non-sterilized disposable PET bottles reduces the time required for media preparation and the fermentation volume by a factor of 2–3 and eliminates much of the cleanup that follows cell harvesting. Disposal of the autoclaved bottles after use reduces the risk of cross-contamination of subsequent cultures.
Another key development includes the availability of specifically labeled proteins for X-ray crystallography or NMR spectroscopy. This protocol also uses disposable fermentation vessels for production of selenomethionine-labeled proteins [1, 22]. It involves a production of in vivo-labeled proteins in optimized minimal media. The method relies on forcing selenomethionine incorporation via inhibition of endogenous methionine biosynthesis by the addition of inhibitory amino acids [23]. Selenium-labeled protein crystals are then used for X-ray diffraction data collection and de novo phasing for structure determination.
The choice of purification and handling procedures plays a critical role in obtaining high-quality protein samples. This standard purification protocol has been designed for His6-tagged proteins and implemented on a semiautomated chromatography platform with minimal human involvement [1, 9]. Purification starts with the preparation of crude extract (or lysate) followed by two chromatography steps. The first affinity chromatography step (IMAC-I) is done on a Ni+2-charged column and immediately followed by buffer exchange on a desalting column. The IMAC-I step produces proteins with purity typically above 80–95 % as judged by SDS-PAGE. After IMAC-I, the His6-tag from the target protein is cleaved with a highly specific recombinant tobacco etch virus (TEV) protease (developed by Dr. B. G. Fox, University of Wisconsin; this protease is fused to a non-cleavable His7-tag and is resistant to auto-inactivation) at a designed cleavage site (ENLYFQ↓S). This cleavage produces a protein with three amino acids (SNA) added to its N-terminus [24, 25]. The second chromatography step, a subtractive IMAC (IMAC-II), is also performed on the Ni+2-charged column. During the IMAC-II process, the cleaved protein elutes in the flow through and is separated from the released His6-tag, undigested protein, persistent endogenous E. coli proteins having affinity for IMAC column, and His7- tagged TEV protease. This step is also followed by buffer exchange. The buffer exchange process is important as it removes low-molecular- weight contaminants and transfers the protein into buffer conditions suitable for downstream steps, such as protease treatment, protein concentration, crystallization, and storage. The protein is then concentrated using centrifugal concentrators. If higher purity protein sample is needed, the buffer exchange step after IMAC-I and IMAC-II or after both processes can be replaced with size-exclusion chromatography (SEC). Batches of different proteins can be purified in parallel using IMAC and buffer exchange or SEC columns. The purification process described above typically produces high-quality protein samples with 95–98 % purity.
One of the key elements of large-scale protein production is the proper long-term storage of protein samples. We recommend that the concentrated protein solutions (20–200 mg/mL) are pipetted dropwise (30–50 μL) directly into liquid nitrogen and the resulting small pellets are collected into plastic tubes and stored in liquid nitrogen or at −80 ºC. These small aliquots can be rapidly thawed at room temperature and used for subsequent crystallizations and biophysical characterizations or in biochemical experiments, while the bulk of the material remains safely stored.
It is important to keep in mind that the high-throughput approach described here involves processing a large number of proteins (or their variants) using standardized protocols that are not optimized for any given protein but work reasonably well for many proteins. Consequently, these protocols may need to be altered for a specific protein that failed the standard high-throughput purification procedure. This also includes efficient protocols for the purification of proteins without affinity tags and stable protein–protein and protein–nucleic acid complexes.
In this chapter, a method for large-scale production of heterologous fusion proteins in E. coli is described. The protocol for the production of native proteins uses 2-L PET bottles as disposable culture vessels. In addition, a modified expression protocol that allows for the production of selenium-labeled proteins in defined minimum medium is included. Finally, a method for the purification of numerous proteins that generates high-purity samples for structural biology and biochemical applications and a technique utilized to prepare protein samples for long-term storage are presented.
2 Materials
Prepare all solutions using ultrapure water (resistance of 18 MΩ in a 1 cm cell; conductivity of less than 5.6 μS/m) and analytical grade reagents. Follow all waste disposal guidelines when disposing waste materials.
2.1 Materials for Media Preparation for the Production of Native Proteins
Ampicillin, sodium salt: Prepare a 100 mg/mL stock by dissolving 1 g of ampicillin in water to a final volume of 10 mL. Store aliquots at −20 °C. 1 mL of 100 mg/mL solution is required per 1 L of culture to achieve a final concentration of 100 μg/mL.
Kanamycin sulfate: Prepare a 30 mg/mL stock by dissolving 300 mg kanamycin sulfate in water to a final volume of 10 mL. Store aliquots at −20 °C. 1 mL of 300 mg/mL solution is required per 1 L of culture to achieve a final concentration of 30 μg/mL.
Luria Bertani (LB) broth: Dissolve 25 g of preformulated LB powder in water to a final volume of 1 L. Add ampicillin and kanamycin sulfate to 100 and 30 μg/mL, respectively (see Note 1).
2.2 Materials for Media Preparation for the Production of Selenomethionine-Labeled Proteins
Ampicillin, kanamycin sulfate, B1, and B12 vitamins, premixed solids in a foil packet (Orion Enterprises, Inc): Prepare a stock solution by dissolving contents of one foil pouch (ampicillin, 2.5 g; kanamycin sulfate, 1.5 g; thiamine (B1), 50 mg; and vitamin B12, 135 mg) in 50 mL of water. Store aliquots at −20 °C, and avoid light during storage. 1 mL of this solution is required per 1 L of culture to achieve a final concentration of 50 μg/mL for ampicillin, 30 μg/mL for kanamycin sulfate, 1 μg/mL for thiamine, and 2.7 μg/mL for vitamin B12.
Trace Metal Supplement, as concentrated solution (Orion Enterprises, Inc): Store at 4 °C, and avoid light. Use 10 mL for 1 L culture to achieve final concentrations of 5 mg EDTA, 430 mg MgCl2 · 6H2O, 5 mg MgSO4, 10 mg NaCl, 1 mg FeSO4 · 7H2O, 1 mg Co(NO3)2 · 6H2O, 11 mg CaCl2, 1 mg ZnSO4 · 7H2O, 0.1 mg CuSO4 · 5H2O, 0.1 mg AlK(SO4)2, 0.1 mg H3BO3, 0.1 mg Na2MoO4 · 2H2O, 0.1 mg Na2SeO3, 0.1 mg Na2WO4 · 2H2O, 0.2 mg NiCl2 · 6H2O.
Vacuum filtration system with a 0.22 μm polyethersulfone membrane such as sterile Stericup® vacuum filtration unit (Millipore Corporation).
50 % Glycerol solution (Sigma Aldrich): Use ultrapure water and a ReagentPlus grade of glycerol to prepare the 50 % solution. Filter the solution using sterile vacuum filtration unit. Use 10 mL of this solution per 1 L of culture to achieve a final glycerol concentration of 0.5 %.
M9 “Pink” medium and non-inhibitory amino acids (NIAAC), premixed solids in a foil packet (Orion Enterprises, Inc): Dissolve one foil pouch (Na2 HPO4, 6.8 g; KH2 PO4, 3.0 g; NaCl, 0.75 g; glucose monohydrate 4.4 g; NH4 Cl, 1.0 g; Na2 SO4, 0.28 g; 11 amino acids (L-glutamate, L-aspartate, L-arginine, L-histidine, L-alanine, L-proline, L-glycine, L-serine, L-glutamine, L-asparagine, L-tryptophan, 200 mg each)) in water to a final volume of 1 L (see Note 2).
Selenomethionine and inhibitory amino acids (IAAC), premixed solids in a foil packet (Orion Enterprises, Inc): Dissolve one foil pouch in 250 mL of water. Use 30 mL for 1 L of cell culture resulting in 150 mg each/L of L-valine, L-isoleucine, L-leucine, L-lysine, L-threonine, L-phenylalanine, and 90 mg/L of selenomethionine.
2.3 Other General Materials for Protein Production
Freshly transformed E. coli BL21(DE3) cells (Stratagene) carrying plasmid pMAGIC and containing a gene for the target protein in vector pMCSG7 [25] or one of its derivatives (see Note 3). Alternatively, a glycerol stock can be used. For details on cloning protocols see Chapter 5 in this book.
Isopropyl-β-D-thio-galactopyranoside (IPTG), premixed solid in a foil packet (Orion Enterprises, Inc). Prepare a 1 M stock by mixing 11.9 g of IPTG (one pouch) with water to a final volume of 50 mL and store the aliquots at −20 °C. 0.5 mL of this solution is required per 1 L of culture to achieve a final concentration of 0.5 mM.
Lysozyme from chicken egg white. Prepare a 50 mg/mL stock by mixing 500 mg of lysozyme with water to a final volume of 10 mL. Store at −20 °C. 1 mL of 50 mg/mL solution is required per 50 mL of a re-suspended culture pellet to achieve a final concentration of 1 mg/mL.
Protease inhibitor cocktail such as Complete Protease Inhibitor Cocktail tablets (Roche). Use one tablet per 8 g of wet cell paste (or 50 mL of resuspended cells).
Sterile tubes for starter cultures such as 5-mL round bottom polypropylene culture tubes.
500-mL polyethylene terephthalate beverage bottles (PET, Continental Glass and Plastics, Inc). Use one bottle per target protein for a small overnight culture. One bottle contains enough culture to inoculate two 1-L cultures [19, 22].
2-L polyethylene terephthalate beverage bottles (PET, Continental Glass and Plastics, Inc.). Use one bottle per liter of culture [19, 22].
Foam tube plugs such as Identi-Plugs® (Jaece Industries).
1-L centrifuge bottles (such as Nalgene® high speed polycarbonate centrifuge bottles with polypropylene-ether screw/blue silicone gaskets).
Incubator/shaker.
Centrifuge.
A spectrophotometer and cuvette that can measure absorbance at 600 nm and 10 mm disposable semi-micro cuvettes.
2.4 Materials for Protein Purification
-
Purification buffers. All buffers must be filtered through a 0.22 μm polyethersulfone filter:
Lysis buffer (50 mM HEPES pH 8.0, 500 mM NaCl, 5 % glycerol, 20 mM imidazole, 10 mM β-mercaptoethanol).
Desalting buffer (50 mM HEPES pH 8.0, 500 mM NaCl, 5 % glycerol, 10 mM β-mercaptoethanol).
Elution buffer (50 mM HEPES pH 8.0, 500 mM NaCl, 5 % glycerol, 250 mM imidazole, 10 mM β-mercaptoethanol).
Crystallization buffer (20 mM HEPES pH 8.0, 250 mM NaCl, 2 mM 1,4-dithio-DL-threitol).
Recombinant tobacco etch virus (TEV) protease carrying a non-cleavable His7-tag [24] and mutated to reduce self-inactivation [26]. Use purified TEV protease as a 5–10 mg/mL solution in 20 mM HEPES pH 8.0, 300 mM NaCl, 10 % glycerol, 2 mM β-mercaptoethanol. For His6-tag cleavage, incubate at a protein: protease mass ratio of 1:50 (see Note 4).
Sonicator.
Centrifuge.
Polyvinylidene fluoride (PVDF) syringe filter (0.45 μM).
ÄKTAxpress chromatography system (GE Healthcare), or the equivalent.
5-mL HisTrap HP column (GE Healthcare) or the equivalent.
53-mL HiPrep 26/10 desalting column (GE Healthcare) or the equivalent.
Superdex 200 HiLoad 26/60 size exclusion chromatography (SEC) column (GE Healthcare) or the equivalent.
96-well deep well blocks such as sterile, 96-well 2-mL square top, round bottom polypropylene plates (BioExpress).
Slide-A-Lyzer™ dialysis cassette (Thermo Scientific) of proper liquid capacity or the equivalent dialysis device.
Centrifugal filter devices.
A spectrophotometer absorbance at 280 nm (such as NanoDrop 1000, for which no cuvette is required)
Cryogenic dewar (such as foam dewar FD-800, Spearlab) with small (10–20 mL) glass beakers.
Plastic cryogenic screw-cap tubes (such as 2.0-mL Nalgene® cryogenic vials).
Conical centrifuge tubes.
3 Methods
It is important to mention that large-scale protein fermentation is one of the many components of the protein production pipeline at a structural genomics center. Before proteins are produced on milligram-scale, target proteins are processed through several stages of the pipeline. First, protein targets are carefully selected, cloned into appropriate expression vectors, and protein expression and solubility is assessed in a small-scale (5 mL). Only the targets with confirmed high-level expression and good solubility are moved into the large-scale fermentation process described here. Failed constructs are salvaged [1].
The incubators, shakers, and sonicators should be disinfected biweekly using 10 % bleach. All disposables should be autoclaved, while non-disposables should be disinfected in 10 % bleach. When disinfecting with 10 % bleach, ensure at least 20 min of contact time. Wear nitrile gloves, safety glasses and a lab coat during these procedures.
3.1 Production of Native Proteins
To prepare a starter culture, inoculate 1 mL of LB medium containing 100 μg/mL ampicillin and 30 μg/mL kanamycin sulfate in a 5-mL plastic tube with a fresh transformant or a glycerol stock of pMCSG7/pMAGIC transformed E. coli BL21(DE3) cells. Place in an incubator and shake at 220 rpm and 37 °C for 4–6 h. Use one tube per protein.
To prepare small overnight culture, add 50 μL of the turbid starter culture to 50 mL of fresh LB medium containing 100 μg/mL ampicillin and 30 μg/mL kanamycin sulfate in a 500-mL PET beverage bottle. Place in an incubator and shake at 150 rpm and 37 °C overnight.
Prepare LB medium in 2-L PET bottles using 990 mL of ultrapure water. Add ampicillin and kanamycin sulfate to a final concentration of 100 and 30 μg/mL, respectively.
Add 10 mL of the saturated overnight culture to each 1 L of fresh LB medium containing 100 μg/mL ampicillin and 30 μg/mL kanamycin sulfate. Shake the bottles in an incubator at 37 ºC at 180 rpm until cells grow to OD600nm 0.8–1.0. Start measuring OD after about 2.0–2.5 h.
Transfer bottles to a pre-cooled 4 °C incubator and shake for 1 h (see Note 5).
Increase incubator temp to 18 ºC. Induce the cells by adding 0.5 mL of 1 M IPTG solution to a final concentration of 0.5 mM.
Grow cells at 18 ºC and shake at 180 rpm overnight.
Pour cell culture in 1-L centrifuge bottles. Centrifuge for 10 min at 7,880 × g (in a rotor such as Sorvall F8S-6x1000y). Remove the supernatant.
Perform all procedures at 4 ºC from here on.
Re-suspend the cells by adding 25 mL of ice-cold lysis buffer and gentle shaking (150 rpm).
Add one tablet of protease inhibitor cocktail per 8 g of wet cell paste.
Transfer the re-suspended cells into a 50-mL tube and bring volume up to 40 mL with lysis buffer (see Note 6).
Add 50 mg/mL lysozyme solution to a final concentration of 1 mg/mL (for example, 0.8 mL of the solution for 40 mL of cell suspension).
Cryo-cool the cell suspension with liquid nitrogen and store at −80 ºC.
3.2 Production of the Selenomethionine Labeled Proteins
Prepare 1-mL starter cultures in LB medium with antibiotics as described in step 1 of Subheading 3.1.
Dissolve 1 pouch of M9 “Pink” salts and NIAAC in 980 mL of ultrapure water in 2-L PET beverage bottles. Add 10 mL of 50 % glycerol, 10 mL of Trace Metal Supplement, 1 mL of ampicillin, kanamycin sulfate, B1, and B12 vitamins stock solution.
To make small overnight cultures, aliquot 50 mL of the prepared M9 “Pink” Medium into 500-mL PET beverage bottles.
Add 50 μL of the turbid starter LB culture to each 50 mL of M9 “Pink” medium. Place the bottles in an incubator and shake at 150 rpm and 37 °C overnight.
Prepare M9 “Pink” medium in 2-L PET bottles described in step 2 of this Subheading 3.1. using 940 mL of ultrapure water (see Note 2).
Add 25 mL of small culture from 500-mL PET bottle to each 2-L bottle of the prepared M9 “Pink” medium.
Shake in an incubator at 37 ºC at 180 rpm until cells grow to OD600nm 1.0–1.4. Start measuring OD after about 2.5–3 h.
Transfer bottles to a precooled 4 °C incubator and shake for 1 h (see Note 5).
Add 30 mL of selenomethionine and IAAC solution per 1 L of culture (see Note 7).
Increase incubator temp to 18 ºC. Induce the cells by adding 0.5 mL of 1 M IPTG solution at a final concentration of 0.5 mM.
Grow cells at 18 ºC and shake at 180 rpm overnight.
Recover the cells by centrifugation, and proceed as described in step 8 of Subheading 3.1.
3.3 Protein Purification
3.3.1 IMAC-I Purification
Lyse the cell suspension. Thaw the cells on ice with gentle mixing and sonicate on ice for 5 min by means of a program or with manual control using a 4-s on and 20-s off mode and power/voltage settings according to the manufacturer’s instructions (see Note 8).
Transfer the cell lysate to 45-mL centrifuge tubes. Centrifuge for 1 h at 29,500 × g in an SS-34 rotor. Alternatively, centrifugation can be performed in the 50-mL tubes for 1 h and 20 min in a conical fixed-angle rotor (such as Sorvall F13–14x50cy). Filter the supernatant into a clean 50-mL tube using 0.45 μm PVDF or polyethersulfone syringe filter to remove residual particulates.
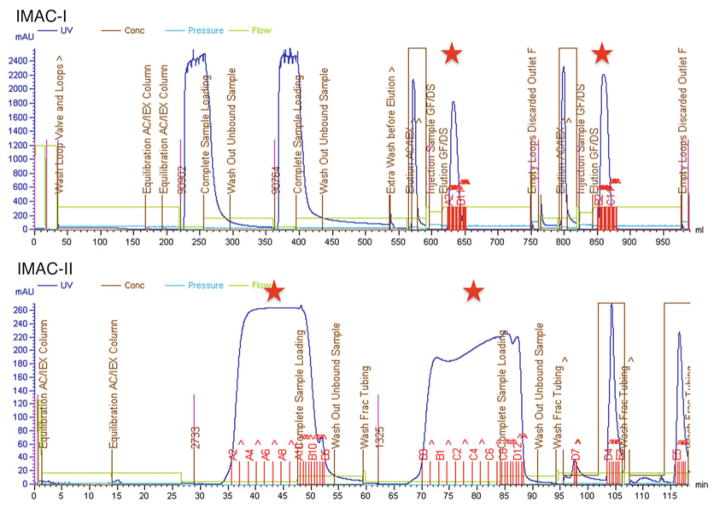
Apply the filtered supernatant with a maximum flow rate of 1 mL/min to a 5-mL HisTrap HP column (see Note 9) that was equilibrated with lysis buffer. Wash the column at 4 mL/min with 30 column volumes (CV) of lysis buffer to remove contaminating endogenous E. coli proteins. Elute the bound His6-target proteins with elution buffer over 5 CV at 4 mL/min. Collect fractions into a 10 mL loop or a 96-well deep-well block (Fig. 1, top). If high-purity protein sample is needed do not use the desalting column in the next step, but proceed to Subheading 3.3.3. Otherwise, see step 4 of this section.
Apply the eluted protein to a 53-mL HiPrep 26/10 desalting column (GE Healthcare) pre-equilibrated with desalting buffer. Wash with 2.5 CV of desalting buffer at 8 mL/min. Collect 1.7-mL fractions into a 96-well deep-well block (see Note 10).
Pool the peak fractions, and measure the volume. Take a 50 μL aliquot of each protein sample. These samples will be later used as “uncut controls” for SDS-PAGE. Estimate the concentration of the partially pure protein spectrophotometrically at 280 nm using an appropriate molar extinction coefficient.
Calculate the amount of TEV protease needed (protein:protease mass ratio 1:50). Add fresh β-mercaptoethanol to the protein pool to replenish the reducing agent. Add the required amount of protease.
Incubate proteins with TEV protease for 1–2 days at 4 ºC. Run SDS-PAGE to determine if the cleavage was successful using the “uncut” samples as a reference. If the His6-tag was cleaved, run IMAC-II (see Note 11).
Fig. 1.
Purification chromatograms of two His6-labeled proteins on IMAC-I (top) and IMAC-II after cleavage with TEV protease (bottom). Red stars indicate collected protein peaks
3.3.2 IMAC-II Purification
Apply the IMAC-I protein pool and TEV protease mixture onto a 5-mL HisTrap HP column (GE Healthcare) pre-equilibrated with lysis buffer with a maximum flow rate of 1 mL/min. Wash the column at 3 mL/min with 8 CV of lysis buffer to elute the cleaved target protein. Next, increase the imidazole concentration to 32.5 mM (5 % elution buffer), and wash the column with 5 CV of that buffer at 4 mL/min to elute any residual target protein that was not washed out in the lysis buffer wash. Finally, elute the uncut His6-tag protein (if any) and the His7-TEV protease with 100 % of elution buffer over 3 CV at 4 mL/min. Collect all peaks from the flow-through, wash, and elution in 1.7-mL fractions into a 96-well deep-well block (Fig. 1, bottom).
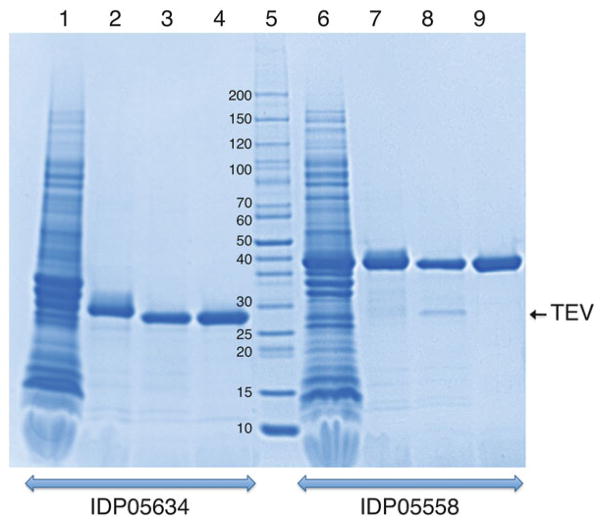
Pool the peak fractions from the flow-through, and wash and measure the volume. Estimate the concentration of the protein spectrophotometrically at 280 nm using appropriate molar extinction coefficient. Run SDS-PAGE to determine sample purity (Fig. 2). If a protein sample of high purity is needed, proceed to the next section (Subheading 3.3.3). Otherwise, see step 3 of this section.
Dialyze against crystallization buffer overnight at 4 ºC in dialysis cassettes. Choose an appropriate dialysis cassette membrane capacity for the nominal molecular weight limit of the protein.
Concentrate to a final volume of ~1 mL using a centrifugal concentrator depending on protein solubility (see Note 12). Measure protein concentration at 280 nm.
Set up crystallizations (see Chapter 12 in this book).
Freeze the remaining protein via drop freezing by dispensing the protein sample as a series of single drops. Use a P200 pipet and aliquot the protein into small beakers filled with liquid nitrogen and placed in a foam dewar. Dispense from a height of approximately 4 in. [27]. Collect the frozen droplets into a pre- chilled plastic cryogenic screw-cap tube and store at −80 ºC (see Note 13).
Fig. 2.
Purification of two His6-labeled proteins monitored by SDS-PAGE (Lonza PAGEr™ Gold 4–20 % gradient precast gel). Lanes 1 and 6: Soluble cell extracts. Lanes 2 and 7: Pooled peak fractions after IMAC-I. Lanes 3 and 8: Cleavage of His6- tag with TEV protease. Lanes 4 and 9: Pooled peak fractions after IMAC-II. Lane 5: Molecular weight standards (kDa). IDP0564 (gi|126699790; amino acid ABC transporter, substrate-binding protein [Clostridium difficile 630]) and IDP05558 (gi|16803797; hypothetical protein lmo1757 [Listeria monocytogenes EGD-e]) are targets from the Center of Structural Genomics of Infectious Diseases (CSGID)
3.3.3 Size-Exclusion Chromatography Purification
If purification of a single oligomeric state is critical, the purity of the sample is not satisfactory, or protein aggregation is suspected, the buffer exchange step after IMAC-I, IMAC-II, or both can be replaced with SEC step. The choice of the running buffer for this step will depend on the condition and destination of the protein sample. In the case where the SEC procedure is the final purification step, it is recommended to use crystallization buffer. If the protein sample needs to undergo His6-tag cleavage with TEV protease, using the desalting buffer is recommended.
Equilibrate the SEC column with at least 2 CV of desalting or crystallization buffer at a flow rate recommended by the manufacturer (for Superdex 200 HiLoad 26/60 SEC column a flow rate of 2.5–3.0 mL/min is used).
Apply the protein sample onto an SEC column (for Superdex 200 HiLoad 26/60 use a flow rate of 1.5–2.0 min/mL). The volume of the samples should be no more than 2 % of the column volume and be free of precipitation (if needed, concentrate to a smaller volume and remove the precipitation by centrifugation or filtration through a 0.22 μm PVDF or polyethersulfone syringe filter). Collect 1.8 mL fractions into a 96-well deep-well block (see Note 14).
Pool the peak fractions from the size-exclusion column. If SEC purification was performed after IMAC-I, follow Subheading 3.3.1, step 6. If SEC purification was performed after IMAC-II, follow Subheading 3.3.2, step 4.
3.3.4 Evaluation of Bacterial Protein Expression in E. coli System
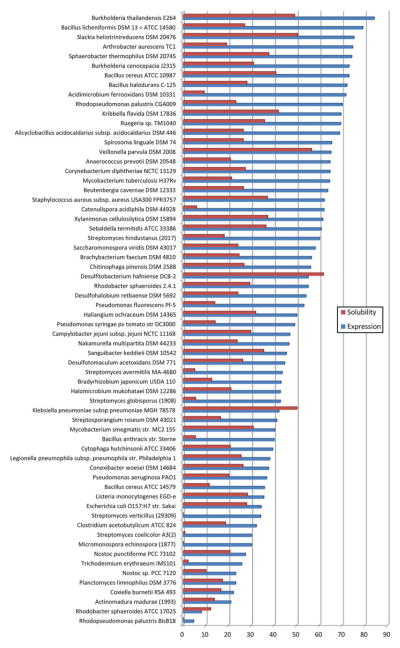
E. coli strains are commonly used as a host for expression of proteins of microbial origin (and many eukaryotic proteins as well). The method is cost effective and remains the first choice for protein expression in structural genomics centers [1, 2, 4]. Over the years, over 20,000 proteins from over 150 different bacterial strains have been produced using the standardized protocols described here. Large-scale expression and solubility levels for a sequence diverse set of proteins from over 60 bacterial strains that were produced in E. coli as an expression host have been analyzed. Results of the analysis are presented in Fig. 3. It was observed that, while good expression levels (40 % or higher) were obtained for proteins from more than 40 of the examined bacterial species, solubility levels for these proteins were generally lower (20–30 %). This method was very successful in expressing over 80 % of target proteins from Gram-negative Burkholderia thailandensis E264, of which almost 50 % were soluble. Interestingly, proteins from Gram-positive Bacillus cereus ATCC 10987 were also expressed at high levels (over 70 %) with 40 % of them soluble, whereas Bacillus anthracis str. Sterne was more problematic, with 40 % of tested proteins expressed and only 5 % soluble. A very limited success was achieved producing proteins from Rhodopseudomonas palustris BisB18, for which only about 5 % of targets were overexpressed and none were soluble. These results suggest that many bacterial species may produce proteins that are less compatible with standard protein production protocols and may require a more customized approach for expression and purification. Alternatively, these difficult proteins may need to be tested in other expression systems like cell-free expression and insect or mammalian cells.
Fig. 3.
Bacterial protein expression and solubility in the E. coli system. The expression and solubility values of more than 1,400 proteins from 25 species are shown on a normalized 0–100 scale. Expression (blue bars) and solubility (red bars) levels are calculated as a percentage of the relative abundance of the target protein in the cell lysate or the soluble fraction, respectively. The highest possible level of expression, when the relative abundance is above 80 %, is set as 100 %
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases Contract (HHSN272200700058C and HHSN272201200026C), the National Institute of Health Grant GM094585, and the US Department of Energy office of Biological and Environmental Research under Contract No. DE-AC02- 06CH11357. This work has been created by UChicago Argonne, LLC, Operator of Argonne National Laboratory (“Argonne”). Argonne, a US Department of Energy Office of Science laboratory, is operated under Contract No. DE-AC02-06CH11357. The US Government retains for itself, and others acting on its behalf, a paid-up nonexclusive, irrevocable worldwide license in said article to reproduce, prepare derivative works, distribute copies to the public, and perform publicly and display publicly, by or behalf of the government.
Footnotes
The use of antibiotics and freshly prepared media alleviates the need for sterilization of media and significantly reduces the labor involved. We have observed that un-inoculated controls exhibited no growth during the time required for protein expression in experimental cultures [19].
Keep the culture volume at exactly or below 1,000 mL for convenient centrifugation in 1-L centrifugation bottles.
Plasmid pMAGIC encodes one rare E. coli tRNA specific for two Arg codons (AGG/AGA) [1]. The Midwest Center for Structural Genomics has developed over 80 expression vectors for large-scale protein production that are optimized for structural genomics applications [14, 25]. Many of these vectors are available to the scientific community from the PSI Biology Materials Repository (http://psimr.asu.edu).
The TEV protease typically used in our procedures was expressed from vector pRK508, which carries a non-cleavable His7-tag (a gift from Dr. D. Waugh, NCI). More recently, we started using a construct expressed from vector pMHT238Δ (developed by Dr. B. G. Fox, University of Wisconsin). This plasmid, in addition to non-cleavable His7-tag, also carries several mutations and a C-terminal truncation that improve TEV protease solubility and produce high levels of the protein (100–130 mg/L of LB medium). This plasmid can be obtained from the PSI Biology Materials Repository [28].
We routinely grow 16 L of cultures at once. Transferring all 16 cultures to an incubator that is kept at 4 °C allows them to cool quickly within 45–60 min to achieve a final culture temperature of 16–18 °C. When a smaller number of cultures are used (such as four to eight 2-L bottles) the incubator can be precooled to 10 °C to achieve the same final effect.
Keep the volume of the cell resuspension below 40 mL to account for an increase of solution volume upon immersing the sonication probe during the lysis step.
We routinely use 90 mg of selenomethionine per 1 L of culture (30 mL of selenomethionine solution per 1 L) with the assumption that methionine residues comprise 2 % of the protein amino acid composition. If the methionine content is higher (5 % or more) the amount of selenomethionine added to the culture needs to be increased to 120 mg/L [29].
We routinely break cells using sonicators from Qsonica equipped with a converter and a dual horn that allows us to process two samples simultaneously. Other lysis techniques such as French press, detergent lysis, or manual shearing should yield comparable results.
For routine purifications, we use an ÄKTAxpress chromatography system (GE Healthcare) and 5-mL HisTrap columns, HiPrep 26/10 desalting columns, and/or Superdex 200 26/60 SEC columns (all columns from GE Healthcare). The ÄKTAxpress system has the capability of purifying four different proteins per purification unit. Each ÄKTAxpress unit has five column positions. We use four of these positions for Ni-affinity columns and one for a desalting or an SEC column. There are five loops available to store peak fractions from the IMAC step before the protein is loaded onto the desalting or the SEC column. If a chromatography system is not available, the IMAC can be performed using manually poured Ni-NTA columns with a peristaltic pump or by gravitational flow.
The buffer exchange step is introduced to prepare the protein sample for a cleavage with TEV protease and a subsequent IMAC-II step. We find that using a desalting column instead of dialysis speeds up the purification process, as the buffer exchange step is included in our ÄKTAxpress program following the IMAC-I step. Specifically, the protein is eluted from a Ni-affinity column using elution buffer into a 10 mL loop and then applied onto a desalting column. To prevent Ni2+ ion leakage from the metal affinity column, 2 mL of 5 mM EDTA solution in the desalting buffer is injected onto the desalting column prior to the injection of the protein. This creates an EDTA zone that sequesters released Ni2+ ions.
We frequently find that His6-tag cleavage is complete after overnight incubation with TEV protease. Sometimes we encounter fusion proteins that are only partially cleaved. This may be due to steric occlusion of the cleavage site. In some cases that problem can be mitigated by using larger amount of the protease and performing the reaction at higher temperature. On occasion, we deal with proteins that do not undergo cleavage of N-terminal His6-tag with TEV protease despite being designed to do so. In such cases, we do not proceed with the IMAC-II step but purify the protein using SEC and set up crystallizations with the uncleaved form. We also typically attempt to re-clone such protein into a vector that has a longer linker between the TEV cleavage site and the N-terminus of the protein or a vector with a C-terminal His6-tag.
Because high protein concentrations are needed for crystallization, we typically concentrate proteins to a concentration of ~1 mM or higher, depending on protein solubility. On average we obtain 50–100 mg of protein from 1 L of culture (thus it is not uncommon to have, for example, a final volume and concentration of 1 mL of 60 mg/mL solution for a 30-kDa bacterial protein).
Long-term protein storage is an important issue. The common practice of freezing proteins in a glycerol solution is problematic because glycerol can possibly interfere with crystallization and may need to be removed by dialysis after thawing. To overcome this problem we have implemented a drop-cryoprotection procedure, in which small aliquots (30–50 μL) of protein solution are dropped directly into small cylindrical containers (10–20 mL beakers) filled with liquid nitrogen. Dropped aliquots form spherical pellets, which sink to the bottom of the container. These pellets are retrieved from the bottom of the container with a pair of forceps, placed into pre-chilled Nalgene® cryogenic vials, and kept at liquid nitrogen temperature or at −80 ºC for long-term storage.
The preparative SEC can be performed on any commercially available purification workstation or with a simple peristaltic pump system. We have developed protocols for ÄKTAxpress and ÄKTAexplorer chromatography systems (GE Healthcare), in which the buffer exchange step after IMAC-I or IMAC-II is replaced with preparative SEC.
References
- 1.Kim Y, Babnigg G, Jedrzejczak R, et al. High-throughput protein purification and quality assessment for crystallization. Methods. 2011;55:12–28. doi: 10.1016/j.ymeth.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gräslund S, Nordlund P, Weigelt J, et al. Protein production and purification. Nat Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincentelli R, Bignon C, Gruez A, et al. Medium-scale structural genomics: strategies for protein expression and crystallization. Acc Chem Res. 2003;36:165–172. doi: 10.1021/ar010130s. [DOI] [PubMed] [Google Scholar]
- 4.Elsliger M-A, Deacon AM, Godzik A, et al. The JCSG high-throughput structural biology pipeline. Acta Crystallorgr Sect F Struct Biol Cryst Commun. 2010;66:1137–1142. doi: 10.1107/S1744309110038212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peti W, Page R, Moy K, et al. Towards miniaturization of a structural genomics pipeline using micro-expression and microcoil NMR. J Struct Funct Genomics. 2005;6:259–267. doi: 10.1007/s10969-005-9000-x. [DOI] [PubMed] [Google Scholar]
- 6.Price WN, II, Handelman SK, Everett JK, et al. Large-scale experimental studies show unexpected amino acid effects on protein expression and solubility in vivo in E. coli. Microb Inform Exp. 2011;1:1–20. doi: 10.1186/2042-5783-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao R, Anderson S, Aramini J, et al. The high-throughput protein sample production platform of the Northeast structural genomics consortium. J Struct Biol. 2010;172:21–33. doi: 10.1016/j.jsb.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Kim Y, Dementieva I, Zhou M, et al. Automation of protein purification for structural genomics. J Struct Funct Genomics. 2004;5:111–118. doi: 10.1023/B:JSFG.0000029206.07778.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lesley SA, Wilson IA. Protein production and crystallization at the joint center for structural genomics. J Struct Funct Genomics. 2005;6:71–79. doi: 10.1007/s10969-005-2897-2. [DOI] [PubMed] [Google Scholar]
- 11.Lesley SA, Kuhn P, Godzik A, et al. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc Natl Acad Sci. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Page R, Peti W, Wilson IA, et al. NMR screening and crystal quality of bacterially expressed prokaryotic and eukaryotic proteins in a structural genomics pipeline. Proc Natl Acad Sci U S A. 2005;102:1901–1905. doi: 10.1073/pnas.0408490102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnelly MI, Zhou M, Millard CS, et al. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr Purif. 2006;47:446–454. doi: 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eschenfeldt W, Maltseva N, Stols L, et al. Cleavable C-terminal His-tag vectors for structure determination. J Struct Funct Genomics. 2010;11:31–39. doi: 10.1007/s10969-010-9082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porath J. Immobilized metal ion affinity chromatography. Protein Expr Purif. 1992;3:263–281. doi: 10.1016/1046-5928(92)90001-d. [DOI] [PubMed] [Google Scholar]
- 16.Burley SK, Joachimiak A, Montelione GT, et al. Contributions to the NIH-NIGMS protein structure initiative from the PSI production centers. Structure. 2008;16:5–11. doi: 10.1016/j.str.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joachimiak A. High-throughput crystallography for structural genomics. Curr Opin Struct Biol. 2009;19:573–584. doi: 10.1016/j.sbi.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y, Bigelow L, Borovilos M, et al. High-throughput protein purification for X-ray crystallography and NMR. Adv Protein Chem Struct Biol. 2008;75:85–105. doi: 10.1016/S0065-3233(07)75003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millard CS, Stols L, Quartey P, et al. A less laborious approach to the high-throughput production of recombinant proteins in Escherichia coli using 2-liter plastic bottles. Protein Expr Purif. 2003;29:311–320. doi: 10.1016/s1046-5928(03)00063-9. [DOI] [PubMed] [Google Scholar]
- 20.Abdullah JM, Joachimiak A, Collart FR. “System 48” high-throughput cloning and protein expression analysis. Methods Mol Biol. 2009;498:117–127. doi: 10.1007/978-1-59745-196-3_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Acton TB, Xiao R, Anderson S, et al. Preparation of protein samples for NMR structure, function, and small-molecule screening studies. Methods Enzymol. 2011;493:21–60. doi: 10.1016/B978-0-12-381274-2.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stols L, Millard CS, Dementieva I, et al. Production of selenomethionine-labeled proteins in two-liter plastic bottles for structure determination. J Struct Funct Genomics. 2004;5:95–102. doi: 10.1023/B:JSFG.0000029196.87615.6e. [DOI] [PubMed] [Google Scholar]
- 23.Van Duyne GD, Standaert RF, Karplus PA, et al. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 24.Kapust RB, Waugh DS. Controlled intracellular processing of fusion proteins by TEV protease. Protein Expr Purif. 2000;19:312–318. doi: 10.1006/prep.2000.1251. [DOI] [PubMed] [Google Scholar]
- 25.Stols L, Gu M, Dieckman L, et al. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 26.Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng J, Davies DR, Wisedchaisri G, et al. An improved protocol for rapid freezing of protein samples for long-term storage. Acta Crystallogr D. 2004;60:203–204. doi: 10.1107/s0907444903024491. [DOI] [PubMed] [Google Scholar]
- 28.Cormier C, Park J, Fiacco M, et al. PSI:Biology-materials repository: a biologist’s resource for protein expression plasmids. J Struct Funct Genomics. 2011;12:55–62. doi: 10.1007/s10969-011-9100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauder MJ, Rutter ME, Bain K, et al. High throughput protein production and crystallization at NYSGXRC. Methods Mol Biol. 2008;426:561–575. doi: 10.1007/978-1-60327-058-8_37. [DOI] [PubMed] [Google Scholar]