Abstract
The TP53 tumor suppressor gene contains a well-studied polymorphism that encodes either proline (P) or arginine (R) at codon 72, and over half of the world’s population is homozygous for R at this codon. The wild-type sequence (wt) p53 peptide, p5365–73, has been identified as a CD8+ T cell-defined tumor antigen for use in broadly applicable cancer vaccines. However, depending on the TP53 codon 72 polymorphism of the recipient, the induced responses to the peptides incorporating R (p5372R) or P (p5372P) can be “self” or “non-self.” Thus, we sought to determine which wt p5365–73 peptide should be used in wt p53-based cancer vaccines. Despite similar predicted HLA-A2-binding affinities, the p5372P peptide was more efficient than the p5372R peptide in HLA-A2 stabilization assays. In vitro stimulation (IVS) of CD8+ T cells obtained from healthy HLA-A2+ donors with these two peptides led to the generation of CD8+ T cell effectors in one-third of the samples tested, at a frequency similar to the responsiveness to other wt p53 peptides. Interestingly, regardless of their p53 codon 72 genotype, CD8+ T cells stimulated with either p5372P or p5372R peptide were cross-reactive against T2 cells pulsed with either peptide, as well as HLA-A2+ head and neck cancer (HNC) cell lines presenting p5372P and/or p5372R peptides for T cell recognition. Therefore, the cross-reactivity of CD8+ T cells for the polymorphic wt p5365–73 peptides, irrespective of their p53 codon 72 polymorphism, suggests that employing either peptide in wt p53-based vaccines can result in efficient targeting of this epitope.
Keywords: Cytotoxic T lymphocytes, Antigen presentation, Tumor immunity
Introduction
Loss of function of the p53 gene is the most common event associated with human cancer, and is usually due to a single missense mutation of one allele, while the remainder of the accumulated p53 protein is wild type (wt) in sequence. Although considered to occur more frequently in human cancers accumulating mutated p53 molecules, the processing and presentation of wt p53 peptides by tumors can lead to elevated presentation of wt p53 peptides that can stimulate cytotoxic T lympocytes (CTL) [1–3]. In this regard, a number of CTL-defined, HLA class I-restricted human wt p53 peptides have been identified for use in p53-based immunotherapy [2, 4–10]. An obstacle to the success of wt p53 immunotherapy, however, is the weak immunogenicity of these “self” tumor peptides and the need to circumvent the tolerance imposed on these antigens. CTL responses to a range of “self” human tumor peptides, including wt p53 peptides, have been improved by amino acid substitution, or optimization, of the parental sequence. Indeed, variant peptides have been shown in vitro to reverse “nonresponsiveness” in PBMC of some HLA-A*0201 (HLA-A2)+ donors to the wt p53264–272 and p53149–157 peptides, thereby expanding the pool of donors likely to respond to vaccines targeting these epitopes [11, 12].
The most common polymorphism in TP53 has been identified at codon 72 and encodes proline (P) or arginine (R). The biological and clinical significance of this polymorphism has been reported [13], and the non-conservative amino acid change at codon 72 is associated with altered electrophoretic mobility of the two polymorphic variants, thus suggesting structural modifications of the p53 protein [14]. One of the HLA-A2-restricted, CTL-defined wt p53 peptides identified for use in cancer immunotherapy is the wt p5365–73 peptide. Consequently, two HLA-A2-resticted, wt p5365–73 peptides, p5372P and p5372R, which express either proline or arginine at residue 8, could be potentially recognized by CTL. The initial studies involving the wt p5365–73 epitope, however, did not take into account the p53 codon 72 polymorphism expressed by the responding CD8+ T cells and/or stimulating wt p5365–73 peptide, which was p5372P. Therefore, it was unclear whether the immune responses generated against this peptide were “self” or “non-self.” As a result, the immunogenicity of the wt p5365–73 peptide was open to further investigation. Because over half of the world’s population is homozygous (R/R) at codon 72, determining whether the p5372P and/or p5372R induces the expansion of CTL equally cross-reactive against both peptides is important to the targeting of this epitope by p53-based cancer vaccines. Furthermore, enhanced binding of one of these polymorphic peptides to HLA-A2 molecules compared to that of the other might make it a more immunogenic wt p5365–73 peptide for use in vaccination. Consequently, we studied whether the p5372P and/or p5372R peptide might be employed as enhanced variant peptides to obviate self-tolerance for individuals without germ line expression of the particular wt p5365–73 polymorphic peptide used in a vaccine. In this report, we investigated the HLA-A2 binding of the p5372P and p5372R peptides, characterized by the CTL raised against each of these peptides, and studied their cross-reactivity against human HLA-A2+ head and neck cancer (HNC) cell lines, which were either homozygous (R/R or P/P) or heterozygous (R/P) at the TP53 codon 72.
Materials and methods
Cells and cell lines
The HLA-A2+ SCCHN cell lines, SCC-4, PCI-13, PCI-30 [15] and UD-SCC-6 (gift of Dr. Henning Bier, University of Dusseldorf), were used; their characteristics and derivation have been published elsewhere [16]. They were cultured in DMEM supplemented with 8% FCS and 1% penicillin/streptomycin solution at 37°C and with 5% CO2. Naturally HPV-16-transformed UPCI:SCC090 (referred to as SCC90) cells were isolated, characterized and described recently [17]. SaOS-2 p53 null cells or the stable mutant p53 cDNA-transfected SaOS-2 cell lines were previously described by Balz et al. [18].
PBMC were isolated from HLA-A2+ healthy normal controls (NCs) by centrifugation over Ficoll-Hypaque gradients (Amersham Pharmacia Biotech, Piscataway, NJ, USA). Healthy control lymphocytes were obtained from leukapheresis products. HLA-A2 expression was determined first by flow cytometry using the anti-HLA-A2 antigen-specific mAb BB7.2 (American Type Culture Collection, Manassas, VA, USA) and an IgG isotype Ab as a negative control, with verification by PCR [19]. Fresh or cryopreserved PBMC were used at a concentration of 5–10 × 106 cells/ml and stored in freezing medium consisting of 90% fetal calf serum (FCS-Life Technologies) + 10% DMSO (Fisher Scientific, Pittsburgh, PA, USA) until further use.
Determination of p53 codon 72 genotype of SCCHN cells and healthy donor PBMC
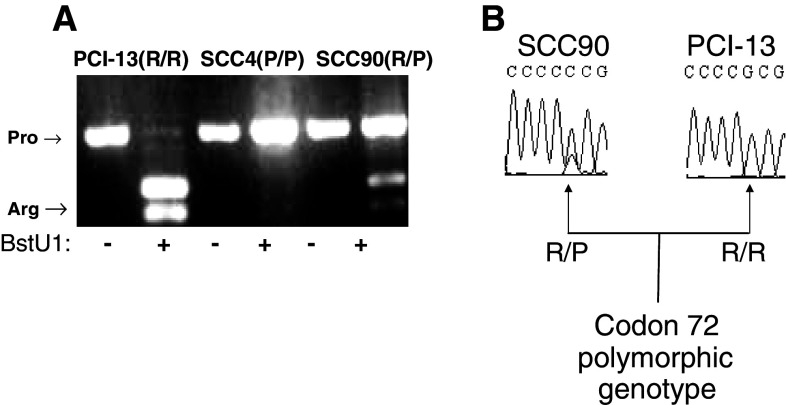
To characterize the distribution of codon 72 genotype, we selected PBMC from 158 HLA-A2+ healthy donors and 55 SCCHN patients. Isolation of genomic DNA from SCCHN patients and PBMC from healthy donors was performed using the QIAGEN DNA Mini Kit (QIAGEN Inc., Valencia, CA, USA), according to the manufacturer’s instructions. PCR-based restriction fragment length polymorphism analysis was used to identify the p53 polymorphism in codon 72 with the primers 5′-ATC TCA AGT CCC CCT TGC CG-3′ and 5′-GCA ACT GAC CGT GCA AGT CA-3′. PCR amplification involved an initial denaturation step at 95°C for 5 min, 40 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 1 min. Then, the PCR product (296 bp fragment) was digested by BstUI (New England Biolabs, Inc., Beverly, MA, USA) overnight at 60°C, which revealed the presence two bands of an arginine-encoding allele (BstU1 sensitive, [20]) or a proline-encoding allele (BstU1 insensitive), as visualized on ethidium bromide-stained agarose gel electrophoresis (Fig. 1a). Direct fluorogenic sequencing analysis (Fig. 1b) of both coding and non-coding strands with subsequent BLAST alignment was used to determine the genotype of TP53 in exons 2 through 11, with particular attention to the sequence of codon 72 of the TP53 exon 4. Using these techniques, we identified homozygous (R/R, two bands; and P/P, one band) and heterozygous (R/P, three bands) tumor cell lines and healthy donors’ PBMC.
Fig. 1.
Genotype analysis in the three different tumor cell lines. DNA was generated from SCCHN cell lines. a Three different tumor cell lines were cultured and harvested as described in “Materials and methods” for PCR amplification with the primers 5′-ATC TCA AGT CCC CCT TGC CG-3′ and 5′-GCA ACT GAC CGT GCA AGT CA-3′. An initial denaturation step was used at 95°C for 5 min, 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. PCR product (296 bp fragment) was digested with BstUI (New England Biolabs, Inc., Beverly, MA, USA) overnight at 60°C, which revealed the presence two bands of an arginine (visible fragment 169 and 127 bp)-encoding allele (BstU1 sensitive, [18]) or a proline-encoding allele (BstU1 insensitive), as visualized on ethidium bromide-stained agarose gel electrophoresis. b These three SCCHN cell lines (SCC-4 not shown) were sent for direct fluorogenic sequencing analysis of both coding and non-coding strands. Subsequent BLAST alignment was used to determine the genotype of the sequence of codon 72 of the TP53 exon 4, and the homozygosity of PCI-13 (R/R) and heterozygosity of SCC-90 (R/P) were confirmed
Peptides
The p5372P (RMPEAAPPV) and p5372R (RMPEAAPRV) peptides were synthesized using f-MOC chemistry by the University of Pittsburgh Peptide Facility and provided at >90% purity as determined by HPLC and confirmed by mass spectrometry. Peptides were stored until use in stock solutions at 1 mg/ml in DMSO.
Tetramers
Lyophilized peptide was used by the NIH Tetramer Core Facility at Emory University to produce the HLA-A2/p5372P and p5372R tetramer linked to allophycocyanin (APC) and phycoerythrin (PE), respectively.
HLA-A2 stabilization assay
For determination of peptide-induced surface HLA-A2 upregulation, T2 cells [21] were incubated for 72 h, then divided and incubated with the indicated HLA-A2-binding peptides for 18 h in AIM-V medium, at a final concentration of 10 μM to 10 nM. Cells were then stained with mAbs against HLA class I monomorphic determinant (W6/32, [22]) or against HLA-A2 (BB7.2, [23]) with similar results. Appropriate fluorescently labeled secondary Abs were added after washing twice. Flow cytometric analysis of at least 50,000 gated events was performed in at least three separate experiments for each indicated curve.
Generation and maturation of DC
DC were generated from plastic adherent human PBMCs and cultured in medium supplemented with IL-4 (1 × 103 U/mL) and GM-CSF (1 × 103 U/mL). On day 6, DC maturation was induced using AIM-V plus 10 ng/ml TNF-α, 10 ng/ml IL-1β and 10 ng/ml IL-6 for 18 h. Both immature and mature DC were phenotyped by flow cytometry, staining for CD80, CD14, CD86, HLA-DR, CD40 and CD83 [4]. DC were incubated for 4 h in the presence of the wt p5365–73 peptides prior to the addition of CTL lines.
IFN-γ ELISPOT assays
ELISPOT assays were performed as described [12]. HLA class I antigen-specific mAb (W6/32) and, as a control, HLA class II antigen-specific mAb (L243) were used for blocking recognition of target HLA-peptide complexes and defining the active T cell subset(s) in the assay system. ELISPOT analyses used IVS cultures of PBMC or enriched populations of CD8+ T cells (CTL lines) obtained from HLA-A2+ healthy donors or SCCHN patients. Briefly, 96-well flat-bottomed plates were coated overnight at 4°C with anti-human IFN-γ mAb (1-D1K: Mabtech) in PBS. The responder cells (PBMC or enriched populations of CD8+ T cells) were added. Controls consisted of unstimulated responder cells, responder cells in the presence of unpulsed APC or APC cells alone. After incubation for 20 h at 37°C, the plates were washed with PBS/0.05% Tween 20, and supplemented with the biotinylated anti-IFN-γ mAb (7-B6-1: Mabtech). After 2 h incubation at 37°C, plates were washed with PBS/0.05% Tween 20, and developed with avidin–peroxidase complex (Vectastain Elite kit). Spots were counted by computer-assisted image analysis (Cellular Technologies).
In vitro stimulation (IVS) of CD8+ T cells with p5372P and p5372R peptides
IVS cultures for stimulation and propagation of CD8+ T cell lines specific for wt p5365–73 were established as described, using autologous DC for the first two stimulations from PBMC, then peptide-loaded T2 cells thereafter [19]. Briefly, DC were pulsed with 10 μg/ml of wt p5372P and p5372R for 4 h at 37°C, and subsequently irradiated (50 Gy), washed and resuspended in AIM-V medium containing 5% (v/v) human AB serum. CD4+ T cells were negatively isolated from non-adherent PBMC using anti-CD4 microbeads (DAKO). The medium was further supplemented with 20 IU/ml of IL-2 (R&D) and 10 ng/ml of IL-7 (R&D); on day 7, the responder cells were harvested and assayed for specificity.
Data analysis
T cell reactivity as measured by the ELISPOT assay was considered positive if the test well had significantly greater spots in triplicate than background wells when using a one-tailed permutation test at α ≤ 0.05. Analysis was performed using Statview statistical software (Abacus Concepts, Inc.).
Results
Frequency of codon 72 polymorphism in the PBMC donor population studied
We first determined the frequency of codon 72 polymorphism in our population of healthy donors to indicate the potential clinical impact of differential immunogenicity of the p5372P or p5372R peptides. Our population frequency of codon 72 genotypes in Western Pennsylvania identified 96 (60.7%) with R/R genotype, 56 (35.4%) with R/P genotype and 6 with P/P genotype in 158 healthy donors, and 41 (74.5%), 8 (14.5%) and 6 (11%), respectively, in 55 HNC patients.
Stabilization of HLA-A2 molecules by wt p5365–73 72P and 72R peptides
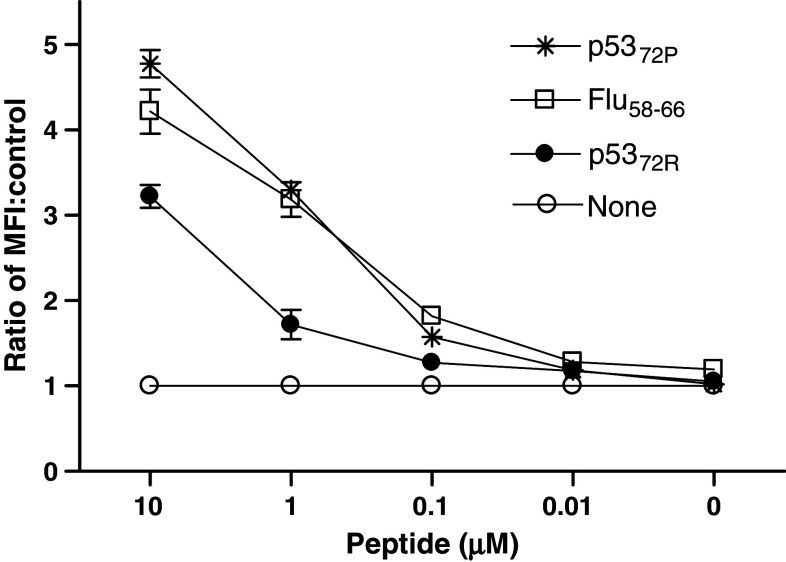
The p5372P and p5372R peptides were predicted by two HLA class I binding algorithms, http://bimas-dcrt.nih.gov/molbio/hla-bind “score >100” and http://www.syfpeithi.de “score >24”, ref. [24], to have similar binding affinities for HLA-A2 molecules. The binding of the p5372P and p5372R peptides HLA-A2 molecules was determined in vitro using the T2 stabilization assay [25]. The ability of these peptides to stabilize surface HLA-A2 expression varied significantly in T2 cells incubated with Flu58–66, p5372P and p5372R peptides (listed in descending order of binding, Fig. 2). Contrary to the predicted similar binding for each peptide, p5372P displayed a consistently higher affinity for HLA-A2 molecules than the p5372R peptide (P = 0.0277).
Fig. 2.
HLA-A2 stabilization assay comparing p53 peptides and influenza. Stabilization of cell surface HLA-A2 molecules was determined after incubation of T2 cells with p5372P and p5372R, or flu peptides at various concentrations (ranging from 10 to 0.01 μg/ml) as described in “Materials and methods”. Flow cytometry staining was performed by incubating 5 × 105 loaded cells with anti-HLA class I mAb W6/32, and then FITC-labeled secondary mAb for 30 min each at 4°C. Mean fluorescence intensity detected by flow cytometry is shown, reflecting results representative of three similar experiments
CD8+ T cells generated against either p5372P or p5372R are cross-reactive and recognize both peptides
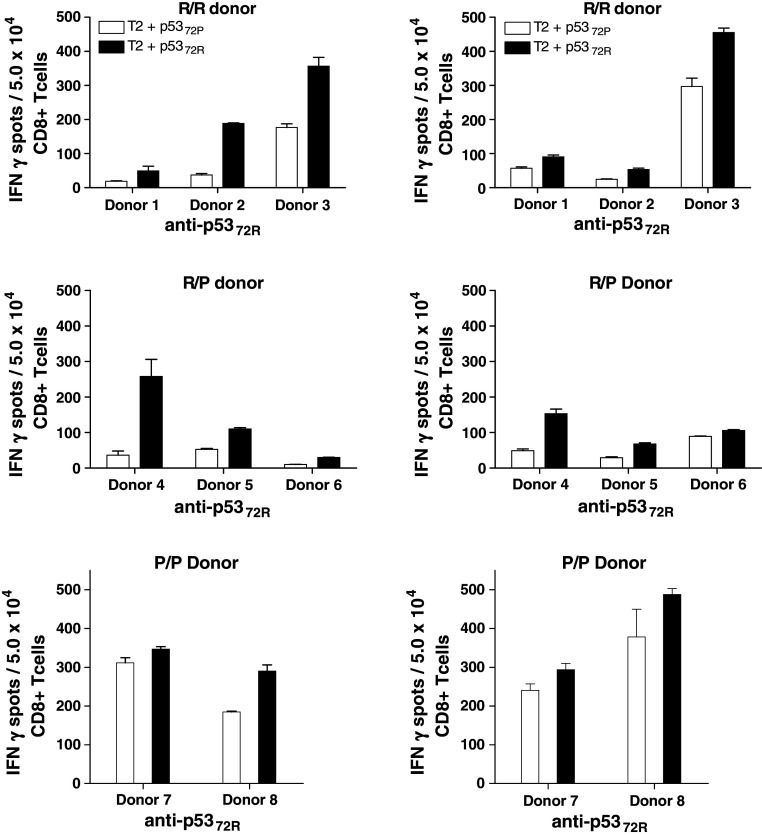
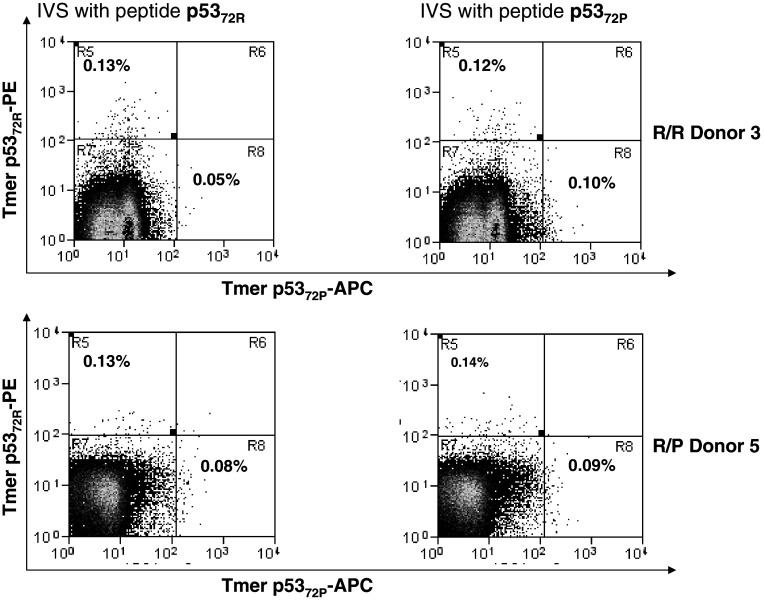
Because the affinity of a given peptide for an HLA class I molecule does not necessarily correlate with its in vitro ability to induce the expansion of CTL [26], CD8+ T cells obtained from 23 HLA-A2+ healthy donors, categorized into three groups according to their genotype (10 R/R, 10 R/P and 3 P/P), were stimulated in vitro with autologous DC pulsed with either the 72P or 72R peptides. After three rounds of IVS, the outgrowing lymphocytes derived from CD8+ T cells from 8 of these 23 donors (34.8%) demonstrated p53-specific CTL reactivity and were characterized for their ability to recognize 72P and 72R-pulsed T2 cells in IFN-γ ELISPOT assays using unpulsed T2 cells and HIV-POL476–484 peptide-pulsed T2 cells as negative controls. Among the R/R and R/P donor PBMC tested, IVS of CD8+ T cells obtained from three donors yielded reactivity in the ELISPOT assay, with consistent cross-reactivity against the p5372P/p5372R peptides for all three. The reactivity was blocked with HLA class I antigen-specific mAb, but not with HLA class II antigen-specific mAb (data not shown). Furthermore, independent of the p53 codon 72 polymorphisms of the stimulating peptide and donor’s PBMC, the induced CTL effectors showed greater recognition of p5372R-pulsed T2 cells (Fig. 3). The outgrowing lymphocytes also were analyzed by flow cytometry for p5372P and p5372R-specific CD8+ T cells using the appropriate HLA-A2/wt p53 p5372P and p5372R peptide tetramers (Fig. 4).
Fig. 3.
Cross-reactivity of CTL derived by IVS using wt p5372P and p5372R-specific T cells against both allelic peptides. CD8+ T cells from HLA-A2+ healthy donors (named as 1–8) were stimulated using IVS with DC loaded either with wt p5372P or p5372R. Reactivity of CTL against peptide-pulsed target cells was tested using IFN-γ ELISPOT assays. This recognition was blocked by HLA class I and HLA-A-specific mAb, but not an HLA class II-specific mAb (not shown). Cross-reactivity of p5372P and p5372R peptide-specific CTL was observed with better recognition of T2 cells loaded with wt p5372R, independent of the peptide used during IVS or the donor genotype. Each bar represents the mean spot number of triplicate experiments ± SD with 5 × 104 CTL per well. Background IFN-γ secretion was measured in response to T2 cells pulsed with HIV peptide or T2 cells alone. Background IFN-γ release was quantified using T2 plus HIV-1 derived peptide. Each data point represents the mean spot number of triplicate determinations ± SD with 104 CTL per well
Fig. 4.
Frequency of wt p5372P and wt p5372R-specific tetramer+ cells in PBMC from healthy donors (HD) after 3-weeks in vitro stimulation (IVS). IVS was performed using autologous DC from HLA-A2+ HD loaded with wt p5372P and p5372R peptides (10µg/ml at 37°C for 4 h). Autologous CD8+ T cells were negatively isolated from PBMC with immunomagnetic beads (Miltenyi Biotech, Germany) and added at 1 × 106 cells/ml to 1 × 105 peptide-pulsed DC in a final volume of 2 ml of culture medium (24-well tissue culture plate). The cells were cultured for 7 days at 37°C with IL-2 (20 units/ml) and IL-7 (5 ng/ml). On day 7, lymphocytes were harvested and restimulated until the completion of 21 days. Two different genotypes (R/R and R/P) were tested for the specificity of PE-labeled HLA-A2-wt p5372R and APC-labeled HLA-A2-wt p5372P tetramer. The PE-labeled HLA-A2-wt p5372R and APC-labeled HLA-A2-wt p5372P tetramer were obtained from the Tetramer Facility of the National Institute of Allergy and Infectious Disease (Atlanta, GA, USA). Specificity was confirmed by staining of the CTL line specific for each peptide after IVS and the lack of staining of irrelevant (MAGE-3271–279 specific or HIV Pol 642A) CTL or HLA-A2− PBMC obtained from normal donors (not shown). Three-color flow cytometry assays (FACScan, BD Biosciences) were performed with fluorescent labeled anti-CD3 and FITC-anti-CD8 Abs (Beckman Coulter) and PE-tetramer or APC-tetramer. Flow cytometry was performed on a CyAn™ flow cytometer (Dako, Ft. Collins, CO, USA) machine, and data analyzed using Summit V4.3 software. Generally, 100,000 events per sample were collected after gating on lymphocytes by forward and side scatter
Only 6/158 healthy donors tested encoded the P/P genotype, a finding consistent with its frequency in the population (3–8%, [27–30]). Of these six P/P donors, three were HLA-A2+ and suitable for this study. One of these did not generate p5372-specific T cells in the IVS cultures. As noted before, IVS of CD8+ T cells obtained from the remaining two HLA-A*0201+ P/P donors with p5372P and p5372R peptides yielded CTL effectors that were cross-reactive against T2 cells loaded with either peptide (Fig. 3).
CD8+ T cells generated against p5372P and p5372R recognize SCCHN cell lines expressing either peptide
We sought to determine whether CD8+ T cells induced by either p5372P or p5372R peptides recognized the naturally processed polymorphic wt p5365–73 peptides presented by human HLA-A2+ tumor cells, a pre-requisite for clinical application in cancer vaccines. CD8+ T cells induced with either p5372P or p5372R were tested for recognition of 3 HLA-A2+ HNC cell lines, PCI-13 (R/R genotype), SCC-90 (R/P genotype) or SCC-4 (P/P genotype) in IFN-γ ELISPOT assays. The peptide-induced effectors, independent of the polymorphisms of the peptide and donor CD8+ T cells used to induce them, recognized all three HNC target cells (Fig. 5a, b). Recognition was blocked by HLA class I antigen, but not HLA class II antigen-specific mAb. More importantly, the CTL showed appreciable cross-reactivity against HNC target cells that encode homozygous (R/R or P/P) alleles at TP53 codon 72, despite the lack of prior exposure of these CTL to the polymorphic peptide presented by the target cell. This observation was confirmed using Saos-2 cells (p53 null) stably transfected with mutant p53 cDNA encoding either the codon 72 P or R polymorphism (Fig. 5c).
Fig. 5.

CTL recognition and cross-reactivity of wt p5372P and p5372R on SCCHN cells. Wt p5372P and p5372R-specific CTL recognition of R/P donor (5A) and R/R donor (5B) of naturally processed peptide expressed by SCCHN cell lines SCC-4 (P/P), PCI-13 (R/R) and SCC-90 (R/P) using IFN-γ ELISPOT assays. Recognition was confirmed using stable transfectants of p53 null Saos-2 cells, which express only one p53 allele (72R or 72P, 5C) in the P/P donor. IFN-γ release was blocked by an anti-HLA class I mAb, but not an HLA class II-specific mAb (not shown). No reactivity was observed using HLA-A2− SCCHN cells (not shown). SCCHN cells were treated with IFN-γ (100 IU/ml for 72 h at 37°C)
Discussion
Since the HLA-A2-restricted, CTL-defined wt p5365–73 peptide [31] incorporates the common p53 codon 72 polymorphism, two polymorphic peptides, p5372P and p5372R, are available for use in cancer vaccines. To clarify their utility in human cancer vaccines, we sought to determine whether a differential in the in vitro immunogenicities for these peptides could be observed when the polymorphism of the peptide and the induced donor CD8+ T cells were taken into account: namely, the extent to which the donor T cell responses to the two polymorphic wt p5365–73 peptides, p5372P and p5372R, were dependent on them being “self” or “non-self.” Interestingly, despite algorithms that predict similar binding to HLA-A2 by either peptide, we observed significantly greater HLA-A2 stabilization by the p5372P peptide.
Using the p5372P and p5372R peptides to stimulate CD8+ T cell cultures, we observed that the CTL were able to cross-react against T2 cells loaded with either peptide. In addition, these T cells also recognized HLA-A2+ SCCHN cells lines, which are either homozygous (R/R or P/P) or heterozygous (R/P) at TP53 codon 72. This was confirmed using Saos-2 (p53 null) cells stably transfected with mutant p53 cDNA that encode either R or P at codon 72. Moreover, we found that IVS of healthy donor HLA-A2+ PBMC led to the generation of CTL that were reactive against the same polymorphoric peptide sequence endogenously encoded by the donor, as well as against the peptide sequence not encoded. Obviously, the important aspect of our findings for clinical utility is the cross-reactivity of the CD8+ T cells recognizing the p5372R epitope when generated by IVS using the p5372P peptide. We confirmed this phenomenon by demonstrating the ability of T cells to recognize both the p5372P and p5372R peptides, either pulsed on T2 cells or naturally presented by HLA-A2+ SCCHN cells. IVS cultures using autologous DC as antigen presenting cells loaded with the peptide of interest were specifically designed to ensure that the alternate, polymorphic peptide was not introduced into the culture either through peptide loading or endogenous antigen processing and presentation. Taken together, these data suggest that in a greater proportion of patients, the p5372P peptide would be more useful than the p5372R peptide for broadly applicable, T cell-mediated immunotherapy of human cancer.
The basis for the cross-reactivity of wt p5365–73 effectors for the p5372P and p5372R peptides requires further investigation, such as exploring the T cell receptor (TCR) vβ usage in various CTL populations, which may be heterogeneous in these IVS cultures after stimulation or vaccination using either polymorphic peptide. It is possible that CTL generated against the exogenous polymorphic wt p5365–73 peptide reacts against and eliminates expanding high-affinity T cells specific for this peptide in the IVS cultures. This phenomenon could potentially skew the outgrowth of low-moderate affinity cross-reactive CTL, which were then detected in our ELISPOT assays after three to four rounds of IVS. Nonetheless, our data clearly provide evidence that either of the (or both) peptides may be used in vivo in clinical vaccine trials for p53 overexpressing malignancies. If one peptide were to be selected for cancer vaccines, it may be desirable to choose the p5372P peptide, based on improved HLA-A2 binding and stabilization. The enhanced stabilization of HLA-A2 antigens by this peptide may provide a stronger stimulus in vivo, which may not translate in vitro into a quantitatively detectable difference in CTL induction efficiency or ELISPOT reactivity, since these assays lack ideal quantitative features necessary to detect such an effect in culture. Due to the lack of a suitable in vivo animal model, we are currently planning a clinical trial in cancer patients to test the hypotheses generated by our findings. The complexity of self–nonself vaccination of mice prevents accurate modeling of endogenous murine, wild-type p53 versus human p53 peptide(s) vaccination, which does not mimic the human situation to a valid degree. This information, if confirmed in expanded studies, would have potential implications for peptide selection in wt p53-based immunotherapy.
Acknowledgments
Supported by PO1 DE12321, P50 CA097190, the Triological Society Research Training Grant, the American College of Surgeons/American Head and Neck Society Career Development Award and the Stout Family Fund for Head and Neck Cancer Research.
References
- 1.Gnjatic S, Cai Z, Viguier M, Chouaib S, Guillet JG, Choppin J. Accumulation of the p53 protein allows recognition by human CTL of a wild-type p53 epitope presented by breast carcinomas and melanomas. J Immunol. 1998;160(1):328–333. [PubMed] [Google Scholar]
- 2.Theobald M, Biggs J, Dittmer D, Levine AJ, Sherman LA. Targeting p53 as a general tumor antigen. Proc Natl Acad Sci USA. 1995;92(26):11993–11997. doi: 10.1073/pnas.92.26.11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Offringa R, Vierboom MP, van der Burg SH, Erdile L, Melief CJ. p53: a potential target antigen for immunotherapy of cancer. Ann N Y Acad Sci. 2000;910:223–233. doi: 10.1111/j.1749-6632.2000.tb06711.x. [DOI] [PubMed] [Google Scholar]
- 4.Chikamatsu K, Nakano K, Storkus WJ, Appella E, Lotze MT, Whiteside TL, et al. Generation of anti-p53 cytotoxic T lymphocytes from human peripheral blood using autologous dendritic cells. Clin Cancer Res. 1999;5(6):1281–1288. [PubMed] [Google Scholar]
- 5.Hald J, Rasmussen N, Claesson MH. Tumour-infiltrating lymphocytes mediate lysis of autologous squamous cell carcinomas of the head and neck. Cancer Immunol Immunother. 1995;41(4):243–250. doi: 10.1007/BF01516999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ropke M, Regner M, Claesson MH. T cell-mediated cytotoxicity against p53-protein derived peptides in bulk and limiting dilution cultures of healthy donors. Scand J Immunol. 1995;42(1):98–103. doi: 10.1111/j.1365-3083.1995.tb03631.x. [DOI] [PubMed] [Google Scholar]
- 7.Barfoed AM, Petersen TR, Kirkin AF, Thor Straten P, Claesson MH, Zeuthen J. Cytotoxic T-lymphocyte clones, established by stimulation with the HLA-A2 binding p5365–73 wild type peptide loaded on dendritic cells in vitro, specifically recognize and lyse HLA-A2 tumour cells overexpressing the p53 protein. Scand J Immunol. 2000;51(2):128–133. doi: 10.1046/j.1365-3083.2000.00668.x. [DOI] [PubMed] [Google Scholar]
- 8.McCarty TM, Yu Z, Liu X, Diamond DJ, Ellenhorn JD. An HLA-restricted, p53 specific immune response from HLA transgenic p53 knockout mice. Ann Surg Oncol. 1998;5(1):93–99. doi: 10.1007/BF02303770. [DOI] [PubMed] [Google Scholar]
- 9.Nijman HW, Van der Burg SH, Vierboom MP, Houbiers JG, Kast WM, Melief CJ. p53, a potential target for tumor-directed T cells. Immunol Lett. 1994;40(2):171–178. doi: 10.1016/0165-2478(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 10.Theobald M, Biggs J, Hernandez J, Lustgarten J, Labadie C, Sherman LA. Tolerance to p53 by A2.1-restricted cytotoxic T lymphocytes. J Exp Med. 1997;185(5):833–841. doi: 10.1084/jem.185.5.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen TR, Buus S, Brunak S, Nissen MH, Sherman LA, Claesson MH. Identification and design of p53-derived HLA-A2-binding peptides with increased CTL immunogenicity. Scand J Immunol. 2001;53(4):357–364. doi: 10.1046/j.1365-3083.2001.00887.x. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann TK, Loftus DJ, Nakano K, Maeurer MJ, Chikamatsu K, Appella E, et al. The ability of variant peptides to reverse the nonresponsiveness of T lymphocytes to the wild-type sequence p53(264–272) epitope. J Immunol. 2002;168(3):1338–1347. doi: 10.4049/jimmunol.168.3.1338. [DOI] [PubMed] [Google Scholar]
- 13.Schneider-Stock R, Mawrin C, Motsch C, Boltze C, Peters B, Hartig R, et al. Retention of the arginine allele in codon 72 of the p53 gene correlates with poor apoptosis in head and neck cancer. Am J Pathol. 2004;164(4):1233–1241. doi: 10.1016/S0002-9440(10)63211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchman VL, Chumakov PM, Ninkina NN, Samarina OP, Georgiev GP. A variation in the structure of the protein-coding region of the human p53 gene. Gene. 1988;70(2):245–252. doi: 10.1016/0378-1119(88)90196-5. [DOI] [PubMed] [Google Scholar]
- 15.Lin CJ, Grandis JR, Carey TE, Gollin SM, Whiteside TL, Koch WM, et al. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007;29(2):163–188. doi: 10.1002/hed.20478. [DOI] [PubMed] [Google Scholar]
- 16.Heo DS, Snyderman C, Gollin SM, Pan S, Walker E, Deka R, et al. Biology, cytogenetics, and sensitivity to immunological effector cells of new head and neck squamous cell carcinoma lines. Cancer Res. 1989;49(18):5167–5175. [PubMed] [Google Scholar]
- 17.Ferris RL, Martinez I, Sirianni N, Wang J, Lopez-Albaitero A, Gollin SM, et al. Human papillomavirus-16 associated squamous cell carcinoma of the head and neck (SCCHN): a natural disease model provides insights into viral carcinogenesis. Eur J Cancer. 2005;41(5):807–815. doi: 10.1016/j.ejca.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 18.Balz V, Scheckenbach K, Gotte K, Bockmuhl U, Petersen I, Bier H. Is the p53 inactivation frequency in squamous cell carcinomas of the head and neck underestimated? Analysis of p53 exons 2–11 and human papillomavirus 16/18 E6 transcripts in 123 unselected tumor specimens. Cancer Res. 2003;63(6):1188–1191. [PubMed] [Google Scholar]
- 19.Sirianni N, Ha PK, Oelke M, Califano J, Gooding W, Westra W, et al. Effect of human papillomavirus-16 infection on CD8+ T cell recognition of a wild-type sequence p53264–272 peptide in patients with squamous cell carcinoma of the head and neck. Clin Cancer Res. 2004;10(20):6929–6937. doi: 10.1158/1078-0432.CCR-04-0672. [DOI] [PubMed] [Google Scholar]
- 20.Birgander R, Sjalander A, Rannug A, Alexandrie AK, Sundberg MI, Seidegard J, et al. p53 polymorphisms and haplotypes in lung cancer. Carcinogenesis. 1995;16(9):2233–2236. doi: 10.1093/carcin/16.9.2233. [DOI] [PubMed] [Google Scholar]
- 21.Salter RD, Cresswell P. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. EMBO J. 1986;5(5):943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parham P, Barnstable CJ, Bodmer WF. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A, B, C, antigens. J Immunol. 1979;123(1):342–349. [PubMed] [Google Scholar]
- 23.Parham P, Bodmer WF. Monoclonal antibody to a human histocompatibility alloantigen, HLA-A2. Nature. 1978;276(5686):397–399. doi: 10.1038/276397a0. [DOI] [PubMed] [Google Scholar]
- 24.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3–4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 25.Filho PA, Lopez-Albaitero A, Xi L, Gooding W, Godfrey T, Ferris RL. Quantitative expression and immunogenicity of MAGE-3 and -6 in upper aerodigestive tract cancer. Int J Cancer. 2009;125(8):1912–1920. doi: 10.1002/ijc.24590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullock TN, Mullins DW, Colella TA, Engelhard VH. Manipulation of avidity to improve effectiveness of adoptively transferred CD8(+) T cells for melanoma immunotherapy in human MHC class I-transgenic mice. J Immunol. 2001;167(10):5824–5831. doi: 10.4049/jimmunol.167.10.5824. [DOI] [PubMed] [Google Scholar]
- 27.Cairey-Remonnay S, Humbey O, Mougin C, Algros MP, Mauny F, Kanitakis J, et al. TP53 polymorphism of exon 4 at codon 72 in cutaneous squamous cell carcinoma and benign epithelial lesions of renal transplant recipients and immunocompetent individuals: lack of correlation with human papillomavirus status. J Invest Dermatol. 2002;118(6):1026–1031. doi: 10.1046/j.1523-1747.2002.01787.x. [DOI] [PubMed] [Google Scholar]
- 28.Rosenthal AN, Ryan A, Al-Jehani RM, Storey A, Harwood CA, Jacobs IJ. p53 codon 72 polymorphism and risk of cervical cancer in UK. Lancet. 1998;352(9131):871–872. doi: 10.1016/S0140-6736(98)07357-7. [DOI] [PubMed] [Google Scholar]
- 29.Shen H, Zheng Y, Sturgis EM, Spitz MR, Wei Q. P53 codon 72 polymorphism and risk of squamous cell carcinoma of the head and neck: a case–control study. Cancer Lett. 2002;183(2):123–130. doi: 10.1016/S0304-3835(02)00117-9. [DOI] [PubMed] [Google Scholar]
- 30.Sourvinos G, Rizos E, Spandidos DA. p53 codon 72 polymorphism is linked to the development and not the progression of benign and malignant laryngeal tumours. Oral Oncol. 2001;37(7):572–578. doi: 10.1016/S1368-8375(00)00139-1. [DOI] [PubMed] [Google Scholar]
- 31.Albers AE, Ferris RL, Kim GG, Chikamatsu K, De Leo AB, Whiteside TL. Immune responses to p53 in patients with cancer: enrichment in tetramer +p53 peptide-specific T cells and regulatory T cells at tumor sites. Cancer Immunol Immunother. 2005;54(11):1072–1081. doi: 10.1007/s00262-005-0670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]