Abstract
Recent advances in genomic research have revealed the existence of a large number of transcripts devoid of protein-coding potential in multiple organisms 1-8. While the functional role for long non-coding RNAs (lncRNAs) has been best defined in epigenetic phenomena such as X inactivation and imprinting, different classes of lncRNAs may have varied biological functions 8-13. We and others have identified a class of lncRNAs, termed ncRNA-activating (ncRNA-a), that function to activate their neighboring genes using a cis-mediated mechanism 5,14-16. To define the precise mode by which such enhancer-like RNAs function, we depleted factors with known roles in transcriptional activation and assessed their role in RNA-dependent activation. Here we report that depletion of the components of the co-activator complex, Mediator, specifically and potently diminished the ncRNA-induced activation of transcription in such a heterologous reporter assay. In vivo, Mediator is recruited to ncRNA-as target genes, and regulates their expression. We show that ncRNA-as interact with Mediator to regulate its chromatin localization and kinase activity toward histone H3 serine 10. Mediator complex harboring disease causing MED12 mutations 17,18 displays diminished ability to associate with activating ncRNAs. Chromosome conformation capture (3C) confirmed the presence of DNA looping between the ncRNA-a loci and its targets. Importantly, depletion of Mediator subunits or ncRNA-as reduced the chromatin looping between the two loci. Our results identify the human Mediator complex as the transducer of activating ncRNAs and highlight the importance of Mediator and activating ncRNAs association in human disease.
To define the transcriptional complex(es) that orchestrate the responsiveness of activating lncRNAs, we used stable cell lines expressing the heterologous TK promoter driving luciferase expression fused to ncRNA-a7 controlled by its own natural promoter and depleted factors known to be involved in transcriptional activation and enhancer function. We also developed stable lines expressing control reporters, in which a DNA fragment devoid of any transcriptional activity was substituted for the ncRNA-a7 genomic region (Fig. 1a). Importantly, depletion of ncRNA-a7 using two different siRNAs specifically decreased the transcription of the reporter construct containing the ncRNA-a7 (Fig. 1b). Next, we depleted factors known to be involved in transcriptional activation (Supplementary Fig. 1a) and examined the responsiveness of cell lines harboring the constructs expressing ncRNA-a7 or control constructs devoid of lncRNAs. Interestingly, only depletion of Mediator subunit, MED12 displayed a differential effect on the transcription of the reporter construct containing the ncRNA-a7 (Fig. 1c). Depletion of other factors was either ineffective in changing transcriptional output (see Cdk9, Cyc T, NIPBL or WDR5) or reduced the transcriptional levels for both constructs (see GTF2B, p300, SMC1) (Fig. 1c).
Figure 1. Mediator confers the ncRNA-as dependent activation of a heterologous reporter.
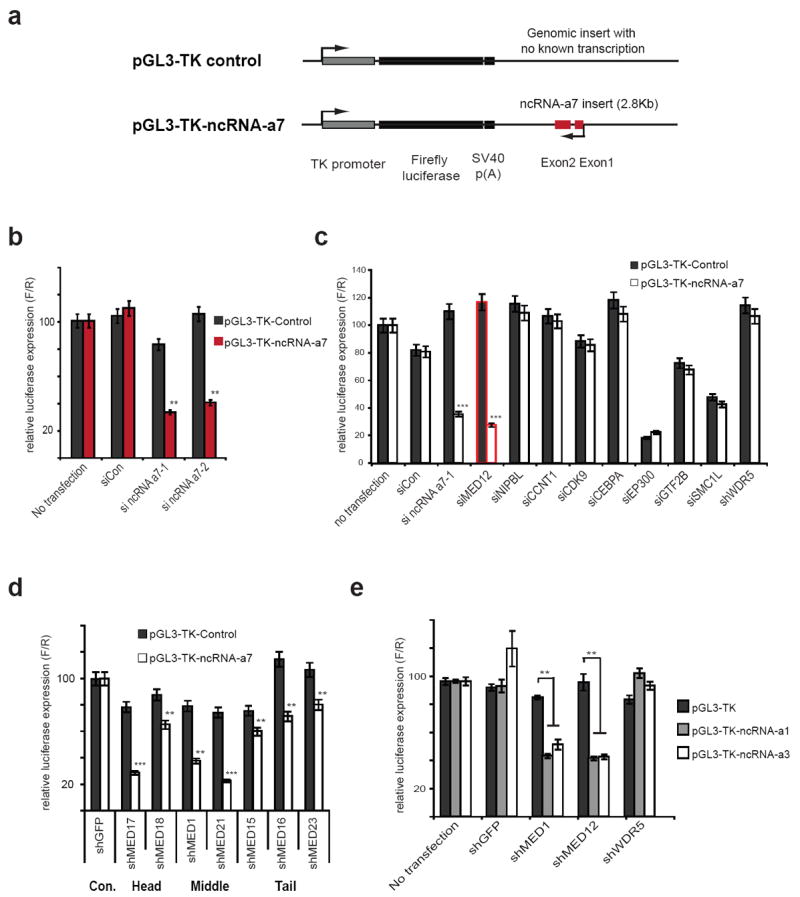
a, Schematic representation of genomic control insert (top) or ncRNA-a7 insert (bottom) in luciferase reporter vector driven by a TK-promoter.
b, Depletion of ncRNA-a7 reduces the luciferase activity in the ncRNA-a7 luciferase reporter cell lines.
c, Depletion of the transcription factors or enhancers in the control or ncRNA-a7 reporter cell lines. The red bar signifies reduced transcription using siRNA against MED12, comparable with siRNA against ncRNA-a7.
d, Depletion of different Mediator subunits using the ncRNA-a7 reporter cell lines.
e, Depletion of the Mediator subunits in ncRNA-a1 or ncRNA-a3 luciferase reporter cell lines. All data shown are mean ±SEM of three independent experiments. **p<0.01, ***p<0.001 by two-tailed Student’s T-test.
Mediator contains a complex and modular subunit composition 19,20. We depleted different components of Mediator corresponding to the head, middle or the leg modules and assessed their effect on the responsiveness of the cell lines harboring the constructs driven by ncRNA-a7 or the control construct. Overall, depletion of other subunits of the Mediator complex also reduced the ncRNA-a7-induced activation, although there was also a small reduction in transcription of the control plasmid with depletion of some of the subunits (Fig. 1d and Supplementary Fig. 1b). Similar to its effects on ncRNA-a7, depletion of the Mediator subunits decreased the RNA-induced activation of two other ncRNA-as, which were previously shown to regulate ECM1 and TAL1 genes (Fig. 1e). Taken together, these results identify the Mediator complex as a transducer of ncRNA-a function.
We next depleted the Mediator subunits and assessed their effect on ncRNA-as targets in vivo. We had previously found that ncRNA-a7 and ncRNA-a3 regulate SNAI1 and TAL1 genes, respectively 5. Moreover, using gene expression arrays, we had observed a profound decrease in AURKA transcription when ncRNA-a7 was depleted 5. We confirmed these results by depleting ncRNA-a7 and ncRNA-a3 which led to a decrease in SNAI1, AURKA and TAL1 transcript levels using real-time PCR (Fig. 2a and b). We next depleted two different subunits of the Mediator complex MED1 and MED12 and assessed the SNAI1, AURKA, and TAL1 transcript levels (Supplementary Fig. 1b). Importantly, depletion of Mediator subunits decreased transcription of all three targets similar to that shown with depletion of ncRNA as (Fig. 2a and b, the affected genes are depicted by a black bar). Interestingly depletion of Mediator subunits did not affect the transcript levels of genes that are not regulated by ncRNA-a (UBE2V1, CSTF, STIL, depicted by a white bar in Fig. 2a and b). However, while ncRNA-a7 transcription was not affected following Mediator depletion, there was a decrease in ncRNA-a3 levels (Fig. 2a and b).
Figure 2. Functional association of Mediator and activating ncRNAs.
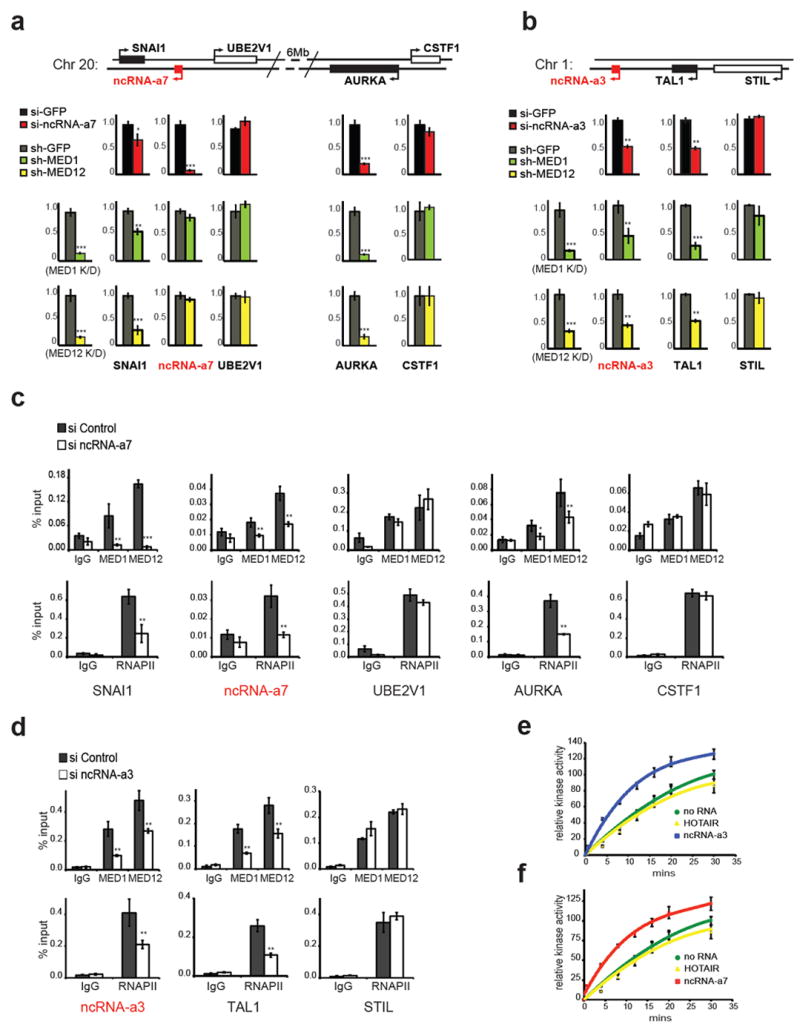
a, Knockdowns of ncRNA-a7 or Mediator subunits decrease SNAI1, AURKA expression. Depletion of ncRNA-a7 diminished SNAI1 and AURKA expression by Real-time PCR (top red bars). Similarly knockdowns of MED1 (green bars) or MED12 (yellow bars) reduce SNAI1 or AURKA expression.
b, Knockdowns of ncRNA-a3 or Mediator subunits decrease TAL1 expression. Expression of TAL1 or control STIL following knockdowns of ncRNA-a3 (red bars), MED1 (green bars) and MED12 (yellow bars) are shown.
c-d, Knockdowns of ncRNA-a7 (c) or ncRNA-a3 (d) reduce the genomic occupancy of MED1, MED12 (top panels) or RNAPII (bottom panels) on SNAI1, AURKA (c) or TAL1 (d) in A549 cells.
e-f, Activating ncRNAs specifically stimulate Mediator kinase activity toward histone H3.1 substrate in vitro. Quantification of kinase assay following addition of ncRNA-a7 (e) or ncRNA-a3 (f). Error bars represent ±SEM (n=3), p<0.01 by two-tailed Student’s T-test.
The mean ±SEM for all results represent three independent experiments. *p<0.05, **p<0.01, ***p<0.001 by two-tailed Student’s T-test.
To ensure that such transcriptional effects following Mediator depletion were direct, we performed chromatin immunoprecipitation (ChIP) using antibodies against the MED12 and MED1 subunits of Mediator. This analysis revealed that the Mediator complex occupied the promoters of ncRNA-a3, ncRNA-a7 and their targets (Fig. 2c and d). Moreover, we found RNA polymerase II (RNAPII) at these promoters consistent with Mediator occupancy (Fig. 2c and d). Importantly, depletion of ncRNA-a7 or ncRNA-a3 decreased the occupancy of Mediator and RNAPII at their target genes without affecting the occupancy at genes not regulated by ncRNA-as, (Fig. 2c and d). These results indicate that the Mediator complex occupies the promoters of genes regulated by ncRNA-as and depletion of ncRNA decreases the occupancy of Mediator at these sites.
The Mediator complex, via its CDK8 module, displays kinase activity toward histone H3 serine 10 (H3S10), a histone modification with strong ties to transcriptional activation 21,22. We next assessed whether the ncRNA-a regulate Mediator kinase activity toward histone H3. Addition of ncRNA-a7 or ncRNA-a3 led to a significant stimulation of the Mediator kinase activity toward histone H3, while the addition of primary let7b ncRNA or ncRNA HOTAIR was devoid of any effect (Fig. 2 e and f, Supplementary Fig. 2a). Such stimulation of Mediator kinase activity by ncRNA-a was specific toward histone H3 as the substrate. We did not observe any stimulation when a GST-CTD (corresponding to the C-terminal domain of RPB1) or cyclin H was used as substrates (Supplementary Fig. 2b and c). Moreover, consistent with a role for ncRNA-a in stimulation of histone H3 phosphorylation, depletion of ncRNA-a7 or ncRNA-a3 led to a specific decrease in H3S10 levels at SNAI1, AURKA and TAL1 (Supplementary Fig. 2d and e).
We next asked whether ncRNA-a could associate with the Mediator complex. We isolated Mediator using stable cell lines expressing Flag-epitope tagged MED12 and examined its interaction with in vitro transcribed ncRNA-a7. An eluate from a stable Flag-GFP cell line was used as a control (Fig. 3a). While the control primary let7 transcript (prilet7) or the ncRNA, HOTAIR, (a 429 nucleotide RNA with a similar base composition as ncRNA-a7) did not specifically interact with the Mediator complex, we detected a robust and specific association between ncRNA-a7 and the Mediator complex (Fig. 3a). Importantly, Mediator also associated with ncRNA-a1 and ncRNA-a3, two other ncRNA-as that were shown to activate transcription of their neighboring genes 5 (Fig. 3b).
Figure 3. Interaction of Mediator and activating ncRNAs is disrupted by FG syndrome mutations of MED12.
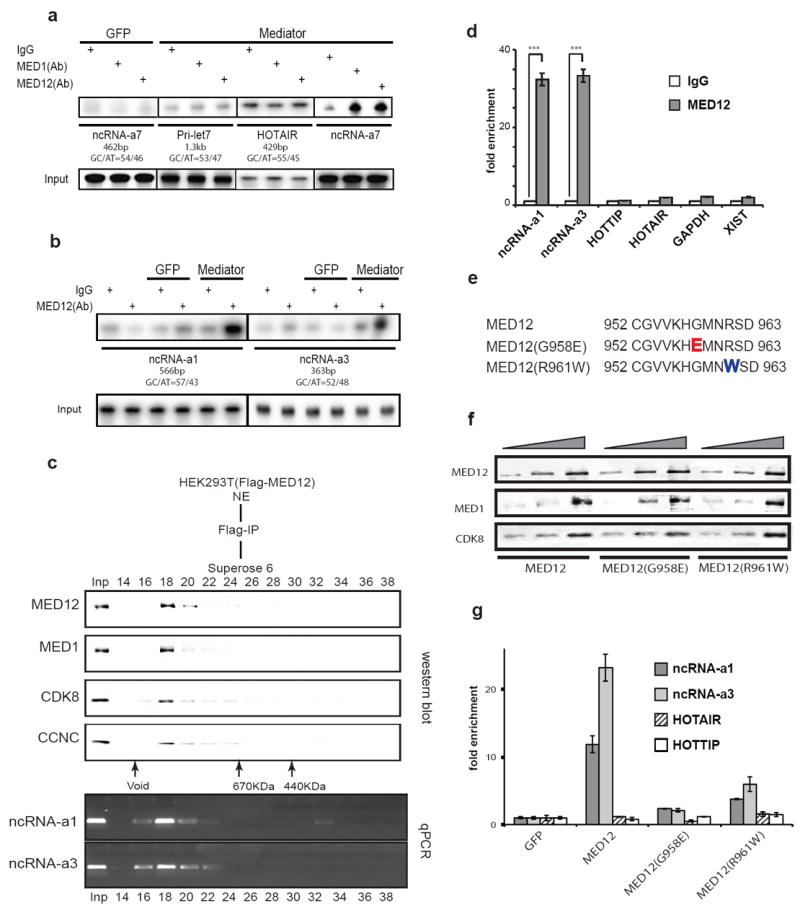
a-b, ncRNA-as associate with Mediator complex in vitro. RNA immune-precipitation (RIP) was performed using protein A dynabeads coupled with IgG, MED1 or MED12 antibodies. Identical results were obtained in three independent experiments.
c, Purification of Mediator protein complex and associated RNAs. The purified FLAG-MED12 affinity elution was subjected to Superose 6 size-exclusion chromatography and the Mediator subunits were detected by Western blot. The Superose 6 fractions (14-38) are shown on top and molecular mass markers (kilodaltons) are at the bottom.
The RNAs in each fraction was extracted and was followed by RT-PCR using specific primers.
d, UV cross-link followed by RIP using HEK293T whole cell lysates. The associated RNAs were analyzed by RT-qPCR with individual specific RNA primers. The mean ±SEM are from three independent experiments. ***p<0.001 by two-tailed Student’s T-test.
e, Protein sequences of MED12 mutants causing FG syndrome (Opitz-Kaveggia syndrome). Part of the protein sequence of MED12 exon 21 (top panel). The mutated sites indicate the G958E mutant (red) and R961W mutant (blue).
f, HEK293T cells were transfected with FLAG-MED12 or mutant constructs. After UV cross-linking, total cell extracts were immunoprecipitated and analyzed by Western blotting.
g, Following UV-RIP of wild type and mutant MED12 samples shown in (f), RT-qPCR was performed using transcript specific primers from the lncRNAs shown. The data is a representative of three independent experiments. The two-tailed Student’s T-test, indicated a p<0.001 for ncRNA-a1 and ncRNA-a3, while there was no significant difference for HOTAIR and HOTTIP.
To assess the association of endogenous ncRNA-a and Mediator, the Flag-MED12 affinity-purified Mediator was fractionated by gel filtration and column fractions were subjected to Western blot analysis and RT-PCR to detect the associated ncRNA-a (Fig 3c). While the HEK293 nuclear extract contained similar levels of ncRNA-a1, ncRNA-a3, HOTAIR and HOTTIP (HEK293 do not express ncRNA-a7, Supplementary Fig. 3a), we could only detect the ncRNA-a1 and ncRNA-a3 in association with the affinity-purified Mediator (Fig. 3c). Indeed, ncRNA-a1 and ncRNA-a3 displayed a co-elution with components of the Mediator complex on gel filtration further supporting their association with Mediator (Fig. 3c). Next, we performed RNA immunoprecipitation following ultraviolet crosslinking (UV-RIP) with anti-MED12 antibodies or IgG, as control. Activating ncRNA-a1 and ncRNA-a3 were significantly enriched in the MED12 UV-RIP, whereas other abundant control mRNAs or non-coding RNAs such as GAPDH, HOTAIR, HOTTIP and XIST were not (Fig. 3d). Two mutations in MED12 proteins have been linked to the development of human genetic disorder Opitz-Kaveggia syndrome (also know as FG syndrome) causing physical anomalies and developmental delays 17,18 (Fig. 3e). Importantly, while such mutations in MED12 do not affect its association with other Mediator subunits, they significantly diminish its association with ncRNA-a1 and ncRNA-a3 as measured by UV-RIP (Fig. 3f and g).
We next asked whether we could detect long-range chromatin looping between the ncRNA-a7 locus and its targets, SNAI1 and AURKA and whether ncRNA-a7 and the Mediator complex have a role in such an association. We performed chromosome conformation capture (3C) to assess the association of ncRNA-a7 with AURKA as well as SNAI1. We also performed a similar analysis between ncRNA-a3 and TAL1 gene. We used anchoring points near the 3’-UTR of ncRNA-a7 or ncRNA-a3 to measure the extent of chromatin looping between the ncRNA-a7 or ncRNA-a3 locus and their targets as shown in Figure 4a, 4d and Supplementary Fig. 3b and c. This analysis revealed a strong association between the ncRNA-a7 locus and the region encompassing the promoter region of SNAI1 (Fig. 4b, upper panel). Moreover, ncRNA-a7 also displayed an interaction with regions near the 5’-end of the AURKA gene that extended into the body of the gene (Fig. 4c, upper panel). There was no detectable association when a control anchor was placed in the genomic region between ncRNA-a7 and AURKA (Fig. 4b and c).
Figure 4. Mediator complex and ncRNA-as promote chromatin looping.
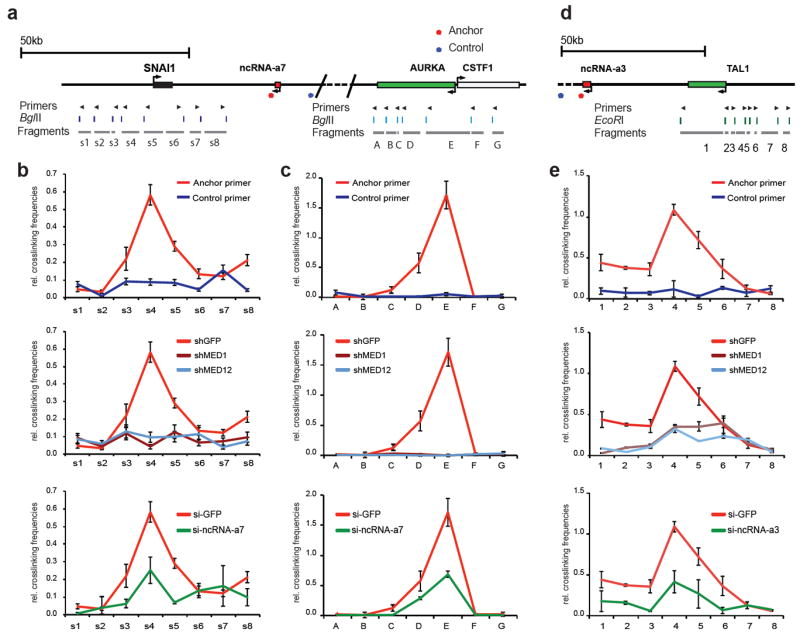
The schematic diagrams represent the genomic locus between the ncRNA-a7, SNAI1 and AURKA loci (a), the ncRNA-a3 and TAL1 (d). The top arrows show the position of primers, the digestion sites are shown in the middle and the s1-8, A-G or 1-8 fragments are presented below.
The looping events between ncRNAs and its targets were detected between ncRNA-a7 and SNAI1 (b), ncRNA-a7 and Aurka (c) and ncRNA-a3 and TAL1(e) using chromosome conformation capture (3C). Depletion of MED1 or MED12 abolished the loop interaction (b,c,e, middle panels). Knockdown of ncRNA-a7 or ncRNA-a3 reduced the chromosomal looping events (b,c,e, lower panels). The interaction frequency between the anchoring points and distal fragments were determined by Real-time PCR and normalized to BAC templates and control anchors. Each error bar represents ±SEM from three independent experiments, p<0.01 by two-tailed Student’s T-test. Representative gel images of the 3C experiments for AURKA are presented in Supplementary Figure 3.
Next we depleted the Mediator subunits MED1 or MED12 and assessed their potential role in mediating the chromosomal looping between ncRNA-a7, SNAI1 and AURKA. Depletion of either MED1 or MED12 completely abrogated the chromosomal looping between the ncRNA-a7 and SNAI1 (Fig. 4b, middle panel). A similar result was obtained following depletion of MED1 or MED12 at the AURKA locus (Fig. 4c, middle panel). Importantly, depletion of ncRNA-a7 had a comparable effect reducing the chromosomal looping between the ncRNA-a7 and both SNAI1 and AURKA loci (Fig. 4b and c, lower panels). Since depletion of ncRNA-a7 using siRNAs resulted in a partial decrease of ncRNA-a7 concentrations (about 60% depletion, Supplementary Fig. 1), the residual ncRNA-a7 may be sufficient to promote smaller levels of DNA looping seen following ncRNA-a7 depletion.
We also performed a similar analysis measuring the extent of DNA looping between the ncRNA-a3 and that of the TAL1 locus. Similar to results obtained with ncRNA-a7, we detected a specific and robust DNA looping between ncRNA-a3 and TAL1 (Fig. 4e). Finally, depletion of Mediator subunits MED1 and MED12 or ncRNA-a3 nearly abolished the DNA looping between ncRNA-a3 and TAL1 (Fig. 4e, middle and lower panels). Notably, Mediator and ncRNA-a association with its target genes extend beyond the transcription start sites to include the more distal sites. This may reflect the engagement of proximal regulatory elements in the ncRNA-a and Mediator chromatin looping (Fig. 4b and 4e). These results suggest a mechanism of action for ncRNA-a and Mediator that may involve additional chromatin contacts beyond that has been observed for distal regulatory elements and transcriptional start sites.
We propose a new mechanism of action for a class of lncRNAs termed ncRNA-activating (ncRNA-a) involved in long-range transcriptional activation through their association with the Mediator complex (Supplementary Fig. 4). It is noteworthy that such Mediator/ncRNA-a-dependent chromatin loops extend beyond the transcription start sites of the target promoters, which may reflect the inclusion of promoter proximal regulatory elements in such associations. Future studies could also assess whether such ncRNA-as–Mediator chromatin loops augment or function exclusively of Mediator-Cohesin complexes23.
Recently, experiments in zebrafish uncovered a fundamental role for lncRNAs in developmental control 24. Moreover, recent reports have linked a number of lncRNAs to human diseases such as cancer and neurological disorders 11,25-27. Importantly, our studies indicated that Mediator complexes containing disease-causing mutant MED12 proteins corresponding to FG syndrome fail to associate with ncRNA-as. These results underscore the significance of Mediator-ncRNA-a association and suggest the loss of such Mediator – ncRNA-a interactions as a possible contributing factor in such developmental disorders.
METHODS
siRNA/DNA transfection and ncRNA Luciferase assays
pGL3-TK-ncRNA-as and pGL3-TK-control constructs were generated as described previously. Firefly luciferase constructs, pRL-CMV and a selectable marker for puromycin resistance were co-transfected in HEK293T cells. Transfected cells were grown in presence of 2.5 ug/ml puromycin (Sigma) for selection. Individual colonies were isolated and screened for luciferase expression. Luciferase assays were performed in 96-well white plates using Dual-Glo luciferase assay system (Promega) according to the manufacturer’s protocol.
On-TargetPlus siRNAs and non-targeting control siRNA were purchased from Dharmacon, Inc. The modified siDNA oligoes were purchased from Integrated DNA Technologies. siRNA or siDNA transfections were performed with Lipofectamine RNAiMAX (Life Technologies, Inc.) according to the manufacturer’s instructions, using 50 nM siRNA/DNAs in a final volume of 2 ml of culture medium without antibiotics. Cells were harvested 48 to 72 h after two-rounds of transfections.
Corresponding ncRNA-as HEK293T cell lines growing in 12-well dishes were transfected with corresponding siRNA oligoes, using Metafectene® Pro (Biontex Laboratories). Twenty four hours post-transfection, the cells were lysed in 200 μl passive lysis buffer (Promega), and firefly and Renilla luciferase activities were measured on 30 μl lysate using the Dual-Luciferase reporter assay kit (Promega). Individual experiments were realized in triplicate, and the ratios presented are the averages of three independent experiments.
siRNA sequences
siControl1: ON-TARGET Non-targeting siRNA #1 (D-001810-01); siControl2: ON-TARGET Non-targeting siRNA #2 (D-001810-02); siGFP: GFP Duplex I (P-002048-01); si ncRNA-a3-1: CUAUGGAAUUUAAGCCCAAUU; si ncRNA-a3-2: GCAAGAUCACGGUGCGUCAUU; si ncRNA-a7-1: CCGAUUUGAGAGAGUGAGAUU; si ncRNA-a7-2: GAAGGGAACCAGUGCUAAAUU; siMed12: UCACUCAUCUCAUGUUAUAUU; siNIPBL: GGGAAUAUGAAGAGCGUGAUU; siCCNT1: GAACAAACGUCCUGGUGAUUU; siCDK9: UGACGUCCAUGUUCGAGUAUU; siCEBPA: ACAAGAACAGCAACGAGUAUU; siEP300: GGACUACCCUAUCAAGUAAUU; siGTF2B: ACAAUCAGACAGUCCUAUAUU; siSMC1L: CAUCAAAGCUCGUAACUUCUU
siDNA sequences
siGFP: T*C*A*C*C*T*T*C*A*C*C*C*T*C*T*C*C*A*C*T;
si ncRNA-a1: C*C*T*G*C*C*C*T*C*T*T*C*C*T*C*C*T*G*T*A;
si ncRNA-a3: G*C*T*C*G*T*C*G*G*G*T*A*C*T*C*T*C*C*A*T*T;
si ncRNA-a7-1: G*T*T*C*C*C*T*T*C*T*G*A*C*G*C*C*T*G*G*T*A;
si ncRNA-a7-2: C*A*T*C*T*T*C*T*C*C*G*T*C*T*C*T*G*C*C*A*C
shRNA infections
pLKO.1 Lentiviral vector containing short hairpin RNAs (shRNAs) or shRNA targeting GFP control (Open Biosystems). The TRC shRNA constructs were purchased from Open Biosystems using Lentiviral packaging system. 24 hours after infection, selection with 2.5 μg/ml puromycin was performed for 48 hours.
shRNA list
shGFP: RHS4459;
MED12 (TRCN0000018576): CGGGTACTTCATACTTTGG;
MED17 (TRCN0000019245): CCGAGCTTGCAGTTATCTATT;
MED18 (TRCN0000053164): GCCAGAAATGGGAGACAAGAA;
MED1 (TRCN0000019800): GCCGAGTTCCTCTTATCCTAA;
MED21 (TRCN0000013429): CCATTGGAGTATTGCAGCAAT;
MED15 (TRCN0000018969): GCTCAGAACCAACCATCAC;
MED16 (TRCN0000022139): CGACATTGACAAGGTCATGAT;
MED23 (TRCN0000019196): CGCAGTTTACACGCTTCCT;
WDR5 (TRCN0000118050): CCAACCTTATTGTCTCAGGAT
Antibodies and recombinant proteins
Pol II (SC-899, Santa Cruz), Med1 (A300-793A, Bethyl labs.), Med12 (A300-774A, Bethyl labs), HistoneH3Ser10p (05-598, Millipore), CDK8 (SC-1521, Santa Cruz), CCNC (Cyclin C, SC-1061, Santa Cruz), Human rHistone H3.1 (M2503S, NEB), Human rCCNH (pka-364, Prospec protein specialists).
RNA Purification, cDNA Synthesis, and Quantitative PCR
Cells were harvested and re-suspended in TRIzol (Invitrogen) and RNA extracted according to the manufacturer’s protocol. cDNA synthesis was done using random primers with Fermentas RevertAid™ first strand cDNA synthesis kit. Quantitative PCR was done using IQ™ SYBR Green super-mixes and CFX96™ Touch Real-Time PCR detection system (Bio-rad). For all quantitative PCR reactions Gapdh was measured for an internal control and used to normalize the data. QPCR primer sequences are listed here:
ncRNA-a1-qfor: GCAAGCGGAGACTTGTCTTT;
ncRNA-a1-qrev: GGCTGGTCTTGAACTCCTGA;
ncRNA-a3-qfor: TTAAGCCCAAGGAATGGAGA;
ncRNA-a3-qrev: AGCGGTGTGGAATAAACTGG;
ncRNA-a7-q1for: ATCCTGGTGGAAAAGGCATC;
ncRNA-a7-q1rev: GCCTGGGAAAAGCTACTTCA;
ncRNA-a7-q2for: CCGTTGGCTCCACAAACCT;
ncRNA-a7-q2rev: CAGTGACAGTAGCAGGCATCCT;
MED17-qfor: CCCAGCGGATAGACTTCAGC;
MED17-qrev: ATTGTTCCTCACTGAGTCCCA;
MED18-qfor: GGGCACCATTAACATGATGGA;
MED18-qrev: TGGTCAAGGAAAGTCTCAGGT;
MED1-qfor: CTGGAACGGCTCCATGCAA;
MED1-qrev: CTTCTCCATGACTTGACGCAC;
MED21-qfor: TTGCACGAACAGCAAAAGACA;
MED21-qrev: TGCGCTTTGTATCTTCTCCAG;
MED15-qfor: ATGGACGTTTCCGGGCAAG;
MED15-qrev: GCATCCTCGATTTGACTGACCA;
MED16-qfor: TGCTGGACATGAACACACTG;
MED16-qrev: AGGGAACCCTGGTTGGGTA;
MED23-qfor: GGGCCTTCAGACAGTTTTGG;
MED23-qrev: AGCTGTGTTCTTTCCCACTCA;
MED12-qfor: CTCCCGATGTTTACCCTCAG;
MED12-qrev: ATGCTCATCCCCAGAGACAG;
NIPBL-qfor: CGGCCCGGATTATAGTCTCT;
NIPBL-qrev: TCAGGTGCCAGCTGTCATTA;
CCNT1-qfor: GCCAATCTGCTTCAGGACA;
CCNT1-qrev: GGGAACTGTGTGAAGGACTGA;
CDK9-qfor: ATACGAGAAGCTCGCCAAGA;
CDK9-qrev: CCTTCTCGTTTTCCATCAGC;
CEBPA-qfor: CAGAGGGACCGGAGTTATGA;
CEBPA-qrev: TTCACATTGCACAAGGCACT;
EP300-qfor: GGGAGGACAAACAGGATTGA;
EP300-qrev: CCAATCTGCTGTCCAGGATT;
GTF2B-qfor: TTCCTGCTTTCGGTGTGTCT;
GTF2B-qrev: TGGATGGTTTGGACATGTGA;
SMC1L1-qfor: TCGGACCATTTCAGAGGTTC;
SMC1L1-qrev: GGTCTTTACCCGCAGGTTG;
WDR5-qfor: AATTCAGCCCGAATGGAGAGT;
WDR5-qrev: GGATATTCCCAGCTTGTGACC;
CSTF1-qfor: ACAGAACCAAAGTGGGCTTGA;
CSTF1-qrev: ACACAGACTGAGGCTTGATTTC;
AURKA-qfor: TTCAGGACCTGTTAAGGCTACA
AURKA-qrev: ATTTGAAGGACACAAGACCCG;
Tal1-qfor: CAGCCTAGTGGCTTGTCCTC;
Tal1-qrev: GGAGCCTGAAATTGAATGGA;
Hotair-qfor: GGTAGAAAAAGCAACCACGAAGC;
Hotair-qrev: ACATAAACCTCTGTCTGTGAGTGCC;
Hottip-qfor: CCTAAAGCCACGCTTCTTTG;
Hottip-qrev: TGCAGGCTGGAGATCCTACT;
STIL-qfor: CCCAACGCCAACTGGAGATTT;
STIL-qrev: AGTCGGATGGTCTTCTCAGTC;
SNAI1-qfor: TCGGAAGCCTAACTACAGCGA;
SNAI1-qrev: AGATGAGCATTGGCAGCGAG;
UBE2V1-qfor: AAGCAAGAGCGACGCAAGAT;
UBE2V1-qrev: CGGAAATTGCGAGGGACTT.
Chromatin Immunoprecipitation
Briefly, 1×107 to 3×107 A549 or MCF7 cells were harvested for each independent IPs. After chemically cross-linking, cells were fixed in 1% formaldehyde 1XPBS with 10% Serum for 15 minutes at room temperature. After quenched with 0.25 M Glycine, rinsed twice with cold 1XPBS, cells were lysed for 15 minutes in cold Chip buffer (150mM NaCl, 1% TritonX-100, 5 mM EDTA; 10 mM Tris-HCl, pH 7.5, 0.5mM DTT) with protease inhibitors on ice for 10 min. DNA was sonicated to between 200- and 350-bp DNA fragments on a Diagenode Bioruptor according to manufacturer’s protocol. The solubilized chromatin was cleared and diluted with lysis buffer (without SDS to bring down SDS concentration to 0.1-0.15 %). 1 ml of the chromatin solution was incubated overnight with 2 to 6 μg of antibody at 4°C. After washed 2 times with Mixed Micelle wash buffer (0.2% SDS, 1% Triton X-100, 5.2 % w/v sucrose, 5 mM EDTA, 20 mM Tris-HCl, pH 8, 150 mM NaCl), 2 time with high-salt buffer (1% Triton X-100, 1 mM EDTA, 50 mM HEPES, pH 8, 0.1% w/v deoxycholate, 500 mM NaCl), 2 time with LiCl buffer (250 mM LiCl, 0.5% NP-40, 0.5% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8), and one time with cold TE buffer (10 mM Tris-HCl, pH 7.6, 1 mM EDTA). Bound chromatin was eluted at 65°C for 10 mins in 60 μl fresh elution buffer twice(TE, 1% SDS) and bring it to 200 μl. Cross-linking samples were reversed overnight at 65°C. After proteinase K treatment (0.5mg/ml), DNA was purified using a QIAquick PCR purification kit (Qiagen).ChIP primers sequences are listed here:
AurkaChip-for: CGAATCCTGCCCAATCTACC;
AurkaChip-rev: GATGGCGAGAAAAGCAAGAG;
ncRNAa7Chip-for: GGATAACCAAAAGCGTAGGAAA;
ncRNAa7Chip-rev: CGGTCTCTAACAACAACAGCTC;
CSTF1-Chip-for: TCCATTTTTCCAGGAGAGAGC;
CSTF1-Chip-rev: CCAGTCGTTTCTGTGGTTTTC;
Tal1-Chip-for: TCCTTCCCCCTTTTCCTTAC;
Tal1-Chip-rev: TGAACGCACTCTCACAATCC;
ncRNA-a3-Cfor: ATGAGAGCCTCGGAAGTTGA;
ncRNA-a3-Crev: CCTTGCATTCCACAGGAAGT;
STIL-Chip-for: AATGTTACCCACCAACCTTCC;
STIL-Chip-rev: CTACCCTGCAAACAGACCTCA;
UBE2V1-Chip-for: TGCTTTTGGGGAGAATGAAG;
UBE2V1-Chip-rev: TCCTCGAACAGGAGACTGGT.
Protein affinity Purification and RNA-Immunopercipitation
The affinity purified mediator complex and the GFP protein control were generated from the flag-tagged MED12 or GFP HEK293T stable cell lines.
RNA probes, internally labeled with [α-32P] CTP, were transcribed using T7 or Sp6 RNA polymerase (Riboprobe System, Promega).
RNA immunopercipitations were performed using Dynabeads Protein A (Invitrogen) according to manufacturer’s recommendations. 2 μg antibody was incubated with each sample, the purified protein complex or the control protein together with labeled RNA probes were incubated with the beads at 4 °C. The immunopercipitated RNAs were extracted by TRIzol (Invitrogen) and resolved on the 6% Novex TBE-Urea gels (Invitrogen).
For UV crosslinking experiments, method is modified from Jeannie T. Lee’s paper 28. Transient transfected HEK293T cells have been UV crosslinked at 254 nm (2000 J/m2) in10 ml ice-cold PBS and collected by scraping. Cells were incubated in lysis solution (0.1% SDS, 0.5% NP40, 0.5% sodium deoxycholate, 400U/ml RNase Inhibitor [Roche], and protease inhibitor at 4°C for 25 min with rotation, followed by DNase treatment (30 U of DNase, 15 min at 37°C).
In vitro transcription of ncRNA-as
In vitro transcription followed the protocol provided by the Riboprobe in vitro transcription systems (promega), the designed oligoes are listed here.
ncRNA-a1-T7FOR: TAATACGACTCACTATAGGGgcgcatctcctacggcctccag;
ncRNA-a1-sp6REV: ATTTAGGTGACACTATAGAAccaagtgtcctgtgtaataggc;
ncRNA-a3-T7FOR: TAATACGACTCACTATAGGGgaagttgagcttcaggcgcggctctt;
ncRNA-a3-sp6REV: ATTTAGGTGACACTATAGAAAaccagcctcagcggtgtggaata;
ncRNA-a7-T7FOR: TAATACGACTCACTATAGGGctgtggcagagacggagaa;
ncRNA-a7-SP6REV: ATTTAGGTGACACTATAGAAacagccttagactgtgaaattttattg;
Hotair-T7FOR: TAATACGACTCACTATAGGGAGGCCCCAAAGAGTCTGATGTT;
Hotair-SP6REV: ATTTAGGTGACACTATAGAACTCAGGTTTTTCCAGCGTTCTC.
Kinase assays
Kinase reactions were carried out with 10 to 50 ng of purified Mediator protein complex and 500 ng human Histone H3.1 recombinant (New England Biolabs) substrates in kinase buffer (25mM Tris pH 8.0, 100 mM KCl, 100 μM ATP, 10 mM MgCl2, 2mM DTT) with the addition of 2.5 μCi [γ-32P]ATP at 24 °C. Samples were resolved on a 4-12% SDS-polyacrylamide gel (Invitrogen) followed by transfer to PVDF membrane (Millipore) and autoradiography.
Chromosome Conformation Capture (3C)
3C assay followed the protocol from Hagège et al. 29, with minor modifications. A549 or MCF7 cells were filtered through a 70 μm to obtain a single cell preparation. 1×107 cells were then fixed in 1% formaldehyde for 30 min at room temperature, followed by 15 mins cross-linking. The reaction was quenched with 0.25 M glycine and cells were collected by centrifugation at 240 g at 4°C. The pellet was re-suspended in 0.5 ml cold lysis buffer (10 mM Tris-HCl, pH 7.5; 10 mM NaCl; 5 mM MgCl2; 0.1 mM EGTA) with freshly added protease inhibitors (Roche) and was lysed on ice for 15 mins. Dounce homogenize the cells on ice with pestle B using 15-20 strokes. The nuclei were collected by centrifugation at 500 g for 10 min at 4°C and were re-suspended in 0.5 ml of 1.2× restriction enzyme buffer (NEB), including 0.3% SDS and were incubated for 1 h at 37°C while shaking at 1200 rpm. 2% (final concentration) Triton X-100 was added to the nuclei and then the samples were incubated for 1 hr at 37°C while shaking. 400 U restriction enzyme was added to the nuclei and the samples were incubated at 37°C over night while shaking. 10 μl of the samples were collected before and after the enzyme reaction to evaluate digestion efficiency. The reaction was stopped by addition of 1.6% SDS (final concentration) and incubation at 65°C for 30 minutes while shaking at 1200 rpm. The sample was then diluted 10 fold with 1.15× ligation Buffer (NEB) with added 1% Triton X-100 and was incubated for 1 h at 37°C while shaking at 900 rpm. 100 U T4 DNA ligase (NEB) were added to the sample and the reaction was carried at 16°C for 4 hrs followed by 30 mins at room temperature. 300 μg of Proteinase K were added to the sample and the reaction was carried at 65°C overnight for de-crosslink. RNA was removed by adding 300 μg of RNAse and incubating the sample for 1 h at 37°C. DNA was purified by twice phenol-chloroform extraction and ethanol precipitation. Purified DNA was then analyzed by conventional or quantitative PCR. As control for ligation products the bac-clones were digested with 10 U of restriction enzyme overnight and then incubated with 10 U T4 DNA-ligase at 16°C overnight. The DNA was extracted by phenol-chloroform and precipitated with ethanol. Purified DNA was then analyzed by conventional or quantitative PCR. For Real time PCR, the ΔCt method was applied for analyzing data, using the bac-clone Ct values as control. Ct values were normalized for each primer pair by setting the Ct value of 100 ng of bac-clone control random ligation matrix DNA at a value of 1. Primer sequences for PCR are listed here:
Backg-Control-Aurka-for: CGTATGAGCTGGCAATAGCA;
Backg-Control-Aurka-rev: GGAGGCATGAGAGCTTGTTC;
Backg-Control-A7-for: ACTGGCTGAGGTTTGTGGAG;
Backg-Control-A7-rev: GGCCGATTTGAGAGAGTGAG;
Backg-Control-Snai1-for: CAAGAGGGGAAGGAGAGGAG;
Backg-Control-Snai1-rev: TCCTAAGTCCCCAGTCTCCA;
ncRNA-a7 anchor primer: CTAGGTCAGGCCCAAAGAGA;
ncRNA-a7 anchor control: ACAGAATAACCCATCCCTTTTCC;
A7-anchorDigTest-for: GCTTAACCCAGTGCAAGCTC;
A7-anchorDigTest-rev: AGTTTGGAGGCTGGGAATCT;
Aurka-N5DigTest-for: CCTGATTCTCACCCTCTTGG;
Aurka-N5DigTest-rev: GAGGCCCCATTCCTTAACAT;
Snai1-N2DigTest-for: GGGAAAATGAGAGCAAACTCC;
Snai1-N2DigTest-rev: AAGGCCAATGTGACAGAGATG;
A3-DigEcoRI-for: CACTTACAGGAACAGAGGACTCA;
A3-DigEcoRI-rev: CAACTTCTAGATCAGCGCCTTC;
3C Aurka-BglII-A: GGGACATAAATGGGAGACTGG;
3C Aurka-BglII-B: TGGGAGGAACTCTTCACCTCT;
3C Aurka-BglII-C: TGGCTCTTTGGTCTTGTCTCT;
3C Aurka-BglII-D: GTTTGGGCCGTTTCTTATTG;
3C Aurka-BglII-E: CAGACCTGATTCTCACCCTCTTG;
3C Aurka-BglII-F: CACCAACTTTTCTTCTCCACCAA;
3C Aurka-BglII-G: CCACAGGCACTACTGTTACCAA;
3C BglII.E-R1: CCAGGGAGGACACAGCATAA;
3C BglII.E-R2: TGGTGTCTGGGTTGAAGTAACA;
3C BglII.E-R3: TCGGCACCTGGTAAACTAGC;
ncRNA-a3 anchor primer: CAACTTCTAGATCAGCGCCTTC;
ncRNA-a3 anchor control: GTCTCTTTCTTTGATTGGAGTGG;
Tal1-1: GGGAATGGCAGAAGACAGAA;
Tal1-2: GTGTCGGGTTTGTTTTTACCTC;
Tal1-3: CTCCATTTTTGCCTCTGTCTCT;
Tal1-4: ATCTTTTCCGTGTAGCTCTTCCT;
Tal1-5: TGGGTTTGATTGCTCAGTACC;
Tal1-6: TCAATTTTGCCTGCCATACA;
Tal1-7: TTAAAAGGGCAGGGAGTTAAAAG;
Tal1-8: CAGATAGCCAGGAAGCAAGG;
Snai1-1: GCTTTGGTCCTGAACTCCTG;
Snai1-2: AACAAACTCACATTAATAAAATATCCAGA;
Snai1-3: CTACAGTCGTGTGCCACCAA;
Snai1-4: ATTGGCAAGAAGGGGAAAAT;
Snai1-5: TCCAACATCCCAGACCTTTC;
Snai1-6: GACCATGAGAAAGCCAGAGG;
Snai1-7: AAGCTCCAGTGGCATGTGAT;
Snai1-8: TGGAGTCCTGGTGTGTTCTG.
Bac clone ID: RP-11-664B18, RP-11-1067D15, RP11-742I1 (Empire Genomics)
Supplementary Material
Acknowledgments
We thank RS laboratory members for support and discussions. We thank Wulan Dang (Blobel lab) and Italo Tempera for suggestions and technical help on 3C experiments. RS was partially supported by cancer core grant from NIH (P30 CA 010815). DJT was supported by a grant from the NCI (R01 CA127364). GAB was supported by 5R37DK058044.
Footnotes
Author Contribution R Shiekhattar, FL and UAO conceived and designed the overall project with help from GAB., MB., DJT., MC. FL and UAO performed the RNAi screens, while FL received advice from GAB to execute the 3C experiments. FL and MC performed the ChIP experiments for Mediator. FL performed all experiments regarding the ncRNA association with Mediator and the kinase assays. FL, MC and RS analyzed the data and wrote the paper.
The authors declare no competing financial interests.
References
- 1.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 3.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 4.Okazaki Y, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 5.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 8.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JT. The X as model for RNA’s niche in epigenomic regulation. Cold Spring Harbor perspectives in biology. 2010;2:a003749. doi: 10.1101/cshperspect.a003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Molecular cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Molecular cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS biology. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Risheg H, et al. A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat Genet. 2007;39:451–453. doi: 10.1038/ng1992. [DOI] [PubMed] [Google Scholar]
- 18.Rump P, et al. A novel mutation in MED12 causes FG syndrome (Opitz-Kaveggia syndrome) Clinical genetics. 2011;79:183–188. doi: 10.1111/j.1399-0004.2010.01449.x. [DOI] [PubMed] [Google Scholar]
- 19.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nature reviews Genetics. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends in biochemical sciences. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Molecular and cellular biology. 2009;29:650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends in genetics : TIG. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sopher BL, et al. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron. 2011;70:1071–1084. doi: 10.1016/j.neuron.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cabianca DS, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149:819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagege H, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nature protocols. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


