Abstract
The increasing population of Aedes aegypti mosquitoes on Madeira Island (Portugal) resulted in the first autochthonous dengue outbreak, which occurred in October 2012. Our study establishes the first genetic evaluation based on the mitochondrial DNA (mtDNA) genes [cytochrome oxidase subunit I (COI) and NADH dehydrogenase subunit 4 (ND4)] and knockdown resistance ( kdr ) mutations exploring the colonisation history and the genetic diversity of this insular vector population. We included mosquito populations from Brazil and Venezuela in the analysis as putative geographic sources. The Ae. aegypti population from Madeira showed extremely low mtDNA genetic variability, with a single haplotype for COI and ND4. We also detected the presence of two important kdr mutations and the quasi-fixation of one of these mutations (F1534C). These results are consistent with a unique recent founder event that occurred on the island of Ae. aegypti mosquitoes that carry kdr mutations associated with insecticide resistance. Finally, we also report the presence of the F1534C kdr mutation in the Brazil and Venezuela populations. To our knowledge, this is the first time this mutation has been found in South American Ae. aegypti mosquitoes. Given the present risk of Ae. aegypti re-invading continental Europe from Madeira and the recent dengue outbreaks on the island, this information is important to plan surveillance and control measures.
Keywords: Aedes aegypti, dengue, knockdown resistance, mtDNA
Invasive mosquito species are a serious threat to the environment and human health. There are several well-known examples of the introduction of non-native mosquito species and the risk they represent. The establishment of Culex quinquefasciatus in Hawaii has endangered the bird fauna of the archipelago ( Fonseca et al. 2000 ). The invasion of the African malaria vector Anopheles gambiae s.l. into Brazil in 1930 was followed by severe malaria epidemics during that decade ( Soper & Wilson 1943 ).
One of the most invasive mosquito species is Aedes aegypti Linnaeus, 1762, which is a major vector of the dengue, urban yellow fever and chikungunya viruses. Originally from Africa, this mosquito has had a very successful expansion history. The early establishment of trade routes between Europe, Africa and the Americas in the XV century and the expansion of the slave trade decisively contributed to the first wave of geographic expansion of this species. It is believed to have invaded the Americas and Europe during the XV-XVII centuries ( Tabachnick 1991 , Lounibos 2002 ). Presumably, Ae. aegypti reached the Asian continent only in the late XIX century ( Smith 1956 ). The evolution of domestic traits in a formerly sylvatic mosquito was critical for its adaptation to artificial water containers ( Tabachnick 1991 ). Ae. aegypti is now well adapted to the domestic environment and its close association with humans allows for long distance dispersal events due to their desiccation-resistant eggs and human host availability.
In the XX century, a vast vector control program resulted in the elimination of Ae. aegypti from most of Central and South America ( Gubler 1989 ). This program was interrupted in the early 1970s and the mosquitoes then began to reinvade areas from which they had been previously eliminated ( Gubler 1989 ). By the 1990s, Ae. aegypti had nearly regained the geographic distribution that it held before elimination was initiated [see Gubler (1998) and references therein]. This recent re-expansion of Ae. aegypti , combined with an intensive urban growth and an increased global movement of people and merchandise, has triggered a dramatic rise in the incidence of dengue over the past 40 years ( Gubler 2011 ).
The most recent example of an epidemic driven by mosquito invasion is the dengue outbreak on Madeira Island (Portugal) that occurred in October 2012, with 1,891 cases (966 laboratory-confirmed) detected at the end of November 2012 ( Sousa et al. 2012 ). This was the first dengue epidemic in Europe since the 1920s and it occurred in a territory where Ae. aegypti was introduced less than a decade ago ( Almeida et al. 2007 ). The mosquito was first identified on Madeira Island in 2005 ( Margarita et al. 2006 ), with no previous records between 1977-1979 ( Capela 1981 ). From then on, a vector control program was implemented to reduce the mosquito’s breeding sites, educate the community and eliminate the pest through the application of insecticides. Specifically, between 2005-2008, pyrethroids (tetramethrin, d-phenothrin, alpha-cypermethrin and Pybutrin®33, containing pyrethrins synergised with piperonyl butoxide) and organophosphate malathion were used against the adult mosquitoes with aerial spraying applications (M Melim, unpublished observations).
However, Ae. aegypti continued to expand throughout the island ( Gonçalves et al. 2008 ). Despite preliminary surveys suggesting a high degree of insecticide resistance ( Seixas 2012 , ECDC 2013 ), no information was yet available on the possible mechanisms underlying the resistance in the local Ae. aegypti population. Emergence of insecticide resistance is a major challenge to vector control. Knockdown resistance ( kdr ) results from mutations in the voltage-gated sodium channel of insects, which is the molecular target of pyrethroids and dichloro-diphenyl-trichloroethane (DDT) ( Soderlund & Knipple 2003 ). Several kdr mutations, such as I1011V, I1011M, V1016G, V1016I and F1534C, have been reported in Ae. aegypti worldwide ( Vontas et al. 2012 )].
Evolutionary and population genetic studies support African Ae. aegypti as the ancestral form of the species and populations outside of Africa belong to one of two major lineages that are associated with West or East Africa ( Brown et al. 2011 , Moore et al. 2013 ). Genetic studies report varying levels of population differentiation at both the local and continental scales, which is a consequence of the major demographic perturbations in Ae. aegypti ’s recent population history ( Urdaneta-Marquez & Failloux 2011 ). These analyses have also been applied to insular populations of Ae. aegypti . In the South West islands of the Indian Ocean, high levels of genetic diversity have been reported, mainly due to historical multiple origins and founder events ( Delatte et al. 2011 ). In the South Pacific archipelagos, the genetic structure of the Ae. aegypti populations was dependent on human population density and recurrent insecticide-based control, which induced repeated population bottlenecks ( Urdaneta-Marquez & Failloux 2011 ). To date, there is no information about the geographic origin of the Ae. aegypti population on Madeira Island. Madeira has strong social and commercial relations with Brazil and Venezuela, mainly due to the presence of large emigrant communities of Madeira descendants in these countries. The intensive movement of people and goods between the island and these dengue-endemic countries makes them potential sources for the introduction of both Ae. aegypti and dengue virus (DENV) into Madeira.
In this study, we conducted a genetic analysis based on the mitochondrial DNA (mtDNA) genes [cytochrome oxidase subunit I (COI) and NADH dehydrogenase subunit 4 (ND4)] and kdr mutations to explore the colonisation history and current population diversity of the Ae. aegypti population of Madeira island. We also included samples from Brazil and Venezuela in the analysis as putative geographic sources. The implications of these results for the control of this invasive species are discussed.
MATERIALS AND METHODS
Mosquito samples and DNA extraction - Samples of Ae. aegypti were collected on Madeira Island and from two populations of Brazil and Venezuela ( Figure , Table I ). Larvae and eggs were collected and reared to adults in an insectary under standard conditions. Adults were kept in dry, individual tubes with silica gel until molecular analyses.
Fig. 1. : map showing the geographic collection sites.
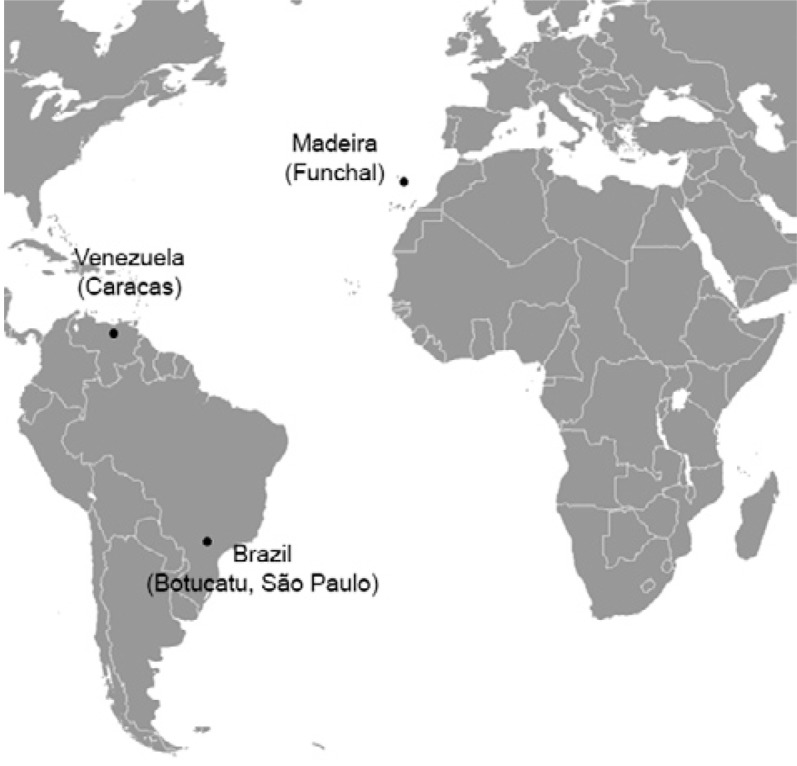
TABLE I. Collection data of the sampled Aedes aegypti populations.
| Localities, country | Locality | Coordinates | Year of collection | Mosquitoes analysed (n) |
|---|---|---|---|---|
| Madeira Island, Portugal | Funchal | 32º40’N 16º55’W | 2009 | 69 |
| São Paulo, Brazil | Botucatu | 22º53’S 48º27’W | 2010 | 32 |
| Caracas, Venezuela | Caracas | 10º29’N 66º54’W | 2013 | 30 |
DNA from the Madeira Island mosquitoes was extracted using a phenol-chloroform protocol modified by Donnelly et al. (1999) . DNA of the individuals from Brazil and Venezuela was extracted using Chelex, as described by Campos et al. (2012) .
Kdr genotyping - Kdr mutations were surveyed in Ae. aegypti from Madeira Island, Brazil and Venezuela. Allele-specific polymerase chain reaction (PCR) was carried out to detect the V1016I mutation in the DDT/pyrethroid insecticide target site of the voltage-gated sodium channel of Ae. aegypti . Primers and PCR conditions are described in Saavedra-Rodriguez et al. (2007) . For the detection of the F1534C mutation, a tetra primer assay, as designed by Harris et al. (2010) , was used. Fragments were visualised in 2% agarose gels after electrophoresis. A subset of samples was selected for sequencing and genotype confirmation. Concerning the F1534C mutation, PCR were carried out using the AaEx31P and AaEx31Q primers described in Harris et al. (2010) , with the same cycling conditions of the tetra primer assay. For the V1016I mutation, the kdr 20f and kdr 21r PCR primers were described in Saavedra-Rodriguez et al. (2007) , with the following cycling conditions: initial denaturation step at 95ºC for 5 min, followed by 33 cycles of 95ºC for 1 min, 56ºC for 1 min, 72ºC for 1 min and a final extension of 72ºC for 10 min. The PCR products were purified with the QIAquick® PCR Purification Kit (Qiagen, Hilden, Germany) and subjected to direct sequencing at STAB VIDA (Portugal). The obtained sequences were aligned and edited using BioEdit v.7.0.9. For each mutation, Hardy-Weinberg equilibrium was tested using the exact probability test in the GenePop v.1.2 software ( Raymond & Rousset 1995 ).
mtDNA analysis - The mitochondrial ND4 gene was amplified using the method described by Paduan and Ribolla (2008), with slight modifications, using the primers ND4f (5’-TGATTGCCTAAGGCTCATGT-3’) and ND4r (5’-TTCGGCTTCCTAGTCGTTCAT-3’). The reaction was performed in a 25 µL volume containing 5 µL of 1x buffer, 2 mM MgCl2, 0.4 mM deoxynucleotide triphosphate (dNTP), 0.2 µM of each primer, 1.5 U of Taq polymerase and 1 µL of DNA template. The PCR cycling conditions included an initial denaturation step at 94ºC for 2 min, followed by 35 cycles of 94ºC for 30 s, 50ºC for 30 s, 72ºC for 1 min and a final extension of 72ºC for 5 min.
Concerning the COI gene, two different fragments were amplified. Initially, a protocol from Paduan and Ribolla (2008) was followed with few modifications. The Fly5IP (5’-GGATTATTAGGATTTATTGT-3’) and Fly10IP (5’-GCCAAATAATGAAATTGTTCT-3’) primers were used. PCR was performed in a 25 µL volume containing 5 µL of 1x buffer, 2 mM MgCl2, 0.4 mM dNTP, 1 µM of each primer, 1.5 U of Taq polymerase and 1 µL of DNA template diluted 1:10. As the COI fragment that was produced was too small to make good genetic comparisons with the GenBank sequences, we used another COI gene amplification protocol to obtain a larger fragment. Thus, we followed the protocol of Paupy et al. (2012), with some modifications. The COI-FOR (5’-TGTAATTGTAACAGCTCATGCA-3’) and COI-REV (5’-AATGATCATAGAAGGGCTGGAC-3’) primers were used. PCR was performed in a 50 µL reaction volume containing 10 µL of 1x buffer, 1.5 mM MgCl2, 0.2 mM dNTP, 0.3 µM of each primer, 1 U of Taq polymerase and 2 µL of DNA template. The PCR cycling conditions included an initial denaturation step at 94ºC for 2 min, followed by 35 cycles of 94ºC for 30 sec, 54ºC for 30 sec, 72ºC for 1 min and a final extension of 72ºC for 5 min. The PCR products were purified using the QIAquick® PCR Purification Kit (Qiagen, Hilden, Germany) and used for direct sequencing at STAB VIDA (Portugal). The obtained sequences were aligned and edited using the BioEdit v.7.0.9 software. These sequences were compared with the reference Ae. aegypti gene sequences deposited in the GenBank.
RESULTS
Kdr genotyping - Genotyping and sequencing of the voltage-gated sodium channel revealed the presence of two kdr mutations. One mutation was located at position 1016 (GTA to ATA) and resulted in the replacement of a valine with an isoleucine (V1016I); this mutation occurred at a low to moderate allelic frequency in the populations studied, ranging from 8% in Madeira to 23% in Venezuela ( Table II ). The second mutation was located at position 1534 (TTC to TGC) and resulted in the replacement of a phenylalanine with a cysteine (F1534C); this mutation occurred at high allelic frequencies in the Madeira (98%) and Venezuela (100%) populations and at a moderate frequency in the Brazil population (23%) ( Table III ). All of the populations were in Hardy-Weinberg equilibrium (exact test, p > 0.05) for the two kdr mutations.
TABLE II. Frequency of the V1016I knockdown resistance mutation in the populations of Madeira, Brazil and Venezuela analysed in this study.
| Population | Mosquitoes analysed (n) | V/V a (n) | V/I b (n) | I/I c (n) | I d (%) |
|---|---|---|---|---|---|
| Madeira | 69 | 59 | 9 | 1 | 0.08 |
| Brazil | 32 | 27 | 4 | 1 | 0.09 |
| Venezuela | 30 | 16 | 14 | 0 | 0.23 |
a : number of individuals homozygous for the susceptible-associated allele; b : number of heterozygous individuals; c : number of individuals homozygous for the resistant-associated allele; d : allele frequency for the resistant-associated allele. Of the total 131 individuals genotyped, 35 were confirmed by sequencing. I: isoleucine; V: valine.
TABLE III. Frequency of the F1534C knockdown resistance mutation in the populations of Madeira, Brazil and Venezuela analysed in this study.
| Population | Mosquitoes analysed (n) | F/F a (n) | F/C b (n) | C/C c (n) | C d (%) |
|---|---|---|---|---|---|
| Madeira | 65 | 0 | 2 | 63 | 0.98 |
| Brazil | 30 | 17 | 12 | 1 | 0.23 |
| Venezuela | 30 | 0 | 0 | 30 | 1 |
a : number of homozygous susceptible individuals; b : number of heterozygous individuals; c : number of homozygous resistant individuals; d : resistance allele frequency in the population. Of the total 125 individuals genotyped, four were confirmed by sequencing. C: cysteine; F: phenylalanine.
mtDNA analysis - Sequences of the COI gene were retrieved from 15 specimens from Funchal, Madeira Island (GenBank accession KF909122). The alignment of 420 bp ( Paduan & Ribolla 2008 ) and 764 bp ( Paupy et al. 2012 ) in individuals revealed the presence of a single haplotype. The COI haplotype found in the population of Ae. aegypti from Madeira matched several sequences, as shown in Table IV . The polymorphic sites of this haplotype (HCOI-Mad) are presented in Supplementary data with references to the published Ae. aegypti (Red eyes strain) sequence of COI (GenBank accession AF390098).
TABLE IV. Geographical distribution of the cytochrome oxidase subunit I (COI) haplotype of Aedes aegypti found in Madeira Island.
| Haplotype | Continent | Countries (localities) | References | Base pairs a | GenBank accessions b |
|---|---|---|---|---|---|
| COI-HMad as in Paduan and Ribolla (2008) | Europe | Portugal (Madeira Island) | Present paper | 420 | - |
| America | Brazil (Porto Velho) | Paduan and Ribolla (2008) | 420 | AY851650 | |
| Puerto Rico | Cook et al. (2006) | 420 | DQ181441 | ||
| Africa | West Africa | Cook et al. (2005) | 420 | AY645261 | |
| West Africa | Cook et al. (2005) | 420 | AY645262 | ||
| Asia | Indonesia, West Timor | Sota and Mogi (2006) | 420 | DQ397892 | |
| COI-HMad as in Paupy et al. (2012) | Europe | Portugal (Madeira Island) | Present paper | 763 | - |
| America | Venezuela, Maracay | Paupy et al. (2012) | 763 | JQ926701 | |
| USA, Florida | Paupy et al. (2012) | 763 | JQ926684 | ||
| Brazil, Foz do Iguaçu | Mousson et al. (2005) | 511 | AJ970965 | ||
| Brazil, Rio Branco | Mousson et al. (2005) | 491 | AJ970975 | ||
| Asia | Vietnam, Hai Phong | Paupy et al. (2012) | 763 | JQ926685 | |
| Thailand, Chiang Mai | Paupy et al. (2012) | 763 | JQ926691 | ||
| Cambodia | Fort et al. (2012) | 763 | HQ688294 | ||
| Vietnam, Ho Chi Minh City | Mousson et al. (2005) | 534 | AJ970968 | ||
| Vietnam, Hanoi | Mousson et al. (2005) | 504 | AJ970967 | ||
| Oceania | French Polynesia, Paea | Mousson et al. (2005) | 534 | AJ970972 | |
| Africa | Madagascar, Toliary | Delatte et al. (2011) | 416 | HQ693081 |
a : number of base pairs matching the COI sequence of Madeira Island; b : list of GenBank accessions, the corresponding references and places of origin for COI sequences matching the haplotypes of Ae. aegypti found in Madeira island.
Mitochondrial DNA [cytochrome oxidase subunit I (COI) and NADH dehydrogenase subunit 4 (ND4)] haplotypes recorded in Aedes aegypti population from Madeira Island.
| ND4 a | 6 | 6 | 6 | 6 | 6 | 6 | 7 | 7 | 7 | 8 | 8 | 8 | 8 | 8 | 9 | 9 | 9 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 3 | 4 | 9 | 5 | 6 | 7 | 1 | 3 | 5 | 8 | 9 | 3 | 7 | 8 | |
| 3 | 5 | 8 | 6 | 2 | 3 | 3 | 2 | 1 | 0 | 7 | 5 | 8 | 1 | 3 | 6 | 4 | |
| Reference sequence | C | A | A | C | C | C | T | C | G | C | G | C | G | A | T | A | A |
| HND4-Mad | T | G | T | T | T | T | C | T | A | T | A | T | A | T | C | C | C |
|
| |||||||||||||||||
| COI a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| 2 | 2 | 2 | 2 | 4 | 5 | 5 | 5 | 6 | 6 | 6 | 6 | 7 | 1 | 1 | 2 | 2 | |
| 0 | 7 | 7 | 8 | 9 | 0 | 1 | 4 | 1 | 6 | 7 | 9 | 8 | 1 | 3 | 7 | 9 | |
| 7 | 3 | 6 | 5 | 8 | 7 | 9 | 0 | 8 | 9 | 5 | 9 | 0 | 0 | 1 | 5 | 0 | |
|
| |||||||||||||||||
| Reference sequence | G | T | C | G | G | G | G | C | A | G | C | T | C | C | C | T | G |
| COI as Paupy et al. (2012) | A | C | T | A | A | A | A | T | G | A | T | C | T | - | - | - | - |
| COI as Paduan and Ribolla (2008) | - | - | - | - | - | - | - | - | - | - | - | - | - | T | T | C | A |
Sequences of the ND4 gene were retrieved from 15 specimens (GenBank accession KF909123). The alignment of 376 bp revealed only one haplotype. The ND4 haplotype matched several sequences previously reported for samples from the Americas ( Table V ). The polymorphic sites of this haplotype (HND4-Mad) are presented in Supplementary data with references to the published Ae. aegypti complete sequence of the ND4 gene (GenBank accession DQ440274).
TABLE V. Geographical distribution of the NADH dehydrogenase subunit 4 (ND4) haplotype of Aedes aegypti found in Madeira island.
| Haplotype | Continent | Countries (localities) | References | Base pairs a | GenBank accessions b |
|---|---|---|---|---|---|
| HND4-Mad | Europe | Portugal (Madeira island) | Present paper | 376 | - |
| America | Perú (Lima) | da Costa-da-Silva et al. (2005) | 360 | DQ177153 | |
| Brazil (Belém, Santos, São Sebastião) | Bracco et al. (2007) | 335 | DQ176842 | ||
| Brazil (Compensa) | Lima and Scarpassa (2009) | 376 | EU650409 | ||
| Brazil (Porto Velho) | Paduan and Ribolla (2008) | 376 | AY906851 | ||
| Brazil, Represa do Cigano | Paupy et al. (2012) | 334 | JQ926726 | ||
| Venezuela (Maracay) | Bracco et al. (2007) | 335 | DQ176842 | ||
| Venezuela | Urdaneta-Marquez et al. (2008) c | 376 | Not available | ||
| Guatemala (Guatemala City) | Bracco et al. (2007) | 335 | DQ176842 | ||
| Mexico | Gorrochotegui-Escalante et al. (2002) | 376 | AF334843 | ||
| USA (Fort Lauderdale) | Bracco et al. (2007) | 335 | DQ176842 | ||
| USA, Florida | Paupy et al. (2012) | 334 | JQ926725 |
a : number of base pairs matching the ND4 sequence of Madeira Island; b : list of GenBank accessions, the corresponding references and places of origin for ND4 sequences matching the haplotypes of Ae. aegypti found in Madeira island; c : sequences supplied by W Black IV.
Several works have reported the presence of NUMTs (nuclear mtDNA) in the Ae. aegypti genome ( Black IV & Bernhardt 2009 , Hlaing et al. 2009 , Behura et al. 2011 ). In our study, no heterozygous sites were detected in any sequences and the observed COI and ND4 haplotypes matched sequences that were previously reported worldwide by several authors and deposited in GenBank (see Tables IV , V for references).
DISCUSSION
This is the first study on the genetic variation of Ae. aegypti in Madeira and, to our knowledge, the first report of the F1534C kdr mutation in the wild mosquito populations of Brazil and Venezuela. Our results showed a very low level of mtDNA diversity, with only one haplotype for COI and ND4, as well as the presence of two important kdr mutations and the quasi-fixation of the F1534C mutation.
Origin of Ae. aegypti from Madeira - The COI haplotype found in the Madeira population was also detected in America, Africa and Asia, while the ND4 haplotype from Madeira was only reported with high frequencies in the American continent ( Tables IV , V ). Overall, these mtDNA haplotypes belong to the West African lineage of Ae. aegypti , as described by Bracco et al. (2007) and Moore et al. (2013) . There are few studies using mtDNA in Ae. aegypti from islands ( Beebe et al. 2005 , Delatte et al. 2011 ). In spite of a general high level of COI diversity in the southwest islands of the Indian Ocean that is associated with old and repeated introductions of Ae. aegypti , a unique haplotype was reported in Reunion Island. The Ae. aegypti population was suggested to be declining on this island due to competition with the recent invader Aedes albopictus ( Delatte et al. 2011 ). Additionally, in mainland Australia, only two COI haplotypes have been identified in a bottlenecked mosquito population restricted to the northern territories, where several dengue outbreaks have occurred since the 1980s ( Beebe et al. 2005 ). The same study analysed a sample from Thursday Island, which is located ca. 39 km off mainland Australia; a distinct COI haplotype was found in this Ae. aegypti population, which exhibited an enhanced vector competence compared with the mainland populations ( Beebe et al. 2005 ). This study provided evidence supporting the hypothesis that island populations may develop unique traits, even with low genetic diversity. The geographic origins of Ae. aegypti populations have epidemiological importance, as several studies have related the origin with traits including vector competence and insecticide resistance ( Failloux et al. 2002 , Lourenço-de-Oliveira et al. 2004 ).
Regarding the kdr mutations associated with insecticide resistance, we observed a very low frequency of the V1016I mutation in Madeira. In samples from Botucatu (Brazil) and Caracas (Venezuela), the frequency of this mutation was low (9%) and moderate (23%), respectively. In Brazil, this mutation has been reported throughout the country, with varying frequencies ranging from 5% in the state of Roraima in the north to 73% in the state of Paraná in the south and this mutation is mainly associated with intensive vector control ( Martins et al. 2009 ). Specifically in the state of São Paulo, Batista (2012) described a significant rise in the frequency of the V1016I mutation from 2001-2011, resulting in a heterogeneous distribution within the state. In the coastal city of Santos, she reported a frequency of 83% for the V1016I mutation, while in the inland city of Bauru, the frequency was limited to 47%. In Venezuela, the frequency of V1016I ranged from 0-14% ( Saavedra-Rodriguez et al. 2007 ). This mutation has also been reported in Cuba, Costa Rica, Panama, Nicaragua ( Saavedra-Rodriguez et al. 2007 ), Mexico ( Saavedra-Rodriguez et al. 2007 , García et al. 2009 ), La Martinique ( Marcombe et al. 2012 ) and the Cayman Islands ( Harris et al. 2010 ), with equal or higher V1016I frequencies than in Madeira.
On the island, a different scenario was observed for the F1534C kdr mutation, with almost complete fixation of the mutant allele. The same result was observed in the sample from Venezuela, with 100% of the mosquitoes homozygous for this mutation. In contrast, the sample from Brazil showed a moderate frequency (23%) of this mutation. To date, there have been a few studies of F1534C in Ae. aegypti populations and these studies have reported moderate to high frequencies of this mutation: 21% in Vietnam, 77% in Thailand and 68% in Cayman Islands ( Kawada et al. 2009 , Harris et al. 2010 , Yanola et al. 2011 ). Moreover, the same mutation was recently described in another invasive mosquito vector, Ae. albopictus , which also demonstrated a high frequency (73%) ( Kasai et al. 2011 ). Thus, more studies are needed to assess the global distribution of the F1534C kdr mutation.
Considering the worldwide distribution of the mtDNA Madeira haplotypes and the frequencies of the kdr mutations recorded on the island, the countries that simultaneously exhibited the same mtDNA haplotypes and comparable frequencies for the two kdr mutations are Brazil and Venezuela. Presently, there are frequent connections between Madeira, Venezuela and Brazil, mainly due to the human migrant population. Approximately 2,800 Brazilians were estimated to live on the island in 2010 ( Chaves-Scarelli 2010 ). Additionally, more than 200,000 people from Madeira live in Venezuela. This is particularly relevant considering that the island population does not exceed 250,000 inhabitants ( OEV 2012 ). Moreover, the DENV serotype 1, which was identified in the recent dengue outbreak on Madeira Island, was associated with viruses circulating in Colombia, Venezuela and northern Brazil ( Alves et al. 2013 ). Based on our genetic data and the present human migrant flows to and from Madeira, the most likely source populations of the Ae. aegypti mosquitoes in Madeira were located in South America, particularly in Brazil and Venezuela. These countries are the most endemic countries for dengue in South America ( WHO 2012 ).
The presence of a single haplotype for both of the sequenced mtDNA loci (COI and ND4) provided strong evidence for a unique recent founder event during the colonisation of Madeira Island by Ae. aegypti mosquitoes harbouring both kdr mutations. Although we cannot fully rule out the possibility of two independent introductions, each harbouring a different kdr mutation, this hypothesis is unlikely given the mtDNA monomorphic haplotype composition. Furthermore, the two kdr mutations may also be present in a single VGSC haplotype. Additional genetic analysis involving more polymorphic markers is required to further refine the geographic origin, colonisation patterns and local genetic structure of this expanding island population.
Implications for vector control - The current study revealed the presence of two kdr mutations in the dengue vector Ae. aegypti from Madeira Island and these mutations were likely previously present in the invading mosquitoes. These mutations may have affected the vector control programs initiated in 2005 and the application of insecticides has been shown to be ineffective ( Seixas 2012 , ECDC 2013 ), leading to the continuous expansion of the mosquito population throughout the southern part of the island and the recent dengue outbreak in 2012. In Brazil, where pyrethroids were introduced for dengue control in 2001 ( Braga & Valle 2007 ), pyrethroid resistance was soon detected ( da-Cunha et al. 2005 ), followed by the subsequent spread of the V1016I kdr mutation throughout most of the country in less than five years ( Martins et al. 2009 , Belinato et al. 2012 ). Nonetheless, the use of insecticides remains the main method for controlling and reducing mosquito populations in emergencies, such as outbreaks. In Madeira, although insecticidal application for vector control was suspended between 2008 and the recent dengue outbreak, the selective pressure for insecticide resistance has been maintained through the use of household insecticides (IASAÚDE 2012). Therefore, the spread of the recently introduced insecticide-resistant Ae. aegypti population is very likely, which could compromise future vector control measures. We recommend the continuous monitoring of Madeira’s Ae. aegypti population, as well as further studies involving samples from future years, to assess the local evolution of the kdr mutations and eventual introduction of the mosquitoes from other geographic regions to the island.
Supplementary data
Mitochondrial DNA [cytochrome oxidase subunit I (COI) and NADH dehydrogenase subunit 4 (ND4)] haplotypes recorded in Aedes aegypti population from Madeira Island.
| ND4 a | 6 | 6 | 6 | 6 | 6 | 6 | 7 | 7 | 7 | 8 | 8 | 8 | 8 | 8 | 9 | 9 | 9 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 3 | 4 | 9 | 5 | 6 | 7 | 1 | 3 | 5 | 8 | 9 | 3 | 7 | 8 | |
| 3 | 5 | 8 | 6 | 2 | 3 | 3 | 2 | 1 | 0 | 7 | 5 | 8 | 1 | 3 | 6 | 4 | |
| Reference sequence | C | A | A | C | C | C | T | C | G | C | G | C | G | A | T | A | A |
| HND4-Mad | T | G | T | T | T | T | C | T | A | T | A | T | A | T | C | C | C |
|
| |||||||||||||||||
| COI a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| 2 | 2 | 2 | 2 | 4 | 5 | 5 | 5 | 6 | 6 | 6 | 6 | 7 | 1 | 1 | 2 | 2 | |
| 0 | 7 | 7 | 8 | 9 | 0 | 1 | 4 | 1 | 6 | 7 | 9 | 8 | 1 | 3 | 7 | 9 | |
| 7 | 3 | 6 | 5 | 8 | 7 | 9 | 0 | 8 | 9 | 5 | 9 | 0 | 0 | 1 | 5 | 0 | |
|
| |||||||||||||||||
| Reference sequence | G | T | C | G | G | G | G | C | A | G | C | T | C | C | C | T | G |
| COI as Paupy et al. (2012) | A | C | T | A | A | A | A | T | G | A | T | C | T | - | - | - | - |
| COI as Paduan and Ribolla (2008) | - | - | - | - | - | - | - | - | - | - | - | - | - | T | T | C | A |
ACKNOWLEDGEMENTS
To William Black IV, for providing the published sequences of the Ae. aegypti mtDNA ND4 gene from Venezuela.
REFERENCES
- Almeida AP, Gonçalves YM, Novo MT, Sousa CA, Melim M, Grácio AJ. Vector monitoring of Aedes aegypti in the autonomous region of Madeira, Portugal. 3311Euro Surveill. 2007;12 doi: 10.2807/esw.12.46.03311-en. [DOI] [PubMed] [Google Scholar]
- Alves MJ, Fernandes PL, Amaro F, Osório H, Luz T, Parreira P, Andrade G, Zé-Zé L, Zeller H. Clinical presentation and laboratory findings for the first autochthonous cases of dengue fever in Madeira Island, Portugal, October 2012. Euro Surveill. 2013;18:3–6. [PubMed] [Google Scholar]
- Batista E. Evolução de mutações no gene do canal de sódio associadas à resistência tipo kdr em populações de Aedes (Stegomyia) aegypti do estado de São Paulo. Faculdade de Saúde Pública/Universidade de São Paulo; São Paulo: 2012. 92 MsD Thesis. [Google Scholar]
- Beebe NW, Whelan PI, van den Hurk A, Ritchie S, Cooper RD. Genetic diversity of the dengue vector Aedes aegypti in Australia and implications for future surveillance and mainland incursion monitoring. Commun Dis Intell Q Rep. 2005;29:299–304. [PubMed] [Google Scholar]
- Behura SK, Lobo NF, Haas B, de Bruyn B, Lovin DD, Shumway MF, Puiu D, Romero-Severson J, Nene V, Severson DW. Complete sequences of mitochondria genomes of Aedes aegypti and Culex quinquefasciatus and comparative analysis of mitochondrial DNA fragments inserted in the nuclear genomes. Insect Biochem Mol Biol. 2011;41:770–777. doi: 10.1016/j.ibmb.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinato TA, Martins AJ, Valle D. Fitness evaluation of two Brazilian Aedes aegypti field populations with distinct levels of resistance to the organophosphate temephos. Mem Inst Oswaldo Cruz. 2012;107:916–922. doi: 10.1590/s0074-02762012000700013. [DOI] [PubMed] [Google Scholar]
- Black WC, IV, Bernhardt SA. Abundant nuclear copies of mitochondrial origin (NUMTs) in the Aedes aegypti genome. Insect Mol Biol. 2009;18:705–713. doi: 10.1111/j.1365-2583.2009.00925.x. [DOI] [PubMed] [Google Scholar]
- Bracco JE, Capurro ML, Lourenço-de-Oliveira R, Sallum MAM. Genetic variability of Aedes aegypti in the Americas using a mitochondrial gene: evidence of multiple introductions. Mem Inst Oswaldo Cruz. 2007;102:573–580. doi: 10.1590/s0074-02762007005000062. [DOI] [PubMed] [Google Scholar]
- Braga IA, Valle D. Aedes aegypti: history of control in Brazil. Epidemiol Serv Saúde. 2007;16:113–118. [Google Scholar]
- Brown JE, McBride CS, Johnson P, Ritchie S, Paupy C, Bossin H, Lutomiah J, Fernandez-Salas I, Ponlawat A, Cornel AJ, Black WC, IV, Gorrochotegui-Escalante N, Urdaneta-Marquez L, Sylla M, Slotman M, Murray KO, Walker C, Powell JR. Worldwide patterns of genetic differentiation imply multiple “domestications” of Aedes aegypti, a major vector of human diseases. Proc Biol Sci. 2011;278:2446–2454. doi: 10.1098/rspb.2010.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M, Spenassatto C, Macoris MLG, Paduan KDS, Pinto J, Ribolla PEM. Seasonal population dynamics and the genetic structure of the mosquito vector Aedes aegypti in São Paulo, Brazil. Ecol Evol. 2012;2:2794–2802. doi: 10.1002/ece3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capela R. Contribution to the study of mosquitoes (Diptera, Culicidae) from the Archipelagos of Madeira and the Salvages. I – Madeira. Arquivos do Museu Bocage A. 1981;1:45–66. [Google Scholar]
- Chaves-Scarelli T. Consulado brasileiro na Ilha da Madeira foi danificado pelas enchentes. 2010 noticias.bol.uol.com.br/internacional/2010/02/23/consulado-brasileiro-na-ilha-da-madeira-foi-danificado-pelas-enchentes.jhtm
- Cook S, Bennett SN, Holmes EC, De Chesse R, Moureau G, de Lamballerie X. Isolation of a new strain of the flavivirus cell fusing agent virus in a natural mosquito population from Puerto Rico. J Gen Virol. 2006;87:735–748. doi: 10.1099/vir.0.81475-0. [DOI] [PubMed] [Google Scholar]
- Cook S, Diallo M, Sall AA, Cooper A, Holmes EC. Mitochondrial markers for molecular identification of Aedes mosquitoes (Diptera: Culicidae) involved in transmission of arboviral disease in West Africa. J Med Entomol. 2005;42:19–28. doi: 10.1093/jmedent/42.1.19. [DOI] [PubMed] [Google Scholar]
- da Costa-da-Silva AL, Capurro ML, Bracco JE. Genetic lineages in the yellow fever mosquito Aedes (Stegomyia) aegypti (Diptera: Culicidae) from Peru. Mem Inst Oswaldo Cruz. 2005;100:539–544. doi: 10.1590/s0074-02762005000600007. [DOI] [PubMed] [Google Scholar]
- da-Cunha MP, Lima JBP, Brogdon WG, Moya GE, Valle D. Monitoring of resistance to the pyrethroid cypermethrin in Brazilian Aedes aegypti (Diptera: Culicidae) populations collected between 2001 and 2003. Mem Inst Oswaldo Cruz. 2005;100:441–444. doi: 10.1590/s0074-02762005000400017. [DOI] [PubMed] [Google Scholar]
- Delatte H, Bagny L, Brengues C, Bouetard A, Paupy C, Fontenille D. The invaders: phylogeography of dengue and chikungunya viruses Aedes vectors on the South West islands of the Indian Ocean. Infect Genet Evol. 2011;11:1769–1781. doi: 10.1016/j.meegid.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Donnelly MJ, Cuamba N, Charlwood JD, Collins FH, Townson H. Population structure in the malaria vector, Anopheles arabiensis Patton, in East Africa. Heredity. 1999;83:408–417. doi: 10.1038/sj.hdy.6885930. [DOI] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control Dengue outbreak in Madeira, Portugal, October-November 2012. ECDC/Mission Report; Stockholm: 2013. 46 [Google Scholar]
- Failloux AB, Vazeille M, Rodhain F. Geographic genetic variation in populations of the dengue virus vector Aedes aegypti. J Mol Evol. 2002;55:653–663. doi: 10.1007/s00239-002-2360-y. [DOI] [PubMed] [Google Scholar]
- Fonseca DM, La Pointe D, Fleischer RC. Bottlenecks and multiple introductions: population genetics of the vector of avian malaria in Hawaii. Mol Ecol. 2000;9:1803–1814. doi: 10.1046/j.1365-294x.2000.01070.x. [DOI] [PubMed] [Google Scholar]
- Fort P, Albertini A, Van-Hua A, Berthomieu A, Roche S, Delsuc F, Pasteur N, Capy P, Gaudin Y, Weill M. Fossil rhabdoviral sequences integrated into arthropod genomes: ontogeny, evolution and potential functionality. Mol Biol Evol. 2012;29:381–390. doi: 10.1093/molbev/msr226. [DOI] [PubMed] [Google Scholar]
- García GP, Flores AE, Fernandez-Salas I, Saavedra-Rodriguez K, Reyes-Solis G, Lozano-Fuentes S, Bond JG, Casas-Martinez M, Ramsey JM, Garcia-Rejon J, Dominguez-Galera M, Ranson H, Hemingway J, Eisen L, Black WC., IV Recent rapid rise of a permethrin knock down resistance allele in Aedes aegypti in Mexico. PLoS Negl Trop Dis. 2009;3: doi: 10.1371/journal.pntd.0000531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves YM, Silva J, Biscoito M. On the presence of Aedes (Stegomyia) aegypti Linnaeus 1762 (Insecta, Diptera, Culicidae) in the Island of Madeira (Portugal) Bol Mus Mun Funchal. 2008;58:53–59. [Google Scholar]
- Gorrochotegui-Escalante N, Gomez-Machorro C, Lozano-Fuentes S, Fernandez-Salas L, Muñoz ML, Farfan-Ale JA, Garcia-Rejon J, Beaty BJ, Black WC., IV Breeding structure of Aedes aegypti populations in Mexico varies by region. Am J Trop Med Hyg. 2002;66:213–222. doi: 10.4269/ajtmh.2002.66.213. [DOI] [PubMed] [Google Scholar]
- Gubler D. Aedes aegypti and Aedes aegypti-borne disease control in the 1990s: top down or bottom up. 1989 doi: 10.4269/ajtmh.1989.40.571. wonder.cdc.gov/wonder/PrevGuid/p0000434/p0000434.asp [DOI] [PubMed]
- Gubler D. Dengue and dengue hemorrhagic fever. 480Clin Microbiol Rev. 1998;11 doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ. Prevention and control of Aedes aegypti-borne diseases: lesson learned from past successes and failures. AsPac J Mol Biol Biotechnol. 2011;19:111–114. [Google Scholar]
- Harris AF, Rajatileka S, Ranson H. Pyrethroid resistance in Aedes aegypti from Grand Cayman. Am J Trop Med Hyg. 2010;83:277–284. doi: 10.4269/ajtmh.2010.09-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlaing T, Tun-Lin W, Somboon P, Socheat D, Setha T, Min S, Chang MS, Walton C. Mitochondrial pseudogenes in the nuclear genome of Aedes aegypti mosquitoes: implications for past and future population genetic studies. 11BMC Genet. 2009;10 doi: 10.1186/1471-2156-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Instituto de Administração da Saúde e Assuntos Sociais Possibilidade de utilização de pesticidas domésticos para a eliminação de Aedes aegypti (fase adulta) 2012 iasaude.sras.gov-madeira.pt/mosquitos/Uploads/Anexos/pesticidas%20uso%20dom%C3%A9stico%20%28documento%205%29.pdf
- Kasai S, Ng LC, Lam-phua SG, Tang CS, Itokawa K, Komagata O, Kobayashi M, Tomita T. First detection of a putative knockdown resistance gene in major mosquito vector Aedes albopictus. Jpn J Infect Dis. 2011;64:217–221. [PubMed] [Google Scholar]
- Kawada H, Higa Y, Komagata O, Kasai S, Tomita T, Yen NT, Loan LL, Sánchez RAP, Takagi M. Widespread distribution of a newly found point mutation in voltage-gated sodium channel in pyrethroid-resistant Aedes aegypti populations in Vietnam. PLoS Negl Trop Dis. 2009;3: doi: 10.1371/journal.pntd.0000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima RS, Scarpassa VM. Evidence of two lineages of the dengue vector Aedes aegypti in the Brazilian Amazon based on mitochondrial DNA ND4 gene sequences. Genet Mol Biol. 2009;32:414–422. doi: 10.1590/S1415-47572009005000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounibos LP. Invasions by insect vectors of human disease. Annu Rev Entomol. 2002;47:233–266. doi: 10.1146/annurev.ento.47.091201.145206. [DOI] [PubMed] [Google Scholar]
- Lourenço-de-Oliveira R, Vazeille M, de Fillippis AM, Failloux AB. Aedes aegypti in Brazil: genetically differentiated populations with high susceptibility to dengue and yellow fever viruses. Trans R Soc Trop Med Hyg. 2004;98:43–54. doi: 10.1016/s0035-9203(03)00006-3. [DOI] [PubMed] [Google Scholar]
- Marcombe S, Mathieu RB, Pocquet N, Riaz M-A, Poupardin R, Sélior S, Darriet F, Reynaud S, Yébakima A, Corbel V, David J-P, Chandre F. Insecticide resistance in the dengue vector Aedes aegypti from Martinique: distribution, mechanisms and relations with environmental factors. PLoS ONE. 2012;7: doi: 10.1371/journal.pone.0030989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margarita Y, Grácio AJ, Lencastre I, Silva AC, Novo MT, Sousa CA. First record of Aedes (Stegomyia) aegypti (Linnaeus, 1762) (Diptera, Culicidae) in Madeira Island - Portugal. Acta Parasitol Port. 2006;13:59–61. [Google Scholar]
- Martins AJ, Lima JB, Peixoto AA, Valle D. Frequency of Val1016Ile mutation in the voltage-gated sodium channel gene of Aedes aegypti Brazilian populations. Trop Med Int Health. 2009;14:1351–1355. doi: 10.1111/j.1365-3156.2009.02378.x. [DOI] [PubMed] [Google Scholar]
- Moore M, Sylla M, Goss L, Burugu MW, Sang R, Kamau LW, Kenya EU, Bosio C, Munoz MDL, Sharakova M, Black WC., IV Dual African origins of global Aedes aegypti s.l. populations revealed by mitochondrial DNA. PLoS Negl Trop Dis. 2013;7: doi: 10.1371/journal.pntd.0002175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousson L, Dauga C, Garrigues T, Schaffner F, Vazeille M, Failloux A-B. Phylogeography of Aedes (Stegomyia) aegypti (L.) and Aedes (Stegomyia) albopictus (Skuse) (Diptera: Culicidae) based on mitochondrial DNA variations. Genet Res. 2005;86:1–11. doi: 10.1017/S0016672305007627. [DOI] [PubMed] [Google Scholar]
- Observatório da Emigração Venezuela Sempre no coração dos portugueses. 2012 observatorioemigracao.secomunidades.pt/np4/2904.html
- Paduan KDS, Ribolla PEM. Mitochondrial DNA polymorphism and heteroplasmy in populations of Aedes aegypti in Brazil. J Med Entomol. 2008;45:59–67. doi: 10.1603/0022-2585(2008)45[59:mdpahi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Paupy C, Le G, Brengues C, Guerra M, Revollo J, Barja Z, Hervé J, Fontenille D. Genetic structure and phylogeography of Aedes aegypti, the dengue and yellow-fever mosquito vector in Bolivia. Infect Genet Evol. 2012;12:1260–1269. doi: 10.1016/j.meegid.2012.04.012. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (v.1.2): population genetics software for exact tests and ecumenicism. Heredity. 1995;86:248–249. [Google Scholar]
- Saavedra-Rodriguez K, Urdaneta-Marquez L, Rajatileka S, Moulton M, Flores AE, Fernandez-Salas I, Bisset J, Rodriguez M, McCall PJ, Donnelly MJ, Ranson H, Hemingway J, Black WC., IV A mutation in the voltage-gated sodium channel gene associated with pyrethroid resistance in Latin American Aedes aegypti. Insect Mol Biol. 2007;16:785–798. doi: 10.1111/j.1365-2583.2007.00774.x. [DOI] [PubMed] [Google Scholar]
- Seixas G. Aedes (Stegomyia) aegypti (Diptera, Culicidae) da ilha da Madeira: origem geográfica e resistência aos insecticidas. Instituto de Higiene e Medicina Tropical/Universidade Nova de Lisboa; Lisboa: 2012. 123 MsD Thesis. [Google Scholar]
- Smith C. The history of dengue in tropical Asia and its probable relationship to the mosquito Aedes aegypti. 243Am J Trop Med Hyg. 1956;59 [PubMed] [Google Scholar]
- Soderlund DM, Knipple DC. The molecular biology of knockdown resistance to pyrethroid insecticides. Insect Biochem Mol Biol. 2003;33:563–577. doi: 10.1016/s0965-1748(03)00023-7. [DOI] [PubMed] [Google Scholar]
- Soper FL, Wilson DB. Anopheles gambiae in Brazil 1930 to 1940. The Rockefeller Foundation; New York: 1943. 262 [Google Scholar]
- Sota T, Mogi M. Origin of pitcher plant mosquitoes in Aedes (Stegomyia): a molecular phylogenetic analysis using mitochondrial and nuclear gene sequences. J Med Entomol. 2006;43:795–800. doi: 10.1603/0022-2585(2006)43[795:ooppmi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Sousa CA, Clairouin M, Seixas G, Viveiros B, Novo MT, Silva AC, Escoval MT, Economopoulou A. Ongoing outbreak of dengue type 1 in the Autonomous Region of Madeira, Portugal: preliminary report. Euro Surveill. 2012;17:8–11. doi: 10.2807/ese.17.49.20333-en. [DOI] [PubMed] [Google Scholar]
- Tabachnick WJ. Evolutionary genetics and arthropod-borne disease: the yellow fever mosquito. Am Entomol. 1991;37:14–26. [Google Scholar]
- Urdaneta-Marquez L, Bosio C, Herrera F, Rubio-Palis Y, Salasek M, Black WC., IV Genetic relationships among Aedes aegypti collections in Venezuela as determined by mitochondrial DNA variation and nuclear single nucleotide polymorphisms. Am J Trop Med Hyg. 2008;78:479–491. [PubMed] [Google Scholar]
- Urdaneta-Marquez L, Failloux A. Population genetic structure of Aedes aegypti, the principal vector of dengue viruses. Infect Genet Evol. 2011;11:253–261. doi: 10.1016/j.meegid.2010.11.020. [DOI] [PubMed] [Google Scholar]
- Vontas J, Kioulos E, Pavlidi N, Morou E, Torre A, Ranson H. Insecticide resistance in the major dengue vectors Aedes albopictus and Aedes aegypti. Pestic Biochem Physiol. 2012;104:126–131. [Google Scholar]
- World Health Organization Global strategy for dengue prevention and control 2012-2020. 2012 apps.who.int/iris/bitstream/10665/75303/1/9789241504034_eng.pdf
- Yanola J, Somboon P, Walton C, Nachaiwieng W, Somwang P, Prapanthadara L. High-throughput assays for detection of the F1534C mutation in the voltage-gated sodium channel gene in permethrin-resistant Aedes aegypti and the distribution of this mutation throughout Thailand. Trop Med Int Health. 2011;16:501–509. doi: 10.1111/j.1365-3156.2011.02725.x. [DOI] [PubMed] [Google Scholar]


