Abstract
Yellow fever virus (YFV), a member of the family Flaviviridae, genus Flavivirus is endemic to tropical areas of Africa and South America and is among the arboviruses that pose a threat to public health. Recent outbreaks in Brazil, Bolivia, and Paraguay and the observation that vectors capable of transmitting YFV are presenting in urban areas underscore the urgency of improving surveillance and diagnostic methods. Two novel methods (RT-hemi-nested-PCR and SYBR®Green qRT-PCR) for efficient detection of YFV strains circulating in South America have been developed. The methods were validated using samples obtained from golden hamsters infected experimentally with wild-type YFV strains as well as human serum and tissue samples with acute disease.
Keywords: Yellow fever virus, molecular diagnostic, RT-hemi-nested-PCR, SYBR®Green qRT-PCR
1. INTRODUCTION
Yellow fever Virus (YFV) is the prototype member of the genus Flavivirus in the family Flaviviridae, and is the etiologic agent of Yellow fever, an ancient infectious disease transmitted by Culicidae mosquitoes in Africa and South America. The YFV genome is single-stranded, positive-sense RNA approximately 11,000 nucleotides in length, encoding a polyprotein that contains three structural (5’-Capsid-C; Pre-Membrane-PrM; and Envelope- E-3’) and seven non-structural (5’-NS1-NS2a-NS2b-NS3-NS4a-NS4b-NS5-3’) proteins (Fauquet et al., 2005).
The International Committee on the Taxonomy of Viruses (ICTV) recognizes 62 different virus species in the family Flaviviridae, many of them transmitted by mosquitoes (Fauquet et al., 2005). YFV and several other flaviviruses including Dengue virus (DENV), West Nile virus (WNV), Japanese encephalitis virus (JEV), Saint Louis encephalitis virus (SLEV) and Rocio virus (ROCV) are associated with human infection in South America (Monath, 1988).
The World Health Organization (WHO) estimates 200,000 annual YFV infections resulting in 30,000 deaths in endemic areas of Africa and South America (Monath, 2001; Robertson et al., 1996; WHO, 2010). In Brazil, the virus is responsible for jungle Yellow fever epidemics and epizootics with increasing morbidity and mortality rates. Viral transmission involves wild mosquito vectors of the Sabethes and Haemagogus genera (mainly the Haemagogus janthinomys species) and arboreal wild vertebrate hosts (mainly monkeys from the genera Allouatta [howlers], Callythrix [marmosets], and Cebus [calucho]). Humans become infected when they enter forest areas to harvest timber, explore for minerals, practice agricultural activities or pursue eco-tourism (Vasconcelos, 2003).
Despite the fact that urban Yellow fever has not been reported in Brazil since the early 1940s, substantial risk exists since Aedes aegypti, the anthropophagic mosquito related to YFV transmission, is present in urban areas of Brazil and other South American countries (Vasconcelos et al., 1999; WHO, 2010). Although RT-PCR assays are commonly employed for detection of several flaviviruses, including DENV, JEV, WNV and SLEV (Coimbra et al., 2008; Hull et al., 2008; Jeong et al., 2011; Lanciotti et al., 1992; Maher-Sturgess et al., 2008; Yeh et al., 2010), protocols for the detection of YFV are less well established. In this article, the development and validation of two molecular methods for detection of YFV are described and their utility with samples obtained from hamsters infected experimentally and infected humans is demonstrated.
2. MATERIAL AND METHODS
2.1 Ethical aspects
All animal serum samples and virus strains used in this study were obtained from the collection of the Department of Arbovirology and Hemorrhagic Fevers at the Instituto Evandro Chagas, a World Health Organization Collaborating Centre for Research and Reference on Arboviruses. Animals used for the experiment were humanely handled in a biosafety level 2 facility and processed in a class II B2 safety cabinet. The study was approved by the Ethics Committee on Animal Research (CEPAN/IEC).Human samples (blood, sera and fresh and paraffin embedded tissues) were sent to the Instituto Evandro Chagas for diagnostic investigation during the 2009 Brazilian Yellow fever epidemic. Informed consent and epidemiological forms were filled out by patients before collection. Furthermore, all samples were included in the national database of the Brazilian Ministry of Health, which controls and enforces all ethical obligations established for the protection of human subjects.
2.2 Hamster specimens
A total of 51 young (2-3 weeks old) hamsters (Mesocricetus auratus) were separated into 17 groups of 3 each. Two hamsters from each group were infected with a YFV strain (Table 1); the third animal served as a negative control. Virus suspensions obtained from infected VERO cells were inoculated intraperitoneally with 6.9 × 106 PFU/mL of each strain. Animals were observed daily until day 3 post infection and then euthanized for sample (blood and viscera) collection. Presence of virus was confirmed by an immunofluorescence assay using an anti-YFV monoclonal antibody (Biomanguinhos, Fiocruz, Brazil). Serum samples were collected daily from 5 groups of infected hamsters to determine the sensitivity of the assays. A total of 200 serum samples were collected from days 0 through 7 post infection.
Table 1.
Brazilian Yellow fever virus strains used for experimental study in hamsters
| Virus strain | Source of isolation | Year of isolation | Passage historyb |
|---|---|---|---|
| BeAR378600 | Haemagogus sp. | 1980 | 1 |
| BeH394880a | human | 1981 | 2 |
| BeH413820a | human | 1983 | 1 |
| BeH422973a | human | 1984 | 1 |
| BeH423602a | human | 1984 | 2 |
| BeH463676a | human | 1987 | 1 |
| BeAR513008 | Sabethes sp | 1992 | 2 |
| BeH521244 | human | 1993 | 1 |
| BeH526722 | human | 1994 | 1 |
| BeH622205 | human | 2000 | 1 |
| BeH622493 | human | 2000 | 1 |
| BeH629290 | human | 2000 | 1 |
| BeAR630785 | Hg. Janthinomys | 2000 | 1 |
| BeAR646536 | Hg. Leucocelaenus | 2001 | 1 |
| BeH655417 | human | 2002 | 1 |
| BeAR678011 | Sa. Chloropterus | 2004 | 2 |
| BeH686174 | human | 2004 | 1 |
YFV strains used for analysis of correlation between genome detection and period post infection
Number of passages in Vero cells after virus isolation in mice
2.3 Sample collection
Four groups of samples were collected (total n=505). Groups 1 and 2 (n=117 each) were used for validation (Table 2). The Group 1 (YF) included blood, sera, fresh liver and paraffin-embedded liver tissues obtained from hamsters infected experimentally with different YFV strains (Table 1). The second group (non YF) group comprised blood, sera, fresh liver and paraffin-embedded liver samples from uninfected hamsters and RNA samples from supernatant of Vero or C6/36 cells infected with different flaviviruses (Table 2).
Table 2.
Samples used for composing the panel for the analyses of molecular methods.
| Group | Type of sample | Source | Diagnostic evidence | No. of samples | Total |
|---|---|---|---|---|---|
| YF | Sera | Infected hamster | Virus isolation, IF positive for YFV | 34 | |
| Whole blood | Infected hamster | Virus isolation, IF positive for YFV | 34 | ||
| Liver fragment | Infected hamster | Virus isolation, IF positive for YFV | 34 | ||
| Paraffin embedded tissue | human liver | Positive Immune Histochemistry for YFV | 15 | 117 | |
| Non YF | sera | non infected hamster | Negative virus isolation, IF negative for flaviviruses including YFV | 17 | |
| blood | non infected hamster | Negative virus isolation, IF negative for flaviviruses including YFV | 17 | ||
| Liver fragment | non infected hamster | Negative virus isolation, IF negative for flaviviruses including YFV | 17 | ||
| Paraffin embedded tissue | human liver | Negative Immune Histochemistry for YFV | 15 | ||
| DENV-1 RNA | C6/36 cell culture | Virus isolation, IF positive for DENV-1 | 15 | ||
| DENV-2 RNA | C6/36 cell culture | Virus isolation, IF positive for DENV-2 | 15 | ||
| DENV-3 RNA | C6/36 cell culture | Virus isolation, IF positive for DENV-3 | 16 | ||
| DENV-4 RNA | C6/36 cell culture | Virus isolation, IF positive for DENV-4 | 1 | ||
| ROCV RNA | VERO cell culture | Virus isolation, positive NT for ROCV | 1 | ||
| SLEV RNA | VERO cell culture | Virus isolation, positive NT for SLEV | 1 | ||
| BSQV RNA | VERO cell culture | Virus isolation, positive NT for BSQV | 1 | ||
| IGUV RNA | VERO cell culture | Virus isolation, positive NT for IGUV | 1 | 117 | |
| Total | 234 | ||||
YFV: Yellow fever virus; DENV-1: Dengue virus type 1; DENV-2: Dengue virus type 2; DENV-3: Dengue virus type 3; DENV-4: Dengue virus type 4; ROCV: Rocio virus; SLEV: Saint Louis Encephalitis virus; BSQV: Bussuquara virus; IGUV: Iguape virus; NT: neutralizing test.
Group 3 was used to determine the sensitivity of the assays during experimental infections of hamsters. This group contained 200 serum samples of hamsters infected with wild type YFV strains collected from days 0 through 7 after infection (Table 1).
Group 4 contained 71 clinical samples (sera and viscera fragments) received during the 2008-2009 Yellow fever epidemic in Rio Grande do Sul, Minas Gerais and Mato Grosso States, Brazil (WHO, 2010). These samples were used to evaluate the clinical performance of the new molecular methods versus virus isolation in Vero cells or newborn mice.
2.4 RNA extraction
Viral RNA from serum, blood, fresh tissue fragments, and paraffin embedded tissues were extracted using the QIAquick RNA extraction kit (Qiagen, Hilden, Germany). Paraffin embedded tissue samples were processed in xylol (Invitrogen, Carlsbad, CA, USA) prior to RNA extraction.
2.5 Primer selection
Primers were selected based on multiple sequence alignment implemented in Geneious 4.5 (Biomatters, Auckland, New Zealand). Alignments included both full-length and partial genome (E and NS5 genes) sequences available from GenBank. Conserved sequences (ranging from 150 nt to 500 nt in length) were selected as candidate regions. Subsequently, selected regions were used as a query in Genbank. All YFV sequences were then chosen for primer design using the Primer Seq software (Lasergene DNA Star Package, Madison, WI, USA). Specific primers were selected according with the lowest E-values (0.021) and highest query coverage score (100%). For the first round of the RT-Hemi-Nested-PCR the following primers were selected on the E gene region: forward (YFV 975F: 5’-CCTACTGGTCTTGGCTGTTGG-3’) and reverse (YFV 1312R: 5’- AATGCTCCCTTTCCCAAATA-3’). For the hemi-nested step the inner forward primer (YFV 995F: 5’-GACAGGGATTTCATTGAGG-3’) was used in combination with the reverse primer YFV 1312R. For the qRT-PCR, primers on the NS5 region were selected (YFVNS5-F: 5’- CATGGTCGATTCATGGGAAAG-3’ and YFVNS5-R: 5’-CGCACAGCTTGTCTTGTCTC-3’). The primers blasted with 100% identity with 102 YFV sequences (GenBank) from around the world, including different geographic regions of Africa, South America (Brazil, Venezuela, Peru and Bolivia) and Central America (Trinidad).
2.6 Reverse Transcription Polymerase Chain Reaction (RT-PCR) and RT-hemi-nested-PCR assays
The RT-PCR method has been previously described by Vasconcelos et al. (2004). RT-PCR reactions contained 5μl (1 to 5 ng) of viral RNA, 200 μM of each forward and reverse primer (Vasconcelos et al., 2004), 1X PCR buffer (50 mM Tris-HCl, 8.3 pH; 75 mM KCL), 50 mM MgCl2, 0.1 mM dithiothreitol (DTT), 1 U/μl of RNAsin RNase inhibitor, 200 μM deoxynucleoside triphosphates (dNTPs), 1.125 U of Platinum Taq DNA polymerase, and 1 U of Superscript II Reverse Transcriptase, and water adjusted for 50 uL. RNA samples, in a one step reaction were reverse transcribed for 1 hour at 42°C followed by 35 PCR cycles of 90°C for 40 seconds, 55°C for 40 seconds and 72°C for 1 minute.
The RT-hemi-nested-PCR was based on the Deubel et al. (1997) protocol to enhance sensitivity of the RT-PCR described above. RT-PCR buffer and cycling conditions were similar to those described above except by the primers sets (see section 2.5). For the hemi-nested PCR, RT-PCR products were diluted 1:100 in nuclease-free water, and the reactions were setup with 5 μL of diluted products, 200 μM of the forward and reverse primers, 1X PCR buffer (50 mM Tris-HCl, 8.3 pH; 75 mM KCL), 50 mM MgCl2, 200 μM deoxynucleoside triphosphates (dNTPs), 1.125 U of Platinum Taq DNA polymerase, and water adjusted to 50 μL. PCR cycling conditions were identical to those described above. Positive (RNA extracted from YFV infected Vero cells) and negative (RNA extracted from uninfected Vero cells and RNAse free water) controls were used. RT-PCR and hemi-nested- PCR products were visualized in a 3% agarose gel and inspected for appropriate size.
2.7 Quantitative real time PCR
For the quantitative real time RT-PCR (qRT-PCR), a commercial kit was used (SuperScript III Platinum SYBR® Green On-step qRT-PCR, Invitrogen, Carlsbad, CA, USA) and the reactions were run on an ABI Prism 7500 Real time PCR System (Applied Biosystem, Carlsbad, CA, USA). The method was adapted from the protocol described by Aquino et al. (2007), using specific primers selected as described above. The reaction was performed in a final volume of 25 μl containing 5 μL of viral RNA (1-2 ng/μL), 0.5 μl of SuperScript III RT Platinum Taq Mix, 0.2 μM of each specific primer, 12.5 μl of 2X SYBR Green fluorescent molecule, and 1 μl of ROX. The amplification was carried out as follows: 50°C for 20 minutes; 95°C for 5 minutes; 45 cycles of 95°C for 15 seconds, 55°C for 40 seconds and 72°C for 30 seconds. The melting curve was calculated during the incubation from 60°C to 90°C with a capture speed of 0.2 °C/seconds. The quantity of the qRT-PCR was expressed in PFU/mL based on the standard curve constructed from amplification values of RNA extracted from Vero E6 cells infected with the YFV strain BE H111 (stock titer 8.9 × 106 PFU/mL) diluted from 10 −1 to 10−7.
2.8 Determination of sensitivity and specificity indexes
Assay sensitivity (S) was determined using serial dilutions (10−1 to 10−7) of supernatant of Vero cells infected with the YFV (strain BE H111) at an initial titer of 8.9 x106 PFU/mL, as well as by testing the group 1 (YF) samples (Table 2). Assay specificity (Sp) for YFV versus other flaviviruses (Group 2; Non YF) was determined using RNA extracted from C6/36 cells infected with DENV 1, 2, and 3 serotypes.
The two new methods (RT-hemi-nested-PCR and qRT-PCR) were compared with the classical RT-PCR (Vasconcelos et al., 2004) and virus isolation methods. The classical RT PCR described by Vasconcelos et al. is the current standard method used in Brazil for clinical diagnosis of YFV infections. This RT-PCR method amplifies a 1200bp region of the envelope gene. The indexes of (S) and (Sp) were calculated using the following formula: S=TP/(TP+FN) × 100 and Sp=TN/(TN+FP) x100, where TP is the number of true positive results; TN is the number true negative results; FP is the number of false-positive results; and FN is the number of false negative results (Table 2).
Assay performance was also assessed by testing 200 serum samples obtained from hamsters infected experimentally with the five YFV strains (Table 1) over days 0 through 7 post-infection for YFV genome (Group 3). Serum samples were tested for the presence of IgM antibodies against YFV using a protocol described previously (Kuno, Gomez, and Gubler, 1987) replacing only the anti-human IgM with the anti-hamster IgM antibody (Mouse anti-hamster IgM, 1mg/mL; RDI, Fitzgerald Industry, Concord, MA, USA).
2.9 Performance evaluation in clinical specimens
The methods were then compared with virus isolation and RT-PCR using 71 clinical samples (sera and tissue fragments, Group 4). Receiver Operating Characteristic curve analysis (ROC) (Fawcett, 2006; Zweig and Campbell, 1993) implemented in the BioStat software v.5.0 (Ayres et al., 2005) was used to determine the best procedure where (S) and (Sp) values closer to 1 and zero, respectively, represent ideal performance.
3. RESULTS
3.1 RT-hemi-nested PCR and qRT-PCR product analyses
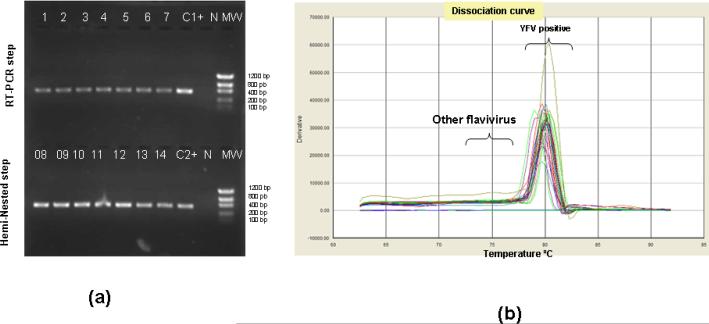
As a measure of specificity, classic assay products were visualized in agarose gels. Melting curve analysis was used in the SYBR® qRT-PCR assay. Amplicon sizes were 390bp for the first round of amplification, and 370bp for the hemi-nested step (Figure 1a). The melting curve for the qRT-PCR assay is shown in Figure 1b.
Figure 1.
Detection of YFV sequences in extracts of infected Vero cells. (a) Size fractionated, ethidium bromide labeled product (4μL) of RT-(lanes 1 to 7) and RT-hemi- nested-PCR (lanes 8 to 14) steps (1% agarose gel). (b) the dissociation curve for the SYBR® qRT-PCR showing the melting temperature for positive YFV genome detection ranging from 79°C to 81°C (mean of 80°C). “C+1 and C+2” stand for positive controls used during the RT-PCR and Hemi-nested-PCR steps, respectively. “NC” stands for negative control (RNA extracted from supernatant of uninfected Vero cells) and “MW” stands for molecular weight marker (100bp DNA ladder, Invitrogen, Carlsbad, CA, USA).
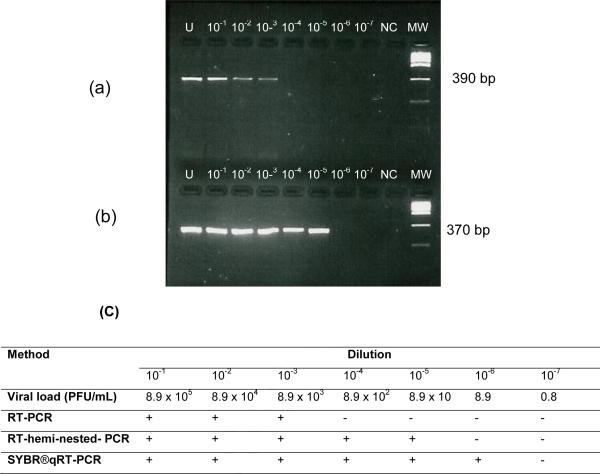
The limits of detection for the RT-PCR, RT-hemi-nested-PCR and qRT-PCR were, respectively, 8.9×103, 8.9×101 and 8.9 PFU/mL (Figure 2).
Figure 2.
Detection of YFV sequences by RT-PCR and RT-hemi-nested-PCR and SYBR® qRT-PCR in serum from an infected hamster (panels a and b) and limits of detection using RT-PCR, RT-hemi-nested-PCR and SYBR® qRT-PCR methods (c). “+” stands for a positive result, “-“ for a negative result, “U” for undiluted sample, “NC” for negative control (RNA extracted from VERO uninfected cells), and “MW” for molecular weight marker (Low DNA mass ladder, Invitrogen, Carlsbad, CA, USA).
3.2 Sensitivity and Specificity
Using the samples from the first and second groups, we calculated the S and Sp indexes and compared them to the “gold standard” virus isolation method. The values determined were: (a) RT-PCR: S: 52.1% (61/117); Sp: 100% (117/117); (b) RT-hemi-nested PCR: S: 70.9% (83/117); Sp: 98.2% (115/117); and (c) qRT-PCR: S: 92.3% (108/117); Sp: 100% (117/117).
Using the third group of samples, we determined that all methods were able to detect YFV during the first two days post infection (Table 3). By day 3, the hemi-nested-PCR and qRT-PCR outperformed RT-PCR. After day 5, qRT-PCR outperformed not only RT-PCR but also virus isolation. At day 6 and 7, hemi-nested PCR also outperformed virus isolation. After day 3, virus isolation outperformed RT-PCR. The RT-PCR protocol was less sensitive (S ≤72%) than hemi-nested-PCR or qRT-PCR (Table 3). Among clinical samples, highest sensitivity was observed in liver samples and lowest in paraffin embedded samples, regardless of the method used. Overall, 100% specificity was observed for all methods except for sera and liver samples tested by the RT-hemi-Nested-PCR method. (Supplementary Table 1).
Table 3.
Performance of RT-PCR, RT-hemi-nested-PCR, qRT-PCR, classical RT-PCR and virus isolation in detection of YFV in infected experimentally hamsters.
| Standard protocol | Current protocol | New protocols |
Serology | |||
|---|---|---|---|---|---|---|
| RT-Hemi-Nested-PCR (a/b) | ||||||
| Days post infection | Virus isolation | RT-PCR (a/b) | RT-PCR step | Nested-PCR step | qRT-PCR (a/b) | MAC-ELISA positive(a/b) |
| Day 0 | 25/25 | 25/25, S=100% 25/25, S=100% | 25/25, S=100% | 25/25, S=100% | 0/25 | |
| Day 1 | 25/25 | 25/25, S=100% 25/25, S=100% | 25/25, S=100% | 25/25, S=100% | 0/25 | |
| Day 2 | 25/25 | 25/25, S=100% 25/25, S=100% | 25/25, S=100% | 25/25, S=100% | 0/25 | |
| Day 3 | 25/25 | 20/25, S=80% | 21/25, S=84% | 25/25, S=100% | 25/25, S=100% | 0/25 |
| Day 4 | 25/25 | 18/25, S=72% | 21/25, S=84% | 24/25, S=96% | 25/25, S=100% | 1/25 |
| Day 5 | 23/25 | 13/25, S=52% | 17/25, S=68% | 21/25, S=88.8% | 22/25, S=96% | 3/25 |
| Day 6 | 10/25 | 2/25, S=8% | 5/25, S=20% | 12/25, S=48% | 11/25, S=44% | 10/25 |
| Day 7 | 1/25 | 0/25, S=0% | 0/25, S=0% | 2/25, S=8% | 4/25, S=16% | 25/25 |
:positive
:tested
High sensitivity
![]() Medium sensitivity
Medium sensitivity
![]() Low sensitivity
Low sensitivity
![]() Very low sensitivity
Very low sensitivity
The number of positive specimens identified with each method for samples obtained during the 2009 YFV outbreak (group 4; n=71) were 9.8% (n=7), 19.7% (n=14), 29.5% (n=21) and 40.8% (n=29) for the standard RT-PCR, RT-PCR step (for the RT-hemi-nested- PCR), hemi-nested PCR step (for the RT-hemi-nested-PCR), and qRT-PCR, respectively. Comparing virus isolation with these new methods revealed high concordance with the qRT PCR showing an index of correlation of 97.2 % (n=69), where 25 were positive and 44 were negative by both methods. The two discordant results (2.8%) were both positive by the qRT PCR and negative by virus isolation into Vero cells or in newborn mice.
3.3 Comparison of test performance
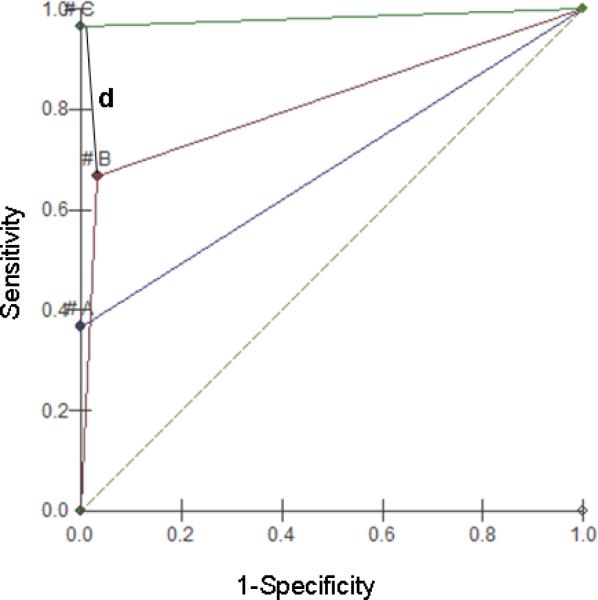
ROC analysis (Figure 3) determined a d value of 0.63 for the RT-PCR (p cohort to S=0.37; p cohort to Sp=1.0), 0.33 for RT-hemi-nested PCR (P cohort to S=0.67; p cohort to Sp= 0.97), and 0.03 for qRT-PCR (p cohort to S=0.67; p cohort to Sp= 0.97).
Figure 3.
Graphic representation of ROC (receiver operating characteristics) test for the RT PCR (A), RT-hemi-nested-PCR (B) e SYBR® qRT-PCR. The best performance for each method is indicated by the line d (distance). In case of methods C and A, the line d coincides with the Y axis (P. cohort to Sp=0). “#” symbol represents the cohort point for each method.
4. DISCUSSION
Yellow fever represents a serious public health concern in Africa and South America. Protocols for genome amplification and sequencing of YFV have been developed (Mutebi et al., 2001; Vasconcelos et al., 2004) and many partial and complete YFV sequences have been generated (Bryant, Holmes, and Barrett, 2007; Bryant et al., 2005). Recently, RT-PCR and other molecular protocols have been established for diagnostic and epidemiological surveillance (Deubel et al., 1997; Domingo et al., 2011; Mendez et al., 2007; Sanchez-Seco et al., 2006; Weidmann et al., 2010). However, none has been fully evaluated for use in endemic regions like South America.
The RT-hemi-nested-PCR and SYBR® qRT-PCR methods described here were developed and tested using well characterized samples from naturally infected humans, experimentally infected hamsters and infected cell lines. This approach enables evaluation of sensitivity, specificity and limit of detection of the assays compared to virus isolation, considered the “gold standard technique”. The current RT-PCR assay used for YFV genome detection in Brazil amplifies a fragment of 1200bp of the viral envelope and was originally adapted from a method described elsewhere (Vasconcelos et al., 2004). Application of that method to local clinical samples obtained during sylvatic Yellow fever outbreaks resulted in a high rate of false-negatives, consistent with low sensitivity indexes (approximately 75% - 80%). Notably, some RT-PCR negative samples were positive by virus isolation (Nunes MR et al., Instituto Evandro Chagas, data not shown). This failure may have resulted from suboptimal primer design due to lack of sequence information representing local strains. Another possibility is that the large size of the amplification product may amplify less efficiently and degrade more readily, thus lowering sensitivity. Therefore, the RT-PCR and RT-hemi-nested PCR protocols were developed with a smaller amplification product making them more sensitive in samples with low viral load such as clinical samples collected during the early phase of infection (less than 12 hours after onset of symptoms), RNA obtained from degraded samples, and paraffin embedded samples. The sensitivity test results demonstrate that both new methods accomplish this goal. Although only samples from South America have been tested in this study, the sequence similarity of the primers to YFV strains from other parts of the world suggests they will amplify any YFV strain. Unfortunately, since the region where the primers are designed is high conserved, the methods cannot differentiate between post vaccine events and wild YF cases.
Whereas SYBR®qRT-PCR performed well in paraffin-embedded tissue samples, detecting signal in 13 of 15 samples, RT-PCR failed to detect signal in any samples and RT- hemi-nested PCR detected signal in only two samples. Whether these findings represent template degradation during processing into paraffin or the extraction of RNA from paraffin, or the presence of some external inhibitor cannot be discerned from our data. Nonetheless, it is clear that qRT-PCR is the method of choice for surveillance of YFV sequences in paraffin embedded tissues.
The RT-hemi-nested PCR assay revealed a specificity of 98.2%. Although every effort to avoid contamination was undertaken (physical separation of RT-PCR and hemi- nested steps, separate pipette sets for each step of the protocol, and UV irradiation between tests), two false-positive results were obtained, presumably due to cross-contamination among samples when assay tubes were opened during the hemi-nested PCR step. The other methods (RT-PCR and qRT-PCR) were 100% specific with no false-positives. Contamination is a problem constantly associated with nested PCR methods and may be an important factor in final method selection. Another factor is sensitivity. The ROC curve test demonstrates the superior performance of qRT-PCR among the three evaluated molecular methods (RT-PCR, RT-hemi-nested PCR and qRT-PCR).
In developing countries cost is another important consideration in assay implementation. Although sensitive, specific assays have been developed with TaqMan™ (Applied Biosystems) and Plexor (Promega) probes, which are relatively expensive. In contrast, the SYBR®qRT-PCR approach offers the advantages of simplicity (one-step qRT PCR), minimal risk of cross-contamination, sensitivity, and relatively low cost.
In conclusion, the new developed methods will be helpful in further YFV epidemics and can be used for the early diagnosis of infections caused by this important human pathogen.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Aaloki Shah for technical assistance and Katrina Ciraldo for editing the manuscript. This study was supported by CNPq (INCT-FHV/CNPq/FAPESPA/CAPES - Grant 573739/2008-0; CNPq Grant 300460/2005-8, and CNPq Grant 302987/2008-8), and IEC/SVS from the Brazilian Government. Work in the CII is supported by awards from the National Institutes of Health including AI057158 (Northeast Biodefense Center-Lipkin) and AI1057158, and the United States Department of Defense.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ayres M, Jr., M.A., Ayres D, Santos A.S.d. BIOSTAT v5.0: Aplicações estatísticas nas áreas das ciências biológicas e médicas. Sociedade Civil Mamirauá/MCT-CNPq/ Conservation International. 2005 [Google Scholar]
- Bryant JE, Holmes EC, Barrett AD. Out of Africa: a molecular perspective on the introduction of yellow fever virus into the Americas. PLoS Pathog. 2007;3:e75. doi: 10.1371/journal.ppat.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant JE, Vasconcelos PF, Rijnbrand RC, Mutebi JP, Higgs S, Barrett AD. Size heterogeneity in the 3′ noncoding region of South American isolates of yellow fever virus. J Virol. 2005;79:3807–21. doi: 10.1128/JVI.79.6.3807-3821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra TL, Santos RN, Petrella S, Nagasse-Sugahara TK, Castrignano SB, Santos CL. Molecular characterization of two Rocio flavivirus strains isolated during the encephalitis epidemic in Sao Paulo State, Brazil and the development of a one-step RT-PCR assay for diagnosis. Rev Inst Med Trop Sao Paulo. 2008;50:89–94. doi: 10.1590/s0036-46652008000200005. [DOI] [PubMed] [Google Scholar]
- Deubel V, Huerre M, Cathomas G, Drouet MT, Wuscher N, Le Guenno B, Widmer AF. Molecular detection and characterization of yellow fever virus in blood and liver specimens of a non-vaccinated fatal human case. J Med Virol. 1997;53:212–7. [PubMed] [Google Scholar]
- Domingo C, Yactayo S, Agbenu E, Demanou M, Schulz AR, Daskalow K, Niedrig M. Detection of Yellow Fever 17D Genome in Urine. J Clin Microbiol. 2011;49:760–2. doi: 10.1128/JCM.01775-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: Eighth Report of the International Committee on the Taxonomy of Viruses. Elsevier; London: 2005. [Google Scholar]
- Fawcett T. An introduction to ROC analysis. Pattern Recognition Letters. 2006;27:861–874. [Google Scholar]
- Hull R, Nattanmai S, Kramer LD, Bernard KA, Tavakoli NP. A duplex real-time reverse transcriptase polymerase chain reaction assay for the detection of St. Louis encephalitis and eastern equine encephalitis viruses. Diagn Microbiol Infect Dis. 2008;62:272–9. doi: 10.1016/j.diagmicrobio.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YE, Jeon MJ, Cho JE, Han MG, Choi HJ, Shin MY, Park HJ, Kim W, Moon BC, Park JS, Park B, Ju YR. Development and field evaluation of a nested RT-PCR kit for detecting Japanese encephalitis virus in mosquitoes. J Virol Methods. 2011;171:248–52. doi: 10.1016/j.jviromet.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Kuno G, Gomez I, Gubler DJ. Detecting artificial anti-dengue IgM immune complexes using an enzyme-linked immunosorbent assay. Am J Trop Med Hyg. 1987;36:153–9. doi: 10.4269/ajtmh.1987.36.153. [DOI] [PubMed] [Google Scholar]
- Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–51. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher-Sturgess SL, Forrester NL, Wayper PJ, Gould EA, Hall RA, Barnard RT, Gibbs MJ. Universal primers that amplify RNA from all three flavivirus subgroups. Virol J. 2008;5:16. doi: 10.1186/1743-422X-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez JA, Parra E, Neira M, Rey GJ. [Detection of yellow fever virus by reverse transcriptase polymerase chain reaction in wild monkeys: a sensitive tool for epidemiologic surveillance]. Biomedica. 2007;27:461–7. [PubMed] [Google Scholar]
- Monath TP. Yellow Fever. In: Monath TP, editor. Arboviruses: ecology and epidemiology. CRC Press; Boca Raton, USA: 1988. pp. 139–241. [Google Scholar]
- Monath TP. Yellow fever: an update. Lancet Infect Dis. 2001;1:11–20. doi: 10.1016/S1473-3099(01)00016-0. [DOI] [PubMed] [Google Scholar]
- Mutebi JP, Wang H, Li L, Bryant JE, Barrett AD. Phylogenetic and evolutionary relationships among yellow fever virus isolates in Africa. J Virol. 2001;75:6999–7008. doi: 10.1128/JVI.75.15.6999-7008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SE, Hull BP, Tomori O, Bele O, LeDuc JW, Esteves K. Yellow fever: a decade of reemergence. JAMA. 1996;276:1157–62. [PubMed] [Google Scholar]
- Sanchez-Seco MP, Rosario D, Hernandez L, Domingo C, Valdes K, Guzman MG, Tenorio A. Detection and subtyping of dengue 1-4 and yellow fever viruses by means of a multiplex RT-nested-PCR using degenerated primers. Trop Med Int Health. 2006;11:1432–41. doi: 10.1111/j.1365-3156.2006.01696.x. [DOI] [PubMed] [Google Scholar]
- Vasconcelos PF. [Yellow Fever]. Rev Soc Bras Med Trop. 2003;36:275–93. doi: 10.1590/s0037-86822003000200012. [DOI] [PubMed] [Google Scholar]
- Vasconcelos PF, Bryant JE, da Rosa TP, Tesh RB, Rodrigues SG, Barrett AD. Genetic divergence and dispersal of yellow fever virus. Brazil. Emerg Infect Dis. 2004;10:1578–84. doi: 10.3201/eid1009.040197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos PFC, Travassos da Rosa APA, Pinheiro FP, Rodrgues SG, Travassos da Rosa ES, Cruz AC, Travassos da Rosa JFS. Aedes aegypti, dengue and re-urbanization of Yellow fever in Brazil and other South American Countries. Past and present, and future perspectives. WHO Dengue Bull. 1999;23:55–66. [Google Scholar]
- Weidmann M, Faye O, Kranaster R, Marx A, Nunes MR, Vasconcelos PF, Hufert FT, Sall AA, Yeh JY, Lee JH, Seo HJ, Park JY, Moon JS, Cho IS, Lee JB, Park SY, Song CS, Choi IS. Improved LNA probe-based assay for the detection of African and South American yellow fever virus strains Fast duplex one-step reverse transcriptase PCR for rapid differential detection of West Nile and Japanese encephalitis viruses. J Clin Virol. 2010;48:187–92. doi: 10.1016/j.jcv.2010.04.013. [DOI] [PubMed] [Google Scholar]
- WHO Yellow Fever. 2010 [Google Scholar]
- Yeh JY, Lee JH, Seo HJ, Park JY, Moon JS, Cho IS, Lee JB, Park SY, Song CS, Choi IS. Fast duplex one-step reverse transcriptase PCR for rapid differential detection of West Nile and Japanese encephalitis viruses. J Clin Microbiol. 2010;48:4010–4. doi: 10.1128/JCM.00582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39:561–77. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.