Abstract
Due to their advantageous materials properties, noble metal nanoparticles are versatile tools in biosensing and imaging. A characteristic feature of gold and silver nanoparticles is their ability to sustain localized surface plasmons that provide both large optical cross-sections and extraordinary photophysical stability. Plasmon Coupling Microscopy takes advantage of the beneficial optical properties and utilizes electromagnetic near-field coupling between individual noble metal nanoparticle labels to resolve subdiffraction limit separations in an all-optical fashion. This Tutorial provides an introduction into the physical concepts underlying distance dependent plasmon coupling, discusses potential experimental implementations of Plasmon Coupling Microscopy, and reviews applications in the area of biosensing and imaging.
1. Introduction – Noble Metal Nanoparticles as Labels in Optical Microscopy
Most cellular processes are complex and require interactions between multiple components well-orchestrated in space and time to generate a desired outcome. It has long been recognized that optical microscopy is – in principle – the method of choice for deciphering the mechanisms of intrinsically dynamic processes in living cells. Monitoring the spatial distribution of selectively labeled components as function of time represents a powerful approach to determine which components interact how in what sequence. The localization accuracy for an individual emitter in optical microscopy is determined by the uncertainty, σ, with which its point-spread-function (PSF), or “image” can be localized in a selected plane:1
| (1) |
where s is the width of the intensity distribution, N the number of collected photons, a the pixel size of the camera, and b the standard deviation of the background. Bright labels in a low background allow for a high localization accuracy for individual emitters and localization precisions of a few nm have been realized. Light emitted by multiple sources interferes, however, so that the wave nature of light sets strict limitations to the level of detail with which multiple labels with identical emission wavelength can be resolved in a conventional light microscope. The lateral spatial resolution of an optical system is given as ~ 0.6λ/NA, where λ is the wavelength of the light and NA is the numerical aperture. In the visible range of the electromagnetic spectrum the diffraction limit is on the order of a few hundreds of nanometers. Unfortunately, this resolution is insufficient to access any molecular details of cellular processes. Dynamic molecular rulers, based on Fluorescence Resonance Energy Transfer (FRET), have traditionally been used to circumvent the diffraction limit in fluorescence microscopy and to monitor the separations between fluorescently labeled species on the order of 1–10 nm.2 Recently true subdiffraction-limit optical imaging with spatial resolution down to ~20 nm has been made possible by fluorescence “nanoscopies” that achieve the separate localization of individual labels at different points in time by switching the dye labels between on- and off-states.3 Fluorescence nanoscopies – like fluorescence microscopy in general – suffer, however, from the limited photophysical stability of organic dyes. After photoexcitation, dyes show a higher reactivity and can undergo a chemical transformation that results in an irreversible loss of fluorescence.2 Photobleaching fundamentally limits the ability to monitor cellular dynamics continuously over extended periods of time with high temporal resolution and, thus, motivates the further improvement of fluorescent labels as well as the development of alternative non-fluorescence based approaches that are not subject to any constraints in observation time.
Due to their large contrast in electron microscopy, Au and Ag nanoparticles (NPs) have long been used as high-contrast labels in electron microscopy.4 But noble metal NPs also have exquisite optical properties and can be imaged in optical scattering or photothermal microscopy without any blinking or bleaching and with no physical limitation in observation time. The time varying electric (E-) field component of light incident on a NP induces a displacement of the conduction band electrons and results in an oscillating charge pile-up on the NP surface (Figure 1a). At the localized surface plasmon (LSP) resonance (LSPR) frequency the surface charge density oscillations form a standing wave so that even weak incident E-fields achieve large charge accumulations. For Au and Ag NPs, the LSPRs lie in the visible range of the electromagnetic spectrum. Since the polarizability, α, of a NP is resonantly enhanced at the LSPR,5 scattering and absorption cross-sections become maximum at this frequency. The strong charge concentration in a small volume on the surface of the NP creates a strong local E-field enhancement at the LSPR, which is of great interest for many non-linear spectroscopies.
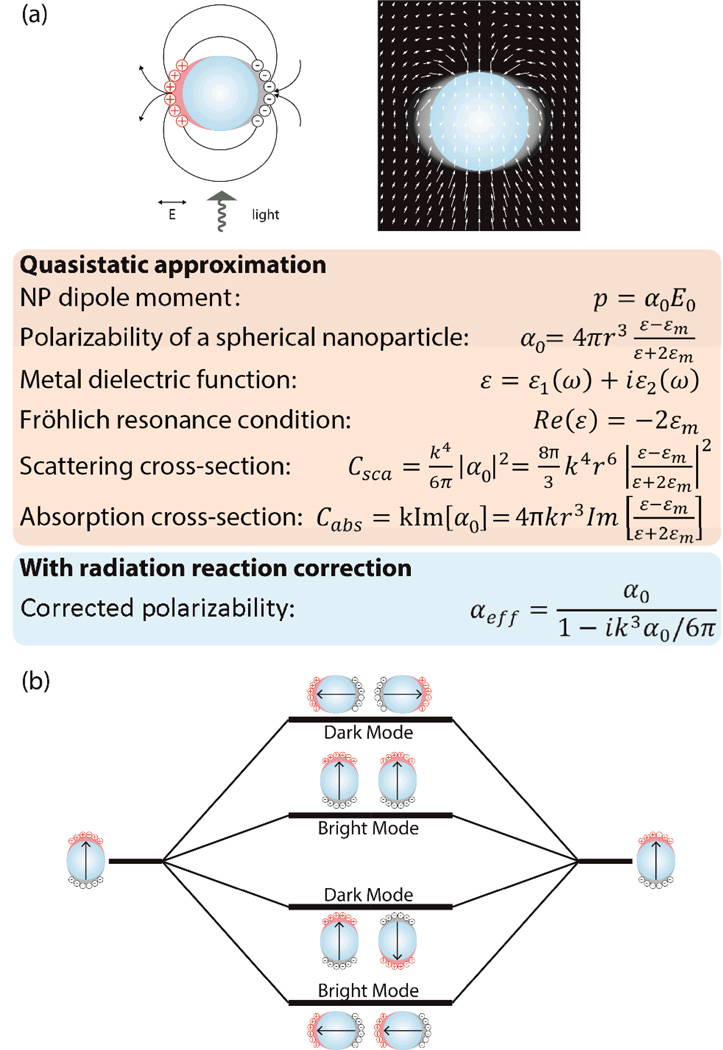
Figure 1.
a) Left: Light incident onto a metal NP creates charge density oscillations on the surface. Right: A plot of the optical powerflow around a noble metal NP at the LSPR frequency shows a rerouting of the incident light towards the NP. Red box: key formulas determining the resonance behavior of the metal NP in the quasistatic approximation (wavenumber k = 2π/λ; εm: dielectric constant of ambient medium). Blue box: corrected effective polarizability including radiation reaction. Adapted with permission from Boriskina and Reinhard Nanoscale, 4, 76 (2012). Copyright 2012 The Royal Society of Chemistry. b) Plasmon hybridization between strongly coupled NPs leads to formation of dark and bright modes, depending on the relative orientation of the NP dipoles (indicated as arrow).
For particles that are much smaller than the wavelength of the incident light it is justified to assume that the phase of the oscillating E-field does not change substantially across the NP diameter, i.e. that the quasistatic approximation applies. Under these conditions the scattering and absorption cross-sections of noble metal NPs, Csca and Cabs, scale as r6 and r3, respectively, where r is the radius of the NP (Figure 1a).5 We note, however, that the polarizability, α0, in Figure 1a does not account for radiation damping, γ, due to light scattering. To avoid a violation of energy conservation, it is therefore necessary to augment the quasistatic approximation with a radiation reaction term, which leads to the corrected polarizability, αeff.6 Light absorption dominates in NPs with very small sizes, but the scattering cross-section increases steeply with size. Individual NPs with a radius of r ≥ 15 nm have sufficiently large scattering cross-sections to facilitate an uncomplicated detection of individual particles in darkfield or total internal reflection microscopy. Both microscopies use excitation geometries that eliminate the excitation beam from detection and, thus, make it possible to selectively collect light that is scattered from the NPs into the direction of the objective. A 40 nm Au NP has a scattering cross-section of ≈6.0×10−12 cm2 at its resonance wavelength of 535 nm, which compares with ≈0.5×10−16 cm2 for a conventional fluorescent dye.7 Even if one takes into account the larger size of the NP and corrects for the fact that one NP corresponds to approximately 1600 dye molecules, the NP still shows a larger cross-section by a factor of ≈75. Considering equation 1, it is immediately clear that one other advantage of NPs is the possibility to collect a large number of photons per time unit, which is beneficial for an accurate localization of the tracked species even at short acquisition times. While the size of optical NP labels is a disadvantage when compared with smaller organic dyes, their extraordinary signal stability and brightness, as well as their field enhancing properties and their multimodal character makes them versatile probes for processes that either directly involve NPs or that occur on length scales of a few tens of nanometers and which can, therefore, tolerate their size. Although the diffraction limit applies to metallic NPs in the same way as for fluorescent labels, oscillating charge distributions in individual NPs create time-dependent electromagnetic fields, which act on the conduction band electrons of other near-by NPs. These fields can then act back on the conduction band electrons in the original NP. Due to the sum of all of these electromagnetic “near-field” interactions, the oscillations in the individual NPs are no-longer independent but coupled. This near-field coupling can be conveniently detected as a shift in the scattering spectra of the NP labels using conventional far-field light microscopy.8 The fundamental concept of plasmon coupling and its application for optically resolving subdiffraction limit separations in a cellular context is the focus of this Tutorial.
The manuscript is structured as follows, in section 2 we will review the fundamental electromagnetic mechanisms underlying plasmon coupling in discrete dimers of noble metal NPs, in section 3 we will review the influence of the configuration in larger clusters. In section 4 we will review the experimental implementation of Plasmon Coupling Microscopy (PCM), and in section 5 we will discuss applications in cellular imaging.
2. Distance Dependent Plasmon Coupling: The Color of Plasmon Rulers
We will start our analysis of the collective resonances in NP clusters with the simplest possible case, the dimer. NP dimers have two natural symmetry axes along which plasmon coupling can occur (Figure 1b). Depending on the polarization of the incident light, the electron density oscillations occur along the long axis (longitudinal mode) or perpendicular to it (two degenerate transverse modes). As we will discuss in more detail below, the treatment of the individual NPs as simple dipoles, p, with p = αE0, where α is the NP polarizability and E0 the incident E-field, is insufficient to describe plasmon coupling quantitatively correct. Nevertheless, the simplified model is still of great value for conceptualizing the relevant electromagnetic coupling mechanisms. In a dimer the two dipoles representing the charge distributions of the individual NPs can be oriented in a parallel or antiparallel fashion. Only the parallel orientations provide “bright” modes for both longitudinal and transverse mode, whereas the antiparallel combinations have no net dipole moment and can, therefore, not couple to the incident light. These modes remain “dark”.9 This coupling model is adequate for small NP sizes, but we point out that in dimers of larger particles modes that are predicted to be dark by this quasistatic model can become bright.10 For the relatively small (20 nm – 60 nm) NPs of interest in most biological contexts, the quasistatic approximation is acceptable and we limit our discussion in the following only to the bright modes, since those will be directly relevant for the optical studies covered in this Tutorial.
Light-induced displacements of conduction band electrons in metal NPs give rise to charge accumulations of opposite sign on opposing surfaces of the particles (Figure 1a). Assuming a semi-classical oscillator model for the plasmon, the restoring force acting on the displacement charge determines the plasmon eigenfrequency of an individual NP. In a dimer these forces can be modulated by electromagnetic interactions between the surface charges on two adjacent NPs.11 In case of the longitudinal dimer mode the individual NP dipoles are arranged in a head-to-tail orientation, whereas in the transverse mode they show a side-by-side orientation (Figure 1b). The dipole orientation for the longitudinal mode creates charge accumulations of opposite signs on both sides of the gap, and the accompanying effective charge screening reduces the net restoring force within the individual NPs. Consequently, the plasmon eigenfrequency of the dimer is reduced relative to that of an individual isolated NP. The reduction in eigenfrequency is equivalent to a spectral red-shift. In contrast, for the bright transverse mode the charge distribution enhances the restoring force and, therefore, results in a spectral blue-shift. We note in passing that the longitudinal plasmon mode has a much higher scattering cross-section than the more weakly coupled transverse modes and, therefore, dominates the dimer spectrum under unpolarized whitelight excitation.
The exact resonance wavelength of a NP or a NP assembly depends also on the refractive index, nr (=εm1/2), of the ambient medium (see Fröhlich resonance condition in Figure 1a).12 In the following we will assume that strong electromagnetic coupling between nanoparticles dominates the spectral NP response. This assumption is justified since the polymer coating assembled on the NP label surface stabilizes the refractive index in the immediate vicinity of the NPs and since refractive index fluctuations in cellular systems – the target application of PCM – are on the order of a few hundredths of refractive index units at most. Changes of this magnitude lead to variations in the resonance wavelength that are relatively modest when compared to those associated with changes of the interparticle separation in the strong electromagnetic coupling regime.13
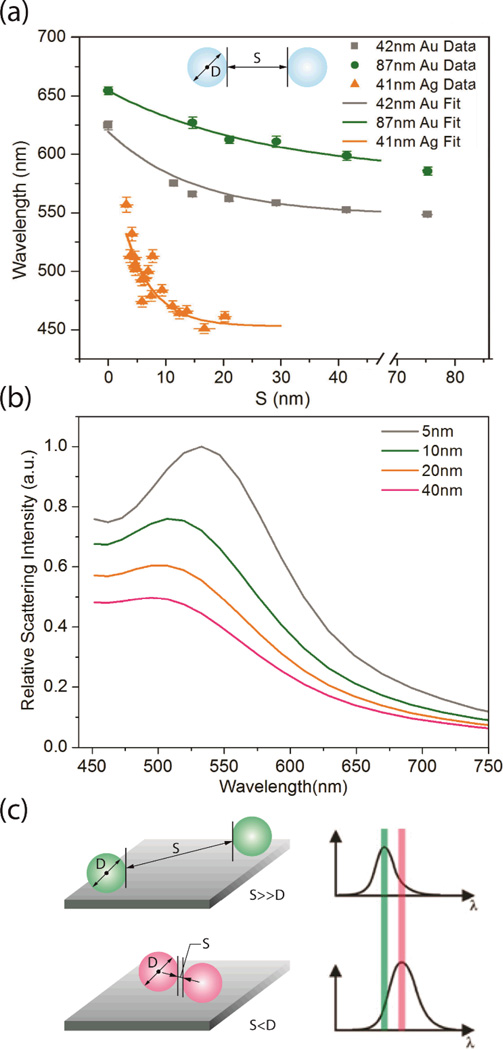
Direct experimental demonstration of the strong distance dependence of plasmon coupling was provided by the spectroscopic characterization of electron beam lithography (EBL) generated NP dimers11,14 and chains of Au NPs8. These and other studies confirmed a monotonic red-shift of the longitudinal plasmon mode with decreasing interparticle separation for strongly coupled NPs. The correlation between plasmon resonance wavelength and interparticle separation obtained with lithographically defined NP dimers forms the foundation for NP based molecular ruler technologies. So-called “Plasmon Rulers” utilize the interparticle separation dependent spectral response of NP dimers to monitor distances and / or distance changes between biopolymer linked NPs at constant refractive index.15–19 Experimental λplas vs. interparticle separation (S) calibration curves have been obtained for DNA tethered dimers assembled from D ≈ 40 nm diameter Au and Ag NPs and for D ≈ 87 nm Au NPs under unpolarized whitelight illumination (Figure 2a). The calibration curves confirm a red-shift of the resonance wavelength as function of increasing average interparticle separation in the flexible linked dimers. This spectral red-shift of the Plasmon Rulers is accompanied by an increase in scattering intensity, as illustrated in the finite-difference time domain (FDTD) simulation for 40 nm Au dimers in Figure 2b.
Figure 2.
a) Experimental calibration curves for 42 nm and 87 nm Au Plasmon Rulers and 41 nm Ag Plasmon Ruler. Adapted with permission from Reinhard et al., Nano Lett., 5, 2246 (2005) and Yang et al., J.Phys.Chem. C 114, 4901 (2010). Copyright 2005 and 2010 American Chemical Society. b) FDTD-simulated scattering spectra for 40 nm Au dimer as function of interparticle separation. c) Principle of Plasmon Coupling Microscopy (PCM): Close contacts between NPs are indicated by a spectral red-shift.
Fluctuations in the size and shape of the NPs used to assemble Plasmon Rulers affect the dimer resonance wavelengths and give rise to some error in the λplas(S) relationship. For a quantitative analysis please refer to ref. 16. This “heterogenous” broadening of the calibration relationship does, however, not impair the ability to detect even very small changes in the interparticle separation of continuously monitored individual Plasmon Rulers. The ability to monitor interparticle separations with Plasmon Rulers has been utilized in biochemical assays, for instance, for detecting i.) DNA hybridization,15,17 ii.) DNA bending upon restriction (EcoRV) enzyme binding,20 iii.) DNA and RNA condensation and cleavage,21,22 or iv.) to image caspase-3 activation23. Other technological applications involved the programmed reversible switching of interparticle separations.24 Most of these assays utilized the large scattering cross-sections of the noble metal NPs and were performed in a parallel fashion, monitoring many individual Plasmon Rulers at the same time. The thus obtained throughput enables the identification of different sub-populations in one ensemble, which was successfully exploited to reveal different transiently stabilized configurations in RNA Plasmon Rulers.21
The experimental Plasmon Ruler calibration curves in Figure 2a confirm that both Au and Ag Plasmon Rulers allow distance measurements of up to at least 1/2 NP diameter and that the dynamic range of the Plasmon Rulers can be tuned through the choice of the size and chemical composition of the NPs used to assemble the dimers. Due to a steeper slope in their λplas vs. S calibration curve, Ag Plasmon Rulers provide a higher sensitivity for detecting small distance changes. Interestingly, the experimentally observed λplas(S) behaviors for all implemented Plasmon Rulers deviate from the expected S−3 dependence for purely dipolar interactions and are, instead, well described by single exponentials. Similar observations were obtained in a series of studies performed with Au or Ag nanodisks nanofabricated on solid supports.14,25,26 The deviation from the dipolar coupling mechanism in the experimental studies has been attributed to a highly inhomogeneous field distribution between the two NPs at short separations.27 The inclusion of plasmon modes of higher order angular momenta (quadrupole etc.) is crucial for an accurate description of the distance dependent plasmon coupling between two strongly coupled NPs, even for relatively small NPs that – as individual particles – are accurately described by the dipole approximation. In strongly coupled dimers the dipolar modes of one NP can couple with higher order angular modes of the second NP, resulting in hybrid modes with contributions from many different angular momenta.9,28 The sum over all of these interactions leads then to an analytical expression that is well approximated by an exponential function.27 For particles with diameters in the range between 20–70 nm it was shown that the ratio of spectral shift, Δλplas, to individual NP resonance wavelength, λ0, is to a first approximation independent of the NP diameter and follows a universal scaling relationship.25 For Au NPs, the universal scaling relationship as function of interparticle separation, S, and particle diameter, D, was determined as:
| (2) |
The Au NP calibration curves in Figure 2a were derived by measuring λplas for DNA assembled from DNA spacers with known number of base pairs and converting the number of DNA spacers into an effective length using a statistical polymer model (worm-like-chain model).16 The Ag Plasmon Ruler calibration curve was obtained by correlating single dimer optical spectroscopy with high resolution transmission electron microscopy (TEM) of surface immobilized Plasmon Rulers.18 The latter approach made it possible to explore plasmon coupling even at very short separations S ≤ 2 nm. These studies revealed that – at least in the case of the Ag Plasmon Rulers – the relative magnitude of the spectral red-shift does not continue to increase at these very short separations. This observation was discussed in the context of quantum plasmonic29 and/or non-local30 electromagnetic effects.18 A similar effect was also observed in one-dimensional clusters of Au NPs assembled via a template guided self-assembly strategy31 and in Ag NP dimers manipulated in a scanning transmission electron microscope32. Although the ability to access the quantum plasmonic regime through chemical assembly strategies will certainly open new vistas, for applications of noble metal NPs in bioimaging, the assumption of classical electromagnetic coupling behavior is justified. The ligand shells required to stabilize NPs in physiological buffers renders the quantum plasmonic regime inaccessible in solution. Consequently, Figure 2a shows the Ag NP plasmon coupling behavior only for the classic electromagnetic coupling regime with S > 2 nm. This classical plasmon coupling behaviour forms the foundation for resolving subdiffraction limit contacts as spectral shift in PCM (Figure 2c).
3. Plasmon Coupling in Clusters: The Effect of the Cluster Configuration
Many-body interactions are of great interest in key biological processes such as receptor clustering in two dimensions (vide infra), and the ability to resolve these interactions with PCM requires a fundamental understanding of plasmon coupling in clusters. In clusters containing more than two NPs, the measured resonance wavelength is no longer determined by the interparticle separation alone but also depends on the number of NPs in the cluster and their geometric arrangement.33,34 In general, the optical properties of NP clusters are strongly size dependent for small cluster sizes but converge in larger clusters. In one- and two-dimensional clusters of close-packed 40 nm diameter Au NPs, a strong red-shift of the peak plasmon resonance was observed with growing size for cluster sizes n≤4 (at constant interparticle separation).31,33 A further increase in cluster size resulted only in small incremental changes of the resonance wavelength. Electromagnetic simulations and experimental SERS spectra for these systems indicated a similar convergence for the E-field intensity. We emphasize that sharp far-field resonances and strong E-field enhancement can be engineered in cluster sizes n > 4 for special configurations that sustain collective cluster resonances, such as Fano resonances.34 These specific structures are, however, of minor relevance in NP labeling experiments due to their stringent symmetry requirements.
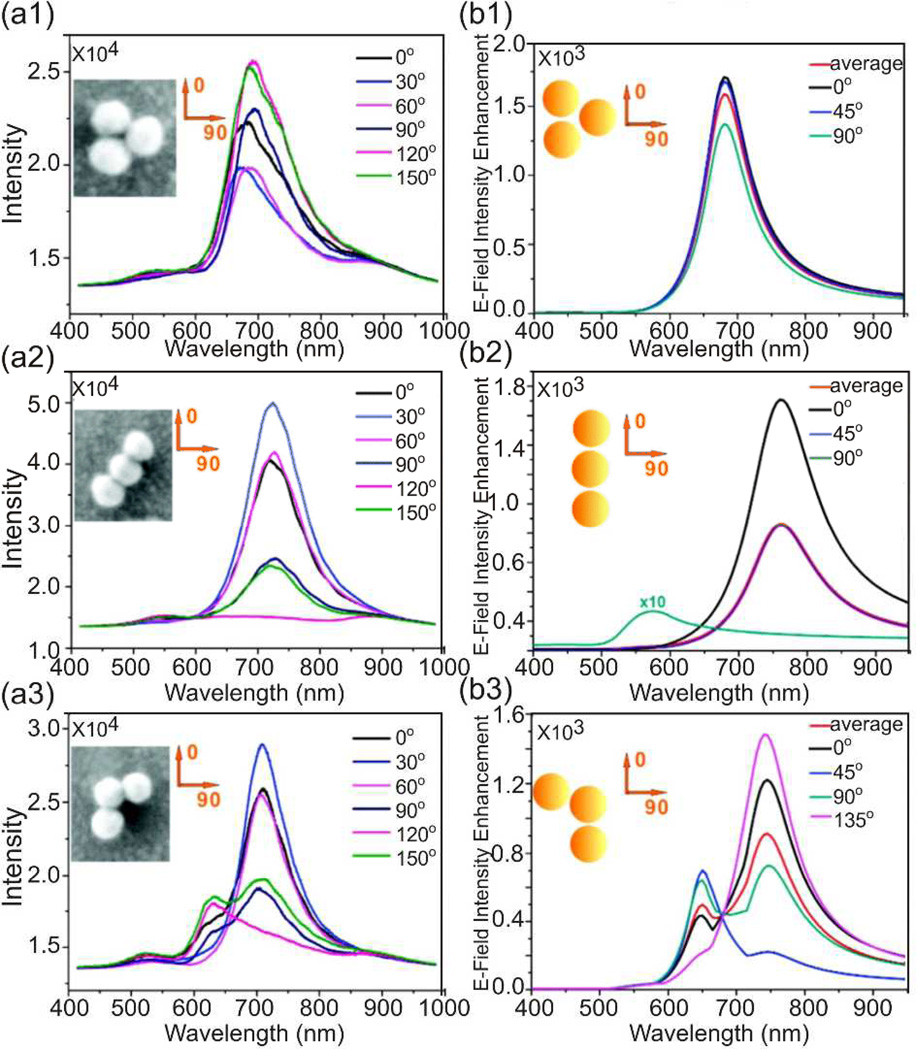
The light polarizing properties of anisotropic NP clusters are of high practical value since they provide additional information about the geometry of NP clusters in the optical microscope.33 This is illustrated for NP trimers with different geometries (D3h, D∞h, C2v) in Figure 3. The figure contains experimental scattering spectra and simulated near-field spectra for different light polarizations. Both the near- and far-field spectra of the highly symmetric D3h configuration show only a weak polarization dependence, whereas the optical response of the anisotropic D∞h configuration, the rod-like cluster, is strongly polarization dependent. Scattering and near-field intensities are much stronger for a light polarization pointing along the long trimer axis. The spectra of the C2v configuration contain two clearly distinct spectral features, whose relative intensities can be modulated by rotation of the light polarization. Overall, Figure 3 illustrates vividly the impact of the geometric cluster configuration on near-and far-field responses and the value of polarization-resolved scattering spectroscopy to retrieve structural information
Figure 3.
Experimental scattering spectra from a1) D3h, a2) D∞h, and a3) C2v trimers for different polarization angles. b1–b3) Simulated E-field intensity enhancement spectra (|E|2/|E0|2) of the same structures. Adopted with permission from Yan et al., J. Phys. Chem. C 115, 4578 (2011). Copyright 2011 American Chemical Society.
4. Implementation of Plasmon Coupling Microscopy (PCM)
After introducing the fundamental concepts of plasmon coupling in noble metal NP dimers and clusters in the previous sections, we will in this section introduce different experimental strategies for its utilization in biosensing and imaging. The spectral and/or light polarization properties collected from a diffraction limited spot in an optical microscope depend on the near-field coupling between the NP labels in this area. The magnitude of the coupling is sensitive to nanoscale interparticle separations and the NP association level. Spatial maps of plasmon coupling, consequently, encode information about the average separation of NP labeled species as function of location, and the strategy of PCM is to collect these information to provide insight into subdiffraction limit separations and separation changes. As we will outline in more detail below, the spectral and/or polarization properties of NP labels can be monitored in conventional widefield microscopy, which makes it possible, for instance, to track plasmon coupling between laterally diffusing NPs35 or to map the local degree of NP clustering simultaneously at many locations across the entire cell surface36, 37. Due to the superb photophysical stability of the NP labels, optical PCM studies can be performed continuously without limitation in observation time. The latter is a great advantage for the investigation of stochastic processes that cannot be induced at defined time points or for slow biological processes. When designing a PCM experiment, one also has to be aware of the limitations of the technique. Since PCM is an elastic light scattering based microscopy, it is primarily intended for cellular studies with low background. In vivo or tissue imaging is difficult to realize due to the intrinsically high scattering background of biological tissue. Before we discuss different experimental strategies that have been applied to quantify plasmon coupling in optical microscopy, we will first review appropriate labeling strategies that allow for a selective tagging of the moiety of interest with an Au or Ag NP.
4.1 NP Immunolabeling
Bioconjugation of NPs is an active research area and a review of all functionalization strategies developed thus far goes beyond the scope of this Tutorial. We refer the interested reader to the excellent review in ref. 38 and focus in the following on functionalizing strategies that we have tested in our Laboratory, instead. Since the analysis of the plasmon resonance wavelength as indicator for spatial clustering, for instance, of receptors in the cell membrane, is only useful when the NP labels are stable and do not show spontaneous self-association in salt buffers, one particular challenge in PCM is the development of NP labels that combine high colloidal stability with high selective binding efficacy for their target. We will provide two examples of labeling strategies that have been demonstrated to fulfill these requirements.
4.1.1. Secondary Antibody Assisted NP Labeling
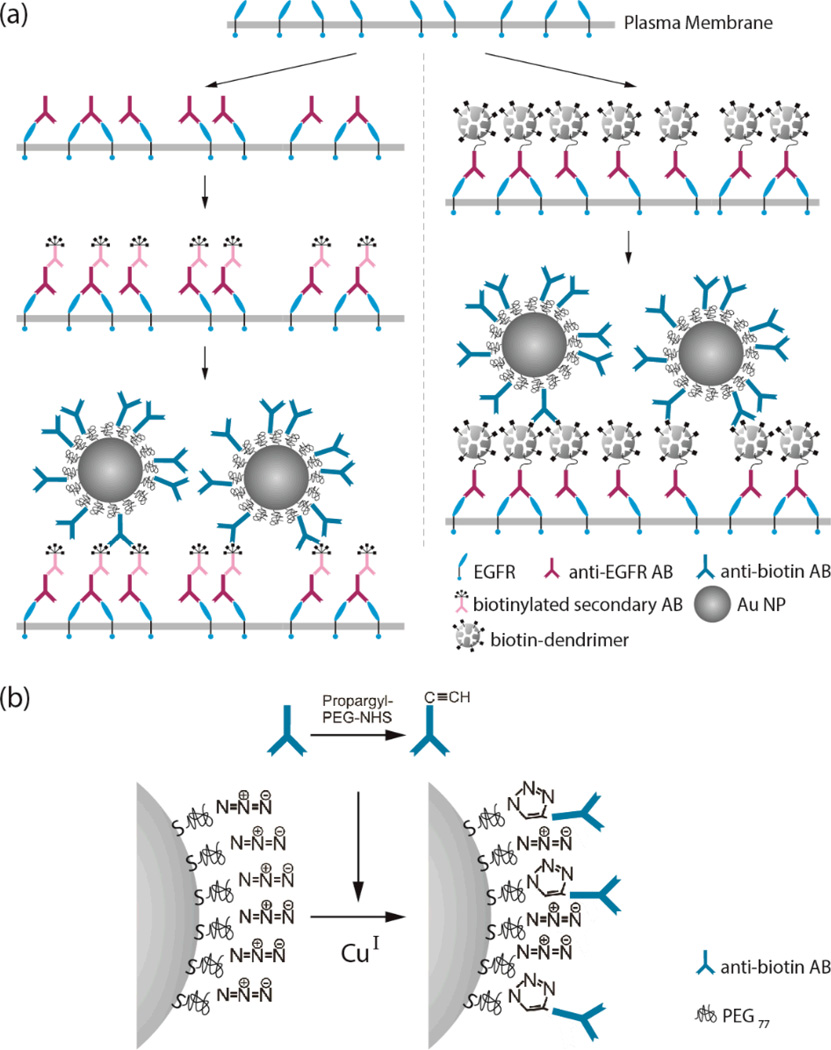
Au NPs with a diameter of 40 nm are frequently used labels in PCM since they provide a good compromise between signal intensity and size. Mammalian cells are coated by a polymeric matrix, which is referred to as glycocalyx. Overcoming this coating can represent a significant challenge for achieving an efficient labeling of cell surface receptors and other functionalities with ~40 nm diameter labels. The NP immunolabeling strategies reviewed in this and the following sections have been demonstrated to achieve a successful targeting of cell surface species with NP labels.39, 40 The left column in Figure 4a outlines a secondary antibody assisted labeling strategy, which was originally developed for the labeling of the epidermal growth factor receptor (EGFR or ErbB1) in the plasma membrane of in vitro cancer cells.39 The combination of primary and secondary antibodies on the cell surface in the secondary antibody assisted labeling procedure creates a spacer that make the targets more accessible for the colloidal NP labels. The binding affinity of the NP label is further increased by multiple biotins bound per secondary antibody. The NPs contain a self-assembled brush of 3.4 kDa polyethylene glycol (PEG) molecules with a thiol terminus to bind to the metal surface and a terminal azide to conjugate to alkyne functionalized anti-biotin antibody or Neutravidin through a CuI catalyzed 1,3-dipolar cycloaddition (Figure 4b).39 We emphasize in passing that the azide-PEG based NP functionalization strategy is not limited to antibodies, but can be expanded to peptides (growth factors etc.), glycopeptides, and other biomolecules of interest.
Figure 4.
a) Secondary antibody (AB) assisted NP labeling strategy (left) and dendrimer enhanced binding approach (right). b) Covalent attachment of antibodies or Neutravidin through 1,3-dipolor cycloaddition. Adopted with permission from Wang et al., ACS Nano 5, 6619 (2011). Copyright 2011 American Chemical Society and Rong and Reinhard, PLoS ONE, e34175 (2012).
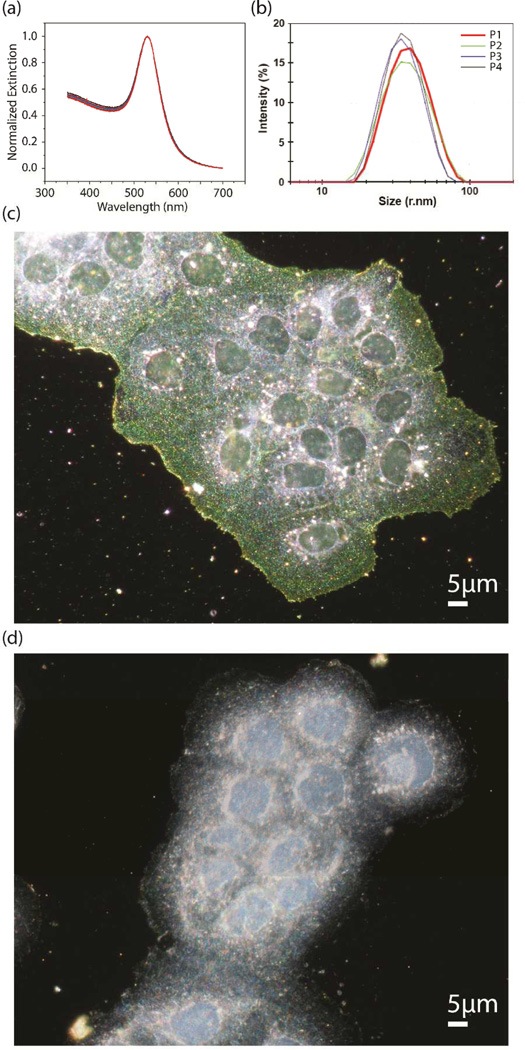
The PEG molecules on the NP surface do not only allow for a facile functionalization of the particle, but they also achieve an efficient steric stabilization of the NPs. UV-Vis spectroscopy (Figure 5a) and dynamic light scattering (DLS) (Figure 5b) have confirmed the stability of the NP immunolabels when incubated with fixed cells in 0.5× phosphate buffered saline (PBS) or living cell in 1× Hank’s buffer.39, 40 The specificity of the labels was validated in a control experiment, in which the cells were incubated without (Figure 5c) or with (Figure 5d) free anti-biotin antibody. The efficient competitive inhibition confirms that NP immunolabel binding is target specific.
Figure 5.
a) UV-Vis spectra of antibody functionalized 40 nm Au NPs in 0.5× PBS recorded over a time interval of 225 min (1 spectum every 15 min) and b) hydrodynamic radii determined by DLS measured before and after incubation of 40 nm Au NPs with living cells in 1X PBS buffer (P1/P2: Neutravidin functionalized NPs before/after incubation with cells; P3/P4: Antibody functionalized NPs before/after incubation with cells). c) Darkfield image of A431 cells labeled with 40 nm Au NPs using a secondary antibody assisted labeling strategy d) Darkfield image of A431 cells labeled under identical conditions in the presence of an excess of free anti-biotin antibody (competitive inhibition). The biotin binding sites are blocked by free anti-biotin antibody from solution, preventing Au NP immunolabeling. Adopted with permission from Wang et al., ACS Nano 5, 6619 (2011) and Yu et al., Anal. Chem. 85, 1290 (2013). Copyright 2011 and 2013 American Chemical Society.
4.1.2. Dendrimer Assisted NP Labeling
The dendrimer assisted labeling strategy outlined in the right column of Figure 4a has been shown to be effective under live cell conditions for which extensive washing or NP co-incubation are not permissible.40 Instead of a combination of primary and secondary biotinylated antibody, this scheme uses primary antibodies that are covalently connected to a biotinylated dendrimer through a flexible linker. Small, but highly branched dendrimers introduce multiple biotins per targeted receptor that are easily accessible and, thus, increase the binding affinity of the NP labels. The Au NP labels can be generated through the synthetic strategies outlined in the previous section.
4.2 Optical Approaches for Quantifying Plasmon Coupling
Different optical techniques can be applied to characterize the spectral properties of NPs spatially resolved with two figures of merits determining the details of the instrumental implementation: temporal and spectral resolution. In most practical realizations one has to compromise between these two figures of merits. In sections 4.2.1–4.2.3 we will review different PCM realizations in the sequence of decreasing spectral but increasing temporal resolution. In section 4.2.4 we will then show how spectrum-based approaches can be augmented to yield light polarization information, as well.
4.2.1 Full Spectral Analysis
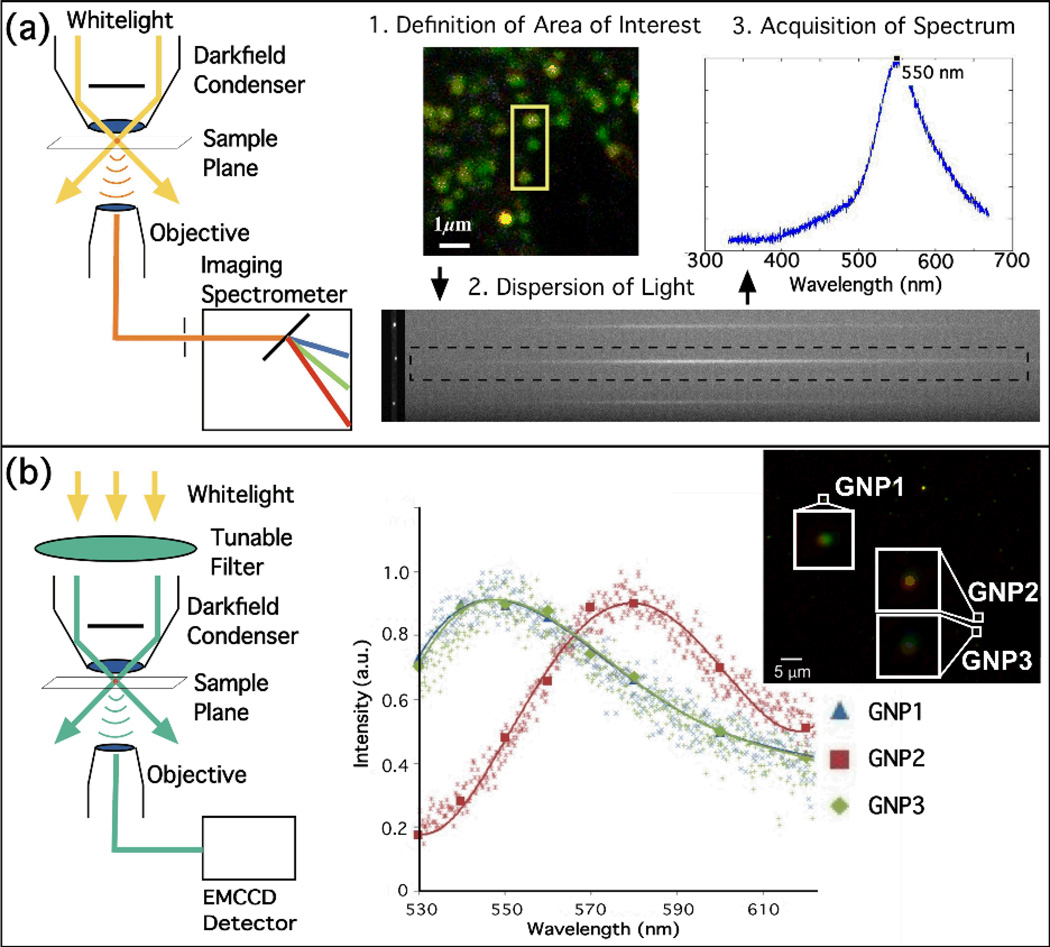
A typical experimental set-up is shown in Figure 6a.39 A spectrometer attached to a darkfield microscope is used to analyze the light scattered from a defined area in the sample plane. The opening of the entrance slit of the spectrometer is adjustable along the horizontal axis and the area of interest along the vertical axis can be controlled via software control. The collected light is then dispersed and collected on an electron multiplying charge-coupled device (EMCCD). While the method provides detailed spectroscopic information of specific locations, it is not suitable for acquiring simultaneous spectral information from many different locations distributed across large areas with high temporal resolution or throughput. Furthermore, the spectral analysis is also not compatible with monitoring the spectra of moving NP labels.
Figure 6.
a) Spatially-resolved spectral analysis of individual NPs and NP clusters using an imaging spectrometer. b) Multispectral Imaging. Left: Set-up, right: comparison of scattering spectra for three Au NPs (GNP1-3) using an imaging spectrometer (small symbols) or multispectral imaging (large symbols and continuous lines).
4.2.2 Multispectral Imaging
To overcome some of the limitations of the full spectral analysis, multispectral imaging schemes have been implemented that compromise between spectral and temporal resolution and that allow for a simultaneous monitoring of plasmon coupling at many arbitrary locations in an extended area.36,41,42 One implementation is shown in Figure 6b, which is described in detail in ref. 41. In a conventional darkfield microscope the entire field of view is subsequently illuminated with different wavelengths and at each wavelength a monochromatic image is recorded on a camera. A rapid switching of the excitation wavelength can be achieved, for instance, using an intense light source in combination with a fast filter switching device or a tunable liquid crystal filter, or using a light emitting diode (LED) illumination array. After the individual images have been corrected for the intensity profile of the excitation source, the monochromatic images can be combined into a composite image, in which each pixel contains the information about the relative intensities on the monitored wavelength channels.41 Multispectral imaging provides spectral information of the entire field of view with reasonable acquisition rates; the collection of multiple complete multispectral images per second is currently routine and significantly higher acquisition rates are technically feasible.42 Figure 6b contains the spectra from multiple Au NPs in one field of view obtained through multispectral imaging as well as through full spectral analysis using an imaging spectrometer. The direct comparison confirms that multispectral imaging and full spectral analysis provide equivalent information. The monitored area in multispectral imaging can be even further increased if the widefield microscope use for data collection is replaced with a lensless imaging systems.43
4.2.3 Ratiometric Imaging
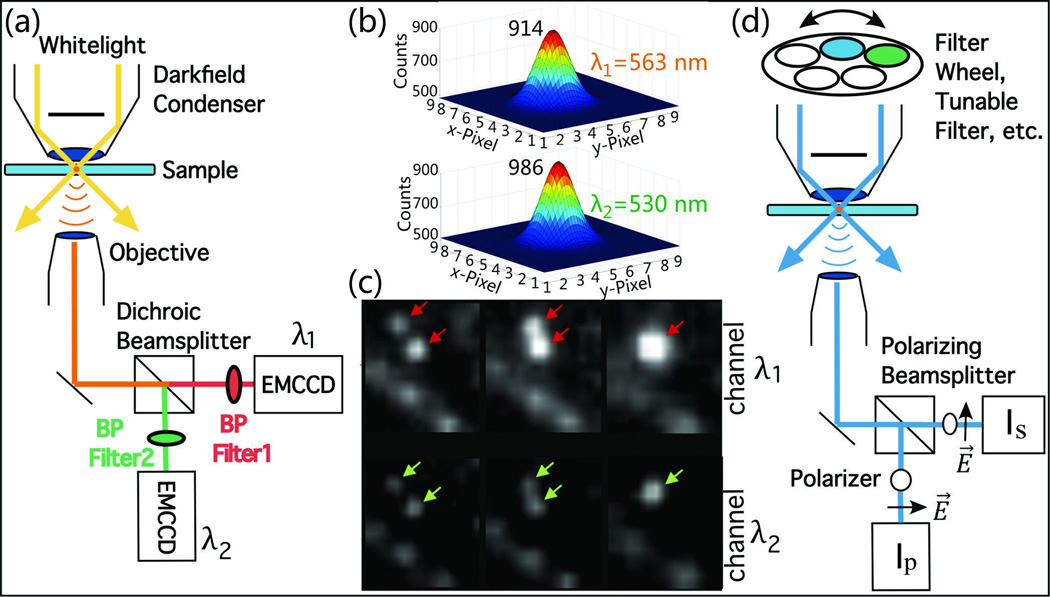
While multispectral imaging allows monitoring of plasmon coupling in a large field of view, the time delays between the individual monochromatic images used to calculate the composite image is problematic for imaging dynamic processes or tracking moving objects. The temporal delay between the individual channels can be avoided through ratiometric imaging approaches where wavelength selective (dichroic) beamsplitters in combination with bandpass filters are used to image the entire field of view simultaneously on multiple detection channels. The most common implementation of this technique for dual color imaging on two wavelength channels λ1 and λ2 is schematically illustrated in Figure 7a.35 The approach generates two images of the same field of view at two defined wavelengths (Figure 7b and c) simultaneously. Fitting the point-spread-functions (PSFs) of individual objects on the monitored wavelength channels, after correction for differences in the transmission properties of the filters and the excitation profile of the light source, provides the integrated intensities Iλ1(n) and Iλ2(n) for a frame n, that can then be used to calculate the intensity ratio, R(n):
| (4) |
Figure 7.
a) Experimental set-up for dual color ratiometric imaging. b) Fitted point-spread-functions (PSFs) for a single gold NPs simultaneously recorded on a 530 nm and 563 nm channel. c) Two Au NPs diffusing on a cell surface simultaneously tracked on two wavelength channels. d) Experimental set-up for polarization resolved PCM. Adapted with permission from Rong et al., Nano Lett. 8, 3386 (2008). Copyright 2008 American Chemical Society.
Although the spectral information encoded in R is limited, it does allow for a ratiometric detection of NP association.35 If the monitored wavelength channels are chosen so that λ1 > λ2, the formation of strongly coupled NPs results in an increase in R.
4.2.4 Polarization Resolved PCM
PCM can be augmented to record both spectral and polarization information. In polarization resolved PCM (PRPCM)22 the light scattered from individual NP labels is collected through an adequate objective and then split into two orthogonal polarization channels using a polarizing beam splitter (Figure 7d). The intensities Is and Ip on the two polarization channels in each frame n are used to calculate the degree of polarization, P, defined in this context as:
| (5) |
P describes the distribution of the detected light on two orthogonal polarization axes, whose direction is defined in lab coordinates. For anisotropic NP clusters with a strongly enhanced scattering cross-section along one direction, e.g. along the long axis of a dimer, P encodes direct information about the orientation of the object in the plane perpendicular to the axis of the objective. For quasi-one-dimensional plasmonic nanostructures, the intensity measured on one of the polarization channels is directly proportional to the square of the cosine of the angle θ, formed by the long particle axis and the respective polarization axis.44 For a dimer with a variable interparticle separation, P depends both on the interparticle separation and the dimer orientation in space.22 However, for a dimer performing a rotation in one plane, only fluctuations in the interparticle separation lead to a change in total scattering intensity, Itot = Is + Ip, which enables to decouple separation from orientation changes. Additional spectral information in PRPCM become available if the sample is illuminated with unpolarized light of alternating excitation wavelength λ1 and λ2 (Figure 5d).45 This can be accomplished with a rapid filter switching device, LED array, alternating laser illumination, etc. The spectral information can be quantified by calculating intensity ratios, R, from the total intensities obtained in two subsequent frames:
| (6) |
where n > 1. The recorded intensities need to be corrected for the different transmission properties and the excitation profile of the light source.
5. PCM in Cellular Imaging and Biosensing
In the following, we will review a number of studies in which PCM was applied to resolve subdiffraction limit contacts in a cellular context. We emphasize that we focus here primarily on case studies performed in our own Laboratory, but refer the interested reader also to the related work by Sokolov and coworkers46, Wax and coworkers13, or El-Sayed and coworkers47. Due to the size of the probes and its dynamic range, plasmon coupling is particularly useful to probe interactions that occur on length scales of a few tens of nanometers, which is of interest in the large scale association (clustering) of transmembrane proteins, membrane domain formation, etc.. Applications of PCM have, consequently, focused on probing the dynamic structures of the plasma membrane, the lateral spatio-temporal heterogeneity of specific cell surface proteins, and the uptake and subsequent trafficking of NPs. All of these applications benefitted from at least one of the advantages of the PCM technique, including its ability to monitor subdiffraction limit interactions between NP tagged species at many locations in a large field of view simultaneously, high temporal resolution, or the extraordinary photophysical stability of the probes which are entirely free of blinking or bleaching. Like with any label-based approach, one important point of consideration in PCM studies is the potential impact of the NP label on the biological system of interest. The use of NP labels is generally unproblematic for investigations using fixed cells or in living cells when the fate of the NPs itself is of interest. In other live cell studies a potential influence of the NPs needs to be evaluated on a case-by-case basis. Strategies to mitigate potential perturbations involve the development of small but bright monovalent NPs with inert surfaces. We also note that a potential perturbation of a ligand or receptor upon nanoconjugation is not bad per se. Instead, it can also provides new opportunities for controlling and manipulating cellular functionalities.48 A successful realization of NP-enabled cellular control requires, however, precise understanding of the underlying mechanisms, and PCM techniques are ideally suited to elucidate them.
5.1 Resolving Transient Interactions between Laterally Diffusing Receptors through Ratiometric NP Tracking
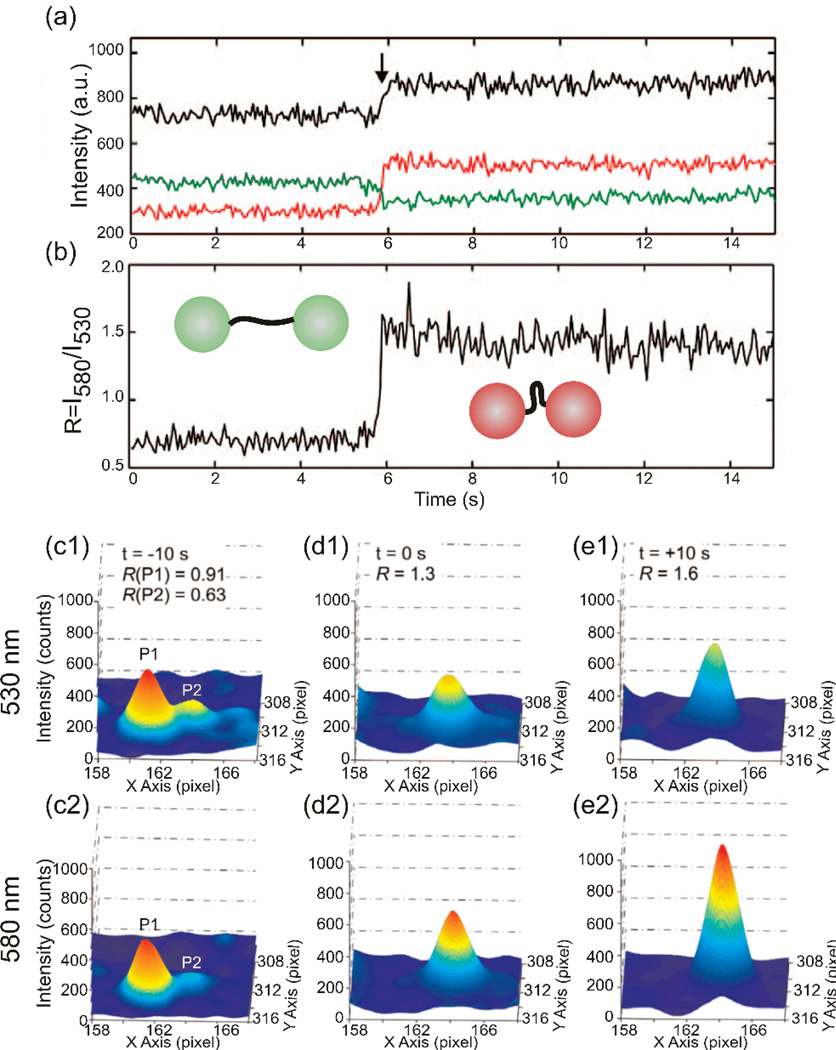
Many cellular processes involve transient interactions that are difficult to detect due to their short-lived nature and because they occur on deeply subdiffraction limit length scales. Although a multicolor tracking approach makes it possible to discern subdiffraction limit separations through spectral separation, the technique fails for identical labels. Consequently, a systematic investigation of interactions between multiple identical groups, as they occur for instance during self-association of proteins, such as ErbB1, in the cellular plasma membrane,49 remains challenging. The ratiometric PCM detection scheme described in section 4.2.3 provides now a potential alternative to overcome this challenge. Rong et al. tested the sensitivity of this approach for detecting the compaction of DNA tethers in Au NP plasmon rulers (Au NP diameter ~40 nm) as well as for detecting the conversion of NP dimers into monomers by enzymatic cleavage of the DNA tether.35 The DNA tethers were compacted from approx. 15 nm to 6 nm through addition of positively charged dendrimers that bind to the negatively charged DNA. According to the experimental Plasmon Ruler calibration relationship (Figure 2a), the resonance wavelength of Plasmon Rulers assembled from 40 nm increases from 568 nm to 621 nm if the interparticle separation is decreased from 15 to 6 nm. The resonance wavelength of an individual NP (dimer with infinite separation) is approximately 537 nm. Rong et al. performed the ratiometric imaging using a dichroic beam splitter with a cutoff wavelength of 560 nm and bandpass filters centered at 530 nm and 580 nm with a spectral width of 10 nm. Figure 8a shows the intensities on the chosen wavelength channels (I580 and I530) and the total intensity for a single Plasmon Ruler as function of time during a compaction event. Figure 8b contains the calculated intensity ratio R = I580 / I530. The induced compaction of the DNA tether is indicated by the sudden increase in total intensity. The intensity on the 580 nm channel increases, whereas the intensity on the 530 nm channel decreases. This anti-correlated intensity change and the resulting increase in R are unique indicators of a spectral red-shift.
Figure 8.
a) Scattering intensities I580nm (red) and I530nm (green), as well as the total intensity (black) of a single Au Plasmon Ruler recorded during its compaction. b) Intensity ratio R plotted as function of time. A spectral red-shift is indicated by an anti-correlated change of I580nm and I530nm and an increase in R. PSFs of two individual Au NPs, P1 and P2, tracked on a 530 nm channel (c1, d1, e1) and 580 nm channel (c2, d2, e2) at three different time points on the plasma membrane of a HeLa cell. The increase in intensity on the 580 nm channel after optical colocalization indicates a spectral red-shift due to strong electromagnetic coupling between P1 and P2. Reproduced with permission from Rong et al., Nano Lett. 8, 3386 (2008). Copyright 2008 American Chemical Society.
After validating the ratiometric detection, Rong et al. applied it to resolve nanometer contacts during optical colocalization of NP labeled integrins that performed a lateral diffusion on the dorsal surface of living HeLa cells.35 Some of the tracked Au NP labels approached each other until they were no longer distinguishable, and the duration of these optical colocalization events showed a broad range of variation. While short periods of colocalization can be coincidental, prolonged colocalization events indicate either a direct association of integrins or a confinement of the labeled integrins to a cell surface compartment with lateral dimensions below the resolution limit of the optical microscope. To differentiate between these two scenarios more information about the separation of the NP labeled integrins are required. This information is available through PCM. Figure 8c–e shows the point-spread-functions for two integrin tethered NPs on the two monitored wavelength channels at three different time points. In Figure 8c, the two NPs are still discernible and for both NPs the intensity is higher on the green than on the red channel, I530 > I580. In Figure 8d, the PSFs of P1 and P2 overlap and the NPs are no longer optically discernible. Concurrent with the optical colocalization, the relative intensity distribution on the two wavelength channels changes. The peak intensity is now higher on the 580 nm than on the 530 nm channel, resulting in an increase in R. In Figure 8e, both the total intensity and the R value reach their maximum. The observed strong spectral shifts in this example indicate short interparticles separations of below one NP diameter, which is consistent with the association of the integrins into a larger cluster.
5.2 Mapping Lateral Membrane Compartmentalization through PCM
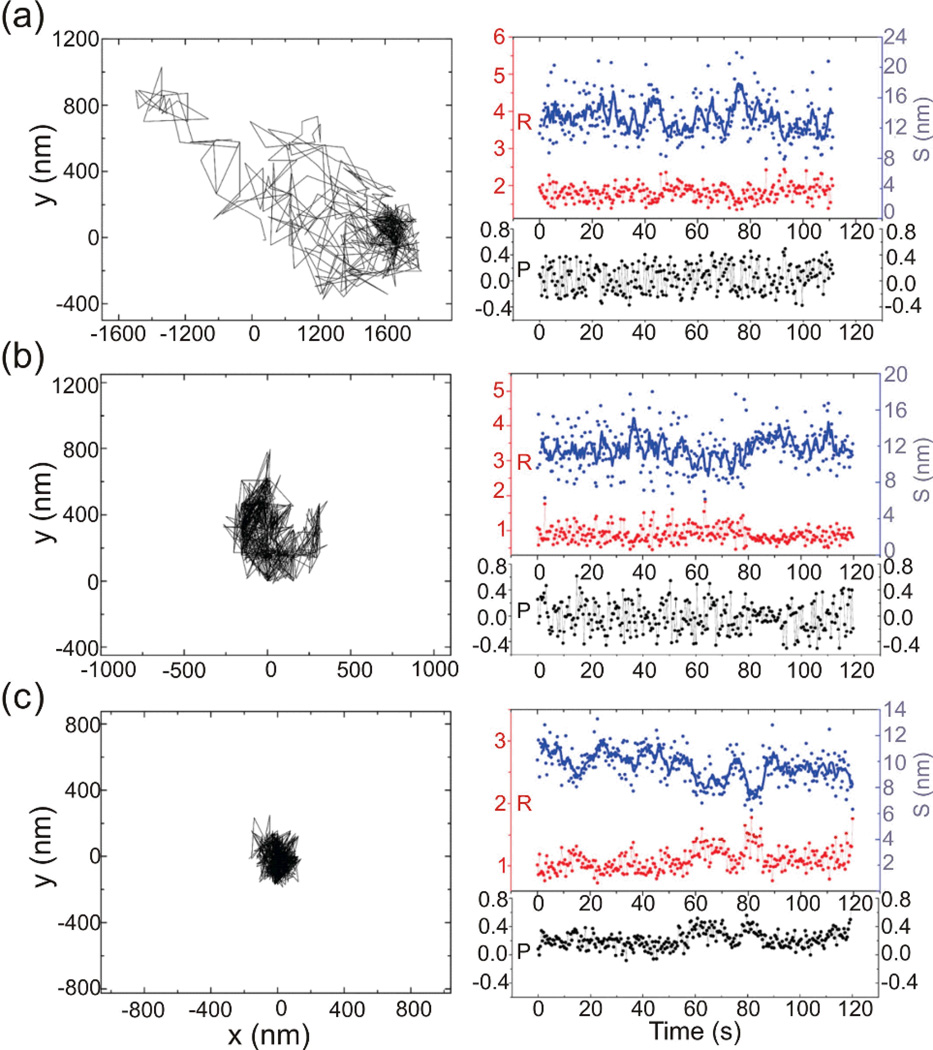
Diffusion studies of individual lipids and proteins in conventional high speed particle tracking have emerged as an important tool for probing the intrinsically dynamic structure of biological membranes and provided key insights into the structural compartmentalization of the plasma membrane.50 PCM augments these approaches now with the ability to simultaneously monitor the separation, S, between individual NP labels once they have approached each other to within approximately one NP diameter and are, thus, no longer optically distinguishable during their lateral diffusion on the plasma membrane. PRPCM provides additional information about the orientation of the long axis in strongly anisotropic clusters, such as NP dimers. This is illustrated in Figure 9, which contains the diffusion tracks and the corresponding P, R, and S values as function of time for three different Plasmon Rulers, PR1-3, as described in ref. 45. The Plasmon Rulers were assembled from ~30 nm diameter Ag NPs using a ~13.7 nm long DNA spacer resulting in an average center-to-center separation of ~44 nm. Under the chosen experimental conditions, the Plasmon Rulers attached spontaneously and non-reversibly to plasma membranes isolated from HeLa cells without the need for any specific binding chemistry.
Figure 9.
Tracking of three individual Ag Plasmon Rulers, PR1-3, on isolated plasma membranes through PRCPM. The Ag dimers were assembled through a DNA-programmed self-assembly strategy and were then monitored on plasma membranes of lysed HeLa cells. Left column: Plasmon Ruler trajectory. Right column: intensity ratios, R (red), degree of light polarization, P (black), and approximate interparticle separation, S (blue), as function of time. Reproduced with permission from Rong et al., Nano Lett. 10, 230 (2010). Copyright 2010 American Chemical Society.
The diffusion trajectory of PR1 was recorded using the PRPCM set-up outlined in Figure 7d. PR1 was tracked using a combination of bandpass filteres centered at 430 nm and 470 nm, PR2-3 were tracked with filters centered at 450 nm and 490 nm. Approximate S values were obtained by converting the R values into separations using an Ag Plasmon Ruler calibration.45 The spatial trajectories in the left column of Figure 9 show that the spatial confinement of the Plasmon Rulers increases from PR1 over PR2 to PR3. The R, S, and P trajectories in the right column were obtained with a frame rate of 5 images per second. The average interparticle separation Sav decreases from 13.4 ± 2.8 nm, over 11.7 ± 2.3 nm to 9.7 ± 1.3 nm for PR1-3.45 The decreasing interparticle separation and the accompanying decrease in the dynamic range in the S trajectories indicate an increasing spatial confinement of the Plasmon Ruler in compartments that are of similar dimensions than the total length of the Plasmon Ruler (DNA spacer + NPs). Although the P and R trajectories were recorded with a relatively low frame rate of 5 Hz in this example, the P trajectories, especially for PR1 and PR2, show a large degree of fluctuations due to transient orientation changes. The observation of resolvable fluctuations in P at this frame rate is a clear indication of significantly hindered rotational motion of the spatially confined Plasmon Rulers. In the case of free rotational motion, the polarization information should have been averaged out at this acquisition rate to result in an average P ≈ 0. For PR3 the frequency and magnitude of the fluctuations in the P trajectory are significantly decreased, but P remains > 0. This observation is consistent with an even stronger spatial confinement and a localization of the Plasmon Ruler in a compartment in which rotational motions between different spatial orientations are separated by high energetic barriers. Overall, the polarization resolved tracking studies in Figure 9 confirm that an increasing spatial confinement is correlated with a decreasing average interparticle separation and decreasing rotational mobility of the Plasmon Rulers. This correlation makes Plasmon Rulers interesting tools for probing membrane compartmentalization on the length scale of the Plasmon Rulers simultaneously at different locations on a cell surface as function of time. This advantage was utilized to detect ErbB1 enriched membrane domains and to monitor their structural dynamics during their lateral diffusion on the surface of living cells.40
5.3 Quantifying Spatial Nanoclustering on Plasma Membranes through Plasmon Coupling
Plasma membrane proteins are often not homogeneously distributed but locally enriched in specific membrane environments.49,50 Especially for proteins that require inter-receptor interactions, spatial clustering represents a potential mechanism for modulating signaling intensity, threshold and response. Quantitative information about the spatial organization of a cell surface protein could, therefore, have similar biomarker relevance as the total expression level.36 As we will discuss in this section, the ability of NP immunolabels to interrogate relative vicinities on the tens of nanometer length scale and to transduce these information into the far-field provides new opportunities for profiling the spatial clustering of the targeted species in addition to its surface expression through an all-optical approach.
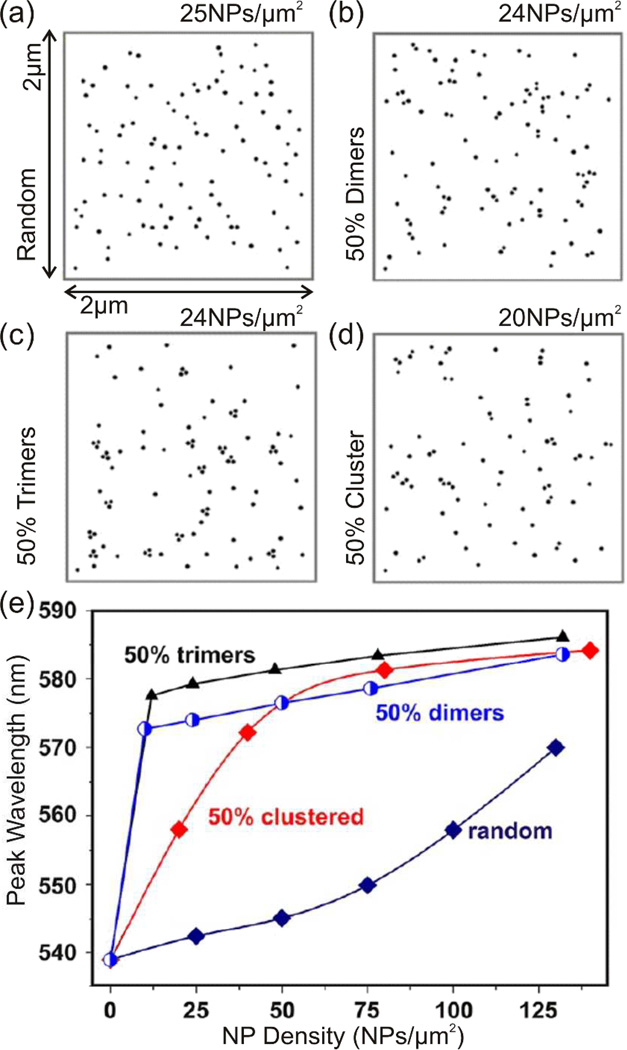
The impact of the NP spatial organization on the collective resonance wavelength, λpl, as function of average NP density, ρ, is shown in Figure 10. The ensemble-averaged resonance wavelengths, λpl(ρ), were obtained from simulated polarization-averaged darkfield scattering spectra. The simulations were performed at defined ρ values for different degrees of NP clustering in an area of 2×2 µm2 using generalized multiple particle Mie theory (GMT).36 Four different clustering configurations were considered in the GMT simulations: random, 50% dimers, 50% trigonal trimers, and 50% random clusters. The latter were generated by randomly placing one fourth of the total number of NPs near distributed NPs with a minimum separation of 5 nm. In case of the 50% dimers and trimers, the interparticle separation was kept constant at 5 nm. The random NP distribution exhibits a very shallow initial increase in λpl with increasing ρ since the probability of the formation of electromagnetically strongly coupled NP pairs or clusters is low at low ρ values. With increasing ρ the average interparticle separation gradually decreases and the effect of plasmon coupling increases. In contrast, configurations enriched in dimers and trimers with constant (and very short) interparticle separations show a much steeper increase in resonance wavelength that converges rapidly with increasing ρ. The 50% clustering configuration has larger average interparticle separations than the clustered dimers and trimers and the slope of its λpl(ρ) curve lies between those of the strongly coupled dimer and trimer configurations and the total random configuration. Overall, Figure 10 confirms clearly a sensitive dependence of the measured spectral response on NP clustering.
Figure 10.
Evaluating the interplay of average Au NP density, ρ, and local clustering on the ensemble-averaged plasmon resonance wavelength, λplas, through GMT simulations. Four different 40 nm NP organization levels were considered for different ρ values: a) random, b) 50% dimers, c) 50% trimers, d) 50% random clusters. e) Peaks resonance wavelength as function of ρ and NP organization. Adapted with permission from Wang et al., Nano Lett. 12, 3231 (2012). Copyright 2012 American Chemical Society.
Although PCM cannot directly image individual NP labeled receptors if their separation lies below the resolution limit of the optical microscope, the spectral response of NP immunolabels provides synergistic information about their clustering. Membrane areas that are locally enriched in the receptor represent high affinity binding sites for NP immunolabels and template the formation of clusters of strongly coupled NPs. These areas are indicated by a spectral red-shift when compared with areas in which the receptors are not clustered and individual NPs dominate the scattering response. Wang et al. utilized the clustering dependent spectral NP response to compare the local ErbB1 and ErbB2 densities on the plasma membrane of selected cancer model cells with different expression levels (A431, SKBR3, MCF7).36 For ErbB family type receptors information about the spatial receptor distribution are especially useful since functionality depends on their organization into dimers and potentially higher oligomers, which is anticipated to be influenced by the local receptor density.4, 36, 49 Wang et al. utilized the secondary antibody based labeling strategy introduced in section 4.1.1 to label ErbB1 and ErbB2. The cells were incubated with increasing concentrations of NP immunolabels to systematically vary the average NP density, ρ. Scattering spectra were then recorded from peripheral stretched membrane areas, where the signal to noise was largest, by averaging the spectral content over the entire opening slit of the spectrometer (section 4.2.1). The cells were subsequently inspected in the SEM to determine ρ. For the highly overexpressed ErbB1 receptor on A431 cells, the experimentally obtained λpl(ρ) relationship reproduces the simulated curve for the 50% random cluster configuration shown in Figure 10e.36 This finding implies an increase in NP clustering throughout the investigated NP density range. In contrast, for ErbB2 on MCF7 and SKBR3, a much faster convergence of the λpl(ρ) relationship against a lower peak resonance wavelength was obtained. The obtained λpl(ρ) relationships confirm less NP immunolabel clustering for MCF7/ErbB2 and SKBR3/ErbB2 than for A431/ErbB1.36 This finding is consistent with overall lower receptor densities and less and smaller receptor enriched membrane compartments for MCF7/ErbB2 and SKBR3/ErbB2.36
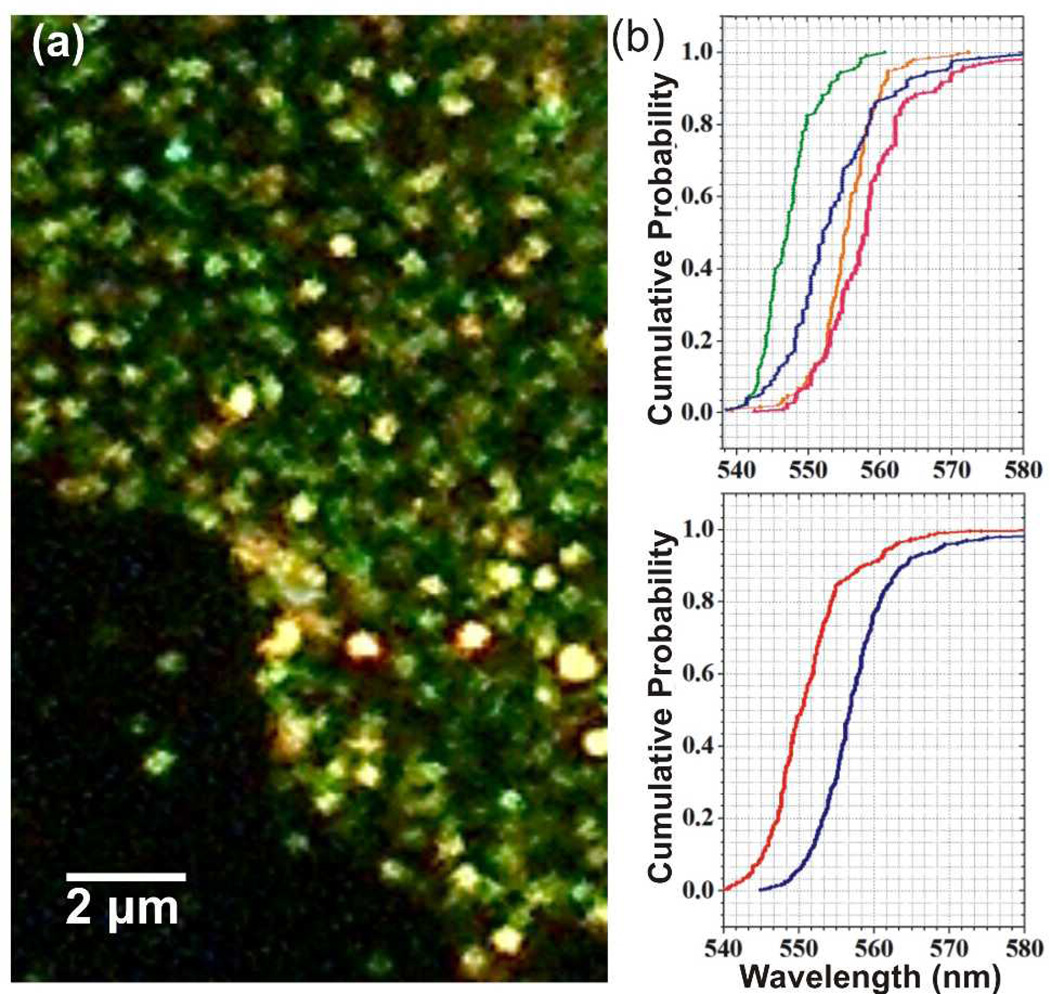
PCM is not limited to an analysis of area-averaged spectral information but can, instead, also be used to detect differences in the spatial organization of the targeted cellular receptors on sub-cellular length scales. This has been utilized, for instance, to compare ErbB1 density and spatial organization on filopodia, (quasi-one-dimensional projections from the cell edge) and the actual cell body.39 Consistent with a higher receptor density, the average plasmon resonance wavelength measured on filopodia was significantly red-shifted when compared to that measured on the actual cell body. The increased NP immunolabel density on the filopodia was subsequently confirmed via SEM. In another study37 both expression and spatial clustering of the cell surface glycoproteins CD24 and CD44 on MCF7 and SKBR3 cells were analyzed. The relative NP labeling intensities were found to be correlated with the receptor expression levels. An optical inspection of the labeled cell surface in the microscope revealed discrete spots on the cell surface with colors ranging from green to orange (Figure 11a) due to clustering of the NP immunolabels on the cell surface. According to a statistical analysis of the scattering spectra of individual spots, CD24 showed a significantly higher degree of clustering than CD44 (Figure 11b, top). Lipid raft disruption through cholesterol sequestration (Figure 11b, bottom) removed the preferential clustering of CD24, indicating an enrichment of CD24 in lipid rafts.
Figure 11.
a) Magnified view of a section of a MCF7 cell after labeling surface expressed CD24 with 40 nm Au NPs. b) top: cumulative distribution plots of λplas after NP labeling of the following cell/receptors: MCF7/CD44 (blue), SKBR3/CD24 (orange), MCF7/CD24 (pink). The distribution for Au NPs in ambient refractive index of nr = 1.40 is included as blue line. bottom: Cumulative distribution plot of λplas for NPs targeted at MCF7/CD24 before (red) and after (blue) treatment with m-β-CD to sequester cholesterol from the membrane. Adapted with permission from Yu et al., Anal. Chem. 85, 1290 (2013). Copyright 2013 American Chemical Society.
5.4 Imaging of NP Uptake and Subsequent Cellular Processing in Living Cells
Although the previous sections focused on the applications of PCM for probing the structure and dynamics of the plasma membrane, the method is not limited to cell surface processes. It is also well suited for investigating the intricate cellular mechanisms orchestrating NP uptake and trafficking. Noble metal NPs are a unique class of cargo for these studies since they can be simultaneously tracked and spectrally analyzed in PCM. It is, therefore, possible to monitor both the spatial trafficking of the uptaken NPs as well as their resonance wavelength, which depends on the local association level of the NPs. This gain of information is important since receptor mediated endocytosis involves the incorporation of NPs into membrane vesicles that can fuse in subsequent trafficking events and, thus, induce a clustering of NPs contained within the vesicles. Furthermore, PCM can be combined with immunofluorescence labeling, which makes it possible to investigate the spatiotemporal distribution of NPs inside cells and to probe their interactions with specific cellular components.
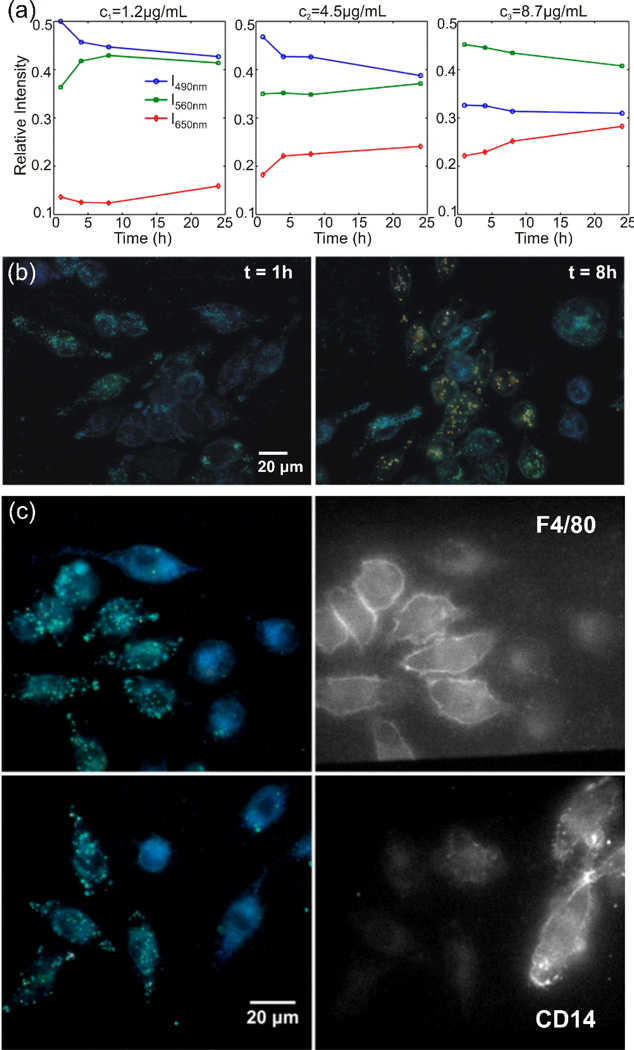
The synergistic gain resulting from a combination of PCM and immunofluorescence microscopy was first utilized in a mechanistic study of the scavenger receptor mediated uptake and trafficking of ~60 nm Ag NPs in the mouse macrophage cell line J774A.1.42 Multispectral imaging (see section 4.2.2) was applied to monitor the plasmon resonance wavelength of the uptaken NPs in hundreds of randomly chosen cells. These studies revealed an average red-shift of the plasmon resonance as function of time and NP concentration (Figure 12a), indicating that NP internalization is associated with a NP clustering within the cells. Optical imaging of the NP association levels on the single cell level showed, however, dramatic cell-to-cell fluctuations in the spatial clustering of the NPs within the cells.42 Even after an incubation of the macrophages with increased Ag NP concentrations for 8h, not all of the cells exhibited a strong intracellular NP association. Instead, a sub-population of cells still contained exclusively individual NPs or small agglomerates that demonstrated vividly blue colors. This is illustrated in Figure 12b, which shows darkfield scattering images of J774A.1 macrophages after exposure to Ag NPs at an effective concentration of 4.5 µg/mL and 8.7 µg/mL for 5 min and subsequent incubation without NPs for 1h or 8h, respectively.
Figure 12.
a) Scattering intensities of silver NPs after uptake by J774A.1 macrophages as function of time on three wavelength channels (490 nm (blue), 560 nm (green), and 650 nm(red)) for three different NP concentrations: 1.2 µg/mL (left), 4.5 µg/mL (middle), and 8.7 µg/mL (right). b) Darkfield images of macrophages exposed to 4.5 µg/mL Ag NPs after 1h (left) and 8.7 µg/mL Ag NPs after 8h (right). c) Correlated darkfield image of NP containing macrophages (left) and fluorescence image (right) after staining with F4/80 (top) or CD14 (bottom) maturity markers. Reproduced with permission from Wang et al., ACS Nano 6, 7122 (2012). Copyright 2012 American Chemical Society.
To elucidate the mechanisms underlying these striking cellular differences, the impact of a panel of pharmacological inhibitors on the intracellular NP association was investigated by Wang et al..42 These studies revealed the existence of two coexisting uptake mechanisms: actin- and clathrin-dependent endocytosis. The clathrin-dependent endocytosis results primarily in an enrichment of non-agglomerated NPs with a strong spectral response in the blue within the cells, while actin-dependent endocytosis is responsible for the formation of compact NP clusters with red-shifted spectral resonances. Wang et al. applied immunofluorescence microscopy to correlate the observed NP association levels with macrophage maturation markers F4/80 (higher concentration on mature macrophages) and CD14 (characteristic of monocytes). These studies revealed that F4/80 is preferentially expressed on cells that show NP association while CD14 is associated with cells containing predominantly monomeric NPs (Figure 12c). Overall, the performed PCM studies – in combination with SEM cell surface characterization – led to the formulation of the following model.42 In one sub-population (monocyte-like macrophages) individual Ag NPs and small NP clusters are directly taken up via a clathrin-dependent but actin-independent pathway. A second sub-population (mature macrophages) accumulates NPs first into larger clusters on the cell surface. Similar to conventional phagocytosis the surface-formed NP clusters are finally engulfed in an actin-dependent step.
6. Perspective
The superb photophysical properties of noble metal NPs and the methods reviewed in this Tutorial for monitoring truly nanoscale separations between them via optical means, make noble metal NPs interesting tools for a broad range of applications in sensing and imaging but also in other materials related fields. The PCM applications demonstrated so far have focused primarily on plasmon ruler tracking and the monitoring of the large-scale association (clustering) of specific cell surface receptors. With the improvement of the NP labels and the development of more homogeneous, smaller, and monovalent NP labels, PCM will be applicable to characterize the spatial distribution of an even broader range of targets beyond the temporal and spatial barriers of FRET. We also envision that PCM based tools will be particularly useful in providing insights into the intricate interactions between nanoscale materials and cellular systems. Many important questions at the nano-bio interface, most importantly, how nanoscale objects can be used to influence cellular processes remain to be characterized.48 Given the wealth of information accessible through PCM tracking of metallic NPs, the technique seems poised to contribute significantly to this important area. Mechanistic information derived from PCM-based imaging approaches will help to mitigate or entirely avoid potential toxic effects associated with nanomaterials and could eventually lead to entire new strategies for controlling cellular activities through nanomaterials.
Key Learning Points.
Optical Properties of Noble Metal Nanoparticles
Distance Dependent Plasmon Coupling
Resolving Subdiffraction Limit Separations in the Optical Microscope through Plasmon Coupling
Experimental Implementation of Plasmon Coupling Microscopy (PCM)
Application of PCM in Biosensing and Imaging
Acknowledgement
We acknowledge the inspiration from many researchers in the area of bioplasmonics whose work we could not cite due to length limitations. We thank the members of the Reinhard Laboratory who have contributed to this work, in particular Hongyun Wang, Guoxin Rong, Jing Wang, Lynell Skewis, Amin Feizpour, Tianhong Chen, Yan Hong and Xinwei Yu. This work was financially supported by the National Institutes of Health through grant 5R01CA138509 and the National Science Foundation through grants 0953121 and 1159552 to BMR. L.W. acknowledges a National Cancer Institute Cancer Nanotechnology Training Center (NCI-CNTC) Graduate Student Traineeship and financial support through NIH R25 CA153955.
Reference
- 1.Thompson RE, Larson DR, Webb WW. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Periasamy A, Day RN, editors. Molecular Imaging: FRET Microscopy and Spectroscopy. New York, USA: Oxford University Press; 2005. [Google Scholar]
- 3.Hell SW. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 4.Yang S, Raymond-Stintz MA, Ying W, Zhang J, Lidke DS, Steinberg SL, Williams L, Oliver JM, Wilson BS. J. Cell.Sci. 2007;120:2763–2773. doi: 10.1242/jcs.007658. [DOI] [PubMed] [Google Scholar]
- 5.Maier SA. Plasmonics: Fundamentals and Applications. New York, USA: Springer; 2007. [Google Scholar]
- 6.Marquier F, Greffet J-J. In: Optical Antennas. Agio M, Alu A, editors. Cambridge, UK: Cambridge University Press; 2013. [Google Scholar]
- 7.Yguerabide J, Yguerabide EE. Anal. Biochem. 1998;262:137–156. doi: 10.1006/abio.1998.2759. [DOI] [PubMed] [Google Scholar]
- 8.Maier SA, Brongersma ML, Kik PG, Atwater HA. Phys. Rev. B. 2002;65:193408. [Google Scholar]
- 9.Nordlander P, Oubre C, Prodan E, Li K, Stockman MI. Nano Lett. 2004;4:899–903. [Google Scholar]
- 10.Rolly B, Stout B, Bonod N. Phys. Rev. B. 2011;84:125420. [Google Scholar]
- 11.Rechberger W, Hohenau A, Leitner A, Krenn JR, Lamprecht B, Aussenegg FR. Optics Commun. 2002;220:137–141. [Google Scholar]
- 12.Jain PK, El-Sayed MA. Nano Lett. 2008;8:4347–4352. doi: 10.1021/nl8021835. [DOI] [PubMed] [Google Scholar]
- 13.Crow MJ, Seekell K, Ostrander JH, Wax A. ACS Nano. 2011;5:8532–8540. doi: 10.1021/nn201451c. [DOI] [PubMed] [Google Scholar]
- 14.Su KH, Wei QH, Zhang X, Mock JJ, Smith DR, Schultz S. Nano Lett. 2003;3:1087–1090. [Google Scholar]
- 15.Sönnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. Nat. Biotech. 2005;23:741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 16.Reinhard BM, Siu M, Agarwal H, Alivisatos AP, Liphardt J. Nano Lett. 2005;5:2246–2252. doi: 10.1021/nl051592s. [DOI] [PubMed] [Google Scholar]
- 17.Chen JIL, Chen Y, Ginger DS. J Am. Chem. Soc. 2010;132:9600–9601. doi: 10.1021/ja103240g. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Wang H, Yan B, Reinhard BM. J Phys Chem. C. 2010;114:4901–4908. doi: 10.1021/jp911858v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Encina ER, Coronado EA. J. Phys. Chem. C. 2010;114:3918–3923. [Google Scholar]
- 20.Reinhard BM, Sheikholeslami S, Mastroianni A, Alivisatos AP, Liphardt J. Proc. Natl. Acad. Sci. USA. 2007;104:2667–2672. doi: 10.1073/pnas.0607826104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skewis LR, Reinhard BM. Nano Lett. 2008;8:214–220. doi: 10.1021/nl0725042. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, Reinhard BM. J Phys Chem. C. 2009;113:11215–11222. doi: 10.1021/jp900874n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jun Y, Sheikholeslami S, Hostetter DR, Tajon C, Craik CS, Alivisatos AP. Proc. Natl. Acad. Sci. USA. 2010;106:17735–17740. doi: 10.1073/pnas.0907367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lermusiaux L, Sereda A, Portier B, Larquet E, Bidault S. ACS Nano. 2012;6:10992–10998. doi: 10.1021/nn304599d. [DOI] [PubMed] [Google Scholar]
- 25.Jain PK, Huang W, El-Sayed MA. Nano Lett. 2007;7:2080–2088. doi: 10.1021/nl071813p. [DOI] [PubMed] [Google Scholar]
- 26.Gunnarsson L, Rindzevicius T, Prikulis J, Kasemo B, Käll M, Zou S, Schatz GC. J Phys Chem. B. 2005;109:1079–1087. doi: 10.1021/jp049084e. [DOI] [PubMed] [Google Scholar]
- 27.Khlebtsov BN, Melnikov AG, Zharov VP, Khlebtsov NG. Nanotechnology. 2006;17:1437–1445. [Google Scholar]
- 28.Sun G, Khurgin JB. Appl. Phys. Lett. 2010;97:263110. [Google Scholar]
- 29.Zuloaga J, Prodan E, Nordlander P. Nano Lett. 2009;9:887–891. doi: 10.1021/nl803811g. [DOI] [PubMed] [Google Scholar]
- 30.de Abajo FJG. J. Phys. Chem. C. 2008;112:17983–17987. [Google Scholar]
- 31.Chen T, Pourmand M, Feizpour A, Cushman B, Reinhard BM. J Phys Chem. Lett. 2013;4:2147–2152. doi: 10.1021/jz401066g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scholl JA, García-Etxarri A, Leen Koh A, Dionne JA. Nano Lett. 2013;13:564–569. doi: 10.1021/nl304078v. [DOI] [PubMed] [Google Scholar]
- 33.Yan B, Boriskina SV, Reinhard BM. J Phys Chem. C. 2011;115:24437–24453. doi: 10.1021/jp207821t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halas NJ, Lal S, Chang W, Link S, Nordlander P. Chem. Rev. 2011;111:3913–3961. doi: 10.1021/cr200061k. [DOI] [PubMed] [Google Scholar]
- 35.Rong G, Wang H, Skewis LR, Reinhard BM. Nano Lett. 2008;8:3386–3393. doi: 10.1021/nl802058q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Yu X, Boriskina SV, Reinhard BM. Nano Lett. 2012;12:3231–3237. doi: 10.1021/nl3012227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu X, Wang J, Feizpour A, Reinhard BM. Anal. Chem. 2013;85:1290–1294. doi: 10.1021/ac303310j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sapsford KE, Algar WR, Gemmill KB, Casey BJ, Oh E, Stewart MH, Medintz IL. Chem. Rev. 2013;113:1904–2074. doi: 10.1021/cr300143v. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Boriskina SV, Wang H, Reinhard BM. ACS Nano. 2011;5:6619–6628. doi: 10.1021/nn202055b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rong G, Reinhard BM. PLoS ONE. 2012;7:e34175. doi: 10.1371/journal.pone.0034175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Rong G, Yan B, Yang L, Reinhard BM. Nano Lett. 2011;11:498–504. doi: 10.1021/nl103315t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, Wu L, Reinhard BM. ACS Nano. 2012;6:7122–7132. doi: 10.1021/nn302186n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei Q, McLeod E, Qi H, Wan Z, Sun R, Ozcan A. Scientific Reports. 2013;3:1699. doi: 10.1038/srep01699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sönnichsen C, Alivisatos AP. Nano Lett. 2005;5:301–304. doi: 10.1021/nl048089k. [DOI] [PubMed] [Google Scholar]
- 45.Rong G, Wang H, Reinhard BM. Nano Lett. 2010;10:230–238. doi: 10.1021/nl903350f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aaron J, Travis K, Harrison N, Sokolov K. Nano Lett. 2009;9:3612–3618. doi: 10.1021/nl9018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian W, Huang X, Kang B, El-Sayed MA. J. Biomed. Opt. 2010;15:046025. doi: 10.1117/1.3477179. [DOI] [PubMed] [Google Scholar]
- 48.Wu L, Yu X, Feizpour A, Reinhard BM. Biomater. Sci. 2014;2:156–166. doi: 10.1039/C3BM60142K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ariotti N, Liang H, Xu Y, Zhang Y, Yonekubo Y, Inder K, Du G, Parton RG, Hancock JF, Plowman SJ. Mol. Cell. Biol. 2010;30:3795–3804. doi: 10.1128/MCB.01615-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, Kasai RS, Kondo J, Fujiwara T. Annu. Rev. Biophys. Biomol. Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]