SUMMARY
Activating mutations in KRAS are among the most frequent events in diverse human carcinomas and are particularly prominent in human pancreatic ductal adenocarcinoma (PDAC). An inducible KrasG12D-driven mouse model of PDAC has established a critical role for sustained KrasG12D expression in tumor maintenance, providing a model to determine the potential for, and the underlying mechanisms of, KrasG12D–independent PDAC recurrence. Here we show that some tumors undergo spontaneous relapse and are devoid of KrasG12D expression and downstream canonical MAPK signaling and instead acquire amplification and overexpression of the transcriptional co-activator Yap1. Functional studies established the role of Yap1 and the transcriptional factor Tead2 in driving KrasG12D–independent tumor maintenance. The Yap1/Tead2 complex acts cooperatively with E2F transcription factors to activate a cell cycle and DNA replication program. Our studies, along with corroborating evidence from human PDAC models, portend a novel mechanism of escape from oncogenic Kras addiction in PDAC.
INTRODUCTION
PDAC remains a largely incurable lethal disease with a median survival of approximately six months (Hidalgo et al., 2010; Vincent et al., 2011). The PDAC genome is characterized by a number of signature mutations involving the KRAS oncogene and the CDKN2A, TP53, and SMAD4 tumor suppressor genes and by significant chromosomal aberrations resulted from telomere dysfunction, centrosome abnormalities, among other mechanisms (Hezel et al., 2006; Jones et al., 2008; Campbell et al., 2010; Biankin et al., 2012). Activating mutations in KRAS are present in the majority of human PDAC cases and genetically engineered mouse (GEM) models have substantiated critical roles of oncogenic Kras in driving tumor initiation and in enabling tumor progression along with deficiencies of P53, Ink4a/Arf, Smad4 and/or Pten tumor suppressors (Aguirre et al., 2003; Guerra et al., 2003; Hingorani et al., 2003; Tuveson et al., 2004; Bardeesy et al., 2006; Hill et al., 2010; Ying et al., 2011; Guerra and Barbacid, 2013). The panoply of signaling pathways engaged by oncogenic Kras provides a basis for its diverse tumor biological roles in proliferation, survival, metabolism and tumor microenvironment remodeling (Pylayeva-Gupta et al., 2011).
The oncogene addiction and tumor maintenance paradigm (Weinstein, 2002; McCormick, 2011; Hanahan and Weinberg, 2011) has rationalized the striking clinical responses achieved with drugs targeting driver oncogenes (Torti and Trusolino, 2011). Despite significant clinical responses to targeted therapies, nearly all tumor remissions are followed by acquired resistance and tumor relapse. Resistance mechanisms vary considerably and include mutations blocking drug-target interaction, genetic alterations sustaining signaling in downstream pathways or alternate survival pathways (Torti and Trusolino, 2011; Berns and Bernards, 2012). The pervasive disease recurrence following targeted therapy has motivated the use of inducible driver oncogene GEM models of cancers to proactively illuminate potential mechanisms of resistance employed by human cancers (Lauchle et al., 2009).
Given the essential roles of oncogenic Kras in both PDAC initiation and maintenance, mutant KRAS and its signaling pathways have been a major focus for the development of disease models for human PDAC (Hingorani et al., 2003; Collisson et al., 2012; Collins et al., 2012; Ying et al., 2012; Eser et al., 2013; Guerra and Barbacid, 2013). To model anti-Ras therapy, we and others have generated an inducible KrasG12D GEM PDAC model and established that extinction of KrasG12D induced rapid tumor regression, highlighting the potential clinical utility of targeting oncogenic KRAS in pancreatic cancer (Collins et al., 2012; Ying et al., 2012).
Despite its critical role in PDAC biology, we sought to determine whether sustained oncogenic Kras suppression would result in tumor relapse and illuminate tumor resistance mechanisms. Employing our previously described doxycycline (doxy)-inducible KrasG12D GEM PDAC model we identified relapse tumors (after KrasG12D extinction induced tumor regression) that lacked transgene expression and instead harbored an activated Yap1/Tead2 transcriptional program enabling KrasG12D -independent tumor cell proliferation that enlists the cooperative actions of the E2F transcription factor. Interestingly, our findings in the mouse model are reinforced by observation in human PDAC showing a prominence of similar transcriptional programs in the quasimesenchymal-subset (QM-subset) of pancreatic cancers, which are notable for lower dependency on oncogenic KRAS relative to other PDAC subsets (Collisson et al., 2011).
RESULTS
Spontaneous pancreatic tumor relapse after KrasG12D inactivation-induced complete clinical regression
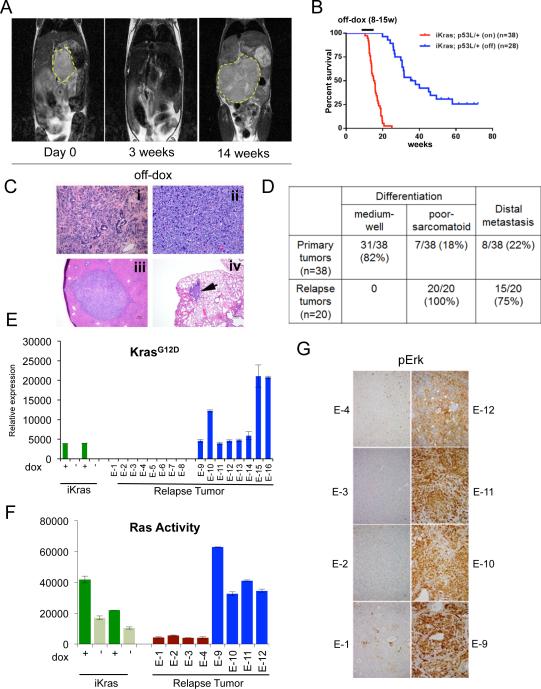
Using mice engineered with a doxy-inducible KrasG12D transgene and conditional p53 null alleles (p48Cre; tetO_LSL-KrasG12D; ROSA_rtTA; p53L/+, designated iKras), we and others established that sustained KrasG12D signaling is essential for pancreatic tumor maintenance (Collins et al., 2012; Ying et al., 2012). To evaluate the potential for recurrence mechanisms following KrasG12D extinction, we utilized MRI imaging to monitor regression of advanced pancreatic tumors measuring at least 8 mm in diameter at time of doxy withdrawal. Consistent with previous findings (Collins et al., 2012; Ying et al., 2012), KrasG12D extinction resulted in complete regression despite significant tumor burdens in all animals (n=28) with virtually no gross tumor detected by MRI imaging at three weeks following doxy withdrawal (Figure 1A). However, 70% of the mice (20/28) escaped from doxy withdrawal with evidence of relapse between 9 to 47 weeks, with a median survival of 36.6 weeks compared to 15.4 weeks for iKras mice maintained on continued doxy treatment (p<0.0001) (Figure 1B). On the morphological level, in contrast to the well differentiated ductal features and predominant CK19 positivity of the doxy-induced PDACs (Figure S1A) (Ying et al., 2012), all recurrent tumors exhibited poorly differentiated or sarcomatoid features, were devoid of acinar (amylase+) or endocrine markers (chromogranin+; CHGA) staining, although some tumors partially retained scattered CK19 ductal marker staining (Figure 1C and S1A). Consistent with the development of more aggressive phenotypes, distal metastases to lung or liver were observed in 75% (15/20) of the animals with recurrent tumors versus 21% (8/38) (p < 0.001) of those carrying primary tumors (Figure 1C and 1D).
Figure 1. Spontaneous pancreatic tumor relapse after complete regression upon KrasG12D inactivation.
(A) Representative MRI scan shows initial tumor regression (3 weeks) but subsequent relapse (14 weeks) after doxy withdrawal. (B) Kaplan-Meier overall survival analysis for iKras; p53L/+ mice after doxy withdrawal. On: mice were fed with doxy. Off: Mice with advanced PDAC were switched to doxy-free water 8-15 weeks after on doxy and observed for relapse. (C) Histopathological characterization of the relapse tumors showing poorly differentiated (i) or sarcomatoid (ii) relapse tumors, with liver (iii) and lung (iv) metastasis (denoted by arrow). (D) Quantitative comparison of histopathological features between primary and relapse tumors. (E) qRT-PCR for KrasG12D transgene shows expression in relapse tumors. Data represented as relative normalized expression. (F) Measurement of Ras activity in relapse tumors. For E and F; Two independent iKras cells were maintained in the presence (+) or absence (-) of doxy for 24 hr and used as controls. Error bars represent SD of duplicate samples. (G) The relapse tumors were stained with antibodies against pErk.
See also Figure S1.
The relapsed tumor lesions were of pancreatic origin as demonstrated by the presence of Cre-mediated p53 deletion in cells cultured from the relapse tumors (Figure S1B). Furthermore, since the iKras PDAC mice do no develop pancreatic tumors in the absence of doxy induction (Ying et al., 2012), we conclude that these doxy-independent pancreatic tumors are bona fide tumor relapses of the original primary tumors, rather than tumors formed de novo in the absence of oncogene induction.
Doxy-independent tumor recurrence in this model could potentially result from doxy-independent activation of the iKras transgene or from Kras independent mechanisms. Indeed, although doxy withdrawal results in loss of KrasG12D expression and downstream signaling in all primary tumors (Ying et al., 2012; Figure 1E and S1C), approximately half of relapse tumors examined exhibited re-expression of the iKras transgene accompanied with canonical downstream signaling; these tumors were designated hereafter as iKras+ relapse tumors (Figure 1E-1G, Figure S1C, tumors E-9 to E-16). The remaining tumors did not express the iKras transgene, or hyper-activated endogenous Kras expression, and exhibited diminished canonical downstream signaling; these tumors were designated hereafter as iKras− relapse tumors (Figure 1E-1G, S1C-S1D; tumors E-1 to E-8). While displaying similar aggressive histopathological features, the iKras+ versus iKras− relapse tumors were readily distinguished molecularly on the basis of mitogen-activated protein kinase (MAPK) pathway activity: In contrast to iKras+ tumors, majority of iKras− tumor lines showed relatively lower phospho-Mek (pMek) and phospho-Erk (pErk) both in vivo and in vitro (Figure 1G and S1C). Furthermore the iKras− tumors did not show compensatory hyper-activation of AKT pathway and levels of phospho-ribosomal protein S6 (pS6) were generally lower relative to iKras+ tumors (Figure S1C). Thus, while oncogenic Kras signaling in the primary tumors tightly controls MAPK pathway activity, recurrence of iKras− tumors results from mechanisms that do not utilize oncogenic Kras or hyper-activated MAPK/AKT signaling. Since the wild-type Kras allele remains intact upon KrasG12D extinction in our model system, the contribution of basal signaling activity from wild-type Kras or other Ras family members during tumor relapse remains to be elucidated.
Yap1 is amplified in iKras− relapse tumors and required for tumor growth
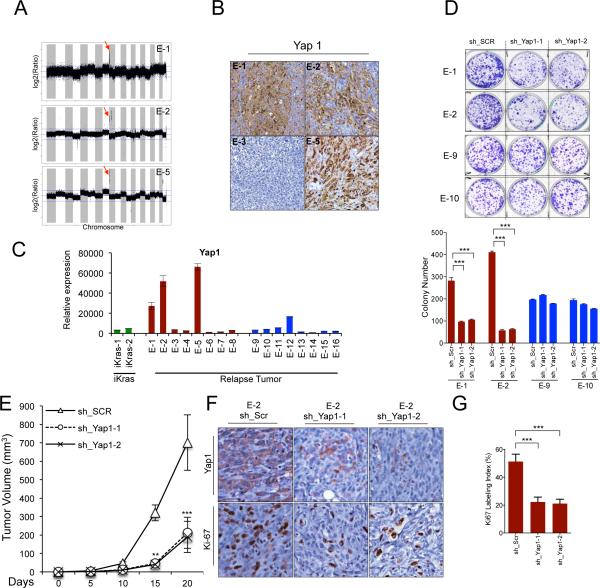
Next, we explored the molecular mechanisms underlying spontaneous tumor relapse after KrasG12D extinction. Array-based comparative genomic hybridization (aCGH) revealed that all iKras+ relapse tumors exhibited focal amplification of the Rosa26 locus rtTA allele providing a likely basis for doxy-independent re-expression of the iKras transgene (Figure S2A; chromosome 6q). In iKras− relapse tumors, the only recurrent genomic alteration was amplification of chromosome 9qA1 region, encompassing 11 genes encoding several metalloproteinases, the transcriptional co-activator Yap1 and the anti-apoptotic genes Birc2 (cIap1) and Birc3 (cIap2). Of these, only Yap1, Birc2 and Birc3 showed a copy number linked increase in gene expression (Figure 2A-2C; Figure S2D). Further, YAP1 protein was found to be elevated in iKras− relapse tumors bearing the 9qA1 amplicon (E-1, E-2 and E-5) whereas iKras− tumors lacking the amplicon showed low levels of Yap1 (Figure 2B-2C; Figure S2C) pointing to additional escape mechanisms (see discussion; and Shao et al.,).
Figure 2. Yap1 is amplified in iKras− relapse tumors and required for tumor growth.
(A) aCGH plots of relapse tumors shows that the 9qA1 locus containing Yap1 is focally amplified (denoted by red arrow) in E-1, E-2 and E-5. Normalized log2 ratio for each probe are plotted. (B) IHC for Yap1 in relapse tumors. Note increased Yap1 expression in 9qA1 amplicon+ tumors, E-1, E-2 and E-5 but not 9qA1 amplicon− tumor E-3. (C) qRT-PCR for Yap1 in relapse tumors. Relative expression levels normalized to reference gene. Error bars represent SD of the mean. (D) Representative wells of the clonogenic growth assay upon knockdown of Yap1 by two independent shRNAs primary cultures in Yap1 amplicon+ tumors (E-1 and E-2) and the iKras+ tumors (E-9 and E-10). Non-targeting shRNA (sh_Scr) was used as control. Quantification of cell growth is shown at the bottom. Error bars represent SD of triplicate wells, ***p < 0.001. (E) Xenograft tumor growth of cell cultures derived from E-2 expressing two independent Yap1 shRNA or control shRNA. Tumor volume was measured at the days indicated, data shown is representative of results from 2 independent experiments (n=5 per group). Error bars represent SD of the mean, **p < 0.01; ***p < 0.001. (F) IHC for proliferation marker Ki-67 and Yap1 in E-2 xenograft tumors expressing Yap1 shRNA described in (E) (G) Quantification of IHC staining for Ki-67 displayed as percentage of cells positive for Ki-67 staining. Error bars represent SD of the mean of 5 random fields, ***p<0.001).
See also Figure S2.
To assess the possible role of Yap1, Birc2 and Birc3 overexpression in driving growth of the iKras− relapse tumors, early passage primary cultures generated from tumors with and without the 9qA1 amplicon were monitored for cell growth following shRNA-mediated knockdown of Yap1, Birc2 or Birc3. Birc2 or Birc3 knockdown had no impact on cell growth relative to control shRNA-expressing cells (Figure S2E-S2F). In contrast, two independent shRNAs targeting Yap1 reduced proliferation in clonogenic assays as well as tumor growth and tumor cell proliferation (Ki-67, Figure 2G) in Yap1 amplified relapse tumors (E-1 and E-2) but exerted no impact on cells without Yap1 amplification (E-9 and E-10) (Figure 2D-2F; Figure S2G-S2H). Interestingly, persistent Yap1 knockdown in xenografts generated from Yap1 shRNA expressing cells resulted in resumption of KrasG12D transgene expression which coincided with a modest increase in MAPK activity and increased tumor cell proliferation, further underscoring the importance of activated Kras in PDAC maintenance (data not shown). Together, these genomic and functional studies strongly support a role for Yap1 amplification-driven overexpression as a mechanism for KrasG12D-independent PDAC recurrence.
Enforced Yap1 expression enables tumor maintenance upon KrasG12D extinction in PDAC
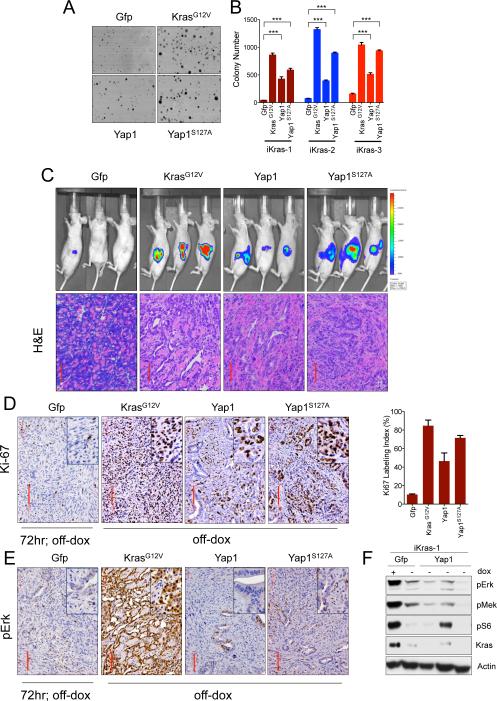
To evaluate whether Yap1 gain-of-function would provide a mechanism for KrasG12D- independent PDAC growth, we engineered three independently derived Kras-dependent iKras tumor lines with wild-type Yap1 or constitutively active Yap1S127A mutant (S127A mutation prevents Yap1 cytoplasmic sequestration by Lats; Zhao et al., 2007) constructs and assayed them for anchorage-independent growth in the absence of doxy (Figure 3A-3B; Figure S3B). As shown in Figure 3A and 3B, Yap1 or Yap1S127A expression along with KrasG12V expressing cells dramatically enhanced anchorage independent growth while GFP-expressing control cells showed profound impairment of cell growth in the absence of doxy. The ability of Yap to drive Kras-independent cell growth aligns with the ability of enforced Yap1 to complement loss of Kras function in human pancreatic cancer cell lines (Shao et al.,).
Figure 3. Enforced Yap1 expression enables tumor maintenance upon KrasG12D extinction in PDAC.
(A) Representative wells of anchorage independent growth assay demonstrating the ability of Yap1 or Yap1S127A to substitute for oncogenic Kras in promoting cell growth of iKras cells. Growth of Gfp infected cells was impaired in the absence of oncogenic Kras. (B) Quantification of anchorage independent growth assay in three independently derived iKras cells (grown offdoxy, KrasG12D off), For each condition, five random fields were counted. Error bars represent SD of the mean, ***p < 0.001. (C) Yap1 mediated bypass of tumor regression (upon inactivation of KrasG12D) in orthotopic xenografts generated from iKras cells used in (A). Mice (n=10 per group) were kept on-doxy for 7 days post-implantation and then switched to off-doxy. Top: Tumor growth off-doxy was visualized by bioluminescent imaging at 4 weeks off-doxy, except for KrasG12V for which image is taken 2 weeks after switching animals off-doxy. Gfp expressing cells regressed upon KrasG12D inactivation. Bottom: H/E of representative tumors is shown, Scale bar: 100μM. (D&E) IHC for Ki-67 (D, quantified on the right) and pErk (E). Gfp expressing tumor 72 hr after doxy was used as a negative control. Note proficient proliferation (as visualized by Ki-67 staining in D) and lack of MAPK activation (as visualized by low pErk staining in E) in the Yap1 expressing tumors. (F) Signaling status of key RAS effectors in short term cultures derived from three independent Yap1 expressing orthotopic tumors described in 3C.
See also Figure S3.
These cell culture-based findings were further substantiated in vivo by the ability of enforced Yap1 expression to substitute for KrasG12D activity in tumor maintenance. Specifically, Yap1, Yap1S127A and KrasG12V expressing iKras tumor cells grown orthotopically (Figure 3C) or subcutaneously (Figure S3A) in nude mice were able to resist tumor regression upon extinction of oncogenic Kras and promote tumor growth and proliferation (as measured by Ki-67 staining, Figure 3D), whereas GFP-expressing control iKras tumor cells fully regressed upon doxy withdrawal (Figure 3C and S3A). Additionally Yap1 or Yap1S127A expressing iKras cells (using two independently derived lines) showed Kras-independent tumor growth when injected into nude mice (Figure S3C-S3D). Furthermore, shRNA-mediated knockdown of Yap1 or YapS127A dramatically suppressed proliferation of short-term cultures from these Yap1-expressing orthotopic tumors (described in Figure 3C), confirming that the growth of bypassed tumors was indeed Yap1-dependent (Figure S3E-S3G). Notably, Yap1 bypassed tumors and early passage derivative cell lines showed lower MAPK activity while compared to KrasG12V bypassed tumors. (Figure 3E-3F; Figure S3A). Together, these results indicate that enforced Yap1 expression can substitute for oncogenic Kras-driven tumor maintenance and associated tumor cell proliferation.
Bypass of KrasG12D extinction by Yap involves interaction with Tead2
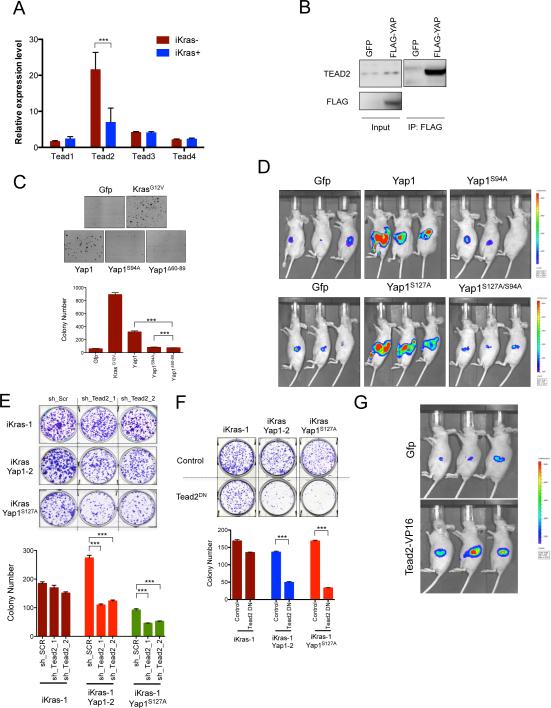
Since Yap1 is a transcriptional co-activator and does not bind directly to DNA, we sought to determine whether transcription factors known to interact with Yap1 signaling might mediate its activity. Smad1, p73, Runx2 and Tead transcription factors are all known to mediate the effects of Yap1 in different contexts (Zhao et al., 2011; Pobbati and Hong, 2013). Interestingly, only Tead2 levels were significantly higher in iKras− relapse tumors compared to the iKras+ ones (Figure 4A) and Tead2 physically interacted with endogenous Yap1 in early passage cultures from primary relapse tumors and in cells derived from Yap1 bypassed tumors (Figure S4A and Figure 4B), implicating Tead2 as a candidate transcription factor mediating Yap1's activity in our system. In line with this hypothesis, the TEAD family of transcription factors are genetically and biochemically validated mediators of YAP1's proliferation-inducing function (Cao et al., 2008; Zhang et al., 2011; Zhao et al., 2011; Schlegelmilch et al., 2011), and this established activity is consistent with the observed robust tumor cell proliferation profile in recurrent tumors (Figure 3C).
Figure 4. Interaction of Yap1 with Tead2 is critical for it's ability to bypass KrasG12D- dependence.
(A) qRT-PCR for expression of Tead family of transcription factors (Tead1-4) in iKras− and iKras+ relapse tumors. Error bars represent SD of the mean, ***p < 0.001. (B) Tead2 interacts with Yap1 in Yap1 (Flag-tagged) expressing cells (described in 3C). Input (25%) is used as a reference. (C) Sustained expression of wild-type Yap1 but not TEAD binding defective Yap1 mutants (YapS94A and Yap1Δ60-89) can promote anchorage independent growth of iKras cells offdoxy. For each condition, five random fields were counted. Error bars represent SD of the mean, ***p < 0.001. (D) Mutation in Tead binding domain of Yap1 (S94A) dramatically decreases the ability of Yap1 or Yap1S127A to substitute for oncogenic Kras in vivo. Representative images shown at 6 weeks off-doxy (n=5 per group). (E) Representative wells (top) of the clonogenic growth assay upon knockdown of Tead2 by two independent shRNAs in Yap1 (or Yap1S127A) expressing cells (described in 3C). Quantification of cell growth is shown below. Error bars represent SD of triplicate wells, ***p < 0.001. (F) Dominant negative Tead2 (Tead2DN) selectively suppresses proliferation of Yap1 (or Yap1S127A) expressing cells but not the KrasG12D expressing iKras cells. Quantification of cell growth is shown below. Error bars represent SD of triplicate wells, ***p < 0.001. (G) Transcriptionally active form of Tead2 (Tead2-VP16) can substitute oncogenic Kras for in vivo tumor growth. Representative images are shown at 6 weeks off-doxy (n=5 per group).
See also Figure S4.
We thus performed a series of experiments to test whether Yap1 exerted its growth effects through Tead2. Mutation in Tead-binding domain (Tead-binding defective Yap1S94A and Yap1Δ60-89; Figure S4B-S4C) completely abolished the ability of Yap1 to drive proliferation and substitute for oncogenic Kras both in vitro (Figure 4C) and in vivo (Figure 4D). Similar results were obtained using the Yap1S127A/S94A double mutant (Figure 4D and S4D). These results strongly suggest that Tead2 is a critical partner of Yap1 in promoting tumor cell growth in the absence of oncogenic Kras. Next, we sought corroborating evidence of the importance of Tead2 in mediating Yap1 function by directly blocking its activity using two complementary approaches.
Firstly, shRNA-mediated knockdown of Tead2 reduced the proliferation of early passage cultures derived from orthotopic Yap1 bypassed tumors while Tead2 knockdown had no effect on the KrasG12D-expressing iKras lines (Figure 4E and S4E). Furthermore, overexpression of a previously characterized dominant-negative Tead2 mutant (Tead2DN; Liu-Chittenden et al., 2012), which harbors a deletion of the C-terminal Yap1-interacting domain while retaining the ability to bind DNA, strongly blocked tumor cell proliferation of Yap1-expressing cells in clonogenic assays (Figure 4F and S4F). In this case as well, growth suppressive activity of Tead2DN is specific to Yap1 expressing cells, as overexpression of Tead2DN did not suppress cell proliferation induced by oncogenic Kras (Figure 4F).
Secondly, we examined whether increasing Tead2 levels could mimic the effect of Yap1 overexpression. Overexpression of full length Tead2 did not promote anchorage independent cell growth or orthotopic tumor growth consistent with a lack of intrinsic transactivation activity of Tead2 (data not shown). However expression of a transcriptionally active form of Tead2, Tead2-VP16 (a fusion protein of the N-terminal region of Tead2 containing the TEA domain and the activation domain of herpes simplex virus VP16; Ota and Sasaki, 2008) in two independent iKras cells promoted orthotopic tumor growth (Figure 4G and S4G), in the absence of Kras expression. Together, these multiple lines of evidence establish a critical role for TEAD2 in mediating Yap1-driven tumor cell growth upon KrasG12D extinction in our model system.
Yap1/Tead2 complex acts cooperatively with E2F transcription factors to activate a cell cycle and DNA replication program
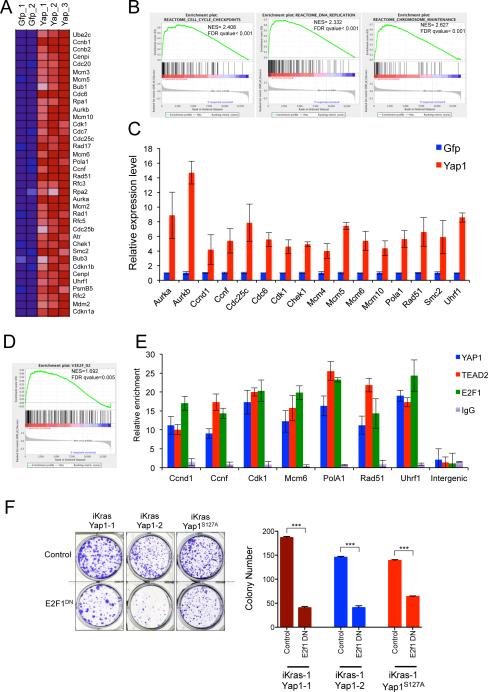
Next, transcriptomic analyses were conducted to elucidate the molecular network of Yap1 actions that facilitate KrasG12D-independent tumor growth. We defined the baseline gene expression upon extinction of oncogenic Kras and subsequently compared this expression profile to that in Yap1 bypassed tumors. To surmise molecular pathways associated with Yap1 overexpression, we performed Gene Set Enrichment Analysis (GSEA) of the expression profiles using gene sets for the canonical pathways in the Molecular Signature Database (MSigDB) (Subramanian et al., 2005). Consistent with our tumor biological observations, GSEA indicated that a significant fraction of KrasG12D-dependent gene sets that are rescued in the Yap1 bypassed tumors related to cell proliferation, DNA synthesis and replication (Figure 5A-5C). We validated several of the differentially expressed genes by qRT-PCR including mitotic kinases (including Aurora kinase A (Aurka), Aurora kinase B (Aurkb), “Budding inhibited by benzimidazoles” (Bub1), cyclins (including Ccna2, Ccnb1, Ccnb2 and Ccnd1), Cell division associated proteins (including Cdc2, Cdc20 and Cdc25c) and DNA replication proteins (including “minichromosome maintenance complex” proteins: Mcm5, Mcm6 and Mcm10).
Figure 5. Yap1/Tead2 cooperate with E2F to promote a cell cycle and DNA replication.
(A) Representative heat maps of the cell cycle and DNA replication genes enriched in Yap1 bypassed tumors compared to control (Gfp, off-doxy for 24 hr). Expression levels shown are representative of log2 values of each replicate. Red signal denotes higher expression relative to the mean expression level within the group and blue signal denotes lower expression relative to the mean expression level within the group. (B) Representative GSEA enrichment plots showing overrepresentation of indicated gene set categories among differentially expressed genes in Yap1 tumors compared with Gfp expressing tumors (24 hr, off-doxy). NES denotes normalized enrichment score. (C) qRT-PCR validation of representative differentially expressed genes in Yap expressing tumors. (D) GSEA enrichment plots showing E2F motif containing gene signatures in the differentially expressed genes in Yap1 expressing tumors compared with control. (E) ChIP showing YAP1 and TEAD2 occupancy at E2F1 bound promoters of several representative genes..No occupancy was seen in the control intergenic region (lacking any putative E2F/TEAD binding sites). IgG served as specificity control for the antibody. Bars represent enrichment at target regions in the promoter relative to the 3’ region of each gene. (F) Dominant negative E2F1 (E2F1DN) suppresses proliferation of Yap1 (or Yap1S127A) expressing cells. Quantification from a representative experiment is shown on the right. Error bars represent SD of triplicate wells, ***p < 0.001.
See also Figure S5.
Our findings are in agreement with the known role of Yap1 in regulating normal cell proliferation through a Tead-mediated transcriptional program (Ota and Sasaki, 2008; Zhao et al., 2008; Schlegelmilch et al., 2011; Zhang et al., 2011; von Gise et al., 2012; Xin et al., 2013). Indeed, several bona fide Yap1 target genes including Ccnd1 and Birc5 were documented to be upregulated in Yap1 bypassed tumors. Together, these analyses support the view that Yap1 enables tumor growth upon KrasG12D extinction through the coordinate activation of genes governing cell cycle and DNA replication.
It has been suggested that Yap/Tead-mediated gene regulation may rely on combinatorial network of transcription factors to drive gene expression thresholds (Nicolay et al., 2011). To determine if Yap/Tead cooperates with a particular transcription factor in a coordinated gene expression program, we performed promoter analysis and identified several transcription factor motifs including E2F, several members of the ATF family (activating transcription factors), and CREB1 (cyclic-AMP response element binding protein 1) enriched in the promoters of differentially expressed genes in Yap1 bypassed tumors (Figure 5D and data not shown). We focused further study on the E2F family of transcription factors since recent studies in Drosophila have emphasized that E2f1 is required for the full activation of specific target genes by fly Yap1 and Tead orthologues, Yki and Sd (Nicolay et al., 2011).
To assess a potential cooperative role of E2F in Yap1/Tead2-mediated bypass of tumor regression, we first sought to determine co-occurrence of TEAD and E2F binding sites among the differentially expressed genes in the Yap1 bypassed tumors. Using the TRANSFAC position frequency matrix, we found significant enrichment for genes containing putative binding sites for both Tead and E2F specifically in the promoters of genes that were upregulated (2-fold upregulated, p-value<0.005) in the Yap1 bypassed tumors (19 out of 241; p-value<0.05). Using chromatin immunoprecipitation (ChIP), we next validated occupancy of E2F1 along with YAP1 (Flag-tagged) /TEAD2 (V5-tagged) at promoters of several representative genes with predicted E2F and TEAD binding sites, and showed no occupancy in an intergenic region lacking any putative E2F/TEAD binding sites (Figure 5E). We further validated occupancy of endogenous Yap1 at three representative loci in early passage cultures derived from Yap1 amplicon positive tumors (E-1 and E-2) but not Yap1 amplicon negative (E-3) tumor (Figure S5). As a specificity control, a nonspecific IgG antibody failed to immunoprecipitate any of the above promoter fragments (Figure 5E). These in silico and ChIP analyses support the view that E2f cooperates with Yap1/Tead2 in coordinating downstream gene expression.
Next, to assess the functional importance of E2F binding, we blocked E2F activity utilizing a dominant negative form of E2F1 (E2F1DN) that lacks the transactivation domain (Adams and Kaelin, 1996). Expression of the dominant negative E2F1 suppressed proliferation of Yap1 expressing cells (Figure 5F), supporting a role for E2F1 activity in enabling Yap1/Tead2-mediated bypass of tumor regression upon KrasG12D extinction.
KrasG12D -independent relapse tumors resemble QM-subtype of human PDAC
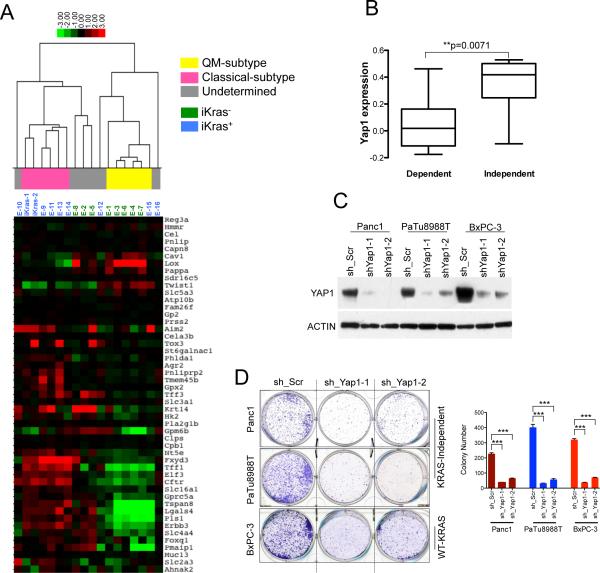
As noted, a subset of human PDAC cell lines become less dependent on oncogenic KRAS (Singh et al., 2009). To assess the potential clinical relevance of our Kras-independent relapse PDAC tumors, we compared the transcriptomic profiling of the relapse tumors with those reported in primary human PDACs and PDAC cell lines (Collisson et al., 2011). As a control, we also performed transcriptomic analysis of doxy-induced iKras PDAC lines. When subjected to unbiased clustering analysis, 5 of 8 iKras− relapsed tumor profiles clustered closely with each other and 5 of 8 iKras+ relapse tumors clustered with the doxy-induced primary PDAC lines (Figure 6A), reinforcing the view that iKras− relapse tumors are molecularly distinct from KrasG12D-dependent tumors.
Figure 6. KrasG12D-independent relapse tumors resemble the quasimesenchymal-subtype of human PDAC.
(A) Hierarchical clustering of murine PDAC iKras cells and the relapse tumors into different PDAC subtypes using PDAssigner genes (Collisson et al., 2011). Subtype analysis found statistically significant association between iKras− relapse tumors and the QM subtype while the iKras+ tumors are associated with the classical subtype (Chi square test, p-value=0.01; Collisson et al., 2011). The subtype identity of the samples (in gray) is not apparent. (B) Gene expressiondata reanalyzed from Collisson et al., 2011 showing YAP1 expression is significantly higher in the Kras-independent lines compared to Kras-dependent human PDAC cells. The y-axis indicates gene expression data expressed as log2 median centered intensity. Boxed bars indicate the medians. (C) Western blots validating the knockdown of YAP1 in the indicated human PDAC cell lines by two independent shRNAs. (D) Representative wells (top) of clonogenic growth of Kras-independent QM human PDAC cells (Panc1 and PaTu8988T) and the Wild-type KRAS cell line BxPC-3 upon YAP1 knockdown. Quantification (bottom) from a representative experiment is shown on the right. Error bars represent SD of triplicate wells, ***p < 0.001.
Human PDAC was recently defined into three subtypes based on transcriptional profiles: classical, quasimesenchymal and exocrine-like, which correspond with distinctive clinical outcome and therapeutic responses (Collisson et al., 2011). 6 of 10 KrasG12D-positive tumors, including the doxy-induced iKras lines and the iKras+ relapse tumors, show expression signatures of classical PDAC, which are highly linked to oncogenic Kras activity. In contrast, 5 of 8 iKras− lines clustered with QM-subtype of human PDAC, which have been reported to show high expression of mesenchymal genes, lower KRAS expression and less KRAS-dependency (Figure 6A; Collisson et al., 2011). The fact that we see a statistically significant association between iKras- relapse tumors and the quasimesenchymal-subtype of human PDAC aligns well with the observation that these human tumors tend to be less KRAS-dependent (Chi square test, p-value=0.01). Interestingly, Shao et al., demonstrated a YAP/FOS mediated epithelial to mesenchymal transition (EMT) program can also drive KRAS-independent tumor growth, further corroborating the relationship between KRAS-independence and Yap1 as well as underscoring the complexity of Yap1 signaling.
To gain insight into whether YAP1 might have a role in driving growth of human Kras-independent PDAC, we first compared its expression in previously established human Kras-dependent and Kras-independent PDAC cells (Singh et al., 2009; Collisson et al., 2011). In line with our murine findings, YAP1 expression was significantly higher in the human KRAS-independent PDAC cells (Figure 6B). Next, to determine whether YAP1 is required for growth of QM KRAS-independent subtype cells, we performed knockdown of YAP1 in two KRAS-independent QM human PDAC cell lines (Panc1 and PaTu8988T) and one wild-type KRAS cell line BxPC-3 (Figure 6C and 6D). shRNA-mediated knockdown of YAP1 strongly suppressed the proliferation of these cells implying that YAP1 is indeed essential for their growth (Figure 6D). Together, our data indicates that the oncogenic Kras-independent relapse tumors tend to resemble the QM-subtype of human PDAC and rely on alternative oncogenic mechanisms including Yap1 for their growth.
DISCUSSION
In this study, we investigated potential resistance mechanisms to oncogenic Kras extinction in the context of significant tumor burden. Following oncogenic Kras extinction and complete tumor regression in all animals, approximately one-third of the animals indeed remained tumor-free for over a period of up to 65 weeks. This observation, together with re-expression of the iKras transgene in approximately half of the mice with tumor recurrence, emphasizes the prominent role of oncogenic Kras in tumor maintenance. Tumor recurrence following complete extinction of oncogenic Kras was not anticipated given the wide spectrum of critical pathways controlled by Kras in cancer. At the same time, our work and that of Shao et al., demonstrate the potential for oncogenic Kras-independent bypass mechanisms involving the Yap1 oncogene, emphasizing that PDAC tumor cells can survive in the absence of oncogenic Kras signaling and acquire alternative mechanisms to foster their own growth, portending the need for anti-Yap1 therapeutic strategies for some tumors in the setting of agents targeting Kras and its signaling pathways.
Our results have important ramifications in anticipating clinical responses in drugs designed directly to target oncogenic Kras. Based on this study and resistance mechanisms discovered in response to other targeted therapies, at least three distinct resistance mechanisms against Kras extinction are possible. First, genomic alterations can act on target itself driving relapse by circumventing target blockade. This is supported by our observation that in approximately half of the relapse tumors, the iKras transgene is amplified. Second, augmentation of key growth signaling pathways through activation of compensatory signaling may induce tumor relapse as shown by Shao et al., that expression of receptor tyrosine kinases bypass the dependency on oncogenic Kras. Our preliminary gene expression data raises the intriguing possibility of activation of multiple RTKs and/or their ligands in Yap1 amplicon negative relapse tumors (E-3, E-4 and E6-E8, data now shown). Third, and most importantly, we and Shao et al., have uncovered a novel mechanism of resistance to Kras inhibition through a Yap1 mediated transcriptional program. Amplification of Yap1 in our study is reminiscent of classic second site suppression events, which substitute for critical functions of oncogenic Kras, particularly tumor cell proliferation, thus allowing Kras-dependent tumors to escape dependency on oncogenic Kras.
Yap1, a transcriptional co-activator and the downstream mediator of Hippo signaling, is regulated post-transcriptionally by either kinase-mediated degradation or cytoplasmic sequestration (Harvey et al., 2013; Zhao et al., 2011). Yap1 is known to be involved in cell proliferation, epithelial to mesenchymal transition, invasion and metastasis. Notably, recurrent amplifications of Yap1 locus 11q22 have been observed in liver carcinoma, oral squamous cell carcinomas, medulloblastomas, and esophageal cancer (Zender et al., 2006; Snijders et al., 2005; Fernandez et al., 2009; Muramatsu et al., 2011). Our findings indicate that Yap1 is sufficient for driving PDAC recurrence upon Kras withdrawal in the iKras model, previously characterized for its dependence on oncogenic Kras signaling for tumor initiation, progression and maintenance. However, Yap1 appears to be insufficient to drive de novo PDAC development since pancreas-specific inactivation of the Mst1/2 kinases and associated Yap1 activation and enhanced cell proliferation fails to initiate tumor formation (George et al., 2012; Gao et al., 2013). Several mechanisms might underlie the differences observed in iKras- tumor recurrence and in pancreas-specific deletion of Mst1/2 including the presence of intact tumor suppressor pathways or absence of transcription factor mediators in the latter study.
In our study, Tead2 is required for Yap1 mediated tumor relapse. Interestingly, Tead2 has been shown to play an important role in stem cell maintenance and self renewal (Cao et al., 2008; Tamm et al., 2011) and thus we speculate that residual surviving tumor cells or tumor stem cells following Kras extinction maintain their viability in Tead2-dependent manner. This rare sub-population of PDAC cells may be enriched for tumor initiating activity and be capable of surviving genetic or pharmacological inactivation of Kras and its surrogates. Such surviving cells could provide a reservoir of relapsed tumor cells to enable acquisition of Kras-independent tumor maintenance events. In support of this hypothesis, we have generated preliminary data showing Tead2 is highly expressed and important for survival of a sub-population of tumor cells that survive KrasG12D extinction (Kapoor and Viale et al., unpublished data). It is not clear, however, whether YAP1 amplification is already present in these rare oncogene-independent cells before KrasG12D ablation or is it acquired after oncogene extinction. Further work using clonal tracking methodologies would be needed to define the relationship between these surviving cells and the relapse tumors seen in our model.
The findings that Yap1 can substitute for oncogenic Kras in advanced PDAC raises the possibility that Yap1 can similarly substitute for activated RAS in other malignancies or in other cellular contexts. In agreement with this supposition, Shao et al., identified Yap1 in a gain of function screen to identify genes that can substitute for Ras signaling in KRAS-dependent human cancer cells. Consistent with the pleiotropic effects of Yap1, both studies converged on overlapping networks, such as ATF and E2Fs, and diverge on distinct transcriptional programs, such as Tead2 (this study) and Fos (Shao et al.,). Our convergent and contrasting findings are consistent with the established fact that Yap1-mediated gene expression program is largely dictated by the cellular context and its interacting transcription factors (Pobbati and Hong, 2013; Zhao et al., 2011).
Interestingly, the iKras− relapse tumors display features similar to the QM subtype of human PDACs associated with poor prognosis (Collisson et al., 2011). Although YAP1 amplified tumors clearly fall into the non-classical subtype, the limited number of tumors prevents us from definitely subgrouping the tumors with YAP1 amplification. However, the significant correlation between iKras status and classical/QM subtypes aligns with previously published results showing that Kras dependency is strongly linked to epithelial differentiation status and that, upon EMT, Kras dependency is reduced in human cancer cells (Singh et al., 2009). Furthermore, the observation by Shao et al., that YAP1 can replace oncogenic Kras in part by regulating an EMT-like program further supports the link between EMT status, Kras-independence and Yap1 expression as well as underscores the complexity of Yap1 signaling.
Future studies should be encouraged to examine the role of Yap1 in the recently defined QM-subset in human PDAC, which is notably less dependent on oncogenic Kras. The continued development of small molecules targeting Yap1 that have delayed tumor progression in mouse models of liver cancer (Liu-Chittenden et al., 2012, Stanger et al., 2012) may prove useful in human PDAC subtypes with elevated Yap1 expression.
EXPERIMENTAL PROCEDURES
Transgenic Mice
TetO_Lox-Stop-Lox-KrasG12D (tetO_LKrasG12D), ROSA26-LSL-rtTA-IRES-GFP (ROSA_rtTA), p48-Cre and Trp53L strains were described previously (Ying et al., 2012). Mice were interbred and maintained on FVB/C57Bl6 hybrid background in pathogen-free conditions at M.D. Anderson Cancer Center. Mice were fed with doxy water (Dox 2g/l in sucrose 20g/l). All manipulations were approved under MD Anderson Cancer Center (MDACC) Institutional Animal Care and Use Committee (IACUC) under protocol number 111113931.
In vivo Imaging
All in vivo imaging was performed at the Small Animal Imaging Facility at MDACC. MRI was performed weekly using with a 4.7T Bruker Pharmascan. For bioluminescent imaging animals were anesthetized with isoflurane, injected intraperitoneally with 3mg of D-Luciferin (Perkin Elmer) and imaged using IVIS Spectrum Imaging System (Perkin Elmer). The Living Image 4.3 software (Perkin Elmer) was used for analysis of the images post acquisition.
Xenograft Studies
All xenograft studies were carried out in NCr nude mice (Taconic) and were approved by the MD Anderson IACUC under protocol number 111113931. Details of the subcutaneous and orthotopic xenograft studies are listed in Extended Experimental Procedures.
Array-CGH Profiling and Analyses
For Array-CGH, genomic DNA processing (from tumor and matched tail DNA), labeling and hybridization to Agilent custom 415K mouse CGH array (Agilent Design ID 025735, NCBI GEO ID:- GPL15058) were performed as per the manufacturer's protocol (Agilent).
Lentivirus Mediated shRNA Knockdown
All lentiviral shRNA clones targeting Yap1, Tead2, Birc2, Birc3 and non-targeting shRNA control were obtained from Sigma Aldrich in the pLKO vector (Moffat et al., 2006). The TRC IDs for the shRNA used in the study are listed in the Extended Experimental Procedures.
Immunohistochemistry and Western Blot Analysis
Tissues were fixed in 10% formalin overnight and embedded in paraffin. Immunohistochemical (IHC) analysis was performed as described earlier (Aguirre et al., 2003). For western blot analysis, cells were lysed on ice using RIPA buffer (Boston BioProducts) supplemented with protease and phosphatase inhibitors (Roche). Primary antibodies used for immunohistochemistry (IHC) and western blot analysis are listed in Extended Experimental Procedures.
Gene Expression Profiling and Subtype Analysis
Gene expression profiling was performed using Affymetrix Gene Chip Mouse Genome 430 2.0 Arrays. Complete gene expression profiles are available at GEO at GSE53169. Details on mRNA expression profiling and data analysis are described in Extended Experimental Procedures.
Statistical Analysis
Tumor volume and tumor free survivals were analyzed using GraphPad Prism. To assess distributional differences of variance across different test groups, the Mann-Whitney test was used. Other comparisons were performed using the unpaired Student t-test. For all experiments with error bars, standard deviation (SD) was calculated to indicate the variation with each experiments and data, and values represent mean ± SD.
Supplementary Material
HIGHLIGHTS.
Spontaneous tumor recurs following oncogenic Kras extinction-induced tumor regression
Yap1 amplification drives Kras-independent tumor relapse in Tead2-dependent manner
Yap1/Tead2 cooperate with E2F in promoting a cell cycle gene expression program
Most Kras-independent tumors resemble the quasimesenchymal-subtype of human PDAC
ACKNOWLEDGEMENTS
We thank the laboratory of Kun-Liang Guan, William Kaelin, Jiandie D. Lin, Hiroshi Sasaki and Marius Sudol for sharing various reagents; Mien-Chie Hung and William Hahn for sharing unpublished data; Douglas Hanahan for helpful suggestions and support for PDAC subtype analysis; Shreya Malu for help with graphical abstract; Giannicola Genovese, Sharmistha Sarkar, Shruti Malu, Simona Colla and other members of the DePinho laboratory for helpful suggestions and technical support. We would also like to thank the Small Animal Imaging Facility, Flow Cytometry and Cellular Imaging Core, Histopathology Core, Sequencing and Non Coding RNA Core Services at The University of Texas, MD Anderson Cancer Center (Cancer Center Support grant, CA16672).
The project was supported by 5U01CA084313 (R.A.D), P01CA117969 (R.A.D), R56DK094865 (R.L.J) from National Institutes of Health, UT Star Award (R.A.D), American-Italian Cancer Foundation Award (G.D), NHS funding to the National Institute for Health Research (NIHR) Biomedical Research Centre for Cancer (A.S), Discovery Award W81XWH-12-1-0459 from Department of Defense (H.Y) and. G.D., H.Y. and T.H. were also supported by funds from the Sheikh Ahmed Center for Pancreatic Cancer Research at the University of Texas MD Anderson Cancer Center. Additional support was provided by the Jane Coffin Childs Postdoctoral fellowship to A.K (61-1512), Damon Runyon Foundation Postdoctoral Fellowship to S.H. and TRIUMPH Postdoctoral fellowship to N.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures and five figures.
REFERENCES
- Adams PD, Kaelin WG., Jr. The cellular effects of E2F overexpression. Current topics in microbiology and immunology. 1996;208:79–93. doi: 10.1007/978-3-642-79910-5_4. [DOI] [PubMed] [Google Scholar]
- Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K, Bernards R. Understanding resistance to targeted cancer drugs through loss of function genetic screens. Drug resistance updates : reviews and commentaries in Antimicrobial and Anticancer chemotherapy. 2012;15:268–275. doi: 10.1016/j.drup.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, Rakshit S, Flannagan KS, Adsay NV, Pasca di Magliano M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122:639–653. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson EA, Trejo CL, Silva JM, Gu S, Korkola JE, Heiser LM, Charles RP, Rabinovich BA, Hann B, Dankort D, et al. A central role for RAF-->MEK-->ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2012;2:685–693. doi: 10.1158/2159-8290.CD-11-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser S, Reiff N, Messer M, Seidler B, Gottschalk K, Dobler M, Hieber M, Arbeiter A, Klein S, Kong B, et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell. 2013;23:406–420. doi: 10.1016/j.ccr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- Fernandez LA, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S, Taylor MD, Kenney AM. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Zhou D, Yang C, Singh T, Penzo-Mendez A, Maddipati R, Tzatsos A, Bardeesy N, Avruch J, Stanger BZ. Hippo signaling regulates differentiation and maintenance in the exocrine pancreas. Gastroenterology. 2013;144:1543–1553. 1553, e1541. doi: 10.1053/j.gastro.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George NM, Day CE, Boerner BP, Johnson RL, Sarvetnick NE. Hippo signaling regulates pancreas development through inactivation of Yap. Mol Cell Biol. 2012;32:5116–5128. doi: 10.1128/MCB.01034-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Barbacid M. Genetically engineered mouse models of pancreatic adenocarcinoma. Molecular Onc. 2013;7:232–247. doi: 10.1016/j.molonc.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, Serrano M, Campuzano V, Barbacid M. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- Hill R, Calvopina JH, Kim C, Wang Y, Dawson DW, Donahue TR, Dry S, Wu H. PTEN loss accelerates KrasG12D-induced pancreatic cancer development. Cancer Res. 2010;70:7114–7124. doi: 10.1158/0008-5472.CAN-10-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancer revelaed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauchle JO, Kim D, Le DT, Akagi K, Crone M, Krisman K, Warner K, Bonifas JM, Li Q, Coakley KM, et al. Response and resistance to MEK inhibition in leukaemias initiated by hyperactive Ras. Nature. 2009;461:411–414. doi: 10.1038/nature08279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F. Cancer therapy based on oncogene addiction. Journal of Surgical Onc. 2011;103:464–467. doi: 10.1002/jso.21749. [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Muramatsu T, Imoto I, Matsui T, Kozaki K, Haruki S, Sudol M, Shimada Y, Tsuda H, Kawano T, Inazawa J. YAP is a candidate oncogene for esophageal squamous cell carcinoma. Carcinogenesis. 2011;32:389–398. doi: 10.1093/carcin/bgq254. [DOI] [PubMed] [Google Scholar]
- Nicolay BN, Bayarmagnai B, Islam AB, Lopez-Bigas N, Frolov MV. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev. 2011;25:323–335. doi: 10.1101/gad.1999211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- Pobbati AV, Hong W. Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther. 2013;14:390–398. doi: 10.4161/cbt.23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RC, Albertson DG. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–4242. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- Stanger BZ. Quit your YAPing: a new target for cancer therapy. Genes Dev. 2012;26:1263–1267. doi: 10.1101/gad.196501.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm C, Bower N, Anneren C. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J Cell Sci. 2011;124:1136–1144. doi: 10.1242/jcs.075796. [DOI] [PubMed] [Google Scholar]
- Torti D, Trusolino L. Oncogene addiction as a foundational rationale for targeted anti-cancer therapy: promises and perils. EMBO Molecular Medicine. 2011;3:623–636. doi: 10.1002/emmm.201100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A, Lin Z, Schlegelmilch K, Honor LB, Pan GM, Buck JN, Ma Q, Ishiwata T, Zhou B, Camargo FD, et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci. 2012;109:2394–2399. doi: 10.1073/pnas.1116136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. Oncogenic Kras Maintains Pancreatic Tumors through Regulation of Anabolic Glucose Metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Pasolli HA, Fuchs E. Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc Natl Acad Sci. 2011;108:2270–2275. doi: 10.1073/pnas.1019603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.