Abstract
During development, the genome adopts specific chromatin states to establish and maintain functionally distinct cell types in a well-controlled environment. A select group of transcription factors have the ability to drive the transition of the genome from a pluripotent to a more specialized chromatin state. The same set of factors can be used as reprogramming factors to reset the already established chromatin state back to pluripotency or directly to an alternative cell type. However, under the suboptimal reprogramming conditions, these factors fall short in guiding the majority of cells to their new fate. In this review, we visit the recent findings addressing the manipulation of chromatin structure to enhance the performance of transcription factors in reprogramming. The main emphasis is on the mechanisms underlying the conversion of somatic cells to pluripotency using OSKM. This review is intended to highlight the windows of opportunities for developing mechanistically-based approaches to replace the phenotypically-guided methods currently employed in reprogramming, in an attempt to move the field of cell conversion towards using next generation technologies.
Introduction
Within adult tissues and organs, fully differentiated cells rarely, if ever, change from one type to another. Somatic cells can be forcibly reprogrammed to pluripotency by nuclear transfer experiments, in which the somatic genome is exposed to a large number of factors found in the egg cytoplasm [1]. Thus it seems remarkable to discover that it can take so few transcription factors to convert cells from one somatic type to another. For instance, MyoD alone can reprogram fibroblasts to myoblasts [2], but more strikingly, the four transcription factors: Oct4, Sox2, Klf4, and c-Myc (OSKM) are able to convert fibroblasts to induced pluripotent stem cells (iPSCs) [3]. However, somatic cells still show a high resistance to such transcription factor-based reprogramming, raising a major obstacle in understanding the molecular mechanism underlying cellular conversion to pluripotency. With this in mind, many studies have recently reported various ways to enhance reprogramming, usually involving a change in the chromatin state of somatic genome to enable a change in cell fate.
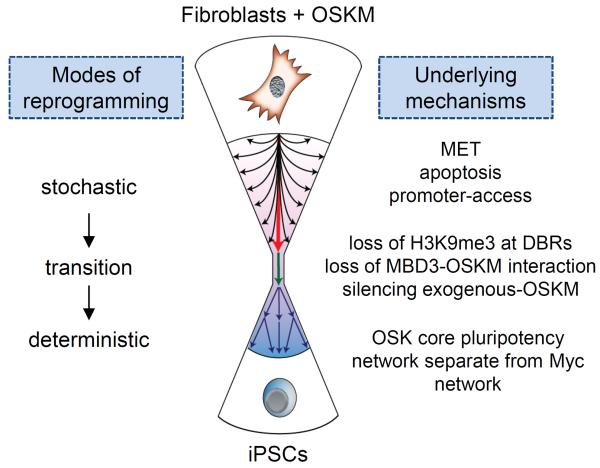
Our understanding so far is that the conversion of somatic cells to pluripotency follows a step-wise process (recently reviewed in [4–6]). However, it has been argued that reprogramming can operate in two modes; a stochastic mode, by which iPS colonies appear with variable latencies, and a deterministic mode in which differentiated cells follow a hierarchal process to pluripotency (Figure 1) [7,8]. Here we will review recent findings of how the different methods to enhance reprogramming fit this model, with a specific emphasis on the chromatin-basis behind both the pliancy and the rigidity of the somatic genome.
Figure 1.
Reprogramming somatic cells to pluripotency is initiated by a stochastic phase followed by a deterministic phase. The ectopic expression of OSKM in fibroblasts drives cells to go through many pathways stochastically (represented by black arrows). Some of these routes represent dead ends and others will lead to successful reprogramming (red arrow). The transition phase (green arrow) is a hallmark of initiating a cascade of deterministic events (blue arrows) resulting in fully reprogrammed iPSCs. The main pathways and processes that define each phase are displayed on the left.
The Pioneer Concept in Reprogramming
A select group of transcription factors, but not others, have the mechanistic ability to reprogram cells. It is intriguing to note that transcription factors involved in the early stages of embryonic development have provided an attractive route for cell fate conversion [9]. Pioneer factors are expressed early in development, and represent a special class of factors that can bind target DNA on nucleosomes [10–13]. This allows pioneer factors to engage silent chromatin and endow the competence for subsequent gene activation [14]. Early in reprogramming, the OSK factors, but not c-Myc, are able to access closed chromatin at distal element and prior to activation of silent genes, including those necessary for pluripotency such as ESRRB and SALL4 [15]. The pioneer activity of Oct4 has been confirmed in the maternal-to-zygotic transition at which Oct4 occupies SOX-POU binding sites before the onset of zygotic transcription [16,17]. The pioneer activity of Oct4 has been carefully assessed in binding to the enhancer elements of NANOG, POU5F1, and MYOD1 genes [18,19]. The concept of pioneer factors expands beyond reprogramming to pluripotency as shown for the case of Ascl1, which can convert fibroblasts to become induced neuronal (iN) cells [20]. Altogether, pioneer factors seem to possess an inherent ability to prime the genome to become susceptible for adapting chromatin states, which are more suited for alternative cell types.
Dissecting OSKM Function in Reprogramming by a Transcription Factor Substitution Approach
Soon after the discovery of OSKM, another set of factors including Oct4, Sox2, Nanog, and Lin28 have been shown to convert somatic cells to pluripotency [21]. Despite these factors being picked as candidates for reprogramming based on their role in pluripotency in ES cells [3,21,22], subsequent studies have attempted to dissect the role of OSKM in reprogramming by using substitutes, and surprisingly showing that they can be replaced with functionally divergent factors. For example the nuclear receptor Nr5a2 and its close family member Nr5a1 are capable of both enhancing reprogramming and replacing Oct4 [23]. The pioneer factors Gata3, Gata4, and Gata6 can replace Oct4, while Sox1 and Sox3 can replace Sox2 if combined with Klf4 and c-Myc [24,25]. The reprogramming activity of Oct4 is conserved among species, as axolotl Oct4, the xenopus Oct91, as well as medaka Pou2 are each able to act together with mouse SKM to generate iPSCs from mouse fibroblasts [26,27]. Oct1 and Oct6 are thought to lack reprogramming capability due to their unstructured linker region between Pou-specific and Pou-homeodomain; the two DNA-binding domains found in the POU (Pit1, Oct1/Oct2, UNC-86) family members [28]. Other Sox family members including Sox7 and Sox17 can be converted into reprogramming factors by engineering a single mutation within the High-Mobility-Group (HMG) DNA-binding domain that promotes co-binding with Oct4 to the canonical Oct-Sox motif [29,30]. Interestingly, Oct4 can switch partners from Sox2 to Sox17 for endoderm specification and binds a compressed version of the Oct-Sox motif at enhancers [31]. Similarly, Sox2 switches partners with another Pou-family member BRN2 (Pou3f2) co-occupying enhancers for neuron progenitors [32]. Sox2 targets yet another distinct network for specifying trophoblast stem cells and marks adult cells in several epithelial tissues [33,34]. Esrrb, an orphan nuclear receptor that is expressed highly in ES cells, has been reported to replace Klf4 [35]. c-Myc was the first factor found to be dispensable for reprogramming and can be replaced by other family members such as n-Myc and l-Myc, as well as by the maternal factor Glis-1 [36–38]. The ability of maternal factors to enhance reprogramming may reflect the potent ability of oocytes to reprogram somatic cells. It is important to note that pioneer factors also appear to interfere with OSKM and counteract their induction to pluripotency, once co-expressed with OSKM [39,40]. Remarkably, these interfering factors extensively co-bind with OSKM [39]. For example, Gata4 can replace Oct4 in reprogramming, yet Gata4 blocks reprogramming if co-expressed with OSKM [24,40]. This indicates that OSKM substitutes engage the somatic genome by binding many overlapping targets, creating alternative, yet conflicting, routes to pluripotency.
Dissecting OSKM Function in Reprogramming by Small Molecule Substitution
Small molecules can also enhance reprogramming and replace the four OSKM factors. Vitamin C enhances the generation of iPS cells, at least partly, due its Tet-dependent induction of DNA demethylation [41–43]. Valporic acid (VPA), suberoylanilide hydroxamic acid (SAHA), trichostatin A (TSA), and butyrate are histone deacetylases (HDAC) inhibitors that can improve reprogramming and replace c-Myc [44,45]. Tranylcypromine, an H3K4 demethylation inhibitor, also promotes iPSC generation in the absence of c-Myc [46]. 5-azacytidine (AZA), a DNA methyltransferase inhibitor, also enhances reprogramming [47,48]. Kenpaullone, like CHIR99021, both inhibit GSK-3β and increase OSKM-based reprogramming and Kenpaullone can replace Klf4 [49]. Tgf-β inhibitor (named 616452), on the other hand, can replace Sox2 and induce reprogramming [50,51]. Forskolin (FSK), a cAMP agonist, can act as a chemical substitutes for Oct4 [52]. The small-molecule combination of VPA, CHIR99021, 616452, and Tranylcypromine enables reprogramming of mouse fibroblasts with only a single gene, Oct4 [46]. And more dramatically, the combination of CHIR99021, 616452, Forskolin and DZNep, an S-adenosylhomocysteine (SAH) hydrolase inhibitor, can replace all four reprogramming factors [52]. Albeit, this cocktail of chemicals can induce pluripotency in mouse with much less efficiency than OSKM, they failed to do so in human [52]. It is important to note that DZNep does not replace any of the factors but instead reduces the global levels of histone methylation associated with heterochromatin including H3K27, H3K9, and H4K20 methylation [53,54]. By these means, DZNep may act at the early stochastic phase of reprogramming as well as the late transition to a deterministic step, as discussed further below. The general view from these studies is that small molecules that induce the loosening of chromatin assist or replace OSKM in reprogramming indicating that OSKM's major role is to open up chromatin of silent genes. However these chemicals target chromatin modifiers that show no cell-type specificity, so they may be useful to induce pluripotency, which is characterized by a general open chromatin state, they won't be useful to directly convert cells to another differentiated cell type.
Stochastic Mode of Reprogramming
Reprogramming is initiated by OSKM binding to the somatic genome at many more sites than will be functional for pluripotency [15]. This unrestrained mode of binding may be simply due to the level of overexpression of the factors, highlighting the importance of OSKM stoichiometry during reprogramming [55–58]. OSKM initially target an extensively overlapped gene network on the somatic genome that is markedly different from the separate OSK core-pluripotency and c-Myc networks found in ES and iPS cells [15,59–61]. Furthermore, the OSKM factors initially bind distal to a large number of genes that both promote “on-target” and impede “off-target” reprogramming [15]. There is a stark difference between OSKM promiscuous binding and Ascl1 “on-target” binding, explaining the higher efficiency of reprogramming to a neuronal fate compared to pluripotency [20]. Although Ascl1 acted as an “on-target” pioneer factor in fibroblasts, it did not do so in keratinocytes, which is in part due to a trivalent chromatin state in fibroblasts that is permissive for Ascl1 occupancy [20]. This argues that somatic cells may be fortuitously primed to be converted to a specific cell type if using the right set of pioneer factor. Regardless, the promiscuous engagement of the somatic genome by OSKM provide a route not only to pluripotency but also to other somatic fates such as neurogenesis and cardiogenesis [62,63].
Other routes induced by OSKM may lead to a dead-end, such as apoptosis, which is considered as a barrier to reprogramming [64–66]. The Mesenchymal-to-Epithelial (MET) transition is one of the early routes that most cells seem to follow when using the OSKM factors [67,68]. The sequential addition approach (Oct4–Klf4 first, then c-Myc and finally Sox2) outperforms the simultaneous introduction of OSKM, but induce EMT first and then MET [69]. Recently, by replacing Oct4 and Sox2 with other lineage specifiers, MET was shown not to be required for reprogramming, but instead a “Seesaw Model” balancing the counteracting mesendodermal and ectodermal lineages was proposed to drive cellular conversion to pluripotency [24,25]. These conclusions were drawn from gene expression analysis in cell populations, where the minority of cells goes through successful conversion to pluripotency. Complimentary studies that worked with purified cell populations have shown that initially cells poised to reprogramming are very similar to those refractory to reprogramming and distinct waves of changes occur progressively during the process [70,71]. These waves include chromatin reorganization, changes in gene expression, cell proliferation, metabolism, DNA-repair and RNA processing [70,71]. Altogether these genes and pathways targeted by OSKM and induced early are not indicative of successful reprogramming. Nevertheless, some of these genes and pathways promote, while the others offset the process of cellular conversion. Indeed, clonal cell populations and single cells studies have confirmed that initially cells go through a stochastic phase before transitioning to more deterministic progression to pluripotency [7,8]. Even turning on pluripotency genes such as Oct4, which is required for pluripotency, are not predictive for successful reprogramming [8]. In summary, the unrestrained nature of OSKM engagement with the somatic genome lead to stochastic genomic responses, which are not necessarily indicative for routes to pluripotency, but are still necessary to increase the probability for cellular conversion.
Enhancing the Stochastic Route to Pluripotency
Initially, OSK are mainly associated with distal elements of silent genes, indicating that promoter accessibility and gene reactivation is a later event in reprogramming [15]. One way that c-Myc may enhance the process is by co-binding with OSK at distal elements and promoting open chromatin as well as promoter accessibility, resulting in a general amplification of gene expression [15,36,37,72–74]. Among genes changing expression early in reprogramming are genes targeted mainly by c-Myc and Klf4 at their promoters and that either gain the open chromatin mark H3K4me2, de novo, or retain this active mark if pre-existing in fibroblasts [15,75]. The ATP-dependent BAF chromatin-remodeling complex has been shown to facilitate the access of Oct4 to promoters and enhance reprogramming even in the absence of c-Myc [76]. The two H3K36me2 demethylases, KDM2A and KDM2B were also shown to enhance reprogramming by cooperating with Oct4 to demethylate promoters and activate gene expression, including that of the micro-RNA gene-cluster 302/367, which is known to induce reprogramming [77–79]. OSK factors can also recruit UTX, an H3K27 demethylase, and WDR5, a core member of the mammalian Trithorax complex that effect H3K4 methylation, to promote open chromatin and enhance reprogramming [80,81]. The general transcription factor, TFIID, that is essential for the assembly of the basal-transcription machinery at promoters to activate gene expression, have also been shown to enhance reprogramming [82]. In conclusion, the OSKM factors are initially wired to an extensive gene network, and hence reprogramming can be stochastically enhanced by stimulating a general and non-selective gene-expression output. This stochastic response can be skewed towards pluripotency if alternative routes are blocked. For example, the down-regulation of H3K79-methyl-transferase DOT1L enhances reprogramming by blocking the mesenchymal fate, from where fibroblasts originate [83]. Blocking apoptosis and senescence, which is the ultimate fate of the majority of cells induced by OSKM, also has a significant boost to reprogramming [64–66].
Deterministic Mode of Reprogramming
In nuclear-transfer experiments, the somatic genome is exposed to an undefined number of reprogramming factors found in the egg's cytoplasm, as well as stripped from factors that endorse the differentiated state found in the somatic cytoplasm. Under these conditions the somatic genome follow deterministic steps, adopting a pluripotent state along the way [84]. So how can a genome that is exposed to only four factors and in the presence of all the endogenous somatic factors, transition to a pluripotent state in a deterministic fashion? The heterogeneous nature of cells going through reprogramming, due to the stochastic events described above, make it challenging to define the deterministic steps taking place in the minority of cells that successfully convert to pluripotency. In an attempt to answer this question, single-cells converting to pluripotency have been tracked morphologically using time-lapse microscopy, to reveal that they follow defined changes including a shrinking in cell size and rapid shift in cell division [85]. To gain more insights into the molecular events that drives successful reprogramming, gene expression dynamics of 48 genes, representing various pathways reported to be involved in reprogramming, have been monitored at single-cells level [8]. This approach has unexpectedly revealed that the reactivation of pluripotency genes such as Oct4 and other genes that promote MET or cell proliferation, despite being necessary, are not predictive of a deterministic pluripotency fate [8]. On the other hand, the reactivation of “top-hierarchy” pluripotency genes such as Nanog and Sox2 are indicative of cells destined for pluripotency [8]. Global genomic and proteomic analysis of sorted cell populations has indicated that the activation of endogenous Nanog and Sox2 is followed by the activation of a cascade of genes including Gdf3, Dppa2, and Dppa3 to stabilize pluripotency [67,70,71,86]. It is intriguing to discover that all of these genes are located within mega-base-scale heterochromatin domains marked by H3K9me3 in fibroblasts that are inaccessible by OSKM early in reprogramming [15]. Quantitative mass-spectrometry has identified that fibroblasts and late reprogramming intermediate (pre-iPSCs), are enriched for H3K9me3, when compared to fully reprogrammed iPSCs [87]. These OSKM-differentially bound regions (DBRs) are conserved in a wide range of cell-types and partly persist in iPS lines, conferring epigenetic memory [15,88–91]. It is worth noting that, the exogenous expression of OSKM must be silenced for transitioning to the deterministic mode of reprogramming [59,86,92]. Failing to silence these over-expressed factors destabilizes pluripotency and occasionally results in partially-reprogrammed cells (pre-iPSCs), that seem to be trapped at this late phase [59,86,93]. By assessing the chromatin occupancy of OSKM in pre-iPSCs, it was shown that c-Myc is associated with promoters in a similar way to that in iPS/ES cells, however OSK are only partly associated with the pluripotency network found in iPS/ESCs [59–61]. Hence, the promiscuous nature of OSK binding to the genome has to be rearranged to “on-target” binding for reprogramming to progress to a deterministic mode. In contrast to nuclear transfer experiments, transcription-factor-based reprogramming requires the genome to be primed by pioneer factors to a “more-pliable” chromatin state before the genome can deterministically transition to a stable pluripotency state.
Enhancing the Deterministic Route to Pluripotency
Modulating the pre-existing chromatin state of the somatic genome is usually the favorable route to enhance reprogramming. One way to induce the transition to a deterministic phase is to remove chromatin barriers that block OSKM access to the top-hierarchy pluripotency genes, which are located within heterochromatin marked by H3K9me3 in fibroblasts [15,88]. Knocking down enzymes that deposit H3K9me3, including SUV39H1/H2 induces OSKM access to heterochromatin and significantly enhances the efficiency and kinetics of programming [15,83,87]. BMP signaling was shown to induce H3K9me3 levels and is responsible for arresting reprogramming in pre-iPSCs, while Vitamin C reduces the levels of H3K9me3 and allow the transition of pre-iPS to fully reprogrammed iPSCs [41,94]. It was also shown that the non-essential amino acid L-proline augments H3K9me3 levels and inhibits converting cells to pluripotency [95]. It is not known how OSKM get access to heterochromatin during the course of reprogramming. It has been reported that Parp1 and Tet2 are recruited to Nanog locus early in reprogramming, and contribute to its subsequent transcriptional induction [96]. However, it is not known whether this takes place prior to or after OSKM access.
Modulating the novel chromatin state as it is modified by OSKM binding may also guide cells to pluripotency. It was reported that depleting the NuRD repressor complex component, MBD3, during reprogramming with OSKM results in a synchronized cell population that deterministically become converted to iPSCs [97]. However, other studies reported only moderate positive effects upon MBD3 knock-down in reprogramming [83,98]. There are several experimental conditions that might account for the discrepancies between these studies. One condition of the former study [97] is that MBD3 was depleted in a clonal secondary-reprogramming system versus the traditionally used primary system. The inter- and intra-clonal efficiencies were accurately measured from separate clones derived from single cells in the secondary system, which differs from the primary method that measures efficiencies by dividing the number of generated iPSC colonies by the number of input cells. This has profound impact on assessing MBD3 effects, for example using the clonal secondary system MBD3 depletion increased the efficiency of reprogramming from 15% to 95% (~7 fold), while in the primary system MBD3 led to an increase from 0.15% to 1.5% (~10 fold) [97,98]. So there is a net 100 fold increase in efficiency of wild type MBD3 just by using the clonal secondary over the primary system. Another difference is the presence of LIF and inhibitors against the GSK3b and MEK kinases in the media for the secondary system [97], which differs from the conditions that are commonly used [83,98]. Surprisingly, depleting MBD3 prior to OSKM induction seems to decrease reprogramming efficiency [83]. A positive effect on reprogramming was only reported when MBD3 was depleted at the same time or, to a larger extent, two days after OSKM induction [97,98]. Nevertheless, MBD3 appears to be recruited by OSKM to promote the formation of heterochromatin, counteracting the reprogramming process [97]. It is intriguing that the re-introduction of MBD3 late in reprogramming had no inhibitory effects on the efficiency, as if MBD3 is only interacting with OSKM at the “off-target” sites early during the process. It is important to note that cells depleted from MBD3 still go through several cell divisions (~7 days), before they get converted to pluripotency [97]. This is different from the deterministic mode of nuclear transfer, in which cells become converted within 48 hrs and without the need to go through cell division [84]. Also of note, the exogenous OSKM expression in MBD3 depleted cells was required for the same duration of time (5–7 days) as in wild type MBD3 in order to generate iPSCs [97]. Altogether, these results indicate that in the absence of MBD3, it would take OSKM at least five days to prime the somatic genome in a synchronous fashion before they get silenced, leaving behind a self-sustaining pluripotency program. So MBD3 does not appear to be enhancing the late deterministic phase but rather synchronizing the early stochastic phase. It is to be determined whether the effect of MBD3 on reprogramming is specific to OSKM or can be expanded to other transcription factors. To dissect the role of MBD3 from culture conditions, it is necessary to re-assess other chromatin modifiers known to enhance reprogramming, under these altered conditions and see if they have the same effects as MBD3.
Perspectives
Despite the short time since the discovery that OSKM can convert somatic cells to pluripotency, we have learned a great deal by using the same concept with alternative transcription factors in a wide range of cell types. Through these studies, the field of reprogramming came to appreciate the dependence of cell identity on the chromatin state of the genome. Manipulating the chromatin state of the genome, however, has been largely based on targeting general chromatin modifiers that show no cell-type specificity. Understanding the cell-type specificity component of the chromatin barriers to reprogramming such as H3K9me3 will greatly contribute to design factors that overcome barriers only required for cell-type-specific conversion. Also, cellular conversion experiments have been heavily reliant on phenotypic-based approaches by mixing factors in their natural protein sequence and observing the cellular output. These factors have not evolved to reprogram cells under culture conditions. It appears time to consider mechanistically-based approaches to engineer reprogramming factors that efficiently convert cells to the specific type of interest. Engineering factors with enhanced reprogramming activity have been attempted by appending transactivation domains from alternative factors such as MyoD and VP16 [99–101]. On top of trans-activation domains, pioneer factors such as FoxA contain other functional domains to interact with nucleosomes, create hypersensitive sites, move on chromatin, and stay bound to mitotic chromatin [13,102–104]. Thus, exploiting the pioneer activity of OSKM by dissecting which functions and domains are essential or missing from the reprogramming factors may help to design better factor for cellular conversion.
Acknowledgment
I thank Prof. Kenneth Zaret for his insightful comments and guidance on preparing the manuscript. A.S. is supported by a US National Institute of Health (NIH) P01 grant (P01GM099134) to K.Z.
References
- 1.Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- 2.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 2013;14:427–39. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papp B, Plath K. Epigenetics of Reprogramming to Induced Pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (**) 8.Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell. 2012;150:1209–22. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using two single-cell approaches, the authors measured gene expression in the minority of cells going through successful reprogramming. The authors revealed an early stochastic phase with varying latencies followed by a late deterministic phase that starts with the activation of top hierarchy pluripotency genes such as Sox2 and Nanog.
- 9.Pasque V, Miyamoto K, Gurdon JB. Efficiencies and mechanisms of nuclear reprogramming. Cold Spring Harb. Symp. Quant. Biol. 2010;75:189–200. doi: 10.1101/sqb.2010.75.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu JK, Bergman Y, Zaret KS. The mouse albumin promoter and a distal upstream site are simultaneously DNase I hypersensitive in liver chromatin and bind similar liver-abundant factors in vitro. Genes Dev. 1988;2:528–41. doi: 10.1101/gad.2.5.528. [DOI] [PubMed] [Google Scholar]
- 11.McPherson CE, Shim EY, Friedman DS, Zaret KS. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell. 1993;75:387–98. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- 12.Cirillo LA, Zaret KS. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell. 1999;4:961–969. doi: 10.1016/s1097-2765(00)80225-7. [DOI] [PubMed] [Google Scholar]
- 13.Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim EY, Clark KL, Burley SK, Zaret KS. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (**) 15.Soufi A, Donahue G, Zaret KS. Facilitators and Impediments of the Pluripotency Reprogramming Factors' Initial Engagement with the Genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors used ChIP-seq to map the initial interaction of OSKM with the somatic genome during reprogramming. OSK act as pioneer factors and engage silent chromatin at distal elements prior to promoter access and gene activation. Mega-base scale heterochromatin regions that are enriched for H3K9me3 are refractory to the initial OSKM binding and impede the efficiency of reprogramming.
- (*) 16.Leichsenring M, Maes J, Mössner R, Driever W, Onichtchouk D. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science. 2013;341:1005–9. doi: 10.1126/science.1242527. [DOI] [PubMed] [Google Scholar]
- (*) 17.Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503:360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]; Studies in refs 16 and 17 show that Oct4 (Pou5f1) act as a pioneer factor and target zygotic genes, prior to activation, at the onset of the maternal-to-zygotic transition.
- 18.You JS, Kelly TK, De Carvalho DD, Taberlay PC, Liang G, Jones PA. OCT4 establishes and maintains nucleosome-depleted regions that provide additional layers of epigenetic regulation of its target genes. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14497–502. doi: 10.1073/pnas.1111309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taberlay PC, Kelly TK, Liu CC, You JS, De Carvalho DD, Miranda TB, Zhou XJ, Liang G, Jones PA. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell. 2011;147:1283–1294. doi: 10.1016/j.cell.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (**) 20.Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, Giresi PG, Ng YH, Marro S, Neff NF, et al. Hierarchical Mechanisms for Direct Reprogramming of Fibroblasts to Neurons. Cell. 2013;155:621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors mapped the association of Ascl1, Brn2 and Myt1l with the genome during the early phase of reprogramming fibroblasts to induced Neurons (iN). The authors revealed that Ascl1 acts as an “on-target” pioneer factor by binding to most of genomic targets in fibroblasts as in iN cells. In contrast, Brn2 and Myt1l binding to cognate targets in fibroblast depends on Ascl1 binding. The authors also revealed that Ascl1 initial binding correlates with a tri-histone modification, pre-existing in fibroblast.
- 21.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Heng J-CD, Feng B, Han J, Jiang J, Kraus P, Ng J-H, Orlov YL, Huss M, Yang L, Lufkin T, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–74. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- (**) 24.Shu J, Wu C, Wu Y, Li Z, Shao S, Zhao W, Tang X, Yang H, Shen L, Zuo X, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–75. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (**) 25.Montserrat N, Nivet E, Sancho-Martinez I, Hishida T, Kumar S, Miquel L, Cortina C, Hishida Y, Xia Y, Esteban CR, et al. Reprogramming of Human Fibroblasts to Pluripotency with Lineage Specifiers. Cell Stem Cell. 2013;13:341–350. doi: 10.1016/j.stem.2013.06.019. [DOI] [PubMed] [Google Scholar]; Studies in refs 24 and 25 show that human and mouse fibroblasts can be reprogrammed to pluripotency using lineage specifiers that are not known to be involved in pluripotency. These studies show that pioneer factors such as Gata3, Gata4, and Gata6 can replace Oct4 in reprogramming. Both studies propose a seesaw model in which pluripotency is induced by balancing the counteracting mesendodermal and ectodermal specification.
- (*) 26.Tapia N, Reinhardt P, Duemmler A, Wu G, Araúzo-Bravo MJ, Esch D, Greber B, Cojocaru V, Rascon CA, Tazaki A, et al. Reprogramming to pluripotency is an ancient trait of vertebrate Oct4 and Pou2 proteins. Nat. Commun. 2012;3:1279. doi: 10.1038/ncomms2229. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reports that different vertebrate Pou2/Oct4 homologues can induce pluripotency in mouse and human fibroblasts. This study highlights the reprogramming ability of Oct4 is conserved between evolutionary-diverged species.
- 27.Jerabek S, Merino F, Schöler HR, Cojocaru V. OCT4: Dynamic DNA binding pioneers stem cell pluripotency. Biochim. Biophys. Acta - Gene Regul. Mech. 2013 doi: 10.1016/j.bbagrm.2013.10.001. pii: S1874-9399(13)00142-9. doi: 10.1016/j.bbagrm.2013.10.001. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- (*) 28.Esch D, Vahokoski J, Groves MR, Pogenberg V, Cojocaru V, Vom Bruch H, Han D, Drexler HCA, Araúzo-Bravo MJ, Ng CKL, et al. A unique Oct4 interface is crucial for reprogramming to pluripotency. Nat. Cell Biol. 2013;3:295–301. doi: 10.1038/ncb2680. [DOI] [PubMed] [Google Scholar]; Using X-ray crystallography, the authors have determined the structure of the POU domain of Oct4 in complex with DNA. The authors revealed that the linker between the Pous and PouHD domains is structured as α-helix and exposed to the protein's surface, in contrast to the unstructured linker of Oct1. This linker region serves as protein-protein interaction interface and is crucial for reprogramming.
- 29.Jauch R, Aksoy I, Hutchins AP, Ng CKL, Tian XF, Chen J, Palasingam P, Robson P, Stanton LW, Kolatkar PR. Conversion of Sox17 into a pluripotency reprogramming factor by reengineering its association with Oct4 on DNA. Stem Cells. 2011;29:940–51. doi: 10.1002/stem.639. [DOI] [PubMed] [Google Scholar]
- 30.Aksoy I, Jauch R, Eras V, Bin ACW, Chen J, Divakar U, Ng CKL, Kolatkar PR, Stanton LW. Sox transcription factors require selective interactions with Oct4 and specific transactivation functions to mediate reprogramming [Internet] Stem Cells. 2013;31:2632–2646. doi: 10.1002/stem.1522. [DOI] [PubMed] [Google Scholar]
- 31.Aksoy I, Jauch R, Chen J, Dyla M, Divakar U, Bogu GK, Teo R, Leng Ng CK, Herath W, Lili S, et al. Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J. 2013;32:938–53. doi: 10.1038/emboj.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lodato MA, Ng CW, Wamstad JA, Cheng AW, Thai KK, Fraenkel E, Jaenisch R, Boyer LA. SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLoS Genet. 2013;9:e1003288. doi: 10.1371/journal.pgen.1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adachi K, Nikaido I, Ohta H, Ohtsuka S, Ura H, Kadota M, Wakayama T, Ueda HRR, Niwa H. Context-Dependent Wiring of Sox2 Regulatory Networks for Self-Renewal of Embryonic and Trophoblast Stem Cells. Mol. Cell. 2013;52:380–392. doi: 10.1016/j.molcel.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K. Sox2+ Adult Stem and Progenitor Cells Are Important for Tissue Regeneration and Survival of Mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng B, Jiang J, Kraus P, Ng J-H, Heng J-CD, Chan Y-S, Yaw L-P, Zhang W, Loh Y-H, Han J, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat. Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- 36.Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 38.Maekawa M, Yamaguchi K, Nakamura T, Shibukawa R, Kodanaka I, Ichisaka T, Kawamura Y, Mochizuki H, Goshima N, Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- 39.Hikichi T, Matoba R, Ikeda T, Watanabe A, Yamamoto T, Yoshitake S, Tamura-Nakano M, Kimura T, Kamon M, Shimura M, et al. Transcription factors interfering with dedifferentiation induce cell type-specific transcriptional profiles. Proc. Natl. Acad. Sci. 2013 doi: 10.1073/pnas.1220200110. doi:10.1073/pnas.1220200110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serrano F, Calatayud CF, Blazquez M, Torres J, Castell JV, Bort R. Gata4 blocks somatic cell reprogramming by directly repressing Nanog. Stem Cells. 2013;31:71–82. doi: 10.1002/stem.1272. [DOI] [PubMed] [Google Scholar]
- 41.Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–9. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–6. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stadtfeld M, Apostolou E, Ferrari F, Choi J, Walsh RM, Chen T, Ooi SSK, Kim SY, Bestor TH, Shioda T, et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 2012;44:398–405. S1–2. doi: 10.1038/ng.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, et al. Butyrate Greatly Enhances Derivation of Human Induced Pluripotent Stem Cells by Promoting Epigenetic Remodeling and the Expression of Pluripotency-Associated Genes. Stem Cells. 2010 doi: 10.1002/stem.402. doi:10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Zhang Q, Yin X, Yang W, Du Y, Hou P, Ge J, Liu C, Zhang W, Zhang X, et al. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011;21:196–204. doi: 10.1038/cr.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 49.Lyssiotis CA, Foreman RK, Staerk J, Garcia M, Mathur D, Markoulaki S, Hanna J, Lairson LL, Charette BD, Bouchez LC, et al. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc Natl Acad Sci U S A. 2009;106:8912–8917. doi: 10.1073/pnas.0903860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, et al. A Small-Molecule Inhibitor of Tgf-β Signaling Replaces Sox2 in Reprogramming by Inducing Nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maherali N, Hochedlinger K. Tgfβ Signal Inhibition Cooperates in the Induction of iPSCs and Replaces Sox2 and cMyc. Curr. Biol. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (*) 52.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, et al. Pluripotent Stem Cells Induced from Mouse Somatic Cells by Small-Molecule Compounds. Science. 2013;341:651–4. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]; This study carried out a chemical compound screen and found a cocktail of small molecules that effectively reprogram mouse fibroblasts into functional iPSCs, replacing all four OSKM factors.
- 53.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RKM, Tan PBO, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, Marquez VE, Jones PA. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 2009;8:1579–88. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Papapetrou EP, Tomishima MJ, Chambers SM, Mica Y, Reed E, Menon J, Tabar V, Mo Q, Studer L, Sadelain M. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12759–64. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carey BW, Markoulaki S, Hanna JH, Faddah DA, Buganim Y, Kim J, Ganz K, Steine EJ, Cassady JP, Creyghton MP, et al. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–98. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 57.Tiemann U, Sgodda M, Warlich E, Ballmaier M, Schöler HR, Schambach A, Cantz T. Optimal reprogramming factor stoichiometry increases colony numbers and affects molecular characteristics of murine induced pluripotent stem cells. Cytometry. A. 2011;79:426–35. doi: 10.1002/cyto.a.21072. [DOI] [PubMed] [Google Scholar]
- 58.Yamaguchi S, Hirano K, Nagata S, Tada T. Sox2 expression effects on direct reprogramming efficiency as determined by alternative somatic cell fate. Stem Cell Res. 2011;6:177–186. doi: 10.1016/j.scr.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 59.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 61.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 2011;13:215–22. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 64.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 68.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 69.Liu X, Sun H, Qi J, Wang L, He S, Liu J, Feng C, Chen C, Li W, Guo Y, et al. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nat. Cell Biol. 2013;15:829–38. doi: 10.1038/ncb2765. [DOI] [PubMed] [Google Scholar]
- (*) 70.Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A Molecular Roadmap of Reprogramming Somatic Cells into iPS Cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (*) 71.Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J. Highly Coordinated Proteome Dynamics during Reprogramming of Somatic Cells to Pluripotency. Cell Rep. 2012 doi: 10.1016/j.celrep.2012.10.014. doi:10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors in refs 70 and 71 employed two complimentary approaches in sorted cell-populations to demonstrate the various pathways and genes involved in early, intermediate and late phases of reprogramming.
- 72.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (*) 73.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (*) 74.Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional Amplification in Tumor Cells with Elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; The two studies in refs 73 and 74 revealed that the overexpression of c-Myc in cell lines and cancer lead to a global amplification of gene expression. Mechanistically, the highly-expressed c-Myc is associated with promoters and enhancers of active genes and induce gene expression through the release of paused RNA Pol-II.
- 75.Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Singhal N, Graumann J, Wu G, Araúzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Schöler HR, Arauzo-Bravo MJ, et al. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 77.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi X, Qin B, Zeng L, Esteban MA, Pan G, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–87. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 79.Liang G, He J, Zhang Y. Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nat. Cell Biol. 2012;14:457–66. doi: 10.1038/ncb2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ang Y-S, Tsai S-Y, Lee D-F, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–97. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y, Krupalnik V, Zerbib M, Amann-Zalcenstein D, Maza I, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–13. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- 82.Pijnappel WWMP, Esch D, Baltissen MPA, Wu G, Mischerikow N, Bergsma AJ, van der Wal E, Han DW, Bruch H vom, Moritz S, et al. A central role for TFIID in the pluripotent transcription circuitry. Nature. 2013;495:516–519. doi: 10.1038/nature11970. [DOI] [PubMed] [Google Scholar]
- (**) 83.Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Mancarci OB, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors took a loss-of-function approach and used shRNA against 22 histone and DNA methylation enzymes to identify chromatin modulators of reprogramming. Knocking down the H3K9 methyltransferase Suv39H1 had the most enhancing effects on reprogramming. The authors also identified the H3K79 methyltransferase Dot1l as a suppressor of reprogramming by promoting the mesenchymal lineage. The authors did not observe a positive effect on reprogramming when knocking down MBD3 as reported in refs 97 and 98.
- 84.Jullien J, Pasque V, Halley-Stott RP, Miyamoto K, Gurdon JB. Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Nat Rev Mol Cell Biol. 2011;12:453–459. doi: 10.1038/nrm3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, Trcka D, Wrana JL. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell. 2012;11:769–82. doi: 10.1016/j.stem.2012.11.008. [DOI] [PubMed] [Google Scholar]
- (*) 87.Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat. Cell Biol. 2013;7:872–82. doi: 10.1038/ncb2768. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors used a quantitative proteomic approach to identify H3K9me3 as as major impediment to reprogramming.
- 88.Soufi A, Zaret KS. Understanding impediments to cellular conversion to pluripotency by assessing the earliest events in ectopic transcription factor binding to the genome. Cell Cycle. 2013;12:1. doi: 10.4161/cc.24663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, et al. Genome-wide Chromatin State Transitions Associated with Developmental and Environmental Cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang T, Wu H, Li Y, Szulwach KE, Lin L, Li X, Chen I-P, Goldlust IS, Chamberlain SJ, Dodd A, et al. Subtelomeric hotspots of aberrant 5-hydroxymethylcytosine-mediated epigenetic modifications during reprogramming to pluripotency. Nat. Cell Biol. 2013 doi: 10.1038/ncb2748. doi:10.1038/ncb2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 93.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- (**) 94.Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Chen J, Wu Y, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 2012;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]; This paper identifies H3K9 methyltransferases and demethylases as downstream effectors to external stimuli, such as BMP and Vitamin C. This study highlights that H3K9me3 as a major barrier to the final phase of reprogramming.
- 95.Comes S, Gagliardi M, Laprano N, Fico A, Cimmino A, Palamidessi A, De Cesare D, De Falco S, Angelini C, Scita G, et al. L-Proline Induces a Mesenchymal-like Invasive Program in Embryonic Stem Cells by Remodeling H3K9 and H3K36 Methylation. Stem Cell Reports. 2013;1:307–321. doi: 10.1016/j.stemcr.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–5. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (**) 97.Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]; By genetically deleting MBD3, the authors showed that reprogramming secondary mouse fibroblast using OSKM is almost 100% efficient and highly synchronized. The authors highlight that OSKM recruit both positive and negative partners for inducing pluripotency and by deleting one single negative influence (MBD3), somatic cells can be deterministically converted to iPS cells.
- (**) 98.Luo M, Ling T, Xie W, Sun H, Zhou Y, Zhu Q, Shen M, Zong L, Lyu G, Zhao Y, et al. NuRD Blocks ReprogAramming of Mouse Somatic Cells into Pluripotent Stem Cells. Stem Cells. 2013;31:1278–86. doi: 10.1002/stem.1374. [DOI] [PubMed] [Google Scholar]; This study shows that knocking down MBD3 enhances the efficiency of reprogramming but not to the same extent as reported in ref 98.
- 99.Wang Y, Chen J, Hu J-L, Wei X-X, Qin D, Gao J, Zhang L, Jiang J, Li J-S, Liu J, et al. Reprogramming of mouse and human somatic cells by high-performance engineered factors. EMBO Rep. 2011;12:373–8. doi: 10.1038/embor.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hirai H, Tani T, Katoku-Kikyo N, Kellner S, Karian P, Firpo M, Kikyo N. Radical acceleration of nuclear reprogramming by chromatin remodeling with the transactivation domain of MyoD. Stem Cells. 2011;29:1349–61. doi: 10.1002/stem.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hammachi F, Morrison GM, Sharov AA, Livigni A, Narayan S, Papapetrou EP, O'Malley J, Kaji K, Ko MSH, Ptashne M, et al. Transcriptional Activation by Oct4 Is Sufficient for the Maintenance and Induction of Pluripotency. Cell Rep. 2012;1:99–109. doi: 10.1016/j.celrep.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 103.Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–9. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Caravaca JM, Donahue G, Becker JS, He X, Vinson C, Zaret KS. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 2013;27:251–60. doi: 10.1101/gad.206458.112. [DOI] [PMC free article] [PubMed] [Google Scholar]