Abstract
Background
The facultatively anaerobic betaproteobacterium Castellaniella defragrans 65Phen utilizes acyclic, monocyclic and bicyclic monoterpenes as sole carbon source under oxic as well as anoxic conditions. A biotransformation pathway of the acyclic β-myrcene required linalool dehydratase-isomerase as initial enzyme acting on the hydrocarbon. An in-frame deletion mutant did not use myrcene, but was able to grow on monocyclic monoterpenes. The genome sequence and a comparative proteome analysis together with a random transposon mutagenesis were conducted to identify genes involved in the monocyclic monoterpene metabolism. Metabolites accumulating in cultures of transposon and in-frame deletion mutants disclosed the degradation pathway.
Results
Castellaniella defragrans 65Phen oxidizes the monocyclic monoterpene limonene at the primary methyl group forming perillyl alcohol. The genome of 3.95 Mb contained a 70 kb genome island coding for over 50 proteins involved in the monoterpene metabolism. This island showed higher homology to genes of another monoterpene-mineralizing betaproteobacterium, Thauera terpenica 58EuT, than to genomes of the family Alcaligenaceae, which harbors the genus Castellaniella. A collection of 72 transposon mutants unable to grow on limonene contained 17 inactivated genes, with 46 mutants located in the two genes ctmAB (cyclic terpene metabolism). CtmA and ctmB were annotated as FAD-dependent oxidoreductases and clustered together with ctmE, a 2Fe-2S ferredoxin gene, and ctmF, coding for a NADH:ferredoxin oxidoreductase. Transposon mutants of ctmA, B or E did not grow aerobically or anaerobically on limonene, but on perillyl alcohol. The next steps in the pathway are catalyzed by the geraniol dehydrogenase GeoA and the geranial dehydrogenase GeoB, yielding perillic acid. Two transposon mutants had inactivated genes of the monoterpene ring cleavage (mrc) pathway. 2-Methylcitrate synthase and 2-methylcitrate dehydratase were also essential for the monoterpene metabolism but not for growth on acetate.
Conclusions
The genome of Castellaniella defragrans 65Phen is related to other genomes of Alcaligenaceae, but contains a genomic island with genes of the monoterpene metabolism. Castellaniella defragrans 65Phen degrades limonene via a limonene dehydrogenase and the oxidation of perillyl alcohol. The initial oxidation at the primary methyl group is independent of molecular oxygen.
Keywords: Monoterpene, Isoprenoids, Biodegradation, Limonene, Phellandrene
Background
Monoterpenes are structurally diverse secondary metabolites of plants. The volatile branched-chain C10 hydrocarbons are major constituents of essential oils and are also known to attract pollinators [1]. Monoterpenes with their characteristic scent possess antimicrobial and anti-herbivore properties and are added to various foods, cosmetics or household products as flavor or antimicrobial agents [2].
Microbes use monoterpenes as carbon and energy source. The capability to transform monoterpenes is widespread among bacteria and fungi [3]. Already in the 1960s, the aerobic metabolism of several monoterpenes (limonene and α-pinene) was described for soil pseudomonads and the fungus Aspergillus niger[4]. Pseudomonads also use the acyclic citronellol [5,6] and Pleurotus sapidus was found to transform the bicyclic Δ3-carene [7]. Limonene may be an intermediate in the metabolism of bicyclic [8] and acyclic monoterpenes [9].
The monocyclic limonene (4-isopropenyl-1-methylcyclohexene) is one of the most common monoterpenes [10]. Several limonene biotransformation pathways with molecular oxygen as co-substrate were described for aerobic microorganisms [11,12]: (I) the oxidation at the primary methyl group to perillyl alcohol and further oxidation; (II) the epoxidation of the ring double bond and formation of a diol; (III) the ring oxidation at the C3 position forming carveol or at the C6 position forming isopiperitenol, and (IV) the epoxidation of the double bond at the isopropenyl group. All these reactions are catalyzed by cytochrome P450 monooxygenases [13]. Alternative initial reactions involve the addition of water to a double bond. The hydration of limonene to α-terpineol was reported for bacterial and fungal species [14]. Anoxic conditions seem to increase the transformation rate [15] and cofactor-independent hydratases were identified to catalyze the reaction [16,17]. The further degradation of α-terpineol via oleuropeic acid or borneol is again dependent on molecular oxygen as co-substrate [18].
A microbial monoterpene mineralization to carbon dioxide in the absence of molecular oxygen was first observed with the enrichment and isolation of denitrifying strains [19-21]. One of these strains, Castellaniella defragrans 65Phen isolated with α-phellandrene, is able to use various acyclic, monocyclic and bicyclic monoterpenes as sole carbon and energy source. Cyclic monoterpenes require a sp2-hybridized C1-atom as precondition for mineralization [22]. Initial metabolite studies showed the formation of geranic acid from β-myrcene in cell-free extracts [23]. A pathway from the acyclic monoterpene β-myrcene to geranic acid was identified. β-Myrcene is enantiospecifically hydrated to (S)-(+)-linalool and further isomerized to geraniol by the linalool dehydratase-isomerase (LDI) [24,25]. The NAD+-dependent geraniol dehydrogenase GeoA and geranial dehydrogenase GeoB oxidize geraniol to geranial and further to geranic acid (published as GeDH and GaDH) [26]. With the development of a genetic system for C. defragrans 65Phen, an in-frame deletion mutant with an inactivated ldi gene showed no growth with the acyclic β-myrcene, but grew like the wild type on limonene or α-phellandrene [27].
In this publication, we report our search for an anaerobic pathway for cyclic monoterpene degradation in Castellaniella defragrans 65Phen. On the basis of the genome, expressed proteins were extracted from α-phellandrene- and acetate-grown cultures and identified by two-dimensional gel electrophoresis coupled to MALDI-TOF-MS as well as membrane protein-enriched LC-ESI-MS/MS. A random transposon mutagenesis with a Mini-Tn5 transposon identified genes essential for the growth on cyclic monoterpenes. Metabolites formed in cultures of several genotypes were identified by GC-MS. The observations were integrated to develop a putative degradation pathway.
Results and discussions
The genome of Castellaniella defragrans
The closed genome of Castellaniella defragrans 65Phen has 3,952,818 bp and an overall G+C content of 68.9%. 3616 protein-coding open reading frames (ORFs), 45 transfer RNA genes and 2 ribosomal RNA operons, comprising 5S, 16S and 23S ribosomal RNA genes, were detected in the genome. C. defragrans 65Phen has complete sets of genes for the citrate cycle, aerobic respiration and denitrification including nitrite reductase nirK and both quinol-dependent nitrite oxide reductase and cytochrome C-dependent nitrite oxide reductase type norB. The degradative pathways matched the observed substrate utilization [21]. The lack of growth on sugars coincided with the absence of a 6-phosphofructokinase, thus the glycolysis pathway was incomplete. The biosynthesis of sugars on anabolic pathways is ensured by a fructose-1,6-bisphosphatase type I. The comparison with other genomes revealed a high similarity to related Alcaligenaceae species, e.g., Bordetella pertussis Tohama I (average nucleotide identity (ANI) 82.1%) or Pusillimonas sp. T7-7 (ANI 80.7%). C. defragrans 65Phen shares 1954 ORFs (54%) with the published genome of Bordetella pertussis Tohama I (Acc. no. NC_002929). In these genes the average amino acid identity was 62%.
An exception was an island of 70 kb DNA located from base 3026577 to 3096437 (Table 1). The island is flanked upstream by a transposable element and downstream by a cluster for the degradation of amino acids. The majority of genes in the island are most similar to betaproteobacterial genes outside the Alcaligenaceae. The island includes the genes for the initial myrcene transformation, ldi for the linalool dehydratase-isomerase, geoA for the geraniol dehydrogenase and geoB for the geranial dehydrogenase, and the previously published contig derived from fosmids (Acc. no. FR669447). The predicted proteins (Tab. 1) resemble proteins of the monoterpene-mineralizing strains, Thauera terpenica 58EuT[20] and Pseudomonas sp. 19-rlim [28], as well as of Azoarcus strains, which have not been tested for monoterpene degradation. Thauera and Azoarcus are Rhodocyclales with a well-established capacity to mineralize aromatic hydrocarbons [29]. Pseudomonas sp. 19-rlim belongs to the gammaproteobacterial Pseudomonadaceae which degrade a wide range of hydrophobic substances [30]. Many predicted proteins in the island were annotated as beta-oxidation pathway-related enzymes and as a transporter of hydrophobic substances.
Table 1.
Genes of the genome island and assigned functions in the metabolism of monoterpenes in Castellaniella defragrans 65Phen
| Protein_id | Proteine detection method a | 2D fold change/LC enrichment level b | Gene annotation |
Related gene product |
|||
|---|---|---|---|---|---|---|---|
| E value | % identity | Organism | Accession no. | ||||
|
CDM25240 |
n.d. |
n.d. |
Hypothetical protein |
4E-60 |
59 |
Thauera terpenica 58Eu |
EPZ16239 |
|
CDM25241 |
LC |
++ |
Acyl-CoA dehydrogenase protein |
0.0 |
82 |
Thauera terpenica 58Eu |
EPZ16240 |
|
CDM25242 |
LC |
0 |
Molybdopterin-binding OR |
3E-70 |
74 |
Thauera terpenica 58Eu |
EPZ16227 |
|
CDM25243 |
n.d. |
n.d. |
Tyrosine/serine phosphatase |
1E-58 |
48 |
Thauera linaloolentis 47Lol |
ENO83508 |
|
CDM25244 |
LC |
0 |
2,4-dienoyl-CoA reductase |
0.0 |
87 |
Thauera terpenica 58Eu |
EPZ16243 |
|
CDM25245 |
n.d. |
n.d. |
NADH:flavin oxidoreductase |
1E-179 |
68 |
Thauera terpenica 58Eu |
EPZ16244 |
|
CDM25246 |
2D/LC |
3.0/++ |
3-hydroxyacyl-CoA dehydrogenase |
5E-167 |
82 |
Azoarcus sp. KH32C |
YP_007598290 |
|
CDM25247 |
n.d. |
n.d. |
IS4 family transposase |
0.0 |
63 |
Thiomonas sp. FB-6 |
WP_018915433 |
|
CDM25248, MrcH |
n.d. |
n.d. |
MaoC-like dehydratase |
2E-36 |
62 |
Azoarcus sp. KH32C |
YP_007598291 |
|
CDM25249, MrcG |
n.d. |
n.d. |
MaoC-like dehydratase |
6E-50 |
67 |
Thauera terpenica 58Eu |
EPZ16257 |
|
CDM25250, MrcF |
LC |
++ |
Perillyl-CoA hydratase |
0.0 |
58 |
Thauera terpenica 58Eu |
EPZ16258 |
|
CDM25251, MrcE |
2D/LC |
18/++ |
4-isopropenyl-2-oxo-cyclohexane-1-carboxyl-CoA hydrolase |
4E-167 |
88 |
Azoarcus sp. KH32C |
YP_007598294 |
|
CDM25252, MrcD |
2D/LC |
3.0/++ |
2-hydroxy-4-isopropenyl-cyclohexane-1-carboxyl-CoA dehydrogenase |
1E-153 |
85 |
Azoarcus sp. KH32C |
YP_007598295 |
|
CDM25253, MrcC |
2D |
2,3 |
2,4-dienoyl reductase |
0.0 |
86 |
Thauera terpenica 58Eu |
EPZ16261 |
|
CDM25254, MrcB |
LC |
++ |
Acyl-CoA dehydrogenase |
0.0 |
90 |
Azoarcus toluclasticus |
WP_018990727 |
|
CDM25255, MrcA |
LC |
++ |
Oxidoreductase, FAD-binding |
0.0 |
73 |
Azoarcus toluclasticus |
WP_018990723 |
|
CDM25256 |
2D/LC |
19/++ |
(R)-specific enoyl-CoA hydratase |
1E-88 |
85 |
Thauera terpenica 58Eu |
EPZ15051 |
|
CDM25257 |
LC |
0 |
Citrate lyase |
6E-141 |
73 |
Thauera terpenica 58Eu |
EPZ15052 |
|
CDM25258 |
LC |
++ |
Acyl-CoA dehydrogenase |
0.0 |
92 |
Thauera terpenica 58Eu |
EPZ15053 |
|
CDM25259 |
LC |
++ |
RND efflux transporter |
0.0 |
62 |
Thauera terpenica 58Eu |
EPZ15054 |
|
CDM25260 |
LC |
++ |
RND efflux transporter |
0.0 |
80 |
Thauera terpenica 58Eu |
EPZ15055 |
|
CDM25261 |
LC |
++ |
RND efflux transporter |
4E-142 |
68 |
Thauera terpenica 58Eu |
EPZ15056 |
|
CDM25262 |
LC |
++ |
RND efflux transporter |
0.0 |
81 |
Thauera terpenica 58Eu |
EPZ15057 |
|
CDM25263 |
LC |
++ |
Acetoacetyl-CoA synthetase |
0.0 |
84 |
Thauera terpenica 58Eu |
EPZ15058 |
|
CDM25264 |
n.d. |
n.d. |
Enoyl-CoA hydratase |
9E-136 |
76 |
Thauera terpenica 58Eu |
EPZ15059 |
|
CDM25265, GeoC |
LC |
++ |
Perillate--CoA ligase |
0.0 |
71 |
Thauera terpenica 58Eu |
EPZ15060 |
|
CDM25266 |
LC |
0 |
Hypothetical protein |
4,3 |
52 |
Fusarium graminearum PH-1 |
XP_382023 |
|
CDM25267, GeoA |
2D/LC |
42/++ |
Geraniol dehydrogenase |
0.0 |
84 |
Thauera terpenica 58Eu |
EPZ14350 |
|
CDM25268 |
LC |
++ |
Hypothetical protein |
8E-117 |
74 |
Thauera terpenica 58Eu |
EPZ14349 |
|
CDM25269 |
n.d. |
n.d. |
Hypothetical protein |
2E-97 |
69 |
Thauera terpenica 58Eu |
EPZ14348 |
|
CDM25270 |
n.d. |
n.d. |
Hypothetical protein |
6E-32 |
66 |
Thauera terpenica 58Eu |
EPZ14347 |
|
CDM25271 |
n.d. |
n.d. |
Thioesterase |
2E-33 |
46 |
Magnetospirillum magneticum |
YP_420191 |
|
CDM25272, LDI |
LC |
++ |
LDI precursor protein |
8E-14 |
25 |
Stereum hirsutum FP-91666 SS1 |
EIM80109 |
|
CDM25273 |
n.d. |
n.d. |
Hypothetical protein |
5E-85 |
43 |
Gordonia paraffinivorans |
WP_006900876 |
|
CDM25274 |
n.d. |
n.d. |
Hypothetical protein |
5E-07 |
30 |
Gordonia paraffinivorans |
WP_006900845 |
|
CDM25275 |
LC |
++ |
Acyl-CoA dehydrogenase |
4E-106 |
54 |
Azoarcus toluclasticus |
WP_018990670 |
|
CDM25276 |
n.d. |
n.d. |
Hypothetical protein |
4E-40 |
40 |
Glaciecola punicea |
WP_006005307 |
|
CDM25277 |
n.d. |
n.d. |
Hypothetical protein |
5E-21 |
58 |
Pseudomonas sp. 19-rlim |
AEO27370 |
|
CDM25278 |
n.d. |
n.d. |
Hypothetical protein |
1E-69 |
62 |
Pseudomonas sp. 19-rlim |
AEO27371 |
|
CDM25279 |
LC |
++ |
Hypothetical protein |
1E-120 |
68 |
Pseudomonas sp. 19-rlim |
AEO27372 |
|
CDM25280 |
n.d. |
n.d. |
MarR transcriptional regulator |
7E-81 |
83 |
Thauera terpenica 58Eu |
EPZ16291 |
|
CDM25281, GeoB |
2D/LC |
15/++ |
Geranial dehydrogenase |
0.0 |
91 |
Thauera terpenica 58Eu |
EPZ16290 |
|
CDM25282 |
LC |
++ |
Acyl-CoA dehydrogenase |
0.0 |
89 |
Thauera terpenica 58Eu |
EPZ16289 |
|
CDM25283 |
n.d. |
n.d. |
LuxR family transcriptional regulator |
0.0 |
59 |
Thauera terpenica 58Eu |
EPZ16271 |
|
CDM25284, CtmG |
LC |
++ |
Hypothetical protein |
5E-35 |
39 |
Azoarcus sp. KH32C |
YP_007598506 |
|
CDM25285, CtmF |
LC |
++ |
NADH:ferredoxin oxidoreductase |
4E-147 |
56 |
Caulobacter sp. AP07 |
WP_007674692 |
|
CDM25286, CtmE |
2D |
6.3/++ |
Ferredoxin, 2Fe-2S |
1E-32 |
50 |
Caulobacter crescentus CB15 |
NP_422318 |
|
CDM25287, CtmD |
n.d. |
n.d. |
Hypothetical protein |
4,1 |
38 |
Ochrobactrum sp. CDB2 |
WP_007881652 |
|
CDM25288, CtmC |
n.d. |
n.d. |
Hypothetical protein |
2,3 |
33 |
Bombus impatiens |
XP_003489707 |
|
CDM25289, CtmB |
2D/LC |
3.4/++ |
Limonene dehydrogenase |
4E-131 |
41 |
Deltaproteobacterium NaphS2 |
WP_006422074 |
|
CDM25290, CtmA |
LC |
++ |
Limonene dehydrogenase |
5E-57 |
30 |
Deltaproteobacterium NaphS2 |
WP_006422074 |
|
CDM25291 |
LC |
++ |
Acetyl-CoA acetyltransferase |
0.0 |
79 |
Thauera terpenica 58Eu |
EPZ16237 |
|
CDM25292 |
LC |
0 |
MarR transcriptional regulator |
2E-76 |
77 |
Thauera terpenica 58Eu |
EPZ16235 |
|
CDM25293 |
n.d. |
n.d. |
Hypothetical protein |
4E-24 |
56 |
Thauera terpenica 58Eu |
EPZ16283 |
|
CDM25294 |
LC |
++ |
Hypothetical protein |
2E-83 |
68 |
Thauera terpenica 58Eu |
EPZ16282 |
|
CDM25295 |
n.d. |
n.d. |
Hypothetical protein |
6E-155 |
76 |
Thauera terpenica 58Eu |
EPZ16281 |
|
CDM25296 |
n.d. |
n.d. |
MarR transcriptional regulator |
5E-94 |
81 |
Thauera terpenica 58Eu |
EPZ16280 |
|
CDM25297 |
n.d. |
n.d. |
Hypothetical protein |
2E-168 |
72 |
Thauera terpenica 58Eu |
EPZ16232 |
|
CDM25298 |
LC |
++ |
Hypothetical protein |
0.0 |
72 |
Thauera terpenica 58Eu |
EPZ16231 |
|
CDM25299 |
LC |
++ |
Acetyl-CoA acetyltransferase |
0.0 |
90 |
Thauera terpenica 58Eu |
EPZ16230 |
|
CDM25300 |
n.d. |
n.d. |
Acetyl-CoA hydrolase/transferase |
0.0 |
74 |
Thauera terpenica 58Eu |
EPZ16229 |
|
CDM25301 |
LC |
++ |
Electron transfer flavoprotein |
7E-133 |
78 |
Thauera terpenica 58Eu |
EPZ16226 |
| CDM25302 | n.d. | n.d. | Electron transfer flavoprotein | 7E-160 | 76 | Azoarcus sp. KH32C | YP_007552025 |
a2D, identified with 2D-SDS-PAGE and MALDI-TOF-MS; LC, identified with LC-ESI-MS/MS.
b++ peptides only identified in α-phellandrene fraction, + increase in α-phellandrene fraction, 0 ratio remained unchanged.
The proteome of monoterpene utilization
The soluble protein fractions of bacteria grown on acetate or on α-phellandrene were analyzed by two-dimensional gel electrophoresis, followed by enzymatic digest and MALDI-TOF mass spectrometry. The enriched membrane protein fractions were analyzed by one-dimensional gel electrophoresis, enzymatic digest and LC-ESI-MS/MS. 234 and 851 individual proteins of C. defragrans 65Phen were identified with 2D-SDS-PAGE separation and MALDI-TOF-MS, and with LC-ESI-MS/MS, respectively. The monoterpene proteome, defined as proteins induced in extracts of α-phellandrene-grown cells in comparison to extracts of acetate-grown cells, included a total of 107 proteins, of which 28 proteins were identified by MALDI-TOF-MS and 97 proteins were identified by LC-ESI-MS/MS, with an overlap of 18 proteins that were identified by both techniques (Additional file 1: Table S1). 32 of these proteins are encoded by genes in the island including the enzymes LDI, GeoA and GeoB. Among the 75 α-phellandrene-induced proteins with a gene location outside of the island, ABC transporter-associated proteins were highly up-regulated.
Growth on oxidized limonene metabolites
The geraniol dehydrogenase GeoA is an allyl-alcohol dehydrogenase with the highest catalytic activity on perillyl alcohol [26]. The high expression of GeoA and GeoB in α-phellandrene-grown cells questioned the utilization of the cyclic monoterpene alcohol by C. defragrans. The strain 65Phen grew on perillyl alcohol, perillyl aldehyde and perillic acid (Figure 1B-D). During active denitrification, perillyl aldehyde accumulated transiently in the culture growing on perillyl alcohol (Figure 1B). Perillic acid accumulated in cultures growing on the alcohol and the aldehyde. When the electron acceptor nitrate was depleted, the cells disproportionated perillyl aldehyde into perillyl alcohol and perillyl acid (Figure 1B, C). During growth on limonene (Figure 1A), the perillyl derivatives were not detected suggesting a rate-limitation in the pathway by the initial limonene-transforming enzyme.
Figure 1.
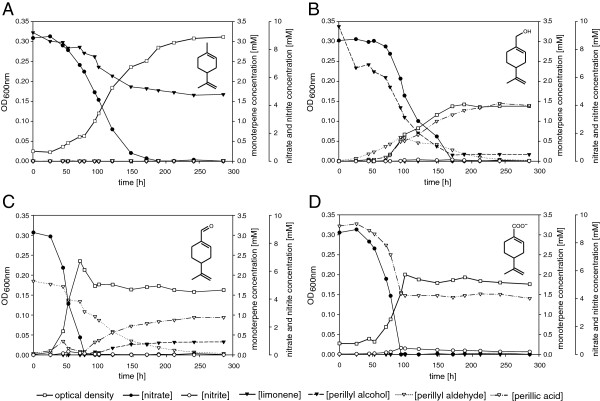
Anaerobic growth of C. defragrans 65Phen on limonene and putative metabolites. Limonene (A), perillyl alcohol (B), perillyl aldehyde (C) and perillic acid (D) were tested as growth substrate, each with a concentration of 3 mM and nitrate limitation (10 mM).
Transposon insertion mutagenesis
To identify the genes involved in monoterpene degradation, a random insertion mutagenesis with a mini-Tn5 transposon was performed according to Larsen et al. [31]. Insertion mutants of C. defragrans 65Phen were obtained on agar plates with a minimal medium and acetate and nitrate as carbon source and electron acceptor, respectively, and rifampicin and kanamycin as selective compounds. These colonies were transferred to acetate-free anoxic plates with nitrate and supplied with limonene via the gas phase. Six of 1000 insertion mutants revealed a lack of growth under these conditions. Our collection of 72 mutant strains covered 46 unique insertion positions in 17 genes. The majority of insertions yielding a loss of function were in ctmA. It was inactivated in 45 mutants at 22 different positions. The adjacent gene ctmB was inactivated thrice at different positions. Also a gene closely located, ctmE, was inactivated at two insertion sites. Five genes were present at least twice in the mutant collection, whereas nine other genes were present once. The five genes coded CDM25082, CDM25239, CDM25260, CDM25338 and CDM25923.
Anaerobic denitrifying growth of selected transposon mutants was tested in liquid culture (Table 2). All transposon mutants denitrified and grew with acetate in liquid culture. In liquid culture on limonene, four strains, the mutants of a putative transcriptional regulator (CDM25239), a putative inner membrane protein (CDM25338), a hypothetical protein (CDM25510) and a plasmid stability protein (CDM24643) grew similarly to the wild type in liquid culture. No colony formation on plates, but growth in liquid medium coincided with a reduced mass transfer limitation of limonene in liquid culture. Three mutant strains showed weak growth and ten mutant strains did not grow on limonene in liquid culture (Table 2). Additional growth experiments showed that the genes ctmA, ctmB or ctmE were not required for growth on perillyl alcohol. This physiology was also observed in aerobic cultures growing in the absence of nitrate (Additional file 2: Figure S1). The three genes are part of an operon-like cluster which was named ctmABCDEFG, for cyclic terpene metabolism-associated genes (Table 1). Also other monoterpenes tested (Table 2) did not support growth of these three mutants.
Table 2.
Growth of Castellaniella defragrans 65Phen transposon insertion mutants in liquid medium
| Inactivated gene | Annotation | Length [b] | Insertion positions [b] |
Growth substrate
*
|
|||
|---|---|---|---|---|---|---|---|
| Acetate | Limonene | β -Myrcene | Further substrates | ||||
| Initial oxidation | |||||||
|
CDM25290 CtmA |
Limonene dehydrogenase, alpha subunit |
1698 |
133 |
+ |
- |
- |
Perillyl alcohol (+), α-phellandrene (-), α-pinene (-), β-pinene (-) |
|
CDM25290 CtmA |
Limonene dehydrogenase, alpha subunit |
1698 |
1188 |
+ |
- |
- |
Perillyl alcohol (+), α-phellandrene (-), α-pinene (-), β-pinene (-) |
|
CDM25289 CtmB |
Limonene dehydrogenase, beta subunit |
1650 |
38 |
+ |
- |
- |
Perillyl alcohol (+), α-phellandrene (-), α-pinene (-), β-pinene (-) |
|
CDM25289 CtmB |
Limonene dehydrogenase, beta subunit |
1650 |
559 |
+ |
- |
- |
Perillyl alcohol (+), α-phellandrene (-), α-pinene (-), β-pinene (-) |
|
CDM25286 CtmE |
Ferredoxin, 2Fe-2S |
324 |
111 |
+ |
- |
- |
Perillyl alcohol (+), α-phellandrene (-), α-pinene (-), β-pinene (-) |
| Ring cleavage and β-oxidation | |||||||
|
CDM23589 |
Electron transfer flavoprotein:ubiquinone oxidoreductase |
1647 |
138 |
+ |
(+) |
(+) |
Perillic acid ((+)) |
|
CDM25250 MrcF |
Perillyl-CoA hydratase |
1239 |
18 |
+ |
- |
- |
α-phellandrene (-) |
|
CDM25253 MrcC |
2,4-dienoyl-CoA reductase |
888 |
841 |
+ |
- |
- |
α-phellandrene (-) |
|
CDM25297 |
Hypothetical protein |
1023 |
34 |
+ |
- |
- |
|
| CDM258488 |
Electron transfer protein |
888 |
385 |
+ |
- |
- |
Perillic acid (-) |
|
CDM25923 |
Enoyl-CoA hydratase |
777 |
461 |
+ |
- |
- |
|
| Methylcitrate cycle | |||||||
|
CDM25082 |
2-methylcitrate dehydratase |
1452 |
262 |
+ |
- |
- |
|
|
CDM25081 |
2-methylcitrate synthase |
1239 |
-18 |
+ |
(+) |
+ |
|
| Other functions | |||||||
|
CDM23676 |
Molybdenum transport system protein |
865 |
47 |
+ |
- |
- |
Perillyl aldehyde (-) |
| CDM25260 | RND efflux transporter, periplasmic component | 1356 | 24 | + | (+) | (+) | Perillic acid ((+)) |
* + growth like wild type; - no growth; (+) decreased growth rate and maximum density compared to wild type.
Metabolite formation from limonene
To demonstrate the in vivo formation of perillyl alcohol from limonene, an in-frame deletion mutant of geoB was generated. The deletion of the alcohol dehydrogenase gene geoA caused a decreased growth rate and biomass formation with several monoterpenes as growth substance. We attribute this residual growth with the expression of a second alcohol dehydrogenase in cells of the in-frame ΔgeoA mutant [27]. The deletion mutant C. defragrans 65Phen ΔgeoB grew on acetate and perillic acid, but lacked the ability to grow on limonene, β-myrcene, perillyl alcohol and perillyl aldehyde (data not shown). To analyze the metabolite formation, the mutant and for comparison the wild type were anaerobically cultured with 3 mM (R)-(+)-limonene and 20 mM acetate as co-substrates in the presence of 20 mM nitrate. To increase the mass transfer, the organic carrier phase 2,2,4,4,6,8,8-heptamethylnonane (HMN) - and with it the two-phase system - was replaced by 0.5% v/v Tween 20 in a homogeneous phase. In the early stationary phase after seven days, hydrophobic compounds were salted out and extracted with isopropanol. The metabolites in the isopropanol phase were analyzed stereospecifically by GC and GC-MS (Figure 2). (R)-(+)-perillyl alcohol as well as (R)-(+)-perillyl aldehyde were not detected in an abiotic control without cells and in the culture of the wild type, but in the deletion mutant ΔgeoB. This in vivo formation of (R)-(+)-perillyl alcohol from (R)-(+)-limonene suggested an oxidation of limonene at the methyl group as initial reaction of the degradation pathway.
Figure 2.
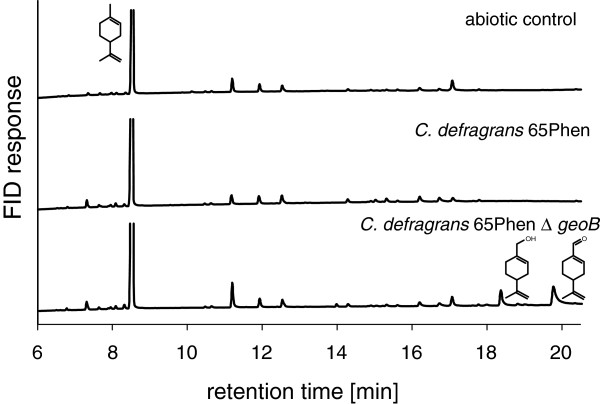
GC chromatogram of metabolite accumulation. Extracts of nitrate-limited cultures were obtained from the primary stationary phase of cultures grown with 20 mM acetate as co-substrate and 3 mM (R)-(+)-limonene.
Hypothetical pathway of limonene degradation based on genome, monoterpene proteome and the physiology of transposon mutants
The genome island of C. defragrans 65Phen is related to genes of T. terpenica 58EuT which also mineralizes cyclic monoterpenes anaerobically [21]. A related gene cluster from Pseudomonas sp. strain 19-rlim has been deposited, but not described (Genbank JN379031). Of these strains, only C. defragrans 65Phen has the cyclic terpene metabolism cluster that contained two FAD-dependent oxidoreductases (ctmA and ctmB), a 2Fe-2S ferredoxin (ctmE) and a ferredoxin reductase (ctmF) together with three hypothetical genes (ctmCD and ctmG) and a putative transcriptional regulator of the luxR family. CtmA, B, E and F were up-regulated proteins in α-phellandrene-grown cells, in comparison to acetate-grown cells. The other proteins of the cluster were not detected. The physiology of the transposon mutants in ctmA, ctmB and ctmE together with the formation of perillyl alcohol in cultures of the ΔgeoB mutant suggests that the Ctm subunits represent a novel enzyme for hydrocarbon activation, catalyzing the oxidation of a methyl group from limonene to the corresponding alcohol. More precisely, a methyl group of an allyl group is oxidized, as it was demonstrated that the carbon-carbon double bond is required for the metabolism [22]. So far, the activation of hydrocarbons in the absence of molecular oxygen is known to be catalyzed by glycine radical enzymes (alkanes, toluene) or molybdenum-containing enzymes (ethylbenzene, cholesterol) [29]. The Ctm enzyme seems to comprise a catalytic core of CtmAB and an electron transfer chain consisting of CtmEF. CtmAB are both annotated as FAD-dependent oxidoreductases, but have a low amino acid identity to each other (27%). The domain structure of both oxidoreductases is similar to those of phytoene dehydrogenases (COG1233, E value of 1e-60) which introduce symmetrically double bonds at phytoene adjacent to existing carbon-carbon double bonds. This process is also an oxidation of the alkyl part of an allylic group. Oxidized dinucleotides like FAD were described as electron acceptors for bacterial phytoene dehydrogenases [32]. Among the COG1233 enzymes, CtmAB showed the largest gene identity to a putative oxidoreductase of the deltaproteobacterial obligate anaerobic sulfate-reducing strain NaphS2 [33]. In contrast, ctmEF is phylogenetically related to genes of the alphabacterial Caulobacter species. Ferredoxins and ferredoxin reductases are well known as electron transfer chain from the NADH/NADPH-pool to cytochrome P450 monooxygenases in aerobic bacteria [13]. Transposon mutants of ctmA, ctmB or ctmE lacked the capability to mineralize the acyclic β-myrcene or the bicyclic α-pinene. Thus, enzymes of the ctm cluster may also be involved in the metabolism of these monoterpenes in C. defragrans 65Phen.
The oxidation of perillyl alcohol involved GeoA, previously identified as geraniol dehydrogenase with a broad substrate spectrum, and GeoB, previously identified as geranial dehydrogenase [26]. The in-frame deletion of geoB revealed in vivo the co-metabolic transformation of limonene to perillyl alcohol and further to perillyl aldehyde. Both GeoA and GeoB were highly expressed in cells grown on α-phellandrene. Another aldehyde dehydrogenase gene (CDM24151) with unknown substrate specificity was also expressed in cells grown on α-phellandrene, but the level of induction was lower than that of GeoB. The gene was located outside the monoterpene island and was related to an aldehyde dehydrogenase of Pusillimonas sp. T7-7 (84% amino acid identity). The activation of perillic acid to a coenzyme A thioester may be catalyzed by the induced CDM26265, an ATP-dependent ligase that we annotated as GeoC due to the gene location next but one near geoA.
The ring cleavage of the cyclic perillyl-CoA resembles the cyclohex-1-ene-1-carboxyl-CoA degradation in anaerobic benzoate degraders or the described monocyclic monoterpene degradation pathways of Pseudomonas putida[34] or Geobacillus (ex. Bacillus) stearothermophilus[35] (Figure 3). We named the genes monoterpene ring cleavage-associated genes (mrc). Perillyl-CoA may be hydrated by MrcF to 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA. Oxidation by the dehydrogenase MrcD may yield 4-isopropenyl-2-oxocyclohexane-1-carboxyl-CoA that may be hydrolysed by MrcE to 4-isopropenylpimelyl-CoA. Related enzymes of the anaerobic benzoate catabolism, BadK, BadH and BadI, catalyze the β-oxidation-like oxidation of cyclohexenecarboxyl-CoA, forming pimelyl-CoA. Among the strains of Azoarcus and Thauera, the enzymes of Azoarcus sp. KH32C are most closely related to MrcDEF. BadK (YP_007598293) catalyzes the hydration of cyclohex-1-ene-1-carboxyl-CoA and has an amino acid identity of 53% to MrcF, affiliating to the enoyl-CoA hydratase/isomerase superfamily. The dehydrogenase BadH (YP_007598295) has a high identity of 85% to MrcD. The formation of pimelyl-CoA is catalzyed by BadI (YP_007598294) which is highly similar to MrcE (88% identity). All three proteins MrcDEF were expressed in cells grown on α-phellandrene. In C. defragrans 65Phen, the genes required for ring cleavage are located in a cluster (mrcABCDEFGH). An insertion mutant of mrcF did not grow on monoterpenes like limonene or α-phellandrene. A second mutant strain with an insertion in the gene mrcC also lacked the capability to grow on limonene. MrcC was annotated as a 2,4-dienoyl reductase.
Figure 3.
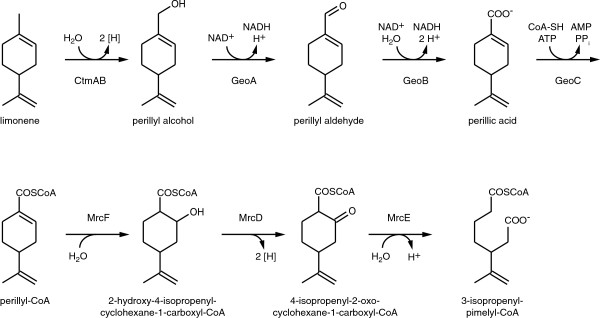
Proposed partial degradation pathway of monocyclic limonene by C. defragrans 65Phen. Enzymes of C. defragrans 65Phen predicted to catalyze reactions of the limonene metabolism: CtmAB, limonene dehydrogenase; GeoA, geraniol dehydrogenase; GeoB, geranial dehydrogenase; GeoC perillate-CoA ligase; MrcF, perillyl-CoA hydratase; MrcD, 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase; MrcE, 4-isopropenyl-2-oxocyclohexane-1-carboxyl-CoA hydrolase (Ctm cyclic terpene metabolism, Mrc monoterpene ring cleavage).
The following degradation of 4-isopropenylpimelyl-CoA may include 3 steps of β-oxidation-like degradation forming three acetyl-CoA and methacrylyl-CoA which may be decarboxylated in a valine-like degradation to propanoyl-CoA. Propanoyl-CoA is connected to the TCA cycle via the methylcitrate cycle. Several enzymes of the valine degradation were up-regulated, but enzymes of the methylcitrate cycle were detected without expression differences between the substrate conditions. However, transposon mutants in 2-methylcitrate synthase and 2-methylcitrate dehydratase showed impaired growth on limonene. Genes assigned to the valine degradation and methylcitrate cycle were located outside of the genome island.
The complete genome of C. defragrans did not show the pathway for the utilization of acyclic monoterpenes as defined by the studies on citronellol in pseudomonads [5]. The atu and liu genes encode two pathways of enzymes required to transform geranyl-CoA to acetyl-CoA and acetoacetate. The liu genes representing the leucine degradation via 3-methylbut-2-enoyl-CoA were present in the genome of C. defragrans 65Phen, but the genes for the acyclic terpene utilization (atu) were not identified. Atu enzymes catalyze the carbon chain cleavage from an acyclic ten-carbon carboxyl-CoA ester to 3-methyl-crotonyl-CoA which is further degraded in the liu pathway to acetyl-CoA and acetoacetate. The tertiary carbon atom undergoes a carbon-carbon cleavage after carboxylation of the methyl group by AtuC/AtuF. Homologues to these pathway-specific genes atuC/atuF were not present in the genome of C. defragrans. The absence of atu genes and the lack of growth on myrcene exhibited by transposon mutants of the limonene degradation pathway is first evidence for a connection between mycrene and limonene metabolism. Other initial reports on the biological formation of cyclic monoterpenes from myrcene [9] and linalool [36] are known, but the molecular basis for this reactions have not been revealed so far.
Conclusions
The degradation of monocyclic monoterpenes in C. defragrans 65Phen is initiated at the methyl group via oxidation by a new enzyme belonging to the phytoene dehydrogenase family (Cog1233). Ferredoxin and ferredoxin reductase are involved in the oxidation of limonene to perillyl alcohol. Expressed proteins and transposon mutants indicate that the further degradation pathway is an oxidation to perillic acid, followed by activation to perillyl CoA thioester and a ring cleavage. Most of the genes for these pathways are located on a genetic island, named the monoterpene island, and seemed to be acquired by horizontal gene transfer from aerobic as well as from anaerobic bacteria.
Methods
Bacterial strains, plasmids and culture conditions
C. defragrans 65Phen-RIF, a strain containing a rifampicin resistance, was used in this study in synonym for C. defragrans 65Phen wild type [27]. Strains of C. defragrans and E. coli as well as plasmids used in this work are listed in Table 3. Additional transposon insertion mutants used in this study are listed in Table 2.
Table 3.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains |
|
|
| C. defragrans |
|
[21] |
| 65Phen-RIF |
RaR |
[27] |
| ΔgeoB |
65Phen-RIF, RaR, ΔgeoB |
This study |
| ctmA::Tn5a |
65Phen-RIF, RaR, KmR, transposon insertion in ctmA at position 133 |
This study |
| ctmA::Tn5b |
65Phen-RIF, RaR, KmR, transposon insertion in ctmA at position 1188 |
This study |
| ctmB::Tn5a |
65Phen-RIF, RaR, KmR, transposon insertion in ctmB at position 38 |
This study |
| ctmB::Tn5b |
65Phen-RIF, RaR, KmR, transposon insertion in ctmB at position 559 |
This study |
| ctmE::Tn5 |
65Phen-RIF, RaR, KmR, transposon insertion in ctmE at position 111 |
This study |
|
E. coli |
|
|
| W20767 |
RP4-2-tet::Mu-1, kan::Tn7 integrant, leu-63::IS10, recA1, creC510, hsdR17, endA1, zbf-5, uidA, (ΔMluI)::pir + thi |
[37] |
| S17-1 |
Thi, pro, hsdR, recA with RP4-2[Tc::Mu-Km::Tn7] |
[38] |
| Plasmids |
|
|
| pRL27 |
Tn5 with KmR, R6K ori, oriT, RP4, tnp |
[31] |
| pCR4-TOPO |
AmR, KmR, lacZα |
Invitrogen |
| pK19mobsacB |
KmR, mob, sacB modified from B. subtilis, lacZα |
[39] |
| pK19mobsacBΔgeoB | KmR, mob, sacB modified from B. subtilis, lacZα, 2000b flanking regions of ΔgeoB | This study |
Preparation of anoxic mineral media and anaerobic cultivation was performed as previously described with small modifications [21]. Media were buffered with 10 mM NaH2PO4 pH 7.2, vitamins were not added and the headspace consisted of N2 gas. Growth experiments were performed in 10 mL media and 300 μL HMN. Inocula were 2% (v/v) of a freshly grown culture. Cultures were incubated at 28°C and shaken at 90 rpm. The optical density was measured directly at 660 nm. Monoterpenes used in this study were purchased from Sigma-Aldrich (Taufkirchen, Germany) with 95 to 97% purity.
Metabolite analysis
For metabolite analysis, triplicates of C. defragrans 65Phen cultures and non-inoculated controls were grown in 500 mL culture flask with 400 mL medium. The mineral medium was autoclaved in the flasks and the headspace was replaced with nitrogen immediately afterwards. After cooling to 21°C, trace minerals, 20 mL HMN, 3 mM monoterpenes (limonene, perillyl alcohol, perillyl aldehyde or perillic acid) and 0.5% inoculum were added. For each measurement 50 μL of the HMN phase and 1 mL of the aqueous phase were sampled. In total, a maximum of 5% (v/v) of each phase was sampled. 1 μL of the organic phase was analyzed by gas chromatography with flame ionization detection (PerkinElmer Auto System XL, Überlingen, Germany). Separation was performed on an Optima-5 column (50 m × 0.32 mm, 0.25 μm film thickness; Macherey-Nagel, Düren, Germany) with the following temperature program: injection port temperature 250°C, detection temperature 350°C, initial column temperature 60°C for 3 min, increasing to 120°C with a rate at 3°C min-1, staying constant for 0.1 min, further increasing to 320°C at 40°C min-1 and hold for 3 min. The split ratio was set to 1:8. All concentrations refer to the aqueous phase. The sample of the aqueous phase was analyzed for optical density at 600 nm and for nitrate and nitrite concentrations as described [19]. Organic acids were separated on a reverse phase HPLC with a Nucleodur C18 Isis column (25 cm × 4.6 mm, 5 μm spheres; Macherey-Nagel, Düren, Germany). The mobile phase consisted of 36% (v/v) acetonitrile and 64% (v/v) 0.05 M ammonium acetate buffer pH 5.0. The flow rate was 0.8 mL min-1 and the effluent was monitored at 217 nm [40].
For additional metabolite analysis, cultures were prepared as described with 0.5% (v/v) Tween 20 replacing the HMN phase. Both arrangements increase the availability of limonene for the cells. 20 mM acetate was added as co-substrate. To salt out metabolites, 5 g KCO3 and 300 μl isopropanol were added to 10 mL culture sampled at the early stationary growth phase. The organic and aqueous phases were separated via centrifugation at 5000 × g for 5 min. 1 μL of the upper phase was analyzed for metabolites and their enantiomer-specificity using a gas chromatograph (PerkinElmer Auto System XL; Überlingen, Germany) equipped with a flame ionization detector. Separation was accomplished on a Hydrodex-β-6TBDM column (25 m × 0.25 mm; Macherey-Nagel, Düren, Germany) by the following temperature program: injection temperature 200°C; detection temperature 230°C, initial column temperature 80°C for 1 min, increasing to 130°C at a rate of 5°C min-1, after 0.5 min further increasing to 230°C at 20°C min-1 and stationary for 2 min. For identification of peaks, a 1 μl sample was analyzed on a Trace GC/MS (Thermo Finnigan, Waltham, USA). Separation was performed on a HP-5 column (25 m × 0.2 mm × 0.33 μm; Agilent, Santa Clara, USA) with the following temperature program: injection port temperature 250°C, initial column temperature 60°C for 6 min, increasing to 120°C at 3°C min-1 further to 320°C at 40°C min-1 and hold for 3 min.
Genome sequencing and annotation
Genomic DNA was extracted as previously described [41]. The genome of Castellaniella defragrans 65Phen was sequenced at the Max Planck Genome Center in Cologne, using the PacBio SMRT system (Pacific Biosciences, Menlo Park, CA). 19858 quality-checked error-corrected reads with at least 4975 bases were de novo assembled to a single contig. The average coverage amounted 91 times and around 4 kb overlapped at each end. Open-reading frames were predicted and annotated by the Rapid Annotations using Subsystems Technology (RAST) pipeline [42]. The G+C content was calculated using Artemis [43]. Putative prokaryotic promoters were predicted with BPROM (http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb) and putative terminators were identified using WebGeSTer [44]. ANI was calculated according to Goris et al.[45]. The complete genome sequence of Castellaniella defragrans 65Phen has been deposited at GenBank under the accession number HG916765.
Label-free quantitative proteome analysis
The soluble and membrane protein enriched proteome of cells grown on acetate and α-phellandrene were compared by 2D-SDS-PAGE and LC-ESI-MS/MS. The cyclic monoterpene α-phellandrene was used as monoterpene growth substrate because of its origin as enrichment substrate for Castellaniella defragrans 65Phen. Anaerobic cultures were grown in a 10 L fermenter with 100 mM nitrate and 10 mM α-phellandrene or 50 mM acetate as previously described [23].
For analyzing the soluble protein fraction, cells were disrupted by sonication in lysis buffer (10 mM Tris pH 7.5, 10 mM EDTA pH 8.0) containing 1.7 mM phenylmethanesulfonylfluoride (PMSF) and cell debris was removed by centrifugation. Proteins (80 μg) were separated on 2D SDS-polyacrylamide gels according to their isoelectric point in the pH range of 3 to 10 and to their molecular mass. Proteins were stained with the fluorescence Sypro Ruby protein gel stain (Invitrogen, Darmstadt, Germany). Spots were detected and quantified on gel images using the software Delta2D (Decodon, Greifswald, Germany). Biological triplicates of each condition were fused and overlaid as dual-channel images and individual spot volumes (%Vol) were calculated as proportion of all proteins on the gel images. Spot ratios which represent an at least 2.5-fold change in spot volume, compared to spots of the acetate samples, were considered. All dominant spots and such spots with ratios >2.5 were automatically excised from the gel (Ettan Spo Picker, GE Healthcare), digested with trypsin and spotted onto a matrix-assisted laser desorption/ionization (MALDI)-target (Ettan Spot Handling Workstation, GE Healthcare). High-throughput MALDI-TOF measurements combined with tandem mass spectrometry were performed on a 4800 MALDI-TOF/TOF Analyser (Applied Biosystems, Darmstadt, Germany). Spectra of a mass range from 900 to 3700 Da were detected during the MALDI-TOF analysis. The three strongest peaks were recorded by the MS/MS analysis. With the GPS Explorer Software (Applied Biosystems) Version 3.6, peaks were indexed and assigned to the corresponding amino acid sequences in the C. defragrans 65Phen database by the Mascot search engine Version 2.1.04 (Matrix Science Ltd, Boston, MA, USA). Proteins of at least 25% sequence coverage, minimum 2 unique assigned peptides and a Mowse score value of 75 or higher were treated as identified.
To analyze the membrane protein-enriched fraction, cells were disrupted with lysis buffer (50 mM Tris pH 7.5, 1 mM PMSF) by sonication. Cell debris was removed by short centrifugation and membranes were pelleted by additional ultracentrifugation (100,000 × g, 1 h, 4°C). Following the protocol established by Eymann et al. [46], the protein pellet was homogenized and washed in high-salt buffer (20 mM Tris pH 7.5, 1 M NaCl), alkaline buffer (0.1 M Na2CO3-HCl pH 11, 0.1 M NaCl) and 50 mM triethylammonium bicarbonate (TEAB) buffer (pH 7.8) with ultracentrifugation (100,000 × g, 1 h, 4°C) after each washing step. The final pellet was resolved in 50 mM TEAB buffer. Samples of 15 μg protein were separated by 1D SDS-PAGE in two technical replicates per condition. After staining with Coomassie Brilliant Blue, each gel lane was divided into 10 slices which were excised and individually analyzed. Proteins were in gel-digested with trypsin, separated by reverse phase chromatography using a nano-Acquity UPLC System (Waters, Milford, MA, USA) and analyzed by MS/MS in a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). Spectra were assigned to the corresponding amino acid sequence of the C. defragrans 65Phen database using Sorcerer-SEQUEST (SEQUEST version 2.7 revision 11, Thermo Scientific) including Scaffold 3_00_08 (Proteome Software Inc., Portland, OR, USA). SEQUEST was searched with a parent ion tolerance of 10 ppm and a fragment ion mass tolerance of 1.00 Da. For protein identification, a stringent SEQUEST filter for peptides was used (Xcorr versus charge state: 1.80 for singly, 2.2 for doubly, and 3.3 for triply charged ions and deltaCn value greater than 0.10) and at least two unique peptides per proteins were required for identification. Protein fractions from both conditions were considered to be different if no peptides were detected in the alternative fraction or if a significant difference was shown by the Scaffold internal t-test analysis (threshold of 95%).
For continuity of the protein abbreviations, GeDH and GaDH [26] were renamed to GeoA and GeoB according to their gene abbreviations.
Deletion mutagenesis
An in frame deletion mutant of geoB was created as described for the ldi gene [27]. The 5'-flanking region (2015 bases) was amplified with the primers GeoB1_XbaI_F (tctagaagagatcgtgaccagctttcc) and GeoB2_NdeI_R (catatgcatcgagggtgtctcctgagt) and the 3'-flanking region (1967 bases) with GeoB3_NdeI_F (catatgtaggatggacggacaccagg) and GeoB4_HindIII_R (aagcttgatgccgacggcgaacttg). Both amplicons were ligated in a pK19mobsacB plasmid via subcloning using a pCR4-TOPO vector (Invitrogen, Darmstadt, Germany). The constructed plasmid pK19mobsacBΔgeoB was transferred in C. defragrans 65Phen by conjugation and colonies were screened for a second recombination event.
Transposon insertion mutagenesis
Transposon insertion mutants of C. defragrans 65Phen were created by biparental conjugation. 2 mL of overnight cultures of C. defragrans 65Phen anaerobically grown on 20 mM acetate and 20 mM nitrate with 150 μg mL-1 rifampicin and of E.coli W20767 (lysogeny broth medium with 50 μg mL-1 kanamycin) carrying the plasmid pRL27 were spin down at 8000 × g for 5 min. The pellets were washed twice and resuspended in 100 μL mineral medium. The optical density at 600 nm of both cell suspensions was adjusted to 1, combined equally (100 μL) and added as one drop on a mineral medium plate (1.5% agar) containing 50 mM acetate without antibiotics. After 24 h incubation at 28°C, cells were resuspended from the plate with 1 mL mineral medium. 100 μL of cell suspension were plated in different dilutions on mineral medium agar plates containing 50 mM acetate, 25 μg mL-1 kanamycin and 150 μg mL-1 rifampicin. The plates were anaerobically incubated in a jar for 4 days at 28°C. Mutants affected in the limonene metabolism were identified by replica plating (replicator stamp, Carl Roth GmbH + Co. KG, Karlsruhe, Germany). The replicon was incubated in an anaerobic jar with a limonene-enriched headspace for at least two days. Putative mutant colonies were transferred twice to new plates with acetate or limonene as carbon source to confirm the phenotype and the insertion position was determined by a direct sequencing approach. The genomic DNA was isolated from a liquid culture using FastDNA Spin Kit for Soil (MP Biomedicals, OH, USA). 2 to 3 μg genomic DNA were applied for the sequencing PCR reaction using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA). The primers tpnRL 17-1 and tpnRL 13-2 [31] were used with following program: 95°C for 5 min, 100 cycles of 96°C for 30 sec, 52°C for 20 sec and 60°C for 4 min. The fragments were sequenced with an ABI Prism 3130xl Genetic Analyzer (Applied Biosystems Life Technologies Corporation, Carlsbad, CA, USA).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JH and JP planed the study. EMD performed the transposon insertion mutagenesis. SM, DB and RS supported the proteome analysis. BH and RR sequenced the genome of Castellaniella defragrans 65Phen. JP analyzed the genome and proteome, created the deletion mutant of geoB, did physiological experiments and metabolite identification. JP and JH analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Up-regulated proteins of cells grown with α-phellandrene. a2D, identified with 2D-SDS-PAGE and MALDI-TOF-MS; LC, identified with LC-ESI-MS/MS. b++ peptides only identified in α-phellandrene fraction, + increase in α-phellandrene fraction, 0 ratio remained unchanged.
Growth of C. defragrans 65Phen transposon mutants. (●) C. defragrans 65Phen wild type, (▽) ctmA::Tn5a, (■) ctmA::Tn5b, (◊) ctmB::Tn5a, (▲) ctmB:Tn5b and (○) ctmE::Tn5 (for details see Tabl. 3) in anoxic incubations with 3 mM limonene (A) and 3 mM perillyl alcohol (B) as well as oxic incubations with 3 mM limonene (C) and 3 mM perillyl alcohol (D) are represented.
Contributor Information
Jan Petasch, Email: jpetasch@mpi-bremen.de.
Eva-Maria Disch, Email: eva-maria.disch@uni-hamburg.de.
Stephanie Markert, Email: stephanie.markert@uni-greifswald.de.
Dörte Becher, Email: dbecher@uni-greifswald.de.
Thomas Schweder, Email: schweder@uni-greifswald.de.
Bruno Hüttel, Email: huettel@mpipz.mpg.de.
Richard Reinhardt, Email: reinhardt@mpipz.mpg.de.
Jens Harder, Email: jharder@mpi-bremen.de.
Acknowledgments
We thank Sarah Moser for measuring the optical density of cultures. Christoph König kindly provided assistance in the genome sequencing. This study was financed by the Max Planck Society.
References
- Kesselmeier J, Staudt M. Biogenic volatile organic compounds (VOC): an overview on emission, physiology and ecology. J Atmos Chem. 1999;33:23–88. [Google Scholar]
- Turek C, Stintzing FC. Stability of essential oils: a review. Compr Rev Food Sci Food Saf. 2013;12:40–53. [Google Scholar]
- Schrader J. In: Flavours and Fragrances: Chemistry, Bioprocessing and Sustainability. 1. Berger RG, editor. Springer-Verlag New York: LLC; 2007. Microbial flavour production. [Google Scholar]
- Dhavalik RS, Bhattach PK. Fermentation of limonene by a soil pseudomonad. Indian J Biochem. 1966;3:144–157. [PubMed] [Google Scholar]
- Forster-Fromme K, Jendrossek D. Identification and characterization of the acyclic terpene utilization gene cluster of Pseudomonas citronellolis. FEMS Microbiol Lett. 2006;264:220–225. doi: 10.1111/j.1574-6968.2006.00454.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya PK, Prema BR, Kulkarni BD, Pradhan SK. Microbiological transformation of terpenes - hydroxylation of alpha-pinene. Nature. 1960;187:689–690. doi: 10.1038/187689b0. [DOI] [PubMed] [Google Scholar]
- Lehnert N, Krings U, Sydes D, Wittig M, Berger RG. Bioconversion of car-3-ene by a dioxygenase of Pleurotus sapidus. J Biotechnol. 2012;159:329–335. doi: 10.1016/j.jbiotec.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Gibbon GH, Pirt SJ. The degradation of alpha-pinene by Pseudomonas PX1. FEBS Lett. 1971;18:103–105. doi: 10.1016/0014-5793(71)80418-0. [DOI] [PubMed] [Google Scholar]
- Esmaeili A, Tavassoli A. Microbial transformation of citral by Penicillium sp. Acta Biochim Pol. 2010;57:265–268. [PubMed] [Google Scholar]
- Geron C, Rasmussen R, Arnts RR, Guenther A. A review and synthesis of monoterpene speciation from forests in the United States. Atmos Environ. 2000;34:1761–1781. [Google Scholar]
- Maróstica MR, Pastore GM. Biotransformation of limonene: a review of the main methabolic pathways. Quim Nova. 2007;30:382–387. [Google Scholar]
- Molina G, Pimentel MR, Pastore GM. Pseudomonas: a promising biocatalyst for the bioconversion of terpenes. Appl Microbiol Biotechnol. 2013;97:1851–1864. doi: 10.1007/s00253-013-4701-8. [DOI] [PubMed] [Google Scholar]
- Schewe H, Mirata MA, Holtmann D, Schrader J. Biooxidation of monoterpenes with bacterial monooxygenases. Process Biochem. 2011;46:1885–1899. [Google Scholar]
- Bicas JL, Fontanille P, Pastore GM, Larroche C. Characterization of monoterpene biotransformation in two pseudomonads. J Appl Microbiol. 2008;105:1991–2001. doi: 10.1111/j.1365-2672.2008.03923.x. [DOI] [PubMed] [Google Scholar]
- Bicas JL, Fontanille P, Pastore GM, Larroche C. A bioprocess for the production of high concentrations of R-(+)-alpha-terpineol from R-(+)-limonene. Process Biochem. 2010;45:481–486. [Google Scholar]
- Cadwallader KR, Braddock RJ, Parish ME, Higgins DP. Bioconversion of (+)-limonene by Pseudomonas gladioli. J Food Sci. 1989;54:1241–1245. [Google Scholar]
- Savithiry N, Cheong TK, Oriel P. Production of alpha-terpineol from Escherichia coli cells expressing thermostable limonene hydratase. Appl Biochem Biotechnol. 1997;63–65:213–220. doi: 10.1007/978-1-4612-2312-2_20. [DOI] [PubMed] [Google Scholar]
- Tadasa K. Intermediates in bacterial-degradation pathway of alpha-terpineol. Agric Biol Chem. 1977;41:2095–2096. [Google Scholar]
- Harder J, Probian C. Microbial-degradation of monoterpenes in the absence of molecular-oxygen. Appl Environ Microbiol. 1995;61:3804–3808. doi: 10.1128/aem.61.11.3804-3808.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss S, Harder J. Thauera linaloolentis sp. nov. and Thauera terpenica sp. nov., isolated on oxygen-containing monoterpenes (linalool, menthol, and eucalyptol) nitrate. Syst Appl Microbiol. 1998;21:365–373. doi: 10.1016/s0723-2020(98)80046-5. [DOI] [PubMed] [Google Scholar]
- Foss S, Heyen U, Harder J. Alcaligenes defragrans sp. nov., description of four strains isolated on alkenoic monoterpenes ((+)-menthene, alpha-pinene, 2-carene, and alpha-phellandrene) and nitrate. Syst Appl Microbiol. 1998;21:237–244. doi: 10.1016/s0723-2020(98)80028-3. [DOI] [PubMed] [Google Scholar]
- Heyen U, Harder J. Cometabolic isoterpinolene formation from isolimonene by denitrifying Alcaligenes defragrans. FEMS Microbiol Lett. 1998;169:67–71. [Google Scholar]
- Heyen U, Harder J. Geranic acid formation, an initial reaction of anaerobic monoterpene metabolism in denitrifying Alcaligenes defragrans. Appl Environ Microbiol. 2000;66:3004–3009. doi: 10.1128/aem.66.7.3004-3009.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodkorb D, Gottschall M, Marmulla R, Lüddeke F, Harder J. Linalool dehydratase-isomerase, a bifunctional enzyme in the anaerobic degradation of monoterpenes. J Biol Chem. 2010;285:30436–30442. doi: 10.1074/jbc.M109.084244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüddeke F, Harder J. Enantiospecific (S)-(+)-linalool formation from beta-myrcene by linalool dehydratase-isomerase. Z Naturforsch C. 2011;66:409–412. doi: 10.1515/znc-2011-7-813. [DOI] [PubMed] [Google Scholar]
- Lüddeke F, Wulfing A, Timke M, Germer F, Weber J, Dikfidan A, Rahnfeld T, Linder D, Meyerdierks A, Harder J. Geraniol and geranial dehydrogenases induced in anaerobic monoterpene degradation by Castellaniella defragrans. Appl Environ Microbiol. 2012;78:2128–2136. doi: 10.1128/AEM.07226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüddeke F, Dikfidan A, Harder J. Physiology of deletion mutants in the anaerobic beta-myrcene degradation pathway in Castellaniella defragrans. BMC Microbiol. 2012;12:192. doi: 10.1186/1471-2180-12-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton RW. Dehydration of the off-flavor chemical 2-methylisoborneol by the R-limonene-degrading bacteria Pseudomonas sp. strain 19-rlim and Sphingomonas sp. strain BIR2-rlima. Biodegradation. 2012;23:253–261. doi: 10.1007/s10532-011-9504-y. [DOI] [PubMed] [Google Scholar]
- Fuchs G, Boll M, Heider J. Microbial degradation of aromatic compounds - from one strategy to four. Nat Rev Microbiol. 2011;9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- Palleroni NJ, Pieper DH, Moore ERB. In: Handbook of Hydrocarbon and Lipid Microbiology. Kenneth NT, editor. Berlin Heidelberg: Springer; 2010. Microbiology of hydrocarbon-degrading Pseudomonas; pp. 1787–1798. [Google Scholar]
- Larsen RA, Wilson MM, Guss AM, Metcalf WW. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol. 2002;178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- Raisig A, Bartley G, Scolnik P, Sandmann G. Purification in an active state and properties of the 3-step phytoene desaturase from Rhodobacter capsulatus overexpressed in Escherichia coli. J Biochem. 1996;119:559–564. doi: 10.1093/oxfordjournals.jbchem.a021278. [DOI] [PubMed] [Google Scholar]
- Galushko A, Minz D, Schink B, Widdel F. Anaerobic degradation of naphthalene by a pure culture of a novel type of marine sulphate-reducing bacterium. Environ Microbiol. 1999;1:415–420. doi: 10.1046/j.1462-2920.1999.00051.x. [DOI] [PubMed] [Google Scholar]
- Speelmans G, Bijlsma A, Eggink G. Limonene bioconversion to high concentrations of a single and stable product, perillic acid, by a solvent-resistant Pseudomonas putida strain. Appl Microbiol Biotechnol. 1998;50:538–544. [Google Scholar]
- Chang HC, Gage DA, Oriel PJ. Cloning and expression of a limonene degradation pathway from Bacillus stearothermophilus in Escherichia coli. J Food Sci. 1995;60:551–553. [Google Scholar]
- Devi JR, Bhattacharyya PK. Fermentation of geraniol, nerol and limonene by a soil pseudomonad, Pseudomonas incognita (linalool strain) Indian J Biochem Biophys. 1977;14:288–291. [PubMed] [Google Scholar]
- Metcalf WW, Jiang WH, Daniels LL, Kim SK, Haldimann A, Wanner BL. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic-engineering - transposon mutagenesis in gram-negative bacteria. Bio Technol. 1983;1:784–791. [Google Scholar]
- Schäfer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19 - selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Ezennia EI, Phillips LR, Wolfe TL, Tabibi SE. Analysis of perillic acid in plasma by reversed-phase high-performance liquid chromatography with ultraviolet detection. J Chromatogr B. 1997;688:354–358. doi: 10.1016/s0378-4347(96)00322-2. [DOI] [PubMed] [Google Scholar]
- Boström KH, Simu K, Hagström A, Riemann L. Optimization of DNA extraction for quantitative marine bacterioplankton community analysis. Limnol Oceanogr Meth. 2004;2:365–373. [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olsen R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Mitra A, Kesarwani AK, Pal D, Nagaraja V. WebGeSTer DB-a transcription terminator database. Nucleic Acids Res. 2011;39:D129–D135. doi: 10.1093/nar/gkq971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Eymann C, Dreisbach A, Albrecht D, Bernhardt J, Becher D, Gentner S, Tam LT, Buttner K, Buurman G, Scharf C, Venz S, Völker U, Hecker M. A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics. 2004;4:2849–2876. doi: 10.1002/pmic.200400907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Up-regulated proteins of cells grown with α-phellandrene. a2D, identified with 2D-SDS-PAGE and MALDI-TOF-MS; LC, identified with LC-ESI-MS/MS. b++ peptides only identified in α-phellandrene fraction, + increase in α-phellandrene fraction, 0 ratio remained unchanged.
Growth of C. defragrans 65Phen transposon mutants. (●) C. defragrans 65Phen wild type, (▽) ctmA::Tn5a, (■) ctmA::Tn5b, (◊) ctmB::Tn5a, (▲) ctmB:Tn5b and (○) ctmE::Tn5 (for details see Tabl. 3) in anoxic incubations with 3 mM limonene (A) and 3 mM perillyl alcohol (B) as well as oxic incubations with 3 mM limonene (C) and 3 mM perillyl alcohol (D) are represented.


