Abstract
Colorectal cancer is the third most common cancer worldwide. Although the transcription factor c-MYC is misregulated in the majority of colorectal tumors, it is difficult to target directly. The deubiquitinase USP28 stabilizes oncogenic factors, including c-MYC; however, the contribution of USP28 in tumorigenesis, particularly in the intestine, is unknown. Here, using murine genetic models, we determined that USP28 antagonizes the ubiquitin-dependent degradation of c-MYC, a known USP28 substrate, as well as 2 additional oncogenic factors, c-JUN and NOTCH1, in the intestine. Mice lacking Usp28 had no apparent adverse phenotypes, but exhibited reduced intestinal proliferation and impaired differentiation of secretory lineage cells. In a murine model of colorectal cancer, Usp28 deletion resulted in fewer intestinal tumors, and importantly, in established tumors, Usp28 deletion reduced tumor size and dramatically increased lifespan. Moreover, we identified Usp28 as a c-MYC target gene highly expressed in murine and human intestinal cancers, which indicates that USP28 and c-MYC form a positive feedback loop that maintains high c-MYC protein levels in tumors. Usp28 deficiency promoted tumor cell differentiation accompanied by decreased proliferation, which suggests that USP28 acts similarly in intestinal homeostasis and colorectal cancer models. Hence, inhibition of the enzymatic activity of USP28 may be a potential target for cancer therapy.
Introduction
The intestine is made up of repetitive units that consist of a differentiated compartment (the villus) and a proliferative compartment (the crypt). Intestinal stem cells are located in the crypt (1, 2) and produce rapidly proliferating daughter cells, the transit amplifying cells, which subsequently differentiate into 2 main epithelial lineages. The absorptive lineage is composed of all enterocytes, while the secretory lineage is composed of goblet cells (secreting protective mucins), enteroendocrine cells (secreting hormones like serotonin or secretin), and Paneth cells (3, 4).
Whether transit amplifying cells differentiate along an absorptive or a secretory lineage is regulated by the Notch pathway (5). Engagement of Notch receptors by Notch ligands, such as Delta or Jagged, induces proteolytic cleavage of the receptor by γ-secretase. The cleaved NOTCH1 receptor (NICD1) translocates into the nucleus, resulting in the formation of an active transcriptional complex composed of RBPJκ (also known as CSL or CBF1) and NICD1. Notch signal activation induces hairy/enhancer of split (Hes) gene expression. Ablation of Notch signaling via genetic deletion of Rbpj results in secretory cell expansion (6). Conversely, in transgenic mice overexpressing NICD1, goblet cells are absent, and the proliferative compartment is expanded (7).
The Wnt signaling pathway is a key regulator of intestinal stem cell homeostasis (8), and 2 of the target genes induced by Wnt signaling, c-MYC and c-JUN, encode transcription factors that have oncogenic potential (9, 10). Consequently, aberrant activation of the adenomatous polyposis coli/β-catenin/T cell factor (APC/β-catenin/TCF) pathway is an initiating event in the majority of human colorectal cancers (11).
c-MYC is a transcription factor with key functions in cell differentiation and cancer development (12–14). In the intestine, c-MYC is required for the altered proliferation and differentiation induced by APC inactivation (15–18). c-MYC is a highly labile protein, and at least 2 ubiquitin ligases, SKP2 and FBW7, can target it for proteasomal degradation (19–21). c-MYC ubiquitination is antagonized by the deubiquitinase USP28, which “piggybacks” on FBW7 and stabilizes c-MYC protein (22). Thus, an E3 ubiquitin ligase and a deubiquitinase, FBW7 and USP28, are together recruited to substrates (22), and a cycle of deubiquitination and ubiquitination controls c-MYC stability.
Genomic data from human cancers suggest that most colorectal cancer mutations converge on c-MYC misregulation (23). Due to its key role in tumorigenesis, much recent research has been directed to finding ways to target c-MYC function (24–29). Dominant-negative approaches targeting c-MYC function impair intestinal tumor formation, and c-Myc heterozygous mice show reduced tumor development in the Apcmin/+ model (16, 17). Transgenic expression of a dominant-negative allele of Myc, OmoMyc, has provided proof of principle that targeting MYC can eliminate tumors with minimal disturbance to normal tissue (30, 31). However, translation of these benefits to human patients will require a small-molecule approach. Recent work has shown that it is possible to selectively inhibit individual USPs using small-molecule inhibitors (32, 33), which suggests that targeting USP28 may be therapeutically feasible. However, whether USP28 has a role in intestinal tumorigenesis is not known.
Here we examined the effect of Usp28 deletion on normal intestinal homeostasis and tumorigenesis, using villin-Cre to delete Usp28 in murine gut epithelium in both WT and Apcmin/+ genetic backgrounds, and Rosa26 CreERT2 to delete Usp28 inducibly in the Apcmin/+ tumor model. We examined the expression of Usp28 mRNA and protein in the murine intestine and in a panel of human colorectal cancers, and correlated Usp28 deletion with a reduction in oncoprotein levels, including 2 novel targets of deubiquitination promoted by USP28, NICD1 and c-JUN. Our data indicate USP28 as a valid candidate for therapeutic inhibition, which may be beneficial in human colorectal cancer patients.
Results
Mice harboring a Usp28 deletion show normal intestinal morphology.
To address the in vivo function of USP28 in intestinal homeostasis, we generated a mouse strain with Usp28 exons 4 and 5 flanked by loxP sites (Usp28fl/fl). Exons 4 and 5 encode part of the catalytic domain, including cysteine 171, which is critical for the deubiquitination activity of USP28 (22). To inactivate USP28 in the intestine, Usp28fl/fl mice were crossed to a strain expressing Cre recombinase under the control of the villin promoter (villin-Cre) (34). The villin-Cre–mediated loss of exons 4 and 5 resulted in a frameshift mutation (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI73733DS1) and prevented the expression of USP28 protein in the gut. These Usp28fl/fl villin-Cre mice (referred to herein as Usp28ΔG) were viable and fertile and showed no gross phenotypic differences compared with their WT littermates.
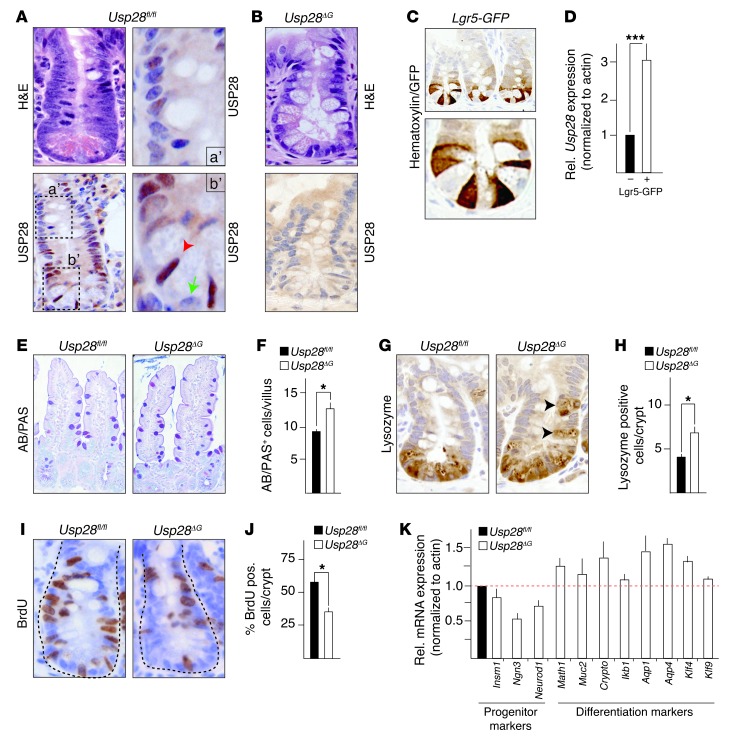
Intestinal fractionation to enrich for either crypts or villi showed significantly more USP28 in protein samples from Usp28fl/fl crypts than in those from villi, whereas USP28 was undetectable in the intestines of Usp28ΔG animals (Supplemental Figure 1B). Immunohistochemistry revealed USP28 protein expression in crypt base columnar (CBC) cells and also several cells within the transit-amplifying cell compartment of the crypt, whereas Paneth cells did not show USP28 staining (Figure 1A). No USP28 was detectable in Usp28ΔG crypts (Figure 1B), confirming the specificity of the antibody. In order to determine whether Usp28 expression was enriched in CBC stem cells, we used an Lgr5-GFP reporter strain and sorted Lgr5+ cells by FACS on the basis of GFP expression. Lgr5+ cells were localized at the crypt base and expressed the stem cell markers Ascl2 and Olfm4 (Figure 1C and Supplemental Figure 1C). Usp28 expression was significantly higher in Lgr5+ versus Lgr5– cells (Figure 1D). Thus, USP28 is highly expressed in CBC intestinal stem and progenitor cells, but appears to be dispensable for intestinal development.
Figure 1. Usp28 is expressed in intestinal crypts and controls intestinal differentiation and proliferation.
(A and B) Expression and localization analysis of USP28 in murine intestine. (A) Usp28fl/fl mice. Boxed regions are shown at higher magnification at right. Bottom right: Usp28 was expressed in crypt base columnar cells (red arrowhead), whereas Paneth cells were devoid of USP28 staining (green arrow). Top right: Usp28 expression gradually decreased within the transit-amplifying cell compartment and was lost in the upper half of the crypt, which contains differentiated cells. (B) Usp28ΔG mice showed no USP28 staining in intestinal tissue. (C) Staining for GFP to label Lgr5+ stem cells in an Lgr5-GFP mouse. (D) Usp28 expression in crypt cells sorted for Lgr5-GFP. (E and F) Representative sections of villi (E) and quantification of goblet cells (AB/PAS+; F), showing increased numbers of goblet cells in Usp28ΔG intestines. n = 10 per group. (G) Representative crypt sections showing mislocalization of Paneth cells (black arrowheads) in Usp28ΔG animals. (H) Quantification of Paneth cells (lysozyme+) per crypt. n = 10 per group. (I and J) Usp28ΔG animals had fewer proliferating cells (BrdU pulse 2.5 hours) within the crypt. n = 10 per group. (K) qRT-PCR of RNA from isolated Usp28ΔG crypt cells showing decreased progenitor and increased differentiation marker gene expression. Expression was normalized to actin and is shown relative to control Usp28fl/fl cells (assigned as 1.0; dashed line). n = 5 per group. Original magnification, ×10 (E); ×20 (C, top); ×40 (A, left; B; C, bottom; G; and I); ×80 (A, right). (D, F, H, J, and K) ***P < 0.0001, *P < 0.001, Student’s t test. Error bars indicate SEM.
USP28 controls intestinal cell differentiation and proliferation.
Immunohistochemical analysis of differentiation markers showed that homozygous Usp28 deletion in the gut epithelium caused a significant increase in goblet cells (Alcian blue/periodic acid–Schiff positive; AB/PAS+) in the villi (Figure 1, E and F). Similarly, Paneth cells (lysozyme+) were found in increased numbers, and were also mislocalized away from the bottom of the crypt (Figure 1, G and H). The number of enteroendocrine cells (chromogranin+) did not differ significantly in the Usp28ΔG animals (data not shown). Therefore, USP28 loss increases differentiation of goblet and Paneth cells, which indicates that USP28 is a negative regulator of secretory fate decisions in the gut.
The number of BrdU-incorporating cells was significantly reduced in Usp28ΔG crypts compared with control Usp28fl/fl mice (Figure 1, I and J), which indicates that USP28 promotes intestinal cell proliferation. Accordingly, expression of the progenitor markers Insm1, Ngn3, and Neurod1 was decreased in Usp28ΔG mice, while a panel of differentiation markers was increased (Figure 1K). Despite the decreased expression of progenitor markers, there was no difference in the number of Olfm4+ stem cells in Usp28ΔG crypts compared with control Usp28fl/fl mice (Supplemental Figure 1, D and E). Thus, USP28 promotes proliferation and inhibits secretory cell differentiation in the intestine.
Mice harboring a Usp28 deletion show lower levels of c-JUN and NICD1.
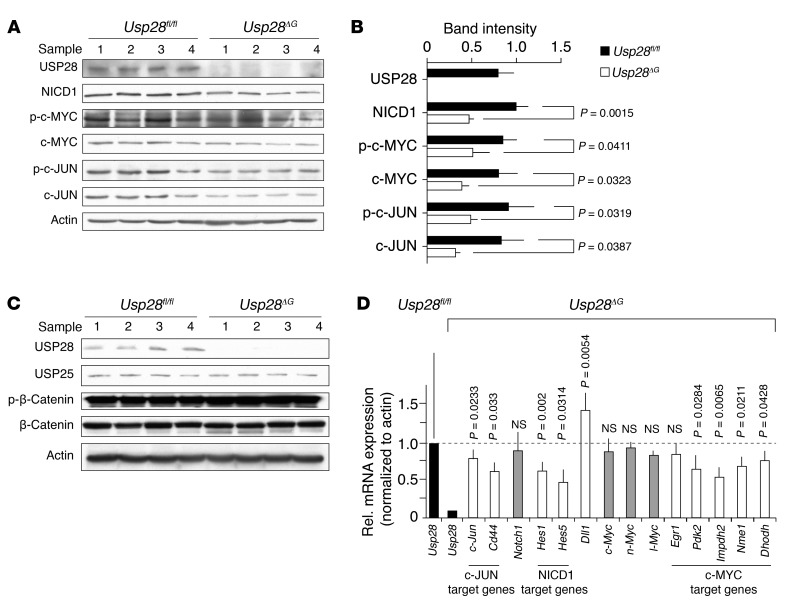
Loss of USP28 in cultured cells destabilizes the c-MYC protein (22). Consistent with this, levels of c-MYC were reduced in Usp28ΔG crypts (Figure 2, A and B). As USP28 is recruited to c-MYC through interaction with the E3 ubiquitin ligase FBW7 (22), we tested whether 2 other FBW7 substrates, c-JUN and NICD1, might also be affected by USP28 deficiency. Indeed, levels of both these proteins were reduced in Usp28ΔG animals (Figure 2, A and B), while the Wnt signaling effector β-catenin and the ubiquitin-specific peptidase USP25, which is closely related to USP28, were not affected (Figure 2C). Consistent with the reduction in c-MYC, c-JUN, and NICD1, quantitative real-time PCR (qRT-PCR) analysis of mRNA from isolated crypt cells showed decreased expression of c-MYC, c-JUN, and NICD1 target genes in Usp28ΔG animals, together with increased expression of Dll1, a gene negatively regulated by Notch signaling (Figure 2D). Expression of Notch1 and 3 Myc genes was unchanged (Figure 2D, gray bars), which indicates that the reduced target gene expression was not due to changes in NICD1 and c-MYC at the mRNA level.
Figure 2. Usp28ΔG intestinal tissue shows decreased c-JUN and NICD1 as well as c-MYC protein.
(A) Western blots of isolated crypt cell lysates from Usp28ΔG intestines show lower USP28, NICD1, phospho– and total c-MYC, and phospho– and total c-JUN proteins compared with Usp28fl/fl controls. (B) Quantification of A. (C) Levels of USP25 and phospho– and total β-catenin were unchanged in crypt cell lysates from Usp28ΔG intestines compared with Usp28fl/fl controls. USP28 and actin are shown as controls. (D) qRT-PCR of RNA from isolated Usp28ΔG crypt cells showing decreased expression of Usp28 (black bars) and c-JUN, NICD1, and c-MYC target genes (white bars) compared with Usp28fl/fl controls, whereas expression of Notch1 and Myc (gray bars) was unaltered. Dll1, which is negatively regulated by Notch signaling, is an example of mRNA increased in Usp28ΔG cells. Expression was normalized to actin and is shown relative to Usp28fl/fl control (assigned as 1.0; dashed line). n = 3 animals per genotype. Error bars indicate SEM.
c-JUN and NICD1 are deubiquitinated and stabilized by USP28.
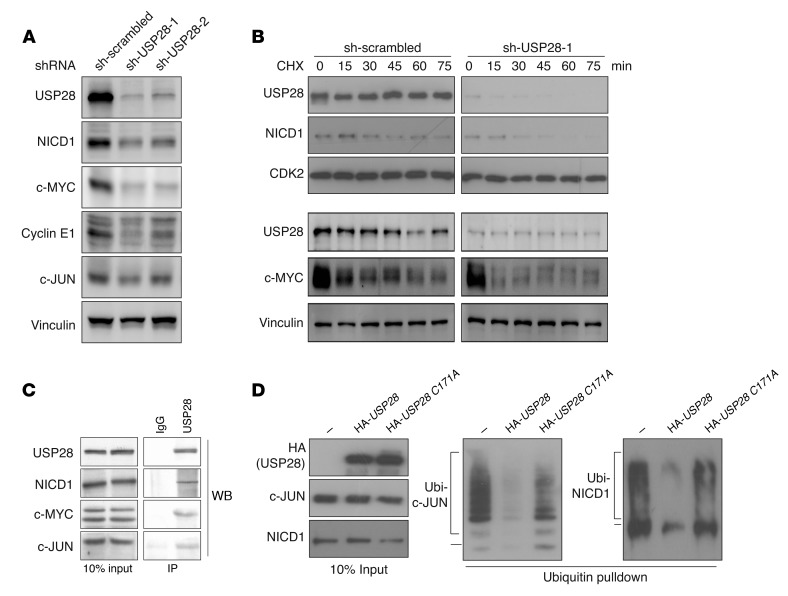
To determine whether the effects on c-JUN and NICD1 are conserved in human cells, we depleted USP28 protein in the human colorectal cancer cell line HCT116 using 2 independent shRNAs. In line with our results in mice, USP28 depletion reduced NICD1 and, to a lesser extent, c-JUN, protein expression as well as levels of the known USP28 substrates c-MYC and cyclin E1 (Figure 3A). When treated with cycloheximide to inhibit protein synthesis, USP28-depleted cells showed a rapid decrease in NICD1 and c-MYC levels, while these proteins were more stable in control cells (Figure 3B and Supplemental Figure 2A). Steady-state levels of c-JUN were also lower in USP28-depleted cells (Supplemental Figure 2B). However, c-JUN protein stability is significantly affected by c-JUN N-terminal phosphorylation (35), and the strong stimulation of c-JUN N-terminal phosphorylation by cycloheximide treatment prevented a clear assessment of c-JUN stability. Thus, USP28 is required for maintaining c-MYC, c-JUN, cyclin E1, and NICD1 protein levels in murine and human intestinal cells, which suggests that c-JUN and NICD1 may also be direct targets of USP28-mediated regulation.
Figure 3. c-JUN and NICD1 are deubiquitinated and stabilized by USP28.
(A) NICD1, c-MYC, cyclin E1, and c-JUN protein levels were decreased in HCT116 cells transiently transfected with 2 independent shRNA constructs against USP28 (sh-USP28-1 and sh-USP28-2) compared with nontargeting control shRNA (sh-scrambled). Vinculin was used as a loading control. Images are representative of 2 independent experiments. (B) Time course of cycloheximide (CHX) treatment of HCT116 cells transiently transfected with scrambled control shRNA or shRNA targeting USP28. Protein stability of USP28, NICD1, and c-MYC was analyzed at the indicated times by Western blot. CDK2 and vinculin were used as controls. Images are representative of 3 independent experiments. (C) Endogenous USP28 coimmunoprecipitated NICD1, c-MYC, and c-JUN. Images are representative of 3 independent experiments. (D) USP28 promoted deubiquitination of c-JUN and NICD1. HeLa cells transfected with c-JUN or NICD1 together with His-tagged ubiquitin, with or without overexpression of USP28 or the catalytically impaired USP28 C171A, were treated 2 days after transfection with MG132 for 3 hours and lysed, after which protein complexes were immunoprecipitated using nickel-NTA beads. Ubiquitinated (Ubi-) complexes were analyzed by Western blot against c-JUN or NICD1. Images are representative of 3 independent experiments.
Consistent with this hypothesis, antibodies directed against USP28 coimmunoprecipitated c-JUN and NICD1 as well as c-MYC from murine intestinal crypt protein extracts (Figure 3C). Conversely, c-JUN and c-MYC were also able to coimmunoprecipitate USP28 (Supplemental Figure 2C). To examine whether USP28 is able to promote deubiquitination of c-JUN and NICD1, we performed ubiquitin pulldown assays in cells expressing HA-tagged USP28, a catalytically impaired mutant (HA-USP28 C171A), or a control vector. Ectopic expression of USP28 deubiquitinated both c-JUN and NICD1, whereas USP28 C171A was less able to do so (Figure 3D). Collectively, these data show that USP28 promotes the deubiquitination of c-JUN and NICD1 and support the notion that these proteins may be direct USP28 substrates. Overexpression of USP28 in HCT116 cells caused only a minor increase in steady-state levels of c-JUN and NICD1 (Supplemental Figure 2D), perhaps because basal USP28 expression is high in these cells, and thus USP28 protein levels may be already saturating.
USP28 is commonly overexpressed in tumors of human colorectal carcinoma patients.
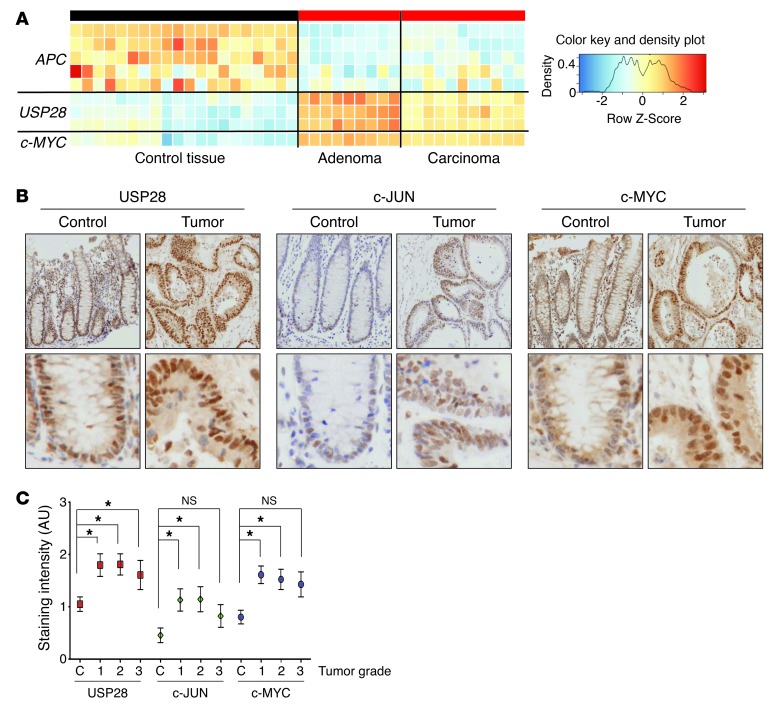
To understand whether USP28 plays a role in human cancers, we analyzed USP28 expression in human colorectal cancer patients using publicly available datasets. USP28 was overexpressed in patient tumor samples compared with normal tissue (Figure 4A). This increase correlated with loss of APC and with strong upregulation of c-MYC expression (Figure 4A), consistent with previous analyses of human colorectal cancers.
Figure 4. USP28 is commonly overexpressed in human colorectal carcinomas.
(A) Heat map of APC, USP28, and c-MYC expression in human intestinal samples from tumors of mucosa and crypt epithelium (adenoma or carcinoma) and the corresponding normal tissue. Data were obtained from Oncomine and Skrzypczak public databases (see Methods). (B) Human colorectal tissue microarray samples from control sections or grade 1 tumor sections stained with antibodies against USP28, c-JUN, and c-MYC. Bottom row shows higher magnifications of the same samples. Original magnification, ×10 (top); ×40 (bottom). (C) Staining intensity was blind-assessed and quantified (0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining) in tumor sections of grade 1–3 and control (C) tissue sections from the tissue microarray (total 80 samples). *P < 0.05.
To investigate the role of USP28 as a tumor promoter in more detail, we evaluated the intensity of USP28, c-JUN, and c-MYC immunostaining on a panel of 70 human intestinal tumor samples. Expression of USP28, c-JUN, and c-MYC all increased in tumors compared with normal tissue (Figure 4B). Notably, similarly increased levels of all 3 proteins were observed in tumor samples, regardless of grade (Figure 4C), which suggests that the upregulation of USP28 occurs early in tumorigenesis. Thus, USP28 is induced in human intestinal tumors, and this correlates with c-JUN and c-MYC overexpression.
USP28 is a c-MYC target.
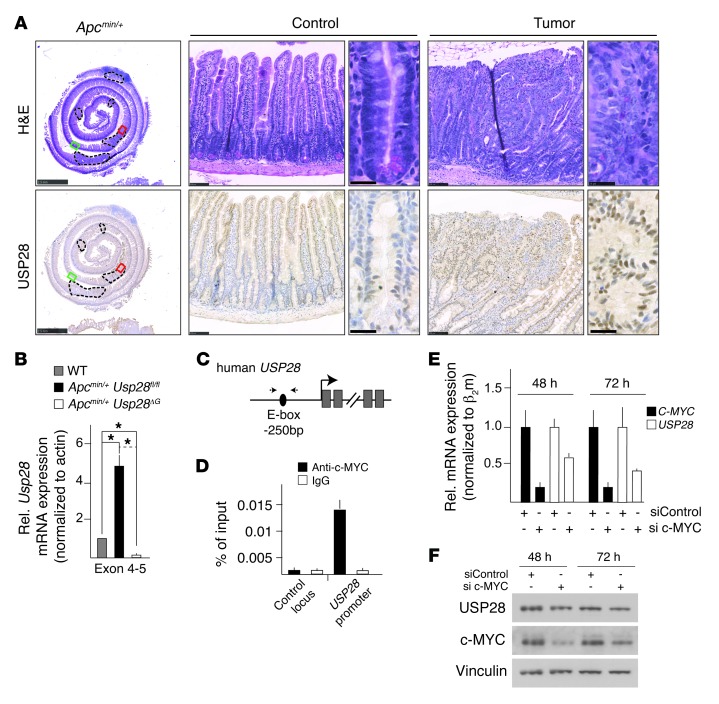
To examine whether the overexpression of USP28 in intestinal tumors is conserved, we analyzed the Apcmin/+ mouse model, which develops intestinal tumors with high penetrance. USP28 protein and mRNA was greatly upregulated in Apcmin/+ tumors compared with normal tissue (Figure 5, A and B). In silico analysis revealed the presence of an E-box (a c-MYC binding site) in the proximal human USP28 promoter (Figure 5C). ChIP analysis in HCT116 cells showed that c-MYC was present at the USP28 promoter (Figure 5D), consistent with published data (www.encodeproject.org), and siRNA-mediated depletion of c-MYC reduced expression of both USP28 mRNA and protein (Figure 5, E and F). Thus, Usp28 is a novel c-MYC target gene that is induced in Apcmin/+ tumors, creating a positive feedback loop stabilizing c-MYC.
Figure 5. USP28 is a c-MYC target gene that is highly expressed in murine as well as human intestinal tumors.
(A) H&E- and USP28-stained sections of the small intestine of an Apcmin/+ animal, including adenomas (dashed outlines). Red and green boxed regions denote tumor and control image areas, respectively. USP28 staining was higher in tumor tissue compared with control crypts and villi. Scale bars: 2.5 mm (gut roll); 100 μm (low-power views); 25 μm (high-power views). (B) qRT-PCR of RNA isolated from Apcmin/+ tumors showed increased Usp28 expression compared with WT tissue. n = 5 per group. Error bars indicate SEM. (C) Human USP28 locus, showing the c-MYC binding site (E-box) upstream of the transcription start site. Small arrows indicate primers used for ChIP in D. (D) ChIP from HCT116 cells. c-MYC efficiently bound to the E-box within the USP28 promoter, but not to an unrelated control locus (see Methods). Data are representative of 3 independent experiments. (E) qRT-PCR validation of c-MYC knockdown and effect on USP28 expression. Error bars indicate SEM. Data are representative of 2 independent experiments. (F) Western blot analysis of HCT116 cells 48 or 72 hours after siRNA transfection. Knockdown of c-MYC decreased USP28 protein levels.
USP28 promotes intestinal tumorigenesis.
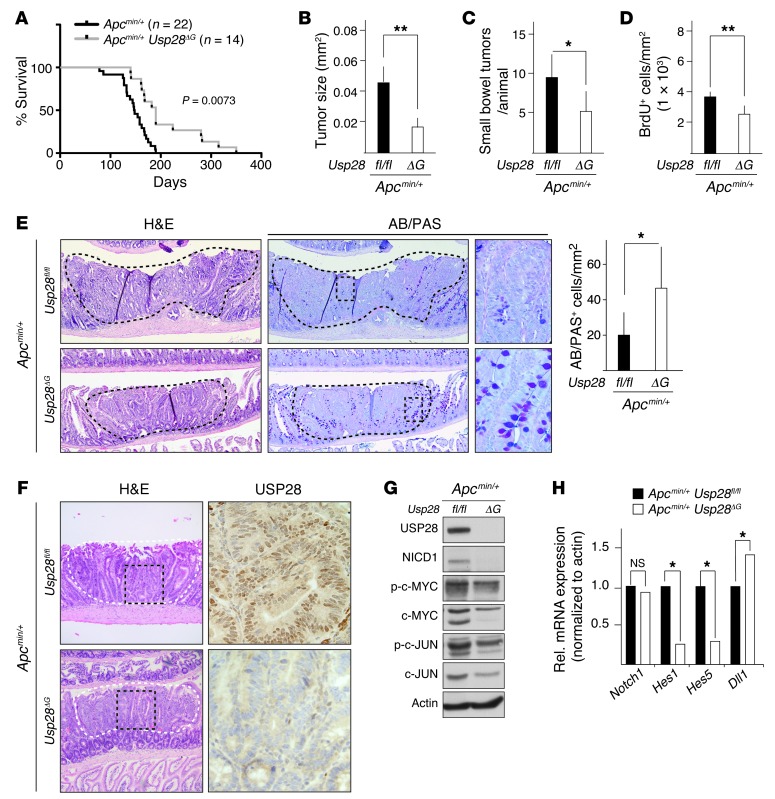
To test whether USP28 contributes to Apcmin/+ tumor formation, we analyzed compound Apcmin/+ Usp28ΔG mice. Usp28 deficiency significantly extended the lifespan of Apcmin/+ animals, from a median survival of 158 days to 190 days (P = 0.0073; Figure 6A). The average tumor size was significantly smaller, and the number of tumors per animal reduced, in Apcmin/+ Usp28ΔG mice (Figure 6, B and C). Apcmin/+ Usp28ΔG tumors had lower rates of cell proliferation, as measured by BrdU incorporation (Figure 6D). They also showed increased differentiation, revealed by AB/PAS staining for goblet cells (Figure 6E), similar to what we observed in the intestines of Usp28ΔG mice. Apcmin/+ Usp28ΔG tumors showed no USP28 staining (Figure 6F and Supplemental Figure 3), which indicated that tumor cells did not escape Usp28 deletion. There were no tumors detected in other organs, and Apcmin/+ Usp28ΔG mice did eventually succumb to intestinal tumors (Supplemental Figure 4, A–C). Therefore, the absence of Usp28 likely improves survival by decreasing proliferation and increasing differentiation within intestinal tumors to slow their progression.
Figure 6. USP28 deficiency ameliorates tumorigenesis in Apcmin/+ animals.
(A) Kaplan-Meier diagram showing increased lifespan of Apcmin/+ Usp28ΔG (n = 14) compared with Apcmin/+ Usp28fl/fl (n = 22) mice. (B) Average tumor size in the small intestine at 18 weeks of age was reduced in Apcmin/+ Usp28ΔG mice. n = 5 per group. (C) Number of tumors in the small intestine and colon at 18 weeks of age is reduced in Apcmin/+ Usp28ΔG mice. n = 5 per group. (D) Quantification of BrdU-incorporating cells per mm2 area of tumor section showing decreased proliferation in Apcmin/+ Usp28ΔG tumors. n = 4 per group. (E) Representative sections of tumors (black dashed outline) of the small intestine stained with H&E or AB/PAS to stain goblet cells. Boxed regions are shown at higher magnification. Quantification of goblet cells is also shown. (F) Sections of tumors (white dashed line) within the small intestine of 18 week-old Apcmin/+ Usp28fl/fl and Apcmin/+ Usp28ΔG animals, stained with H&E (left) and USP28 antibody (right, enlarged). Black dashed box indicates enlarged area. (G) Lysates from isolated small intestinal Apcmin/+ tumors show decreases in the indicated proteins in Usp28ΔG tumors when analyzed by western blotting. Actin was used as loading control. (H) qRT-PCR of RNA isolated from small intestinal tumors showing expression of genes positively (Hes1, Hes5) and negatively (Dll1) regulated by NICD1. n = 5 per group. Data represent means. Original magnification, ×5 (E, left and middle, and F, left); ×40 (E and F, right). (B–E and H) Where shown, error bars indicate SEM. *P < 0.05, **P < 0.005.
Consistent with this, Usp28-deficient tumors showed lower expression levels of c-JUN, c-MYC, and NICD1 compared with Apcmin/+ Usp28fl/fl controls (Figure 6G). Notch signaling is a key regulator of goblet cell differentiation, and while Notch1 mRNA levels were unchanged in Usp28ΔG mice, the altered expression of the NICD1 target genes Hes1, Hes5, and Dll1 (Figure 6H) reflected a decrease in Notch signaling. These results suggest that the absence of USP28 impairs intestinal tumor formation by controlling the protein levels of substrates involved in proliferation and differentiation.
Acute USP28 loss reduces tumor burden and extends lifespan.
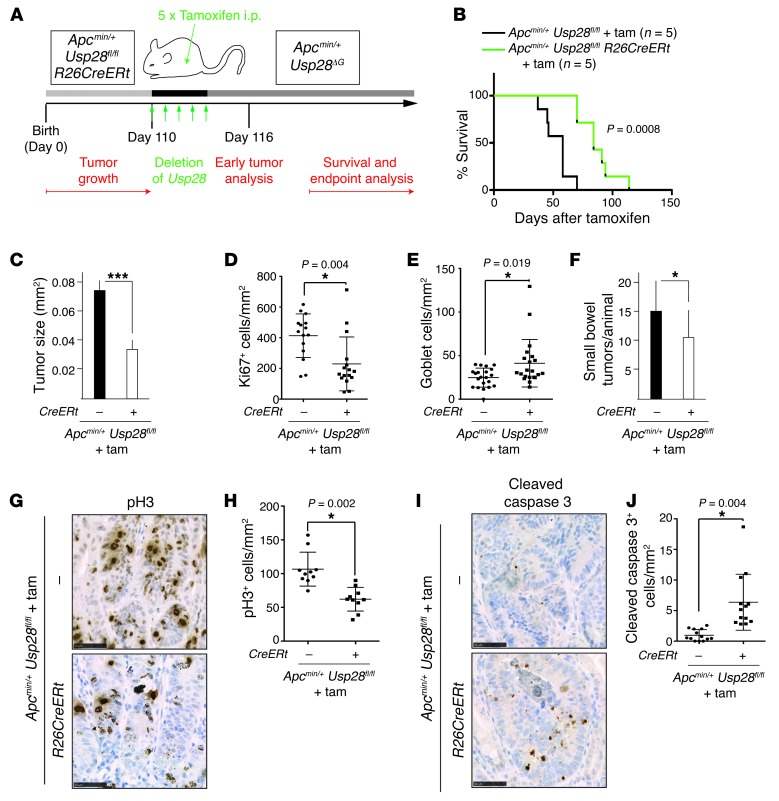
The above results indicated that loss of USP28 in the gut epithelium inhibits tumor development in the Apcmin/+ model. As any systemic therapy targeting USP28 would inactivate the protein in all tissues, we asked whether global inactivation of USP28 would also impair intestinal cancer development. Usp28Δ/Δ mice are viable and fertile (C. Schülein-Völk and M. Eilers, unpublished observations), which indicates that germline deletion of Usp28 is well tolerated systemically. We used the Rosa26 Cre-ERT2 line, which enables systemic gene inactivation through administration of tamoxifen (36). The inducible nature of the system also enabled us to test whether USP28 is required for maintenance of established tumors in adult mice. Apcmin/+ Usp28fl/fl Rosa26 Cre-ERT2 mice were allowed to develop tumors, and at 110 days, Usp28 was deleted (Figure 7A and Supplemental Figure 5A). Ubiquitous acute deletion of Usp28 was sufficient to extend the survival of mice with established tumors to a median of 12 weeks after tamoxifen injection, a 50% increase compared with controls (Figure 7B). Average tumor size was significantly reduced in tamoxifen-injected Apcmin/+ Usp28fl/fl Rosa26 Cre-ERT2 mice at the experimental endpoint (P = 0.0002; Figure 7C). Consistent with this, cellular proliferation in tumors was significantly decreased upon USP28 depletion, and numbers of differentiated goblet cells were moderately increased (Figure 7, D and E, and Supplemental Figure 5B). Tumor number was also somewhat reduced (P = 0.0046; Figure 7F), which suggests that acute loss of USP28 is sufficient to eradicate or prevent some tumors. When examined 2 days after acute Usp28 deletion, tumors exhibited signs of regression, including decreased phospho–histone 3 staining for mitotic cells, and increased cleaved caspase 3 staining, indicative of apoptosis (Figure 7, G–J). Thus, systemic loss of USP28 suppresses Apcmin/+-mediated intestinal hyperplasia, and may be beneficial for the treatment of established intestinal tumors. Taken together, our data indicate an important role for USP28 in intestinal tumor development and suggest inhibition of USP28 as a potential strategy for cancer therapy.
Figure 7. Inducible Usp28 deletion reduces tumor burden and improves survival in Apcmin/+ animals.
(A) Inducible Usp28 deletion. Cre expression was induced by 5 consecutive days of i.p. tamoxifen injection at 110 days of age, when intestinal tumors were already established. R26CreERt, Rosa26 Cre-ERT2. (B) Kaplan-Meier diagram showing survival of tamoxifen-injected Apcmin/+ Usp28fl/fl and Apcmin/+ Usp28fl/fl Rosa26 Cre-ERT2 mice. n = 5 per genotype. (C) Average tumor size in the small intestine at endpoint. n = 5 per group. (D) Quantification of Ki67+ cells per mm2 in tumors at endpoint. (E) AB/PAS+ goblet cells per mm2 in tumors at endpoint. (F) Number of tumors in the small intestines of mice from A postmortem. n = 5 per group. (G) Acute loss of Usp28 reduced cell proliferation in tumors. Representative immunohistological sections of tumors from tamoxifen-treated Apcmin/+ Usp28fl/fl and Apcmin/+ Usp28fl/fl Rosa26 Cre-ERT2 mice, 2 days after acute Usp28 deletion, stained for phospho–histone 3 (pH3). Scale bars: 50 μm. (H) pH3+ cells per mm2 in tumors 2 days after acute Usp28 deletion. (I) Apcmin/+ tumors stained with cleaved caspase 3, showing increased cell death after acute Usp28 deletion. Scale bars: 50 μm. (J) Cleaved caspase 3+ cells per mm2 in tumors 2 days after acute Usp28 deletion. (C–F, H, and J) Error bars indicate SEM. Symbols in D, E, H, and J indicate individual tumors. ***P < 0.001, *P < 0.05.
Discussion
Oncogenic proteins, such as the transcription factors c-MYC and c-JUN, play important roles in the development of intestinal cancer and other tumor types (14, 18, 37, 38). c-MYC has attracted particular attention, since it is universally deregulated in colorectal tumors, and since its function in these tumors is essential and nonredundant (15). Cancer treatments targeting c-MYC would thus severely restrict the potential for drug resistance. However, transcription factors are difficult to inhibit pharmacologically, fueling the search for alternative ways to target these proteins.
Here we demonstrated that loss of the deubiquitinase USP28 reduced the levels of 3 key oncogenic factors: c-MYC, c-JUN, and NICD1. By inhibiting proliferation and promoting differentiation of tumor cells, loss of USP28 suppressed tumor formation in the Apcmin/+ mouse model. USP28 is a cysteine protease, and this class of enzymes can be specifically inhibited by small molecules (39), which suggests that targeting USP28 is a useful strategy for the development of new cancer therapies.
USP28 in intestinal homeostasis.
Previous work has demonstrated that USP28 can bind to its substrates c-MYC and cyclin E1 via the E3 ligase FBW7 (22). Consistent with this model, USP28 was found to promote the deubiquitination of 2 additional SCF(FBW7) substrates, c-JUN and NICD1. Consistent with their opposite effects on shared substrates, USP28 and FBW7 had opposite effects on intestinal cell proliferation and differentiation (Figure 1 and ref. 40). Usp28ΔG mice showed increased numbers of differentiated goblet and Paneth cells, thus biasing cells toward a secretory fate. Goblet cell number is controlled by Notch signaling, since constitutively expressed NICD1 decreases the number of goblet cells, whereas treatment with a γ-secretase inhibitor, which prevents NICD1 production, increases goblet cell number (6). Most likely, therefore, the function of USP28 toward NICD1 underlies its role in secretory fate determination.
Similarly, c-MYC, c-JUN, and cyclin E1 play key roles in cellular proliferation, and alteration in the protein levels of these substrates likely contributes to the role of USP28 in intestinal proliferation. The FBW7/USP28 system thus controls the abundance of a number of proteins with important functions in intestinal cell proliferation and differentiation.
USP28 in tumorigenesis.
The targets of USP28 have well-characterized roles in tumor formation. c-MYC is required for neoplasia after APC inactivation, and c-JUN is required for oncogenesis in the Apcmin/+ mouse model (15, 16, 37). Cyclin E1, a key target of c-MYC activation, is required for the chromosomal instability phenotype of colorectal cancers with FBW7-inactivating mutations (41). Preventing NICD production with a γ-secretase inhibitor is able to reduce proliferation and increase goblet cell differentiation within Apcmin/+ tumors, just as in our Usp28 deletion model (6). Activation of Notch signaling is also known to occur in both human and mouse tumors and strongly promotes Apcmin/+ tumorigenesis, reducing differentiation and increasing proliferation within tumors and also shortening mouse survival (42). The destabilization and thus reduced protein levels of these oncogenic factors is likely to cause the tumor-suppressive effect of USP28 loss.
The rapid tumor progression of the Apcmin/+ Fbw7ΔG mouse model (40), and the increased expression of Usp28 mRNA in Apcmin/+ tumors, is reflected in human colorectal cancers. Apc loss in human colorectal cancer correlated with USP28 overexpression and increased c-MYC levels (Figure 4). Thus, the findings of the Apcmin/+ Usp28ΔG mouse model might be applicable to human cancers. This is especially important, as inducible deletion of Usp28 in Apcmin/+ mice with established tumors attenuated tumor development (Figure 7).
USP28 was originally described as a regulator of p53-mediated apoptosis (43). However, we found no evidence of p53 deregulation in Usp28ΔG mice. No change in p53 protein was detected, either by immunohistochemistry or by Western blot, in Usp28ΔG versus Usp28fl/fl intestines (Supplemental Figure 6, A and B). Usp28ΔG crypts also did not show elevated staining for cleaved caspase 3, and expression of p53 target genes was unchanged (Supplemental Figure 6, C and D). p53 is commonly mutated in human colon cancers (44), but based on these data, the tumor-suppressive effect of inhibiting USP28 is predicted to be independent of p53 status.
The observation that loss of USP28 was able to restrain tumor progression is also encouraging, given the current inability to target c-MYC–driven cancers using drugs. Proof-of-principle experiments using a dominant-negative transgene have unequivocally established that targeting MYC will have high therapeutic efficacy for many tumors (30, 31, 45), but small molecules that directly inhibit c-MYC are not available. The bromodomain protein inhibitor JQ1 inhibits MYC transcription and suppresses hematologic malignancies, although it is not yet optimized for in vivo delivery (46, 47). An alternative strategy is to target MYC stability. As an enzyme that stabilizes the c-MYC protein, USP28 is a good candidate for small-molecule inhibition; moreover, it has the advantage of simultaneously decreasing the activity of other oncogenic factors in addition to c-MYC. Recent studies have shown that small-molecule deubiquitinase inhibitors are effective in cell lines (32, 48, 49), further underlining USP28 inhibition as a promising approach to cancer treatment.
Methods
Mouse lines.
Exons 4 and 5 of the Usp28 locus were flanked by loxP sites. A neomycin cassette flanked by Frt sites was introduced into intron 3 and used as a positive selection marker. The neomycin cassette was excised after successful targeting by crossing mice harboring the targeted allele with Flp deleter mice. A thymidine kinase cassette served as a negative selection marker. Cre-mediated deletion of exons 4 and 5 results in loss of function by removing the exons encoding part of the peptidase domain (including the catalytic C171 in exon 5) and generating a frameshift to downstream exons.
villin-Cre, Apcmin/+, and Rosa26 Cre-ERT2 mice have been described previously (34, 50, 51).
Histological analysis and quantification.
Mice were injected i.p. with 100 mg/kg BrdU (Sigma-Aldrich) 2.5 hours prior to sacrifice. Mice were euthanized by cervical dislocation, and the small intestines were dissected out. The intestines were cut longitudinally into pieces of similar size, opened out and fixed overnight in 10% neutral buffered formalin, briefly washed with PBS and transferred into 70% ethanol, rolled, processed, and embedded into paraffin. Sections were cut at 4 μm for H&E staining, AB/PAS staining, immunohistochemistry, and immunofluorescence. Antibodies against USP28 (Sigma-Aldrich), c-JUN (BD Biosciences), c-MYC (Roche), BrdU (Roche), Ki67 (Abcam), p53 (Vector Laboratories), cleaved caspase 3 (R&D Systems), pH3 (Cell Signaling Technology), GFP (Abcam), and lysozyme (DAKO) were used.
To quantify BrdU+ cells per crypt, 100 full crypts or villi were scored from at least 3 mice per genotype. Data are represented as percentage of control ± SEM (considering each control littermate BrdU+ cell number as 100%). To quantify goblet cells, AB/PAS+ cells were quantified from 100 ileal villi from at least 5 mice per genotype; data are represented as mean ± SEM. To quantify Paneth cells, lysozyme+ cells were quantified from 100 crypts from at least 3 mice per genotype; data are represented as mean ± SEM.
Western blot analysis and qRT-PCR of the intestine.
Immunoblots were carried out as previously described (40). Antibodies against USP28 (Sigma-Aldrich, or Ptglab 17707-1-AP), USP25 (Sigma-Aldrich), p53 (NEB), c-JUN (BD Biosciences), phospho–c-JUN(S63) (Cell Signaling Technology), active NICD1 (Abcam), c-MYC (Millipore, OP31), phospho–c-MYC(T58/S62) (Santa Cruz), β-catenin (Sigma-Aldrich), phospho–β-catenin (S37/T41) (Millipore), and β-actin (Sigma-Aldrich) were used.
For qRT-PCR analysis, total mRNA was isolated from dissected ileum as previously described (40). Results (normalized to β-actin) are presented as fold induction relative to control mice. See Supplemental Table 1 for primers used for qRT-PCR.
Apcmin/+ model of intestinal tumorigenesis.
Apcmin/+ mice were generated in Usp28ΔG backgrounds. Mice were monitored and culled when they showed symptoms of sickness; control Apcmin/+ littermates were culled as controls. Tumor number, incidence, and area were determined in at least 5 mice per genotype, and intestinal tissue was used for histological analysis of USP28, c-MYC, c-JUN, active NICD1, H&E, and AB/PAS stainings. Tumors from mice of the same genotype were pooled, and RNA and protein was extracted and used for qRT-PCR and Western blot analyses.
Tamoxifen treatment.
Apcmin/+Usp28fl/flRosa26 Cre-ERT2 and Apcmin/+ Usp28fl/fl animals were given 100 μg/g BW tamoxifen dissolved in peanut oil i.p., as previously described (51), for 5 consecutive days. Mice were monitored and culled when they showed symptoms of sickness, and overall survival was documented.
Cell lines.
HEK293T, Phoenix-ampho, HCT116, and HeLa cells were provided by Cancer Research UK Cell Services. Cell lines were cultured in DMEM with 10% heat-inactivated FBS and 1% penicillin and streptomycin at 37°C in a 5% CO2/95% humidity incubator except where indicated.
Expression vectors.
The HA-USP28, Flag-tagged USP28, and mutant HA-USP28 C171A constructs were described previously (22).
Western blot analysis of cell line extracts.
Immunoblots were carried out as previously described (40). Briefly, cells were washed with cold PBS and subsequently resuspended in RIPA buffer (Cell Signaling Technology) supplemented with complete protease inhibitor cocktail (Sigma-Aldrich). The resuspended pellet was sonicated for 10 seconds, and lysates were cleared by centrifugation for 15 minutes at 4°C.
Antibodies against USP28 (Sigma-Aldrich or Ptglab 17707-1-AP), c-JUN (BD Biosciences or Cell Signaling Technology), active Notch1 (Abcam or Cell Signaling Technology), c-MYC (Santa Cruz; Millipore, OP31; or Abcam, ab32072), cyclin E1 (Santa Cruz), HA (Covance, 16B12), vinculin (Sigma-Aldrich, V9131), Cdk2 (Santa Cruz, M2/sc-163), and β-actin (Sigma-Aldrich) were used.
siRNA transfection.
siRNAs targeting MYC (L-003282-00-0020) and a nontargeting siRNA pool (D-001206-14-05) were purchased from Thermo Scientific (Dharmacon). Transfection was performed according to the manufacturer’s protocol (Lipofectamine RNAiMAX; Invitrogen). Cells were harvested 48 or 72 hours after transfection. For protein analysis, cells were washed with cold PBS and directly lysed in RIPA buffer. For gene expression analysis, RNA was extracted with peqGOLD (peqlab) according to the manufacturer’s instructions, and the first strand was synthesized with M-MLV Reverse Transcriptase (Promega) and random hexamer primers (Roche). Results, normalized to β2m, were presented as fold regulation over siControl-treated cells. Error bars indicate SD of technical triplicates. Primer sequences were as follows: β2m, 5′-GTGCTCGCGCTACTCTCTC and 5′-GTCAACTTCAATGTCGGAT; MYC, 5′-CCTACCCTCTCAACGACAGC and 5′-CTCTGACCTTTTGCCAGGAG; USP28, 5′-ACTCAGACTATTGAACAGATGTACTGC and 5′-CTGCATGCAAGCGATAAGG.
ChIP.
ChIP was performed as described previously (52). Briefly, HCT116 cells were crosslinked with 1% formaldehyde, cells were lysed, and chromatin was fragmented by sonication. Chromatin was immunoprecipitated with anti-MYC antibodies (Santa Cruz, N-262, sc-764) or control rabbit IgG (Sigma-Aldrich, I5006). Crosslinks were reverted, and DNA was isolated by phenol/chloroform extraction and ethanol precipitation. Primers used for qRT-PCR amplified a fragment containing a conserved consensus E-box at –250 bp from the transcriptional start site of human USP28 (5′-CCGAACAGTTCTGCGTGCT and 5′-CACCGGCTGTGAAGCTGA), or a negative control region on chromosome 11 (5′-TTTCTCACATTGCCCCTGT and 5′-TCAATGCTGTACCAGGCAAA). Percentage of binding relative to input was calculated using the 2ΔCt method, with ΔCt defined as Ctinput – CtIP. Error bars show SD for technical triplicates.
Cycloheximide assay.
HCT116 cells were transfected using Lipofectamine (Invitrogen) with pRetroSuper-puro vector containing scrambled shRNA or shRNA against USP28 (22). Transfected cells were selected with 2 μg/ml puromycin (InvivoGen). To measure protein stability, protein synthesis was blocked by addition of 100 μg/ml cycloheximide (Sigma-Aldrich), and cells were collected at different time points after drug addition. Whole cell extracts were obtained by boiling cell suspensions in SDS sample buffer.
Coimmunoprecipitation of endogenous proteins from murine crypt cells.
Coimmunoprecipitation was carried out as previously described (53). In short, mouse primary crypt material was prepared as described above for Western blot and qRT-PCR of intestine, and then prepared in PLB buffer (1% Triton X-100, 2 mM EDTA, 1 mM DTT, 5% glycerol in PBS) supplemented with complete protease inhibitor cocktail (Sigma-Aldrich). The resuspended pellet was sonicated for 10 seconds, and lysates were cleared by centrifugation for 15 minutes at 4°C. Next, lysates were precleared with IgG-coupled agarose beads (Sigma-Aldrich) for 2 hours at 4°C. Immunoprecipitations of endogenous complexes were carried out overnight at 4°C with anti-Usp28 (Sigma-Aldrich) or, alternatively, with anti–c-Jun (Santa Cruz) or anti–c-Myc (Santa Cruz) in combination with a 50% protein A/G–agarose mix slurry (Sigma-Aldrich). Immunoprecipitates were washed 5 times with lysis buffer, and sample proteins were separated by SDS-PAGE and subsequently transferred onto nitrocellulose membranes and blotted with the indicated antibodies.
Ubiquitination assay.
Cells were transfected with the indicated plasmids. 24 hours after transfection, cells were treated with proteasome inhibitor (Merck Biosciences, MG132) for 3 hours. After proteasome inhibitor treatment, cells were washed with cold PBS and subsequently subjected to in vivo ubiquitination assays. His-ubiquitin was affinity purified with nickel-NTA-agarose beads, as described previously (54), and sample proteins were separated by SDS-PAGE and subsequently transferred onto nitrocellulose membranes and blotted with the indicated antibodies.
Human colorectal tumor analysis.
Human Tumor Micro-Array slides containing 70 colon cancer samples and 10 controls were ordered from US Biomax (order no. CO811; http://www.biomax.us/tissue-arrays/Colon/CO811). For USP28, c-MYC, and c-JUN immunohistochemistry quantification, all samples were blind scored (from 0, lowest staining intensity, to 3, highest staining intensity) by 4 independent observers. Mean ± SD of the score was calculated for each tumor sample (for USP28, c-MYC, and c-JUN) using Graphpad Prism5.
Computational analyses.
Human USP28, c-MYC, and APC expression data were downloaded from Oncomine (https://www.oncomine.org), and expression data from Skrzypczak (Skrzypczak Colorectal 2) were used to correlate the expression of APC, USP28, and c-MYC in normal and tumor samples. Expression data were available for 20 tumors and 20 normal colorectal samples from the Skrzypczak Colorectal 2 dataset.
Statistics.
Statistical evaluation was performed by 2-tailed Student’s unpaired t test. Data are presented as mean ± SEM. A P value of 0.05 or less was considered statistically significant.
Study approval.
Experiments in mice were carried out with the approval of the London Research Institute’s Ethical Review Committee and under the guidance of the Biological Resources Unit.
Supplementary Material
Acknowledgments
We thank B. Vogelstein (Johns Hopkins University School of Medicine, Baltimore, Maryland, USA) for providing cell lines; M. Frye, M. Kumar, I. Malanchi, and P. Meier for critical reading of the manuscript; and the Experimental Histopathology Laboratory as well as Richard Mitter (Bioinformatics and Biostatistics Lab, London Research Institute) for expert technical assistance. M.E. Diefenbacher is funded by a postdoctoral fellowship (DFG-GEPRIS DI 1679/1-1). S.M. Blake is funded by FEBS and EMBO Long-Term Fellowships. N. Popov is funded by a grant from the Deutsche Forschungsgemeinschaft (PO 1458/3-1). L.A. Jaenicke was funded by a Boehringer Ingelheim Fonds fellowship. The generation and initial analysis of the Usp28 knockout mice was supported by grants from Bayer and the Wilhelm-Sander-Stiftung to M. Eilers. The Mammalian Genetics Laboratory and London Research Institute are funded by Cancer Research UK.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(8):3407–3418. doi:10.1172/JCI73733.
Nikita Popov’s present address is: University of Würzburg, Comprehensive Cancer Center Mainfranken, Würzburg, Germany.
References
- 1.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. Stem Cells: A unifying theory for the crypt. Nature. 2013;495(7439):53–54. doi: 10.1038/nature11958. [DOI] [PubMed] [Google Scholar]
- 3.Scoville DH, Sato T, He XC, Li L. Current view: intestinal stem cells and signaling. Gastroenterology. 2008;134(3):849–864. doi: 10.1053/j.gastro.2008.01.079. [DOI] [PubMed] [Google Scholar]
- 4.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469(7330):415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sancho E, Batlle E, Clevers H. Signaling pathways in intestinal development and cancer. Annu Rev Cell Dev Biol. 2004;20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805. [DOI] [PubMed] [Google Scholar]
- 6.van Es JH, et al. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435(7044):959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 7.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435(7044):964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 8.de Lau W, Barker N, Clevers H. WNT signaling in the normal intestine and colorectal cancer. Front Biosci. 2007;12:471–491. doi: 10.2741/2076. [DOI] [PubMed] [Google Scholar]
- 9.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653(1):1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 10.Grigoryan T, Wend P, Klaus A, Birchmeier W. Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of β-catenin in mice. Genes Dev. 2008;22(17):2308–2341. doi: 10.1101/gad.1686208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87(2):159–170. doi: 10.1016/S0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 12.Heinen CD, et al. The APC tumor suppressor controls entry into S-phase through its ability to regulate the cyclin D/RB pathway. Gastroenterology. 2002;123(3):751–763. doi: 10.1053/gast.2002.35382. [DOI] [PubMed] [Google Scholar]
- 13.Heath VJ, Gillespie DA, Crouch DH. Inhibition of adipocyte differentiation by cMyc is not accompanied by alterations in cell cycle control. Biochem Biophys Res Commun. 2000;269(2):438–443. doi: 10.1006/bbrc.2000.2316. [DOI] [PubMed] [Google Scholar]
- 14.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sansom OJ, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446(7136):676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 16.Athineos D, Sansom OJ. Myc heterozygosity attenuates the phenotypes of APC deficiency in the small intestine. Oncogene. 2010;29(17):2585–2590. doi: 10.1038/onc.2010.5. [DOI] [PubMed] [Google Scholar]
- 17.Ignatenko NA, et al. Role of c-Myc in intestinal tumorigenesis of the ApcMin/+ mouse. Cancer Biol Ther. 2006;5(12):1658–1664. doi: 10.4161/cbt.5.12.3376. [DOI] [PubMed] [Google Scholar]
- 18.He TC, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281(5382):1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 19.Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol. 2004;14(20):1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 20.Minella AC, Clurman BE. Mechanisms of tumor suppression by the SCF(Fbw7). Cell Cycle. 2005;4(10):1356–1359. doi: 10.4161/cc.4.10.2058. [DOI] [PubMed] [Google Scholar]
- 21.Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popov N, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9(7):765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 23.Muzny DM, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frenzel A, Zirath H, Vita M, Albihn A, Henriksson MA. Identification of cytotoxic drugs that selectively target tumor cells with MYC overexpression. PLoS One. 2011;6(11):e27988. doi: 10.1371/journal.pone.0027988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kessler JD, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335(6066):348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadiminty N, et al. MicroRNA let-7c suppresses androgen receptor expression and activity via regulation of Myc expression in prostate cancer cells. J Biol Chem. 2012;287(2):1527–1537. doi: 10.1074/jbc.M111.278705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toyoshima M, et al. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci U S A. 2012;109(24):9545–9550. doi: 10.1073/pnas.1121119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bidwell GL, et al. Thermally targeted delivery of a c-Myc inhibitory polypeptide inhibits tumor progression and extends survival in a rat glioma model. PLoS One. 2013;8(1):e55104. doi: 10.1371/journal.pone.0055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, et al. Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature. 2012;483(7391):608–612. doi: 10.1038/nature10927. [DOI] [PubMed] [Google Scholar]
- 30.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455(7213):679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soucek L, et al. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27(5):504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. Conformational stabilization of ubiquitin yields potent and selective inhibitors of USP7. Nat Chem Biol. 2013;9(1):51–58. doi: 10.1038/nchembio.1134. [DOI] [PubMed] [Google Scholar]
- 33.Chauhan D, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22(3):345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39(3):186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 35.Musti AM, Treier M, Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275(5298):400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 36.Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2(4):292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nateri AS, Spencer-Dene B, Behrens A. Interaction of phosphorylated c-Jun with TCF4 regulates intestinal cancer development. Nature. 2005;437(7056):281–285. doi: 10.1038/nature03914. [DOI] [PubMed] [Google Scholar]
- 38.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3(11):859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 39.Hernandez AA, Roush WR. Recent advances in the synthesis, design and selection of cysteine protease inhibitors. Curr Opin Chem Biol. 2002;6(4):459–465. doi: 10.1016/S1367-5931(02)00345-9. [DOI] [PubMed] [Google Scholar]
- 40.Sancho R, Jandke A, Davis H, Diefenbacher ME, Tomlinson I, Behrens A. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology. 2010;139(3):929–941. doi: 10.1053/j.gastro.2010.05.078. [DOI] [PubMed] [Google Scholar]
- 41.Rajagopalan H, et al. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428(6978):77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 42.Fre S, et al. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci U S A. 2009;106(15):6309–6314. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126(3):529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 44.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sodir NM, Swigart LB, Karnezis AN, Hanahan D, Evan GI, Soucek L. Endogenous Myc maintains the tumor microenvironment. Genes Dev. 2011;25(9):907–916. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Issaenko OA, Amerik AY. Chalcone-based small-molecule inhibitors attenuate malignant phenotype via targeting deubiquitinating enzymes. Cell Cycle. 2012;11(9):1804–1817. doi: 10.4161/cc.20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010;70(22):9265–9276. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 50.Luongo C, Moser AR, Gledhill S, Dove WF. Loss of Apc+ in intestinal adenomas from Min mice. Cancer Res. 1994;54(22):5947–5952. [PubMed] [Google Scholar]
- 51.Hameyer D, et al. Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol Genomics. 2007;31(1):32–41. doi: 10.1152/physiolgenomics.00019.2007. [DOI] [PubMed] [Google Scholar]
- 52.von Eyss B, et al. The SNF2-like helicase HELLS mediates E2F3-dependent transcription and cellular transformation. EMBO J. 2012;31(4):972–985. doi: 10.1038/emboj.2011.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahmoudi T, et al. The kinase TNIK is an essential activator of Wnt target genes. EMBO J. 2009;28(21):3329–3340. doi: 10.1038/emboj.2009.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davies CC, Chakraborty A, Cipriani F, Haigh K, Haigh JJ, Behrens A. Identification of a co-activator that links growth factor signalling to c-Jun/AP-1 activation. Nat Cell Biol. 2010;12(10):963–972. doi: 10.1038/ncb2098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.