Abstract
We review here recent advances in our understanding of sexual reproduction in fungal pathogens that commonly infect humans, including Candida albicans, Cryptococcus neoformans/gattii, and Aspergillus fumigatus. Where appropriate or relevant, we introduce findings on other species associated with human infections. In particular, we focus on rapid advances involving genetic, genomic, and population genetic approaches that have reshaped our view of how fungal pathogens evolve. Rather than being asexual, mitotic, and largely clonal, as was thought to be prevalent as recently as a decade ago, we now appreciate that the vast majority of pathogenic fungi have retained extant sexual, or parasexual, cycles. In some examples, sexual and parasexual unions of pathogenic fungi involve closely related individuals, generating diversity in the population but with more restricted recombination than expected from fertile, sexual, outcrossing and recombining populations. In other cases, species and isolates participate in global outcrossing populations with the capacity for considerable levels of gene flow. These findings illustrate general principles of eukaryotic pathogen emergence with relevance for other fungi, parasitic eukaryotic pathogens, and both unicellular and multicellular eukaryotic organisms.
Many pathogenic fungi, including Cryptococcus, Candida, and Aspergillus, have sexual or parasexual cycles. Some unions involve closely related individuals; others involve diverse individuals and the capacity for significant gene flow.
A variety of fungal species are now recognized as being major human pathogens, responsible for causing life-threatening infections worldwide (Brown et al. 2012). Some species appear to have evolved to grow commensally, or as pathogens, on animal hosts while other species exist primarily in different ecological niches and cause opportunistic infections of humans, notably in those with suppressed immune systems (Tekaia and Latge 2005; Sharpton et al. 2009; Brown et al. 2012). A key aspect of the success of many of these fungal pathogens is their ability to produce infectious propagules by which they can enter and colonize a human host. Most human pathogens have traditionally been considered to reproduce by asexual means, thought to result in largely clonal populations. However, in this review we focus on advances involving a combination of experimental, genomic, and population genetic approaches that have reshaped our view of how fungal pathogens reproduce, with consequences for our understanding of how they might be evolving. We consider diverse fungi, focusing on basidiomycete Cryptococcus species, hemiascomycete Candida species, and euascomycete Aspergillus species, causal agents of the diseases cryptococcosis, candidiasis, and aspergillosis, respectively (Brown et al. 2012).
SEXUAL REPRODUCTION OF Cryptococcus neoformans
The sexual cycles of Cryptococcus neoformans and Cryptococcus gattii were described to occur under laboratory conditions nearly four decades ago by June Kwon-Chung (1975, 1976a,b). Her studies revealed a bipolar mating system involving cells of two mating types, α and a. However, the observations that the vast majority of the clinical and environmental isolate population are of mating type α (>95% for serotype D, >99.9% for serotype A, >95% for C. gattii) posed a conundrum (Kwon-Chung and Bennett 1978; Lengeler et al. 2000; Yan et al. 2002; Cogliati 2013). This was further compounded by the facts that (1) many isolates were sterile under lab conditions, (2) some lineages (serotype A VNII) are exclusively of one mating type, and (3) in some lineages isolates of a mating type are present but geographically restricted (serotype A VNB) (Litvintseva et al. 2003, 2006, 2011; Nielsen et al. 2003; Bui et al. 2008; Litvintseva and Mitchell 2012). Taken together, this led to a prevailing view that the a-α sexual cycle might be uncommon, geographically restricted, or absent in nature. Over the past decade, robust genomic, genetic, and population genetic studies have provided considerable insight into how, when, and where Cryptococcus undergoes sexual reproduction, and have revealed extant heterothallic (a-α opposite sex) and homothallic (α-α unisexual) reproduction cycles that contribute to genetic recombination and both the preservation and the generation of genetic diversity (Hull and Heitman 2002; Heitman 2010; Heitman et al. 2013).
a-α Opposite Sex Bisexual Reproduction Results in a Dimorphic Transition to Hyphal Development
The Cryptococcus a-α bipolar sexual cycle involves fusion of yeast cells of opposite mating type, formation of a dikaryon that undergoes a dimorphic transition to hyphae, and culminates in the formation of basidia in which nuclear fusion and meiosis occur (Hull and Heitman 2002; McClelland et al. 2004). The four postmeiotic nuclei undergo repeated rounds of mitosis and budding from the basidium surface to produce four long chains of spores. The initial steps in mating involve pheromone production, secretion, and sensing by the two opposite mating partners (Davidson et al. 2000). This interaction has features of anisogamy in some lineages, in which the a cells form enlarged, round cells, whereas the α cells produce long conjugation tubes that fuse with the enlarged a partner (Wang et al. 2000; Nielsen et al. 2003). Under some conditions, and in some lineages, conjugation tubes and enlarged cells are less morphologically apparent, or both partners appear to produce conjugation tubes. Cell–cell fusion results in migration of the α nucleus into the a partner, and the paired nuclei migrate together and coordinately divide in the hyphae that are produced (McClelland et al. 2004). Concomitantly, mitochondria from the α parent are thought to be destroyed, leading to uniparental mitochondrial inheritance from the a parent (Xu et al. 2000). The dikaryotic hyphae produced during sexual reproduction have hallmark features of other basidiomycetes: the generation and fusion of clamp cells to produce fused clamp connections (a process that ensures faithful segregation of both parental nuclei), formation of blastospores (yeast cells produced by budding from the dikaryotic hyphae), under some conditions, formation of haustoria branches, and finally, formation of the fruiting structures, the basidia. In contrast to the yeast S. cerevisiae, nuclear fusion does not occur immediately following cell–cell fusion, and is instead delayed until basidia form, wherein nuclear fusion and meiosis occur in response to unknown signals (Lee and Heitman 2012).
Monokaryotic Fruiting Resembles Hyphal Sexual Development
A process that morphologically resembles a-α sexual reproduction, (formation of hyphae, basidia, spores) but involves the solo culture of only one individual isolate, termed monokaryotic fruiting, was described by a series of investigators, including Jean Shadomy, Keith Erke, June Kwon-Chung, and Brian Wickes and Jeff Edman (Shadomy and Utz 1966; Lurie et al. 1971; Shadomy and Lurie 1971; Erke and Schneidau 1973; Erke 1976; Kwon-Chung 1978; Wickes et al. 1996). Shadomy and Erke independently first observed clinical isolates that in solo culture produced largely monokaryotic hyphae with unfused clamps. However, terminal or subterminal cells were observed to be dikaryotic. This hyphal developmental process was more completely described by Wickes et al. as occurring when haploid α cells were cultured on a very low nitrogen synthetic medium (filamentation agar), leading to a dimorphic transition from yeast to monokaryotic hyphae with unfused clamp connections, basidia, and rare spores (Wickes et al. 1996). Because the process occurred when isolates were cultured all on their own, without a partner of opposite mating type, it was hypothesized that this pathway involved an asexual transition to hyphal growth and sporulation that did not involve ploidy changes, meiosis, or recombination (Wickes et al. 1996; Wickes 2002).
Although the initial reports focused on α strains that undergo monokaryotic fruiting, it was later appreciated that several isolates of a mating type are also competent to undergo this developmental pathway (Hull and Heitman 2002; Tscharke et al. 2003; Lin et al. 2006). Subsequently, detailed genetic studies revealed that this hyphal developmental pathway is a quantitative trait, controlled by as many as ∼20 genes in the genome, the most prominent of which was found to be the MAT locus for which the α allele made a greater contribution to promote hyphal growth when compared to the a allele (Lin et al. 2006). In addition to nitrogen limitation, higher temperatures may also promote the formation of hyphae associated with monokaryotic fruiting under some conditions, or with some isolates (Fu et al. 2013).
Unisexual Reproduction Is a Novel Form of Self-Fertility
A paradigmatic advance was the discovery that monokaryotic fruiting represents a novel form of selfing sexual reproduction, which has been termed unisexual reproduction or same sex mating (Lin et al. 2005). Appropriate conditions lead to a transition from yeast to hyphal growth of solo incubated lab strains, and this involves monokaryotic hyphae, unfused clamp connections, basidia, and spores. Multiple lines of evidence prove conclusively that this is a true sexual cycle that includes meiosis. First, ploidy changes commonly occur during the process, and diploid blastospores can be recovered from hyphae produced by haploid isolates (Lin et al. 2005; Feretzaki and Heitman 2013a). Second, diploid isolates are enhanced for unisexual reproduction (Lin et al. 2005), and several hyperfilamentous isolates recovered from a genetic screen for altered unisexual reproduction were diploid (Feretzaki and Heitman 2013a). Third, key meiotic genes (DMC1, SPO11) are required for sporulation and spore viability, and this includes both Spo11, which inflicts the DNA DSB that provoke meiotic recombination, and Dmc1, which is the protein that repairs these double strand breaks (Lin et al. 2005; Feretzaki and Heitman 2013a). Finally, recombination occurs at a high frequency during unisexual reproduction of a genetically marked diploid strain, on par with the frequency of meiotic recombination during a-α sexual reproduction (Lin et al. 2005). The karyogamy gene KAR7 is required for both bisexual and unisexual reproduction, providing compelling evidence that nuclear fusion occurs during both developmental cascades (Lee and Heitman 2012). Laboratory conditions (darkness, room temperature, low CO2, desiccation, low nitrogen, or presence of inositol) that stimulate bisexual reproduction of Cryptococcus similarly support unisexual reproduction of well-validated haploid serotype D strains (XL280, JEC21) (Bahn et al. 2005; Idnurm and Heitman 2005; Xue et al. 2007).
Importantly, population genetics studies also provide evidence that unisexual reproduction also occurs in nature for both C. neoformans and C. gattii. First, the discovery and characterization of both αADα and αAAα isolates from the environment and clinical or veterinary isolates provides support that unisexual reproduction occurs in nature from fusion of two α cells or endoreplication to generate hybrid diploid or homozygous diploid intermediates (Lin et al. 2007, 2009; Bui et al. 2008). Second, a series of studies provides evidence that recombination is observed in populations that are exclusively of α mating type (Bui et al. 2008; Hiremath et al. 2008; Saul et al. 2008; Chowdhary et al. 2011). Taken together, these laboratory and population genetics studies provide evidence that unisexual reproduction may be a predominant mode of sexual reproduction for both C. neoformans and C. gattii, and may be a route by which infectious propagules are produced.
Unisexual reproduction can involve cell–cell fusion between genetically divergent isolates of the same mating type, leading to recombination and admixture of genetic information from both parents. Alternatively, unisexual reproduction can also involve two seemingly genetically identical genomes that are brought together into the diploid state either via cell–cell fusion (possibly even between mother and daughter cell) or via endoreplication (involving karyogamy or independent of karyogamy) (Lin et al. 2009; Feretzaki and Heitman 2013a,b). In the most extreme examples of selfing, a central question is, why undergo sexual reproduction if there is no genetic diversity to reshuffle? The null hypothesis is that there is no evolutionary benefit to this form of selfing, and any cost might be minimal or simply tolerated. On the other hand, the frequent and independent origins of homothallism throughout the fungal kingdom suggest there may be benefits conferred by this novel mode of sexual reproduction. Unisexual reproduction involves a dimorphic transition from yeast to hyphal growth, and recent studies reveal that unisexual strains have an enhanced capacity to explore their environment to forage for nutrients compared to isolates that are asexual (Phadke et al. 2013). Other recent studies reveal that unisexual reproduction can generate phenotypic and genotypic diversity de novo, and that much of this diversity is attributable to aneuploidy (Ni et al. 2013). Although a prevailing view is that aneuploidy is uniformly deleterious, in fungi, a large number of studies have revealed that aneuploidy promotes adaptive evolutionary trajectories and underlies morphogenic switches (Yona et al. 2012; Tan et al. 2013). Similar pathways may operate in animals as well, such as the frequent changes in ploidy and aneuploidy observed in hepatocytes (Duncan et al. 2010). In the specific case of aneuploid progeny produced by unisexual reproduction, these isolates can exhibit azole resistance and virulence equivalent to the euploid parental strain even in the harsh milieu of experimentally infected mice (Ni et al. 2013). It is likely that other adaptive benefits may be conferred by unisexual reproduction that will be brought to light by future studies (Roach et al. 2014).
Insights on Sexual Reproduction from the Vantage Point of Genetic Hybrids
The discovery and analysis of hybrid, diploid Cryptococcus isolates reveal that both opposite sex and unisexual reproduction occur in nature, and provide insights into which isolates are undergoing hybridization, possible benefits of hybridization, and species boundaries. These isolates include those between the different serotypes (AD, AB, BD) and within (AA, DD, BB), and provide several key insights. First, the finding of aADα and αADa hybrids with two opposite mating types provides indirect evidence of opposite sex mating that must occur in nature (Lengeler et al. 2001; Xu et al. 2002; Li et al. 2012). Second, the discovery of diploid aADα hybrids found globally that descend from the haploid serotype A VNB lineage that is geographically restricted to South Africa and Botswana (Litvintseva et al. 2003) reveals that hybridization can broaden the environmental niche for otherwise provincial species and subtypes (Litvintseva et al. 2007). This may result from hybrid fitness, as the hybrids are more resistant to UV and more fit at higher growth temperatures. The ability of these hybrids to survive in distinct environmental niches, such as pigeon guano, compared to the restricted niche of the VNB parental isolates (Mopane trees) may also contribute. Third, the discovery of hybrid serotype A isolates that are αAAα provides evidence that these may represent both an intermediate and a product of sexual reproduction (in addition to haploid recombinant progeny), and also provide evidence of unisexual reproduction in nature (Bui et al. 2008; Lin et al. 2009). This is echoed by the discovery and characterization of αADα hybrids (Lin et al. 2007).
Although C. neoformans had been thought to be a predominantly haploid organism, diploid isolates represent ∼10% of the clinical and environmental isolates examined (Lin et al. 2009). A fundamental question is by which mechanisms are these diploids formed? There are two general routes: (i) endoreplication, and (ii) pathways involving karyogamy with or without cell–cell fusion. Endoduplication yields homozygous diploids, and identified αAAα, aAAa, αDDα, aDDa homozygous isolates may have been generated by this process (Cogliati et al. 2001; Lin et al. 2007; Li et al. 2012). Other heterozygous diploids (αAAa, αDDa, αADa, aADα, and αADα) are clearly hybrid isolates that must have been generated by cell–cell and nuclear fusion (Cogliati et al. 2001; Litvintseva et al. 2007; Lin et al. 2009). Diploidization can occur either early or late during both opposite-sex and unisexual reproduction. During opposite-sex reproduction, nuclear fusion typically occurs late, in the basidium, just before meiosis. However, nuclear fusion can also occur early during opposite-sex mating, giving rise to diploid isolates that form a hyphal monokaryon with unfused clamp connections, which go on to produce basidia and undergo meiosis. This is a well-established lab method to isolate and study diploids in the lab (Hull and Heitman 2002; Hull et al. 2002; Idnurm 2010). a/α diploid strains are thermally dimorphic; the diploid cell grows as yeast at 37°C, but, at 24°C, the diploid cell produces monokaryotic hyphae and completes sexual reproduction (Sia et al. 2000). Nuclear fusion also occurs either early or late during unisexual reproduction, giving rise to either haploid or diploid monokaryotic hyphae. In the case of early nuclear fusion this enhances hyphal growth; in the case of late nuclear fusion, this likely occurs in the basidium just before meiosis, similar to opposite-sex sexual reproduction (Feretzaki and Heitman 2013a).
Evidence from Population Genetics Studies on Sexual Reproduction
Population genetic analysis can be used to detect signatures of sexual recombination as the random association of alleles among individuals within a population. Polymorphic alleles within a genome can be generated via amplification (e.g., DNA fingerprinting and amplified fragment length polymorphisms [AFLP]) or sequencing (e.g., multi-locus sequence polymorphisms [MLST] and whole genome sequencing [WGS]), and there are various approaches to detect statistically significant evidence of clonality or recombination (for examples, see Burt et al. 1996; Carter et al. 2007; Simwami et al. 2011). As fungi can reproduce both sexually and asexually, with either mode favored under different conditions, most fungal species comprise a mosaic of clonal lineages and sexually recombining clusters, with any given population potentially dominated by one or the other type. This can complicate population studies but can also provide important insights into the expansion and spread of fungi, in which migration, propagation, and recombination work together to drive evolution and adaptation. Understanding these can help us to unravel how Cryptococcus has evolved to become the important global pathogen that it is today, and may allow us to predict the likelihood of future changes in its virulence, dispersal, and range.
The evolution of a species is driven at the population level, but it can be difficult to define exactly what constitutes a population and at what stage divergent populations are considered different species. Cryptococcus neoformans was formerly described as one species, first with two varieties, var. neoformans and var. gattii (Kwon-Chung et al. 1982), which were subsequently joined by a third, var. grubii (Franzot et al. 1999). The recognition of considerable differences in genetic makeup, epidemiology, ecology, host range, and disease manifestation led to the elevation of C. neoformans var. gattii to full species status as Cryptococcus gattii (Kwon-Chung et al. 2002). Serological and molecular analyses have since determined that there are distinctly different serotypes and genotypes within both C. neoformans and C. gattii (Chen et al. 1996; Meyer et al. 2003, 2009; Bovers et al. 2008), and it has been argued that some of these may also represent different species (Ngamskulrungroj et al. 2009). Diploid and aneuploid interspecies and intervarietal hybrids have also been reported (Boekhout et al. 2001; Cogliati et al. 2001; Lengeler et al. 2001; Lin et al. 2007), and subgenotypes have been further defined within some Cryptococcus genotypes (Fraser et al. 2005; Chen et al. 2008; Byrnes et al. 2009, 2011; Day et al. 2011). It is clear that the population structure of the pathogenic Cryptococcus species is complex and is subject to ongoing, dynamic changes. Although only two species are currently recognized, as many as six to nine distinct taxa likely populate the pathogenic species complex (VNI, VNII, VNB, VNIV, VGI, VGII, VGIIIa, VGIIIb, and VGIV).
At the global level, C. neoformans var. grubii (serotype A; molecular genotypes VNI, VNII, and VNB) causes the vast majority of infections. Within this variety, molecular genotype VNI is the most prevalent and is found on every continent except Antarctica, as well as inhabited subcontinents and islands (Cogliati 2013). Weathered pigeon guano is a major environmental reservoir and is thought to be central to its transmission from the environment to humans (Khan et al. 1978; Gallo et al. 1989; Stenderup et al. 1989; Khosravi 1997; Haag-Wackernagel and Moch 2004). As the common pigeon (Columba livia) is also found globally, and C. neoformans var. grubii can grow and mate on media supplemented with pigeon guano (Nielsen et al. 2007), it is generally assumed that widespread dissemination has co-occurred with pigeons following migratory and trade routes (Littman and Borok 1968). Population analyses of C. neoformans var. grubii using MLST found a very low level of genetic diversity, with the majority of global strains represented by very few genotypes, and an extreme bias of mating type α strains, consistent with recent, clonal propagation and widespread dispersal to most areas of the world. However, studies conducted at more local levels have found evidence of recombination among isolates sourced from trees in Africa (Litvintseva and Mitchell 2012) and India (Hiremath et al. 2008) and infected animals in Australia (Bui et al. 2008). The presence of ancestral genotypes in African populations originating from native mopane (Colophospermum mopane) trees suggests that contemporary global strains associated with pigeon guano originated in Africa (Litvintseva et al. 2011; Simwami et al. 2011; Litvintseva and Mitchell 2012), and the jump in niche to guano enabled a massive amplification and expansion of the species' range.
There is evidence that host shift and range expansion appear to likewise shape modern outbreaks of cryptococcosis caused by C. gattii. Phylogenetic analysis indicates molecular genotype VGII is the ancestral C. gattii genotype (Bovers et al. 2008; Ngamskulrungroj et al. 2009), and population studies show that recombination occurs within this genotype on global (Ngamskulrungroj et al. 2009; Carter et al. 2011) and local (Campbell et al. 2005a; Carriconde et al. 2011) scales. However, this genotype also comprises some highly clonal derived lineages, including subgenotypes VGIIa, VGIIb, and VGIIc that are responsible for large, ongoing outbreaks of cryptococcosis in Vancouver Island, Canada, and in the Pacific Northwest of the United States. These temperate regions are unusual for C. gattii, which generally causes infection only in warmer regions, apart from a single isolate obtained from a sputum sample from a patient in Seattle in the early 1970s (Kidd et al. 2004; Byrnes et al. 2009, 2010). The ecological niche for VGII is not known; however, recent evidence using coalescent analysis suggests it may have emerged from the Amazon rainforest (Hagen et al. 2013). Isolates from the outbreaks are exclusively mating type α, and amplification in local niches may be via asexual propagation or α-α mating (Fraser et al. 2005).
The other major C. gattii genotype that is found globally is VGI (Cogliati 2013). The origin of this genotype is enigmatic. Although it occurs at a relatively high frequency in Australia and can be readily sourced from certain Eucalyptus tree species (Ellis and Pfeiffer 1990a,b), the level of genetic diversity across Australia is very limited (Campbell and Carter 2006), which is inconsistent with an ancestral origin in this region. In addition, although both mating types have been found (Halliday et al. 1999), fertility is low (Campbell et al. 2005b), there is no evidence for sexual recombination except on extremely local scales comprising single tree hollows (Campbell et al. 2005a; Saul et al. 2008; Carter et al. 2011), and dispersal even among trees in close proximity appears to be minimal (Halliday and Carter 2003). It is therefore possible that, as with pigeon guano in C. neoformans, Eucalyptus trees are not the realized ecological niche of C. gattii VGI, but are the site of a successful host jump, with subsequent large-scale amplification.
In summary, population studies of Cryptococcus indicate a capacity for novel strains to emerge, colonize new hosts, and rapidly amplify in the environment. If this occurs in association with susceptible hosts, as in the case of C. neoformans and the HIV/AIDS population, or with an increased propensity to cause disease, as has occurred with C. gattii VGIIa in the Pacific Northwest, large outbreaks of cryptococcosis can ensue. This has parallels with other fungal pathogen-host systems, including chytridiomycosis in frogs, white-nose disease in bats, and die-back disease in jarrah forests, in which a combination of high virulence, rapid reproduction, long-lived environmental stages and the capacity to infect multiple hosts can combine to cause massive outbreaks and drive host species to extirpation and extinction (Fisher et al. 2012). Cryptococcus manifests all of these attributes, and with ongoing globalization and climate change, capacity for pathogens to spread and evolve may be further enhanced (Garcia-Solache and Casadevall 2010). The biology and population structure of Cryptococcus predicts that it will not be a question of whether further outbreaks will occur, but when, and how large they will be.
Sexual Reproduction Links to Virulence
Sexual reproduction is linked to virulence of Cryptococcus via the production of infectious spores, in that α isolates can be more virulent than congenic a isolates, and possibly also via the production of a novel cell type (Titan or giant cells) observed in the lungs of infected animals or patients (Okagaki et al. 2010; Zaragoza et al. 2010). Infections by Cryptococcus are caused by inhalation of infectious propagules, which are thought to be spores, desiccated yeast cells, or both, all of which are small enough to fit down into the alveoli of the lung. Spores have been documented to be bona fide infectious propagules in a series of studies, including via inhalation and direct intracerebral routes of infection (Zimmer et al. 1984; Sukroongreung et al. 1998; Giles et al. 2009; Velagapudi et al. 2009). Moreover, when spores encounter alveolar macrophages, they can be directly phagocytosed, in contrast to yeast cells, which require opsonization (Giles et al. 2009; Velagapudi et al. 2009). Mating type has also been associated with virulence in animal model studies, and found to be linked to α mating type in some serotypes and strain backgrounds, but not in others (Kwon-Chung et al. 1992; Nielsen et al. 2003, 2005a,b; Barchiesi et al. 2005; Zhai et al. 2013; Zhu et al. 2013). Finally, the discovery of dramatically enlarged giant or titan cells in the lungs of infected animals, in which the ploidy has increased from haploid to octaploid or even higher, may produce a novel cell type that evades phagocytosis and plays roles in latency, immune evasion, or dissemination (Okagaki et al. 2010; Zaragoza et al. 2010). How these cells of higher ploidy are produced is not yet understood at a mechanistic level, but may involve some type of unisexual reproduction occurring in vivo (Zaragoza and Nielsen 2013).
PARASEXUAL REPRODUCTION OF Candida albicans
Mating by Candida albicans and related species is highly similar, but not identical, to that of its preeminent relative Saccharomyces cerevisiae, which has served as a general model and reference point for elucidating mating systems in other fungi. Some of the differences are attributable to the rewiring of regulatory networks, but others are more biological and appear to be related to the pathogenic life style of members of the Candida clade.
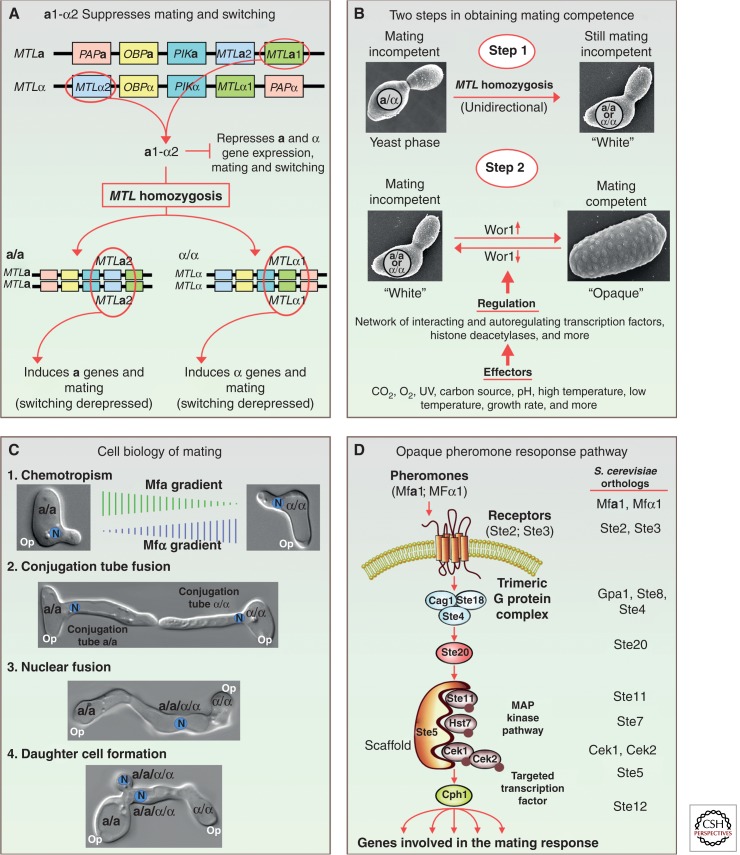
For S. cerevisiae to become mating-competent, diploids (a/α), which predominate in nature, undergo meiosis, and the progeny, which are a or α at the mating type locus (MTL), and thus are released from a1-α2 repression, are immediately mating competent. C. albicans, the species that will be highlighted in the Candida clade, and its closely related species, are also diploid in nature, but must undergo homozygosis at the mating type locus (MTL) to mate, remaining diploid with either an a/a or α/α MTL genotype (Fig. 1A). Other members of the Candida clade are haploid and some clearly must undergo meiosis (Butler et al. 2009; Reedy et al. 2009). But although a/a and α/α strains of C. albicans are relieved of a1-α2 repression, they are still not mating competent. Homozygosis is only the first step in attaining mating competence. Homozygosis occurs in vitro primarily by the loss of one chromosome 5 homolog, which harbors the MTL locus (Hull and Johnson 1999), followed by duplication of the retained homolog (Wu et al. 2005). In nature, mitotic crossing over may be the major mechanism generating MTL homozygous isolates (Wu et al. 2007). In C. albicans, release from a1-α2 repression by loss of MTLa1 or MTLα2, derepresses MTLa2 or MTLα1, respectively, which encode an activator of cell-type-specific mating genes (Fig. 1A). But to achieve mating-competence in C. albicans (Miller and Johnson 2002; Lockhart et al. 2003a) and related species (Pujol et al. 2004; Porman et al. 2011), a/a and α/α cells must then undergo a second complex phenotypic transition, from a “white” traditional yeast-like morphology to a unique opaque morphology (Fig. 1B), a transition discovered more than 25 years ago (Slutsky et al. 1987). This transition, like mating, is also suppressed by the a1-α2 complex (Miller and Johnson 2002). Thus homozygosis releases C. albicans switching from a1-α2 repression. This transition involves changes in virtually every architectural and physiological aspect of the cell and involves the regulation of more than 5% of the genes in the C. albicans genome (Lan et al. 2002; Lohse and Johnson 2010). Switching is regulated by a putative master switch gene, WOR1 (Huang et al. 2006; Srikantha et al. 2006; Zordan et al. 2006), which in turn is regulated by a network of interacting transcription factors (Zordan et al. 2006; Lohse and Johnson 2010; Hernday et al. 2013). Switching is also regulated by histone deacetylases (Fig. 1B) (Klar et al. 2001; Srikantha et al. 2001; Hnisz et al. 2009), alternative regulatory pathways, including one that involves the MAP kinase pathway, the target transcription factor Cph1 (Ramirez-Zavala et al. 2013), and the mediator complex (Zhang et al. 2013). The white-opaque transition occurs spontaneously, but is affected by levels of CO2 and O2 (Ramirez-Zavala et al. 2008; Huang et al. 2009), environmental insults such as UV irradiation (Morrow et al. 1989), low and high temperatures (Slutsky et al. 1987), and the carbon source, in particular N-acetyl glucosamine (Fig. 1B) (Huang et al. 2009). Recently, it has been shown that a/α cells of C. albicans can also undergo switching under select conditions, but are mating incompetent (Xie et al. 2013), and that rare haploid strains of C. albicans must switch to mate (Hickman et al. 2013).
Figure 1.
Mating in Candida albicans. Mating in Candida albians involves both homozygosis from a/α to a/a or α/α, and a switch from the white to opaque phenotype. (A) Homozygosis from a/α to a/a or α/α results in the loss of the a1-α2 corepressor complex, which represses both mating and switching. (B) Attaining mating competence is a two-step process, involving first homozygosis at the mating type locus, and a second phenotypic switch from the white to opaque phenotype. The latter involves a network of inter-regulatory and autoregulatory transcription factors and chromatin modifications, affected by a number of environmental conditions. (C) The cell biology of mating between opaque a/a and α/α cells is similar to that of S. cerevisiae, but involves long conjugation tubes and the formation of an a/a/α/α daughter cell. (D) The pheromone response pathway for mating activation has been conserved in the hemiascomycetes, so that all components, pheromone to the major targeted transcription factor, are orthologs of those components in the S. cerevisiae response pathway.
In the mating process, a/a cells release the a-pheromone Mfa1 (Bennett et al. 2003; Lockhart et al. 2003b; Panwar et al. 2003) and α/α cells produce the α-pheromone Mfα1 (Daniels et al. 2006). As in S. cerevisiae, pheromones play two direct roles in the mating process: first as inducers of the opaque cell-mating response, and, second, as a chemotropic attractant (Fig. 1C). In the mating response, cells with the MTL configuration a/a respond to Mfα1 by up-regulating genes involved in the signal transduction pathway, in the processing and release of pheromones, and in the cellular changes involved in the mating process such as polarization and evagination (shmooing) (Bennett et al. 2003; Daniels et al. 2003; Lockhart et al. 2003b; Panwar et al. 2003; Zhao et al. 2005). Interestingly, pheromones also up-regulate a set of genes involved in hyphae formation (Zhao et al. 2005), which may explain why C. albicans can form such long conjugation tubes with some features shared with hyphae (Zhao et al. 2005; Chapa et al. 2011). These genes are not up-regulated in the response of S. cerevisiae to pheromone, which involves formation of only small mating evaginations, probably because S. cerevisiae lacks many of these hypha-related genes (Zhao et al. 2005). Mating projections of opaque a/a and α/α cells, on the other hand, may have to travel long distances to find each other. The pheromones of C. albicans also act as chemotropic agents over long distances, as demonstrated by orientation studies of seeded opaque cells in white cell biofilms (Fig. 1C) (Daniels et al. 2006). In environments in which gradients can form and be protected from diffusion and mechanical perturbations, a/a and α/α conjugation tubes move up opposing pheromone gradients, fusing at their tips (Fig. 1C). The nuclei move into the tubes and fuse near the position of tip fusion (Fig. 1C) (Daniels et al. 2003; Bennett and Johnson 2005). The first daughter cell forms next to the junction on the a/a side (Daniels et al. 2003). The tetraploid nucleus divides and one tetraploid a/a/α/α daughter nucleus enters the daughter cell (Fig. 1C). The pheromone-induced mating response pathway includes the alternative pheromones (Mfα1, Mfa1), their respective receptors (Ste2, Ste3), the trimeric G-protein complex (composed of Cag1, Ste18, and Ste4), the MAP kinase cascade (composed of Cst20, Ste11, Hst7, and Cek1/Cek2), the scaffold for the cascade (Cst5), and the target transcription factor Cph1 (Fig. 1D) (Chen et al. 2002; Magee et al. 2002; Yi et al. 2008, 2011a; Cote et al. 2011). These are all orthologs of the components in the S. cerevisiae pathway (Fig. 1D), suggesting conservation of this pheromone response pathway for mating in the hemiascomycetes.
Although there is the suggestion that C. albicans has many of the genes necessary for a full sexual cycle, including meiosis (Tzung et al. 2001), the latter has not been observed. Instead, tetraploids undergo stochastic chromosome loss to return to the diploid or near diploid state via a parasexual process (Bennett et al. 2003; Forche et al. 2008; Berman and Hadany 2012). Similarly, it is assumed that the very rare occurrence of haploids (Hickman et al. 2013) is a result of a similar mechanism. Genes for mating systems have also been demonstrated in a variety of other species, both haploid and diploid, in the Candida clade, with small variations (Butler et al. 2009; Reedy et al. 2009; Sai et al. 2011). For instance, Candida lusitaniae lacks α2 in the MTLα locus, Candida guilliermondii lacks α2 in the MTLα locus and a1 in the MTLa locus, and C. parapsilosis lacks a1 in the MTLa locus, yet they contain all of the nonsex genes and the rest of the a and α genes found in the C. albicans MAT locus (Butler et al. 2009; Reedy et al. 2009; Sai et al. 2011). Even with such deletions, some of these Candida species, such as C. lusitaniae, undergo mating and a complete sexual cycle, including meiosis (Francois et al. 2001; Reedy et al. 2009).
But what may prove to be the most unique aspect of mating, in at least the subgroup of the Candida clade that includes C. albicans, is the role of the white-opaque transition. The pheromone released by MTL-homozygous opaque cells also induces mating-incompetent white cells of opposite mating type to form a biofilm architecturally similar to that formed by a/α cells (Yi et al. 2008, 2011a; Daniels et al. 2013). This MTL-homozygous biofilm facilitates chemotropism (Daniels et al. 2006) and mating (Park et al. 2013) between seeded minority opaque a/a and α/α cells (Daniels et al. 2006). It has been hypothesized that the white-opaque transition may have evolved to facilitate mating in a host (Soll 2013). Although the white biofilm is architecturally similar to the a/α biofilm, it does not exhibit the pathogenic traits of impermeability to low and high molecular weight molecules, impenetrability by human polymorphonuclear leukocytes, and drug resistance (Yi et al. 2011b). In stark contrast, MTL-homozygous white cell biofilms are permeable, penetrable, and drug susceptible, traits consistent with a microenvironment that will allow pheromone gradients to form, but retard diffusion, protect against mechanical perturbations, and permit conjugation tube extension during chemotropism. Both the opaque mating response and the white biofilm response are coordinately induced by the same pheromones, a necessity if the latter is to facilitate the former (Soll 2013). Furthermore, if there is a paucity of opaque cells, white cells will self-induce the formation of white cell biofilms by a paracrine system in which white a/a cells release α-pheromone and opaque α/α cells release a-pheromone for self-induction (Yi et al. 2011b).
SEXUAL REPRODUCTION OF Aspergillus fumigatus
The majority of Aspergillus species are only known to reproduce by asexual means. The opportunistic pathogen Aspergillus fumigatus was typical of these in that for most of its documented history it was presumed to be a purely asexual organism (Samson et al. 2009). However, accumulating evidence from population genetic, genomic, and experimental work strongly indicated that A. fumigatus might have a cryptic (i.e., covert, hidden, and not yet identified [Heitman 2010]) sexual cycle, with the species exhibiting many signs of “clandestine” sexual activity (Gow 2005). Population genetic analyses had revealed evidence of genetic divergence and possible recombination within worldwide populations (Varga and Toth 2003; Paoletti et al. 2005; Pringle et al. 2005; Bain et al. 2007; O’Gorman et al. 2009). Genomic analysis had shown the presence of >200 known “sex-related” genes in the genomes of representative isolates, and importantly, these genes appeared functional, with no evidence of point or frameshift mutations (Galagan et al. 2005; Fedorova et al. 2008; Dyer and O’Gorman 2012). Finally, experimental work had revealed the presence of complementary MAT1-1 and MAT1-2 isolates within worldwide populations with a nearly even distribution of mating types, consistent with a heterothallic (obligate outcrossing) sexual breeding system encountered in many filamentous ascomycete fungi (Paoletti et al. 2005). The latter study also showed that mating-type and pheromone signaling genes associated with sex and mating were expressed under laboratory conditions in both single and mixed mating-type cultures.
Sexual Cycle in Aspergillus fumigatus
A major breakthrough was then reported in 2009 with the discovery that a complete heterothallic sexual cycle could be induced in vitro for A. fumigatus when isolates of opposite MAT1-1 and MAT1-2 mating type were crossed in a barrage formation on oatmeal agar at 30°C in the dark (O’Gorman et al. 2009). The developmental process took 6–12 mo to complete and led to the production of cleistothecia (enclosed sexual fruiting bodies), which contained asci within which were formed sexual spores (ascospores) (Fig. 2). These ascospores exhibited high temperature resistance, for example, being able to survive heat shock at 70°C for 90 min (as is characteristic of many Neosartorya species [Samson et al. 2007]), which has recently been correlated with the presence of a relatively thick ascospore cell wall (Kwon-Chung and Sugui 2013). Ascospores germinated to form colonies that showed evidence of genetic recombination based on analysis of mating type and DNA fingerprint markers (O’Gorman et al. 2009). The newly discovered teleomorph (sexual state) was named Neosartorya fumigata. This discovery was of great significance as it indicated that A. fumigatus should be able to generate increased genetic diversity via the sexual cycle to allow evolution in response to environmental change. In the context of an opportunistic pathogen this might involve evolution of resistance to antifungal agents (see below) and greater virulence. The sexual cycle also offered a valuable genetic tool to study the inheritance and genetic basis of traits of interest.
Figure 2.
Sexual reproduction in Aspergillus species. Micrograph showing larger, ornamented lenticular ascospores (white arrow) of A. lentulus relative to smaller, globose less-heat-resistant conidia (black arrow). Scale bar, 5 µm. (Image courtesy of C. O’Gorman and S. Swilaiman.)
The discovery of a sexual cycle triggered much subsequent work. Szewczyk and Krappmann (2010) reported that conserved MAT1-1, MAT1-2, and nsdD regulators of mating in filamentous ascomycete species were also required for sexual development in A. fumigatus (Szewczyk and Krappmann 2010). Alvarez-Perez et al. (2010) reported a possible link between the MAT1-1 genotype and increased virulence in a collection of Spanish isolates. A major advance in the use of the sexual cycle as a genetic tool was then made by Sugui et al. (2011), who identified a ‘‘supermater’’ pair of isolates that exhibited high sexual fertility relative to other isolates tested and could complete the sexual cycle and produce viable ascospores within 4 wk (although highest ascospore viability was not reached until 20 wk of incubation). The supermater isolates, AFB62 (MAT1-1) isolated from a case of invasive aspergillosis and AFIR928 (MAT1-2) from the environment (O’Gorman et al. 2009), were further characterized and shown to be highly virulent in two different murine models and to have a high recombination frequency. Thus, they provide an excellent tool for genetic studies of A. fumigatus (Sugui et al. 2011).
Identification of Sexual Cycles in Other ‘‘Asexual’’ Pathogenic Aspergillus Species
Although A. fumigatus is the principal causal agent of aspergillosis worldwide, other species of Aspergillus such as A. lentulus, A. flavus, A. terreus, and A. niger are also known to cause infections in immunocompromised hosts (Balajee et al. 2005; Dagenais and Keller 2009). As with A. fumigatus, all of these species were once presumed asexual organisms. However, by genetic analysis of the presence of mating-type genes and careful manipulation of laboratory conditions, following on from the discovery of sex in A. fumigatus, it has since proved possible to induce heterothallic sexual cycles in all of these species except for A. niger (Horn et al. 2009; Dyer and O’Gorman 2011, 2012; Arabatzis and Velegraki 2013; Swilaiman et al. 2013), indicating that these opportunistic pathogens also have the opportunity to evolve by sexual recombination. However, it is interesting to note that all of these species are “fastidious” with respect to their demands for environmental conditions to induce sex relative to many other nonpathogenic Aspergillus species, possibly linked to an increased ability to reproduce prolifically by asexual means (Kwon-Chung and Sugui 2009; Samson et al. 2009).
Significance of Sexual Fertility for Aspergillus Pathogen Population Biology
Although a sexual cycle has now been demonstrated in A. fumigatus and other aspergilli capable of causing opportunistic disease, one outstanding question regards the degree of sexual fertility within natural populations. It has been suggested that many fungal species, particularly well-adapted pathogenic species, might be undergoing a slow decline in sexual fertility as a step toward the evolution of asexuality (Dyer and Paoletti 2005). In the case of A. fumigatus, O’Gorman et al. (2009) initially demonstrated sexual fertility of isolates only from a population from Dublin, Ireland and the fertility status of worldwide isolates remained unknown (O’Gorman et al. 2009). However, since then, sexual fertility (in terms of ability to produce cleistothecia with viable ascospores) has been demonstrated for many clinical isolates from the United States and other parts of Europe (Szewczyk and Krappmann 2010; Sugui et al. 2011; Camps et al. 2012; C O’Gorman and P Dyer, unpubl.) consistent with A. fumigatus representing a globally distributed single species capable of sexual reproduction (Pringle et al. 2005). In contrast, many crosses of the emerging agent of aspergillosis Neosartorya (Aspergillus) udagawae either failed to produce cleistothecia or produced ascospores that did not germinate, suggesting a fertility decline in this species (Sugui et al. 2010). Similarly, the majority of attempted crosses between MAT1-1 and MAT1-2 isolates of A. lentulus failed to produce cleistothecia, despite the selection of highly fertile tester isolates, again consistent with a slow decline in sexual fertility in this species (Swilaiman et al. 2013).
Significance of Sex in Aspergillus fumigatus for the Evolution and Spread of Resistance to Antifungal Drugs
There has been a worrying increase in the incidence of resistance to azole and other antifungal drugs in A. fumigatus over the past 20 years. This has followed a greatly increased deployment of antifungal drugs as a result of the increase in cases of aspergillosis in humans, itself linked to an increase in the number of susceptible, immunocompromised patients (Snelders et al. 2008, 2011). It is possible that the sexual cycle of A. fumigatus might be an important factor contributing to the evolution and spread of such antifungal-resistant strains of the fungus because of genetic recombination and dispersal of wind-borne ascospores, which might have a greater ability to survive harsh environmental conditions than asexual conidia. Previous work with the fungal plant pathogen Tapesia yallundae demonstrated that sexual recombination could give rise to ascospore progeny showing a greater than threefold increased resistance in azole fungicide resistance than either parent of a cross (Dyer et al. 2000), while spread of disease by wind-borne ascospores has also been reported for various plant pathogenic fungi (Camps et al. 2012), and the increased heat resistance of ascospores might allow survival in ecological niches in which high temperatures might be encountered, such as composting vegetation (Swilaiman et al. 2013).
The molecular epidemiology of the most frequent form of resistance to azole antifungals by A. fumigatus in Europe, involving TR34/L98H mutations in the region of the target CYP51A gene, was studied by Camps et al. (2012). Evidence was found that sexual reproduction might have contributed to early genetic diversification of a pool of resistant isolates, and that sexual crossing was possible between different CSP (putative cell-surface-protein)-type populations of the fungus, meaning that the resistance genotype might spread further in the future. Intriguingly, it was also found that sexual recombination led to the production of a novel microsatellite marker, presumably caused by meiotic slippage, which might confound the use of such markers to monitor the spread of isolates (Camps et al. 2012). Sexual reproduction and gene flow has also been shown to be possible, albeit infrequent, between closely related fungal species, for example, in the plant pathogenic Ophiostoma species (Paoletti et al. 2006). This is of concern because there is the prospect that antifungal resistance might spread between closely related Aspergillus species, and there have been claims of abortive crossing between A. fumigatus and other Neosartorya species (Takada et al. 1986). However, attempted crosses between highly fertile isolates of A. fumigatus and A. lentulus failed to produce cleistothecia, demonstrating reproductive isolation between these sibling species. This was of medical significance because most isolates of A. lentulus exhibit increased natural resistance to several azole and echinocandin antifungal agents relative to A. fumigatus, so fortunately there would appear to be little risk of gene flow between the species (Swilaiman et al. 2013).
CONCLUSION
Based on the data presented for Cryptococcus, Candida and Aspergillus species it is clear that rather than being purely asexual and mitotic in nature, reproduction in fungal human pathogens instead appears to be much more varied. There have been the discoveries of typical fungal sexual cycles that had previously been overlooked in Aspergillus species, or more unusual systems to promote nuclear fusion and gene flow as seen in the unisexual mating of Cryptococcus and diploid mating in Candida species. The fact that these diverse fungal taxa have maintained methods to promote genetic variation, which might also confer other selective benefits, illustrates the general principles that we should not underestimate the potential for the continued evolution and emergence of fungal pathogens, a finding of relevance for other parasitic unicellular and multicellular eukaryotic organisms.
Footnotes
Editors: Arturo Casadevall, Aaron P. Mitchell, Judith Berman, Kyung J. Kwon-Chung, John R. Perfect, and Joseph Heitman
Additional Perspectives on Human Fungal Pathogens available at www.perspectivesinmedicine.org
REFERENCES
- Alvarez-Perez S, Jose LB, Alba P, Garcia ME 2010. Mating type and invasiveness are significantly associated in Aspergillus fumigatus. Med Mycol 48: 273–277 [DOI] [PubMed] [Google Scholar]
- Arabatzis M, Velegraki A 2013. Sexual reproduction in the opportunistic human pathogen Aspergillus terreus. Mycologia 105: 71–79 [DOI] [PubMed] [Google Scholar]
- Bahn YS, Cox GM, Perfect JR, Heitman J 2005. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr Biol 15: 2013–2020 [DOI] [PubMed] [Google Scholar]
- Bain JM, Tavanti A, Davidson AD, Jacobsen MD, Shaw D, Gow NA, Odds FC 2007. Multilocus sequence typing of the pathogenic fungus Aspergillus fumigatus. J Clin Microbiol 45: 1469–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee SA, Gribskov JL, Hanley E, Nickle D, Marr KA 2005. Aspergillus lentulus sp. Nov., a new sibling species of A fumigatus. Eukaryot Cell 4: 625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchiesi F, Cogliati M, Esposto MC, Spreghini E, Schimizzi AM, Wickes BL, Scalise G, Viviani MA 2005. Comparative analysis of pathogenicity of Cryptococcus neoformans serotypes A, D and AD in murine cryptococcosis. J Infect 51: 10–16 [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Johnson AD 2005. Mating in Candida albicans and the search for a sexual cycle. Annu Rev Microbiol 59: 233–255 [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Uhl MA, Miller MG, Johnson AD 2003. Identification and characterization of a Candida albicans mating pheromone. Mol Cell Biol 23: 8189–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J, Hadany L 2012. Does stress induce (para)sex? Implications for Candida albicans evolution. Trends Genet 28: 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhout T, Theelen B, Diaz M, Fell JW, Hop WC, Abeln EC, Dromer F, Meyer W 2001. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology 147: 891–907 [DOI] [PubMed] [Google Scholar]
- Bovers M, Hagen F, Kuramae EE, Boekhout T 2008. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet Biol 45: 400–421 [DOI] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC 2012. Hidden killers: Human fungal infections. Sci Transl Med 4: 165rv113. [DOI] [PubMed] [Google Scholar]
- Bui T, Lin X, Malik R, Heitman J, Carter D 2008. Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange in unisexual, α mating type populations. Eukaryot Cell 7: 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A, Carter DA, Koenig GL, White TJ, Taylor JW 1996. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc Natl Acad Sci 93: 770–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459: 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ III, Bildfell RJ, Frank SA, Mitchell TG, Marr KA, Heitman J 2009. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. J Infect Dis 199: 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ III, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, et al. 2010. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog 6: e1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ III, Li W, Ren P, Lewit Y, Voelz K, Fraser JA, Dietrich FS, May RC, Chaturvedi S, Chaturvedi V, et al. 2011. A diverse population of Cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PLoS Pathog 7: e1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LT, Carter DA 2006. Looking for sex in the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res 6: 588–598 [DOI] [PubMed] [Google Scholar]
- Campbell LT, Currie BJ, Krockenberger M, Malik R, Meyer W, Heitman J, Carter D 2005a. Clonality and recombination in genetically differentiated subgroups of Cryptococcus gattii. Eukaryot Cell 4: 1403–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LT, Fraser JA, Nichols CB, Dietrich FS, Carter D, Heitman J 2005b. Clinical and environmental isolates of Cryptococcus gattii from Australia that retain sexual fecundity. Eukaryot Cell 4: 1410–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps SM, Rijs AJ, Klaassen CH, Meis JF, O’Gorman CM, Dyer PS, Melchers WJ, Verweij PE 2012. Molecular epidemiology of Aspergillus fumigatus isolates harboring the TR34/L98H azole resistance mechanism. J Clin Microbiol 50: 2674–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriconde F, Gilgado F, Arthur I, Ellis D, Malik R, van de Wiele N, Robert V, Currie BJ, Meyer W 2011. Clonality and α-a recombination in the Australian Cryptococcus gattii VGII population—An emerging outbreak in Australia. PLoS ONE 6: e16936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D, Saul N, Campbell L, Bui T, Krockenberger M 2007. Sex in natural populations of Cryptococcus gattii. In Sex in fungi: Molecular determination and evolutionary implications (ed. Heitman J, Kronstad JW, Taylor JW, Casselton LA), pp. 477–488 ASM, Washington, DC [Google Scholar]
- Carter D, Campbell L, Saul N, Krockenberger M 2011. Sexual reproduction of Cryptococcus gattii: A population genetics perspective. In Cryptococcus: From human pathogen to model yeast (ed. Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Cassadevall A), pp. 299–311 ASM, Washington, DC [Google Scholar]
- Chapa YLB, Lee S, Regan H, Sudbery P 2011. The mating projections of Saccharomyces cerevisiae and Candida albicans show key characteristics of hyphal growth. Fungal Biol 115: 547–556 [DOI] [PubMed] [Google Scholar]
- Chen J, Chen J, Lane S, Liu H 2002. A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol Microbiol 46: 1335–1344 [DOI] [PubMed] [Google Scholar]
- Chen J, Varma A, Diaz MR, Litvintseva AP, Wollenberg KK, Kwon-Chung KJ 2008. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg Infect Dis 14: 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SC, Brownlee AG, Sorrell TC, Ruma P, Nimmo G 1996. Identification by random amplification of polymorphic DNA of a common molecular type of Cryptococcus neoformans var. neoformans in patients with AIDS or other immunosuppressive conditions. J Infect Dis 173: 754–758 [DOI] [PubMed] [Google Scholar]
- Chowdhary A, Hiremath SS, Sun S, Kowshik T, Randhawa HS, Xu J 2011. Genetic differentiation, recombination and clonal expansion in environmental populations of Cryptococcus gattii in India. Env Microbiol 13: 1875–1888 [DOI] [PubMed] [Google Scholar]
- Cogliati M 2013. Global molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii: An atlas of the molecular types. Scientifica (Cairo) 2013: 675213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati M, Esposto MC, Clarke DL, Wickes BL, Viviani MA 2001. Origin of Cryptococcus neoformans var. neoformans diploid strains. J Clin Microbiol 39: 3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote P, Sulea T, Dignard D, Wu C, Whiteway M 2011. Evolutionary reshaping of fungal mating pathway scaffold proteins. mBio 2: e00230–00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagenais TR, Keller NP 2009. Pathogenesis of Aspergillus fumigatus in invasive Aspergillosis. Clin Microbiol Rev 22: 447–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels KJ, Lockhart SR, Staab JF, Sundstrom P, Soll DR 2003. The adhesin Hwp1 and the first daughter cell localize to the a/a portion of the conjugation bridge during Candida albicans mating. Mol Biol Cell 14: 4920–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels KJ, Srikantha T, Lockhart SR, Pujol C, Soll DR 2006. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J 25: 2240–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels KJ, Park YN, Srikantha T, Pujol C, Soll DR 2013. Impact of environmental conditions on the form and function of Candida albicans biofilms. Eukaryot Cell 12: 1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Moore TD, Odom AR, Heitman J 2000. Characterization of the MFα pheromone of the human fungal pathogen Cryptococcus neoformans. Mol Microbiol 38: 1017–1026 [DOI] [PubMed] [Google Scholar]
- Day JN, Hoang TN, Duong AV, Hong CT, Diep PT, Campbell JI, Sieu TP, Hien TT, Bui T, Boni MF, et al. 2011. Most cases of cryptococcal meningitis in HIV-uninfected patients in Vietnam are due to a distinct amplified fragment length polymorphism-defined cluster of Cryptococcus neoformans var. grubii VN1. J Clin Microbiol 49: 658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, Grompe M 2010. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467: 707–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer PS, O’Gorman CM 2011. A fungal sexual revolution: Aspergillus and Penicillium show the way. Curr Opin Microbiol 14: 649–654 [DOI] [PubMed] [Google Scholar]
- Dyer PS, O’Gorman CM 2012. Sexual development and cryptic sexuality in fungi: Insights from Aspergillus species. FEMS Microbiol Rev 36: 165–192 [DOI] [PubMed] [Google Scholar]
- Dyer PS, Paoletti M 2005. Reproduction in Aspergillus fumigatus: Sexuality in a supposedly asexual species? Med Mycol 43: S7–S14 [DOI] [PubMed] [Google Scholar]
- Dyer PS, Hansen J, Delaney, Lucas JA 2000. Genetic control of resistance to the sterol 14α-demethylase inhibitor fungicide prochloraz in the cereal eyespot pathogen Tapesia yallundae. Appl Environ Microbiol 66: 4599–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis DH, Pfeiffer TJ 1990a. Ecology, life cycle, and infectious propagule of Cryptococcus neoformans. Lancet 336: 923–925 [DOI] [PubMed] [Google Scholar]
- Ellis DH, Pfeiffer TJ 1990b. Natural habitat of Cryptococcus neoformans var. gattii. J Clin Microbiol 28: 1642–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erke KH 1976. Light microscopy of basidia, basidiospores, and nuclei in spores and hyphae of Filobasidiella neoformans (Cryptococcus neoformans). J Bacteriol 128: 445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erke KH, Schneidau JD Jr 1973. Relationship of some Cryptococcus neoformans hypha-forming strains to standard strains and to other species of yeasts as determined by deoxyribonucleic acid base ratios and homologies. Infect Immun 7: 941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova ND, Khaldi N, Joardar VS, Maiti R, Amedeo P, Anderson MJ, Crabtree J, Silva JC, Badger JH, Albarraq A, et al. 2008. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet 4: e1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Heitman J 2013a. Genetic circuits that govern bisexual and unisexual reproduction in Cryptococcus neoformans. PLoS Genet 9: e1003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Heitman J 2013b. Unisexual reproduction drives evolution of eukaryotic microbial pathogens. PLoS Pathog 9: e1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484: 186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ 2008. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol 6: e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois F, Noel T, Pepin R, Brulfert A, Chastin C, Favel A, Villard J 2001. Alternative identification test relying upon sexual reproductive abilities of Candida lusitaniae strains isolated from hospitalized patients. J Clin Microbiol 39: 3906–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzot SP, Salkin IF, Casadevall A 1999. Cryptococcus neoformans var grubii: Separate varietal status for Cryptococcus neoformans serotype A isolates. J Clin Microbiol 37: 838–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, et al. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437: 1360–1364 [DOI] [PubMed] [Google Scholar]
- Fu J, Morris IR, Wickes BL 2013. The production of monokaryotic hyphae by Cryptococcus neoformans can be induced by high temperature arrest of the cell cycle and is independent of same-sex mating. PLoS Pathog 9: e1003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, et al. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Gallo MG, Cabeli P, Vidotto V 1989. Presence of pathogenic yeasts in the feces of the semi-domesticated pigeon (Columba livia, Gmelin 1789, urban type) from the city of Turin. Parassitologia 31: 207–212 [PubMed] [Google Scholar]
- Garcia-Solache MA, Casadevall A 2010. Global warming will bring new fungal diseases for mammals. mBio 1: e00061-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles SS, Dagenais TRT, Botts MR, Keller NP, Hull CM 2009. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect Immun 77: 3491–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NA 2005. Fungal genomics: Forensic evidence of sexual activity. Curr Biol 15: R509–R511 [DOI] [PubMed] [Google Scholar]
- Haag-Wackernagel D, Moch H 2004. Health hazards posed by feral pigeons. J Infect 48: 307–313 [DOI] [PubMed] [Google Scholar]
- Hagen F, Ceresini PC, Polacheck I, Ma H, van Nieuwerburgh F, Gabaldon T, Kagan S, Pursall ER, Hoogveld HL, van Iersel LJ, et al. 2013. Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PLoS ONE 8: e71148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday CL, Carter DA 2003. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var gattii isolates from Australia. J Clin Microbiol 41: 703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday CL, Bui T, Krockenberger M, Malik R, Ellis DH, Carter DA 1999. Presence of α and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia. J Clin Microbiol 37: 2920–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J 2010. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe 8: 86–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Sun S, James TY 2013. Evolution of fungal sexual reproduction. Mycologia 105: 1–27 [DOI] [PubMed] [Google Scholar]
- Hernday AD, Lohse MB, Fordyce PM, Nobile CJ, DeRisi JL, Johnson AD 2013. Structure of the transcriptional network controlling white-opaque switching in Candida albicans. Mol Microbiol 90: 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD, Wang YM, Su CH, Bennett RJ, Wang Y, et al. 2013. The “obligate diploid” Candida albicans forms mating-competent haploids. Nature 494: 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiremath SS, Chowdhary A, Kowshik T, Randhawa HS, Sun S, Xu J 2008. Long-distance dispersal and recombination in environmental populations of Cryptococcus neoformans var. grubii from India. Microbiology 154: 1513–1524 [DOI] [PubMed] [Google Scholar]
- Hnisz D, Schwarzmuller T, Kuchler K 2009. Transcriptional loops meet chromatin: A dual-layer network controls white-opaque switching in Candida albicans. Mol Microbiol 74: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn BW, Ramirez-Prado JH, Carbone I 2009. Sexual reproduction and recombination in the aflatoxin-producing fungus Aspergillus parasiticus. Fungal Genet Biol 46: 169–175 [DOI] [PubMed] [Google Scholar]
- Huang G, Wang H, Chou S, Nie X, Chen J, Liu H 2006. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci 103: 12813–12818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Srikantha T, Sahni N, Yi S, Soll DR 2009. CO2 regulates white-to-opaque switching in Candida albicans. Curr Biol 19: 330–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Heitman J 2002. Genetics of Cryptococcus neoformans. Annu Rev Genet 36: 557–615 [DOI] [PubMed] [Google Scholar]
- Hull CM, Johnson AD 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285: 1271–1275 [DOI] [PubMed] [Google Scholar]
- Hull CM, Davidson RC, Heitman J 2002. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1α. Genes Dev 16: 3046–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A 2010. A tetrad analysis of the basidiomycete fungus Cryptococcus neoformans. Genetics 185: 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Heitman J 2005. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol 3: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan ZU, Pal M, Randhawa HS, Sandhu RS 1978. Carriage of Cryptococcus neoformans in the crops of pigeons. J Med Microbiol 11: 215–218 [DOI] [PubMed] [Google Scholar]
- Khosravi AR 1997. Isolation of Cryptococcus neoformans from pigeon (Columba livia droppings in northern Iran). Mycopathologia 139: 93–95 [DOI] [PubMed] [Google Scholar]
- Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, Macdougall L, Boekhout T, Kwon-Chung KJ, Meyer W 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci 101: 17258–17263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar AJ, Srikantha T, Soll DR 2001. A histone deacetylation inhibitor and mutant promote colony-type switching of the human pathogen Candida albicans. Genetics 158: 919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ 1975. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67: 1197– 1200 [PubMed] [Google Scholar]
- Kwon-Chung KJ 1976a. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68: 821–833 [PubMed] [Google Scholar]
- Kwon-Chung KJ 1976b. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68: 943–946 [PubMed] [Google Scholar]
- Kwon-Chung KJ 1978. Heterothallism vs. self-fertile isolates of Filobasidiella neoformans (Cryptococcus neoformans). In The black and white yeasts, Proceedings of the IV International Conference on Mycoses, pp. 204–213 Pan American Health Organization Scientific Publication No. 356, Washington, DC [Google Scholar]
- Kwon-Chung KJ, Bennett JE 1978. Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am J Epidemiol 108: 337–340 [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Sugui JA 2009. Sexual reproduction in Aspergillus species of medical or economical importance: Why so fastidious? Trends Microbiol 17: 481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Sugui JA 2013. Aspergillus fumigatus—What makes the species a ubiquitous human fungal pathogen? PLoS Pathog 9: e1003743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Bennett JE, Rhodes JC 1982. Taxonomic studies on Filobasidiella species and their anamorphs. Antonie van Leeuwenhoek 48: 25–38 [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Edman JC, Wickes BL 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun 60: 602–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Boekhout T, Fell JW, Diaz M 2002. Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae). Taxon 51: 804–806 [Google Scholar]
- Lan CY, Newport G, Murillo LA, Jones T, Scherer S, Davis RW, Agabian N 2002. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc Natl Acad Sci 99: 14907–14912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Heitman J 2012. Function of Cryptococcus neoformans KAR7 (SEC66) in karyogamy during unisexual and opposite-sex mating. Eukaryot Cell 11: 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Wang P, Cox GM, Perfect JR, Heitman J 2000. Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Proc Natl Acad Sci 97: 14460–14555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Cox GM, Heitman J 2001. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect Immun 69: 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Averette AF, Desnos-Ollivier M, Ni M, Dromer F, Heitman J 2012. Genetic diversity and genomic plasticity of Cryptococcus neoformans AD hybrid strains. G3 (Bethesda) 2: 83–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Hull CM, Heitman J 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434: 1017–1021 [DOI] [PubMed] [Google Scholar]
- Lin X, Huang J, Mitchell T, Heitman J 2006. Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATα allele enhances filamentation. PLoS Genet 2: e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J 2007. αADα hybrids of Cryptococcus neoformans: Evidence of same-sex mating in nature and hybrid fitness. PLoS Genet 3: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Patel S, Litvintseva AP, Floyd A, Mitchell TG, Heitman J 2009. Diploids in the Cryptococcus neoformans serotype A population homozygous for the α mating type originate via unisexual mating. PLoS Pathog 5: e1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman ML, Borok R 1968. Relation of the pigeon to cryptococcosis: Natural carrier state, heat resistance and survival of Cryptococcus neoformans. Mycopathol Mycol Appl 35: 329–345 [DOI] [PubMed] [Google Scholar]
- Litvintseva AP, Mitchell TG 2012. Population genetic analyses reveal the African origin and strain variation of Cryptococcus neoformans var. grubii. PLoS Pathog 8: e1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Marra RE, Nielsen K, Heitman J, Vilgalys R, Mitchell TG 2003. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot Cell 2: 1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG 2006. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 172: 2223–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Lin X, Templeton I, Heitman J, Mitchell T 2007. Many globally isolated AD hybrid strains of Cryptococcus neoformans originated in Africa. PLoS Pathog 3: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Carbone I, Rossouw J, Thakur R, Govender NP, Mitchell TG 2011. Evidence that the human pathogenic fungus Cryptococcus neoformans var. grubii may have evolved in Africa. PLoS ONE 6: e19688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart SR, Daniels KJ, Zhao R, Wessels D, Soll DR 2003a. Cell biology of mating in Candida albicans. Eukaryot Cell 2: 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart SR, Zhao R, Daniels KJ, Soll DR 2003b. α-Pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot Cell 2: 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Johnson AD 2010. Temporal anatomy of an epigenetic switch in cell programming: The white-opaque transition of C. albicans. Mol Microbiol 78: 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie HI, Shadomy HJ, Still WJ 1971. An electron microscopic study of Cryptococcus neoformans (Coward strain). Sabouraudia 9: 15–16 [PubMed] [Google Scholar]
- Magee BB, Legrand M, Alarco AM, Raymond M, Magee PT 2002. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol Microbiol 46: 1345–1351 [DOI] [PubMed] [Google Scholar]
- McClelland CM, Chang YC, Varma A, Kwon-Chung KJ 2004. Uniqueness of the mating system in Cryptococcus neoformans. Trends Microbiol 12: 208–212 [DOI] [PubMed] [Google Scholar]
- Meyer W, Castaneda A, Jackson S, Huynh M, Castaneda E, IberoAmerican Cryptococcal Study G. 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis 9: 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, Esposto MC, Fisher M, Gilgado F, Hagen F, Kaocharoen S, et al. 2009. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol 47: 561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MG, Johnson AD 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110: 293–302 [DOI] [PubMed] [Google Scholar]
- Morrow B, Anderson J, Wilson J, Soll DR 1989. Bidirectional stimulation of the white-opaque transition of Candida albicans by ultraviolet irradiation. J Gen Microbiol 135: 1201–1208 [DOI] [PubMed] [Google Scholar]
- Ngamskulrungroj P, Gilgado F, Faganello J, Litvintseva AP, Leal AL, Tsui KM, Mitchell TG, Vainstein MH, Meyer W 2009. Genetic diversity of the Cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PLoS ONE 4: e5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Feretzaki M, Li W, Floyd-Averette A, Mieczkowski P, Dietrich FS, Heitman J 2013. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLoS Biol 11: e1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect Immun 71: 4831–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Litvintseva AP, Mylonakis E, Malliaris SD, Benjamin DK Jr, Giles SS, Mitchell TG, Casadevall A, Perfect JR, et al. 2005a. Cryptococcus neoformans α strains preferentially disseminate to the central nervous system during coinfection. Infect Immun 73: 4922–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Marra RE, Hagen F, Boekhout T, Mitchell TG, Cox GM, Heitman J 2005b. Interaction between genetic background and the mating-type locus in Cryptococcus neoformans virulence potential. Genetics 171: 975–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, De Obaldia AL, Heitman J 2007. Cryptococcus neoformans mates on pigeon guano: Implications for the realized ecological niche and globalization. Eukaryot Cell 6: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Gorman CM, Fuller H, Dyer PS 2009. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457: 471–474 [DOI] [PubMed] [Google Scholar]
- Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chretien F, Heitman J, Dromer F, Nielsen K 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLOS Pathogens 6: e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar SL, Legrand M, Dignard D, Whiteway M, Magee PT 2003. MFα1, the gene encoding the α mating pheromone of Candida albicans. Eukaryot Cell 2: 1350–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M, Rydholm C, Schwier EU, Anderson MJ, Szakacs G, Lutzoni F, Debeaupuis JP, Latge JP, Denning DW, Dyer PS 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr Biol 15: 1242–1248 [DOI] [PubMed] [Google Scholar]
- Paoletti M, Buck KW, Brasier CM 2006. Selective acquisition of novel mating type and vegetative incompatibility genes via interspecies gene transfer in the globally invading eukaryote Ophiostoma novo-ulmi. Mol Ecol 15: 249–262 [DOI] [PubMed] [Google Scholar]
- Park YN, Daniels KJ, Pujol C, Srikantha T, Soll DR 2013. Candida albicans forms a specialized “sexual” as well as “pathogenic” biofilm. Eukaryot Cell 12: 1120–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadke SS, Feretzaki M, Heitman J 2013. Unisexual reproduction enhances fungal competitiveness by promoting habitat exploration via hyphal growth and sporulation. Eukaryot Cell 12: 1155–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porman AM, Alby K, Hirakawa MP, Bennett RJ 2011. Discovery of a phenotypic switch regulating sexual mating in the opportunistic fungal pathogen Candida tropicalis. Proc Natl Acad Sci 108: 21158–21163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle A, Baker DM, Platt JL, Wares JP, Latge JP, Taylor JW 2005. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59: 1886–1899 [PubMed] [Google Scholar]
- Pujol C, Daniels KJ, Lockhart SR, Srikantha T, Radke JB, Geiger J, Soll DR 2004. The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot Cell 3: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Zavala B, Reuss O, Park YN, Ohlsen K, Morschhauser J 2008. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog 4: e1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Zavala B, Weyler M, Gildor T, Schmauch C, Kornitzer D, Arkowitz R, Morschhauser J 2013. Activation of the Cph1-dependent MAP kinase signaling pathway induces white-opaque switching in Candida albicans. PLoS Pathog 9: e1003696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reedy JL, Floyd AM, Heitman J 2009. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol 19: 891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach KC, Feretzaki M, Sun S, Heitman J 2014. Unisexual reproduction. Adv Genet 85: 255–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai S, Holland LM, McGee CF, Lynch DB, Butler G 2011. Evolution of mating within the Candida parapsilosis species group. Eukaryot Cell 10: 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Hong S, Peterson SW, Frisvad JC, Varga J 2007. Polyphasic taxonomy of Aspergillus section Fumigati and its teleomorph Neosartorya. Stud Mycol 59: 147–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Varga J, Dyer PS 2009. Morphology and reproductive mode of Aspergillus fumigatus. In Aspergillus fumigatus and Aspergillosis (ed. Latge JP, Steinbach WJ), pp. 7–13 ASM, Washington, DC [Google Scholar]
- Saul N, Krockenberger M, Carter D 2008. Evidence of recombination in mixed-mating-type and α-only populations of Cryptococcus gattii sourced from single eucalyptus tree hollows. Eukaryot Cell 7: 727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadomy HJ, Lurie HI 1971. Histopathological observations in experimental cryptococcosis caused by a hypha-producing strain of Cryptococcus neoformans (Coward strain) in mice. Sabouraudia 9: 6–9 [DOI] [PubMed] [Google Scholar]
- Shadomy HJ, Utz JP 1966. Preliminary studies on a hypha-forming mutant of Cryptococcus neoformans. Mycologica 58: 383–390 [PubMed] [Google Scholar]
- Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, Jordar VS, Maiti R, Kodira CD, Neafsey DE, Zeng Q, et al. 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res 19: 1722–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia RA, Lengeler KB, Heitman J 2000. Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet Biol 29: 153–163 [DOI] [PubMed] [Google Scholar]
- Simwami SP, Khayhan K, Henk DA, Aanensen DM, Boekhout T, Hagen F, Brouwer AE, Harrison TS, Donnelly CA, Fisher MC 2011. Low diversity Cryptococcus neoformans variety grubii multilocus sequence types from Thailand are consistent with an ancestral African origin. PLoS Pathog 7: e1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR 1987. “White-opaque transition”: A second high-frequency switching system in Candida albicans. J Bacteriol 169: 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelders E, van der Lee HA, Kuijpers J, Rijs AJ, Varga J, Samson RA, Mellado E, Donders AR, Melchers WJ, Verweij PE 2008. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med 5: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelders E, Melchers WJ, Verweij PE 2011. Azole resistance in Aspergillus fumigatus: A new challenge in the management of invasive aspergillosis? Future Microbiol 6: 335–347 [DOI] [PubMed] [Google Scholar]
- Soll DR 2013. The evolution of alternative biofilms in an opportunistic fungal pathogen: An explanation for how new signal transduction pathways may evolve. Infect Genet Evol: S1567–1348 [DOI] [PubMed] [Google Scholar]
- Srikantha T, Tsai L, Daniels K, Klar AJ, Soll DR 2001. The histone deacetylase genes HDA1 and RPD3 play distinct roles in regulation of high-frequency phenotypic switching in Candida albicans. J Bacteriol 183: 4614–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, Gerstein M, Yi S, Snyder M, Soll DR 2006. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell 5: 1674–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenderup J, Flensted K, Jorgensen C, Sorensen AH, Hansen NC, Siersted HC 1989. Occurrence of the yeast, Cryptococcus (Cr) neoformans, in pigeon droppings. Ugeskr Laeger 151: 2974–2975 [PubMed] [Google Scholar]
- Sugui JA, Vinh DC, Nardone G, Shea YR, Chang YC, Zelazny AM, Marr KA, Holland SM, Kwon-Chung KJ 2010. Neosartorya udagawae (Aspergillus udagawae), an emerging agent of aspergillosis: How different is it from Aspergillus fumigatus? J Clin Microbiol 48: 220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui JA, Losada L, Wang W, Varga J, Ngamskulrungroj P, Abu-Asab M, Chang YC, O’Gorman CM, Wickes BL, Nierman WC, et al. 2011. Identification and characterization of an Aspergillus fumigatus “supermater” pair. mBio 2: e00234–00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukroongreung S, Kitiniyom K, Nilakul C, Tantimavanich S 1998. Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans. Med Mycol 36: 419–424 [PubMed] [Google Scholar]
- Swilaiman SS, O’Gorman CM, Balajee SA, Dyer PS 2013. Discovery of a sexual cycle in Aspergillus lentulus, a close relative of A. fumigatus. Eukaryot Cell 12: 962–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, Krappmann S 2010. Conserved regulators of mating are essential for Aspergillus fumigatus cleistothecium formation. Eukaryot Cell 9: 774–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada M, Udagawa S, Norizuki K 1986. Isolation of Neosartorya fennelliae and interspecific pairings between N. fennelliae, N spathulata, and Aspergillus fumigatus. Trans Mycol Soc Jpn 27: 415–423 [Google Scholar]
- Tan Z, Hays M, Cromie GA, Jeffery EW, Scott AC, Ahyong V, Sirr A, Skupin A, Dudley AM 2013. Aneuploidy underlies a multicellular phenotypic switch. Proc Natl Acad Sci 110: 12367–12372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekaia F, Latge JP 2005. Aspergillus fumigatus: Saprophyte or pathogen? Curr Opin Microbiol 8: 385–392 [DOI] [PubMed] [Google Scholar]
- Tscharke RL, Lazera M, Chang YC, Wickes BL, Kwon-Chung KJ 2003. Haploid fruiting in Cryptococcus neoformans is not mating type α-specific. Fungal Genet Biol 39: 230–237 [DOI] [PubMed] [Google Scholar]
- Tzung KW, Williams RM, Scherer S, Federspiel N, Jones T, Hansen N, Bivolarevic V, Huizar L, Komp C, Surzycki R, et al. 2001. Genomic evidence for a complete sexual cycle in Candida albicans. Proc Natl Acad Sci 98: 3249–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J, Toth B 2003. Genetic variability and reproductive mode of Aspergillus fumigatus. Infect Genet Evol 3: 3–17 [DOI] [PubMed] [Google Scholar]
- Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J 2009. Spores as infectious propagules of Cryptococcus neoformans. Infect Immun 77: 4345–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Perfect JR, Heitman J 2000. The G-protein β subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol Cell Biol 20: 352–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes BL 2002. The role of mating type and morphology in Cryptococcus neoformans pathogenesis. Int J Med Microbiol 292: 313–329 [DOI] [PubMed] [Google Scholar]
- Wickes BL, Mayorga ME, Edman U, Edman JC 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: Association with the α-mating type. Proc Natl Acad Sci 93: 7327–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Pujol C, Lockhart SR, Soll DR 2005. Chromosome loss followed by duplication is the major mechanism of spontaneous mating-type locus homozygosis in Candida albicans. Genetics 169: 1311–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Lockhart SR, Pujol C, Srikantha T, Soll DR 2007. Heterozygosity of genes on the sex chromosome regulates Candida albicans virulence. Mol Microbiol 64: 1587–1604 [DOI] [PubMed] [Google Scholar]
- Xie J, Tao L, Nobile CJ, Tong Y, Guan G, Sun Y, Cao C, Hernday AD, Johnson AD, Zhang L, et al. 2013. White-opaque switching in natural MTLa/α isolates of Candida albicans: Evolutionary implications for roles in host adaptation, pathogenesis, and sex. PLoS Biol 11: e1001525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Ali RY, Gregory DA, Amick D, Lambert SE, Yoell HJ, Vilgalys RJ, Mitchell TG 2000. Uniparental mitochondrial transmission in sexual crosses in Cryptococcus neoformans. Curr Microbiol 40: 269–273 [DOI] [PubMed] [Google Scholar]
- Xu J, Luo G, Vilgalys RJ, Brandt ME, Mitchell TG 2002. Multiple origins of hybrid strains of Cryptococcus neoformans with serotype AD. Microbiology 148: 203–212 [DOI] [PubMed] [Google Scholar]
- Xue C, Tada Y, Dong X, Heitman J 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1: 263–273 [DOI] [PubMed] [Google Scholar]
- Yan Z, Li X, Xu J 2002. Geographic distribution of mating type alleles of Cryptococcus neoformans in four areas of the United States. J Clin Microbiol 40: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Sahni N, Daniels KJ, Pujol C, Srikantha T, Soll DR 2008. The same receptor, G protein, and mitogen-activated protein kinase pathway activate different downstream regulators in the alternative white and opaque pheromone responses of Candida albicans. Mol Biol Cell 19: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Sahni N, Daniels KJ, Lu KL, Huang G, Srikantha T, Soll DR 2011a. Self-induction of a/a or α/α biofilms in Candida albicans is a pheromone-based paracrine system requiring switching. Eukaryot Cell 10: 753–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Sahni N, Daniels KJ, Lu KL, Srikantha T, Huang G, Garnaas AM, Soll DR 2011b. Alternative mating type configurations (a/α versus a/a or α/α) of Candida albicans result in alternative biofilms regulated by different pathways. PLoS Biol 9: e1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona AH, Manor YS, Herbst RH, Romano GH, Mitchell A, Kupiec M, Pilpel Y, Dahan O 2012. Chromosomal duplication is a transient evolutionary solution to stress. Proc Natl Acad Sci 109: 21010–21015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Nielsen K 2013. Titan cells in Cryptococcus neoformans: Cells with a giant impact. Curr Opin Microbiol 16: 409–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Garcia-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodriguez-Tudela JL, Casadevall A 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog 6: e1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B, Zhu P, Foyle D, Upadhyay S, Idnurm A, Lin X 2013. Congenic strains of the filamentous form of Cryptococcus neoformans for studies of fungal morphogenesis and virulence. Infect Immun 81: 2626–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Liu Z, Myers LC 2013. Differential regulation of white-opaque switching by individual subunits of Candida albicans mediator. Eukaryot Cell 12: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Daniels KJ, Lockhart SR, Yeater KM, Hoyer LL, Soll DR 2005. Unique aspects of gene expression during Candida albicans mating and possible G1 dependency. Eukaryot Cell 4: 1175–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Zhai B, Lin X, Idnurm A 2013. Congenic strains for genetic analysis of virulence traits in Cryptococcus gattii. Infect Immun 81: 2616–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer BL, Hempel HO, Goodman NL 1984. Pathogenicity of the basidiospores of Filobasidiella neoformans. Mycopathologia 85: 149–153 [DOI] [PubMed] [Google Scholar]
- Zordan RE, Galgoczy DJ, Johnson AD 2006. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci 103: 12807–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]