Abstract
Several human skin diseases and disorders are associated with two groups of fungi, the dermatophytes and Malassezia. Although these skin-related problems are not generally life threatening, they are among the most common diseases and disorders of mankind. These fungi are phylogenetically divergent, with the dermatophytes within the Ascomycota and Malassezia within Basidiomycota. Genome analysis indicates that the adaptations to the skin environment are different in these two groups of fungi. Malassezia are dependent on host lipids and secrete lipases and phospholipases that likely release host fatty acids. The dermatophytes encode multiple enzymes with potential roles in modulating host interactions: polyketide synthases, nonribosomal peptide synthetases, LysM, proteases, kinases, and pseudokinases. These two fungal groups have maximized their interactions with the host using two very different mechanisms.
Dermatophytes and Malassezia are present on the skin of most people and cause disorders, such as dandruff, eczema, ringworm, and nail infections. Genome sequencing has provided insights into their virulence and survival.
Skin fungi are associated with many of humanity’s most common disorders, including dandruff, atopic dermatitis/eczema, ringworm, athlete’s foot, jock itch, and nail infections (onychomycosis). These diseases are attributed to two sets of fungi, ascomycete dermatophytes, including the genera Trichophyton, Microsporum, and Epidermophyton, and basidiomycete fungi in the genus Malassezia. Research on dermatophytes and Malassezia has lagged behind research on fungi associated with human mortality, although most of the world’s people constantly deal with fungal skin disease, whether acute or chronic.
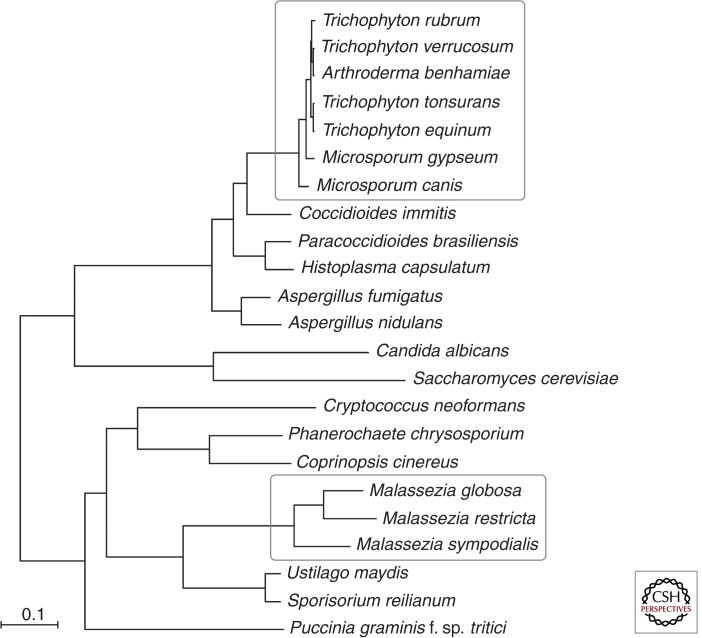
Genome sequencing has led to new hypotheses for the molecular means by which these fungi inhabit human skin and cause disease. Not only are these two sets of skin fungi from distant branches of the fungal phylogenetic tree (Fig. 1), but they also use different sets of proteins to adapt to mammalian skin (Table 1). The dermatophytes appear to use a plethora of proteases to devour keratin, LysM proteins to hide from the host immune system, kinases and pseudokinases to modulate cell metabolism, and a wealth of polyketide synthases and nonribosomal peptide synthases to generate metabolites for unknown purposes. Malassezia are found on the skin of warm-blooded animals and are the most numerous fungus on many human skin sites. Their phylogenetically closest relatives are plant pathogens such as Ustilago maydis (Fig. 1), but Malassezia secrete a set of proteins with activities similar to secreted proteins of another skin-inhabiting fungus, Candida albicans. Most Malassezia species are unable to synthesize fatty acids, a deficiency not known in other free-living fungi. Malassezia compensate by using many secreted hydrolases (lipases and phopholipases C) to provide fatty acids from host lipids.
Figure 1.
Phylogenetic relationship of dermatophytes and Malassezia to other fungi. The phylogenetic relationship of selected fungi was estimated from 616 single-copy orthologs present in all species; protein sequences were aligned using MUSCLE (Edgar 2004), and a phylogeny of the concatenated alignment was inferred using RAxML (version 7.3.3, with model PROTCATWAG and 1000 bootstrap replicates) (Stamatakis 2006). All nodes in the best tree were highly supported by 100% of bootstrap replicates. Dermatophyte and Malassezia species groups are each highlighted by a white box.
Table 1.
Significant observations from the dermatophyte and Malassezia genomes
| Dermatophytes | Malassezia |
|---|---|
| Whole-genome phylogeny consistent with rRNA phylogeny–dermatophytes are closely related to Coccidioides immitis | Phylogeny indicates nearest relatives are plant fungi, such as U. maydis, cause of corn smut |
| Each genome is haploid, containing about 23–24 Mb of DNA | Haploid genomes of 8–9 Mb, among the smallest for free-living fungi |
| Little repetitive DNA | Little repetitive DNA |
| Consistent with their ecological niche, fewer CAZymes to degrade plant material than closely related Ascomycetes | Fewer CAZymes than close relative U. maydis |
| Enriched for genes encoding endo- and exoproteases, consistent with their ecological niche | Enriched for genes encoding lipases, phospholipases C, acid sphingomyelinases, and aspartyl proteases, consistent with ecological niche |
| Enriched for secondary metabolism genes, which may produce molecules that contribute to the interactions with the host immune system | Shortage of secondary metabolite genes, with Malassezia globosa and Malassezia sympodialis each having one polyketide synthase gene and one nonribosomal peptide synthetase gene |
| Two copies of fatty acid synthase genes | No fatty acid synthase gene; among free-living fungi, only Malassezia are known to lack fatty acid synthase gene |
| Enriched for genes encoding kinases, including pseudokinases, which may provide diverse regulation of signaling | Fewer kinase genes; tyrosine protein kinase genes absent |
| Enriched for LysM genes, which may mask fungal cell surfaces and limit exposure to the host immune system | No LysM genes |
Other skin fungi are responsible for emerging diseases that are altering the biosphere. Many amphibian species are threatened with extinction, and a chytrid fungus, growing on frog skin, may be primarily responsible (Cheng et al. 2011). More recently, bat populations have plummeted because of a Geomyces fungal infection of bats, which colonizes the facial and wing skin (Blehert et al. 2009). Although these wildlife-killing fungi will not be covered in this review, understanding of one skin fungal pathogen may lead to hypotheses relevant to others.
We will describe the dermatophytes and Malassezia, along with some of the diseases with which they are associated. We will provide a perspective on the phylogeny, ecology, and genomics of these organisms.
HUMAN MICROBIOME ANALYSIS OF FUNGI FROM SKIN AND OTHER BODY SITES
The human body is home to a plethora of microorganisms, many of which are commensal nonpathogenic residents, including bacteria, archaea, viruses, and fungi. These human-associated microbes are essential in establishing and maintaining human health and disease. Enabled by significant advances in next-generation sequencing technologies, the microbiome of several body sites, including skin, has been characterized (Grice and Segre 2011; Cui et al. 2013; Huffnagle and Noverr 2013). The majority of these studies have taken a bacteriocentric view, only highlighting the diversity of bacterial species (Parfrey et al. 2011).
On human skin, microbial diversity is not limited to bacteria. Fungi, viruses, and mites also contribute to the human skin microbiome. The first large-scale sequencing analysis characterized fungal (“mycobiome”) diversity on the skin of 10 healthy adult volunteers (HVs) (Findley et al. 2013). A total of 14 skin sites representing a range of physiological characteristics—dry, moist, and oily—were sampled via the swab method. Findley et al. used both a culture-dependent and a culture-independent approach to collect microbiota at each body site. Two molecular markers for fungi, the 18S rRNA gene and the internal transcribed spacer 1 (ITS1) region, as well as the 16S rRNA gene for bacteria, were sequenced and used to classify these communities (Khot et al. 2009). The next-generation sequencing technology used in this study resulted in more than five million sequences, whereas approximately 140 isolates were recovered from the culturing method (Findley et al. 2013).
Sequence analysis revealed that Malassezia is the most abundant fungus on many human skin sites, as previously shown for the scalp (Gemmer et al. 2002). These authors cultured three of the human-associated Malassezia species: M. globosa, M. restricta, and M. sympodialis. Sequencing uncovered an additional eight Malassezia species residing on skin, as well as an unclassified Malassezia spp. The ear and the forehead harbored mainly M. restricta, whereas the remaining sites were a mixture of M. globosa and M. sympodialis.
In contrast to extensive bacterial diversity found at all human skin sites tested (Grice et al. 2009), the skin fungal diversity is more site-dependent. Sites on the back and head are the most stable with the lowest diversity, whereas the proximal arm sites display intermediate diversity. The foot sites are not very stable, change over time, and are the most diverse, with at least 40 genera colonizing each foot site (Findley et al. 2013). The longitudinal analysis of six of the 10 HVs revealed that feet microbiota change over time, which is to be expected, because sampling occurred during the summer when feet are frequently exposed to the environment. Taken together, these data suggest that Malassezia species are abundant on human skin, the feet are highly diverse, and the topography of fungi differs from that of bacteria in that the physiological characteristics of skin do not define fungal communities; rather, the body site location is important.
Studies that explore dermatophytes are of interest to dermatologists, because foot fungal infections are a health concern. One study explores foot disease in soccer athletes using a culture-dependent approach (Purim et al. 2005). The major genera observed in this population of athletes were primarily the dermatophytes, Trichophyton rubrum (40%), and Trichophyton mentagrophytes (36.4%), whereas Candida and other fungi made up the remaining 24%. Findley et al. (2013) also observed Trichophyton as the only dermatophyte in the fungal survey study and only on the feet of HVs. Genera such as Microsporum and Epidermophyton were not observed.
Malassezia have historically been described as found on the skin of warm-blooded animals. Since the advent of DNA-based methods to characterize microbial communities, there are several reports of Malassezia or Malassezia-like organisms. The samples include beetle guts (Zhang et al. 2003), nematodes (Renker et al. 2003), sponges (Gao et al. 2008), coral (Amend et al. 2012), and airborne samples (Pitkaranta et al. 2008). A better understanding of Malassezia adaptation to mammalian skin may arise from comparisons with Malassezia-like organisms from other biological sources.
DERMATOPHYTES
Dermatophytes are a group of pathogenic fungi that cause mostly superficial diseases on humans and other mammals (Kwon-Chung and Bennett 1992; White et al. 2008; Achterman and White 2012a,b). The diseases that result from a dermatophyte infection are known as tineas. The location of the disease on the body further defines the disease, so that tinea pedis are dermatophyte infections of the feet, tinea cruris of the genitals, tinea corporis of the torso, and tinea capitis of the head. There are at least 40 species of dermatophytes that infect humans, and many of these fungi can cause disease in more than one body location. Still the most prevalent cause of tinea pedis is T. rubrum, and the most prevalent causes of tinea capitis are Trichophyton tonsurans and Microsporum canis.
Dermatophytes are ascomycetes with septate hyphae, most closely related to Coccidioides immitis within the Onygenales (Kwon-Chung and Bennett 1992; Graser et al. 2008). There are three genera of dermatophytes, Trichophyton, Microsporum, and Epidermophyton. Although the species were historically divided into these genera by morphology and physical attributes, recent analysis by rRNA sequencing indicates that the dermatophytes as a whole are a cohesive group, with no clear distinction between the three genera. The closest relatives to any one Microsporum species might be two Trichophyton species (Graser et al. 2008). Thus, no comparisons should be made between genera without consulting the rRNA-based phylogenetic tree.
DISEASES OF DERMATOPHYTES
The dermatophytes cause significant disease in humans, horses, pets, and other animals. It is often argued that dermatophyte infections are the most common human infection in the world (not just the most common fungal infection), as tinea pedis is a common problem in the developed world, and tinea capitis is a common problem in the developing world. Dermatophyte infections are responsible for at least half a billion dollars in health-care costs (Achterman et al. 2011). It is also important to remember that dermatophytes colonize 30%–70% of humans without causing disease.
There are three types of dermatophytes, unrelated to the three genera (White et al. 2008). Some dermatophyte species have only been found on humans and are assumed to be human-specific. Such species are termed anthropophilic and include T. rubrum and T. tonsurans. There are species that are found on humans and on other animals, and these species are referred to as zoophilic. These zoophilic species include Microsporum canis, Trichophyton equinum, Trichophyton verrucosum, and Arthroderma benhamiae. Geophilic species, including M. gypseum, are found in soil and occasionally infect animals.
Each of the species in these three types of dermatophytes can cause a spectrum of disease. However, in general, geophilic species are accidental infections, and there has been no evolutionary selection to maximize host/pathogen interactions. Therefore, these geophilic species generally cause acute, severe disease (Kwon-Chung and Bennett 1992). Anthropophilic species have usually interacted with the human host for a very long time and, thus, the interactions between these species and the host immune system have evolved together and can be subtle. Therefore, these species usually are associated with long-term, chronic, but limited disease and tissue destruction. Finally, the zoophilic species interact somewhere between the acute geophiles and the chronic anthropophiles.
Another aspect of the three types of dermatophytes is their ability to complete a sexual cycle. In general, the sexual stages of the geophiles have been identified, and the sexual stages of the anthropophiles have not. Specifically, sexual stages are known for M. gypseum, M. canis, and A. benhamiae, but not for T. tonsurans or T. rubrum. In fact, recent work based on the genome sequencing has identified strains containing each of two mating types in M. gypseum (Li et al. 2010), yet no isolate representing a second mating type in T. rubrum has as of yet been identified. This suggests that this most common dermatophyte has predominantly one mating type in clinical infections, implying that mating is not occurring for this species during most infections, or if it is, it may be unisexual. Although most T. rubrum infections from around the world might be the result of a single strain or clone, microsatellite analysis suggests the existence of a second subpopulation of predominantly African origin (Graser et al. 2007). The collection and analysis of additional globally diverse isolates, particularly from Africa, could assist a search for the second mating type in T. rubrum.
Research into the virulence and pathogenicity of dermatophytes has been slow, in part because dermatophyte infections do not have significant mortality. There are occasionally reports of disseminated dermatophyte infections, but these individuals usually have other underlying conditions. A fundamental question in dermatophyte research is how the dermatophytes interact with the host immune system. Major dermatophyte antigens have been defined, including Tri r 2 and Tri r 4 (Gao and Takashima 2004). In active disease, the dermatophytes grow, cause tissue destruction, and cause inflammation, essentially a war of attrition between the fungus and the host. When a dermatophyte infection is under control, it is not clear if the fungus has been eliminated or if it is still present but controlled by the host immune system. Dermatophyte infections are difficult to cure entirely and frequently recur in the same site of initial infection. Hence, the question of reactivation or new infections is important to our understanding of these infections.
Some epidemiology has been conducted to investigate the major dermatophyte infections in a specific area or group of patients. Yet only recently have the precise molecular tools, including genome sequencing (see below), been available to clearly determine if a strain is indeed a new infection or a reactivation. As described above, this question is central to dermatophyte biology. Dermatophytes are treated with a variety of antifungal drugs (Borgers et al. 2005); the most common are the ergosterol biosynthesis inhibitors (EBIs), including azoles and terbinafine (Lamisil). These drugs inhibit steps in the biosynthesis of ergosterol, the fungal equivalent of cholesterol, the major sterol component of the plasma membrane. Azoles target the enzyme Erg11, lanosterol demethylase, whereas terbinafine targets Erg1, squalene epoxidase (Marr et al. 1998). Most of the azoles that are used to treat dermatophytes are imidazoles, available over the counter. Prescription triazole azoles that are commonly used to treat invasive fungal infections are generally not used to treat dermatophyte infections. Terbinafine is available over the counter as a topical medication and by prescription to treat recalcitrant nail infections; however, this drug can show significant liver toxicity, so the prescribing physician and the patient must carefully decide when to use this drug orally (Gupta and Cooper 2008).
Curiously, despite the availability of imidazole azole drugs over the counter, and the subsequent sporadic use of these medicines by those infected, there are only a few case reports of azole drug–resistant infections (Ghannoum et al. 2006; Ghannoum et al. 2008). Azole drug resistance is common in other pathogenic fungi including C. albicans, Cryptococcus neoformans, and Aspergillus fumigatus, under conditions where the drugs are used regularly, sporadically, and at varying strengths (Pfaller 2012). In addition, the azoles are static drugs that inhibit growth but do not kill the fungal cells—excellent conditions for the development of resistance. Therefore, it is curious that the dermatophytes have not developed widespread resistance despite the overuse and misuse of azoles. Understanding how the dermatophytes respond to azole drugs is of wide interest in the use of these drugs to treat all pathogenic fungi.
The three most important questions relating to dermatophytes are thus: (1) is an infection a reactivation or a new infection, (2) why do dermatophytes not develop azole resistance, and (3) how do the fungus and the immune response interact to create a balance that can result in either clinical disease, latent and controlled colonization, or the elimination of the fungus (Table 1)?
GENOME ANALYSIS OF DERMATOPHYTES
The first step in addressing the most intriguing questions relating to the dermatophytes was to obtain and compare the genome sequences of different species. These were completed and published in 2011 and 2012 (Burmester et al. 2011; Martinez et al. 2012). The species that were sequenced include T. rubrum, T. tonsurans, T. equinum, M. canis, M. gypseum, A. benhamiae, and T. verrucosum. These species were selected because they represent (1) all three dermatophyte types (geophilic, zoophilic, and anthropophilic); (2) mating-competent species as well as species with no known sexual phase; and (3) acute and chronic infections. The strains selected to represent each species to be sequenced were representative and isolated relatively recently. The complete genomes confirmed many aspects of the dermatophyte biology: (1) The whole-genome phylogeny is consistent with the rRNA-based phylogeny, (2) the strains were haploid, (3) the genomes were approximately the same size (23–24 Mb), and (4) the genomes had few repeat sequences.
An analysis of these genomes was not sufficient to characterize a set of genes that direct a species to be geophilic, zoophilic, or anthropophilic, as the genes specific to these categories were open reading frames (ORFs) without functional annotation. Neither did the genome analysis identify differences correlating with mating competence. Each strain exhibited its own mating locus, and using that information, the other mating locus could be identified in other strains from mating-competent species, but not from species with no known mating. However, each of the genomes contains all of the genes usually necessary for mating of a species, and thus does not give any indication as to why some species are mating competent and others are not. Analysis of the genomes did not reveal any features that clearly correlated with pathogenicity, including how the species might cause acute or chronic disease. Although duplications of some ergosterol pathway genes were identified, this did not clearly explain why drug resistance is rare in the dermatophytes. Further study of how these pathogens respond to host and drug pressure is needed to address these questions.
The genomes provided insights into several areas of pathogenicity in dermatophytes that should guide further study (Table 1). These observations were the result of comparing the genomes of the seven dermatophyte species with the genomes of their close relatives, C. immitis, and the three other dimorphic fungi, Histoplasma, Blastomyces, and Paracoccidioides, as well as Aspergillus fumigatus and other fungal pathogens. One of the first observations is that the dermatophyte genomes have a reduced number of genes that are involved in degradation of plant carbohydrates. This class of plant-degrading carbohydrate-active enzymes, or CAZymes, is present in most species that grow on plants. The fact that the genes are lost in fungi adapted to growth on human tissue instead of plant tissue is not unexpected; however, directly testing growth on substrates, including those produced by plants, would help correlate phenotype with genotype and identify differences between the species.
Comparing the genomes also revealed that dermatophytes have amplified a number of gene families with potential roles in infection. Dermatophytes have a higher number of genes encoding enzymes that could degrade human proteins, including exopeptidases and endopeptidases. Several types of proteases are enriched, including subtilisins, deuterolisins, and metallopeptidases.
In addition to proteases, the dermatophytes have an increased number of genes encoding kinases and pseudokinases. Presumably, an increase in these signaling factors may indicate that the dermatophytes are able to respond to diverse conditions, as might be expected from constant interaction between the fungus and the host immune system. It is particularly interesting that the dermatophytes also have an increased number of genes encoding pseudokinases, also involved in signaling. Pseudokinases do not have the conserved amino acids necessary for kinase function, but these proteins can function in signaling by interacting with functional kinases. The types of interactions that could involve pseudokinases may include (1) competing with kinases for substrates, (2) competing with kinases to act as a substrate for upstream kinases, or (3) interacting with kinases in a multisubunit complex with allosteric regulation (Reese and Boyle 2012). Signaling systems can also contain phosphatases and pseudophosphatases, but genes encoding these types of molecules were not amplified or lost in the dermatophytes compared to other fungi.
Genes encoding secondary metabolites are also present at higher number in dermatophytes and are highly variable between the different species. These metabolites are the products of polyketide synthesis or nonribosomal protein synthesis. Many of these metabolites are toxins, with antimicrobial and immunosuppressive activities. It is interesting to speculate that dermatophytes increased the number of these genes so that the metabolites could be used in the interactions between the fungal cells and the host, perhaps as immune suppressors to control the immune response to fungal infections. Alternatively, the toxins might control superinfection with other microbes, which can lead to “trench foot,” the result of dermatophyte infection and bacterial superinfection.
Dermatophytes also contain very high numbers of proteins with LysM domains, which in other fungi can bind to various carbohydrate moieties including chitin. The LysM domain was originally identified in bacteria in which proteins containing the domain appear to be secreted and bind to and mask bacterial peptidoglycan from the host immune system. Recently, LysM-containing proteins have been identified in plant fungi, where they are thought to bind to and mask chitin in the fungal cell wall from recognition by the plant immune system (Koharudin et al. 2011; Marshall et al. 2011). The LysM-containing proteins defined in the dermatophytes are the first LysM genes described from a human fungal pathogen. Although many pathogenic fungi have a few proteins with LysM domains, the dermatophytes have amplified this gene family to 15 to 48 members. The LysM-containing proteins in the dermatophytes all have a GH18 chitin-binding domain. This supports the hypothesis that the dermatophyte LysM proteins bind to and mask chitin to protect the fungal cell from the host immune system, allowing latent colonization by the dermatophyte.
Analysis of these genomes has increased our understanding of dermatophytes and raised new questions for further study: (1) What is the function of the many dermatophyte-specific or species-specific genes with no known function (this problem is not unique to the dermatophytes), (2) what is the role of the additional secondary metabolism gene clusters unique to the dermatophytes, (3) what is the role of the additional kinases and the unique pseudokinases in the interactions between the fungus and the host, and (4) what is the role of the proteins containing LysM domains—do they mask the fungus from the host immune system, and to what surface markers does the unique LysM domain bind on the fungal cell surface (Table 1)?
Malassezia
The genus Malassezia belongs to the subphylum Ustilaginomycotina (Begerow et al. 2000; Xu et al. 2007; Guého-Kellermann et al. 2010) of Basidiomycota, but within this group, their position is not yet clear. The order Malasseziales has been proposed in the Exobasidiomycetes (Begerow et al. 2000; Guého-Kellermann et al. 2010), but other researchers placed them in Ustilaginomycetes (Matheny et al. 2007). Consequently, Hibbett et al. (2007) treated them as insertae cedis in their outline of a phylogenetically supported fungal taxonomy. Based on a multigene analysis of many species of basidiomycetous yeasts, a new class, Malasseziomycetes, was proposed in Ustilaginomycotina, indicating the isolated taxonomic position of these yeasts (Wang et al. 2014).
Complex media are needed for isolation, cultivation, and culture-based enumeration of Malassezia yeasts. Dixon’s agar (DA) is used according to its original formula but more commonly in a modified version (modified Dixon agar, mDA) (Midgley 1989). A second widely used medium is Leeming and Notman agar (LNA), which allows isolation and maintenance of all known Malassezia yeasts. Recipes for these media can be found in Guého-Kellermann et al. (2010). Optimal growth temperature is 32°C–34°C. Samples from human or animal sources should be transferred quickly to these media and incubated in a moist environment for at least 2 wk. Different Malassezia species show different growth rates and plating efficiencies, so culture-based methods are not recommended for identification or enumeration of resident Malassezia.
Species can be recognized using ribosomal DNA (rDNA) sequence analysis, especially that of the ITS 1 + 2 regions and the D1/D2 domains of the large subunit (LSU) rDNA (Guillot and Guého 1995; Gemmer et al. 2002; Gupta et al. 2004). Based on this molecular recognition of species, it became possible to differentiate them by phenotypic means to some extent (Guého et al. 1996, 1998; Guillot et al. 1996; Mayser et al. 1997; Crespo Erchiga and Guého 2005; Guého-Kellermann et al. 2011). Discrimination of closely related species (e.g., M. sympodialis, Malassezia caprae, and Malassezia equine) by these phenotypic methods remains challenging. More recently, matrix-assisted laser desorption-ionization time-of-flight mass spectroscopy (MALDI-TOF MS) allowed the identification of all recognized species without extensive phenotypic characterization and showed full concordance with rDNA-based identifications (Kolecka et al. 2014).
Our knowledge on the biodiversity of Malassezia yeasts has greatly expanded during the last two decades. Because the recognition that the lipid-dependent Malassezia furfur, as formerly recognized, contained multiple species (Guého et al. 1996), 13 lipid-dependent species are currently recognized in addition to the lipophilic, but not lipid dependent, Malassezia pachydermatis (Cabanes et al. 2010; Guého-Kellermann et al. 2010, 2011). The phylogenetic position of M. pachydermatis is not yet settled, as it clusters at different positions based on the gene fragments studied.
SKIN DISEASES AND Malassezia
Dandruff and Seborrheic Dermatitis
Dandruff and seborrheic dermatitis (D/SD) are common disorders of the scalp and occasionally the eyebrows, moustache, and beard (Warner et al. 2001). Dandruff is characterized by flaking and pruritis, and it is classified as SD when accompanied by visible redness and extension beyond the scalp to other areas of the face. D/SD is the most common Malassezia-associated dermal disorder, occurring in up to 50% of healthy humans and up to 75%–90% of immunocompromised subjects, especially AIDS patients (Farthing et al. 1985; Warner et al. 2001). Dandruff has received increased attention because of the following: its common occurrence, its link to loss of self-esteem, its negative social image (Warner et al. 2001), and the publication of the dandruff-associated fungal genomes (M. globosa and M. restricta). The resurgence of interest in Malassezia in D/SD supports that dandruff and seborrheic dermatitis are manifestations of the same disorder differing only in severity (Priestley and Savin 1976; Kligman 1979; McGrath and Murphy 1991; Danby et al. 1993; Bulmer and Bulmer 1999; Faergemann 2000b; Pierard-Franchimont et al. 2000; Warner et al. 2001).
D/SD most commonly first appears in adolescents and young adults, likely because of the increase in sebaceous activity during puberty (Ramasastry et al. 1970; Cotterill et al. 1972). The number of scalp Malassezia has been reported to be higher in patients with seborrheic dermatitis or dandruff than in normal controls (McGinley et al. 1975), but the evidence is controversial and contradicted by other reports (Gaitanis et al. 2012). The differences are likely caused by the difficulty in accurately enumerating Malassezia from human skin (Kligman 1979; Weiss et al. 1991; Gupta et al. 2001b; Gemmer et al. 2002; Sugita et al. 2002). D/SD is most commonly treated by shampoos containing antifungal ingredients, indicating the role of fungi. However, as M. globosa and M. restricta are present on all human scalps, the presence of fungus alone is not sufficient to cause D/SD. It has been reported that D/SD is not caused by overgrowth of Malassezia but by an abnormal host immune response (Bergbrant and Faergemann 1989; Midgley 2000). However, D/SD sufferers do not have higher total antibody levels than controls, and there are several reports that an aberrant immune response is not involved (Midgley and Hay 1988; Bergbrant and Faergemann 1989; Parry and Sharpe 1998). A Malassezia lipase has been shown to split triglycerides into irritant fatty acids that can induce scaling (Troller 1971; McGrath and Murphy 1991; Parry and Sharpe 1998) or can release arachidonic acid, which is involved in the inflammation of skin (Faergemann and Fredriksson 1979; Binder and Jonelis 1983; Riciputo et al. 1996). Further research may reveal to what extent the D/SD-promoting role of Malassezia is mediated by the immune system, by the enzymatic activity of the fungus, or by additional features such as secondary metabolites (Faergemann and Fredriksson 1979; Hay and Graham-Brown 1997; Watanabe et al. 2001).
The Malassezia most closely associated with D/SD are M. globosa (Kligman 1979; Crespo Erchiga et al. 1999; Nakabayashi et al. 2000; Gupta et al. 2001b) and M. restricta (Kligman 1979; Crespo Erchiga et al. 1999; Gemmer et al. 2002). However, some authors have also reported M. furfur, M. sympodialis, Malassezia obtuse, and Malassezia slooffiae (Nakabayashi et al. 2000).
Atopic Dermatitis
Atopic dermatitis (AD), also designated atopic eczema, is a chronic inflammatory skin disease marked by pruritus (often intense) and characteristic eczematous lesions with erythema, fine scaling, and thickening of the epidermis. The disease is complex with several subgroups and frequently associated with allergic rhinitis, asthma, and immunoglobulin E (IgE)-mediated food reactions (Bieber 2008). Genetic factors, such as those leading to an impaired skin barrier, play an important role in the development of AD. However, environmental factors, such as lifestyle and exposure to microbes, most likely contribute to the increased frequency of the disease (Bieber 2010). The prevalence of AD has rapidly increased, estimated today at 15%–30% in children and 2%–10% in adults, with the highest prevalence in industrialized countries.
Many studies have found that Malassezia is a microbial trigger that exacerbates AD, especially when the disorder is localized to the head and neck, sebum-rich areas frequently colonized by Malassezia. Some studies have examined the prevalence and the species composition of Malassezia yeasts in AD. Sandström Falk et al. (2005) sampled skin on the upper back and found that M. sympodialis was the species most commonly isolated from both AD patients and healthy controls and also found a difference in species distribution on lesional versus nonlesional skin in AD patients; nonlesional skin was most frequently colonized by M. globosa, whereas M. sympodialis was most commonly found on lesional skin. Other studies in Japan have preferentially found M. globosa as the major cutaneous fungus in AD (Cho et al. 2013), which might reflect geographical differences in the distribution of Malassezia species.
Approximately 50% of adult patients with AD have allergen-specific IgE and T-cell reactivity and/or positive atopy patch test reactions to Malassezia, leading to the hypothesis that in AD Malassezia act as allergens rather than infectious agents (Scheynius and Crameri 2010). These reactions are rarely found in other allergic diseases, indicating a specific link between AD and Malassezia (Casagrande et al. 2006). Allergen-specific IgE against Malassezia can also be detected in children associated with their AD severity in infancy (Kekki et al. 2013). Thirteen allergens have so far been identified in Malassezia (Scheynius and Crameri 2010), and at least 12 genes encoding different classes of the proteins have been confirmed to be present in both the M. sympodialis and M. globosa genomes (Gioti et al. 2013). Interestingly, several of the identified allergens are proteins of unknown function with no sequence homology to characterized proteins, whereas others are homologous to host proteins, suggesting the possibility of cross-reactive immune responses. In fact, using recombinant Malassezia allergens, new subgroups of AD patients have been discovered, who, in addition to IgE reactivity to Malassezia allergens, have an autoimmune IgE-mediated reactivity against self-antigens (Scheynius and Crameri 2010). One example is T-cell-mediated cross-reactivity between the M. sympodialis allergen Mala s 13 and the human homologous protein thioredoxin (Balaji et al. 2011). Such cross-reactivity might contribute to the pathogenesis of AD by perpetuating skin inflammation in patients with AD sensitized to Malassezia. Another mechanism by which Malassezia can interact with the host is the release of extracellular exosomelike nanovesicles carrying allergens (Gehrmann et al. 2011). These nanovesicles, designated MalaEx, can induce inflammatory cytokine responses with a significantly higher IL-4 production in AD patients compared to healthy controls. These findings suggest that MalaEx might be potential therapeutic targets.
Pityriasis Versicolor
Pityriasis versicolor (PV) is a chronic superficial fungal disease characterized by round to oval lesions on the trunk and upper arms. The lesions vary in color and can be hypo- (white) or hyper- (pink, tan, brown, or black) pigmented. Large lesions exhibit minor flaking, but only at the border. PV is regarded as a cosmetic disorder, as most patients are asymptomatic or have mild pruritus (Faergemann 2000a). PV is associated with warm, damp weather, as it is more prevalent in the summer months, and it is more prevalent in tropical locations than in temperate regions. However, endogenous host factors are also involved, including malnutrition (Stein 1983), oral contraceptives (Borelli et al. 1991), systemic corticosteroids or immunosuppressants, and hyperhidrosis (Burke 1961; Faergemann and Bernander 1979).
PV is most common in adolescents and young adults, when sebaceous gland activity is highest (Michalowski and Rodziewicz 1963; Wyre and Johnson 1981; Congly 1984; Terragni et al. 1991). PV may occur when the Malassezia that normally colonize the skin change morphology from the round yeast form to a pathological hyphal form that invades the stratum corneum. The most common Malassezia species cultured from PV are M. globosa (Crespo Erchiga et al. 2000; Nakabayashi et al. 2000) and M. sympodialis (Gupta et al. 2001a,b). Other Malassezia, including M. slooffiae and M. furfur, are less common but occasionally isolated. The ability to grow well at higher temperatures may explain why M. furfur is more frequently isolated from PV in warmer climates than M. globosa (Midgley 2000; Guého-Kellermann et al. 2011).
Other Disorders
Malassezia can cause folliculitis and has been implicated in psoriasis (Gaitanis et al. 2012). M. furfur may also be implicated in invasive infections, especially in low-birth-weight neonates that receive intravenously lipid supplementation (Gaitanis et al. 2012). Children and adults with serious immune suppression caused by gastrointestinal disease may be at risk (Gaitanis et al. 2012); for treatment guidelines, see Arendrup et al. (2013). Part of the older literature on such infections, however, may refer to other lipid-dependent Malassezia species.
PATHOGENESIS
Genome sequences from M. globosa, M. restricta (Xu et al. 2007), and M. sympodialis (Gioti et al. 2013) have provided insights into hypotheses for virulence and survival on host skin. These fungi are the only known free-living fungi to lack fatty acid synthase (Xu et al. 2007), suggesting that they obtain lipids from the host skin. The lack of a fatty acid synthase is a trait shared with some skin bacteria in the genus Corynebacterium (Tauch et al. 2005).
Malassezia encode many secreted aspartyl proteases, lipases, phospholipases C, and acid sphingomyelinases, a trait shared with C. albicans (Xu et al. 2007; Park et al. 2013), which also grows on human skin. This similarity suggests that these enzymes have a role in providing nutrients. However, the dermatophytes do not share this trait. Phylogenetic analysis of lipase and aspartyl protease genes show examples of multiple M. globosa genes related to a single gene from the phylogenetically close relative, U. maydis, implying gene duplications or losses from the common ancestor of these fungi (Xu et al. 2010). There is some indication that these enzymes have a role in pathogenesis. Scalp gene expression at the mRNA level has been demonstrated for lipase and phospholipase genes of M. globosa (Xu et al. 2007) and M. restricta (Lee et al. 2013). Increased expression of M. globosa lipase genes was demonstrated in scalp skin scales from seborrheic dermatitis patients (Patiño-Uzcátegui et al. 2011). Oleic acid, a product of M. globosa lipase activity on scalp, can induce dandruff-like flaking in human subjects (DeAngelis et al. 2007). Higher levels of phospholipase were observed from M. pachydermatis isolates from dogs with dermatitis and otitis than from healthy dogs (Cafarchia and Otranto 2004). The latter study suggests that some Malassezia strains may be more pathogenic than others, a conclusion demonstrated with atopic dermatitis patients and strains with different rRNA sequences (Cho et al. 2013). Insights into virulence may arise from the complete genome sequence of strains of different pathogenicity.
There are additional ways by which Malassezia might affect human skin, resulting in disease. Tryptophan metabolism produces a variety of indoles that are potent agonists of the aryl alcohol receptor. These indoles (malassezin, indolo[3,2-b] carbazole, pityriacitrin, and indirubin) were found in extracts of skin from SD and PV patients but not in healthy controls (Magiatis et al. 2013). The first three of these materials were produced by M. furfur strains isolated from SD patients when these strains were cultivated in l-tryptophan agar (Gaitanis et al. 2008). Control strains from healthy patients grew poorly and produced lower amounts of indoles. Genomics may not readily provide insight into the pathways of indole production, as many indoles can be formed from tryptophan and indole pyruvic acid in the absence of cells, suggesting that a tryptophan aminotransferase may be the only enzyme necessary for the synthesis of these indoles (Zuther et al. 2008). However, additional enzymes may be used in vivo, and Hort et al. (2009) have identified some candidate M. furfur genes with a role in indole synthesis.
These indoles are potent activators of the aryl hydrocarbon receptor. Indirubin is synergistic with Toll-like receptor ligands in the up-regulation of a gene regulated by the aryl hydrocarbon receptor (Vlachos et al. 2012). The indoles appear to modulate the response of monocyte-derived dendritic cells to Toll-like receptor ligands, resulting in less production of costimulatory markers CD80, CD83, and CD86, as well as MHC II and the inflammatory cytokines IL-6 and IL-12. These results suggest that Malassezia can depress the immune response through indoles. Other indole activities include the lowered host pigment production mediated by malassezin, likely through apoptosis of melanocytes (Gaitanis 2012). Together, these indole derivatives may be responsible for hypopigmentation and lack of inflammation associated with PV (Gaitanis et al. 2012).
In another example of a Malassezia secondary metabolite that interacts with animal cells, several Malassezia species are recognized by a C-type lectin receptor, Mincle, found on activated macrophages (Yamasaki et al. 2009). Binding induces the production of the cytokines MIP-2, TNF-α, KC, and IL-10. The Mincle ligand was recently isolated from M. pachydermatis and characterized as a glycolipid comprising 10-hydroxystearic acid, d-mannose, and l-mannitol (Ishikawa et al. 2013).
With the discovery of mating-type genes in Malassezia (Xu et al. 2007), there has been interest in whether Malassezia has a sexual cycle and, if so, to what extent sex impacts virulence and population dynamics. There are several indications that Malassezia may be sexual. A mating locus has been found in all three Malassezia genome sequences that have been reported; a population study of M. sympodialis has provided evidence for recombination; and many meiosis and mating signaling genes are present (Gioti et al. 2013). However, despite repeated attempts, no one has yet demonstrated mating in Malassezia (Gioti et al. 2013).
SUMMARY
For the dermatophytes and for Malassezia, the completion of genome sequences represents a turning point, facilitating molecular analysis of how these fungi survive and cause disease. With the recent development of transformation protocols for dermatophytes (Grumbt et al. 2011a,b), the role of a gene can be tested. Dermatophytes and Malassezia have adapted to human skin, an environment that is relatively amenable to sample recovery, facilitating rapid progress in studying host–fungal interactions. Dermatophytes and Malassezia have a large impact on human health, as they are found on the skin of most people and are frequently associated with disease. With greater attention to the human microbiome, we will learn how these organisms may contribute to health and disease.
Footnotes
Editors: Arturo Casadevall, Aaron P. Mitchell, Judith Berman, Kyung J. Kwon-Chung, John R. Perfect, and Joseph Heitman
Additional Perspectives on Human Fungal Pathogens available at www.perspectivesinmedicine.org
REFERENCES
- Achterman RR, White TC 2012a. Dermatophyte virulence factors: Identifying and analyzing genes that may contribute to chronic or acute skin infections. Int J Microbiol 2012: 358305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achterman RR, White TC 2012b. A foot in the door for dermatophyte research. PLoS Pathog 8: e1002564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achterman RR, Smith AR, Oliver BG, White TC 2011. Sequenced dermatophyte strains: Growth rate, conidiation, drug susceptibilities, and virulence in an invertebrate model. Fungal Genet Biol 48: 335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amend AS, Barshis DJ, Oliver TA 2012. Coral-associated marine fungi form novel lineages and heterogeneous assemblages. ISME J 6: 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendrup MC, Boekhout T, Akova M, Meis JF, Cornely OA, Lortholary O, Arikan-Akdagli S, Cuenca-Estrella M, Dannaoui E, van Diepeningen AD, et al. 2013. ESCMID/ECMM joint clinical guideline for the diagnosis and management of rare invasive yeast infections. Clin Microbiol Infect 10.1111/1469-0691.12360 [DOI] [PubMed] [Google Scholar]
- Balaji H, Heratizadeh A, Wichmann K, Niebuhr M, Crameri R, Scheynius A, Werfel T 2011. Malassezia sympodialis thioredoxin-specific T cells are highly cross-reactive to human thioredoxin in atopic dermatitis. J Allergy Clin Immun 128: 92–99 [DOI] [PubMed] [Google Scholar]
- Begerow D, Bauer R, Boekhout T 2000. Phylogenetic placements of ustilaginomycetous anamorphs as deduced from nuclear LSU rDNA sequences. Mycol Res 104: 53–60 [Google Scholar]
- Bergbrant IM, Faergemann J 1989. Seborrhoeic dermatitis and Pityrosporum ovale: A cultural and immunological study. Acta Derm Venereol 69: 332–335 [PubMed] [Google Scholar]
- Bieber T 2008. Atopic dermatitis. New Engl J Med 358: 1483–1494 [DOI] [PubMed] [Google Scholar]
- Bieber T 2010. Atopic dermatitis. Ann Dermatol 22: 125–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder RL, Jonelis FJ 1983. Seborrheic dermatitis in neuroleptic-induced parkinsonism. Arch Dermatol 119: 473–475 [PubMed] [Google Scholar]
- Blehert DS, Hicks AC, Behr M, Meteyer CU, Berlowski-Zier BM, Buckles EL, Coleman JT, Darling SR, Gargas A, Niver R, et al. 2009. Bat white-nose syndrome: An emerging fungal pathogen? Science 323: 227. [DOI] [PubMed] [Google Scholar]
- Borelli D, Jacobs PH, Nall L 1991. Tinea versicolor: Epidemiologic, clinical, and therapeutic aspects. J Am Acad Dermatol 25: 300–305 [DOI] [PubMed] [Google Scholar]
- Borgers M, Degreef H, Cauwenbergh G 2005. Fungal infections of the skin: Infection process and antimycotic therapy. Curr Drug Targets 6: 849–862 [DOI] [PubMed] [Google Scholar]
- Bulmer AC, Bulmer GS 1999. The antifungal action of dandruff shampoos. Mycopathologia 147: 63–65 [DOI] [PubMed] [Google Scholar]
- Burke RC 1961. Tinea versicolor: Susceptibility factors and experimental infection in human beings. J Invest Dermatol 36: 389–401 [DOI] [PubMed] [Google Scholar]
- Burmester A, Shelest E, Glockner G, Heddergott C, Schindler S, Staib P, Heidel A, Felder M, Petzold A, Szafranski K, et al. 2011. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol 12: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanes FJ, Vega S, Castella G 2010. Malassezia cuniculi sp. nov., a novel yeast species isolated from rabbit skin. Med Mycol 49: 40–48 [DOI] [PubMed] [Google Scholar]
- Cafarchia C, Otranto D 2004. Association between phospholipase production by Malassezia pachydermatis and skin lesions. J Clin Microbiol 42: 4868–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casagrande BF, Fluckiger S, Linder MT, Johansson C, Scheynius A, Crameri R, Schmid-Grendelmeier P 2006. Sensitization to the yeast Malassezia sympodialis is specific for extrinsic and intrinsic atopic eczema. J Invest Dermatol 126: 2414–2421 [DOI] [PubMed] [Google Scholar]
- Cheng TL, Rovito SM, Wake DB, Vredenburg VT 2011. Coincident mass extirpation of neotropical amphibians with the emergence of the infectious fungal pathogen Batrachochytrium dendrobatidis. Proc Natl Acad Sci 108: 9502–9507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho O, Saito M, Tsuboi R, Kato H, Nishikawa A, Nakajima S, Sugita T 2013. Relationships among the genotypes of Malassezia globosa colonizing patients with atopic dermatitis, the clinical severity of the disease, and the level of specific IgE antibodies. J Clin Exp Dermatol Res 4: 197 [Google Scholar]
- Congly H 1984. Pityriasis versicolor in a 3-month-old boy. Can Med Assoc J 130: 844–845 [PMC free article] [PubMed] [Google Scholar]
- Cotterill JA, Cunliffe WJ, Williamson B, Bulusu L 1972. Age and sex variation in skin surface lipid composition and sebum excretion rate. Br J Dermatol 87: 333–340 [DOI] [PubMed] [Google Scholar]
- Crespo Erchiga V, Guého E 2005. Superficial diseases caused by Malassezia species. In Topley & Wilson’s microbiology and microbial infections (ed. Merz W, Hay R), pp. 202–219 Wiley, New York [Google Scholar]
- Crespo Erchiga V, Ojeda Martos A, Vera Casaño A, Crespo Erchiga A, Sanchez-Fajardo F, Guého-Kellermann E 1999. Mycology of pityriasis versicolor. J Mycol Méd 9: 143–148 [Google Scholar]
- Crespo Erchiga V, Ojeda Martos A, Vera Casano A, Crespo Erchiga A, Sanchez Fajardo F 2000. Malassezia globosa as the causative agent of pityriasis versicolor. Br J Dermatol 143: 799–803 [DOI] [PubMed] [Google Scholar]
- Cui L, Morris A, Ghedin E 2013. The human mycobiome in health and disease. Genome Med 5: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danby FW, Maddin WS, Margesson LJ, Rosenthal D 1993. A randomized, double-blind, placebo-controlled trial of ketoconazole 2% shampoo versus selenium sulfide 2.5% shampoo in the treatment of moderate to severe dandruff. J Am Acad Dermatol 29: 1008–1012 [DOI] [PubMed] [Google Scholar]
- DeAngelis YM, Saunders CW, Johnstone KR, Reeder NL, Coleman CG, Kaczvinsky JR Jr, Gale C, Walter R, Mekel M, Lacey MP, et al. 2007. Isolation and expression of a Malassezia globosa lipase gene, LIP1. J Invest Dermatol 127: 2138–2146 [DOI] [PubMed] [Google Scholar]
- Edgar RC 2004. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faergemann J 2000a. Management of seborrheic dermatitis and pityriasis versicolor. Am J Clin Dermatol 1: 75–80 [DOI] [PubMed] [Google Scholar]
- Faergemann J 2000b. Seborrheic dermatitis (dandruff), pp. 197–202 Marcel Dekker, New York [Google Scholar]
- Faergemann J, Bernander S 1979. Tinea versicolor and Pityrosporum orbiculare: A mycological investigation. Sabouraudia 17: 171–179 [PubMed] [Google Scholar]
- Faergemann J, Fredriksson T 1979. Tinea versicolor with regard to seborrheic dermatitis. An epidemiological investigation. Arch Dermatol 115: 966–968 [PubMed] [Google Scholar]
- Farthing CF, Staughton RC, Rowland Payne CM 1985. Skin disease in homosexual patients with acquired immune deficiency syndrome (AIDS) and lesser forms of human T cell leukaemia virus (HTLV III) disease. Clin Exp Dermatol 10: 3–12 [DOI] [PubMed] [Google Scholar]
- Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, NIH Intramural Sequencing Center Comparative Sequencing Program, Kong HH, et al. 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498: 367–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanis G, Magiatis P, Stathopoulou K, Bassukas ID, Alexopoulos EC, Velegraki A, Skaltsounis AL 2008. AhR ligands, malassezin, and indolo[3,2-b]carbazole are selectively produced by Malassezia furfur strains isolated from seborrheic dermatitis. J Invest Dermatol 128: 1620–1625 [DOI] [PubMed] [Google Scholar]
- Gaitanis G, Magiatis P, Hantschke M, Bassukas ID, Velegraki A 2012. The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev 25: 106–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Takashima A 2004. Cloning and characterization of Trichophyton rubrum genes encoding actin, Tri r2, and Tri r4. J Clin Microbiol 42: 3298–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Li B, Zheng C, Wang G 2008. Molecular detection of fungal communities in the Hawaiian marine sponges Suberites zeteki and Mycale armata. Appl Environ Microbiol 74: 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrmann U, Qazi KR, Johansson C, Hultenby K, Karlsson M, Lundeberg L, Gabrielsson S, Scheynius A 2011. Nanovesicles from Malassezia sympodialis and host exosomes induce cytokine responses—Novel mechanisms for host–microbe interactions in atopic eczema. PLoS ONE 6: e21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmer CM, DeAngelis YM, Theelen B, Boekhout T, Dawson TL Jr 2002. Fast, noninvasive method for molecular detection and differentiation of Malassezia yeast species on human skin and application of the method to dandruff microbiology. J Clin Microbiol 40: 3350–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum M, Isham N, Sheehan D 2006. Voriconazole susceptibilities of dermatophyte isolates obtained from a worldwide tinea capitis clinical trial. J Clin Microbiol 44: 2579–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum MA, Wraith LA, Cai B, Nyirady J, Isham N 2008. Susceptibility of dermatophyte isolates obtained from a large worldwide terbinafine tinea capitis clinical trial. Br J Dermatol 159: 711–713 [DOI] [PubMed] [Google Scholar]
- Gioti A, Nystedt B, Li W, Xu J, Andersson A, Averette AF, Munch K, Wang X, Kappauf C, Kingsbury JM, et al. 2013. Genomic insights into the atopic eczema-associated skin commensal yeast Malassezia sympodialis. mBio 4: e00572-00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser Y, Frohlich J, Presber W, de Hoog S 2007. Microsatellite markers reveal geographic population differentiation in Trichophyton rubrum. J Med Microbiol 56: 1058–1065 [DOI] [PubMed] [Google Scholar]
- Graser Y, Scott J, Summerbell R 2008. The new species concept in dermatophytes—A polyphasic approach. Mycopathologia 166: 239–256 [DOI] [PubMed] [Google Scholar]
- Grice EA, Segre JA 2011. The skin microbiome. Nature Rev Microbiol 9: 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Program NCS, Bouffard GG, Blakesley RW, Murray PR, et al. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324: 1190–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumbt M, Defaweux V, Mignon B, Monod M, Burmester A, Wostemeyer J, Staib P 2011a. Targeted gene deletion and in vivo analysis of putative virulence gene function in the pathogenic dermatophyte Arthroderma benhamiae. Eukaryot Cell 10: 842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumbt M, Monod M, Staib P 2011b. Genetic advances in dermatophytes. FEMS Microbiol Lett 320: 79–86 [DOI] [PubMed] [Google Scholar]
- Guého E, Midgley G, Guillot J 1996. The genus Malassezia with description of four new species. Antonie van Leeuwenhoek 69: 337–355 [DOI] [PubMed] [Google Scholar]
- Guého E, Boekhout T, Ashbee HR, Guillot J, Van Belkum A, Faergemann J 1998. The role of Malassezia species in the ecology of human skin and as pathogens. Med Mycol 36: 220–229 [PubMed] [Google Scholar]
- Guého-Kellermann E, Velegraki A, Begerow D 2010. Biodiversity, phylogeny and ultrastructure. In Malassezia and the skin (ed. Boekhout T, Guého-Kellermann E, Mayser P, Velegraki A), pp. 17–63 Springer, Berlin [Google Scholar]
- Guého-Kellermann E, Batra R, Boekhout T 2011. The genus Malassezia Baillon. In The yeasts: A taxonomic study (ed. Kurtzman C, Fell JW, Boekhout T), pp. 1807–1832 Elsevier, Amsterdam [Google Scholar]
- Guillot J, Guého E 1995. The diversity of Malassezia yeasts confirmed by rRNA sequence and nuclear DNA comparisons. Antonie van Leeuwenhoek 67: 297–314 [DOI] [PubMed] [Google Scholar]
- Guillot J, Gueho E, Lesourd M, Midgley G, Chevrier G, Dupont B 1996. Identification of Malassezia species: A practical approach. Masson, Paris [Google Scholar]
- Gupta AK, Cooper EA 2008. Update in antifungal therapy of dermatophytosis. Mycopathologia 166: 353–367 [DOI] [PubMed] [Google Scholar]
- Gupta AK, Kohli Y, Faergemann J, Summerbell RC 2001a. Epidemiology of Malassezia yeasts associated with pityriasis versicolor in Ontario, Canada. Med Mycol 39: 199–206 [DOI] [PubMed] [Google Scholar]
- Gupta AK, Kohli Y, Summerbell RC, Faergemann J 2001b. Quantitative culture of Malassezia species from different body sites of individuals with or without dermatoses. Med Mycol 39: 243–251 [DOI] [PubMed] [Google Scholar]
- Gupta AK, Boekhout T, Theelen B, Summerbell R, Batra R 2004. Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and large-subunit regions of ribosomal DNA. J Clin Microbiol 42: 4253–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RJ, Graham-Brown RA 1997. Dandruff and seborrhoeic dermatitis: Causes and management. Clin Exp Dermatol 22: 3–6 [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lucking R, et al. 2007. A higher-level phylogenetic classification of the Fungi. Mycol Res 111: 509–547 [DOI] [PubMed] [Google Scholar]
- Hort W, Lang S, Brunke S, Mayser P, Hube B 2009. Analysis of differentially expressed genes associated with tryptophan-dependent pigment synthesis in M. furfur by cDNA subtraction technology. Med Mycol 47: 248–258 [DOI] [PubMed] [Google Scholar]
- Huffnagle GB, Noverr MC 2013. The emerging world of the fungal microbiome. Trends Microbiol 21: 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Itoh F, Yoshida S, Saijo S, Matsuzawa T, Gonoi T, Saito T, Okawa Y, Shibata N, Miyamoto T, et al. 2013. Identification of distinct ligands for the C-type lectin receptors Mincle and Dectin-2 in the pathogenic fungus Malassezia. Cell Host Microbe 13: 477–488 [DOI] [PubMed] [Google Scholar]
- Kekki OM, Scheynius A, Poikonen S, Koskinen A, Kautiainen H, Turjanmaa K 2013. Sensitization to Malassezia in children with atopic dermatitis combined with food allergy. Pediatr Allergy Immun 24: 244–249 [DOI] [PubMed] [Google Scholar]
- Khot PD, Ko DL, Fredricks DN 2009. Sequencing and analysis of fungal rRNA operons for development of broad-range fungal PCR assays. Appl Environ Microbiol 75: 1559–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kligman AM 1979. Perspectives and problems in cutaneous gerontology. J Invest Dermatol 73: 39–46 [DOI] [PubMed] [Google Scholar]
- Koharudin LM, Viscomi AR, Montanini B, Kershaw MJ, Talbot NJ, Ottonello S, Gronenborn AM 2011. Structure-function analysis of a CVNH-LysM lectin expressed during plant infection by the rice blast fungus Magnaporthe oryzae. Structure 19: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolecka A, Khayhan K, Arabatzis M, Velegraki A, Kostrzewa M, Andersson A, Scheynius A, Cafarchia C, Iatta R, Montagna MT, et al. 2014. Efficient identification of Malassezia yeasts by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Br J Dermatol 170: 332–341 [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Bennett JE 1992. Medical mycology. Lea & Febiger, Philadelphia [Google Scholar]
- Lee YW, Lee SY, Lee Y, Jung WH 2013. Evaluation of expression of lipases and phospholipases of Malassezia restricta in patients with seborrheic dermatitis. Ann Dermatol 25: 310–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Metin B, White TC, Heitman J 2010. Organization and evolutionary trajectory of the mating type (MAT) locus in dermatophyte and dimorphic fungal pathogens. Eukaryot Cell 9: 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiatis P, Pappas P, Gaitanis G, Mexia N, Melliou E, Galanou M, Vlachos C, Stathopoulou K, Skaltsounis AL, Marselos M, et al. 2013. Malassezia yeasts produce a collection of exceptionally potent activators of the Ah (Dioxin) receptor detected in diseased human skin. J Invest Dermatol 133: 2023–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr KA, Lyons CN, Rustad TR, Bowden RA, White TC 1998. Rapid, transient fluconazole resistance in Candida albicans is associated with increased mRNA levels of CDR. Antimicrob Agents Chemother 42: 2584–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R, Kombrink A, Motteram J, Loza-Reyes E, Lucas J, Hammond-Kosack KE, Thomma BP, Rudd JJ 2011. Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol 156: 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez DA, Oliver BG, Graser Y, Goldberg JM, Li W, Martinez-Rossi NM, Monod M, Shelest E, Barton RC, Birch E, et al. 2012. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBio 3: e00259-00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny PB, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson RH, Hughes KW, Hofstetter V, Ammirati JF, Schoch CL, et al. 2007. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol Phylogenet Evol 43: 430–451 [DOI] [PubMed] [Google Scholar]
- Mayser P, Haze P, Papavassilis C, Pickel M, Gruender K, Gueho E 1997. Differentiation of Malassezia species: Selectivity of cremophor EL, castor oil and ricinoleic acid for M. furfur. Br J Dermatol 137: 208–213 [DOI] [PubMed] [Google Scholar]
- McGinley KJ, Leyden JJ, Marples RR, Kligman AM 1975. Quantitative microbiology of the scalp in non-dandruff, dandruff, and seborrheic dermatitis. J Invest Dermatol 64: 401–405 [DOI] [PubMed] [Google Scholar]
- McGrath J, Murphy GM 1991. The control of seborrhoeic dermatitis and dandruff by antipityrosporal drugs. Drugs 41: 178–184 [DOI] [PubMed] [Google Scholar]
- Michalowski R, Rodziewicz H 1963. Pityriasis versicolor in children. Br J Dermatol 75: 397–400 [DOI] [PubMed] [Google Scholar]
- Midgley G 1989. The diversity of Pityrosporum (Malassezia) yeasts in vivo and in vitro. Mycopathologia 106: 143–153 [DOI] [PubMed] [Google Scholar]
- Midgley G 2000. The lipophilic yeasts: State of the art and prospects. Med Mycol 38: 9–16 [PubMed] [Google Scholar]
- Midgley G, Hay RJ 1988. Serological responses to Pityrosoporum (Malassezia) in seborrhoeic dermatitis demonstrated by ELISA and Western blotting. Bull Soc Fr Mycol Med 17: 267–276 [Google Scholar]
- Nakabayashi A, Sei Y, Guillot J 2000. Identification of Malassezia species isolated from patients with seborrhoeic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Med Mycol 38: 337–341 [DOI] [PubMed] [Google Scholar]
- Parfrey LW, Walters WA, Knight R 2011. Microbial eukaryotes in the human microbiome: Ecology, evolution, and future directions. Front Microbiol 2: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Do E, Jung WH 2013. Lipolytic enzymes involved in the virulence of human pathogenic fungi. Mycobiology 41: 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry ME, Sharpe GR 1998. Seborrhoeic dermatitis is not caused by an altered immune response to Malassezia yeast. Br J Dermatol 139: 254–263 [DOI] [PubMed] [Google Scholar]
- Patiño-Uzcátegui A, Amado Y, Cepero de Garcia M, Chaves D, Tabima J, Motta A, Cardenas M, Bernal A, Restrepo S, Celis A 2011. Virulence gene expression in Malassezia spp from individuals with seborrheic dermatitis. J Invest Dermatol 131: 2134–2136 [DOI] [PubMed] [Google Scholar]
- Pfaller MA 2012. Antifungal drug resistance: Mechanisms, epidemiology, and consequences for treatment. Am J Med 125: S3–13 [DOI] [PubMed] [Google Scholar]
- Pierard-Franchimont C, Hermanns JF, Degreef H, Pierard GE 2000. From axioms to new insights into dandruff. Dermatology 200: 93–98 [DOI] [PubMed] [Google Scholar]
- Pitkaranta M, Meklin T, Hyvarinen A, Paulin L, Auvinen P, Nevalainen A, Rintala H 2008. Analysis of fungal flora in indoor dust by ribosomal DNA sequence analysis, quantitative PCR, and culture. Appl Environ Microbiol 74: 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestley GC, Savin JA 1976. The microbiology of dandruff. Br J Dermatol 94: 469–471 [DOI] [PubMed] [Google Scholar]
- Purim KS, Bordignon GP, Queiroz-Telles F 2005. Fungal infection of the feet in soccer players and non-athlete individuals. Rev Iberoam Micol 22: 34–38 [DOI] [PubMed] [Google Scholar]
- Ramasastry P, Downing DT, Pochi PE, Strauss JS 1970. Chemical composition of human skin surface lipids from birth to puberty. J Invest Dermatol 54: 139–144 [DOI] [PubMed] [Google Scholar]
- Reese ML, Boyle JP 2012. Virulence without catalysis: How can a pseudokinase affect host cell signaling? Trends Parasitol 28: 53–57 [DOI] [PubMed] [Google Scholar]
- Renker C, Alphei JR, Buscot FO 2003. Soil nematodes associated with the mammal pathogenic fungal genus Malassezia (Basidiomycota: Ustilaginomycetes) in Central European forests. Biol Fert Soils 37: 70–72 [Google Scholar]
- Riciputo RM, Oliveri S, Micali G, Sapuppo A 1996. Phospholipase activity in Malassezia furfur pathogenic strains. Mycoses 39: 233–235 [DOI] [PubMed] [Google Scholar]
- Sandström Falk MH, Tengvall Linder M, Johansson C, Bartosik J, Bäck O, Särnhult T, Wahlgren CF, Scheynius A, Faergemann J 2005. The prevalence of Malassezia yeasts in patients with atopic dermatitis, seborrhoeic dermatitis and healthy controls. Acta Derm Venereol 85: 17–23 [DOI] [PubMed] [Google Scholar]
- Scheynius A, Crameri C 2010. Malassezia in atopic eczema/dermatitis. In Malassezia and the skin (ed. Boekhout T, Guého-Kellermann E, Mayser P, Velegraki A), pp. 212–228 Springer, Berlin [Google Scholar]
- Stamatakis A 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Stein DH 1983. Superficial fungal infections. Pediatr Clin N Am 30: 545–561 [DOI] [PubMed] [Google Scholar]
- Sugita T, Takashima M, Shinoda T, Suto H, Unno T, Tsuboi R, Ogawa H, Nishikawa A 2002. New yeast species, Malassezia dermatis, isolated from patients with atopic dermatitis. J Clin Microbiol 40: 1363–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauch A, Kaiser O, Hain T, Goesmann A, Weisshaar B, Albersmeier A, Bekel T, Bischoff N, Brune I, Chakraborty T, et al. 2005. Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. J Bacteriol 187: 4671–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terragni L, Lasagni A, Oriani A, Gelmetti C 1991. Pityriasis versicolor in the pediatric age. Pediatr Dermatol 8: 9–12 [DOI] [PubMed] [Google Scholar]
- Troller JA 1971. Model system for the investigation of dandruff. J Soc Cosmet Chem 22: 187–198 [Google Scholar]
- Vlachos C, Schulte BM, Magiatis P, Adema GJ, Gaitanis G 2012. Malassezia-derived indoles activate the aryl hydrocarbon receptor and inhibit Toll-like receptor-induced maturation in monocyte-derived dendritic cells. Br J Dermatol 167: 496–505 [DOI] [PubMed] [Google Scholar]
- Wang Q-M, Theelen B, Groenewald M, Bai F-Y, Boekhout T 2014. Moniliellomycetes and Malasseziomycetes, two new classes in Ustilaginomycotina. Persoonia (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner RR, Schwartz JR, Boissy Y, Dawson TL Jr 2001. Dandruff has an altered stratum corneum ultrastructure that is improved with zinc pyrithione shampoo. J Am Acad Dermatol 45: 897–903 [DOI] [PubMed] [Google Scholar]
- Watanabe S, Kano R, Sato H, Nakamura Y, Hasegawa A 2001. The effects of Malassezia yeasts on cytokine production by human keratinocytes. J Invest Dermatol 116: 769–773 [DOI] [PubMed] [Google Scholar]
- Weiss SJ, Schoch PE, Cunha BA 1991. Malassezia furfur fungemia associated with central venous catheter lipid emulsion infusion. Heart Lung 20: 87–90 [PubMed] [Google Scholar]
- White TC, Oliver BG, Graser Y, Henn MR 2008. Generating and testing molecular hypotheses in the dermatophytes. Eukaryot Cell 7: 1238–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyre HW Jr, Johnson WT 1981. Neonatal pityriasis versicolor. Arch Dermatol 117: 752–753 [PubMed] [Google Scholar]
- Xu J, Saunders CW, Hu P, Grant RA, Boekhout T, Kuramae EE, Kronstad JW, Deangelis YM, Reeder NL, Johnstone KR, et al. 2007. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc Natl Acad Sci 104: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Boekhout T, DeAngelis Y, Dawson T, Saunders CW 2010. Genomics and pathophysiology: Dandruff as a paradigm. In Malassezia and the skin (ed. Boekhout T, Guého-Kellermann E, Mayser P, Velegraki A), pp. 253–270 Springer, Berlin [Google Scholar]
- Yamasaki S, Matsumoto M, Takeuchi O, Matsuzawa T, Ishikawa E, Sakuma M, Tateno H, Uno J, Hirabayashi J, Mikami Y, et al. 2009. C-type lectin Mincle is an activating receptor for pathogenic fungus, Malassezia. Proc Natl Acad Sci 106: 1897–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Suh SO, Blackwell M 2003. Microorganisms in the gut of beetles: Evidence from molecular cloning. J Invertebr Pathol 84: 226–233 [DOI] [PubMed] [Google Scholar]
- Zuther K, Mayser P, Hettwer U, Wu W, Spiteller P, Kindler BL, Karlovsky P, Basse CW, Schirawski J 2008. The tryptophan aminotransferase Tam1 catalyses the single biosynthetic step for tryptophan-dependent pigment synthesis in Ustilago maydis. Mol Microbiol 68: 152–172 [DOI] [PubMed] [Google Scholar]