Abstract
MYC is a noncanonical transcription factor that binds to thousands of genomic loci and affects >15% of the human transcriptome, with surprisingly little overlap between MYC-bound and -regulated genes. This discordance raises the question whether MYC chooses its targets based on their individual biological effects (“a la carte”) or by virtue of belonging to a certain group of genes (on a “prix fixe” basis). This review presents evidence for a prix fixe, posttranscriptional model whereby MYC initially deregulates a select number of microRNAs. These microRNAs then target a broad spectrum of genes based solely on the presence in their 3′ UTRs (untranslated regions) of distinct “seed” sequences. Existing evidence suggests that there are significant microRNA components to all key MYC-driven phenotypes, including cell-cycle progression, apoptosis, metabolism, angiogenesis, metastasis, stemness, and hematopoiesis. Furthermore, each of these cell-intrinsic and -extrinsic phenotypes is likely attributable to deregulation of multiple microRNA targets acting in different, yet frequently overlapping, pathways. The habitual targeting of multiple genes within the same pathway might account for the robustness and persistence of MYC-induced phenotypes.
MYC affects the expression of more than 15% of human genes, either directly or indirectly. In the process, it the process, it regulates several microRNAs that are involved in MYC-driven phenotypes, including cell-cycle progression, apoptosis, and cancer.
V-MYC was first discovered as an oncogenicity factor of several acutely transforming avian myelocytomatosis retroviruses and subsequently found to have a cellular homolog c-MYC (thereafter referred to simply as MYC) (Sheiness and Bishop 1979). In the early 1990s, great strides were made in characterizing its subcellular localization and biochemical properties. It was found to be a nuclear phosphoprotein tightly bound to chromatin (Abrams et al. 1982). It later became apparent that MYC preferentially binds to the E-box motif in the genomic DNA through its carboxyl terminus as a heterodimer with Max (Blackwell et al. 1990; Prendergast and Ziff 1991), whereas its amino terminus possesses an intrinsic transactivation activity when fused to the GAL4 DNA-binding domain (Kato et al. 1990). Curiously, full-length MYC has never been purified or produced in quantities sufficient for rigorous analyses. Nevertheless, it seemed at the time that identification of MYC target genes would be fairly straightforward, and that the identity of its key targets would explain MYC-driven phenotypes in a way that proapoptotic (e.g., Puma and Noxa) and antiproliferation (e.g., p21) targets account for the major tumor suppressive effects of p53 (Lowe et al. 2004). These hopes for clarity never materialized (see Conacci-Sorrell et al. 2014).
As more and more cell types were tested, the number of MYC targets rose vertiginously. A hubsite (www.myccancergene.org) was created in the early 2000s to keep the researchers abreast of new developments. Per its last update (September 2003), the database contained 1697 genes. Many more genes have since been identified. Even if one limits the analysis to just one cell line, the number of genes whose expression is influenced by MYC is staggering. By some estimates, MYC regulates >15% of the human transcriptome (Eilers and Eisenman 2008), which is commonly referred to as the “MYC signature.”
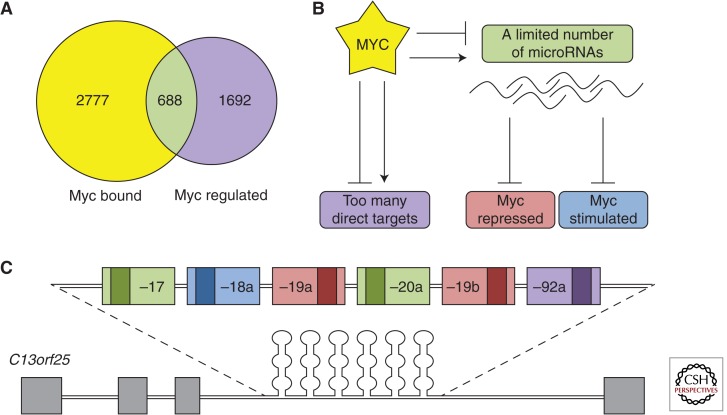
One could certainly argue that not all genes comprising the MYC signature are its direct targets, and if one were to catalog MYC-binding sites in the DNA, the “true” targets would emerge. Such analysis was performed in several cell types, and the first part of the prediction certainly held true. Out of thousands of MYC signature genes, only a small fraction contained experimentally confirmed MYC-binding sites. Strikingly, the majority of MYC-bound genes showed little evidence of regulation by MYC, and a consensus has emerged that “… only a minority of loci to which MYC and Max are bound in vivo correspond to MYC-regulated protein-coding genes” (Adhikary and Eilers 2005). Given the minimal overlap between MYC-bound and -regulated genes (Fig. 1A), it is fair to ask what exactly an MYC-target gene is and whether MYC chooses them on individual merit (“a la carte”) or by virtue of belonging to a certain group of genes (on a “prix fixe” basis). Although many papers in the field emphasize important functional relationships between MYC and a handful of key targets, it is worth reviewing evidence in favor of the prix fixe model.
Figure 1.
Gene regulation by MYC: a la carte or prix fixe? (A) Comparison of MYC-bound versus MYC-regulated genes in P493-6 cells. (B) Model of MYC-mediated gene regulation through the microRNA pathway. (C) The miR-17–92 cluster, which accounts for most posttranscriptional MYC targets.
One possible scenario is that MYC regulates several key transcription factors, which broaden the MYC signature by pooling together different subsets of targets. In principle, MYC could trans-activate genes x, y, and z, each with their own cohort of target genes. For example, Id2 was reported to be a direct transcriptional target of both c- and N-MYC (Lasorella et al. 2000), leading the investigators to conclude that Id2 mediates MYC-induced signaling, in particular, through the Rb-E2F pathway. That same pathway could be affected through the binding of MYC to the E2F2 promoter (Sears et al. 1997). To be sure, there is a considerable overlap between MYC and E2F targets (Ogawa et al. 2002). Yet, this indirect mode of regulation by proxy is inconsistent with the rapidity of MYC-induced changes in gene expression. Several reports show that gene deregulation by MYC occurs within 4–12 hours, and it is doubtful that two cycles of transcription and one cycle of translation could occur within this time frame. Indeed, published studies from the Eick group show that the majority of MYC-affected genes are regulated in the absence of new protein synthesis (Schuhmacher et al. 2001).
An intriguing alternative to the mechanism described above is the recent “amplification model” in which MYC neither deliberately chooses its targets on individual basis nor delegates this task to “subordinate” transcription factors, but instead broadly enhances transcription of genes poised to be transcribed. This is thought to occur via the recruitment of the P-TEFb antipausing complex by MYC and the promotion of transcription elongation (Lin et al. 2012; Nie et al. 2012; also see Rahl and Young 2014).
As thought-provoking as these transcriptional studies are, currently there is little evidence that the amplification model accounts for the transforming activity of MYC. Furthermore, there are also examples of MYC-target genes that are not at all regulated at the level of messenger RNA (mRNA) synthesis (either transcription initiation or elongation). In 1989 and 1990, it was reported that the plasminogen activator inhibitor 1 gene is regulated by MYC at the level of mRNA turnover (Prendergast and Cole 1989; Prendergast et al. 1990). Ten years later, our laboratory made a surprising discovery that although MYC strongly represses expression of the antiangiogenic factor thrombospondin-1, it does not significantly affect the activity of the thbs1 promoter. Instead, it negatively regulates thrombospondin-1 mRNA stability, as determined in experiments with actinomycin D, an inhibitor of de novo transcription (Janz et al. 2000). Furthermore, the Eick and Dang groups found numerous discrepancies between nuclear run-off rates and mRNA steady-state levels, leading them to conclude that “genes such as RFC4 and MCM4 are likely to be direct targets of MYC, but that posttranscriptional mechanisms may contribute to accumulation of mRNA at later time points” (Schuhmacher et al. 2001; Fan et al. 2010). Thus, alternatives to transcription-based mechanisms of gene regulation by MYC must exist. The discovery of microRNAs in mammalian cells and their tight connection to MYC proved to be a breakthrough in the field.
Since their discovery in 1993 (Lee et al. 1993; Wightman et al. 1993), microRNAs, a subclass of short double-stranded RNAs, have emerged as major players affecting mRNA turnover and translation in a sequence-specific manner. They are synthesized as long primary transcripts (pri-microRNAs) with loop/hairpin structures and undergo cleavage by the RNase III endonuclease Drosha. The cleaved products (termed pre-microRNAs) are exported from the nucleus into the cytoplasm where the Dicer RNase III endonuclease cleaves the loop off the hairpin, thus generating two complementary single-stranded molecules. One of them, called the guide strand, gets incorporated in the RISC-loading complex (RLC). RLC, which at minimum is composed of Dicer, TRBP, and Ago2, guides the microRNA to the target mRNA (Winter et al. 2009). Although it is sometimes incorporated into the RLC instead of the conventional guide strand, the complementary strand (miR*) usually undergoes degradation (Zamore and Haley 2005).
The outcome of these events depends on the degree of homology between the microRNA and the mRNA. In rare cases in which the microRNA–mRNA complementarity is complete, Argonaute can mediate the cleavage and rapid degradation of target mRNA. In the majority of cases, nucleotides 2–7 of the microRNA, termed the “seed sequence,” are responsible for target recognition through homology with the cognate mRNA (Bartel 2009). Additional contributing factors include target accessibility and the free energy of the entire duplex (Miranda et al. 2006). However, base pairing of the nonseed nucleotides appears to vary greatly between targets, and seed sequence interactions are, in many cases, sufficient to result in translation repression and mRNA destabilization (Zamore and Haley 2005). Thus, the microRNA binding sites in mRNAs are barely longer than MYC-binding sites in the genomic DNA and equally abundant. In fact, common algorithms predict that most microRNAs have hundreds, if not thousands, of targets. Thus, they are ideally suited to mediate broad effects of MYC on gene expression should they be subject to regulation by MYC (Fig. 1B).
Several papers published in the summer of 2005 showed that such connections indeed exist. First, the Mendell and Dang laboratories showed that MYC transcriptionally up-regulates the human C13orf25 locus (O’Donnell et al. 2005) encoding the primary transcript for the miR-17–92 cluster. This cluster encodes six distinct microRNAs, miR-17, -18a, -19a, -20a, -19b, and -92a (Fig. 1C), which collectively down-regulate a number of target genes including E2F1, a protein that drives both proliferation and apoptosis. The investigators proposed that microRNA-mediated down-regulation of E2F1 by MYC alleviates apoptosis and enhances overall tumor growth (O’Donnell et al. 2005). Indeed, in the accompanying study by He et al. (2005), miR-17–92 was shown to cooperate with MYC in inducing neoplastic transformation of B-cell progenitors.
Moreover, a subsequent study from the Mendell and Thomas-Tikhonenko laboratories showed that, in addition to MYC-activated microRNAs (which could, in principle, account for a wealth of MYC-repressed genes), there exist MYC-repressed microRNAs (which could account for equally numerous MYC-activated genes) (Chang et al. 2008). Interestingly, in a B-lymphoma cell line, MYC-repressed (but not MYC-stimulated) genes were highly enriched for predicted MYC-regulated microRNA seed sequences (Psathas et al. 2013). This suggested that gene repression by MYC was effected in large part (and more so than gene activation) through microRNA-mediated mechanisms.
In the following pages, we will present evidence that microRNA-dependent gene regulation by MYC accounts for many of its “target” genes and, most importantly, for key MYC-driven phenotypes. We will preferentially cite and discuss papers that place microRNA-driven events in the context of MYC-associated events and, thus, this should not be construed as a comprehensive survey of cancer-related microRNAs; for that, we refer the reader to other review articles (Croce 2009; Sotillo and Thomas-Tikhonenko 2011). Additionally, most of the papers cited herein focus on phenotypes driven by MYC overexpression, which is often observed in cancer. Although the same regulatory pathways are likely to be in place in normal cellular contexts, how endogenous MYC regulates microRNA expression remains a subject for further studies.
CELL CYCLE AND MITOTIC DIVISION
Precise regulation of cell-cycle progression ensures genetic fidelity during mitotic division. These regulatory decisions incorporate diverse extracellular and cell-intrinsic signals. If DNA damage is sensed or replication is incomplete, cells will enter cell-cycle arrest or apoptosis to avert the propagation of mutations. Aberrant regulation of the cell cycle, either through mutations or deregulated gene expression, is a hallmark of cancer development. To prevent this, cells have evolved extensive cell-cycle checkpoints that verify whether each phase has concluded properly and completely before progression to the next phase. The balance between cell-cycle progression and arrest is dependent chiefly on the equilibrium of cyclins, cyclin-dependent kinases (CDKs), and cyclin-dependent kinase inhibitors (CDKIs) (Malumbres and Barbacid 2009). The establishment of multiple, independent checkpoints preclude a single alteration to the checkpoint machinery from driving uncontrolled division. However, deregulation of a single factor that controls multiple aspects of the cell cycle in concert could alter this balance.
Of all the biological processes that MYC regulates, cell-cycle progression and mitotic division have been studied most extensively and conclusively. Rat-1 fibroblasts with homozygous deletion of MYC were viable, but significantly impaired in mitosis (Mateyak et al. 1997) and this defect could be rescued by other MYC-family members (Landay et al. 2000), but by virtually no other genes, with the possible exception of SHMT2 (Nikiforov et al. 2002). Early efforts to characterize this regulation focused on transcriptional mechanisms; as a result, CDK-4 and the CDK tyrosine phosphatase Cdc25A were identified as MYC targets (Galaktionov et al. 1996; Hermeking et al. 2000). However, reconstitution of the cyclin D-CDK-4/-6 complex in MYC null cells minimally affected cell-cycle regulation (Mateyak et al. 1999). MYC must therefore regulate the cell cycle at multiple independent points and through multiple targets. This idea is consistent with a microRNA-dependent mechanism.
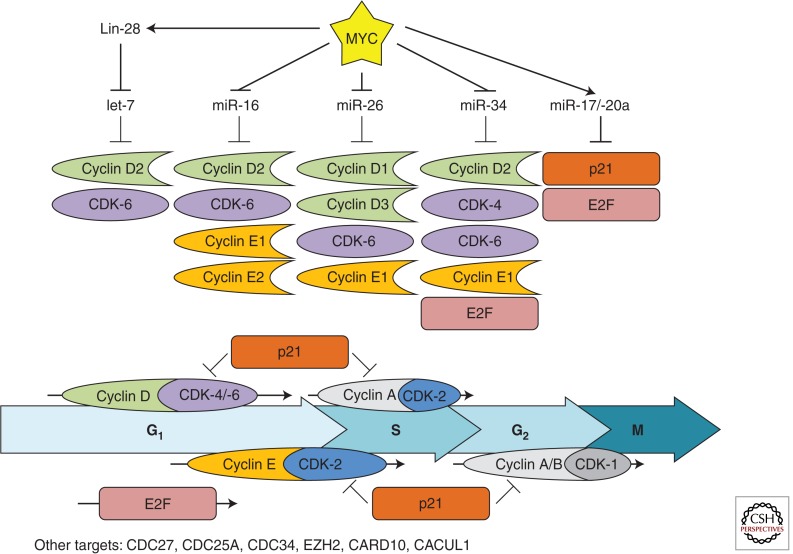
Indeed, 5 years ago we found that, in MYC-driven lymphomas, restoring expression of MYC-repressed microRNAs conferred significant growth disadvantage in competition assays (Chang et al. 2008). Briefly, murine B-lymphoma cells were retrovirally infected with constructs expressing MYC-repressed microRNAs and green fluorescent protein (GFP), and equal numbers of noninfected- and microRNA-expressing cells were subcutaneously injected into severe combined immunodeficiency mice. We observed a significant underrepresentation of cells expressing MYC-repressed microRNAs, as measured by enumerating GFP-positive cells. This was in contrast to the very mild effects of CDK-4 deficiency on general cell proliferation (save for specific endocrine cell types) (Malumbres et al. 2004). How do MYC-repressed microRNAs affect cell division so potently? As one would expect, certain MYC-repressed microRNAs (miR-34a, miR-15a/-16, miR-26, and let-7) have been shown to specifically target positive regulators of the cell cycle (Fig. 2).
Figure 2.
MYC-regulated microRNAs and their cell-cycle-related targets. The MYC-stimulated cyclin D1/2/3 (green), cyclin E1/2 (orange), and CDK-4/-6 (purple), and the MYC-repressed p21 (red) are shown. E2F (pink) is both MYC-stimulated and -repressed. Each stage of the cell cycle regulated by these targets is represented and the coordinated regulation of these targets promotes cell-cycle progression.
miR-34
Overexpression of miR-34 family members has been shown to cause cell-cycle arrest in multiple cell lines (He et al. 2007; Tarasov et al. 2007; Sun et al. 2008). This phenotype is, at least in part, mediated through simultaneous repression of CCND1, CCNE2, CDK-4, and CDK-6, positive regulators of cell-cycle progression (Matsushime et al. 1992; Kato et al. 1993; Quelle et al. 1993; Bates et al. 1994; Meyerson and Harlow 1994; Lauper et al. 1998). Of note, miR-34 was more effective at driving cell-cycle arrest than knockdown of the individual targets, supporting a model of cooperativity. Furthermore, miR-34a has been shown to down-regulate E2F protein levels in human colon cancer cells, although a direct targeting mechanism has yet to be established (Tazawa et al. 2007).
miR-15a/-16
The miR-16 family of microRNAs is comprised of miR-15a/b, miR-16, miR-195, miR-424, and miR-427. Levels of the miR-15a and miR-16 microRNAs, encoded by the DLEU2 locus, fluctuate dynamically during cell-cycle progression (Rissland et al. 2011) and are integral to cell-cycle regulation. The deregulation of these microRNAs has been implicated in multiple cancers: DLEU2 is often found to be deleted in B-cell chronic lymphocytic leukemias (B-CLL) (Calin et al. 2002) and is down-regulated in B-cell lymphomas by MYC through the recruitment of HDAC3 (Zhang et al. 2012). When restored, miR-16 family members inhibit the G1/S transition by targeting several positive regulators of the cell cycle. Specifically, CCND1, CCND3, CCNE1, and CDK-6 were directly targeted by miR-16 in various cell lines (Linsley et al. 2007; Liu et al. 2008). Additionally, CARD10, CDC27, and CACUL1 were down-regulated after transfection of miR-16 into Dicer-deficient HCT116 cells (Linsley et al. 2007). Importantly, the cell-cycle arrest phenotype required the targeting of multiple cell-cycle regulators to achieve the same degree of arrest driven by miR-16.
miR-26
Several groups have established miR-26 as a regulator of the cell cycle. In vitro, overexpression of miR-26 in hepatocellular carcinoma (HCC) cells (which express low levels of miR-26) led to cell-cycle arrest, in part, through repression of CCND2 and CCNE2 (Kota et al. 2009). Furthermore, the investigators showed the therapeutic efficacy of miR-26 overexpression in a MYC-driven mouse model of HCC (Felsher and Bishop 1999). This efficacy is likely due to the array of cell-cycle regulators targeted by miR-26. In addition to CCND2 and CCNE2, miR-26 directly repressed CCNE1, CDK-6, and EZH2, and induced cell-cycle arrest in cell lines of various origin (Sander et al. 2008; Zhu et al. 2012).
let-7
Whereas the regulation of most microRNAs by MYC is transcriptional (Chang et al. 2008), MYC-mediated repression of let-7 is unconventional. MYC stimulates the expression of Lin-28b (Chang et al. 2009), which binds the pri-let-7 hairpin and prevents processing by Drosha in the nucleus and Dicer in the cytoplasm (Heo et al. 2008; Newman et al. 2008; Viswanathan et al. 2008). Inhibition of let-7 leads to increased proliferation (Legesse-Miller et al. 2009) and concordantly, overexpression of mature let-7 results in cell-cycle arrest (Johnson et al. 2007). The mechanisms driving these phenotypes involve let-7 directly repressing several positive regulators of the cell cycle including CCND2, CDK-6, CDC25A, and CDC34 (Johnson et al. 2007; Legesse-Miller et al. 2009). Additionally, targeting of high mobility group protein A2 (Hmga2) by let-7 is known to limit anchorage-independent growth, most likely by a cell-cycle-dependent mechanism (Mayr et al. 2007).
miR-17–92
In contrast to MYC-repressed microRNAs, the MYC-stimulated miR-17, miR-20a (Ota et al. 2004), and miR-106a/b (O’Donnell et al. 2005; Zhao et al. 2012) directly target CDKN1A (p21), a negative regulator of the cell cycle (Ivanovska et al. 2008). In human mammary epithelial cells, individual transfection of these microRNAs resulted in cell-cycle progression similar to anti-CDKN1A small interfering RNA (Ivanovska et al. 2008). This regulation is functionally important during transformation; repression of CDKN1A by miR-17 or miR-20 was necessary to avoid oncogene-induced senescence in Ras-expressing fibroblasts (Hong et al. 2010). The E2F family of transcription factors that regulate cell-cycle progression is also targeted by miR-17 and miR-20a (O’Donnell et al. 2005; Sylvestre et al. 2007). In a reciprocal relationship, E2F family members stimulate expression of the miR-17–92 cluster creating a negative feedback loop (Sylvestre et al. 2007; Woods et al. 2007). MYC, therefore, both stimulates (miR-17/-20a) and represses (miR-34a) microRNAs that target E2F. The cumulative effect of these relationships on cell-cycle progression may be context-dependent and requires further exploration; but, overall, MYC represses microRNAs that prevent cell-cycle progression and induces microRNAs that target cell-cycle inhibitors.
APOPTOSIS
Cellular stresses resulting in DNA damage can lead not only to cell-cycle arrest but also to apoptosis—if the surveillance machinery deems the damage irreparable. The apoptotic signaling cascade is efficient, irreversible, and poised to kill a single cell for survival benefit to the organism. Regulation of apoptosis is complex and depends on the antagonistic balance between pro- and antiapoptotic factors. Despite robust MYC-driven effects on cell-cycle progression, MYC overexpression alone is apparently insufficient to transform cells, most likely because MYC has intrinsic proapoptotic activities, both p53 dependent and independent (Shortt and Johnstone 2012).
Mechanisms of p53-dependent apoptosis by MYC are well understood and generally involve activation by MYC of p19Arf (Zindy et al. 1998). Arf then counteracts Mdm2, an E3 ubiquitin ligase that normally targets p53 for proteasomal degradation (Haupt et al. 1997; Honda et al. 1997; Kubbutat et al. 1997). Additionally, MYC is known to directly activate the tp53 promoter (Reisman and Rotter 1993) as well as affect the expression (Ceballos et al. 2005) and phosphorylation of Mdm2 (Rogulski et al. 2005). In contrast, mechanisms of p53-independent apoptosis by MYC are not well understood. In some cases, they might incorporate p53-independent effects of p19Arf (Qi et al. 2004; Boone et al. 2011). However, the Ink4a locus, which encodes Arf, is frequently deleted, particularly in B-cell lymphomas. Other key mediators of MYC-driven p53-independent apoptosis are Noxa (Nikiforov et al. 2007; Nawrocki et al. 2008; Qing et al. 2012) and Bim, a well-recognized direct MYC target (Hemann et al. 2005).
Induction of apoptosis is detrimental for the transforming activity of MYC and in mouse models slows down tumorigenesis by necessitating additional antiapoptotic events such as Bcl-2/Bcl-xL overexpression (Strasser et al. 1990; Pelengaris et al. 2002). Thus, one would surmise that genetic selection in tumors would yield a mechanism that uncouples proliferation and apoptosis. Indeed, Hemann et al. (2005) showed that a common mutant MYC allele found in Burkitt lymphoma does just that. Specifically, Thr-58 (or alternatively, Pro-57) mutants retain the ability to stimulate proliferation and activate p53, but cannot up-regulate Bim and thus do not induce apoptosis in the mouse lymphoma models. In Burkitt lymphoma, p53 inactivation and these MYC mutations (themselves associated with low Bim levels) were mutually exclusive (Hemann et al. 2005). However, the mechanism by which MYC activates, or fails to activate, apoptosis remained to be identified (Dang et al. 2005). Could this uncoupling mechanism be microRNA-dependent?
It is not presently known whether Thr-58 MYC mutants are more adept at regulating microRNAs. However, in our recent paper, we observed that overexpression of MYC due to the strengthening of the PI3K-Akt axis and ensuing inhibition of GSK-3β resulted in a more pronounced microRNA deregulation (Chung et al. 2012). Because GSK-3β is the enzyme that phosphorylates Thr-58, it could be argued that nonphosphorylation of this residue (because of either mutation or GSK-3β inhibition) would cause more robust up-regulation of miR-17–92 and more robust down-regulation of miR-34a and miR-15/-16 (Fig. 3 illustrates how this would affect apoptosis).
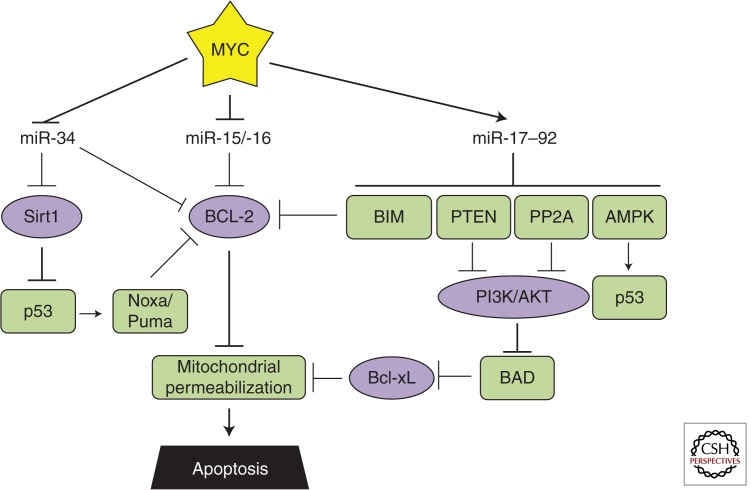
Figure 3.
MYC-regulated microRNAs and apoptosis. Relevant MYC-repressed (green) and -stimulated (purple) targets are shown. The coordinated regulation of these targets inhibits apoptosis.
miR-17–92
As mentioned above, the MYC-stimulated miR-17–92 cluster of microRNAs was identified as an oncogene by virtue of its ability to repress apoptosis in a murine B-cell lymphoma model (He et al. 2005). Subsequent analyses have identified the two dominant effectors responsible for the decreased apoptosis, Bim and PTEN (Xiao et al. 2008). Bim, which inhibits the antiapoptotic Bcl-2, is down-regulated by miR-17–92 during normal development to promote survival of B-cell progenitors (Ventura et al. 2008). However, in adult mice, constitutive overexpression of miR-17–92 leads to lymphoproliferative disease, and heterozygous deletion of the proapoptotic Bim and PTEN partially mimicked this effect of miR-17–92 (Xiao et al. 2008). Subsequently, two other studies identified the miR-19 family of microRNAs as the key oncogenic component of miR-17–92 in Eμ-MYC lymphomas, both converging on PTEN as the functionally important target (Mu et al. 2009; Olive et al. 2009). Its importance stems from the ability of PTEN to inhibit PI3K and subsequent Akt phosphorylation, an event that promotes survival in a signaling cascade involving BAD, Bcl-xL, and mitochondrial membrane permeabilization (Yang et al. 1995; Datta et al. 1997; Billen et al. 2008).
One year later, a screen to identify the oncogenenic component of miR-17–92 in T-cell acute lymphoblastic leukemia also implicated miR-19 (Mavrakis et al. 2010). The investigators transduced IL-3-dependent lymphocytes with individual members of the cluster before induction of apoptosis by IL-3 withdrawal. miR-19 was the only cluster member whose overexpression allowed for escape from apoptosis. In addition to Bim and PTEN, the investigators identified PP2A (Ppp2r5e) and AMPK (Prkaa1) as being directly targeted by miR-19. The PP2A phosphatase acts on phospho-Akt and was shown to contribute to miR-19-associated antiapoptotic phenotype. In contrast, the investigators were unable to observe an antiapoptotic effect after AMPK knockdown alone, despite its known roles in p53 activation. Perhaps the coordinated down-regulation of all four miR-19 targets is needed to confer the greatest survival advantage.
miR-15a/-16
Bcl-2 inhibits mitochondrial membrane permeabilization and is a central node in apoptotic signaling. Not surprisingly, multiple MYC-microRNA regulatory pathways converge on Bcl-2. In addition to its activation through the miR-17–92/Bim/Bcl-2 axis, MYC represses the miR-15a/-16 cluster that targets Bcl-2 directly. Thus, there are multiple MYC-regulated microRNAs that could increase endogenous Bcl-2 levels. Nevertheless, at least in MYC-driven B-lymphoid malignancies, ectopic reexpression of Bcl-2 enhances neoplastic growth (Strasser et al. 1990), suggesting that transcriptional modulation of miR-17–92 and -15a/-16 levels may not be sufficient to evade apoptosis and copy number variation in the corresponding genes might be required for a highly malignant phenotype.
Indeed, in chronic lymphocytic leukemia, Bcl-2 is up-regulated following miR-15a/-16 loss (Calin et al. 2002) and deletion of the DLEU2 locus in mice also results in CLL-like disease (Klein et al. 2010). Consistent with these findings, transfection of miR-15a/-16 expression constructs into a leukemia-derived cell line-induced apoptosis (Cimmino et al. 2005), at least in part because of Bcl-2 repression. The investigators also identified a strong negative correlation between Bcl-2 and miR-15a/-16 expression in 26 CLL samples providing physiological relevance. Interestingly, miR-15a/-16 is also reported to target p53, suggesting a complex interplay between genetic and microRNA-mediated events in CLL (Fabbri et al. 2011).
miR-34
MYC-mediated repression of miR-34 can promote or inhibit apoptosis in a context-dependent manner. Several studies established the apoptosis-inducing ability of miR-34 in a variety of human cancer cell lines (lung, breast, colon, osteosarcoma) as well as mouse embryo fibroblasts (MEFs) (Chang et al. 2007; Raver-Shapira et al. 2007; Tarasov et al. 2007). This is thought to occur downstream of p53, which transcriptionally regulates the miR-34 family members (He et al. 2007). Additionally, miR-34 induces apoptosis by repressing the Sirt1 deacetylase (Yamakuchi et al. 2008), which inactivates p53 through deacetylation of lysine-382. This results in decreased expression of the p53 targets Puma and Noxa, both of which repress Bcl-2. miR-34 also directly targets Bcl-2 (Bommer et al. 2007).
Counter to these proapoptotic roles, miR-34 also harbors antiapoptotic activities in certain contexts. This is because miR-34 directly targets MYC for repression (Kong et al. 2008; Christoffersen et al. 2010) and in cells with the intact MYC-ARF-MDM2-p53 pathway, miR-34 overexpression actually reduced MYC levels and p53 stability (Sotillo et al. 2011). This correlated with decreased sensitivity to chemotherapeutic agents. Similar chemoprotective effects of miR-34 family members have been described in other tumor types as well (Catuogno et al. 2013). Hence, as manipulation of microRNA levels is being developed into therapies, the genetic context of the malignancy must be considered very carefully.
METABOLISM
With each division, a cell must double its biomass and genetic material. The accelerated rate of cell division in cancers results in extraordinary metabolic demand and therefore requires metabolic adaptation. Otto Warburg first observed the altered cellular metabolism of cancers 80 years ago. Relative to normal tissue, glucose uptake and lactate production were increased in rapidly dividing cancers and this aerobic glycolysis is now known as the Warburg effect (Vander Heiden et al. 2009). In support of this model, several oncogenes, including MYC, are capable of reprogramming the cell's metabolic pathways.
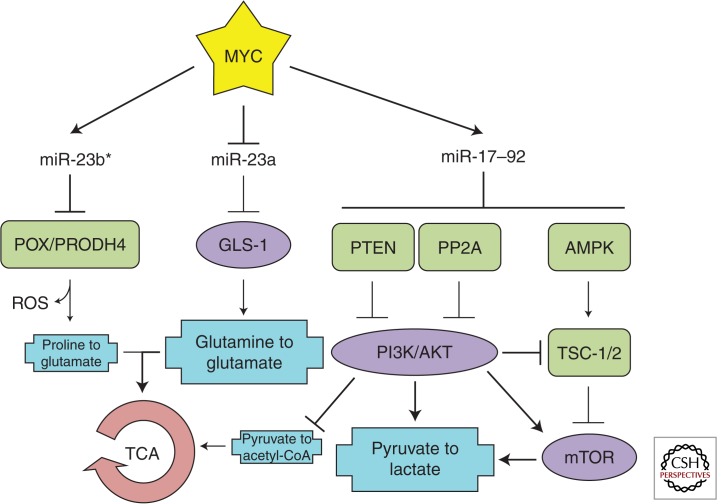
With regard to the Warburg effect, MYC promotes multiple aspects of aerobic glycolysis. This conversion involves the transport of glucose into the cytoplasm (via glucose transport proteins, or GLUTs), enzymatic conversion of glucose to pyruvate (via HK2, PFK1, PFK2, and PKM2), and, finally, using pyruvate to make lactate (via PDK1 and LDHA) instead of acetyl-CoA, which enters the TCA cycle. To be sure, MYC transcriptionally regulates several of these genes: GLUT1, HK2, PKM2, PDK1, and LDHA (Dang et al. 2009). However, it has not been established whether (typically mild) up-regulation of these genes by MYC is physiologically relevant. Given that regulation of metabolism is complex and involves multiple feedback and feed-forward loops, one can argue that there might exist a more systemic, pathway-wide approach to metabolome reprogramming (Fig. 4).
Figure 4.
MYC-regulated microRNAs and metabolic alterations. Relevant MYC-repressed (green) and -stimulated (purple) targets are shown. Important metabolic processes (blue) are depicted. The coordinated regulation of these targets promotes aerobic glycolysis and uses glutamine for the tricarboxylic acid cycle (TCA) cycle.
miR-17–92
In addition to conventional transcriptional regulation by MYC (e.g., of GLUT1) (Osthus et al. 2000), regulation of aerobic glycolysis has an miR-17–92 component. As mentioned above, the MYC-stimulated miR-19a/b directly represses PTEN and PP2A leading to increased Akt phosphorylation (Mu et al. 2009; Olive et al. 2009; Mavrakis et al. 2010). Phosphorylation of Akt stimulates glycolysis through multiple mechanisms: by activating PFK1 and PFK2, increasing expression of several GLUTs, and stimulating mTORC1 (reviewed in Robey and Hay 2009), a major facilitator of glycolysis and cell growth (Yecies and Manning 2011). Furthermore, miR-19 also directly represses AMPK (Mavrakis et al. 2010), an inhibitor of mTOR activity (Bolster et al. 2002). Thus, as far as the promotion of aerobic glycolysis is concerned, the main strength of MYC lies not only in its repertoire of direct targets, but in cunning use of microRNAs to stimulate multiple components of the Akt and mTOR pathways.
miR-23a/b-23b*
In addition to the systemic effects of miR-17–92, some MYC-regulated microRNAs could still work by a one-target-at-a-time mechanism. A consequence of aerobic glycolysis is reduced levels of acetyl-CoA entering the TCA cycle. In the absence of this substrate, cancer cells must find alternative energy sources to power the mitochondria. Glutamine catabolism (to glutamate and, subsequently, α-ketoglutarate) can fulfill this requirement (Daye and Wellen 2012). In fact, several MYC-overexpressing cancer cell lines display glutamine dependency (Wise et al. 2008; Qing et al. 2012). Although glutaminase (GLS-1), the enzyme responsible for the conversion of glutamine to glutamate, is not a direct MYC target, MYC has found a way to stimulate its expression by repressing miR-23a and miR-23b, which directly target the GLS-1 3′ UTR (Gao et al. 2009). The fact that glutamine deprivation slowed down proliferation more than glucose deprivation highlights the importance of this MYC-microRNA pathway in cancer.
The contribution of the miR-23b cluster to metabolomic changes could be even more complex. While investigating the repression of GLS-1, Gao and colleagues established that MYC binds the miR-23b promoter and repressed its expression (Gao et al. 2009). In contrast, Liu and colleagues found that MYC up-regulated miR-23b* (Liu et al. 2012). The mechanism behind this differential regulation of opposite strands remains unclear. The investigators showed that stimulation of miR-23b* by MYC resulted in altered proline metabolism. miR-23b* directly inhibited POX/PRODH, the enzyme responsible for the catabolism of proline to Δ1-pyrroline-5-carboxylate. How exactly this shift away from glutamate and toward proline biosynthesis affects the energy requirements of cancer cells has not been established. It does, however, reinforce the importance of the MYC-miR-23a/b-GLS-1 regulatory pathway in maintaining sufficient glutamate levels for the TCA cycle. Additionally, there are clear implications for cell survival. POX/PRODH activity results in the creation of reactive oxygen species (ROS), which can trigger apoptosis (Donald et al. 2001). By stimulating miR-23b* and repressing POX/PRODH, MYC simultaneously alters proline/glutamate metabolism and limits the ROS by-product.
METASTASIS AND ANGIOGENESIS
As tumors grow beyond the size of several cubic millimeters, even metabolic reprogramming is unable to satisfy the increased energy demands. Angiogenic growth then becomes necessary to provide greater access to oxygen and nutrients. Angiogenesis also provides a conduit for metastatic spread, alleviating the spatial limitations of the current tumor microenvironment. Tumors induce angiogenesis by secreting proangiogenic factors (such as vascular endothelial growth factor, or VEGF) that elicit new vascular growth from preexisting endothelial cells. Opposing this are several endogenous antiangiogenic factors that function by directly inhibiting proangiogenic factors or initiating antiangiogenic signaling (Bergers and Benjamin 2003). Oncogenic drivers of tumorigenesis shift this equilibrium toward angiogenesis, allowing unfettered tumor growth (Rak and Yu 2004).
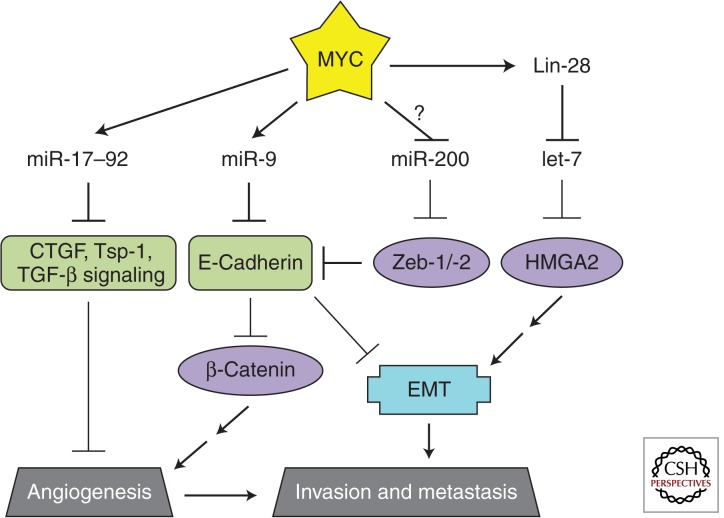
Metastasis, too, is a complex process involving multiple stages: epithelial to mesenchymal transition (EMT), invasion of tumor cells into the surrounding tissue, entry into the vasculature, translocation through the circulatory system, extravasation, formation of micrometastases, and, finally, expansion into macrometastases. At each step, barriers exist that must be overcome. Chief among them is E-cadherin, a transmembrane protein that plays an important role in cell adhesion of adjacent epithelial cells. As such, it acts as a barrier to cell migration and invasion, two components of metastasis (Chambers et al. 2002). Additionally, E-cadherin represses β-catenin signaling and, as a result, expression of the proangiogenic VEGF (Gottardi et al. 2001; Wong and Gumbiner 2003; Skurk et al. 2005; Ceteci et al. 2007). Deregulation of MYC has been implicated in both angiogenesis and metastasis with MYC-regulated microRNAs playing an integral role (Fig. 5).
Figure 5.
MYC-regulated microRNAs during angiogenesis and metastasis. Relevant MYC-repressed (green) and -stimulated (purple) targets are shown. The EMT (blue) promotes metastasis. The coordinated regulation of these targets promotes angiogenesis and metastasis.
miR-9
Recently, the MYC-stimulated miR-9 was shown to directly repress E-cadherin (Ma et al. 2010). Using overexpression and inhibition approaches, Ma and colleagues showed the proangiogenic and prometastatic properties of miR-9 in breast cancer models. miR-9 overexpression in previously nonmetastatic breast cancer cells led to micrometastases in mice. The primary tumors were 10-fold more vascularized and allowed for increased invasion and subsequent metastases. Conversely, inhibition of miR-9 using a “microRNA sponge” (which outcompetes endogenous targets for binding to miR-9) decreased lung metastases from implanted highly metastatic mouse mammary cancer cells. MYC and its family member N-MYC directly stimulated miR-9 expression in breast cancer and neuroblastoma models, respectively. Thus, by repressing E-cadherin, the MYC-stimulated miR-9 promotes several aspects in the development of metastatic disease: angiogenesis, EMT, invasion, and formation of micrometastases. Perhaps not surprisingly, a strong correlation between N-MYC, miR-9, tumor grade, and metastatic status was observed in human neuroblastomas providing clinical relevance (reviewed in Khew-Goodall and Goodall 2010).
let-7
The EMT is coordinated by several factors (Snail, Twist, Slug) that are regulated by HMGA2, a chromatin remodeling protein. In two back-to-back studies, HMGA2 was characterized as a target of the let-7 family of microRNAs (Lee and Dutta 2007; Mayr et al. 2007). The first study showed that mutation of let-7 binding sites in the HMGA2 3′ UTR increased oncogenic transformation as measured by soft agar assay (Mayr et al. 2007). Similarly, expression of HMGA2 without its 3′ UTR partially relieved let-7-mediated growth inhibition in lung cancer cell lines (Lee and Dutta 2007). Investigation into the upstream regulation of let-7 and the downstream effectors of HMGA2 unveiled a MYC-repressed, antimetastatic-microRNA program in breast cancer cell lines (Dangi-Garimella et al. 2009). Specifically, upstream signaling events converge on MYC to sequentially stimulate LIN28B, repress let-7, and therefore derepress HMGA2. HMGA2 derepression resulted in up-regulation of Snail, increased invasiveness in vitro, and increased bone metastases in vivo. Additionally, LIN28B was shown to promote metastasis of colon (King et al. 2011) and ovarian cancers (Helland et al. 2011); in the latter case, MYCN seems to be directly responsible for LIN28B deregulation.
miR-200
Another well-established microRNA regulator of the EMT is miR-200. The miR-200 family of microRNAs directly targets the 3′ UTRs of both Zeb-1 and Zeb-2, transcription factors that inhibit E-cadherin expression (Bracken et al. 2008; Gregory et al. 2008; Korpal et al. 2008; Park et al. 2008). Knockdown of miR-200 in epithelial cells resulted in an EMT that was dependent on up-regulation of Zeb-1 and Zeb-2. Conversely, overexpression of miR-200 in mesenchymal cells led to a mesenchymal to epithelial transition. Invasive breast cancer cell lines have been shown to have lost miR-200 expression and miR-200 correlates with E-cadherin levels in ovarian cancers. Although data demonstrating a causal link between MYC and the miR-200/Zeb-1/EMT pathway has yet to be established, a screen for MYC-regulated microRNAs in stem cells indicated that miR-200 is repressed by MYC in murine lymphomas (Lin et al. 2009; further discussed in the Stemness section).
miR-17–92
The proangiogenic properties of miR-17–92 were first identified in a study investigating the role of MYC in angiogenesis. Introduction of MYC into Ras-transformed colonocytes (RasMyc) increased tumor growth threefold over the parental Ras transformed cells, and analysis of the tumors revealed that RasMyc tumors were highly vascularized (Dews et al. 2006). This difference was, in part, mediated by miR-17–92 through the repression of the antiangiogenic factors Tsp-1 and CTGF. miR-19a/b and miR-18a directly target the THBS1 (Sundaram et al. 2011) and CTGF (Ernst et al. 2010; Fox et al. 2013) 3′ UTRs, respectively. The angiogenic effects of miR-17–92 expression were also imposed through the repression of the TGF-β signaling pathway. Direct targeting of TGFBR2 by miR-17/-20a and SMAD4 by miR-18a resulted in decreased responsiveness to TGF-β (Dews et al. 2010). Tsp-1, CTGF, and clusterin were among the TGF-β responsive genes whose expression was reduced in the presence of exogenous miR-17–92. Thus, the miR-17–92 cluster is a potent activator of angiogenesis; it directly represses antiangiogenic factors (Tsp-1 and CTGF) while also targeting the antiangiogenic TGF-β signaling pathway to exert indirect repression (clusterin, Tsp-1, and CTGF). A similar pattern of regulation is driven by NMYC in neuroblastoma (Mestdagh et al. 2010). Of note, TGF-β signaling can induce the EMT (reviewed in Katsuno et al. 2013). MYC therefore trades invasion for angiogenesis by inducing miR-17–92, and perhaps balances the repression of TGF-β signaling by stimulating miR-9 and repressing let-7.
In contrast to these proangiogenic properties, a study examining the effects of miR-17–92 overexpression in endothelial cells established a cell-intrinsic antiangiogenic role for the cluster, in particular, miR-92 (Doebele et al. 2010). How the context (tumor-initiated paracrine signaling vs. cell intrinsic effects) of miR-17–92 expression affects angiogenesis requires further investigation.
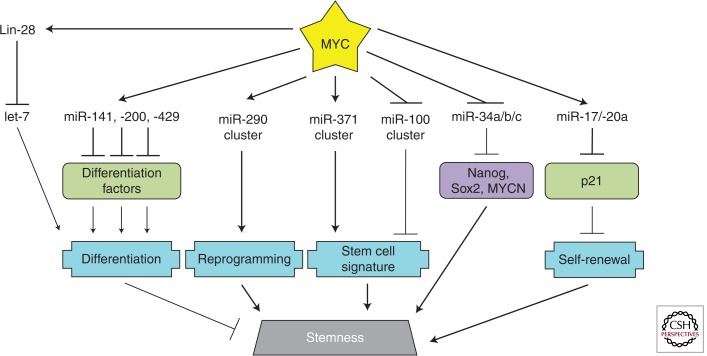
STEMNESS
Of all biological phenomena that MYC regulates, stemness is the most recently identified and least well characterized (see Chappell and Dalton 2013). The stemness-promoting potential of MYC was most notably described by Takahashi and Yamanaka in their seminal paper describing four factors (Oct4, Sox2, Klf4, and MYC) used to transcriptionally induce pluripotent stem cells (Takahashi and Yamanaka 2006). Although only two of the Yamanaka factors, Oct4 and Sox2, were indispensable for this transition (Yu et al. 2007), the stemness-driving potential of MYC came into focus. Rather than using Klf4 and MYC, Yu and colleagues substituted Nanog and (the MYC-stimulated) Lin-28; this naturally led to the suggestion that MYC-mediated stemness was conferred through the repression of the let-7 family of microRNAs. A direct link between MYC, Lin-28, let-7, and stemness has yet to be established; however, let-7 has been implicated in the balance of stemness and differentiation (Melton et al. 2010; Zhong et al. 2010). Could other MYC-regulated microRNAs be relevant as well? (see Fig. 6.)
Figure 6.
MYC-regulated microRNAs in stem cells. Relevant MYC-repressed (green) and -stimulated (purple) targets are shown. Important processes in stem cell biology (blue) are depicted. The coordinated regulation of these targets promotes stem-cell-like properties.
miR-290 Cluster
The microRNAome of stem cells is dissimilar from that of differentiated cells. Up to 70% of microRNAs in mouse embryonic stem cells (ESCs) are derived from the miR-290 cluster (Marson et al. 2008). Within this cluster are the miR-291–3p, miR-294, and miR-295 microRNAs (all of the same seed family) that have been established as positive regulators of the cell cycle (Wang et al. 2008). In an effort to understand the role of these microRNAs in stem cell maintenance/reprogramming, it was shown that introduction of Oct4, Sox2, Klf4, and miR-294 (rather than MYC) could reprogram MEFs (Judson et al. 2009). Both c-MYC and N-MYC were found to bind the promoter of the miR-290 cluster and MYC was required to transcriptionally activate the cluster during reprogramming of MEFs. This implicated the miR-290 cluster as a powerful downstream effector of MYC and provided the first concrete link between MYC, microRNAs, and stemness. The investigators did, however, observe differences among the induced pluripotent stem (iPS) cells from the miR-294 and MYC-transformed MEFs, possibly caused by other MYC-regulated microRNAs.
miR-141, miR-200, and miR-429
Shortly thereafter, the MYC-microRNA-stem cell connection was strengthened in a screen designed to identify microRNAs regulated by MYC exclusively in ESCs (Lin et al. 2009). The investigators compared MYC-overexpressing ESCs to three other conditions: ESC-derived-induced hematopoietic stem/progenitor cells (HSPs), HSP-derived tumors from transplanted mice, and, finally, MYC-knockdown ESCs. This approach allowed ESC-specific MYC-regulated microRNAs to be distinguished from those microRNAs regulated by MYC in many other cell contexts. In ESCs, miR-141, miR-200, miR-338, and miR-429 were shown to be directly activated by MYC (in contrast to MYC-driven tumors in which they are repressed). In functional assays, transfection of miR-141, miR-200, and miR-429 microRNAs result in reduced differentiation of the ESCs. Conversely, inhibitors targeting these microRNAs enhanced differentiation. Through transfection with synthetic microRNAs and microarray profiling, the investigators identified a host of microRNA-targeted genes involved in differentiation pathways. This model, in which MYC regulates several microRNAs to target multiple genes within a single pathway, provides a robust regulatory network that can efficiently drive phenotypes.
miR-371 and miR-100 Clusters
The human miR-371 cluster of microRNAs, the murine miR-290 cluster homolog, is also activated by MYC. This relationship was initially identified in hepatoblastomas, a rare malignant neoplasm arising from liver precursor cells (Cairo et al. 2010). MYC has been implicated in a subclass of particularly aggressive and undifferentiated hepatoblastomas. Also characterized was the repression of the miR-100/let-7a-2/miR-125b-1 cluster by MYC. Activation of the miR-371 cluster together with repression of the miR-100 cluster constituted a MYC-driven stem-cell-like microRNA signature. This signature was also observed in aggressive HCCs. Considering the link between MYC, the miR-290 cluster, and stemness in murine ESCs, it is possible that MYC deregulation in human liver precursor cells could result in up-regulation of miR-371 and the undifferentiated phenotype of these aggressive hepatoblastomas. Emerging evidence suggests that tumors are initiated and maintained by a population of cancer stem cells (CSCs) (discussed in Gupta et al. 2009). In support of this, the investigators show that reversal of the MYC-driven stem-cell-like microRNA signature in HCC cell lines led to decreased colony growth in soft agar assays and smaller tumors when injected into immunodeficient mice (Cairo et al. 2010).
miR-34
After the initial discovery that Yamanaka factors could induce reprogramming of somatic cells into iPS cells, questions remained regarding how to increase the efficiency and rate of reprogramming. Five groups simultaneously established p53 as a major roadblock to reprogramming (Hong et al. 2009; Kawamura et al. 2009; Li et al. 2009; Marion et al. 2009; Utikal et al. 2009). Inactivation of p53 significantly enhanced the generation of iPS cells by the Yamanaka factors compared to the factors alone; however, self-renewal and subsequent differentiation of these iPS cells was negatively affected (Hong et al. 2009; Marion et al. 2009). Activation of p21 and miR-34 are two of the mechanisms by which p53 inhibits reprogramming (Hong et al. 2009; Li et al. 2009; Choi et al. 2011). p21 effects were mediated mainly through cell proliferation whereas miR-34 directly targets Nanog, Sox2, and N-MYC, all of which are important in maintaining stemness (Choi et al. 2011). miR-34−/− iPS cells had similar self-renewal and differentiation capabilities relative to wild-type iPS cells, but were generated with increased efficiency (Choi et al. 2011). In contrast to p53, MYC indirectly represses p21 (through stimulation of miR-17–92, as discussed in the section on cell cycles above, and below in the context of CSCs). It also directly represses miR-34. The ability of MYC to counteract these p53-mediated reprogramming roadblocks likely contributes to its ability to drive stemness.
miR-17–92
Another line of evidence supports a role for MYC in the maintenance of CSCs. In mixed lineage leukemia (MLL), the MYC-stimulated miR-17 and miR-20a microRNAs target p21. Although the effects of this regulation on the cell cycle had been established (Ivanovska et al. 2008), another group explored how p21 loss affected leukemia stem cells (Wong et al. 2010). Overexpression of miR-17 and miR-20a in a mouse model of acute myeloid leukemia led to leukemic cells expressing increased c-Kit and decreased Mac-1, characteristic of leukemia stem cells. This phenomenon was attributed to repression of p21 and, subsequently, increased self-renewal of CSCs. Thus, during normal development, MYC plays a fundamental role in stemness and tissue homeostasis; however, its deregulation can result in cancer development through the maintenance of CSCs.
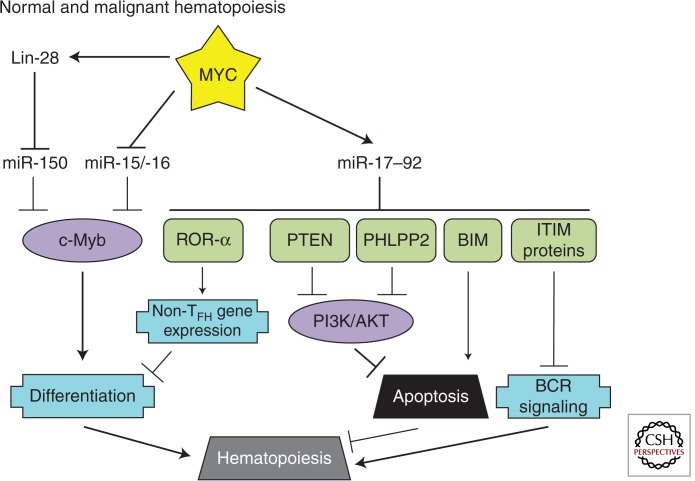
NORMAL AND MALIGNANT HEMATOPOIESIS
In addition to the phenotypes discussed above, MYC has been implicated in a variety of developmental processes. This is especially evident in lymphocyte development in which MYC functionally interacts with other transcription factors such as c-Myb (Fig. 7).
Figure 7.
MYC-regulated microRNAs in normal and malignant hematopoiesis. Relevant MYC-repressed (green) and -stimulated (purple) targets are shown. Important processes in hematopoiesis (blue) are depicted.
miR-150
c-Myb is of critical importance during early B-cell development as well as during the activation of mature B cells. miR-150, in contrast, is expressed specifically in mature B cells and directly targets c-Myb to repress this developmental factor once B-cell differentiation has occurred. Altering the normal expression of miR-150 or c-Myb results in B-cell differentiation defects (Xiao et al. 2007). The first evidence supporting a role for MYC in the miR-150/c-Myb regulatory pathway was the identification of miR-150 as a MYC-repressed microRNA (Chang et al. 2008). Interestingly, the processing of pri-miR-150, like pri-let-7, is repressed by the MYC-stimulated LIN28 (Chang et al. 2009). Therefore, in addition to its role in normal development, deregulation of MYC and the resultant block in miR-150 maturation is a driver of MLL-associated leukemia (Jiang et al. 2012). Another MYC-repressed cluster, miR-15a/-16, also targets c-Myb (Chung et al. 2008) demonstrating the important role of microRNAs in sustaining c-Myb levels (Zhao et al. 2009).
miR-17–92
The miR-17–92 microRNA cluster is another important regulator of lymphocyte development. As discussed in the Apoptosis section, overexpression of miR-17–92 in B cells leads to lymphoproliferative disorder caused by the repression of proapoptotic Bim (Xiao et al. 2008). During normal development, this repression is necessary for the pro-B- to pre-B-cell transition. Recently, several studies have identified additional mechanisms by which miR-17–92 regulates lymphocyte development and shown how aberrant miR-17–92 expression can have a causal role in lymphomagenesis.
Two recently published studies established that mice with a T-cell-specific deletion of miR-17–92 show defects in follicular helper T-cell (TFH cell) differentiation. As mediators of immune responses, mice with compromised TFH cell differentiation have reduced germinal-center formation, antibody production, and response to viral infection. In contrast, T-cell-specific miR-17–92 overexpression promoted TFH cell differentiation.
Multiple miR-17–92 targets are responsible for these phenotypes. In one study, the investigators observed that miR-17–92 null TFH cells expressed a subset of TFH cell-inappropriate genes due to up-regulation of ROR-α, a transcription factor normally targeted by all miR-17–92 cluster members. Reducing ROR-α expression in the miR-17–92 null T cells partially rescued TFH cell differentiation (Baumjohann et al. 2013). In the other study, miR-17–92 was shown to target the PHLPP2 and PTEN phosphatases. Both PHLPP2 and PTEN target the PI3K signaling pathway, which is important during T-cell activation and differentiation (Kang et al. 2013). Of note, repression of PHLPP2 and PTEN by miR-17–92 is also relevant in B-lymphomagenesis. Mice with B-cell-specific overexpression of miR-17–92 develop lymphomas. Furthermore, PHLPP2 and PTEN were identified as miR-17–92 targets in PAR-CLIP experiments designed to interrogate specific microRNA–mRNA interactions (Jin et al. 2013). These results show that a microRNA-target relationship can be required for normal development, but can also be oncogenic when deregulated.
Deregulation of phosphatases appears to be a common theme in miR-17–92-mediated signaling. We have recently shown that among its direct targets are several immunoreceptor tyrosine inhibitory motif-containing proteins (such as CD22 and FCGR2B) that normally recruit phosphatases of the SHIP/SHP family to the B-cell receptor (BCR). Consistent with this finding, either MYC or miR-17–92 expression was necessary to maintain phosphorylation of SYK and BLNK on ligation of the BCR. Further downstream, amplification of the BCR response by miR-17–92 resulted in enhanced calcium signaling and elevated levels of Myc itself constituting a feed-forward loop. Additionally, miR-17–92 levels were limiting to the BCR response of diffuse large B-cell lymphoma (DLBCL) cell lines and elevated in the BCR subtype of primary DLBCL (Psathas et al. 2013).
CONCLUDING REMARKS
MYC is an atypical transcription factor capable of both stimulating and repressing the expression of thousands of genes. Although in the P493-6 system both miR-17–92 and LIN28B are induced quite robustly, for the vast majority of protein-coding genes, the magnitude of regulation is often modest relative to conventional transcription factors (Dang et al. 2006; Eilers and Eisenman 2008). In this regard, MYC is strikingly similar to microRNAs, which are also capable of exerting modest repression on hundreds of target genes. Thus, the discovery that MYC regulates several microRNAs provided a new cornerstone in MYC biology and a mechanistic link between the two classes of gene regulators (O’Donnell et al. 2005; Chang et al. 2008).
In general, investigations of microRNA–target interactions fall into two broad categories. One is based on the assumption that in a given cell type, each microRNA has one or few essential targets that account for its effects on cell phenotypes. This concept held true for more than 20 years in the Caenorhabditis elegans system (Lee et al. 1993; Wightman et al. 1993) and has significant experimental support from mouse genetics studies. For example, in the context of normal B-cell differentiation, MYC-regulated miR-150 controls this process chiefly by targeting c-Myb (Xiao et al. 2007). During MYC-induced B-lymphomagenesis, miR-17–92 acts as an onco-miR (He et al. 2005) and its oncogenic effects could be attributed, to a large extent, to down-regulation of the PTEN tumor suppressor gene by miR-19 (Mu et al. 2009; Olive et al. 2009).
The latter approach posits that miR-controlled cell phenotypes are attributable to deregulation of multiple targets acting in different, frequently overlapping pathways and simultaneously exerting both activating and inhibitory effects. Although less common, this approach is better suited to the analysis of microRNA clusters and “promiscuous” transcription factors such as MYC. Because MYC regulates dozens of microRNAs and each microRNA, in turn, can target hundreds of mRNAs, the sum effects of MYC deregulation are inevitably complex.
The MYC-driven phenotypes described herein all have microRNA-regulated components. The robust nature of these phenotypes owes a great deal to microRNA-mediated targeting of multiple genes within the same pathway, which makes MYC effects potent, persistent, and pervasive.
ACKNOWLEDGMENTS
We thank Dr. Elena Sotillo-Pineiro, Dr. Patrick Viatour (Children's Hospital of Philadelphia), Dr. Chi Dang (University of Pennsylvania), and Dr. Joshua Mendell (University of Texas Southwestern) for many valuable comments on the manuscript. The work in the authors' laboratory was funded by National Institutes of Health grants CA122334, CA115299, and CA009140.
Footnotes
Editors: Chi V. Dang and Robert N. Eisenman
Additional Perspectives on MYC and the Pathway to Cancer available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Abrams HD, Rohrschneider LR, Eisenman RN 1982. Nuclear location of the putative transforming protein of avian myelocytomatosis virus. Cell 29: 427–439 [DOI] [PubMed] [Google Scholar]
- Adhikary S, Eilers M 2005. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, Bonetta L, MacAllan D, Parry D, Holder A, Dickson C, Peters G 1994. CDK6 (PLSTIRE) and CDK4 (PSK-J3) are a distinct subset of the cyclin-dependent kinases that associate with cyclin D1. Oncogene 9: 71–79 [PubMed] [Google Scholar]
- Baumjohann D, Kageyama R, Clingan JM, Morar MM, Patel S, de Kouchkovsky D, Bannard O, Bluestone JA, Matloubian M, Ansel KM, et al. 2013. The microRNA cluster miR-17∼92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat Immunol 14: 840–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergers G, Benjamin LE 2003. Tumorigenesis and the angiogenic switch. Nat Rev Cancer 3: 401–410 [DOI] [PubMed] [Google Scholar]
- Billen LP, Kokoski CL, Lovell JF, Leber B, Andrews DW 2008. Bcl-XL inhibits membrane permeabilization by competing with Bax. PLoS Biol 6: e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H 1990. Sequence-specific DNA binding by the c-Myc protein. Science 250: 1149–1151 [DOI] [PubMed] [Google Scholar]
- Bolster DR, Crozier SJ, Kimball SR, Jefferson LS 2002. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J Biol Chem 277: 23977–23980 [DOI] [PubMed] [Google Scholar]
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al. 2007. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol 17: 1298–1307 [DOI] [PubMed] [Google Scholar]
- Boone DN, Qi Y, Li Z, Hann SR 2011. Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Proc Natl Acad Sci 108: 632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ 2008. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res 68: 7846–7854 [DOI] [PubMed] [Google Scholar]
- Cairo S, Wang Y, de Reyniès A, Duroure K, Dahan J, Redon MJ, Fabre M, McClelland M, Wang XW, Croce CM, et al. 2010. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci 107: 20471–20476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. 2002. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci 99: 15524–15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catuogno S, Cerchia L, Romano G, Pognonec P, Condorelli G, de Franciscis V 2013. miR-34c may protect lung cancer cells from paclitaxel-induced apoptosis. Oncogene 32: 341–351 [DOI] [PubMed] [Google Scholar]
- Ceballos E, Munoz-Alonso MJ, Berwanger B, Acosta JC, Hernandez R, Krause M, Hartmann O, Eilers M, Leon J 2005. Inhibitory effect of c-Myc on p53-induced apoptosis in leukemia cells. Microarray analysis reveals defective induction of p53 target genes and upregulation of chaperone genes. Oncogene 24: 4559–4571 [DOI] [PubMed] [Google Scholar]
- Ceteci F, Ceteci S, Karreman C, Kramer BW, Asan E, Gotz R, Rapp UR 2007. Disruption of tumor cell adhesion promotes angiogenic switch and progression to micrometastasis in RAF-driven murine lung cancer. Cancer Cell 12: 145–159 [DOI] [PubMed] [Google Scholar]
- Chambers AF, Groom AC, MacDonald IC 2002. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2: 563–572 [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26: 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, et al. 2009. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci 106: 3384–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Chappell J, Dalton S 2013. Roles for MYC in the establishment and maintenance of pluripotency. Cold Spring Harb Perspect Med 3: a014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C, et al. 2011. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol 13: 1353–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen RC, Oren M, Lund AH 2010. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ 17: 236–245 [DOI] [PubMed] [Google Scholar]
- Chung EY, Dews M, Cozma D, Yu D, Wentzel EA, Chang TC, Schelter JM, Cleary MA, Mendell JT, Thomas-Tikhonenko A 2008. c-Myb oncoprotein is an essential target of the dleu2 tumor suppressor microRNA cluster. Cancer Biol Ther 7: 1758–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EY, Psathas JN, Yu D, Li Y, Weiss MJ, Thomas-Tikhonenko A 2012. CD19 is a major B cell receptor-independent activator of MYC-driven B-lymphomagenesis. J Clin Invest 122: 2257–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci 102: 13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Conacci-Sorrell M, McFerrin L, Eisenman RN 2014. An overview of MYC and its interactome. Cold Spring Harb Perspect Med 4: a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM 2009. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10: 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, O’Donnell KA, Juopperi T 2005. The great MYC escape in tumorigenesis. Cancer Cell 8: 177–178 [DOI] [PubMed] [Google Scholar]
- Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F 2006. The c-Myc target gene network. Semin Cancer Biol 16: 253–264 [DOI] [PubMed] [Google Scholar]
- Dang CV, Le A, Gao P 2009. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res 15: 6479–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM, Minn AJ, Rosner MR 2009. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J 28: 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME 1997. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91: 231–241 [DOI] [PubMed] [Google Scholar]
- Daye D, Wellen KE 2012. Metabolic reprogramming in cancer: Unraveling the role of glutamine in tumorigenesis. Semin Cell Dev Biol 23: 362–369 [DOI] [PubMed] [Google Scholar]
- Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, et al. 2006. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet 38: 1060–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dews M, Fox JL, Hultine S, Sundaram P, Wang W, Liu YY, Furth E, Enders GH, El-Deiry W, Schelter JM, et al. 2010. The myc-miR-17∼92 axis blunts TGF-β signaling and production of multiple TGF-β-dependent antiangiogenic factors. Cancer Res 70: 8233–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, Urbich C, Hofmann WK, Zeiher AM, Dimmeler S 2010. Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood 115: 4944–4950 [DOI] [PubMed] [Google Scholar]
- Donald SP, Sun XY, Hu CA, Yu J, Mei JM, Valle D, Phang JM 2001. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res 61: 1810–1815 [PubMed] [Google Scholar]
- Eilers M, Eisenman RN 2008. Myc’s broad reach. Genes Dev 22: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A, Campos B, Meier J, Devens F, Liesenberg F, Wolter M, Reifenberger G, Herold-Mende H, Lichter P, Radlwimmer B 2010. De-repression of CTGF via the miR-17–92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene 29: 3411–3422 [DOI] [PubMed] [Google Scholar]
- Fabbri M, Bottoni A, Shimizu M, Spizzo R, Nicoloso MS, Rossi S, Barbarotto E, Cimmino A, Adair B, Wojcik SE, et al. 2011. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA 305: 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Zeller K, Chen YC, Watkins T, Barnes KC, Becker KG, Dang CV, Cheadle C 2010. Time-dependent c-Myc transactomes mapped by Array-based nuclear run-on reveal transcriptional modules in human B cells. PLoS ONE 5: e9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM 1999. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell 4: 199–207 [DOI] [PubMed] [Google Scholar]
- Fox JL, Dews M, Minn AJ, Thomas-Tikhonenko A 2013. Targeting of TGF-β signature and its essential component CTGF by miR-18 correlates with improved survival in glioblastoma. RNA 19: 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaktionov K, Chen X, Beach D 1996. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature 382: 511–517 [DOI] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JR, et al. 2009. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458: 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardi CJ, Wong E, Gumbiner BM 2001. E-cadherin suppresses cellular transformation by inhibiting β-catenin signaling in an adhesion-independent manner. J Cell Biol 153: 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601 [DOI] [PubMed] [Google Scholar]
- Gupta PB, Chaffer CL, Weinberg RA 2009. Cancer stem cells: Mirage or reality? Nat Med 15: 1010–1012 [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M 1997. Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299 [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. 2005. A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. 2007. A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helland A, Anglesio MS, George J, Cowin PA, Johnstone CN, House CM, Sheppard KE, Etemadmoghadam D, Melnyk N, Rustgi AK, et al. 2011. Deregulation of MYCN, LIN28B and LET7 in a molecular subtype of aggressive high-grade serous ovarian cancers. PLoS ONE 6: e18064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, Cleveland JL, Tansey WP, Lowe SW 2005. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 436: 807–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN 2008. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 32: 276–284 [DOI] [PubMed] [Google Scholar]
- Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, O’Connell BC, Mateyak MK, Tam W, Kohlhuber F, et al. 2000. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci 97: 2229–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 420: 25–27 [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S 2009. Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature 460: 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Lai M, Chen M, Xie C, Liao R, Kang YJ, Xiao C, Hu WY, Han J, Sun P 2010. The miR-17–92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res 70: 8547–8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, et al. 2008. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol 28: 2167–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz A, Sevignani C, Kenyon K, Ngo CV, Thomas-Tikhonenko A 2000. Activation of the myc oncoprotein leads to increased turnover of thrombospondin-1 mRNA. Nucleic Acids Res 28: 2268–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S, Chen P, He C, You D, Zhang S, et al. 2012. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell 22: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin HY, Oda H, Lai M, Skalsky RL, Bethel K, Shepherd J, Kang SG, Liu WH, Sabouri-Ghomi M, Cullen BR, et al. 2013. MicroRNA-17 ∼ 92 plays a causative role in lymphomagenesis by coordinating multiple oncogenic pathways. EMBO J 32: 2377–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. 2007. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67: 7713–7722 [DOI] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R 2009. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 27: 459–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Liu WH, Lu P, Jin HY, Lim HW, Shepherd J, Fremgen D, Verdin E, Oldstone MB, Qi H, et al. 2013. MicroRNAs of the miR-17∼92 family are critical regulators of TFH differentiation. Nat Immunol 14: 849–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato GJ, Barrett J, Villa-Garcia M, Dang CV 1990. An amino-terminal c-Myc domain required for neoplastic transformation activates transcription. Mol Cell Biol 10: 5914–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ 1993. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev 7: 331–342 [DOI] [PubMed] [Google Scholar]
- Katsuno Y, Lamouille S, Derynck R 2013. TGF-β signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol 25: 76–84 [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khew-Goodall Y, Goodall GJ 2010. Myc-modulated miR-9 makes more metastases. Nat Cell Biol 12: 209–211 [DOI] [PubMed] [Google Scholar]
- King CE, Cuatrecasas M, Castells A, Sepulveda AR, Lee JS, Rustgi AK 2011. LIN28B promotes colon cancer progression and metastasis. Cancer Res 71: 4260–4268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Mbesi-Impiombato A, Califano A, Migliazza A, Bhagat G, et al. 2010. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 17: 28–40 [DOI] [PubMed] [Google Scholar]
- Kong YW, Cannell IG, de Moor CH, Hill K, Garside PG, Hamilton TL, Meijer HA, Dobbyn HC, Stoneley M, Spriggs KA, et al. 2008. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci 105: 8866–8871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y 2008. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283: 14910–14914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. 2009. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137: 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH 1997. Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- Landay M, Oster SK, Khosravi F, Grove LE, Yin X, Sedivy J, Penn LZ, Prochownik EV 2000. Promotion of growth and apoptosis in c-myc nullizygous fibroblasts by other members of the myc oncoprotein family. Cell Death Differ 7: 697–705 [DOI] [PubMed] [Google Scholar]
- Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A 2000. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature 407: 592–598 [DOI] [PubMed] [Google Scholar]
- Lauper N, Beck AR, Cariou S, Richman L, Hofmann K, Reith W, Slingerland JM, Amati B 1998. Cyclin E2: A novel CDK2 partner in the late G1 and S phases of the mammalian cell cycle. Oncogene 17: 2637–2643 [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A 2007. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21: 1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Legesse-Miller A, Elemento O, Pfau SJ, Forman JJ, Tavazoie S, Coller HA 2009. let-7 Overexpression leads to an increased fraction of cells in G2/M, direct down-regulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. J Biol Chem 284: 6605–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M 2009. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN 2009. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J 28: 3157–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, et al. 2007. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol 27: 2240–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X 2008. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res 36: 5391–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, Phang JM 2012. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci 109: 8983–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G 2004. Intrinsic tumour suppression. Nature 432: 307–315 [DOI] [PubMed] [Google Scholar]
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. 2010. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 12: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M 2009. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer 9: 153–166 [DOI] [PubMed] [Google Scholar]
- Malumbres M, Sotillo R, Santamaria D, Galan J, Cerezo A, Ortega S, Dubus P, Barbacid M 2004. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell 118: 493–504 [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA 2009. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460: 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Adachi S, Sedivy JM 1997. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ 8: 1039–1048 [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Sedivy JM 1999. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol 19: 4672–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushime H, Ewen ME, Strom DK, Kato JY, Hanks SK, Roussel MF, Sherr CJ 1992. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell 71: 323–334 [DOI] [PubMed] [Google Scholar]
- Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, et al. 2010. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol 12: 372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP 2007. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315: 1576–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R 2010. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Bostrom AK, Impens F, Fredlund E, Van Peer G, De Antonellis P, von Stedingk SK, Ghesquiere B, Schulte S, Dews M, et al. 2010. The miR-17–92 microRNA cluster regulates multiple components of the TGF-β pathway in neuroblastoma. Mol Cell 40: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Harlow E 1994. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol 14: 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I 2006. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217 [DOI] [PubMed] [Google Scholar]
- Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D’Andrea A, Sander C, Ventura A 2009. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev 23: 2806–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki ST, Carew JS, Maclean KH, Courage JF, Huang P, Houghton JA, Cleveland JL, Giles FJ, McConkey DJ 2008. Myc regulates aggresome formation, the induction of Noxa, and apoptosis in response to the combination of bortezomib and SAHA. Blood 112: 2917–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM 2008. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA, Chandriani S, O’Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD 2002. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol 22: 5793–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA, Riblett M, Tang WH, Gratchouck V, Zhuang D, Fernandez Y, Verhaegen M, Varambally S, Chinnaiyan AM, Jakubowiak AJ, et al. 2007. Tumor cell-selective regulation of NOXA by c-MYC in response to proteasome inhibition. Proc Natl Acad Sci 104: 19488–19493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296: 1132–1136 [DOI] [PubMed] [Google Scholar]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L 2009. miR-19 is a key oncogenic component of mir-17–92. Genes Dev 23: 2839–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV 2000. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem 275: 21797–21800 [DOI] [PubMed] [Google Scholar]
- Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M 2004. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res 64: 3087–3095 [DOI] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22: 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan GI 2002. Suppression of Myc-induced apoptosis in β cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell 109: 321–334 [DOI] [PubMed] [Google Scholar]
- Prendergast GC, Cole MD 1989. Posttranscriptional regulation of cellular gene expression by the c-myc oncogene. Mol Cell Biol 9: 124–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast GC, Ziff EB 1991. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science 251: 186–189 [DOI] [PubMed] [Google Scholar]
- Prendergast GC, Diamond LE, Dahl D, Cole MD 1990. The c-myc-regulated gene mrl encodes plasminogen activator inhibitor 1. Mol Cell Biol 10: 1265–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psathas JN, Doonan PJ, Raman P, Freedman BD, Minn AJ, Thomas-Tikhonenko A 2013. The Myc-miR-17–92 axis amplifies B-cell receptor signaling via inhibition of ITIM proteins: A novel lymphomagenic feed-forward loop. Blood 122: 4220–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR 2004. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature 431: 712–717 [DOI] [PubMed] [Google Scholar]
- Qing G, Li B, Vu A, Skuli N, Walton ZE, Liu X, Mayes PA, Wise DR, Thompson CB, Maris JM, et al. 2012. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell 22: 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelle DE, Ashmun RA, Shurtleff SA, Kato JY, Bar-Sagi D, Roussel MF, Sherr CJ 1993. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev 7: 1559–1571 [DOI] [PubMed] [Google Scholar]
- *.Rahl PB, Young RA 2014. MYC and transcripton elongation. Cold Spring Harb Perspect Med 4: a020990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak J, Yu JL 2004. Oncogenes and tumor angiogenesis: The question of vascular “supply” and vascular “demand.” Semin Cancer Biol 14: 93–104 [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M 2007. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 26: 731–743 [DOI] [PubMed] [Google Scholar]
- Reisman D, Rotter V 1993. The helix-loop-helix containing transcription factor USF binds to and transactivates the promoter of the p53 tumor suppressor gene. Nucleic Acids Res 21: 345–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Hong SJ, Bartel DP 2011. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol Cell 43: 993–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey RB, Hay N 2009. Is Akt the “Warburg kinase?”—Akt-energy metabolism interactions and oncogenesis. Semin Cancer Biol 19: 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulski K, Li Y, Rothermund K, Pu L, Watkins S, Yi F, Prochownik EV 2005. Onzin, a c-Myc-repressed target, promotes survival and transformation by modulating the Akt-Mdm2-p53 pathway. Oncogene 24: 7524–7541 [DOI] [PubMed] [Google Scholar]
- Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Moller P, Stilgenbauer S, Pollack JR, Wirth T 2008. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood 112: 4202–4212 [DOI] [PubMed] [Google Scholar]
- Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, et al. 2001. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res 29: 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Ohtani K, Nevins JR 1997. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol 17: 5227–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheiness D, Bishop JM 1979. DNA and RNA from uninfected vertebrate cells contain nucleotide sequences related to the putative transforming gene of avian myelocytomatosis virus. J Virol 31: 514–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortt J, Johnstone RW 2012. Oncogenes in cell survival and cell death. Cold Spring Harb Perspect Biol 4: a009829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurk C, Maatz H, Rocnik E, Bialik A, Force T, Walsh K 2005. Glycogen-synthase kinase 3β/β-catenin axis promotes angiogenesis through activation of vascular endothelial growth factor signaling in endothelial cells. Circ Res 96: 308–318 [DOI] [PubMed] [Google Scholar]
- Sotillo E, Thomas-Tikhonenko A 2011. Shielding the messenger (RNA): MicroRNA-based anticancer therapies. Pharmacol Ther 131: 18–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo E, Laver T, Mellert H, Schelter JM, Cleary MA, McMahon S, Thomas-Tikhonenko A 2011. Myc overexpression brings out unexpected antiapoptotic effects of miR-34a. Oncogene 30: 2587–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Bath ML, Cory S 1990. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 348: 331–333 [DOI] [PubMed] [Google Scholar]
- Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X 2008. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett 582: 1564–1568 [DOI] [PubMed] [Google Scholar]
- Sundaram P, Hultine S, Smith LM, Dews M, Fox JL, Biyashev D, Schelter JM, Huang Q, Cleary MA, Volpert OV, et al. 2011. p53-responsive miR-194 inhibits thrombospondin-1 and promotes angiogenesis in colon cancers. Cancer Res 71: 7490–7501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P 2007. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem 282: 2135–2143 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev E, Menssen A, Meister G, Hermeking H 2007. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 6: 1586–1593 [DOI] [PubMed] [Google Scholar]
- Tazawa H, Tsuchiya N, Izumiya M, Nakagama H 2007. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci 104: 15472–15477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K 2009. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460: 1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. 2008. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI 2008. Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R 2008. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40: 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862 [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S 2009. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234 [DOI] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci 105: 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AS, Gumbiner BM 2003. Adhesion-independent mechanism for suppression of tumor cell invasion by E-cadherin. J Cell Biol 161: 1191–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P, Iwasaki M, Somervaille TC, Ficara F, Carico C, Arnold C, Chen CA, Cleary ML 2010. The miR-17–92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res 70: 3833–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM 2007. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem 282: 2130–2134 [DOI] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K 2007. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131: 146–159 [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K 2008. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol 9: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ 2008. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci 105: 13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ 1995. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell 80: 285–291 [DOI] [PubMed] [Google Scholar]
- Yecies JL, Manning BD 2011. mTOR links oncogenic signaling to tumor cell metabolism. J Mol Med (Berl) 89: 221–228 [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Ntosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Haley B 2005. Ribo-gnome: The big world of small RNAs. Science 309: 1519–1524 [DOI] [PubMed] [Google Scholar]
- Zhang X, Chen X, Lin J, Lwin T, Wright G, Moscinski LC, Dalton WS, Seto E, Wright K, Sotomayor E, et al. 2012. Myc represses miR-15a/miR-16–1 expression through recruitment of HDAC3 in mantle cell and other non-Hodgkin B-cell lymphomas. Oncogene 31: 3002–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Kalota A, Jin S, Gewirtz AM 2009. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood 113: 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZN, Bai JX, Zhou Q, Yan B, Qin WW, Jia LT, Meng YL, Jin BQ, Yao LB, Wang T, et al. 2012. TSA suppresses miR-106b-93–25 cluster expression through downregulation of MYC and inhibits proliferation and induces apoptosis in human EMC. PLoS ONE 7: e45133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Li N, Liang S, Huang Q, Coukos G, Zhang L 2010. Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J Biol Chem 285: 41961–41971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang JR, Zeng C, Zhuang SM 2012. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res 40: 4615–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 12: 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]