Abstract
Although p53 has long been known as the “guardian of the genome” with a role in tumor suppression in many tissues, the discovery of two p53 ancestral genes, p63 and p73, more than a decade ago has triggered a considerable amount of research into the role of these genes in skin development and diseases. In this review, we primarily focus on mechanisms of action of p53 and p63, which are the best-studied p53 family members in the skin. The existence of multiple isoforms and their roles as transcriptional activators and repressors are key to their function in multiple biological processes including the control of skin morphogenesis, regeneration, tumorigenesis, and response to chemotherapy. Last, we provide directions for further research on this family of genes in skin biology and pathology.
p53, the “guardian of the genome,” and two ancestral genes—p63 and p73—play key roles in skin development, regeneration, tumorigenesis, and response to chemotherapy.
The p53 gene family is derived from a common ancestor that appeared very early during evolution in the metazoan sea anemone more than a billion years ago (Belyi et al. 2010). This ancestor gene duplicated in early vertebrates resulting in the appearance of the gene that is closely related to p53, whereas its second duplication in bony fish led to the appearance of the p63 and p73 genes (Belyi et al. 2010). During evolution, the p63 and p73 genes showed a marked increase in the number of introns resulting in the appearance of their numerous isoforms in vertebrates. In contrast, the structure of the p53 gene did not change dramatically (Belyi et al. 2010). The expansion of the p53 gene family in vertebrates, whose role was originally linked to the protection of germ cells against DNA damage in early metazoans, resulted in a diversity of novel functions including the control of cell proliferation, differentiation, tissue development, stem cell regulation, and tumor suppression (Flores et al. 2005; Belyi et al. 2010; Allocati et al. 2012).
During the last two decades, tremendous progress has been achieved in the delineation of the functions of the p53 family of genes in mammals (Lane and Levine 2010). In particular, the structure and role for the p63 gene as a master regulator of the development and maintenance of the stratified epithelia as well as its involvement in the control of aging and tumor growth were shown (reviewed in Koster and Roop 2007; Crum and McKeon 2010; Melino 2011; Vanbokhoven et al. 2011; Su et al. 2013). Furthermore, the roles of the distinct p63 and p73 isoforms in the control of cell proliferation, differentiation, and apoptosis in the skin were dissected by using a number of genetically engineered mouse models and in vitro analyses.
Consistent with the role of p53 in genome surveillance, p53 mutations are found in more than half of human tumors including squamous cell carcinomas, and missense p53 mutations lead to aggressive cancer phenotypes (Muller and Vousden 2013). In contrast to p53, mutations of the p63 gene cause numerous ectodermal dysplasia syndromes also associated with craniofacial and limb abnormalities (for review, see Rinne et al. 2007; Vanbokhoven et al. 2011). Unlike p53 and p63, mutations in the p73 gene are extremely rare; however, p73 is inactivated in cancers epigenetically, that is, via hypermethylation of its promoter (Maas et al. 2013).
In this review, we summarize our current knowledge of the role of the p53 gene family in the control of skin development, regeneration, tumorigenesis, and response to chemotherapy. We also discuss the distinct roles for different p63 and p73 isoforms in the regulation of skin regeneration and tumor growth. Finally, we discuss directions for further research in these exciting areas of skin biology and pathology.
THE p53 FAMILY AND ITS ISOFORMS
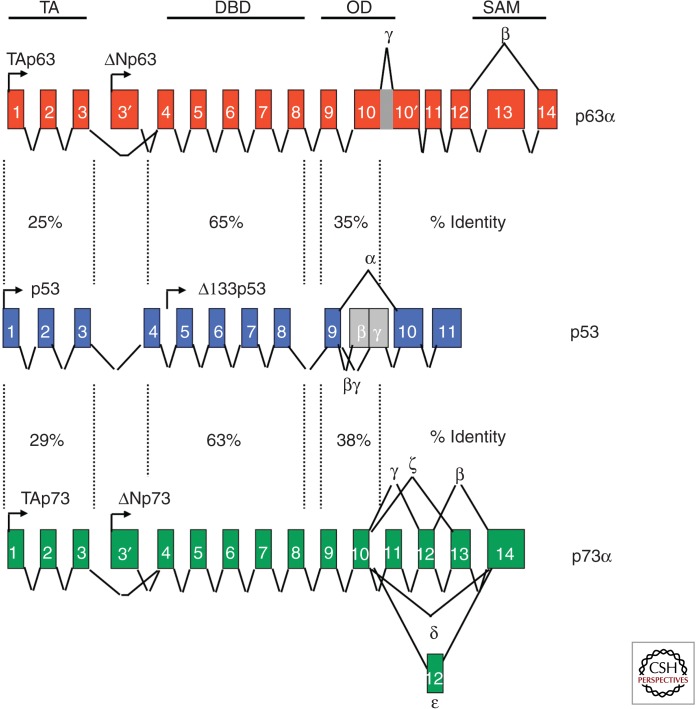
All three family members, p53, p63, and p73, share structural similarities in three main domains: the transactivation domain (TA), the DNA-binding domain (DBD), and the oligomerization domain (OD) (Fig. 1). All three proteins are transcription factors, and each domain of the three plays a critical role in the downstream transactivation of target genes. The OD is necessary for the formation of functional tetramers that bind to DNA via the DBD. The acidic TA domain is needed for transcriptional activity of the family members.
Figure 1.
p53 family gene organization and isoforms. The p53 family members have four main domains: the transactivation domain (TA), the DBD, the oligomerization domain (OD), and the sterile alpha motif domain (SAM; present in a subset of the isoforms). For each gene, full length (TA) and amino-terminal spliced isoforms exist (ΔN). Multiple carboxy-terminal spliced isoforms exist as shown (α, β, δ, etc.).
Over the last decade or so, data from multiple laboratories indicates that all three family members have isoforms that perform overlapping and unique functions. The isoforms that have been identified for p63 and p73 can be categorized into two groups that are driven by distinct promoters. One set contains an acidic TA domain and is known as the TA isoforms. The other set of isoforms lack this domain. In addition to these amino-terminal variants of p63 and p73, carboxy-terminal spliced isoforms of each TA and ΔN isoforms of p63 and p73 exist. These are known as α, β, δ, ε, γ, ζ (Fig. 1) (Su et al. 2013). The alternatively spliced isoforms differ in their ability to transactivate downstream target genes that induce biological activities, such as cell-cycle arrest and apoptosis (Yang et al. 1998). Importantly, some of the isoforms share more structural similarity to p53 and have been found to transactivate the same target genes such as p21, bax, NOXA, PUMA, and Perp (Yang et al. 1998; Su et al. 2013). p53 has also been found to have multiple isoforms driven by two promoters with carboxy-terminal spliced isoforms (Bourdon et al. 2005): p53, p53β, p53γ, Δ133p53, Δ133p53β, and Δ133p53γ (Fig. 1). Similar to the ΔN isoforms of p63 and p73, the Δ133 isoforms lack an acidic TA domain.
The existence of multiple isoforms of all p53 family members leads to great complexity in biological function. This is because the isoforms are not ubiquitously expressed, but are only expressed in specific tissues. A prime example is ΔNp63α, which is highly expressed in the basal layer of the epidermis (Mills et al. 1999; Yang et al. 1999; Koster and Roop 2007). This isoform is essential for terminal differentiation of all epithelial tissues and has been best studied in the skin, as discussed below (Koster et al. 2004; Koster and Roop 2007). The function of other isoforms of p53 and p73 in epidermal morphogenesis has not been as well studied, but they may also play different roles or form heterotetramers with the isoforms of p63, leading to various types of tetramers with varying transactivation ability that induce a certain complement of target genes depending on spatial and temporal cues.
p53, p63, AND p73 IN EPIDERMAL AND HAIR FOLLICLE MORPHOGENESIS
p63 and Control of Epidermal Development
The process of skin development results in the formation of a stratified epidermis with self-renewing ability and a number of skin appendages (hair follicles, nails, glands) (Blanpain and Fuchs 2009; Chuong and Richardson 2009; Dhouailly 2009). The p63 transcription factor serves as a master regulator of epidermal development and differentiation (Fig. 2) (Koster and Roop 2007; Mikkola 2007). In mice, development of the epidermis begins after the onset of ΔNp63 expression in cells of the embryonic ectoderm that express epithelial keratins K8 and K18, which is followed by the appearance of basal epidermal keratins K5/K14 at E9.5 (Byrne et al. 1994; Koster and Roop 2007; Blanpain and Fuchs 2009). Between E9.5 and E12.5, K5/K14-positive epidermal progenitors expand laterally via symmetric cell division to cover the rapidly growing embryo (Weiss and Zelickson 1975; Lechler and Fuchs 2005). The onset of epidermal stratification in mice occurs at about E12.5 when basal keratinocytes begin to form the intermediate epidermal layer between the basal layer and periderm via asymmetric cell division (Poulson and Lechler 2012). The intermediate layer is subsequently transformed into the spinous layer (Byrne et al. 1994), in which keratinocytes occasionally proliferate and begin to express suprabasal keratins K1/K10 at about E13.5–E14.5, whereas expression of K5/K14 is down-regulated (Byrne et al. 1994).
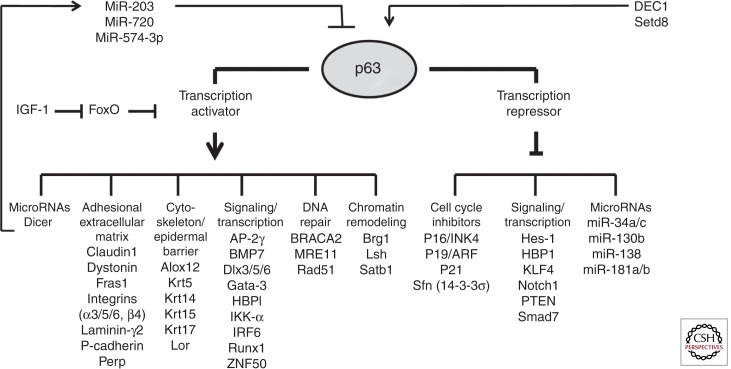
Figure 2.
Transcriptional targets of p63 in epidermal keratinocytes. p63 functions as a transcriptional activator and repressor and regulates expression of distinct groups of genes in epidermal keratinocytes. In turn, p63 expression in keratinocytes is controlled by microRNAs, the DEC1 transcription factor, and Setd8 histone methyltransferase (see references in the text).
By day E14.5–E15.5 of mouse embryonic development, keratinocytes of the upper spinous layer accumulate lamellar granules enriched in lipids and lipid-processing enzymes and form the granular epidermal layer (Byrne et al. 2003; Candi et al. 2005). The final step of the differentiation process of epidermal cells is the formation of the cornified layer, which occurs in the mouse embryo between E17.5–E18.5 (Byrne et al. 2003). Cells of the granular epidermal layer undergo cornification, which represents a unique form of cell death and includes formation of the cytoplasmic cornified envelope, a submembranous structure, in which intermediate filaments, proteins, lipids, fragments of degraded organelles, and nuclear remnants are cross-linked (Simpson et al. 2011).
Genetic p63 ablation in mice results in the failure of stratification of the epidermis and other squamous epithelia, lack of formation of epidermal appendages (hair follicles, glands, teeth), and severe abnormalities in the development of the limbs and external genitalia (Mills et al. 1999; Yang et al. 1999; Ince et al. 2002). The p63 isoforms play distinct roles in the control of epidermal development; the ΔNp63 isoforms are much more abundant in the epidermis compared to TAp63, which is strongly expressed in basal epidermal keratinocytes and is markedly down-regulated in the spinous epidermal layer (Laurikkala et al. 2006; Romano et al. 2009, 2012; LeBoeuf et al. 2010; Shalom-Feuerstein et al. 2011). TAp63 is also expressed in response to stresses such as wound healing (Su et al. 2009b). ΔNp63 plays a major role in mediating the effects of p63 on epidermal development, whereas TAp63 keeps stem cells within the skin in quiescence to avoid early depletion of these cells and suppresses tumorigenesis in postnatal epidermis (Su et al. 2009b; Romano et al. 2012; Chakravarti et al. 2014).
p63 is a master regulator of epidermal development and operates as a transcriptional activator and repressor regulating the expression of several groups of genes involved in the control of cell proliferation, adhesion, cytoskeletal organization, DNA repair, transcription, and chromatin remodeling (Fig. 2) (for review, see Koster and Roop 2007; Vanbokhoven et al. 2011; Botchkarev et al. 2012; Su et al. 2013). p63 induces in epidermal keratinocytes the expression of the genes encoding basal epidermal keratins 5/14, distinct cell adhesion molecules (P-cadherin, integrin-α3, Perp, dystonin, etc.), cell matrix regulators (Fras-1), transcription factors (AP-2γ, IKK-α, IRF6, HBP1, Runx1, ZNF750), and DNA repair machinery (BRACA2, MRE11, Rad51) (Koster et al. 2004; Ihrie et al. 2005; Koster and Roop 2007; Lin et al., 2009; Shalom-Feuerstein et al. 2011; Vanbokhoven et al. 2011; Sen et al. 2012). p63 transcriptional activity is also regulated and inhibited by the FoxO transcription factor, which directly binds to p63 protein and prevents its interaction with gene promoters. In basal epidermal cells, the levels of intranuclear FoxO are negatively regulated by the IGF-1 signaling that stimulates the nuclear export of FoxO (Gunschmann et al. 2013).
As a transcriptional repressor, p63 maintains proliferation of basal epidermal keratinocytes by directly repressing expression of antiproliferative target genes, including 14-3-3s, p16/Ink4a, p19/Arf, p21, and PTEN (Westfall et al. 2003; Watt et al. 2008; Su et al. 2009a; Leonard et al. 2011; Ferone et al. 2012). In addition to cell-cycle-associated genes, p63 inhibits the expression of nonkeratinocyte genes (i.e., mesodermal genes), as well as a number of other genes such as distinct components of the Notch (Notch1, Hes1) and bone morphogenetic protein (BMP) signaling pathways (Smad7) in epidermal keratinocytes (Nguyen et al. 2006; De Rosa et al. 2009; Shalom-Feuerstein et al. 2011). Repressive effects of p63 on the expression of nonepidermal genes are mediated, at least in part, by Smad7 inhibition, which activates the BMP signaling pathway and promotes expression of epidermal genes (De Rosa et al. 2009).
There are several indications that many effects of p63 on gene expression in epidermal keratinocytes are mediated by its interactions with epigenetic regulatory machinery (Botchkarev et al. 2012; Frye and Benitah 2012; Zhang et al. 2012). For instance, p63 directly regulates expression of several genes encoding the genome organizer and AT-rich-binding protein Satb1, as well as ATP-dependent chromatin remodelers Lsh and Brg1 in epidermal keratinocytes (Fessing et al. 2011; Keyes et al. 2011; Mardaryev et al. 2014). Both Brg1 and Satb1 play important roles in mediating a p63-regulated program of higher-order chromatin remodeling within the epidermal differentiation complex (EDC) locus. Brg1 regulates relocation of the EDC away from the nuclear periphery into the nuclear interior and propinquity with nuclear speckles. This is followed by Satb1-regulated arrangement of chromatin conformation within the central domain of the locus, which contains a large number of genes activated during terminal keratinocyte differentiation (Fessing et al. 2011; Mardaryev et al. 2014). In turn, Lsh regulates chromatin remodeling and mediates ΔNp63-dependent proliferation and survival of Keratin 15-positive stem cells in the skin (Keyes et al. 2011).
p63 also localizes to the genes that show binding of histone acetyltransferase complex containing ING5, but not ING5 + meCpG (Mulder et al. 2012). ΔNp63 also cooperates with HDAC1/2 to directly repress p16/INK4a in epidermal keratinocytes, whereas HDAC1/2 independently deacetylate p53 and suppress its ability to induce apoptosis and activate target genes, including p21 (LeBoeuf et al. 2010). Conversely, p63 gene expression in epidermal keratinocytes is positively regulated by the histone methyltransferase Setd8, as well as by the DEC1 transcription factor (Driskell et al. 2011; Qian et al. 2011).
The p63-dependent transcriptional program is also regulated by distinct microRNAs, whereas selected microRNA genes represent an important class of p63 targets in basal epidermal keratinocytes (reviewed in Botchkareva 2012). Global effects of p63 on microRNAs are realized via direct regulation and transactivation of the Dicer promoter by TAp63 (Su et al. 2010) and the DiGeorge syndrome critical region gene 8 (DGCR8) promoter by ΔNp63 (Chakravarti et al. 2014). In addition, p63 negatively regulates expression of several microRNAs of the miR-34 family, which show reciprocal expression patterns with p63 in the epidermis (Antonini et al. 2010). Furthermore, p63 directly inhibits expression of miR-138, miR-181a/b, and miR-130b in primary human keratinocytes (Rivetti di Val Cervo et al. 2012). In turn, p63 expression in epidermal keratinocytes is regulated by miR-203, miR-720, and miR-574-3p, which are expressed in cells within the suprabasal epidermal layer and restrict p63 expression to basal cells (Yi et al. 2008; Chikh et al. 2011).
p53/p63 and Regulation of Hair Follicle Development and Cycling
Hair follicle development is governed by the signaling exchanges between the epidermal keratinocytes committed to hair-follicle-specific differentiation and the mesenchymal cells of the follicular papilla (Hardy 1992; Millar 2002; Schmidt-Ullrich and Paus 2005; Blanpain and Fuchs 2009; Lee and Tumbar 2012). Hair follicle morphogenesis results in formation of the hair bulb in which hair matrix keratinocytes proliferate and differentiate into six distinct cell lineages: the cuticle, Huxley and Henle layers of the inner root sheath, as well as the medulla, cortex, and cuticle of the hair shaft (Hardy 1992; Millar 2002; Schmidt-Ullrich and Paus 2005). In addition to the hair bulb, epidermal progenitors generate the sebaceous gland, which secretes sebum via the duct penetrating the upper part of the follicular epithelium and extrudes it into the hair follicle infundibulum (Niemann and Horsley 2012). Postnatally, the hair follicle undergoes cyclic transformations with periods of active growth (anagen), involution (catagen), and resting (telogen) (Stenn and Paus 2001).
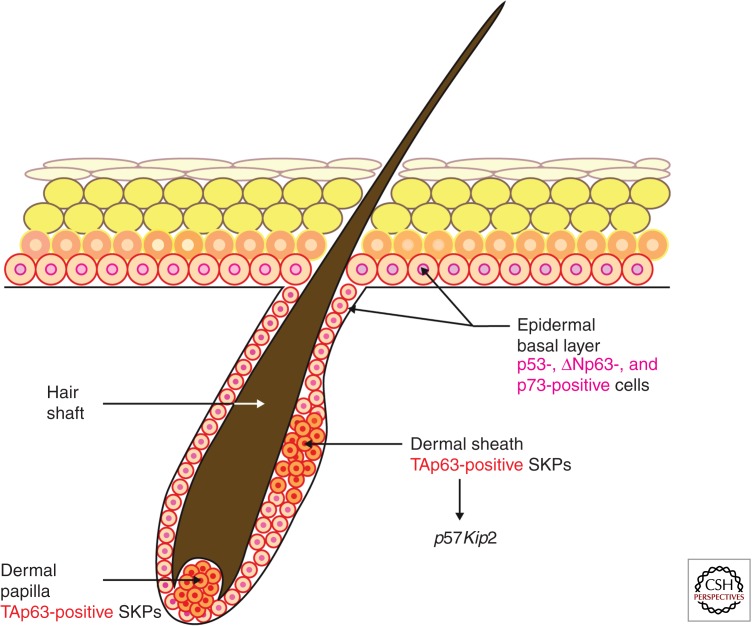
ΔNp63 is expressed in the epithelial cells of the developing hair follicle as well as in the bulge, outer root sheath, and hair matrix of the postnatal hair follicles, whereas TAp63 is expressed in cells of the dermal sheath and follicular papilla (Su et al. 2009b; Romano et al. 2010). TAp63 keeps skin-derived precursor cells (SKPs), which reside in the dermis in the postnatal hair follicle, in quiescence to protect them from premature depletion via transcriptional regulation of the cyclin-dependent kinase inhibitor p57Kip2 (Su et al. 2009b). ΔNp63 is also involved in regulation of hair follicle stem cell maintenance and self-renewal, although its overexpression in the outer root sheath results in the switch of hair-follicle-specific differentiation toward epidermal differentiation and depletion of bulge stem cells (Romano et al. 2010). TAp73 might also play a role in the control of bulge stem cell activity because its conditional ablation results in failure of the hair follicles to enter anagen phase (Rufini et al. 2012).
p53 is also expressed in the outer root sheath and hair matrix of the postnatal hair follicles, and its expression is increased during the hair follicle transition from anagen to catagen (Botchkarev et al. 2001). p53 regulates apoptosis in hair matrix keratinocytes during catagen (Botchkarev et al. 2001), and BRCA1 operates upstream of p53, suppressing DNA damage in hair matrix cells and resulting in p53 stabilization (Sotiropoulou et al. 2013). The roles of p63 and p73 in the control of apoptosis in hair matrix keratinocytes during hair follicle involution remain to be defined, although ΔNp63 might antagonize the proapoptotic function of p53, which is evident from the fact that ΔNp63-overexpressing mice show a markedly delayed transition from anagen to catagen (Romano et al. 2010).
Taken together, these data show that different members of p53 family play distinct, yet partially overlapping, functions in the control of epidermal and hair follicle development, and their effects on epithelial or mesenchymal cell populations in the skin are largely isoform specific.
THE ROLES OF p63 AND ITS ISOFORMS IN WOUND HEALING
The TA and ΔN isoforms of p63 have distinct roles in wound healing. The expression of the isoforms differs in adult skin. Whereas ΔNp63 is expressed throughout the basal layer of the epidermis (Koster and Roop 2007), TAp63 is undetectable under normal conditions and is up-regulated on stress (Su et al. 2009b). One example of stress in which TAp63 is detected is in the wound healing response (Su et al. 2009b). This distinct expression pattern gives rise to different roles of these isoforms in various biological processes from cancer to wound healing as has been shown using mouse models, and is discussed further below.
Using a TAp63 conditional knockout mouse model, the roles of TAp63 in the wound healing response have been defined. Mice with a germline deletion of TAp63 (TAp63−/−) developed alopecia, a severe blistering disease giving rise to spontaneous wounds, and wound healing defects evidenced by a failure to completely heal wounds created on the epidermis. These defects are reminiscent of those seen in human patients with Hay–Wells syndrome, in which patients with mutations in p63 develop skin abrasions, dermatitis, and alopecia (McGrath et al. 2001). In contrast, mice deficient for TAp63 in K14-expressing cells only, generated by intercrossing TAp63 conditional knockout mice (TAp63fl/fl) with K14cre-expressing mice, displayed no such wound healing defect. These data suggested a role for TAp63 in a K14 negative progenitor cell that is critical for the wound healing and hair regeneration. Indeed, TAp63 was shown to be critical to maintain dermal-derived stem cells known as SKPs (Biernaskie et al. 2009) in quiescence through transcriptional regulation of the cyclin-dependent kinase inhibitor, p57KIP2. Consequently, in mice deficient in TAp63, SKP cells hyperproliferate and are subsequently depleted, and wound healing is impaired (Fig. 3) (Su et al. 2009b). Thus, TAp63 plays a key role in maintaining quiescence of dermal stem cells critical for wound healing. When regulation of TAp63 is lost, these stem cells are prematurely depleted leading to deficits in wound healing.
Figure 3.
TAp63 and ΔNp63 are expressed in distinct cell types within the skin. TAp63 is expressed in SKPs within the dermal papilla and dermal sheath (TAp63 positive). TAp63 maintains these cells in quiescence by transcriptional regulation of p57Kip2. p53, ΔNp63, and p73 isoforms are expressed in the basal layer of the epidermis.
The role of ΔNp63 in wound healing was also shown using genetically engineered mice. In this case, the mice were engineered to express a ΔNp63 small hairpin RNA (shRNA) under the control of an inducible-epidermal-specific promoter to silence ΔNp63 in the basal layer of the epidermis. After induction of ΔNp63 shRNA, the mice displayed skin fragility characterized by skin erosions. In a wound-healing model, the ΔNp63 knockdown mice showed severely impaired healing with a failure of the epidermis to terminally differentiate over the wound site. Importantly, ΔNp63 was found to function by its ability to transactivate two key genes, Fras1, required for basement membrane integrity, and IKK-α, required for terminal differentiation and formation of the spinous layer. Thus, ΔNp63 orchestrates a transcriptional program critical for epithelial integrity and morphogenesis (Fig. 3). Taken together, these data indicate that p63 isoforms play distinct roles in wound healing and epidermal homeostasis.
THE p53 FAMILY IN SKIN CANCER
p53 Mutations in Skin Cancer
A variety of skin cancers harbor p53 mutations including basal cell carcinomas (BCCs), squamous cell carcinomas (SCC), precursors of SCCs known as actinokeratosis, and melanoma (Hodis et al. 2012). The p53 mutations observed in these cancers and precancerous lesions are UV induced and frequently C–T transitions at dipyrimidine sites. The most frequently detected mutations in both BCCs and SCCs are located at codons 196, 248, and 282 of p53. Phosphorylation of the p53 protein at specific residues, like Serine 389, has been found to be of particular importance in the UV response (Bruins et al. 2004). Mouse models genetically engineered with a serine to alanine mutation at codon 389 (p53 S389A) showed increased sensitivity to UV-induced skin tumors (Bruins et al. 2004). Additional mouse models with mutations in the p53-binding domain combined with a KrasG12D mutation also showed increased susceptibility to skin tumors (Caulin et al. 2007). Mouse models for each of the common p53 mutations in skin cancers have not yet been developed; however, these models will likely shed light on unknown functions of p53 and its family members in skin cancer.
p63 in Skin Cancer
The expression patterns of p63 in cutaneous SCCs have been well studied. In many studies of human cutaneous SCCs, p63 levels are high and now used as diagnostic markers (Dotto et al. 2006). A large majority of these studies have been performed using pan-p63 antibodies that bind in the DBD of the protein and do not distinguish between the TA and ΔN isoforms. Although specific isoforms were not detected in these studies, it is well known and accepted that the ΔNp63α is the most highly expressed in basal epithelial cells and detected in these tumors, indicating the potential role of ΔNp63 α as an oncogene. Consistent with this function, ΔNp63α is down-regulated in response to UV, allowing p53, TAp63, and TAp73 to be freed from inhibitory transcriptional complexes with ΔNp63. This, in turn, results in downstream TA of target genes that protect the cell (Liefer et al. 2000).
Mouse models have also shed light on the roles of p63 in SCC formation. In a mouse model heterozygous for both p53 and p63 (p53+/−;p63+/−), 50% of mice spontaneously develop head and neck SCC or cutaneous SCC (Flores et al. 2005). Moreover, 100% of these tumors undergo loss of heterozygosity and lose the wild-type allele of p63 (Flores et al. 2005). Loss of heterozygosity of p53 was not detected in this model. Because of the complexity of p63 and the existence of multiple isoforms, TAp63 knockout mice have been generated. Both TAp63−/− and TAp63+/− mice develop spontaneous cutaneous SCCs at high frequency that are highly metastatic (Su et al. 2010). These findings were further validated in human SCC patient samples in which high-grade, metastatic tumors lose expression of TAp63 and its downstream target genes. These findings indicate that TAp63 is a critical suppressor of SCC metastasis and acts through transcriptional regulation of Dicer and miR130b.
p73 in Skin Cancer
p73 resides on chromosome 1p36.33, a region frequently lost in multiple tumor types. p73 may also play a role in skin cancer. Although infrequently mutated, p73 mutations have been identified in Merkel cell carcinoma (MCC) (Van Gele et al. 2000). p53 mutations have been identified in MCC indicating that these two p53 family members may play a role in the suppression of this tumor type. In mouse models heterozygous for p53 and p73 (p53+/−;p73+/−), mice spontaneously develop cutaneous SCC, further indicating that p73 may play a role in skin cancer (Flores et al. 2005). To date, no extensive research on p73’s role in skin tumorigenesis has been performed and further investigation is warranted.
p53 AND SKIN RESPONSE TO CHEMOTHERAPY
Chemotherapeutic drugs alter mitotic cell division and metabolism in the hair follicles, actively generating hairs via induction of DNA damage in proliferating hair matrix keratinocytes (Paus et al. 2013). DNA damage sites are detected by the MRN (MRE11-RAD50-NBS1) complex (primary damage sensor), followed by activation of ATM and ATR kinases, which phosphorylate the histone H2AX (Jackson and Bartek 2009). γ-H2AX foci serve as sites for further recruitment of the 53BP1 and BRAC1, whereas ATM and ATR kinases operate as transducers of the response from the DNA damage sensors to a number of proapoptotic effectors including p53 (Jackson and Bartek 2009).
p53 and ΔNp63 are expressed in proliferating hair matrix keratinocytes (Botchkarev et al. 2000, 2001; Romano et al. 2010). Whereas the role of ΔNp63 in the response of hair follicles to chemotherapy remains to be defined, several lines of evidence show an essential role for p53 in the control of chemotherapy-induced hair loss. First, massive apoptosis in hair matrix keratinocytes induced by cyclophosphamide in mice in vivo or in human hair follicles ex vivo is preceded by a marked increase in p53 protein levels in hair matrix keratinocytes (Botchkarev et al. 2000; Bodo et al. 2007). Second, p53 knockout mice show complete resistance to hair loss induced by cyclophosphamide in vivo (Botchkarev et al. 2000). Finally, other p53 transcriptional targets, such as Fas, Bax, or IGFBP-3, are also up-regulated in hair matrix keratinocytes of mice treated with cyclophosphamide (Botchkarev et al. 2000), and Fas knockout mice show significant protection against chemotherapy-induced hair loss (Sharov et al. 2004).
In contrast to hair matrix keratinocytes, epithelial stem cells residing in the bulge and dermal papilla fibroblasts show relative protection against DNA damage induced by chemotherapy or ionizing radiation (Blanpain et al. 2011; Paus et al. 2013). Both cell populations are slow cycling and survive chemotherapy, and their protection against DNA damage is mediated, at least in part, by high levels of expression of the antiapoptotic Bcl2 protein that blocks p53-dependent apoptosis (Lindner et al. 1997; Sotiropoulou et al. 2010). In addition to Bcl2, protection of bulge stem cells against ionizing radiation is mediated by high activity of the DNA repair machinery including DNA-PK (Sotiropoulou et al. 2010). Interestingly, ΔNp63 is capable of inhibiting DNA damage induced by chemotherapy (Li et al. 2012). Thus, because ΔNp63 is also expressed in the bulge stem cells of telogen hair follicles and likely inhibits the proapoptotic effects of p53 in hair matrix keratinocytes during physiological hair follicle involution (Romano et al. 2010), it may play a role in the protection of bulge stem cells and hair matrix keratinocytes against DNA damage after chemotherapy.
CONCLUSIONS
Over the last decade, a wealth of knowledge has accumulated on the function of the p53 family of genes in multiple systems from stem cell maintenance and renewal to the suppression of cancer and metastasis. The functions of this family of genes are now well understood in the skin and include critical roles for p63 and its isoforms in epidermal morphogenesis and maintenance. Importantly, we know that the p53 family plays important roles in responses to stresses like DNA damage, wound healing, and tumor suppression. Functions for p73 and its isoforms in the skin are not as well known.
Given that the p53 family of genes has been shown to interact extensively in the DNA damage response, it will be important to understand how they form complexes in various contexts to regulate the ever-growing list of downstream transcriptional target genes. Performing chromatin immunoprecipitation using antibodies for the various isoforms of the p53 family coupled with next-generation sequencing (ChIP-Seq) in discrete stem cell compartments within the skin and during development or after various stresses will be key to understanding transcriptional regulation by this family of genes. Recently, roles for p63 and p73 in metabolism have been identified (Flores and Lozano 2012; Rufini et al. 2012; Su et al. 2012) and understanding how the p53 family influences the cellular metabolism of skin cells will be an area of new research.
The next decade will bring additional insights to the functions of p53 family genes in multiple contexts and, in particular, skin biology. This knowledge will be important for the generation of therapies for patients afflicted with skin diseases harboring p63 mutations and also aid in the design of targeted therapies for patients with tumors harboring alterations in the p53 family of genes.
Footnotes
Editors: Anthony E. Oro and Fiona M. Watt
Additional Perspectives on The Skin and Its Diseases available at www.perspectivesinmedicine.org
REFERENCES
- Allocati N, Di Ilio C, De Laurenzi V 2012. p63/p73 in the control of cell cycle and cell death. Exp Cell Res 318: 1285–1290 [DOI] [PubMed] [Google Scholar]
- Antonini D, Russo MT, De Rosa L, Gorrese M, Del Vecchio L, Missero C 2010. Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Invest Dermatol 130: 1249–1257 [DOI] [PubMed] [Google Scholar]
- Belyi VA, Ak P, Markert E, Wang H, Hu W, Puzio-Kuter A, Levine AJ 2010. The origins and evolution of the p53 family of genes. Cold Spring Harb Perspect Biol 2: a001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernaskie J, Paris M, Morozova O, Fagan BM, Marra M, Pevny L, Miller FD 2009. SKPs derive from hair follicle precursors and exhibit properties of adult dermal stem cells. Cell Stem Cell 5: 610–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E 2009. Epidermal homeostasis: A balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 10: 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Mohrin M, Sotiropoulou PA, Passegue E 2011. DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell 8: 16–29 [DOI] [PubMed] [Google Scholar]
- Bodo E, Tobin DJ, Kamenisch Y, Biro T, Berneburg M, Funk W, Paus R 2007. Dissecting the impact of chemotherapy on the human hair follicle: A pragmatic in vitro assay for studying the pathogenesis and potential management of hair follicle dystrophy. Am J Pathol 171: 1153–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VA, Komarova EA, Siebenhaar F, Botchkareva NV, Komarov PG, Maurer M, Gilchrest BA, Gudkov AV 2000. p53 is essential for chemotherapy-induced hair loss. Cancer Res 60: 5002–5006 [PubMed] [Google Scholar]
- Botchkarev VA, Komarova EA, Siebenhaar F, Botchkareva NV, Sharov AA, Komarov PG, Maurer M, Gudkov AV, Gilchrest BA 2001. p53 involvement in the control of murine hair follicle regression. Am J Pathol 158: 1913–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VA, Gdula MR, Mardaryev AN, Sharov AA, Fessing MY 2012. Epigenetic regulation of gene expression in keratinocytes. J Invest Dermatol 132: 2505–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkareva NV 2012. MicroRNA/mRNA regulatory networks in the control of skin development and regeneration. Cell Cycle 11: 468–474 [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP 2005. p53 isoforms can regulate p53 transcriptional activity. Genes Dev 19: 2122–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruins W, Zwart E, Attardi LD, Iwakuma T, Hoogervorst EM, Beems RB, Miranda B, van Oostrom CT, van den Berg J, van den Aardweg GJ, et al. 2004. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol Cell Biol 24: 8884–8894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne C, Tainsky M, Fuchs E 1994. Programming gene expression in developing epidermis. Development 120: 2369–2383 [DOI] [PubMed] [Google Scholar]
- Byrne C, Hardman M, Nield K 2003. Covering the limb—Formation of the integument. J Anat 202: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G 2005. The cornified envelope: A model of cell death in the skin. Nat Rev Mol Cell Biol 6: 328–340 [DOI] [PubMed] [Google Scholar]
- Caulin C, Nguyen T, Lang GA, Goepfert TM, Brinkley BR, Cai WW, Lozano G, Roop DR 2007. An inducible mouse model for skin cancer reveals distinct roles for gain- and loss-of-function p53 mutations. J Clin Invest 117: 1893–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D, Su X, Cho MS, Bui NH, Coarfa C, Venkatanarayan A, Benham AL, Flores Gonzalez RE, Alana J, Xiao W, et al. 2014. Induced multipotency in adult keratinocytes through down-regulation of ΔNp63 or DGCR8. Proc Natl Acad Sci 111: E572–E581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikh A, Matin RN, Senatore V, Hufbauer M, Lavery D, Raimondi C, Ostano P, Mello-Grand M, Ghimenti C, Bahta A, et al. 2011. iASPP/p63 autoregulatory feedback loop is required for the homeostasis of stratified epithelia. EMBO J 30: 4261–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong CM, Richardson MK 2009. Pattern formation today. Int J Dev Biol 53: 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crum CP, McKeon FD 2010. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu Rev Pathol 5: 349–371 [DOI] [PubMed] [Google Scholar]
- De Rosa L, Antonini D, Ferone G, Russo MT, Yu PB, Han R, Missero C 2009. p63 suppresses non-epidermal lineage markers in a bone morphogenetic protein-dependent manner via repression of Smad7. J Biol Chem 284: 30574–30582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhouailly D 2009. A new scenario for the evolutionary origin of hair, feather, and avian scales. J Anat 214: 587–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto JE, Glusac EJ 2006. p63 is a useful marker for cutaneous spindle cell squamous cell carcinoma. J Cutaneous Pathol 33: 413–417 [DOI] [PubMed] [Google Scholar]
- Driskell I, Oda H, Blanco S, Nascimento E, Humphreys P, Frye M 2011. The histone methyltransferase Setd8 acts in concert with c-Myc and is required to maintain skin. EMBO J 31: 616–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferone G, Thomason HA, Antonini D, De Rosa L, Hu B, Gemei M, Zhou H, Ambrosio R, Rice DP, Acampora D, et al. 2012. Mutant p63 causes defective expansion of ectodermal keratinocytes and impaired FGF signalling in AEC syndrome. EMBO Mol Med 4: 192–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessing MY, Mardaryev AN, Gdula MR, Sharov AA, Sharova TY, Rapisarda V, Gordon KB, Smorodchenko AD, Poterlowicz K, Ferone G, et al. 2011. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J Cell Biol 194: 825–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores ER, Lozano G 2012. The p53 family grows old. Genes Dev 26: 1997–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, McKeon F, Jacks T 2005. Tumor predisposition in mice mutant for p63 and p73: Evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7: 363–373 [DOI] [PubMed] [Google Scholar]
- Frye M, Benitah SA 2012. Chromatin regulators in mammalian epidermis. Semin Cell Dev Biol 23: 897–905 [DOI] [PubMed] [Google Scholar]
- Gunschmann C, Stachelscheid H, Akyuz MD, Schmitz A, Missero C, Bruning JC, Niessen CM 2013. Insulin/IGF-1 controls epidermal morphogenesis via regulation of FoxO-mediated p63 inhibition. Dev Cell 26: 176–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Keyes WM, Papazoglu C, Zuber J, Li W, Lowe SW, Vogel H, Mills AA 2009. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol 11: 1451–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy MH 1992. The secret life of the hair follicle. Trends Genet 8: 55–61 [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. 2012. A landscape of driver mutations in melanoma. Cell 150: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrie RA, Marques MR, Nguyen BT, Horner JS, Papazoglu C, Bronson RT, Mills AA, Attardi LD 2005. Perp is a p63-regulated gene essential for epithelial integrity. Cell 120: 843–856 [DOI] [PubMed] [Google Scholar]
- Ince TA, Cviko AP, Quade BJ, Yang A, McKeon FD, Mutter GL, Crum CP 2002. p63 coordinates anogenital modeling and epithelial cell differentiation in the developing female urogenital tract. Am J Pathol 161: 1111–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes WM, Pecoraro M, Aranda V, Vernersson-Lindahl E, Li W, Vogel H, Guo X, Garcia EL, Michurina TV, Enikolopov G, et al. 2011. ΔNp63α is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell 8: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Roop DR 2007. Mechanisms regulating epithelial stratification. Annu Rev Cell Dev Biol 23: 93–113 [DOI] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR 2004. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev 18: 126–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D, Levine A 2010. p53 research: The past thirty years and the next thirty years. Cold Spring Harb Perspect Biol 2: a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I 2006. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development 133: 1553–1563 [DOI] [PubMed] [Google Scholar]
- LeBoeuf M, Terrell A, Trivedi S, Sinha S, Epstein JA, Olson EN, Morrisey EE, Millar SE 2010. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal keratinocytes. Dev Cell 19: 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Fuchs E 2005. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437: 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Tumbar T 2012. Hairy tale of signaling in hair follicle development and cycling. Semin Cell Dev Biol 23: 906–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard MK, Kommagani R, Payal V, Mayo LD, Shamma HN, Kadakia MP 2011. ΔNp63α regulates keratinocyte proliferation by controlling PTEN expression and localization. Cell Death Differ 18: 1924–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen J, Yi Y, Li C, Zhang Y 2012. DNA damage down-regulates ΔNp63α and induces apoptosis independent of wild type p53. Biochem Biophys Res Commun 423: 338–343 [DOI] [PubMed] [Google Scholar]
- Liefer KM, Koster MI, Wang XJ, Yang A, McKeon F, Roop DR 2000. Down-regulation of p63 is required for epidermal UV-B-induced apoptosis. Cancer Res 60: 4016–4020 [PubMed] [Google Scholar]
- Lin YL, Sengupta S, Gurdziel K, Bell GW, Jacks T, Flores ER 2009. p63 and p73 transcriptionally regulate genes involved in DNA repair. PLoS Genet 5: e1000680. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lindner G, Botchkarev VA, Botchkareva NV, Ling G, van der Veen C, Paus R 1997. Analysis of apoptosis during hair follicle regression (catagen). Am J Pathol 151: 1601–1617 [PMC free article] [PubMed] [Google Scholar]
- Maas AM, Bretz AC, Mack E, Stiewe T 2013. Targeting p73 in cancer. Cancer Lett 332: 229–236 [DOI] [PubMed] [Google Scholar]
- Mardaryev AN, Gdula MR, Yarker JL, Emelianov VN, Poterlowicz K, Sharov AA, Sharova TY, Scarpa JA, Chambon P, Botchkarev VA, et al. 2014. p63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. Development 141: 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JA, Duijf PH, Doetsch V, Irvine AD, de Waal R, Vanmolkot KR, Wessagowit V, Kelly A, Atherton DJ, Griffiths WA, et al. 2001. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet 10: 221–229 [DOI] [PubMed] [Google Scholar]
- Melino G 2011. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ 18: 1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola ML 2007. p63 in skin appendage development. Cell Cycle 6: 285–290 [DOI] [PubMed] [Google Scholar]
- Millar SE 2002. Molecular mechanisms regulating hair follicle development. J Invest Dermatol 118: 216–225 [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398: 708–713 [DOI] [PubMed] [Google Scholar]
- Mulder KW, Wang X, Escriu C, Ito Y, Schwarz RF, Gillis J, Sirokmány G, Donati G, Uribe-Lewis S, Pavlidis P, et al. 2012. Diverse epigenetic strategies interact to control epidermal differentiation. Nature Cell Biol 14: 753–763 [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH 2013. p53 mutations in cancer. Nat Cell Biol 15: 2–8 [DOI] [PubMed] [Google Scholar]
- Niemann C, Horsley V 2012. Development and homeostasis of the sebaceous gland. Semin Cell Dev Biol 23: 928–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BC, Lefort K, Mandinova A, Antonini D, Devgan V, Della Gatta G, Koster MI, Zhang Z, Wang J, Tommasi di Vignano A, et al. 2006. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev 20: 1028–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus R, Haslam IS, Sharov AA, Botchkarev VA 2013. Pathobiology of chemotherapy-induced hair loss. Lancet Oncol 14: e50–e59 [DOI] [PubMed] [Google Scholar]
- Poulson ND, Lechler T 2012. Asymmetric cell divisions in the epidermis. Int Rev Cell Mol Biol 295: 199–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Jung YS, Chen X 2011. ΔNp63, a target of DEC1 and histone deacetylase 2, modulates the efficacy of histone deacetylase inhibitors in growth suppression and keratinocyte differentiation. J Biol Chem 286: 12033–12041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne T, Brunner HG, van Bokhoven H 2007. p63-associated disorders. Cell Cycle 6: 262–268 [DOI] [PubMed] [Google Scholar]
- Rivetti di Val Cervo P, Lena AM, Nicoloso M, Rossi S, Mancini M, Zhou H, Saintigny G, Dellambra E, Odorisio T, Mahe C, et al. 2012. p63-microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci 109: 1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano RA, Ortt K, Birkaya B, Smalley K, Sinha S 2009. An active role of the ΔN isoform of p63 in regulating basal keratin genes K5 and K14 and directing epidermal cell fate. PLoS One 4: e5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano RA, Smalley K, Liu S, Sinha S 2010. Abnormal hair follicle development and altered cell fate of follicular keratinocytes in transgenic mice expressing ΔNp63α. Development 137: 1431–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano RA, Smalley K, Magraw C, Serna VA, Kurita T, Raghavan S, Sinha S 2012. ΔNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development 139: 772–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufini A, Niklison-Chirou MV, Inoue S, Tomasini R, Harris IS, Marino A, Federici M, Dinsdale D, Knight RA, Melino G, et al. 2012. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev 26: 2009–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Ullrich R, Paus R 2005. Molecular principles of hair follicle induction and morphogenesis. Bioessays 27: 247–261 [DOI] [PubMed] [Google Scholar]
- Sen GL, Boxer LD, Webster DE, Bussat RT, Qu K, Zarnegar BJ, Johnston D, Siprashvili Z, Khavari PA 2012. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev Cell 22: 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom-Feuerstein R, Lena AM, Zhou H, De La Forest Divonne S, Van Bokhoven H, Candi E, Melino G, Aberdam D 2011. ΔNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ 18: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA, Siebenhaar F, Sharova TY, Botchkareva NV, Gilchrest BA, Botchkarev VA 2004. Fas signaling is involved in the control of hair follicle response to chemotherapy. Cancer Res 64: 6266–6270 [DOI] [PubMed] [Google Scholar]
- Simpson CL, Patel DM, Green KJ 2011. Deconstructing the skin: Cytoarchitectural determinants of epidermal morphogenesis. Nat Rev Mol Cell Biol 12: 565–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulou PA, Candi A, Mascre G, De Clercq S, Youssef KK, Lapouge G, Dahl E, Semeraro C, Denecker G, Marine JC, et al. 2010. Bcl-2 and accelerated DNA repair mediates resistance of hair follicle bulge stem cells to DNA-damage-induced cell death. Nat Cell Biol 12: 572–582 [DOI] [PubMed] [Google Scholar]
- Sotiropoulou PA, Karambelas AE, Debaugnies M, Candi A, Bouwman P, Moers V, Revenco T, Rocha AS, Sekiguchi K, Jonkers J, et al. 2013. BRCA1 deficiency in skin epidermis leads to selective loss of hair follicle stem cells and their progeny. Genes Dev 27: 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenn KS, Paus R 2001. Controls of hair follicle cycling. Physiol Rev 81: 449–494 [DOI] [PubMed] [Google Scholar]
- Su X, Cho MS, Gi YJ, Ayanga BA, Sherr CJ, Flores ER 2009a. Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf. EMBO J 28: 1904–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Paris M, Gi YJ, Flores ER 2009b. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell 5: 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, Leung ML, El-Naggar A, Creighton CJ, Suraokar MB, et al. 2010. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 467: 986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Gi YJ, Chakravarti D, Chan IL, Zhang A, Xia X, Tsai KY, Flores ER 2012. TAp63 is a master transcriptional regulator of lipid and glucose metabolism. Cell Metab 16: 511–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Chakravarti D, Flores ER 2013. p63 steps into the limelight: Crucial roles in the suppression of tumorigenesis and metastasis. Nat Rev Cancer 13: 136–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanbokhoven H, Melino G, Candi E, Declercq W 2011. p63, a story of mice and men. J Invest Dermatol 131: 1196–1207 [DOI] [PubMed] [Google Scholar]
- Van Gele M, Kaghad M, Leonard JH, Van Roy N, Naeyaert JM, Geerts ML, Van Belle S, Cocquyt V, Bridge J, Sciot R, et al. 2000. Mutation analysis of P73 and TP53 in Merkel cell carcinoma. Br J Cancer 82: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, Frye M, Benitah SA 2008. MYC in mammalian epidermis: How can an oncogene stimulate differentiation? Nat Rev Cancer 8: 234–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LW, Zelickson AS 1975. Embryology of the epidermis: Ultrastructural aspects: 1. Formation and early development in the mouse with mammalian comparisons. Acta Derm Venereol 55: 161–168 [PubMed] [Google Scholar]
- Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA 2003. The ΔNp63 α phosphoprotein binds the p21 and 14-3-3 σ promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol 23: 2264–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 2: 305–316 [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398: 714–718 [DOI] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E 2008. A skin microRNA promotes differentiation by repressing “stemness.” Nature 452: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Bardot E, Ezhkova E 2012. Epigenetic regulation of skin: Focus on the Polycomb complex. Cell Mol Life Sci 69: 2161–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]