Abstract
Background
Evidence supports the possibility of a role of the Y chromosome in prostate cancer, but controversy exists.
Methods
A novel analysis of a computerized population-based resource linking genealogy and cancer data was used to test the hypothesis of a role of the Y chromosome in prostate cancer predisposition. Using a statewide cancer registry from 1966 linked to a computerized genealogy representing over 1.2 million descendants of the Utah pioneers, 1,000 independent sets of males, each set hypothesized to share the same Y chromosome as represented in genealogy data, were tested for a significant excess of prostate cancer.
Results
Multiple Y chromosomes representing thousands of potentially at-risk males were identified to be associated to have a significant excess risk for prostate cancer.
Conclusions
This powerful and efficient in silico test of an uncommon mode of inheritance has confirmed evidence for Y chromosome involvement in prostate cancer.
Keywords: Y chromosome, prostate cancer, UPDB
INTRODUCTION
Evidence suggests that genes present on the Y chromosome may be involved in increased risk for prostate cancer; however, the Y chromosome has received little attention in conventional genetic studies of prostate cancer. Investigation of the Y chromosome is challenging due to lack of recombination and the high content of repetitive and ampliconic sequences; this results in exclusion of the Y chromosome from most genome sequencing projects 1. The Y chromosome is thought to harbor almost no genes; some rodent groups 2; 3 have lost the Y chromosome and some marsupials have degraded Y chromosomes 4. There is a sole documented human Mendelian hearing loss disorder exhibiting linkage to the Y chromosome 5.
Y haplogroups are geographically specific, so that males from different ethnic groups have different Y lineages and potentially different predisposition to prostate cancer. It is well recognized that the incidence of prostate cancer is higher in African-American populations than in Caucasians, which is higher than in Japanese men 6; 7. Further, cytogenetic studies in primary prostate tumors demonstrate that loss of the Y chromosome is the most common chromosomal aberration observed 8.
The Y chromosome is haploid and does not recombine over much of its length. Consequently, classical linkage mapping studies are not possible. Association studies of haplotypes constructed from genetic markers (short tandem repeats i.e., STRs, or single nucleotide polymorphisms i.e., SNPs) have been performed. To date the results of studies of the Y chromosome in prostate cancer cases and controls representing various ethnic groups have been conflicting. Prostate cancer incidence was reported to vary across Y chromosome lineages in a study of Japanese men 9. No statistically significant differences in haplogroup frequencies were identified in a study of Y chromosomal markers in Korean prostate cancer cases and controls 10. A rare Y lineage associated with an increased risk of prostate cancer was reported in an analysis of 5 binary Y-chromosome markers in Swedish prostate cancer cases and controls 11; however, this was not confirmed in an independent data set. Significant risk- and protective effects were identified in a study that analyzed STRs at Yp11.2 in Portuguese cases and controls; testis-specific Y-encoded protein (TSPY) was proposed as a candidate gene 12. In a study of 4 STRs on the Y-chromosome in Malaysian cases and controls significant risk- and protective haplotypes were identified 13. In a larger study of 34 binary Y chromosome markers in approximately 4,000 cases and 4,000 controls inherited Y-chromosome variation was suggested to play a limited role in prostate cancer in European populations 14.
These previous studies have not clarified the role of the Y chromosome in prostate cancer. One reason for lack of clarity may be insufficiently informative study design. These published analyses of Y chromosomes were performed in unrelated men with prostate cancer, most of whom likely have different Y chromosomes that are associated with differing risks. A more informative design would identify and analyze sets of men who share a specific Y chromosome for association with increased prostate cancer risk. Such a study requires a large population with informative genealogy so that large groups of men sharing the same Y chromosome can be identified, and so that a statistically reliable and powerful test for an excess of prostate cancer can be made for the specific Y chromosomes.
In this study, a population resource for Utah, the Utah Population Database (UPDB) was analyzed to identify large groups of men sharing the same Y chromosome. Prostate cancer risk in each independent Y-chromosome group was estimated in order to identify those specific Y chromosomes with a significant excess of prostate cancer cases.
MATERIALS AND METHODS
The Utah Population Database (UPDB)
The UPDB is a population-based resource containing computerized genealogy records for the European-Americans that settled Utah in the mid 1800s and their modern day descendants. The database originated in the early 1970s 15, and has been used extensively for successful gene identification studies (NF1, BRCA1, BRCA2, p16, APC). Genealogy data added since the 1970s consists of vital statistics data on trios (e.g. mother, father and child from a birth certificate). The database has been record-linked to the Utah Cancer Registry (UCR), which is part of the national SEER cancer surveillance effort, and contains data on every independent primary tumor occurring in the State of Utah since 1966, when the contribution of cancer data to the UCR became mandated by state law.
The UPDB includes over 6.5 million individuals whose records have been linked to over 400,000 cancer records, birth and death certificates, inpatient hospital data, and more 16; 17. There are approximately 1.25 million individuals in the UPDB that have genealogy data for parents, all 4 grandparents, and at least 6 of their 8 great grandparents. Restriction to these individuals with high quality and quantity genealogy data (12 of 14 immediate ancestors) is implied for all further discussion. Within this set of over 1.25 million individuals there are a total of 87,037 individuals diagnosed with cancer; 18,291 of them have been diagnosed with prostate cancer. Each male in the UPDB was assigned to a cohort based on 5-year birth year range and birthplace (Utah or not) for estimation of prostate cancer disease rates. Cohort-specific rates of prostate cancer were estimated by dividing the number of UPDB prostate cancer cases by the total number of UPDB males, by cohort.
All males without a father in the genealogy data (founders) were identified; if they had any male descendants the founder was assigned a unique, sequential Y chromosome id (YID); each of their male descendants, and all of his male descendants, and so forth, were assigned this same YID, effectively identifying each independent Y chromosome segregating in the UPDB. This resulted in the identification of 257,252 YIDs for which there were at least 2 males who shared each Y chromosome (a father and son pair constitute the smallest YID group). The largest YID group included 2,264 males. All YIDs were assumed to be distinct based on genealogy data.
Risk for Prostate cancer
Using cohort-specific prostate cancer rates estimated internally from the UPDB, any group of males identified in UPDB can be tested to determine whether there is a significant excess of prostate cancer observed. For any YID founder in the genealogy we can consider 3 sets of male descendants, the first set includes all of the male descendants of the founder, whether descended through male or female lineages; this subset includes the other two subsets. The second subset is those male descendants who share the Y chromosome of the founder, and the third subset is the male descendants who do not share his Y chromosome. The observed number of prostate cancer cases among the male descendants of each founder was counted, and the expected number of prostate cancer cases among the male descendants of each founder was calculated by multiplying the number of descendants in each cohort by the cohort-specific rate of prostate cancer, and summing over all cohorts. This same method was used to calculate the expected number of prostate cancer cases among the two mutually exclusive subsets of male descendants of each founder. RR’s were calculated as the ratio of the observed to expected number of cases. A two-tailed significance test for the null hypothesis of relative risk = 1.0 was performed. The number of observed cases was assumed to follow a Poisson distribution with mean and standard deviation equal to the expected number of cases. Confidence intervals for the relative risks were estimated by Agresti’s method 18.
Randomization test
An excess of observed prostate cancers among males sharing the same Y chromosome is suggestive of a high-risk Y chromosome; however, it is not sufficient to consider a significant excess among the Y-sharing males in a YID group as conclusive of a Y chromosome effect. Because of the confounding of prostate cancer and maleness, autosomal sharing could be totally, or partially, responsible for what might appear to be Y chromosome sharing. This is obvious when considering, for example, a high-risk prostate pedigree in which most offspring are males. It would not be possible to differentiate between autosomal and Y sharing as being responsible for prostate cancers in such a pedigree.
Since any excess risk for prostate cancer observed in the Y-sharing males may be partly, or entirely, due to autosomal sharing, any excess risk hypothesized to arise from the shared Y chromosome must be assessed against the autosomal risk background of all descendants of each YID founder. A randomization test was used to establish whether each Y-sharing group was significantly different from a cohort-matched, but randomly selected, subset of all of the descendants of the Y-founder. This approach implicitly takes into account the prostate cancer risk in the ‘all descendants’ and the ‘non-Y sharing’ group.
For each YID group, the total number of Y-sharing descendants in each cohort was counted. Then, descendants were chosen at random, without replacement, from the set of all male descendants of the founder, with the restriction that the cohort counts and total counts matched the configuration of the YID group. This was repeated for a total of 10,000 replicates. For each replicate, the number of prostate cancer cases among the replicate sampled set of descendants was counted. These 10,000 counts of cases determined the null distribution. The number of replicates for which the case count exceeded the actual case count in the YID group was used to estimate the empirical statistical significance of an excess of prostate cancer for each YID group. A significance threshold of p < 0.05 was used; each dataset represents an independent experiment.
Institutional Review Board approval was in place for this study. Analysis was performed without use of personal identifiers.
RESULTS
To ensure power to assess prostate cancer risk we considered the 1,000 YIDs (groups of Y-chromosome-sharing males) with the largest total male membership in the UPDB. These YID groups ranged in size from 168 to 2,379 males who share a Y chromosome. Each of these 1,000 YID groups had at least two prostate cancer cases observed among all Y-sharing male descendants; the maximum number of prostate cancer cases observed in any YID group was 59.
Table I shows summary data for the 100 YID groups with the most significant excess of prostate cancer (ranked by p value for the randomization test for excess prostate cancer cases) selected from the largest 1,000 YID groups. Table I includes summary data for each YID, including the number of male descendants of the founder male counted 3 ways: total male descendants (# males), number of male descendants who do not share the Y chromosome of the founder (# non-YID males), and number of male descendants sharing the Y chromosome of the founder (# YID males). Table I also shows the number of prostate cancer cases observed in each of the 3 groups of males, followed by the empirical p-value observed in the randomization test.
Table I.
Test for excess risk for prostate cancer by YID for the 100 YIDs with the most significant randomization test results.
| Rank | # Males |
# Non- YID males |
# YID males |
All PRCA Obs |
Non- YID Obs |
YID Obs |
Emp p-value |
Rank | # Males |
# Non- YID males |
# YID males |
All PRCA Obs |
Non- YID Obs |
YID Obs |
Emp p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2515 | 2295 | 220 | 34 | 15 | 19 | 0.0000 | 51 | 6216 | 5871 | 345 | 57 | 44 | 13 | 0.0336 |
| 2 | 1460 | 1289 | 171 | 20 | 7 | 13 | 0.0001 | 52 | 3229 | 3049 | 180 | 42 | 32 | 10 | 0.0340 |
| 3 | 2829 | 2519 | 310 | 32 | 16 | 16 | 0.0002 | 53 | 2254 | 2001 | 253 | 25 | 15 | 10 | 0.0347 |
| 4 | 3418 | 3061 | 357 | 27 | 12 | 15 | 0.0003 | 54 | 2457 | 2197 | 260 | 22 | 13 | 9 | 0.0353 |
| 5 | 3694 | 3404 | 290 | 34 | 23 | 11 | 0.0006 | 55 | 10595 | 9790 | 805 | 127 | 94 | 33 | 0.0359 |
| 6 | 4403 | 3966 | 437 | 39 | 23 | 16 | 0.0010 | 56 | 3689 | 3468 | 221 | 30 | 24 | 6 | 0.0359 |
| 7 | 1165 | 992 | 173 | 13 | 4 | 9 | 0.0011 | 57 | 6632 | 6397 | 235 | 49 | 42 | 7 | 0.0368 |
| 8 | 4981 | 4633 | 348 | 74 | 50 | 24 | 0.0017 | 58 | 7875 | 7678 | 197 | 70 | 62 | 8 | 0.0372 |
| 9 | 1107 | 861 | 246 | 16 | 3 | 13 | 0.0020 | 59 | 1669 | 1431 | 238 | 24 | 12 | 12 | 0.0380 |
| 10 | 2695 | 2519 | 176 | 22 | 13 | 9 | 0.0021 | 60 | 1647 | 1470 | 177 | 18 | 10 | 8 | 0.0394 |
| 11 | 7349 | 7138 | 211 | 65 | 52 | 13 | 0.0025 | 61 | 4822 | 4151 | 671 | 31 | 17 | 14 | 0.0395 |
| 12 | 2941 | 2569 | 372 | 27 | 10 | 17 | 0.0026 | 62 | 2183 | 1996 | 187 | 23 | 16 | 7 | 0.0398 |
| 13 | 2496 | 2305 | 191 | 37 | 25 | 12 | 0.0035 | 63 | 606 | 391 | 215 | 7 | 0 | 7 | 0.0401 |
| 14 | 7720 | 7299 | 421 | 61 | 47 | 14 | 0.0037 | 64 | 15927 | 15459 | 468 | 135 | 118 | 17 | 0.0408 |
| 15 | 5240 | 4716 | 524 | 71 | 45 | 26 | 0.0048 | 65 | 14093 | 13035 | 1058 | 201 | 157 | 44 | 0.0417 |
| 16 | 2403 | 2145 | 258 | 26 | 12 | 14 | 0.0053 | 66 | 2224 | 1869 | 355 | 27 | 10 | 17 | 0.0437 |
| 17 | 5329 | 4967 | 362 | 82 | 60 | 22 | 0.0058 | 67 | 2989 | 2767 | 222 | 30 | 17 | 13 | 0.0438 |
| 18 | 3452 | 3023 | 429 | 38 | 21 | 17 | 0.0062 | 68 | 5745 | 5506 | 239 | 43 | 36 | 7 | 0.0446 |
| 19 | 3308 | 3138 | 170 | 34 | 25 | 9 | 0.0086 | 69 | 1117 | 865 | 252 | 12 | 3 | 9 | 0.0453 |
| 20 | 1968 | 1653 | 315 | 17 | 7 | 10 | 0.0097 | 70 | 3427 | 3108 | 319 | 38 | 25 | 13 | 0.0454 |
| 21 | 4006 | 3633 | 373 | 42 | 26 | 16 | 0.0098 | 71 | 2891 | 2614 | 277 | 33 | 22 | 11 | 0.0463 |
| 22 | 1547 | 1319 | 228 | 20 | 8 | 12 | 0.0103 | 72 | 942 | 754 | 188 | 3 | 0 | 3 | 0.0488 |
| 23 | 3670 | 3471 | 199 | 30 | 21 | 9 | 0.0103 | 73 | 10154 | 9953 | 201 | 103 | 96 | 7 | 0.0499 |
| 24 | 5230 | 4589 | 641 | 47 | 25 | 22 | 0.0115 | 74 | 2464 | 2266 | 198 | 21 | 14 | 7 | 0.0507 |
| 25 | 751 | 555 | 196 | 13 | 2 | 11 | 0.0115 | 75 | 6898 | 6549 | 349 | 49 | 37 | 12 | 0.0515 |
| 26 | 1487 | 1277 | 210 | 27 | 14 | 13 | 0.0123 | 76 | 2104 | 1883 | 221 | 12 | 5 | 7 | 0.0529 |
| 27 | 1548 | 1338 | 210 | 13 | 6 | 7 | 0.0155 | 77 | 995 | 790 | 205 | 7 | 0 | 7 | 0.0538 |
| 28 | 1986 | 1742 | 244 | 12 | 5 | 7 | 0.0156 | 78 | 4226 | 4012 | 214 | 43 | 32 | 11 | 0.0543 |
| 29 | 2447 | 2205 | 242 | 18 | 11 | 7 | 0.0160 | 79 | 2363 | 2139 | 224 | 25 | 15 | 10 | 0.0558 |
| 30 | 13511 | 12848 | 663 | 128 | 106 | 22 | 0.0169 | 80 | 10971 | 10652 | 319 | 101 | 88 | 13 | 0.0564 |
| 31 | 4344 | 4050 | 294 | 35 | 24 | 11 | 0.0179 | 81 | 6260 | 6071 | 189 | 67 | 59 | 8 | 0.0570 |
| 32 | 984 | 812 | 172 | 22 | 8 | 14 | 0.0196 | 82 | 1422 | 1217 | 205 | 8 | 4 | 4 | 0.0584 |
| 33 | 3613 | 3287 | 326 | 48 | 32 | 16 | 0.0196 | 83 | 6826 | 6555 | 271 | 84 | 71 | 13 | 0.0585 |
| 34 | 1387 | 1199 | 188 | 12 | 4 | 8 | 0.0206 | 84 | 1062 | 886 | 176 | 11 | 4 | 7 | 0.0604 |
| 35 | 2398 | 2150 | 248 | 32 | 17 | 15 | 0.0207 | 85 | 4069 | 3528 | 541 | 39 | 21 | 18 | 0.0619 |
| 36 | 1180 | 969 | 211 | 7 | 1 | 6 | 0.0208 | 86 | 8608 | 8274 | 334 | 66 | 54 | 12 | 0.0642 |
| 37 | 1043 | 872 | 171 | 12 | 4 | 8 | 0.0210 | 87 | 4623 | 4122 | 501 | 69 | 48 | 21 | 0.0644 |
| 38 | 4808 | 4490 | 318 | 60 | 44 | 16 | 0.0218 | 88 | 4363 | 3967 | 396 | 42 | 29 | 13 | 0.0654 |
| 39 | 2552 | 2259 | 293 | 32 | 20 | 12 | 0.0218 | 89 | 3671 | 3446 | 225 | 36 | 29 | 7 | 0.0657 |
| 40 | 3500 | 3262 | 238 | 46 | 32 | 14 | 0.0266 | 90 | 4005 | 3727 | 278 | 43 | 30 | 13 | 0.0658 |
| 41 | 2516 | 2321 | 195 | 16 | 12 | 4 | 0.0270 | 91 | 1979 | 1805 | 174 | 22 | 15 | 7 | 0.0675 |
| 42 | 1371 | 1194 | 177 | 19 | 11 | 8 | 0.0273 | 92 | 2644 | 2338 | 306 | 16 | 9 | 7 | 0.0682 |
| 43 | 2080 | 1783 | 297 | 24 | 9 | 15 | 0.0279 | 93 | 8010 | 7782 | 228 | 76 | 69 | 7 | 0.0683 |
| 44 | 6094 | 5824 | 270 | 58 | 48 | 10 | 0.0288 | 94 | 2014 | 1789 | 225 | 21 | 11 | 10 | 0.0702 |
| 45 | 1975 | 1781 | 194 | 22 | 12 | 10 | 0.0297 | 95 | 1989 | 1689 | 300 | 29 | 14 | 15 | 0.0703 |
| 46 | 10389 | 9192 | 1197 | 94 | 63 | 31 | 0.0316 | 96 | 2318 | 2098 | 220 | 31 | 18 | 13 | 0.0706 |
| 47 | 2879 | 2687 | 192 | 28 | 19 | 9 | 0.0328 | 97 | 8226 | 7332 | 894 | 100 | 65 | 35 | 0.0708 |
| 48 | 5831 | 5498 | 333 | 59 | 48 | 11 | 0.0328 | 98 | 5819 | 5535 | 284 | 31 | 24 | 7 | 0.0729 |
| 49 | 2165 | 1895 | 270 | 17 | 6 | 11 | 0.0330 | 99 | 4484 | 4228 | 256 | 48 | 37 | 11 | 0.0741 |
| 50 | 6652 | 6310 | 342 | 55 | 43 | 12 | 0.0331 | 100 | 2747 | 2567 | 180 | 22 | 16 | 6 | 0.0748 |
The randomization test considers whether the excess of prostate cancers observed in Y-sharing males is significantly greater than that observed in non Y-sharing males, thus a YID with excess prostate cancers among the Y sharing males, even if not a significant excess, would have a significant result if there were many fewer prostate cancers observed in the entire pedigree. An example of this is YID 28 in Table I where 12 overall prostate cancer cases were observed among male descendants of the founder (p=0.19); 7 among Y chromosome sharers (6.35 expected; p = 0.84), and 5 were observed in non Y chromosome sharers (11.3 expected; p =0.07). The randomization test empirical p value for the RR ratio was 0.03. Prioritization of Y chromosomes for study will rank such Y chromosomes lower than those with a significant excess of prostate cancer among Y chromosome sharers.
With a nominal cutoff of p<0.05 one would expect to see 50 false positives out of 1000 independent experiments; 73 of the YID groups summarized in Table I showed a significant excess of prostate cancer cases observed among the YID sharing descendants of the founder compared to all descendants (empirical p < 0.05). This suggests that there are Y chromosomes associated with increased risk for prostate cancer that is independent of risk is conferred by the autosomes. Figure 1 shows an example high-risk prostate pedigree with significant evidence for an excess of cases among Y chromosome sharing males. This example pedigree is the pedigree with rank 32 in Table I.
Figure 1.
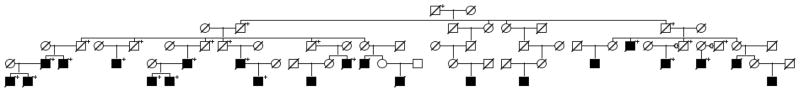
Example pedigree with significant excess of prostate cancer among Y chromosome-sharing males.
High-risk prostate cancer Y chromosome pedigree 32 (from Table I) showing only descendants leading to prostate cancer cases. Those male descendants who share the founder’s Y chromosome are marked with “+”. Only prostate cancer cases diagnosed since the Utah Cancer Registry began in 1966 are known; males in upper generations remain unknown for prostate cancer status.
Characteristics of Y chromosome associated prostate cancer
It is of interest whether prostate cancer cases that appear to be due to Y chromosome variants differ in characteristics of the prostate cancer. The available cancer characteristics for all of the Y sharing prostate cancer cases who were descendants in the 73 YID Y-chromosomes with a significant excess of prostate cancer (n=951 cases) were compared to those characteristics measured for all prostate cancer cases in UPDB (n=18,291). The results for age at diagnosis, BMI, survival months, percent of cases with high grade at diagnosis, and percent of cases with distant stage at diagnosis are shown in Table II.
Table II.
Clinical characteristics of the prostate cancer cases in all Y chromosome-sharing prostate cancer cases belonging to the 73 YID groups with a significant excess of prostate cancer.
| Group | n | mean age | mean BMI | survival months | % high grade | % distant stage |
|---|---|---|---|---|---|---|
| High-risk Y cases | 951 | 69.3 | 26.8 | 101.6 | 26 | 4.8 |
| All prostate cases | 18,291 | 70.5 | 26.9 | 94.6 | 27 | 5.1 |
DISCUSSION
A role of the Y chromosome in prostate cancer risk seems likely given published evidence. In light of the confounding of prostate cancer with male sex and the difficulty of sequencing the Y chromosome, it is understandable that Y chromosome predisposition genes would rarely have been searched for, or identified. Considering the evidence supporting the existence of multiple prostate cancer predisposition genes on autosomal chromosomes (from both linkage and association studies), as well as the likely existence of environmental risk factors, and the potential over-diagnosis of prostate cancer based on PSA screening, it is no surprise that it has been difficult to appropriately test the Y chromosome hypothesis.
Here analysis of a unique population-based genealogical resource linked to 50 years of statewide cancer data has identified specific Y chromosomes shared by multiple males with known cancer status. This resource has allowed a test of whether some Y chromosomes are associated with an excess risk of prostate cancer. Many Y chromosomes are well represented in the UPDB, with from hundreds, and up to thousands, of males sharing the same Y chromosome. This analysis has provided strong evidence of Y chromosome involvement in prostate cancer, and has identified a powerful resource of individuals and pedigrees for efficiently examining these high-risk Y chromosomes to identify and characterize the predisposing genes or variants.
The hypothesis of a Y chromosome contribution to prostate cancer risk has support from many studies. Identification of specific Y chromosomes associated with increased risk is difficult, and was only possible here because the UPDB has decades of linked genealogy and cancer data. Nevertheless, even with genealogy and cancer data in extended pedigrees it is not always possible to discriminate between the possibilities of autosomal versus Y chromosome contribution. In the simple example of a family with a preponderance of sons, autosomal and Y chromosome inheritance could lead to the same pedigree pattern. For this reason we performed a randomization test for Y chromosome status; this test provided significant evidence for the independent role of the Y chromosome for the observed effects.
This analysis of 1,000 Y chromosomes suggests that approximately 73/100 or 7.3% of Y chromosomes are associated with high risk for prostate cancer. Analysis of a subset of the prostate cancer cases from the largest YID groups with the most significant excess of prostate cancer suggests few clinically significant differences in the prostate cancer characteristics compared with all prostate cancer cases in the UPDB, although most of the differences are likely statistically significant given the overall sample size for Utah prostate cancer cases (Table II). We did not precisely estimate penetrance of the hypothesized Y chromosome variant given the censoring of prostate cancer diagnosis data prior to 1966 and the presence of multiple males in each YID group who are still too young to have been diagnosed. A rough estimate of 11% penetrance is obtained when only considering those male descendants born after 1866 and before 1940 in the 73 high risk YID groups considered here.
Since the Utah genealogy data only extends to the mid 1700s, some of the YID groups could potentially represent the same Y chromosome; lack of genealogy data would prevent such identification. In addition some genealogy may not be correctly represented in the UPDB. In future studies, the coalescence of YIDs that appear to be independent, but are not, could be determined by sufficiently informative genotyping or sequencing.
Most of the males identified as sharing the same Y chromosome by their representation in genealogy data are expected to share. In decades of study of Utah high-risk pedigrees, the genealogy data in the UPDB has been used for the ascertainment and study of pedigrees. Pedigree analysis with genetic markers (which allow identification of non-paternity or other incompatibilities) has almost universally confirmed the validity of the genealogy data with very few misrepresentations. This may be due in part to the fact that non-paternity rates in Utah have been reported to be low compared to US figures of 1.5% 19, as well as to the significant attention given to the correct construction of Latter-Day Saint (LDS or Mormon) genealogies.
While prostate cancer is an obvious phenotype to begin investigation of Y-chromosome-associated risk, this innovative study design can be applied to many different phenotypes represented in the Utah resources. Initial focus on those other cancers that also show evidence of Y chromosome losses in tumors is warranted. Loss of the Y chromosome has been noted for many cancers in addition to prostate cancer, including: male breast cancer (63% loss) 20, head and neck tumors (69% loss) 21, urothelial bladder cancer (23% loss) 22, hepatocellular cancer (90% loss) 23, pancreatic cancer (67% loss) 24–26, esophageal squamous cell carcinoma (100% loss) 27, and hematological disorders (10% loss) 28. The analysis of informative Y chromosome groups in the UPDB can increase understanding of a role for the Y chromosome for these cancers, as well as for many other phenotypes of interest. Initial disease association studies for Y chromosome association with risk can be performed efficiently, without data collection and with no subject recruitment.
This analysis has also identified Y chromosomes that appear to be associated with a significant deficit of prostate cancer; data not shown. It is possible that study of these “low-risk” Y chromosomes might allow identification of protective genes or variants for resistance to prostate cancer. Identification of such resistance (or protective) genes for disease could be as valuable as the identification of high-risk genes in terms of advancing our knowledge of prostate cancer genetics. However, the UPDB data are much more powerful for identifying a significant excess, than a significant deficit, for cancer. Data for cancers diagnosed before 1966, or outside Utah, are censored in the UPDB, and incorrect genealogy data typically leads to record linking failure. Thus data quality issues might more easily contribute to the conclusion of a significant deficit of observed cancers in the absence of such an effect. The hypothesis of inherited resistance to prostate cancer is provocative and will be pursued, but is likely not possible as a purely insilico study such as this one.
Association analysis of Y chromosome haplotypes, followed with sequence analysis of regions of interest, could allow identification of the genes or variants on the Y chromosome responsible for the increased risk for prostate cancer observed for some Y chromosomes. Identification of Y chromosome haplotypes or variants associated with increased risk for prostate cancer would expand understanding of the genetics of prostate cancer and potentially permit meaningful counseling and personalized screening for men identified to be at risk. Identification of specific Y-haplotypes associated with increased risk would support a very different sort of risk prediction scenario than individual genetic testing. One high-risk Y chromosome can represent many men. For example, in the Utah database there is a single Y chromosome shared by over 2,000 men born since the 1700’s. Y-haplotype data is relatively inexpensive and straightforward to generate, and risk estimates from a single test could be useful to many individuals.
CONCLUSIONS
Insilico analysis of an existing population-based genealogy linked to cancer records has shown significant evidence for specific Y chromosomes that are associated with increased risk for prostate cancer. This efficient approach using an existing genealogical resource can be extended to consider Y chromosome involvement for other phenotypes, and can be extended to consideration of other modes of inheritance.
Acknowledgments
Research supported by the U.S. Department of Defense Prostate Cancer Research Program of the Office of the Congressionally Directed Medical Research Programs, Grant Number W81XWH-11-1-0342 awarded to Lisa Cannon-Albright; a subcontract from Johns Hopkins University with funds provided by grant R01 CA89600 from the NIH National Cancer Institute (to L.A. Cannon Albright). The project was also supported by Award Number P30CA042014 from the National Cancer Institute, and the Utah Cancer Registry, which is funded by Contract No. HHSN261201000026C from the National Cancer Institute’s SEER Program with additional support from the Utah State Department of Health and the University of Utah. Partial support for all data sets developed within the Utah Population Database (UPDB) was provided by Huntsman Cancer Institute and the University of Utah and the Huntsman Cancer Institute’s Cancer Center Support grant, P30 CA42014 from National Cancer Institute. RAS acknowledges the Keith and Susan Warshaw Fund, the Maurice Warshaw Fund, the C. Scott Watkins Fund, and the Tennity Family Fund in support of this research.
References
- 1.Hughes JF, Rozen S. Genomics and genetics of human and primate y chromosomes. Annual review of genomics and human genetics. 2012;13:83–108. doi: 10.1146/annurev-genom-090711-163855. [DOI] [PubMed] [Google Scholar]
- 2.Just W, Baumstark A, Suss A, Graphodatsky A, Rens W, Schafer N, Bakloushinskaya I, Hameister H, Vogel W. Ellobius lutescens: sex determination and sex chromosome. Sexual development: genetics, molecular biology, evolution, endocrinology, embryology, and pathology of sex determination and differentiation. 2007;1:211–221. doi: 10.1159/000104771. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa Y, Nishida-Umehara C, Matsuda Y, Sutou S, Suzuki H. X-chromosomal localization of mammalian Y-linked genes in two XO species of the Ryukyu spiny rat. Cytogenetic and genome research. 2002;99:303–309. doi: 10.1159/000071608. [DOI] [PubMed] [Google Scholar]
- 4.Graves JA. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 5.Wang QJ, Lu CY, Li N, Rao SQ, Shi YB, Han DY, Li X, Cao JY, Yu LM, Li QZ, et al. Y-linked inheritance of non-syndromic hearing impairment in a large Chinese family. Journal of medical genetics. 2004;41:e80. doi: 10.1136/jmg.2003.012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.SEER Stat Fact Sheets: Prostate. SEER NCI NIH; 2012. [Google Scholar]
- 7.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 8.Brothman AR, Maxwell TM, Cui J, Deubler DA, Zhu XL. Chromosomal clues to the development of prostate tumors. The Prostate. 1999;38:303–312. doi: 10.1002/(sici)1097-0045(19990301)38:4<303::aid-pros6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 9.Ewis AA, Lee J, Naroda T, Sano T, Kagawa S, Iwamoto T, Shinka T, Shinohara Y, Ishikawa M, Baba Y, et al. Prostate cancer incidence varies among males from different Y-chromosome lineages. Prostate cancer and prostatic diseases. 2006;9:303–309. doi: 10.1038/sj.pcan.4500876. [DOI] [PubMed] [Google Scholar]
- 10.Kim W, Yoo TK, Kim SJ, Shin DJ, Tyler-Smith C, Jin HJ, Kwak KD, Kim ET, Bae YS. Lack of association between Y-chromosomal haplogroups and prostate cancer in the Korean population. PloS one. 2007;2:e172. doi: 10.1371/journal.pone.0000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindstrom S, Adami HO, Adolfsson J, Wiklund F. Y chromosome haplotypes and prostate cancer in Sweden. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:6712–6716. doi: 10.1158/1078-0432.CCR-08-0658. [DOI] [PubMed] [Google Scholar]
- 12.Carvalho R, Pinheiro MF, Medeiros R. Localization of candidate genes in a region of high frequency of microvariant alleles for prostate cancer susceptibility: the chromosome region Yp11.2 genetic variation. DNA and cell biology. 2010;29:3–7. doi: 10.1089/dna.2009.0905. [DOI] [PubMed] [Google Scholar]
- 13.Nargesi MM, Ismail P, Razack AH, Pasalar P, Nazemi A, Oshkoor SA, Amini P. Linkage between prostate cancer occurrence and Y-chromosomal DYS loci in Malaysian subjects. Asian Pacific journal of cancer prevention: APJCP. 2011;12:1265–1268. [PubMed] [Google Scholar]
- 14.Wang Z, Parikh H, Jia J, Myers T, Yeager M, Jacobs KB, Hutchinson A, Burdett L, Ghosh A, Thun MJ, et al. Y chromosome haplogroups and prostate cancer in populations of European and Ashkenazi Jewish ancestry. Human genetics. 2012;131:1173–1185. doi: 10.1007/s00439-012-1139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skolnick MH. The Utah genealogical database: a resource for genetic epidemiology. In: Cairns J, Lyon JL, Skolnick M, editors. Banbury Report 4: Cancer Incidence in Defined Populations. New York, NY: Cold Spring Harbor Laboratories; 1980. p. xi.p. 458. [Google Scholar]
- 16.Utah Population Database - Data Overview. Huntsman Cancer Institute, University of Utah; 2013. [Google Scholar]
- 17.Utah Population Database - Overview. Huntsman Cancer Institute, University of Utah; 2013. [Google Scholar]
- 18.Agresti A. Categorical data analysis. New York: Wiley; 1990. [Google Scholar]
- 19.Jorde LB. Inbreeding in the Utah Mormons: an evaluation of estimates based on pedigrees, isonymy, and migration matrices. Annals of human genetics. 1989;53:339–355. doi: 10.1111/j.1469-1809.1989.tb01803.x. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs PA, Maloney V, Cooke R, Crolla JA, Ashworth A, Swerdlow AJ. Male breast cancer, age and sex chromosome aneuploidy. British journal of cancer. 2013;108:959–963. doi: 10.1038/bjc.2012.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kujawski M, Jarmuz M, Rydzanicz M, Szukala K, Wierzbicka M, Grenman R, Golusinski W, Szyfter K. Frequent chromosome Y loss in primary, second primary and metastatic squamous cell carcinomas of the head and neck region. Cancer letters. 2004;208:95–101. doi: 10.1016/j.canlet.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Minner S, Kilgue A, Stahl P, Weikert S, Rink M, Dahlem R, Fisch M, Hoppner W, Wagner W, Bokemeyer C, et al. Y chromosome loss is a frequent early event in urothelial bladder cancer. Pathology. 2010;42:356–359. doi: 10.3109/00313021003767298. [DOI] [PubMed] [Google Scholar]
- 23.Park SJ, Jeong SY, Kim HJ. Y chromosome loss and other genomic alterations in hepatocellular carcinoma cell lines analyzed by CGH and CGH array. Cancer genetics and cytogenetics. 2006;166:56–64. doi: 10.1016/j.cancergencyto.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 24.Wallrapp C, Hahnel S, Boeck W, Soder A, Mincheva A, Lichter P, Leder G, Gansauge F, Sorio C, Scarpa A, et al. Loss of the Y chromosome is a frequent chromosomal imbalance in pancreatic cancer and allows differentiation to chronic pancreatitis. International journal of cancer Journal international du cancer. 2001;91:340–344. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1014>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 25.Zeng X, Zhao DC, Liu TH, Wu SF, Gao J. Detection of Y chromosome loss by fluorescence in situ hybridization in pancreatic cancer. Zhonghua bing li xue za zhi Chinese journal of pathology. 2004;33:523–526. [PubMed] [Google Scholar]
- 26.Long PP, Hruban RH, Lo R, Yeo CJ, Morsberger LA, Griffin CA. Chromosome analysis of nine endocrine neoplasms of the pancreas. Cancer genetics and cytogenetics. 1994;77:55–59. doi: 10.1016/0165-4608(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 27.Yamaki H, Sasano H, Ohashi Y, Shizawa S, Shineha R, Satomi S, Nagura H. Alteration of X and Y chromosomes in human esophageal squamous cell carcinoma. Anticancer research. 2001;21:985–990. [PubMed] [Google Scholar]
- 28.Zhang LJ, Shin ES, Yu ZX, Li SB. Molecular genetic evidence of Y chromosome loss in male patients with hematological disorders. Chinese medical journal. 2007;120:2002–2005. [PubMed] [Google Scholar]


