SUMMARY
Cue1p is an integral component of yeast endoplasmic reticulum (ER)-associated degradation (ERAD) ubiquitin ligase (E3) complexes. It tethers the ERAD ubiquitin-conjugating enzyme (E2), Ubc7p, to the ER and prevents its degradation, and also activates Ubc7p via unknown mechanisms. We have now determined the crystal structure of the Ubc7p-binding region (U7BR) of Cue1p with Ubc7p. The U7BR is a unique E2-binding domain that includes three α-helices that interact extensively with the ‘backside’ of Ubc7p. Residues essential for E2 binding are also required for activation of Ubc7p and for ERAD. We establish that the U7BR stimulates both RING-independent and dependent ubiquitin transfer from Ubc7p. Moreover, the U7BR enhances ubiquitin-activating enzyme (E1)-mediated charging of Ubc7p with ubiquitin. This is the first example where an essential component of E3 complexes both binds to E2 and enhances E2 loading with ubiquitin. These findings provide new insights into mechanisms of stimulating ubiquitination.
INTRODUCTION
Endoplasmic reticulum (ER)-associated degradation (ERAD) is the primary means for the removal of misfolded, misassembled, or degradation-regulated secretory and membrane proteins (Brodsky and Skach, 2011; Smith et al., 2011). ERAD substrates are polyubiquitinated by a multi-step enzymatic cascade and degraded by the 26S proteasome. First, the C-terminal carboxyl group of ubiquitin is linked by a thioester bond to the active-site cysteine of ubiquitin-activating enzyme (E1). Ubiquitin is then transferred to the catalytic cysteine of a ubiquitin-conjugating enzyme (E2) via a trans-thiolation reaction. RING finger ubiquitin ligases (E3s) function as substrate recognition factors and most often mediate the directed transfer of ubiquitin from E2 to the ε-amino group of a lysine on the substrate, or on ubiquitin itself, generating a polyubiquitin chain (Deshaies and Joazeiro, 2009; Metzger et al., 2012). The sites of E1 and RING finger binding on E2s substantially overlap, so E2s must dissociate from ligase domains to be ‘reloaded’ with ubiquitin (Eletr et al., 2005; Olsen and Lima, 2013; Zheng et al., 2000).
The degradation of most known ERAD substrates in Saccharomyces cerevisiae requires one of two RING finger proteins, Hrd1p (Der3p) or Doa10p (Bays et al., 2001; Deak and Wolf, 2001; Swanson et al., 2001). These function as components of the HRD1 and DOA10 E3 complexes, respectively (Carvalho et al., 2006). Essential to both complexes is Cue1p, a transmembrane protein that recruits the ERAD E2, Ubc7p, to the ER (Biederer et al., 1997). Cue1p is also required for the stability of Ubc7p (Biederer et al., 1997; Gardner et al., 2001; Ravid and Hochstrasser, 2007). When Cue1p is absent or under-represented relative to Ubc7p, Ubc7p forms thioester-linked ubiquitin chains on its catalytic cysteine that target it for proteasomal degradation (Ravid and Hochstrasser, 2007). Cue1p’s role in ERAD expands beyond this since a stable, ER membrane-anchored form of Ubc7p still requires Cue1p (Bazirgan and Hampton, 2008; Kostova et al., 2009). In vitro, Cue1p stimulates RING-dependent and independent ubiquitination by Ubc7p (Bazirgan and Hampton, 2008; Kostova et al., 2009). Until now, the molecular mechanisms of activation of Ubc7p by Cue1p have been unknown.
In addition to its N-terminal transmembrane domain, Cue1p has a CUE domain (Shih et al., 2003), to which no functionally significant ubiquitin binding has been ascribed until recently (Bagola et al., 2013), and a C-terminal Ubc7p-binding region (U7BR; Figure 1A; Kostova et al., 2009). We previously identified the U7BR as the minimal region needed for binding to Ubc7p and for degradation of ERAD substrates when Ubc7p is stabilized by tethering it to the ER membrane by addition of a transmembrane domain. The U7BR also stimulates RING-dependent ubiquitination in vitro with Ubc7p (Kostova et al., 2009). Thus, it is the minimal region required for the activity of Cue1p, with other regions enhancing activity.
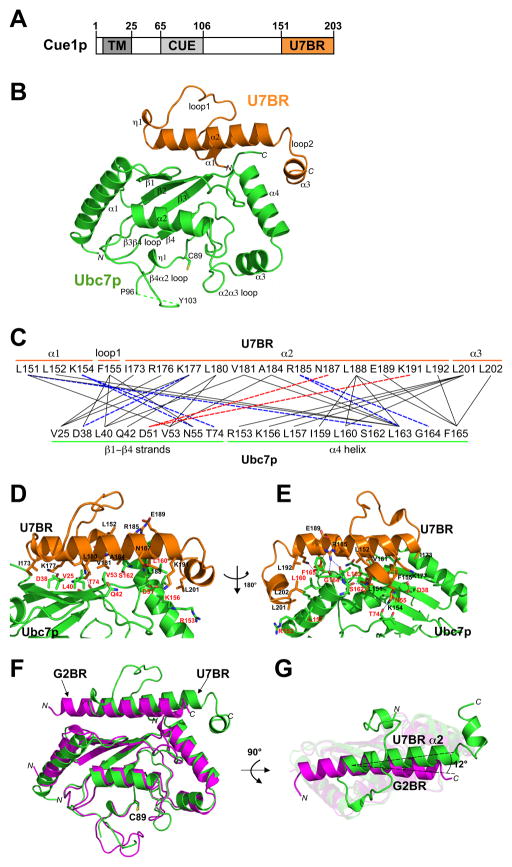
Figure 1.
Crystal structure of the Ubc7p:U7BR complex. (A) Schematic of Cue1p showing its transmembrane (TM), ubiquitin-binding (CUE), and Ubc7p-binding region (U7BR) domains. (B) Ribbon diagram of the Ubc7p:U7BR structure. Helices, strands, and loops are illustrated as spirals, arrows, and tubes, respectively. The N- and C-termini and secondary structure elements are labeled. The structure includes residues 2–96 and 103–165 of Ubc7p, residues 151–203 of Cue1p (U7BR), one 2-(bis-(2-hydroxy-ethyl)-amino)-2hydroxymethyl-propane-1,3-diol (Bis-Tris) molecule, two diethylene glycol molecules (a degradation product of PEG), and 136 water molecules. Amino acids 97–102 in the β4α2 loop of Ubc7p are indicated with a dashed line. There are five (GSAAA) and one (M) extra residues at the N-termini of Ubc7p and U7BR, respectively, resulting from removal of the GST tag and expression of the fragment, respectively. (C) Summary of the Ubc7p:U7BR interactions: solid black lines indicate van der Waals interactions (cutoff distance = 4.0 Å) and dashed lines indicate hydrogen bonds or salt bridges (cutoff distance = 3.5 Å) as seen from one side (red; corresponding to 1D) or the other side (blue; corresponding to 1E) of the U7BR. (D, E) View of the Ubc7p:U7BR interface from opposite directions (180° rotation). Amino acid side chains involved in the electrostatic and van der Waals interactions are shown as sticks. Those labeled in red are from Ubc7p and those in black from the U7BR. (F) Superposition of the structures of Ubc7p:U7BR (in green, this work) and Ube2g2:G2BR (in magenta, PDB 3H8K) on the basis of Cα positions in E2. (G) As in (F), viewing from the top of the G2BR helix (related to the view in panel F by a 90° rotation). See also Supplemental Figure 1.
We now report the crystal structure of Ubc7p in complex with the U7BR. The Ubc7p:U7BR structure reveals an extensive interface between the U7BR, which forms a novel multi-helix E2-binding domain, and the ‘backside’ of Ubc7p. Further, we assess the consequences of U7BR binding and uncover multiple functional outcomes, including stimulation of E1-dependent ubiquitin loading, as well as RING finger-dependent and independent ubiquitin transfer, likely through allosteric effects. Unique among known E2 interactions with E3 complex components, the loading of Ubc7p with ubiquitin is enhanced by binding to Cue1p, suggesting the potential for E2s to be reloaded with ubiquitin when bound to a component of an E3 complex.
RESULTS
Crystal structure of the Ubc7p:U7BR complex
To address the mode of interaction and activation of Ubc7p by the U7BR of Cue1p, they were co-crystallized (Table 1 and Figure 1B). The structure reveals one Ubc7p:U7BR complex in the asymmetric unit and includes residues 2–96 and 103–165 of Ubc7p and residues 151–203 of Cue1p (the U7BR). Ubc7p (Figure 1B, green) has a typical UBC-like fold. Notably, residues 97–102 of the β4α2 loop are disordered without observed electron density. This region lies within an extended loop also found in Ubc7p’s mammalian ortholog, Ube2g2, the related Ube2g1, and Cdc34p and its mammalian orthologs (Ju et al., 2009).
Table 1.
Data collection and refinement statistics
| DATA COLLECTION | |
| Wavelength (Å) | 1.000 |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 46.81, 48.46, 95.73 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 40-1.81 (1.87-1.81)a |
| Number of unique reflections | 20415 (1899) |
| Rmerge (%)b | 6.3 (49.4) |
| I/σ | 27.0 (3.4) |
| Completeness (%) | 98.7 (93.7) |
| Redundancy | 7.5 (6.4) |
|
REFINEMENT
| |
| Resolution | 33.47-1.81 (1.92-1.81) |
| Rwork (%)c | 18.45 (21.37) |
| Rfree (%)d | 22.47 (24.85) |
| No. of Atoms | |
| Protein | 1829 |
| Water | 136 |
| Ligands | 28 |
| B factors | |
| Protein | 29.2 |
| Water | 36.3 |
| Ligands | 49.2 |
| R.m.s.d | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.974 |
| Ramachandran plot | |
| Most favored region (%) | 90.4 |
| Additional allowed region (%) | 9.0 |
| Generously allowed region (%) | 0.5 |
| Disallowed region (%) | 0 |
Values in parentheses are for the highest resolution shell.
Rmerge = Σ|(I − <I>)|/σ(I), where I is the observed intensity.
Rwork = Σhkl | |Fo| − |Fc| |/Σhkl |Fo|, calculated from working dataset.
Rfree is calculated from 4.5% of data randomly chosen and not included in refinement.
The U7BR of Cue1p (Figure 1B, orange) covers the surface of Ubc7p composed of the β-sheet and α4 helix, on the ‘backside’ of the E2. This surface is removed from the catalytic cysteine (C89) and the shared E1- and RING-binding regions of the E2 on the α1 helix, β3β4 loop, and β4α2 loop (Brzovic et al., 2003; Dominguez et al., 2004; Olsen and Lima, 2013; Zheng et al., 2000). The U7BR-interacting surface of Ubc7p is also distinct from regions of E2 that contact thioester-linked ubiquitin, as recently identified in structures of RINGs or a U-box in complex with E2~Ub (Dou et al., 2012; Plechanovova et al., 2012; Pruneda et al., 2012).
The U7BR structure consists of a 310-helix (η1) and a long α-helix (α2) in the central part of the peptide. η1 is linked to a short helix at the N-terminus (α1), while α2 is linked to a short helix at the C-terminus (α3) via loop 1 and loop 2, respectively. No structure in the Protein Data Bank (PDB) shows high structural homology with the U7BR, and all proteins showing overall sequence similarity with the U7BR are from fungi (Supplemental Figure 1A).
The Ubc7p:U7BR interface is largely hydrophobic with stabilizing electrostatic interactions
The extensive Ubc7p:U7BR interface buries a surface of 2064 Å2. The residues involved in intermolecular contacts are summarized in Figure 1C. Hydrophobic interactions dominate the interface, which can be approximately divided into two regions. In one region, five residues (I173, K177, L180, V181, and A184) from the N-terminal half of helix α2, two residues (L151 and L152) from α1, and one residue (F155) from loop 1 of the U7BR surround three residues (V25, L40 and V53) from the β1–β3 strands of Ubc7p. In the other region, six residues (R185, L188, E189, L192, L201, and L202) from α2, loop2, and α3 of the U7BR stack with residues from the α4 helix (R153, K156, L157, L160, L163 and F165) of Ubc7p (Figures 1D and 1E). Surrounding the hydrophobic interactions, there are eight hydrogen bonds and salt bridges, six on one side of the α2 helix of U7BR and two on the other side (Figure 1C; dashed blue and red lines, respectively). Residues L151 and K154 from the α1 helix of the U7BR form hydrogen bonds with S162, Q55, and T74 from Ubc7p, which anchors the α1 helix onto the surface of Ubc7p and stabilizes loop1 of U7BR.
The U7BR is structurally distinct from the gp78 G2BR
Ubiquitination by Ube2g2 is enhanced by the Ube2g2-binding region (G2BR) of the RING E3, gp78 (Das et al., 2009). The 27-amino acid G2BR consists of a single helix that binds the backside of Ube2g2 (Das et al., 2009). Relative to their E2s, the central α2 helix of the 53-amino acid U7BR differs in orientation from the G2BR by only 12° (Figures 1F and 1G), and these two helices are ~40% similar (Supplemental Figure 1A). However, the N- and C-terminal ends of these helices are offset relative to their E2s by at least one complete turn at each end (Figures 1F and 1G). The other two helices of the U7BR, which are required for interaction with Ubc7p and for function (see below; Kostova et al., 2009), further distinguish the two binding domains. Hence, the footprints of U7BR and G2BR on the surface of E2 are different in both location and shape (Supplemental Figures 1B and 1C).
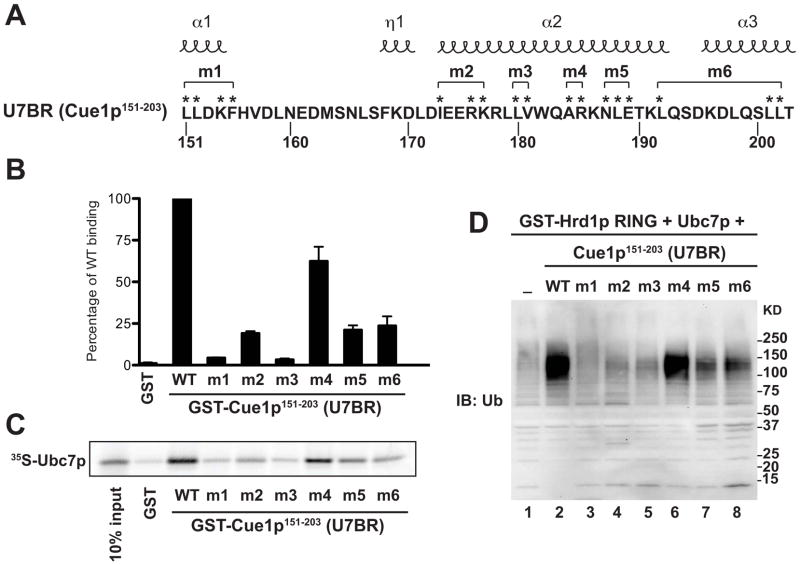
A role for Ubc7p:U7BR contact residues in binding to Ubc7p and activation of ubiquitination in vitro
To validate the Ubc7p:U7BR structure, we analyzed groups of mutations in U7BR residues that contact Ubc7p (Figure 2A, brackets indicate residues mutated together). When U7BR alanine mutants (U7BRm1-U7BRm6; alanine 184 of U7BRm4 was mutated to glycine) are expressed as GST fusion proteins, all show a significant decrease in binding to Ubc7p expressed in vitro (Figures 2B and 2C). Interestingly, U7BRm4, with its only significant change being an arginine to alanine substitution in the α2 helix, also shows diminished binding to Ubc7p.
Figure 2.
Ubc7p contact residues are important for in vitro binding and ubiquitination. (A) The amino acid sequence of the U7BR. Asterisks mark mutated Ubc7p contact residues. Those grouped by brackets (m1-m6) were mutated to alanine together, with the exception of A184 of m4, which was mutated to glycine. Helices above the sequence denote the location of the indicated structural elements. (B) Quantification of the in vitro binding of equimolar amounts of GST or GST-Cue1p151-203 wild-type or the indicated mutant to in vitro transcribed and translated 35S-labeled Ubc7p. Binding was calculated as a percentage of wild-type (WT) levels by quantifying the signal for each mutant using ImageQuant and normalizing to the wild-type signal (100%). Graphed values are the average of three independent experiments and error bars represent standard error. (C) SDS-PAGE showing a representative binding experiment from (B). 10% of the input 35S-Ubc7p used in the binding assays is also shown. (D) In vitro auto-ubiquitination reactions with Glutathione-Sepharose-bound GST-Hrd1p RING (amino acids 321–551), mouse E1, ubiquitin, Cue1p151-203 wild-type or mutants (lanes 2–8), and Ubc7p, both of which were cleaved from N-terminal GST fusions. Lane 1 contains the same reaction mix only without Cue1p151-203. After the reaction, bead-bound proteins were eluted, resolved by SDS-PAGE, and analyzed by immunoblotting.
The U7BR stimulates the in vitro autoubiquitination of Hrd1p by Ubc7p (Kostova et al., 2009), so we evaluated whether U7BR mutations affect this. As shown (Figure 2D), the U7BR-stimulated autoubiquitination of a GST-fusion containing the C-terminal RING domain of Hrd1p (GST-Hrd1p321-551) was significantly reduced by most of the mutants. U7BRm4, despite a loss of ~40% of Ubc7p binding, had a less pronounced effect on ubiquitination. Thus, U7BR residues that contact Ubc7p are required for in vitro binding and for the Ubc7p-dependent activation of ubiquitination by Hrd1p, and decreased binding and loss of ubiquitination correlate well. Notably, mutations in each of the three α-helices (α1-3) severely diminish binding and ubiquitination, demonstrating the significance of each.
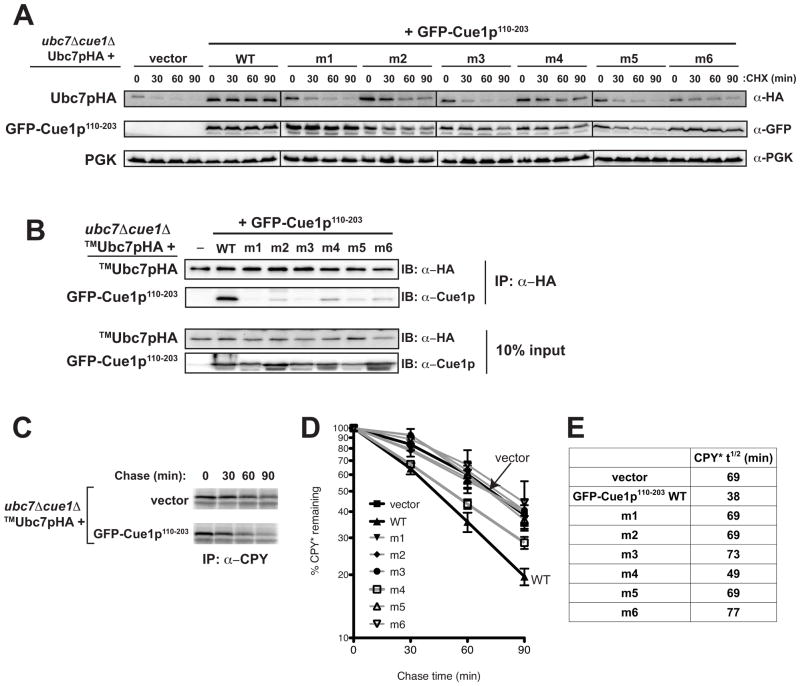
U7BR mutants do not bind Ubc7p in vivo, resulting in Ubc7p instability
To further validate Ubc7p contact residues, we examined U7BR mutants in vivo. Ubc7p is highly unstable in the absence of Cue1p (Biederer et al., 1997; Gardner et al., 2001; Ravid and Hochstrasser, 2007) and the U7BR is insufficient to stabilize Ubc7p from proteasomal degradation (Kostova et al., 2009). Therefore, we used a longer, N-terminal GFP-tagged fragment of Cue1p (GFP-Cue1p110-203, Figure 1A) that efficiently stabilized Ubc7p (Figure 3A, α-HA, compare vector and WT), allowing for Ubc7p stability to serve as an additional functional read-out for binding of U7BR mutants to Ubc7p.
Figure 3.
U7BR contact residues are essential in vivo for Ubc7p’s stability, binding to Cue1p, and ERAD. (A) Degradation of Ubc7pHA and GFP-Cue1p110-203 were assessed by cycloheximide (CHX) chase at the indicated time points using a ubc7Δ cue1Δ strain containing a chromosomal CPY* allele transformed with a low copy plasmid encoding Ubc7pHA and low copy plasmids encoding vector (pRS314) or the indicated GFP-Cue1p110-203 wild-type- or mutant-expressing plasmid. Protein stability was analyzed by SDS-PAGE and immunoblotting with anti-HA and anti-GFP antibodies, respectively. PGK was used as a loading control. (B) The binding of transmembrane anchored TMUbc7pHA to the indicated GFP-Cue1p110-203 wild-type or U7BR mutant was assessed by co-immunoprecipitation of TMUbc7pHA from equivalent amounts of detergent-solubilized protein lysates using anti-HA affinity matrix. Bound proteins were eluted, analyzed by SDS-PAGE, and TMUbc7pHA and GFP-Cue1p110-203 wild-type and indicated mutants were visualized by immunoblotting with anti-HA and anti-Cue1p antibodies, respectively. 10% of each input lysate was also analyzed. (C) The degradation of CPY* was analyzed at the indicated time points by 35S pulse-chase analysis using the indicated strain carrying vector (pRS314) or wild-type GFP-Cue1p110-203. (D) The average of three independent pulse-chase experiments of the GFP-Cue1p110-203 wild-type and mutant strains from (B). Error bars represent the standard error. (E) The half-life (t1/2, in min) of CPY* in each of the strains was calculated from the graphs in (D).
By cycloheximide chase, Ubc7pHA is significantly less stable when co-expressed with any of the U7BR mutants than with ‘wild-type’ GFP-Cue1p110-203 (Figure 3A, α-HA). Most of the GFP-Cue1p110-203 U7BR mutant proteins are themselves relatively stable over the same time frame (Figure 3A, α-GFP). The exception is the m5 mutation, which is significantly less stable than wild-type, making it difficult to conclusively assess its function in vivo. In agreement with its intermediate in vitro binding, the m4 mutant retains some capacity to stabilize the E2.
To assess in vivo binding, we analyzed the association between GFP-Cue1p110-203 and TMUbc7pHA, an ER membrane-anchored form of Ubc7p, which remains stable independent of Cue1p expression (Kostova et al., 2009) (Figure 3B). TMUbc7pHA efficiently co-immunoprecipitated wild-type GFP-Cue1p110-203. In contrast, U7BR mutants all exhibited greatly reduced binding to TMUbc7pHA, mirroring the pattern seen in vitro. Although the GFP-Cue1p110-203 m4 mutant retains some in vivo binding to Ubc7p, it is greatly reduced compared to wild-type. Therefore, the inability of the U7BR mutants to bind to Ubc7p in vivo provides an explanation for the instability of Ubc7pHA when co-expressed with these mutants.
Ubc7p contact residues are essential for degradation of ERAD substrates
To analyze the effect that mutating Ubc7p contact residues within the U7BR has on ERAD, we examined the degradation of the prototypical Hrd1p substrate, CPY*, by pulse-chase analysis (Figures 3C–3E). To dissociate the functions of Cue1p in stabilizing and recruiting Ubc7p to the ER membrane from its role in ERAD activation, we again used TMUbc7pHA. CPY* degradation is impaired in cells lacking Cue1p (t1/2 = ~69 min) and its degradation is accelerated in cells expressing wild-type GFP-Cue1p110-203 (t1/2 = ~38 min) to a rate similar to that seen with full-length Cue1p (data not shown). Cue1p-dependent degradation of CPY* was not restored in strains expressing U7BR mutants. The only exception was the m4 GFP-Cue1p110-203 mutant, which again shows an intermediate phenotype and a partial restoration of CPY* turnover (t1/2 = ~49 min).
To determine the general importance of the U7BR to ERAD, we assessed whether the U7BR was essential for degradation of the Doa10p substrate Ste6p* (Huyer et al., 2004). This required the use of full-length Cue1p, since efficient Doa10p substrate degradation requires the Cue1p transmembrane domain (Supplemental Figure 2A, and M.B.M. and A.M.W, unpublished observations). Cue1p lacking the C-terminal α2 and α3 helices of the U7BR (Cue1p1-173) or mutated in the U7BR α1 helix (Cue1p1-203 m1) both significantly impaired Ste6p* degradation (Supplemental Figure 2B). Collectively, these findings definitively establish U7BR binding to Ubc7p, through multiple structural elements, as being critical to the in vivo ERAD function of the E2.
U7BR binding alters loops surrounding the catalytic cysteine of Ubc7p
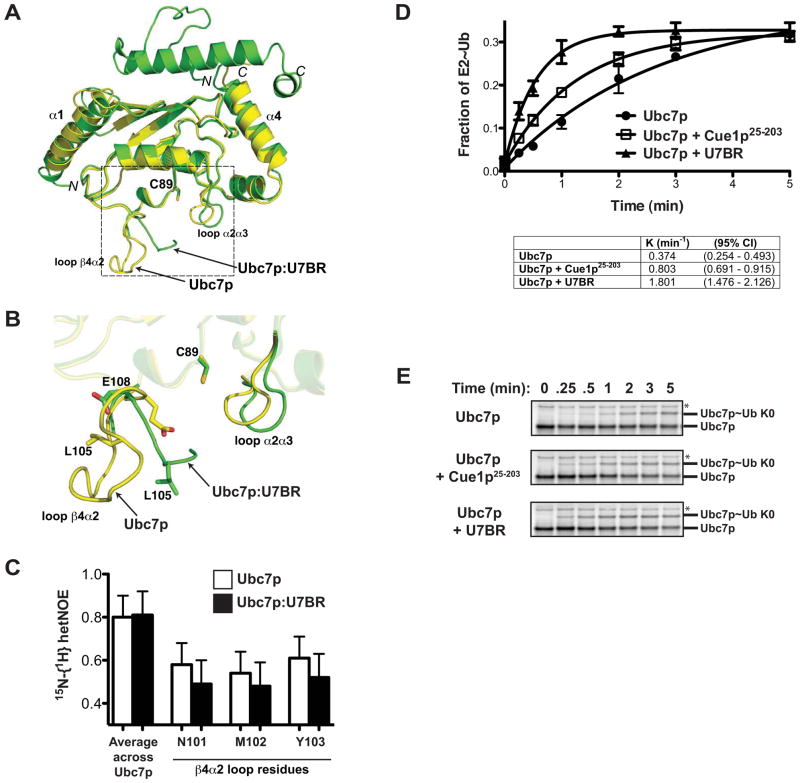
To assess potential mechanisms whereby the U7BR might activate Ubc7p, the Ubc7p:U7BR structure (this work) and the U7BR-free (apo) Ubc7p structure [PDB entry 2UCZ (Cook et al., 1997)] were compared. U7BR binding does not significantly change the overall structure of Ubc7p (Figure 4A). However, there are notable changes in the loops that bracket the catalytic cysteine (C89, Figure 4B). The α2α3 loop of Ubc7p appears to move away from C89 when bound by U7BR. Additionally, while the β4α2 loop of Ubc7p is found in a single conformation in the apo Ubc7p crystal structure, residues 97–102 within this loop are completely disordered with no observed electron density in the Ubc7p:U7BR complex. The limited resolved portions of the β4α2 loop (residues 95–96 and 103–108) exhibit elevated mobility (with an average B value of 72 compared to an average of 29 across all protein atoms) and appear to shift several residues, including E108, away from C89 but re-orient the distal end of the loop toward it. Although crystal lattice contacts may have played a role in stabilizing this loop in a conformation that is favored by its hydrophobic core (residues 96, 100, 105, and 109) in the apo structure, the lack of electron density in the Ubc7p:U7BR structure suggests this region might be more dynamic when U7BR is bound.
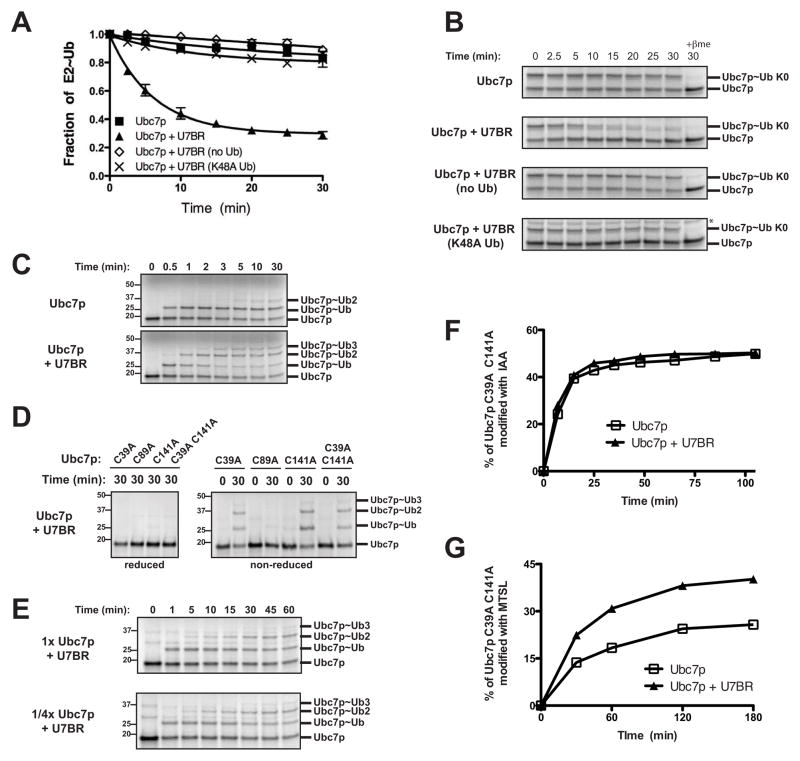
Figure 4.
The U7BR stimulates Ubc7p thioester formation. (A) Superposition of the structures of Ubc7p (in yellow, PDB 2UCZ) and Ubc7p:U7BR (in green, this work) on the basis of Cα positions in E2. The backbone root-mean-square deviation between the free and bound Ubc7p is 1.86 Å over all residues and decreases to 0.99 Å when residues 96–108 are excluded (calculated using program LSQKAB from the CCP4 suite). The rectangle highlights the region enlarged in (B). (B) A close-up view of the framed region in (A) that undergoes conformational changes upon U7BR binding. (C) 15N-{1H} heteronuclear NOE (hetNOE) values for residues of Ubc7p in the β4α2 loop (N101, M102, and Y103; see also Supplemental Figure 3A) compared to the average hetNOE for all residues of Ubc7p. The experiments were repeated for Ubc7p:U7BR peptide complex at concentrations ranging from 0.1 to 0.2 mM. Lower hetOE values indicate increased dynamics. Graphed is the average hetNOE value of two independent experiments. Error bars represent standard error from the mean. (D) Quantification of the rate of K0 ubiquitin loading of in vitro transcribed and translated 35S-labeled Ubc7p alone, or with cleaved Cue1p151-203 (U7BR) or Cue1p25-203. Graphed is the average fraction of E2~Ub for three independent experiments. Error bars represent the standard error; rate constant (K) and 95% confidence index (CI) are shown for each. See also Supplemental Figure 3B. (E) SDS-PAGE showing a representative K0 ubiquitin loading experiment from (D). The asterisk indicates a Promega E. coli T7 S30 Extract System lot-dependent nonspecific band.
To directly assess the dynamics of the β4α2 loop, 15N-{1H} heteronuclear NOE (hetNOE) NMR experiments were performed. Although issues of protein stability precluded assignment of all residues of Ubc7p, we assigned peaks corresponding to several residues in the β4α2 loop (N101, M102, and Y103) via mutational analysis (Supplemental Figure 3A). The average hetNOE value for each of these residues is significantly lower than apo Ubc7p as a whole, indicating that the β4α2 loop is more dynamic than the rest of Ubc7p (Figure 4C). U7BR binding to Ubc7p does not change the overall hetNOE value for Ubc7p, however, the β4α2 loop residues all trend towards a further decrease in hetNOE values, consistent with an even more dynamic state when the U7BR is bound. Although each of these differences falls within the standard error, the trend is consistent with the interpretation of the structural data as reflecting greater loop dynamics when the U7BR is bound to Ubc7p. It is worth noting that this hetNOE experiment measures molecular dynamics on a pico- to nano-second time scale, and any difference in motion over a longer time scale will elude this analysis.
The U7BR accelerates Ubc7p ubiquitin thioester formation
When the G2BR of gp78 binds to Ube2g2, its β4α2 and α2α3 loop impinge on the catalytic cysteine, the side chain of which is also reoriented (Das et al., 2009). These changes correlate with decreased loading of Ube2g2 with ubiquitin. Consistent with differences between bound and free Ubc7p versus Ube2g2 in these regions, the rate of E1-dependent Ubc7p~Ub thioester formation (using ‘K0’ ubiquitin that is unable to form chains) is dramatically accelerated (~5-fold) in the presence of wild-type, but not binding defective, U7BR (Figures 4D and 4E, compare Ubc7p to Ubc7p + U7BR, and Supplemental Figure 3B). The entire cytoplasmic domain of Cue1p (Cue1p25-203) also significantly increases (~2-fold) the rate of formation of Ubc7p~Ub (Figures 4D and 4E, compare Ubc7p to Ubc7p + Cue1p25-203). Whether the acceleration mediated by the U7BR or the complete cytoplasmic domain of Cue1p is most relevant in vivo remains to be determined. For example, in vivo constraints on Cue1p imposed by its transmembrane domain or by binding of polyubiquitinated ERAD substrates are not present in vitro, where there could also be unanticipated interactions that affect loading. Regardless, the significant acceleration of Ubc7p~Ub thioester formation by both the U7BR and the full cytoplasmic domain likely contributes to the in vivo activating role of Cue1p.
The U7BR accelerates the rate of transfer of thioester-linked ubiquitin from the Ubc7p catalytic cysteine to free or thioester-linked ubiquitin
The accelerated rate of ubiquitin loading, in conjunction with the dynamics and structural changes, is suggestive of a more ‘open’ environment around the catalytic cysteine of Ubc7p when the U7BR is bound. This might be predicted to increase the susceptibility of the E2~Ub thioester to nucleophilic attack. This was directly assessed by examining the rate of discharge of the Ubc7p~Ub (K0) thioester to lysines of wild-type ubiquitin free in solution in the absence of a RING domain. The Ubc7p~Ub thioester alone shows negligible discharge (Figures 5A and 5B). However, consistent with alterations surrounding the active site, the transfer of ubiquitin from Ubc7p~Ub is stimulated by addition of wild-type U7BR, but not by a binding-defective mutant (Figures 5A and 5B and Supplemental Figure 5A). Discharge in the presence of the U7BR is facilitated by addition of wild-type, but not K48A, ubiquitin (Figures 5A and 5B), confirming that the Ubc7p~Ub thioester is specifically more accessible to K48 of acceptor ubiquitins. This is consistent with the role of K48-linked polyubiquitin chains in proteasomal degradation.
Figure 5.
The U7BR increases the accessibility of the active site cysteine of Ubc7p to modification. (A) The discharge rate of the 35S-Ubc7p thioester was analyzed in the presence of excess wild-type ubiquitin alone, the U7BR alone, or both the U7BR and either wild-type or K48A ubiquitin. Ubc7p was first loaded with K0 ubiquitin, followed by apyrase treatment to deplete ATP and thereby inactivate E1 prior to the addition of ubiquitin or the U7BR. Three independent experiments for each set of conditions were quantified and the average was graphed as the fraction of E2~Ub remaining at each time point. Error bars represent the standard error. See also Supplemental Figure 5A. (B) SDS-PAGE showing a representative experiment from (A). The asterisk indicates a Promega E. coli T7 S30 Extract System lot-dependent non-specific band. (C) 35S-Ubc7p was loaded with ubiquitin and analyzed at the indicated times points as in Figure 4D except wild-type ubiquitin was used and the reaction was carried out at 24°C to maximize ubiquitin loading and chain formation. (D) 35S-Ubc7p cysteine mutants were loaded with wild-type ubiquitin as in (C) and analyzed at 0 and 30 min. Samples were reduced where indicated by the addition of 10% β-mercaptoethanol. (E) 35S-Ubc7p was loaded with wild-type ubiquitin and analyzed at the indicated times points as described in (C), except using 0.5 pmol 35S-Ubc7p (‘1x Ubc7p’) or 0.125 pmol 35S-Ubc7p (‘1/4x Ubc7p‘). (F, G) 20 μM Ubc7pC39A C141A was reacted with 2 mM iodoacetamide (F) or MTSL (G) in the presence or absence of the U7BR and analyzed by mass spectrometry at the indicated time points. The experiment was repeated twice with similar results and a representative quantification showing the percentage of Ubc7pC39A C141A modified at each time point is shown.
An additional predicted consequence of a U7BR-dependent increase in the accessibility of C89 would be the increased susceptibility of the ubiquitin thioester to nucleophilic attack by a lysine of ubiquitin that is thioester-linked to another Ubc7p molecule. We analyzed this by assessing the formation of Ubc7p C89-linked chains during loading experiments with wild-type ubiquitin. The addition of the U7BR rapidly stimulates the modification of Ubc7p with multiple ubiquitins (Figure 5C), which are reducible (Figure 5D, left panel), indicating that they are thioester-linked to cysteines. To verify that these chains are formed on C89, the other two cysteines of Ubc7p were mutated to alanine without effect (Figure 5D, right panel, Ubc7pC39A C141A). Interestingly, decreasing the concentration of Ubc7p in the reaction by 75% without changing the concentration of free, acceptor ubiquitin had no effect on the rate of formation of C89-linked ubiquitin chains (Figure 5E). This indicates that chain formation does not occur as a random event and implies a direct interaction between Ubc7p~Ub molecules in the presence of the U7BR, which facilitates the formation of ubiquitin chains linked to C89 of Ubc7p.
The increased susceptibility of Ubc7p~Ub to nucleophilic attack is also stimulated by Cue1p25-203 (Supplemental Figures 4A–4C), suggesting it, too, increases the accessibility of Ubc7p~Ub to lysines of ubiquitin.
The U7BR increases the accessibility of the catalytic cysteine of Ubc7p to modification
To directly assess the effect of the U7BR on the accessibility of Ubc7p C89, its rate of alkylation was examined using Ubc7p containing only this cysteine (Ubc7pC39A C141A). When the small, commonly used, alkylating agent iodoacetamide was used, there was no difference in the rate of alkylation of C89 with or without the U7BR bound (Figure 5F). However, when a considerably bulkier alkylating agent was employed (MTSL, C12H21NO3S2), the presence of the U7BR enhanced the rate of E2 modification (Figure 5G), consistent with greater accessibility of C89. Together with the enhanced rate of loading and the RING-independent discharge stimulated by the U7BR, these findings provide strong evidence that the rearrangement and seemingly increased dynamic nature of the β4α2 loop and movement of the α2α3 loop represent functionally significant allosteric alterations around the Ubc7p active site cysteine.
The U7BR increases the affinity of Ubc7p for a RING finger domain
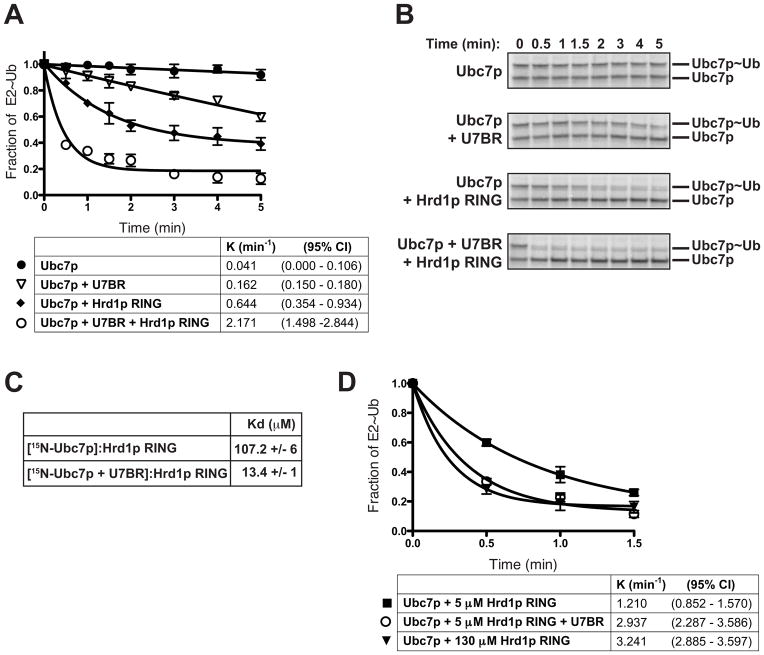
Binding of the gp78 G2BR to Ube2g2 induces allosteric changes that increase the affinity of the E2 for RING finger domains and thus enhances RING-dependent ubiquitination (Das et al., 2009). U7BR binding to Ubc7p also stimulates RING-dependent ubiquitination (Figure 2D and Kostova et al., 2009), so we asked whether similar affinity changes occur. To assess this, the rate of ubiquitin discharge from the Ubc7p~Ub thioester to free ubiquitin was analyzed in the presence or absence of excess purified Hrd1p321-551, containing its RING finger domain. The Hrd1p RING is active and stimulates transfer of thioester-linked ubiquitin from Ubc7p~Ub (Figures 6A and 6B). Consistent with the longer time course shown in Figure 5A, the U7BR alone stimulates a relatively slower rate of Ubc7p~Ub discharge. Strikingly, the U7BR and RING are synergistic in promoting single-turnover ubiquitin discharge, resulting in an ~3-fold increase in rate compared to RING alone. A U7BR mutant that does not bind to Ubc7p fails to synergize with the Hrd1p RING (Supplemental Figure 5A). Stimulation of discharge of Ubc7p~Ub also occurs when the RING domain of Doa10p is used (Supplemental Figure 5D), and when Cue1p25-203 is used with the Hrd1p RING (Supplemental Figures 5B and 5C). Hrd1p RING domain proteins consisting of either 8 amino acids N-terminal (Hrd1p341-551) or 2 amino acids C-terminal (Hrd1p321-401) to the RING finger each stimulate Ubc7p discharge with the U7BR in a manner identical to the longer RING fragment (data not shown). This is consistent with an allosteric effect mediated through the backside binding of the U7BR.
Figure 6.
The U7BR increases Ubc7p affinity for the Hrd1p RING finger domain. (A) Quantification of the rate of discharge of thioester-linked ubiquitin was analyzed as described in Figure 5A at the indicated time points in the presence of wild-type ubiquitin (80 μM) alone, or plus the U7BR (4 μM), plus the Hrd1p RING (5 μM), or plus both U7BR and RING. Data are graphed as the fraction of E2~Ub remaining at each time point from the average of three independent discharge experiments. Error bars represent the standard error; rate constant (K) and 95% confidence index (CI) are shown for each condition. (B) SDS-PAGE showing a representative experiment from (A). (C) Kd values for 15N-Ubc7p alone or bound to the U7BR for the Hrd1p RING were calculated by NMR. Error reflects the standard error from mean. Primary NMR data used to calculate these values is shown in Supplemental Figures 6A–6C; values were calculated over all peaks that shifted upon binding to the Hrd1p RING. (D) Quantification of 35S-Ubc7p discharge, performed as in (A), using the indicated Hrd1p RING concentrations. Three independent experiments were quantified and graphed as in (A). Error bars represent the standard error.
To directly assess whether the U7BR is functioning by increasing the affinity of the E2 for the RING domain, we measured the affinity of Ubc7p for the Hrd1p RING by NMR. The dissociation constant (Kd) of Ubc7p for the RING was 107.2 (±6) μM, similar to that reported for other E2-RING domain pairs (Deshaies and Joazeiro, 2009). Strikingly, the Kd of Ubc7p for the RING decreases to 13.4 (±1) μM when the U7BR is present, an ~8 fold increase in the affinity (Figure 6C and Supplemental Figures 6A–6C). We find a similar U7BR-mediated increase in the affinity of Ubc7p for Doa10p (Supplemental Figure 5E).
To assess the extent to which the increased rate of Ubc7p~Ub discharge is attributable to this change in affinity, we re-analyzed the rate of discharge at Hrd1p RING concentrations greater than the Kd for Ubc7p (~130 μM). Under these conditions, ubiquitin is discharged from Ubc7p~Ub at an enhanced rate, regardless of the presence of the U7BR (Figure 6D). Therefore, the increased affinity of Ubc7p for RING finger domains can account for the in vitro stimulation of RING-dependent discharge of Ubc7p~Ub by the U7BR, independent of changes that affect the accessibility of the catalytic cysteine.
DISCUSSION
The U7BR of Cue1p, a critical yeast ERAD E3 complex component, is a unique multi-helix E2-interacting domain that binds to an extended region on the backside of Ubc7p, away from both the overlapping site of RING and E1 interactions and the catalytic cysteine (C89) (Eletr et al., 2005; Olsen and Lima, 2013; Zheng et al., 2000). The U7BR activates Ubc7p by multiple mechanisms whose relative in vivo importance are difficult to assess, but together have the potential to greatly enhance the rate of ubiquitination. Analogous to the allosteric effect of the G2BR on the binding of Ube2g2 to RING fingers (Das et al., 2009), the U7BR increases the affinity of Ubc7p for its cognate RING fingers. Based on the degree of stimulation, this is likely to be a critical factor in Ubc7p activation. As the sites of RING finger and U7BR binding to E2 are distinct, this affinity increase is likely also due to allosteric mechanisms.
The U7BR additionally increases the availability of C89 of Ubc7p, as suggested by structural changes in the loops surrounding C89 and its enhanced susceptibility to alkylation when the U7BR is bound. Interestingly, the extended β4α2 loop is significantly more dynamic than the rest of Ubc7p even without the U7BR. The precise role of this in ubiquitin transfer requires further study. These U7BR-induced active site alterations correlate with both an acceleration in ubiquitin loading by E1 and enhanced transfer of thioester-linked ubiquitin (Ubc7~Ub) to K48 of acceptor ubiquitins. Importantly, the Cue1p-dependent enhancement of ubiquitin loading implies that Ubc7p can be loaded by E1 while Cue1p is bound, providing what we believe to be the first example of a stable ubiquitin ligase component (Carvalho et al., 2006) that can enhance the loading of an E2 with ubiquitin.
In vivo, we find Cue1p in excess of Ubc7p (our unpublished observations). This is consistent with findings that Ubc7p expressed in excess of Cue1p in vivo is unstable due to autoubiquitination and proteasomal degradation (Ravid and Hochstrasser, 2007), suggesting that the large majority of Ubc7p will be Cue1p bound, and therefore the mechanisms of E2 activation presented herein would be predicted to be relevant in vivo. Cue1p not only ensures that Ubc7p is stable and properly localized to the HRD1 and DOA10 ERAD E3 complexes in the ER (Carvalho et al., 2006), but also activates the E2 for ubiquitination of ERAD substrates.
The formation of thioester-linked ubiquitin chains on C89 of Ubc7p was previously reported (Bazirgan and Hampton, 2008; Ravid and Hochstrasser, 2007), as was the formation of such chains on its mammalian ortholog, Ube2g2 (Li et al., 2007). In vitro, the U7BR also stimulates formation of C89-linked chains (Figure 5C). In vivo, these target Ubc7p for degradation in the absence of stoichiometric amounts of Cue1p. By localizing Ubc7p to the ER and enhancing its interaction with E3, Cue1p may facilitate the transfer of such chains to substrates, explaining why they are not detectable when Cue1p and Ubc7p are present at endogenous levels (Ravid and Hochstrasser, 2007). The in vitro formation of C89-linked chains occurs in a Ubc7p concentration-independent manner (Figure 5E), suggesting a physical interaction between Ubc7p molecules. Yeast two-hybrid data for Ubc7p is also suggestive of dimerization (Chen et al., 1993). As there is no in vitro evidence for dimerization or higher order oligomerization of Ubc7p (Bazirgan and Hampton, 2008; data not shown), this implies that thioester-linked Ubc7p (Ubc7p~Ub) is required for these homotypic interactions.
While Cue1p25-203 retains all of the functions we ascribe to the U7BR, regions N-terminal of the U7BR may affect Ubc7p in a distinct manner, in accordance with previous observations (Bazirgan and Hampton, 2008; Kostova et al., 2009). This is consistent with a role for the CUE domain of Cue1p in the targeting of some substrates (Bagola et al., 2013). We have, however, thus far been unsuccessful in crystallizing Cue1p fragments longer than the U7BR, either alone or in complex with Ubc7p.
Several other proteins contact E2s on their backside (Wenzel et al., 2011). Ubiquitin itself binds with low affinity to a region on E2s that overlaps where the U7BR binds Ubc7p and stimulates ubiquitin chain formation (Brzovic et al., 2006). SUMO (small ubiquitin-related modifier), NIP45, and the SUMO E3 Nup358/RanBP2 all contact the backside of the SUMO E2, Ubc9 (Prudden et al., 2009; Reverter and Lima, 2005; Sekiyama et al., 2010).
Most comparable to the U7BR is the G2BR of gp78 (Das et al., 2009). Both induce allosteric changes around C89, but the G2BR inhibits rather than stimulates E1-mediated E2 loading, suggesting a need for dissociation of the G2BR for Ube2g2 to be reloaded with ubiquitin (Das et al., 2009). The Rad6-binding domain (R6BD) of the Rad18 RING E3 (Hibbert et al., 2011) interacts with its E2, Rad6, on the latter’s backside, but in a significantly different orientation. The α-helical R6BD competes with ubiquitin binding and decreases ubiquitination. The RING finger E3 AO7/RNF25 has a secondary binding domain that binds to the backside of its E2 and imparts functionally distinct effects (our unpublished observations). The peroxisomal protein, Pex22p, binds its E2, Pex4p, (Williams et al., 2011) on a different face of the E2 than the U7BR, but also stimulates E3-independent auto-ubiquitination by E2, through unknown mechanisms. Also, Ubc13~Ub forms heterodimers with the E2-like molecule, Mms2, through a distinct face and stimulates formation of K63-linked chains (Eddins et al., 2006). There is also evidence for E2 homodimerization via unknown domains (David et al., 2010) and herein we provide functional evidence for such dimerization.
It is likely that RING-independent binding sites on E2s are prevalent and heterogeneous and that the functional consequences of these interactions will be similarly varied. Among those described to date, the U7BR stands out as an extreme example of a domain that properly localizes the E2 in the cell and enhances ubiquitination through multiple distinct mechanisms.
EXPERIMENTAL PROCEDURES
Crystallization, Data Collection, and Structure Determination
The initial screening for crystallization conditions of the Ubc7p:U7BR complex was carried out with a Hydra II Plus One robot system (Matrix Technologies Corporation), using the sitting drop vapor diffusion method at 19±1°C. The optimization of crystallization conditions was set up at 4°C to reduce the nucleation rate and crystallization speed, thus improving the diffraction quality of the crystals. Crystals suitable for X-ray diffraction experiments were grown by mixing 1 μL protein solution (12 mg/mL in 150 mM NaCl, 50 mM Tris-HCl, pH 7.5) and 1 μL reservoir solution (25% w/v PEG3350 and 0.2 M NH4CH2COOH in 0.1 M Bis-Tris buffer, pH 6.5), and the 2 μL droplets were equilibrated against 70 μl reservoir solution. The rod-shaped crystals grew to full size (~0.08 mm × 0.08 mm × 0.15 mm) in three days. They were flash-cooled in liquid nitrogen after a brief soak in the reservoir solution premixed with 25% (wt/vol) PEG400 as a cryoprotectant. X-ray diffraction data were collected from a single crystal at beamline BM-22 of SER-CAT at the Advanced Photon Source, Argonne National Laboratory, and processed using the HKL3000 program suite (Otwinowski and Minor, 1997).
The Ubc7p:U7BR structure was determined by molecular replacement using phenix.automr of the PHENIX program suite (Adams et al., 2010). The crystal structure of yeast Ubc7p (Cook et al., 1997) was used as the search model. The U7BR structure was built into the difference electron density map, first using phenix.autobuild and then by manual interpretation. The structure was refined with phenix.refine of PHENIX and model building and adjustment was done with COOT (Emsley and Cowtan, 2004). About 4.5% of the reflections were randomly selected for cross validation. Water, Bis-Tris, and diethylene glycol molecules were included at the last stage of refinement on the basis of difference electron density (Fo-Fc, above 3s) and verified with omit maps. The refined structure was validated using the PROCHECK (Laskowski et al., 1993) and WHATIF(Hooft et al., 1996) programs. Figures were generated with PyMol (DeLano Scientific LLC).
E2 loading and discharge
E2s were in vitro transcribed and translated from pET3a plasmids in the Escherichia coli T7 S30 Extract System using [35S]Methionine and used in loading and discharge experiments as previously described (Das et al., 2009), with the following exceptions. For loading experiments, 5 μM wild-type or K0 ubiquitin and ~2 μM cleaved Cue1p151-203 (when indicated) were used and reactions were incubated at the indicated temperature. For discharge reactions, E2 was loaded as described above, except without Cue1p151-203. Reactions were quenched with 10 U/mL apyrase (Sigma) for 5 min at room temperature (Ozkan et al., 2005). Discharge was initiated by the addition of 80 μM wild-type or K48A ubiquitin (unless otherwise indicated; added ubiquitins serve as nucleophilic acceptors for ubiquitin discharge from E2) in 50mM Tris-HCl pH 7.5 with or without Cue1p151-203 (4 μM) or Hrd1p321-551 (5 μM unless otherwise indicated). Reactions were incubated at 20°C. For both loading and discharge reactions, samples were run on 10% Bis-Tris gels in 1X NuPAGE® MES SDS running buffer (Invitrogen). After gels were dried, they were visualized using a Storm Phosphorimager. The fraction of E2~Ub was quantified with ImageQuant™ Software and data analysis was carried out using Prism software (GraphPad).
NMR Spectroscopy
For NMR titration experiments, pET3a-GB1-His6-Thrombin-Ubc7p was expressed, refolded in 4 M urea, and purified by Ni-NTA purification. The GB1 tag was then cleaved by Thrombin protease, yielding purified Ubc7p. Ubc7p was incubated with GST-Cue1p151-203 prior to cleavage of the GST tag by PreScission protease and purification on the S-300 sizing column, yielding the Ubc7p:U7BR complex. All NMR samples were dialyzed in 50mM Tris pH 7.2, 2mM TCEP. Ubc7p and Ubc7p:U7BR (50–75 μM) were titrated with up to 3x concentration equivalents of purified Hrd1p321-551 or Doa10p1-112 RING fragments and the changes in amide shifts of Ubc7p were monitored by 1H-15N HSQC experiments, which were plotted and fitted against protein to ligand concentrations to obtain Kd values.
For 15N-{1H} heteronuclear NOE (hetNOE) experiments, pET3a-Ubc7p wild-type (pMM43) or mutants (pMM154, 155, and 156) were expressed, refolded in 4 M urea, and purified by ion exchange followed by gel-filtration. A U7BR peptide synthesized at the Keck Biotechnology Resource Center was used to form the Ubc7p:U7BR complex. Before acquiring hetNOE and HSQC spectra for assignments, all samples were co-dialyzed in 50 mM Tris pH 7.2, 50 mM Arg, 50 mM Glu and 2 mM TCEP. The NMR sample concentrations ranged from 0.1 to 0.2 mM. The hetNOE were taken as the ratio of peak intensities observed for experiments with and without 3.0 seconds of 1H-presaturation during the recycle delay acquired in an interleaved manner. Experimental errors for the relaxation time measurements were estimated based on the observed root-mean-square deviation between peak intensities measured for replicate experiments.
Alkylation and Mass Spectroscopy
Ubc7pC39A C141A (~20 μM) alone or bound to U7BR was treated with 2 mM MTSL (Toronto Research Chemicals) or iodoacetamide (Sigma) and incubated at room temperature (MTSL) or on ice in the dark (iodoacetamide). At the indicated times, samples were removed and analyzed by ESI mass spectrometry to determine the fraction of Ubc7pC39A C141A modified.
Supplementary Material
HIGHLIGHTS.
U7BR of Cue1p binds on the ‘backside’ of Ubc7p, away from its active site.
Binding via the backside is required for Ubc7p function in vivo and in vitro.
U7BR-mediated changes near Cys89 activate Ubc7p for ubiquitination.
The U7BR markedly increases the affinity of Ubc7p for RING finger domains.
Acknowledgments
We thank Stanley Lipkowitz and Yien Che Tsai for helpful discussion and comments regarding this manuscript. X-ray diffraction data were collected at the 22-BM synchrotron beamline of the Southeast Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source, Argonne National Laboratory. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Supplemental Experimental Methods
Additional experimental methods can be found in the Supplemental Information.
Accession Numbers
Coordinates and structure factors of the Ubc7p:U7BR complex were deposited in the Protein Data Bank under accession code 4JQU.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagola K, von Delbruck M, Dittmar G, Scheffner M, Ziv I, Glickman MH, Ciechanover A, Sommer T. Ubiquitin binding by a CUE domain regulates ubiquitin chain formation by ERAD E3 ligases. Mol Cell. 2013;XX:XXX–XXX. doi: 10.1016/j.molcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Bazirgan OA, Hampton RY. Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J Biol Chem. 2008;283:12797–12810. doi: 10.1074/jbc.M801122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Skach WR. Protein folding and quality control in the endoplasmic reticulum: Recent lessons from yeast and mammalian cell systems. Curr Opin Cell Biol. 2011;23:464–475. doi: 10.1016/j.ceb.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D, 3rd, Fukuda M, Ohta T, Klevit R. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci U S A. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Cook WJ, Martin PD, Edwards BF, Yamazaki RK, Chau V. Crystal structure of a class I ubiquitin conjugating enzyme (Ubc7) from Saccharomyces cerevisiae at 2.9 angstroms resolution. Biochemistry. 1997;36:1621–1627. doi: 10.1021/bi962639e. [DOI] [PubMed] [Google Scholar]
- Das R, Mariano J, Tsai YC, Kalathur RC, Kostova Z, Li J, Tarasov SG, McFeeters RL, Altieri AS, Ji X, et al. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell. 2009;34:674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem. 2010;285:8595–8604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak PM, Wolf DH. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J Biol Chem. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Bonvin AM, Winkler GS, van Schaik FM, Timmers HT, Boelens R. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure. 2004;12:633–644. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gardner RG, Shearer AG, Hampton RY. In vivo action of the HRD ubiquitin ligase complex: mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol Cell Biol. 2001;21:4276–4291. doi: 10.1128/MCB.21.13.4276-4291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbert RG, Huang A, Boelens R, Sixma TK. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc Natl Acad Sci U S A. 2011;108:5590–5595. doi: 10.1073/pnas.1017516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- Ju T, Bocik W, Majumdar A, Tolman JR. Solution structure and dynamics of human ubiquitin conjugating enzyme Ube2g2. Proteins. 2009;78:1291–1301. doi: 10.1002/prot.22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostova Z, Mariano J, Scholz S, Koenig C, Weissman AM. A Ubc7p-binding domain in Cue1p activates ER-associated protein degradation. J Cell Sci. 2009;122:1374–1381. doi: 10.1242/jcs.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallog. 1993;26:283–291. [Google Scholar]
- Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 2012;125:531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SK, Lima CD. Structure of a Ubiquitin E1-E2 Complex: Insights to E1-E2 Thioester Transfer. Mol Cell. 2013;49:884–896. doi: 10.1016/j.molcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci U S A. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Perry JJ, Arvai AS, Tainer JA, Boddy MN. Molecular mimicry of SUMO promotes DNA repair. Nat Struct Mol Biol. 2009;16:509–516. doi: 10.1038/nsmb.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M. Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol. 2007;9:422–427. doi: 10.1038/ncb1558. [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiyama N, Arita K, Ikeda Y, Hashiguchi K, Ariyoshi M, Tochio H, Saitoh H, Shirakawa M. Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45. Proteins. 2010;78:1491–1502. doi: 10.1002/prot.22667. [DOI] [PubMed] [Google Scholar]
- Shih SC, Prag G, Francis SA, Sutanto MA, Hurley JH, Hicke L. A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J. 2003;22:1273–1281. doi: 10.1093/emboj/cdg140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochem J. 2011;433:31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, van den Berg M, Panjikar S, Stanley WA, Distel B, Wilmanns M. Insights into ubiquitin-conjugating enzyme/co-activator interactions from the structure of the Pex4p:Pex22p complex. EMBO J. 2011;31:391–402. doi: 10.1038/emboj.2011.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.