Abstract
DNA methylation is an epigenetic mechanism for gene silencing. In Arabidopsis, MET1 is the primary DNA methyltransferase that maintains CG DNA methylation. Plants having an overall reduction of MET1 activity, caused by a met1 mutation or a constitutively expressed MET1 antisense gene, display genome hypomethylation, inappropriate gene and transposon transcription, and developmental abnormalities. However, the effect of a transient reduction in MET1 activity caused by inhibiting MET1 expression in a restricted set of cells is not known. For this reason, we generated transgenic plants with a MET1 antisense gene fused to the DEMETER (DME) promoter (DME:MET1 a/s). Here we show that DME is expressed in leaf primordia, lateral root primoridia, in the region distal to the primary root apical meristem, which are regions that include proliferating cells. Endogenous MET1 expression was normal in organs where the DME:MET1 a/s was not expressed. Although DME promoter is active only in a small set of cells, these plants displayed global developmental abnormalities. Moreover, centromeric repeats were hypomethylated. The developmental defects were accumulated by the generations. Thus, not maintaining CG methylation in a small population of proliferating cells flanking the meristems causes global developmental and epigenetic abnormalities that cannot be rescued by restoring MET1 activity. These results suggest that during plant development there is little or no short-term molecular memory for reestablishing certain patterns of CG methylation that are maintained by MET1. Thus, continuous MET1 activity in dividing cells is essential for proper patterns of CG DNA methylation and development.
Keywords: DEMETER, DNA methylation, FWA, METHYLTRANSERASE1, transposon
INTRODUCTION
Eukaryote genomes are covalently modified on both the DNA and the associated histones (Henderson and Jacobsen, 2007). CG DNA methylation plays a critical role in genome stabilization. DNA methylation is also important for the regulation of genomic imprinting in mammals and flowering plants (Bender, 2004; Chan et al., 2005; Scott and Spielman, 2006; Zhu et al., 2008).
Arabidopsis is an excellent model system to study DNA methylation and subsequent epigenetic inheritance owing to the viability of DNA methyltransferase mutants. In Arabidopsis, the MET1 gene is responsible for maintaining most of the CG DNA methylation. Plants with loss-of-function met1 mutations or MET1 antisense transgenes exhibit global DNA hypomethylation, diverse phenotypes such as homeotic transformation of floral organs, and ectopic gene and transposon expression (Finnegan et al., 1996; Kankel et al., 2003; Ronemus et al., 1996; Saze et al., 2003; Xiao et al., 2003). These results show that DNA methylation is a critical regulatory epigenetic mark for genome integrity and plant development.
Loss-of-function met1 mutations and MET1 antisense transgenic plants uniformly reduce DNA methyltransferase activity in all or most plant cells (Finnegan et al., 1996; Kankel et al., 2003; Ronemus et al., 1996; Saze et al., 2003; Xiao et al., 2003). However, little is known about the effect of a transient reduction of MET1 activity caused by inhibiting MET1 gene expression in a small subset of cells. Is there a short-term molecular memory that will allow patterns of DNA methylation to be reset after MET1 activity is restored in the rest of the plant? Or will the loss of DNA methylation even for a small number of cell divisions lead to irreversible global epigenetic changes outside the zone of MET1 inhibition? To address these questions, we reduced MET1 activity in tissues where cells are actively dividing by spatially and temporally controlling expression of a MET1 antisense gene with the DEMETER (DME) promoter.
DME is a member of a family of helix-hairpin-helix DNA glycosylases in Arabidopsis (Choi et al., 2002). DME excises 5-methylcytosine, which is replaced by cytosine by the base excision DNA repair pathway (Gehring et al., 2006; Morales-Ruiz et al., 2006). Thus, DME DNA glycosylase functions as an active DNA demethylase. The maternal DME allele, expressed in the central cell of the female gametophyte, demethylates and activates maternal allele expression of two Polycomb group protein genes, MEDEA (MEA) (Choi et al., 2002; Gehring et al., 2006) and FERTILIZATION-INDEPENDENT SEED2 (FIS2) (Jullien et al. 2006), as well as a homeodomain transcription factor gene, FLOWERING WAGENINGEN (FWA) (Kinoshita et al., 2004). This causes imprinted expression of these genes in the endosperm. Imprinting is critical to reproduction and seeds that inherit a maternal null dme allele abort their development and are not viable (Choi et al., 2002). However, dme-1, a weak allele, allows for rare maternal mutant allele transmission and the formation of homozygous dme-1 plants, which display sporadic developmental abnormalities, including reduced or increased flower organ number, fused organs, and abnormal leaf and stem morphology (Choi et al., 2002). This result suggested that DME expression might be important for vegetative plant development.
Using a DME:GUS reporter gene, we show here that the DME promoter is active in regions with dividing cells that express cyclin-dependent protein kinases (Doerner et al., 1996; Ferreira et al., 1994). By generating DME:MET1 a/s transgenic plants, we show that transient disruption of MET1-mediated DNA methylation, caused by inhibition of MET1 gene expression in a small subset of cells, has global effects on patterns of DNA methylation and plant development.
MATERIALS AND METHODS
Plant materials
Wild type, met1-6, DME:MET1 a/s, and DME:GUS plants were in the Columbia glabrous background. Plants were grown under long day condition (16 h light/8 h dark) at 23°C. Genotyping of met 1-6 was performed as described previously (Xiao et al., 2003).
Generation of DME:GUS and DME:GFP reporter genes
Previously, we constructed DME:GUS and DME:GFP translation gene fusions in the pBI101.1 vector consisting of a DME promoter (2.3 kb of 5′-flanking sequences), 1922 base pairs of the first DME exon (148 amino acids with a nuclear localization signal), and a GUS cDNA or GFP cDNA, respectively (Choi et al., 2002). The DME:GUS and DME:GFP transgenes used in this manuscript is the same as DME:GUS and DME:GFP transgenes in Choi et al. (2002) except that their respective promoters are truncated, with 555 base pairs of DME 5′-flanking sequences and 1922 base pairs of the first DME exon are present.
Construction of a DME:MET1 a/s gene
We generated transgenic plants with a MET1 antisense gene (DME:MET1 a/s) by inserting a 2.5 kb MET1 sequence (+2192 to +4659 base pairs from the transcription initiation site, BgIII site was added in the 5′-flanking end of each primer) in the antisense orientation into a BamHI site located between the DME promoter (555 base pairs 5′-flanking sequences plus 1922 base pairs of the first DME exon) and the GFP cDNA. However, stop codons in the MET1 antisense portion of the mRNA would prevent GFP from being translated. Primers for amplification of the MET1 cDNA from the Colombia glabrous genomic DNA are; 5′-GCA GAT CTG GGA TGG TGA GAG TCT AGG-3′ (BglasMET2202f) and 5′-GGA GAT CTG GGT TGG TGT TGA GGA GAC-3′ (BglasMET4668r).
Analysis of GUS activity
Seedlings were harvested at 1, 4 and 14 days after germination and stained for GUS activity as described previously (Yadegari et al., 2000). Histochemical localization of GUS staining was observed using a Zeiss Lumiar V12 microscope. Patterns of -555DME:GUS gene expression were analyzed in a minimum of 5 independent transgenic lines.
RNA preparation and RT-PCR
Seedlings and rosette leaves were harvested at two and four weeks after germination, respectively. Total RNAs from seedlings, rosette leaves, cauline leaves, pistils, and stamens were isolated using a Trizol Kit (Invitrogen). cDNAs were synthesized using a RETROscript Kit (Ambion).
DNA preparation and McrBC treatment
Genomic DNAs from rosette leaves, cauline leaves, pistils, and stamens were isolated using CTAB (Gehring et al., 2006) and 2 ug of DNA was treated with 20 unit of McrBC as describe by the manufacturer (New England Biolabs). The 60 ng of McrBC-digested DNA was used for PCR amplification. Primer sets for amplification of 180-bp repeat are; 5′-ACC ATC AAA GCC TTG AGA AGC A-3′ (JP1623) and 5′-CCG TAT GAG TCT TTG TCT TTG TAT CTT CT-3′ (JP1624).
RESULTS
DME promoter is active in cells flanking shoot and root meristems
Previously we reported DME promoter activity in the central cell of the female gametophyte within the ovule using green fluorescent protein (GFP) and β-glucuronidase (GUS) reporter genes under the control of DME promoter (DME:GFP and DME:GUS, respectively) (Choi et al., 2002). DME:GUS expression was not detected in the male gametophyte and stamen (Choi et al., 2002). The DME promoter used in these experiments consisted of 2.3 kb of 5′-flanking sequences plus 1.9 kb of the first DME exon. These constructs produced fusion polypeptides consisting of 148 N-terminal amino acids of DME, which includes a nuclear localization signal, followed by the C-terminal GUS or GFP protein, respectively. To dissect cis-acting regulatory sequences in the DME promoter, we truncated the DME promoter and constructed a DME:GUS transgene with 555 base pairs of DME 5′-flanking sequences plus 1.9 kb of the first DME exon, which were ligated to the GUS reporter gene (Fig. 1A) as described in “Materials and Methods”.
Fig. 1.
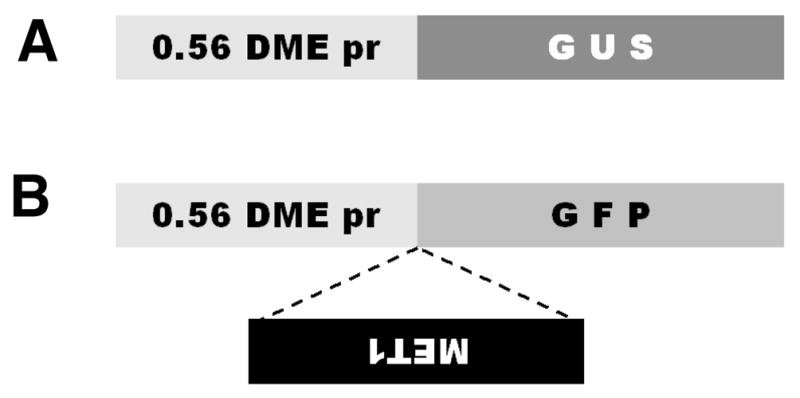
Generation of DME:GUS construct and DME:MET1 a/s transgene by using DME:GFP construct. DME promoter region includes 555 base pairs of DME 5′ flanking sequences plus 1922 base pairs of the first DME exon, which encode 148 amino acids of DME spanning a putative nuclear localization signal. (A) DME:GUS construct for checking DME expression pattern. 0.56 DME pr, 0.56 kb (555 base pairs) DME promoter. (B) Nucleotides +2192 to +4659 from the transcription start site of MET1 was cloned and ligated in an antisense direction between 0.56 kb DME promoter and GFP translation start site.
GUS staining was detected in the nucleus of the central cell of the female gametophyte in DME:GUS transgenic lines (Fig. 2A) and not in the male gametophyte or stamen (Fig. 2B). These results suggest that cis-acting regulatory sequences for central cell-specific expression are located between −555 and +1922 in the DME gene.
Fig. 2.
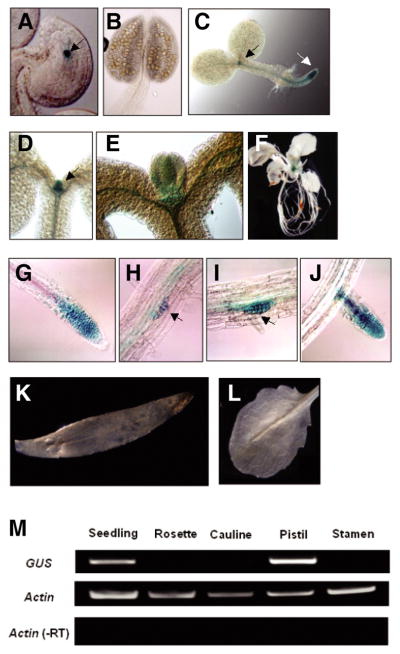
Spatiotemporal DME expression during plant development. Transgenic plants containing GUS reporter gene fused with DME promoter region were used to confirm where DME is expressed during plant development. (A) Strong GUS signal was detected in the central cell nucleus (arrow) of the female gametophyte, but not in male gametophytes or stamens (B). (C) In 1-day seedling, GUS was detected primarily in proliferating cells flanking the shoot apical meristem and root apical meristem regions (black and white arrows, respectively). (D–E) Strong GUS signal is restricted in young leaf primordia emerging from the shoot apical meristem in 4-day seedlings. (F) Whole mount view of transgenic plants before bolting. DME promoter is active only in the undifferentiated tissue. (G) GUS was expressed in primary roots adjacent to the meristems. (H–J) GUS was also expressed in secondary roots adjacent to the meristems. (K–L) GUS expression was not detected in mature tissues such as cauline or rosette leaves, respectively. (M) GUS RNA was detected mainly from seedling tissues and pistils that contain undifferentiated proliferating cells and central cell, respectively.
In addition to expression in the central cell, we also detected DME promoter activity during sporophyte development of DME:GUS transgenic plants. GUS staining was detected in leaf primordia in 1-day (Fig. 2C) and 4-day (Figs. 2D and 2E) seedlings, as well as at primary (Figs. 2C and 2G) and secondary (Figs. 2H–2J) roots distal to their respective root apical meristems. Cells in these regions have dividing cells that express cyclin-dependent protein kinases (Doerner et al., 1996; Ferreira et al., 1994). Thus, the DME promoter is active in leaf and root primorida which contain proliferating cells.
By contrast, DME:GUS expression was not detected in mature rosette leaves (Figs. 2F and 2L), cauline leaves (Fig. 2K) or floral organs (data not shown) with the exception of the central cell (Fig. 2A). Similar patterns of GUS staining were observed in 5 independently isolated DME:GUS transgenic lines, as well as in lines bearing a DME:GUS gene with 2.3 kb of 5′-flanking sequences (data not shown). Thus, DME promoter activity is highly restricted to discrete regions of the seedling where rapidly proliferating cells emerge from shoot and root meristems.
The level of GUS RNA in DME:GUS transgenic seedlings, rosette leaves, cauline leaves, pistils, and from stamens was measured by semi-quantitative RT-PCR. GUS RNA was detected in the seedling and pistil, which contain shoot/root apex regions and central cell, respectively, and was not detected in rosette leaves, cauline leaves, or stamens (Fig. 2M). Thus, the GUS staining patterns are correlated with the presence of GUS RNA, and likely reflect DME promoter-mediated transcription of the GUS reporter gene. These results suggest that 555 base pairs of DME 5′ flanking sequence plus 1922 base pairs of the first DME exon are responsible for GUS transcription in discreet bands of highly proliferating cells that flank shoot and root meristems.
Plants with a DME:MET1 a/s transgene display pleiotropic mutant phenotypes
We induced passive DNA demethylation by generating transgenic plants that express MET1 antisense sequences under the control of the DME promoter (DME:MET1 a/s) (Fig. 1B). Expression of the antisense MET1 transgene is predicted to block the endogenous MET1 expression through an RNAi mechanism (Matzke and Birchler, 2005). Because the same DME promoter is used for DME:GUS and DME:MET1 a/s expression, we expect that suppression of MET1 expression will occur specifically in regions delineated by the DME:GUS reporter described above (Fig. 2).
To reveal the effect of suppressing MET1 expression in regions where the DME promoter is active, we generated 109 DME:MET1 lines. 74 T1 lines, which were designated NF-1 to NF-74 (Fig. 3B), appeared to develop normally when compared to wild type control plants (Fig. 3A). However, in 35 T1 lines we detected alterations in the development. Delayed flowering time (Fig. 3B), aberrant floral and vegetative structures, and decreased fertility (data not shown) were observed in 26 lines which were designated LF-1 to LF-26. Moreover, 9 other T1 lines displayed numerous developmental abnormalities such as altered phyllotaxy, fasciated stems, and a dwarf stature (data not shown).
Fig. 3.
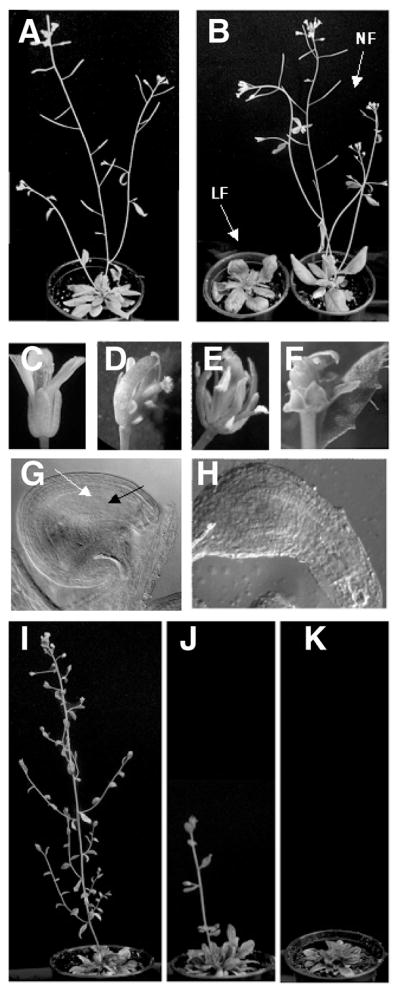
Pleiotropic mutant phenotypes of DME:MET1 a/s transgenic plants. (A) Wild type plants. (B) 26 out of total 109 DME::MET1 a/s transgenic T1 lines displayed late flowering phenotype (LF) and 74 transgenic lines normally flowered (NF). (C) Wild type flower. (D–F) Pleiotropic floral defects were observed in the T2-LF generations. (G) Wild type ovule. Mature embryo sac is shown including egg cell nucleus (black arrow) and central cell nucleus (white arrow). (H) Abnormal elongated ovule in T2-LF lines. Among 8 T3-LF plants, 3 plants successively flowered, although the growth rate deliberately differed depending on lines (I and J). Rest of five plants failed to undergo the transition to flowering and showed greatly delayed senescence (K).
To understand the heritability of mutant phenotypes, four LF T1 lines (LF-1, LF-2, LF-3, and LF-4) and two NF T1 lines (NF-1 and NF-2) lines were self-pollinated and their development was analyzed in subsequent T2 and T3 generations. Whereas control wild type flowers developed normally (Fig. 3C), in T2 LF lines, in addition to late flowering, we observed reduced floral organ number and bending pistils (Fig. 3D), increased number of stamens and defective pollen grain formation (Fig. 3E), and fascinated pistils (Fig. 3F). T2 LF lines also displayed ovules with abnormal shapes and aberrant female gametophytes (Fig. 3H) compared to wild type ovules (Fig. 3G). In general, phenotypes became more severe with each succeeding generation. For example, T2 LF plants displayed greatly reduced fertility and produced almost no T3 seeds. Among 8 T3 plants that were produced, 3 were able to flower (Figs. 3I and 3J), whereas 5 failed to undergo the transition to flowering and showed greatly delayed senescence (Fig. 3K). Thus, the DME:MET1 a/s transgene causes dramatic mutant phenotypes that are heritable.
From the results above, we concluded that the developmental abnormalities observed in whole stages of development in the DME:MET1 a/s lines are due to suppression of MET1 expression in the limited, defined regions where the DME promoter is active (Figs. 2 and 3).
DME:MET1 a/s plants revealed global hypomethylation
CG DNA methylation can be analyzed by McrBC treatment followed by PCR amplification. McrBC is a methyl-cytosinespecific restriction endonuclease, which digests only 5-methylcytosine-containing DNA when two methylated 5′-PuC sites are separated by approximately 40–80 non-defined base pairs (Sutherland et al., 1992). Thus, successful PCR amplification after McrBC digestion indicates the lack of DNA methylation (Lippman et al., 2003; Rabinowicz et al., 2003).
To determine if the DME:MET1 a/s transgene causes genome DNA hypomethylation, we measured the level of DNA methylation at the highly repeated 180 base pair centromeric repeat regions (Lister et al., 2008). We observed very little PCR amplification after McrBC digestion of the centromeric repeats using DNAs isolated from wild type and control normal flowering DME:MET1 a/s lines (Fig. 4), which is consistent with their being highly methylated. By contrast, strong PCR amplification was observed after McrBC digestion using DNAs from late flowering DME:MET1 a/s lines as well as a met1-6 control line (Fig. 4), which is consistent with hypomethylation of the 180 base pair centromeric repeat. These results suggest that expression of the DME:MET1 a/s transgene in cells flanking the shoot and root meristem can cause global DNA hypomethylation throughout the plant.
Fig. 4.
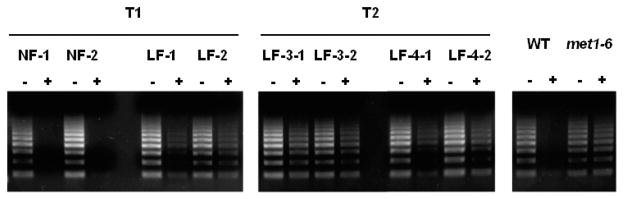
Global genomic hypomethylation at 180 base pair centromeric repeat region in DME:MET1 a/s transgenic plants. After McrBC digestion of genomic DNAs, the 180 base pair centromeric repeat region was amplified by PCR using primers shown in “Materials and Methods”. If DNA methylation is depleted, successful amplification is expected. Since 180 base pair centromeric repeat region is a repetitive sequence, ladder shape band was detected in late-flowering transgenic lines. NF, normal flowering transgenic line; LF, late flowering transgenic line.
DISCUSSION
To elucidate the importance of continuous maintenance of DNA methylation, we sought to express antisense MET1 sequences for a relatively short period of time in proliferating cells that flank the meristems. This was achieved by constructing DME:MET1 a/s transgenic lines. As revealed by analysis of DME:GUS transgene expression and RT-PCR experiments, the DME promoter is active in leaf and lateral root primordia, as well as the region that is distal to the root apical meristem (Fig. 2). These regions typically have proliferating cells that contribute to leaf and root morphogenesis (Barkoulas et al., 2007). Cells in the region of DME promoter activity and antisense MET1 expression presumably suppress endogenous MET1 expression. However, as cells exit the region of DME promoter activity (Fig. 2), the MET1 wild type allele should be expressed. Thus, we were able to see the outcome of inhibiting MET1 expression in a restricted set of cells, which is different from previous studies that reported on plants where MET1 function was globally reduced throughout the plant by met1 loss-of-function mutations or constitutively expressed MET1 antisense gene (Finnegan et al., 1996; Kankel et al., 2003; Ronemus et al., 1996; Saze et al., 2003).
We found that that a very brief loss of MET1 activity is sufficient to cause many developmental defects including late flowering, aberrant floral and vegetative structures, decreased fertility, altered phyllotaxy, fasciated stems, and dwarf stature (Fig. 3 and data not shown). Moreover, global DNA hypomethylation was observed at highly repeated centromeric sequences in late flowering DME:MET1 a/s lines (Fig. 4). Once the syndrome of late flowering and DNA hypomethylation was established, it was stably inherited in the next generations (Fig. 2). Thus, even a brief hiatus of MET1 expression alters DNA methylation which has dramatic effects on plant development that can persist for many generations. This reflects, in part, that the DME promoter is active in proliferating cells (Fig. 2).
Maintenance of the majority of CG DNA methylation in the plant genome depends primarily on MET1 (Cokus et al., 2008; Lister et al., 2008; Zilberman et al., 2007). Without maintenance MET1 activity, a cell with a fully methylated genome only passes on methylated strand to the first daughter cells upon duplication. Subsequent divisions further dilute the ratio of methylated versus hypomethylated DNA, resulting in daughter cells with mainly hypomethylated DNA within a few cell divisions.
DME is transcribed in the central cell, where it establishes imprinting of the MEA, FIS2, and FWA genes by demethylating their respective maternal alleles (Choi et al., 2002; Gehring et al., 2006; Jullien et al., 2006; Kinoshita et al., 2004; Xiao et al., 2003). DME-like polypeptides are broadly expressed and demethylate at the 5′- and 3′-boundaries of over 100 genes (Lister et al., 2008; Ortega-Galisteo et al., 2008; Penterman et al., 2007). Here we show that the DME promoter is active in proliferating cells that flank the shoot and root meristems (Fig. 2). Moreover, plants homozygous for a weak dme mutant allele, dme-1, display sporadic developmental abnormalities, including alterations in floral organ number, organ fusion, and aberrant leaf and stem morphology (Choi et al., 2002). We speculate that DME-mediated DNA demethylation in primordia flanking the shoot meristems is required to prevent these sporadic developmental abnormalities.
Taken together, our results show that a subtle reduction of MET1 activity in a restricted region of proliferating cells results in a global epigenetic alteration, which for the most part is irreversible, and cannot be recovered merely by restoring of MET1 activity. Thus, there may be no efficient mechanism that constitutes a memory for patterns of MET1-mediated DNA methylation.
Acknowledgments
We would like to thank Dr. Tzung-Fu Hsieh for critical comments on this manuscript. This work was support by Korea Research Foundation (KRF-2005-070-C00129) and Korea Science and Engineering Foundation (R01-2007-000-10706-0) to Y.C. Brain Korea 21 project supported this work to H.O. This work was also supported by National Institute of Health (GM069415) and United States Department of Agriculture (2005-02355) to R.L.F.
References
- Barkoulas M, Galinha C, Grigg SP, Tsiantis M. From genes to shape: regulatory interactions in leaf development. Curr Opin Plant Biol. 2007;10:660–666. doi: 10.1016/j.pbi.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Bender J. DNA Methylation and Epigenetics. Annu Rev Plant Biol. 2004;55:41–68. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner P, Jorgensen JE, You R, Steppuhn J, Lamb C. Control of root growth and development by cyclin expression. Nature. 1996;380:520–523. doi: 10.1038/380520a0. [DOI] [PubMed] [Google Scholar]
- Ferreira PC, Hemerly AS, Engler JD, van Montagu M, Engler G, Inze D. Developmental expression of the arabidopsis cyclin gene cyc1At. Plant Cell. 1994;6:1763–1774. doi: 10.1105/tpc.6.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES. Reduced DNA methylation in Arabidopsis results in abnormal plant development. Proc Natl Acad Sci USA. 1996;93:8449–8454. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Jullien PE, Kinoshita T, Ohad N, Berger F. Maintenance of DNA methylation during the Arabidopsis life cycle Is essential for parental imprinting. Plant Cell. 2006;18:1360–1372. doi: 10.1105/tpc.106.041178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated SIngle-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, Martinez-Macias RR, Ariza RR, Roldan-Arjona T. DEMETER and REPRESSOR OF SILENCING1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz PD, Palmer LE, May BP, Hemann MT, Lowe SW, McCombie WR, Martienssen RA. Genes and transposons are differentially methylated in plants, but not in mammals. Genome Res. 2003;13:2658–2664. doi: 10.1101/gr.1784803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL. Demethylation-induced developmental pleiotropy in Arabidopsis. Science. 1996;273:654–657. doi: 10.1126/science.273.5275.654. [DOI] [PubMed] [Google Scholar]
- Saze H, Scheid OM, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- Scott RJ, Spielman M. Genomic imprinting in plants and mammals: how life history constrains convergence. Cytogenet Genome Res. 2006;113:53–67. doi: 10.1159/000090815. [DOI] [PubMed] [Google Scholar]
- Sutherland D, Coe L, Raleigh EA. McrBC: A Multisubunit GTP-dependent restriction endonuclease. J Mol Biol. 1992;225:327–348. doi: 10.1016/0022-2836(92)90925-a. [DOI] [PubMed] [Google Scholar]
- Xiao W, Gehring M, Choi Y, Margossian L, Pu H, Harada JJ, Goldberg RB, Pennell RI, Fischer RL. Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell. 2003;5:891–901. doi: 10.1016/s1534-5807(03)00361-7. [DOI] [PubMed] [Google Scholar]
- Yadegari R, Kinoshita T, Lotan O, Cohen G, Katz A, Choi Y, Katz A, Nakashima K, Harada JJ, Goldberg RB, et al. Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell. 2000;12:2367–2381. doi: 10.1105/tpc.12.12.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JQ, Liu JH, Liang XW, Xu BZ, Hou Y, Zhao XX, Sun QY. Heat stress causes aberrant DNA methylation of H19 and Igf-2r in mouse blastocysts. Mol Cells. 2008;25:211–215. [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]


