Abstract
While genetic determinants strongly influence HDL cholesterol (HDLc) levels, most genetic causes underlying variation in HDLc remain unknown. We aimed to identify novel rare mutations with large effects in candidate genes contributing to extreme HDLc in humans, utilizing family-based Mendelian genetics. We performed next-generation sequencing of 456 candidate HDLc-regulating genes in 200 unrelated probands with extremely low (≤10th percentile) or high (≥90th percentile) HDLc. Probands were excluded if known mutations existed in the established HDLc-regulating genes ABCA1, APOA1, LCAT, cholesteryl ester transfer protein (CETP), endothelial lipase (LIPG), and UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GALNT2). We identified 93 novel coding or splice-site variants in 72 candidate genes. Each variant was genotyped in the proband’s family. Family-based association analyses were performed for variants with sufficient power to detect significance at P < 0.05 with a total of 627 family members being assessed. Mutations in the genes glucokinase regulatory protein (GCKR), RNase L (RNASEL), leukocyte immunoglobulin-like receptor 3 (LILRA3), and dynein axonemal heavy chain 10 (DNAH10) segregated with elevated HDLc levels in families, while no mutations associated with low HDLc. Taken together, we have identified mutations in four novel genes that may play a role in regulating HDLc levels in humans.
Keywords: high density lipoprotein, Mendelian genetics, lipids, high density lipoprotein metabolism, genetics, lipoproteins
Heritability estimates of 47–76% for plasma HDL cholesterol (HDLc) levels suggest that genetic variation plays a pivotal role in HDL metabolism (1–3). However, despite major advances in family- and population-based association studies (2, 4), most genetic causes of extreme HDLc levels in humans remain unknown. Cohen et al. (5) reported mutations in the established HDLc genes ABCA1, APOA1, and LCAT in only 12.4% of individuals with low HDLc (<5th percentile). Similarly, we reported that mutations in the known HDLc-regulating genes ABCA1, APOA1, and LCAT were found in 28.7% of unrelated individuals with low HDLc levels (≤10th percentile) (6), consistent with other published data (7, 8). Furthermore, mutations with an HDLc-increasing effect in the cholesteryl ester transfer protein (CETP), endothelial lipase (LIPG), and UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GALNT2) genes were found in only 14.6% of probands with high HDLc (≥90th percentile) (9). Additionally, only ∼10–15% of the inter-individual variation in lipid levels is explained by common genetic variation (10, 11).
One significant limitation of identifying novel mutations that result in large changes to HDLc is that they are frequently extremely rare (i.e., <0.1% in the general population), which precludes testing for associations. Thus assessing the segregation of genetic variations with apparent Mendelian forms of extreme HDLc in families is a useful and well-validated method to examine whether novel rare mutations associate with HDLc phenotypes (6, 9, 12–15).
Recent advances in sequencing technology allow parallel sequencing of many genes and even whole exomes or genomes across populations. However, a major challenge to this approach is identifying those few mutations that associate with large changes in HDLc, or any other complex trait, from the many additional identified unrelated sequence variations. We propose here that novel mutations that underlie extreme HDLc levels can be distinguished from other sequence variations found by next-generation sequencing by initially assessing their segregation in families with extreme HDLc levels.
Here, we sequenced the exons and exon/intron boundaries of 456 genes that potentially influence HDLc levels in 200 unrelated individuals, including 80 with low HDLc (LHDL; ≤10th percentile) and 120 with high HDLc (HHDL; ≥90th percentile). We then employed a series of data analysis filters to identify rare novel sequence variants that are enriched in either the extreme high or low HDLc phenotype and are expected to have large impacts on protein function. Finally, we performed segregation analyses of these sequence variations across family members of the probands, recruiting a total of 59 families (constituting 685 individuals), and identified mutations in four novel genes that may play a role in elevated plasma HDLc levels.
MATERIALS AND METHODS
Probands
We identified 80 unrelated Dutch Caucasian probands with LHDL and 120 unrelated probands of Dutch Caucasian ancestry with HHDL, based on age- and sex-specific Lipid Research Clinic data and as described previously (6, 16) with no other abnormal lipid measures. We also studied 685 family members of 59 probands with mutations (59 pedigrees). Study protocols were approved by the ethics committees of the Academic Medical Center, Amsterdam and the University of British Columbia, Vancouver. All subjects provided written informed consent. Lipoprotein measurements were performed on fresh plasma as described (17). Cholesterol and triglyceride levels were determined in total plasma and plasma at d < 1.006 g/ml obtained after preparative ultracentrifugation and before and after precipitation with dextran manganese. Other covariables such as age, sex, BMI, medical history, alcohol intake, and smoking history were available for all individuals.
DNA sequencing and data analysis
Genomic coordinates for the exons of the 456 genes were compiled by querying the Ensembl database (v56) using the perl application program interface. Proband DNA was enriched for each exon and at least 50 bp of adjacent intron sequence using SureSelect bead technology (Agilent, Santa Clara, CA). Captured sequences from a pool of five proband DNA samples (24 pools for HHDL samples and 16 pools for LHDL samples) were then pooled in equimolar amounts and sequenced together (thus sequencing of 200 individuals consisted of 40 sequencing runs) using next-generation paired-end read sequencing (Illumina, San Diego, CA) (18). For each sequencing run of five pooled DNA samples, we obtained an average 246 ± 37-fold sequencing coverage per sample pool, or 49 ± 7-fold coverage per proband DNA sample.
Sequence changes in sample pools were identified by alignment of sequence data to the human genome (NCBI build 36.1) using the CASAVA v1.7 software (Illumina). Identified sequence changes were characterized using the Ensembl v56 database and the perl application program interface as synonymous, missense, nonsense, or splice site variants.
Sequence changes of interest were confirmed by standard fluorescent dye terminator chemistry sequencing (Beckman Coulter Genomics, Danvers, MA and SeqWright, Houston, TX) and analyzed using Sequencher v4.7 (Gene Codes Corporation, Ann Arbor, MI). For each sequence change, standard Sanger sequencing was performed on each of the original proband DNAs comprising the five pooled samples to identify the specific carrier individual. In all cases, data analyses were performed by scientists blinded to the phenotypes of the sequenced samples. Exon-probe and primer sequences are available upon request.
Segregation analysis of mutations in families and statistical analyses
A total of 685 family members of 59 probands with sequence changes of interest were genotyped using standard Sanger sequencing techniques described above for 93 SNPs in 72 genes. Only the variant found in a specific proband was genotyped in the proband’s family. After genotyping, only variants where genotypes from six or more family members were available, and of these family members, at least three mutation carriers identified were included in the statistical analyses for segregation.
Linear regression was performed on each variant for HDLc level, fitting age, sex, and BMI as covariables. The β-statistic indicating the effect-size and direction of the mutation on HDLc was used to recode alleles to H (for increasing HDL) and L (for decreasing HDL). Family-based analysis was performed on the recoded alleles using within- and between-family-based association tests for each gene, noting that most genes were represented by a single variant, using the family-based-associated test function of PLINK and permuting the data 1,000,000 times to obtain the empirical P values for significance (19). Each gene was considered an independent test, as different samples and families were used for the analysis (different variants were genotyped in different families), and thus not amenable to multiple-testing correction. An empirical P value of <0.05 was deemed significant.
RESULTS
Selection of probands for sequencing
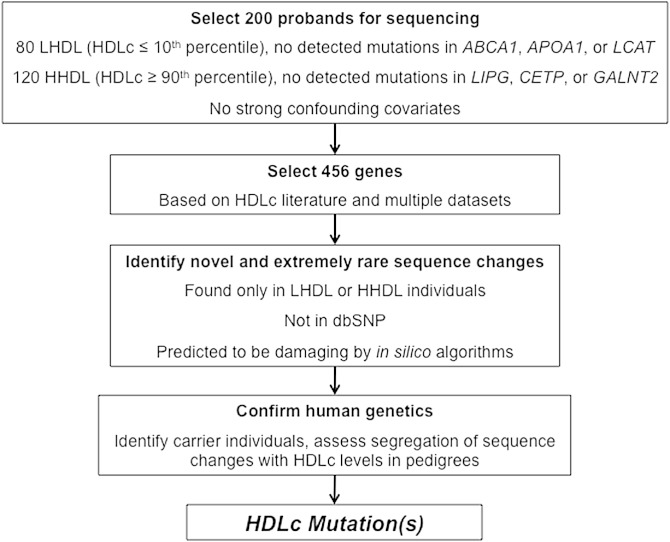
From an initial cohort of 178 unrelated Dutch probands (6), 80 probands with HDLc ≤10th percentile and no known coding or splicing mutations in ABCA1, APOA1, or LCAT were selected as part of low-HDLc screening cohort (LHDL; Table 1). Separately, from an initial cohort of 171 unrelated Dutch probands (9), 120 probands with HDLc ≥90th percentile with no known coding or splicing mutations in CETP, LIPG, or GALNT2 were selected as part of high-HDLc screening cohort (HHDL; Table 1). No other major lipid abnormalities or other confounding factors like severely elevated BMI, diabetes mellitus, hypertension, extensive medical history or medication use, or excessive alcohol, smoking, hormone replacement therapy, or other drug use were reported by these probands. An overview of the study design is shown in Fig. 1.
TABLE 1.
Basic demographic and clinical characteristics of the low and high HDL probands who underwent sequencing
| Low HDLc | High HDLc | |
| Number of probands | 80 | 120 |
| Number of males (%) | 52 (76.2%) | 61 (50.8%) |
| Age (yrs) | 52.0 (14.2) | 51.9 (14.7) |
| Total cholesterol | 4.46 (1.49) | 5.86 (1.05) |
| Triglycerides | 1.67 (0.87) | 0.84 (0.44) |
| HDLc | 0.70 (0.19) | 2.24 (0.45) |
| HDLc percentile | 4.7 (1.7) | 95.1 (1.2) |
| Number with HDLc percentile <5 (%) | 66 (82.5%) | 96 (80.0%) |
| LDL cholesterol | 2.99 (1.38) | 3.23 (0.93) |
| BMI (kg/m2) | 25.9 (2.9) | 23.6 (2.9) |
| Number with cardiovascular disease (%) | 35 (43.8%) | 1 (0.8%) |
Values are average (standard deviation or percent). Lipids are mmol/l.
Fig. 1.
Overview of the study design.
Selection of genes for sequencing
We manually curated and prioritized 456 genes for sequencing from multiple datasets that included genes or sets of genes identified from, or implicated in: A) genetic regions with suggestive linkage (Log of odds score ≥2.0) with HHDL or LHDL in families (data not shown); B) HDLc regulation reported in published genome-wide association studies (4, 20); C) significant expression changes in an in vitro APOA1 siRNA screen in HepG2 cells (21); D) in silico liver centric Bayesian network analysis for five well-characterized genes related to HDL metabolism [APOA1, ABCA1, CETP, scavenger receptor class B member 1 (SCARB1), and LIPG] (22); E) significant expression changes in Apoa1 knockout mice (23); and F) direct and indirect literature support for roles in HDL regulation or metabolism not reported in genome-wide assiciation studies (GWAS), in addition to paralogs of select genes with literature support (supplementary Tables I, II).
In the union of these gene lists from A to F, we selected 450 genes for sequencing. We also included ABCA1, APOA1, LCAT, CETP, LIPG, and GALNT2 as internal controls to identify any additional mutations not detected by our previous sequencing efforts in the 200 selected probands (6, 9). Supplementary Table III lists the total 456 genes sequenced in the 200 probands.
Validation of sequencing data
To validate that sequence variations were readily detected in sample pools, where each sample pool constituted five proband DNA samples, we assessed the presence of known and presumably benign sequence variation from previous standard sequencing of ABCA1, APOA1, and LCAT in LHDL probands, and CETP, LIPG, and GALNT2 in HHDL probands (6, 9). Of 97 SNPs that were either noncoding or synonymous sequence variations previously detected in these probands, all 97 were readily detected in the sample pools, including 29 of 97 SNPs (29.9%) where the minor allele was present in no more than one proband per sample pool, indicating that sequence variation present in single individuals is readily detected from pooled sample sequence data (supplementary Table IV).
We also identified additional mutations not previously detected by standard sequencing, including six within the LHDL probands (APOA1 L202P in one proband, LCAT V371M in one proband, L338H in two probands, and T147I in two probands) (6), and four within the HHDL probands (LIPG N396S in three probands and G196R in one proband) (9). These mutations were inadvertently missed during our standard sequencing of control genes in the extreme HDLc probands, but were confirmed by reviewing the initial standard sequencing chromatograms, demonstrating again that rare mutations are readily detected by our experimental approach.
No CETP, LIPG, or GALNT2 mutations were found in LHDL probands, and no ABCA1, APOA1, or LCAT mutations were found in HHDL probands. Our final discovery cohorts therefore constituted 74 probands for LHDL and 116 for HHDL.
Identification of novel sequence changes
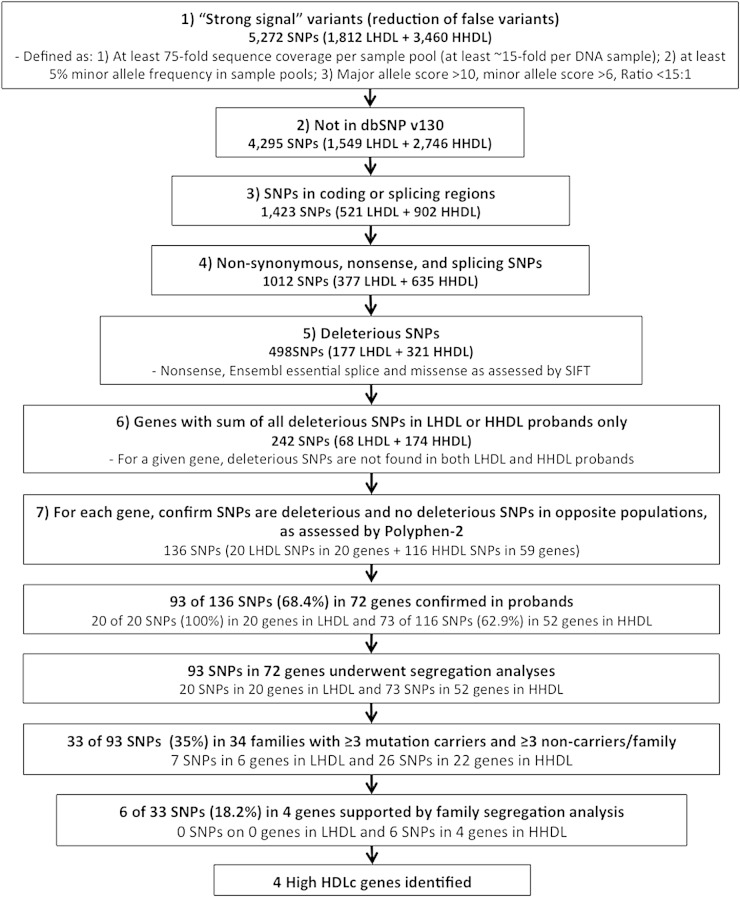
As we could detect known SNPs and mutations in known HDLc-regulating genes in these probands, we next searched for novel mutations in the remaining 450 genes to identify novel sequence variants that potentially underlie extreme HDLc levels (Fig. 2). To reduce the identification of false variant calls, the following quality control filters were applied: A) at least 75-fold sequence coverage per sample pool of five probands (at least ∼15-fold per proband DNA); B) a minor allele frequency >0.05 in sample pools; C) detection in pooled high quality sequence data: major allele BaCON score >10, minor allele BaCON score >6, and BaCON score ratio <15:1 (the BaCON score is generated by Illumina and represents the discrepancy of variants. A score of ≤6 indicates variants below the threshold of detection) (24); and D) found only in LHDL or HHDL sample pools (Fig. 2).
Fig. 2.
Filters used to identify novel SNPs most likely to underlie extreme HDLc levels in sequenced probands and process to identify novel SNPs that significantly associate with reduced or elevated HDLc in families.
As we anticipated that mutations underlying large changes in HDLc levels would be extremely rare in the general population (i.e., <0.1% or unique to individual families), we next prioritized sequence changes that were absent from database of SNPs (dbSNPs) (build 130). We then prioritized those sequence changes that were predicted to result in nonsense mutations, splicing defects in the first or last 2 bp of an intron (Ensembl essential splice), or nonsynonymous amino acid substitutions predicted to be damaging according to the sorting intolerant from tolerant (SIFT) algorithm (25). We next prioritized those genes where all remaining sequence variation in a given gene was found in LHDL or HHDL probands only. Finally, all remaining sequence variation was confirmed to be “possibly damaging” or “probably damaging” by the PolyPhen-2 algorithm (26), and sequence data in opposite phenotype sample pools was reviewed to confirm that no potentially deleterious sequence variations were present (i.e., SNPs predicted to be deleterious by PolyPhen-2 but not SIFT). Using this approach, we identified a total of 136 variants with a putative role in HDLc regulation, 20 sequence changes in 20 genes from LHDL probands, and 116 sequence changes in 59 genes from the HHDL probands (Fig. 2).
Confirmation of the presence of the 136 identified variants was performed by standard Sanger sequencing in the original probands (Fig. 2). Of the 136 variants identified, 93 changes in 72 genes were confirmed (68%). The changes and genes are shown in supplementary Table V.
Segregation of novel sequence changes with HDLc phenotypes
To assess the likelihood of the 93 confirmed variants underlying the extreme HDLc levels in families, we genotyped each mutation in all available family members of the originating proband. A total of 685 individuals from 59 families (representing 59 separate pedigrees) were genotyped for specific mutations found only in the respective proband. To provide sufficient statistical power for further analyses, a given variant was required to be present in at least three family members, and absent in at least three additional family members in the same pedigree. Only 33 of the 93 variants passed this filter. For these 33 variants, the average size of each pedigree was 22 ± 16 and consisted of a total of 627 family members in 34 pedigrees. This indicates that some of the 60 remaining novel variants identified cannot account for the extreme HDLc phenotype observed in a given family, as insufficient affected family members would be carriers, while others would be bona fide HDLc modulating mutations for which insufficient statistical power was available to assess segregation.
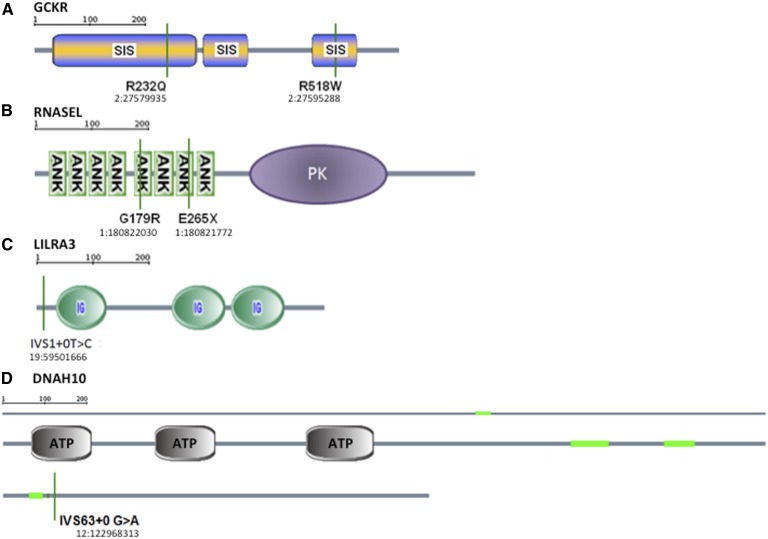
Nevertheless, mutations in several genes were identified to segregate significantly with extreme HDLc levels, as determined by family-based association analysis. For LHDL, we did not identify any novel mutations that significantly segregated with HDLc percentile. For HHDL, we identified new mutations in glucokinase regulatory protein (GCKR), RNase L (RNASEL), leukocyte immunoglobulin-like receptor 3 (LILRA3), and dynein axonemal heavy chain 10 (DNAH10) (Table 2; Fig. 3). For GCKR, the mutation R232Q was found in two unrelated probands, while R518W was found in a third; both mutations fall within sugar isomerase domains. For RNASEL, a G179R mutation was found in one proband while E265X was found in five unrelated probands; these mutations occur in the fifth and seventh ankyrin domains, respectively. For LILRA3, an IVS1+0T>C mutation was found in two unrelated probands, which is predicted to disrupt splicing within the N-terminal coding sequence (Fig. 3). Finally, for DNAH10, one mutation was identified near the C terminus, IVS63+0 G>A, an essential splice site mutation in one proband. Of these probands, only those with pedigrees with three or more carriers and three or more noncarriers underwent statistical analyses. In all genes, carriers of the mutations had significantly elevated HDLc percentiles when compared with noncarrier family members. Table 2 summarizes the HDLc phenotypes of mutation carriers and noncarriers in families.
TABLE 2.
Segregation of mutations in extreme HDL genes
| Gene | Mutations (number of probands) | HDLp Mutation Carriers | HDLp Family Controls |
| GCKR | R232Q(2)+R518W(1) | 85.1 (11.4) (21) | 77.2 (22.1) (14); 0.004; 12.7 |
| LILRA3 | IVS1+0T>C(1) | 86.4 (10.9) (7) | 51.7 (30.4) (21); 0.036; 34.9 |
| RNASEL | G179R(1)+E265X(1) | 90.6 (11.1) (16) | 71.3 (27.0) (33); 0.01; 20.1 |
| DNAH10 | IVS63+0 G>A(1) | 95.8 (0.5) (4) | 41.8 (33.1) (4); 0.04; 59.8 |
HDLp = HDLc percentile; values are average (standard deviation) (n); empirical P value following 1,000,000 permutations (in bold); beta.
Fig. 3.
Predicted mutation effects on GCKR (A), RNASEL (B), LILRA3 (C), and DNAH10 (D). The chromosomal and base pair location of each mutation, based on Human Genome build 18 (hg18) is shown. SIS, sugar isomerase domain; ANK, ankyrin domain; PK, protein kinase domain; Ig, immunoglobulin domain; ATP, ATPase domain; green, coiled-coil domain.
DISCUSSION
Here we combined next-generation sequencing of 456 genes having known or putative roles in HDL metabolism with Mendelian family-based analyses of 59 different families and 685 total family members. As a result, we identified mutations in four genes, GCKR, RNASEL, LILRA3, and DNAH10, that significantly segregate with increased plasma HDLc in families. Our goal was to identify novel and extremely rare nonsynonymous, nonsense, and splice mutations with large functional impacts on HDLc, most of which are not captured by current genome-wide association strategies. Although extreme HDLc in the absence of other lipid abnormalities was the only phenotype considered here, clearly this strategy could be applicable to identifying mutations for other extreme lipid traits.
Overall, six mutations in seven probands (with 120 family members) in four novel genes that are in coding regions or exon/intron boundaries, not present in dbSNP (build 130), predicted damaging by in silico algorithms showed significant segregation with elevated HDLc in families. We previously reported that in the initial cohort of 171 unrelated HHDL probands, we found 22 with LIPG mutations (12.9%, inclusive of those mutations described here), 1 with a CETP mutation (0.6%), 4 with GALNT2 mutations (2.3%), 2 with APOC3 mutations (2.3%) (27), and 2 with SCARB1 mutations (2.3%) (9, 28, 29). Taken together, we have identified to date a total of 43 of 171 HHDL probands with mutations (25.1%), making this the highest frequency of mutations identified to date in a HHDL cohort. Of note, the RNASEL E265X mutation was found in 5 of 171 HHDL probands (2.9%), raising the possibility of testing for associations with HDLc levels in large populations with this mutation.
Because our goal was to identify only those mutations that were novel, likely deleterious, and exclusive to one phenotype, nonsynonymous mutations predicted to be benign by at least one in silico analysis were discounted here. Indels were also excluded, as >75% of these sequence changes detected from next-generation sequencing data using algorithms available to us were not detected by Sanger sequencing. We also excluded variants present in dbSNP at low frequency, although this likely excluded a subset of bona fide HDLc-modulating mutations. This step also importantly excluded common sequence variation that was unlikely to underlie large changes in HDLc levels. Mutations that were suppressed by strong mutations of the opposite phenotype were also excluded. For example, we previously showed that the HDLc-elevating phenotypes of LIPG N396S and SCARB1 S112F mutations may be suppressed by ABCA1 mutations IVS24+1G>C and V2091I, respectively (9, 28). Finally, we also did not assess the association of common polymorphisms in these 456 genes with HDLc phenotypes, although it is worth exploring in concert with additional populations. Clearly, false negatives and positives will inevitably arise from our initial approach, reflecting the challenges of understanding the genetics of complex traits, and requiring subsequent analysis strategies for mutation discovery. Evolving bioinformatics tools and statistical methods will also continually afford identification of additional mutations with potential functional consequences.
In addition to the novel mutations identified here, we identified both APOC3 and SCARB1 mutations (4, 27–29) that have previously been associated with both loss-of-function and elevated plasma HDLc in humans, thus offering validation for our approach. We also identified additional mutations in APOA1 (L202P), LCAT (V371M, L338H, and T147I), and LIPG (N396S and G196R) that were not detected in the original Sanger sequencing, providing further validation. We note that only 93 of 136 (68.4%) prioritized sequence changes predicted by next-generation sequencing were confirmed in the Sanger sequencing, possibly reflecting that our initial quality filters were not stringent enough.
While GCKR, LILRA3, and DNAH10 have been implicated by GWAS to modulate HDLc (4, 20), nonsynonymous mutations segregating with an elevated HDLc trait have not been reported in these genes. Our observations support previous studies that implicate novel genes in HDLc regulation and validate the use of next-generation sequencing and family-based segregation approaches to identify novel mutations.
GCKR was included in our 456 gene list because a variant in GCKR showed genome-wide significant association with HDL in published GWAS (20, 30, 31). We identified two missense mutations, R232Q and R518W, in three unrelated probands (R232Q was found in two probands). GCKR is a regulatory protein that inhibits glucokinase (GK) in liver and pancreatic islet cells. It participates in the modulation of GK activity and location by binding free cytoplasmic GK. As glucose levels decline, GCKR moves GK into the nucleus, where it is held in reserve in an inactive form (32). As glucose and insulin levels rise, GK is released from GCKR and moves back to the cytoplasm. It is localized widely, with highest expression in the liver, testes, ovaries, and adipocytes (Illumina human body map). The identification of GCKR as an HDLc gene suggests a novel link between genes influencing HDLc levels and glucose metabolism.
LILRA3 was included in our 456 gene list because a SNP was previously identified showing genome-wide significant association with elevated plasma HDLc levels (4, 33). We identified one mutation, IVS1+0T>C, an essential splice site mutation, in two probands in this gene. Genotyping did not identify three or more carriers in the pedigree of one proband. However, the mutation segregated significantly with elevated HDLc in the pedigree of the second proband. LILRA3, also known as CD85 antigen-like family member E (CD85E), is expressed predominantly on monocytes and B cells and at lower levels on dendritic cells and natural killer cells. It acts as a soluble receptor for class I major histocompatibility complex antigens, binding both classical and nonclassical human leukocyte antigen class I molecules (34). While its specific role in regulating plasma HDLc is not known, its presence is associated with chronic inflammatory conditions such as rheumatoid arthritis, suggesting that it may affect HDLc levels indirectly through regulation of chronic inflammation (35).
DNAH10 was included in our 456 gene list because it was supported by GWAS (20). We identified one mutation in one proband with HHDL, IVS63+0G>A, an essential splice site mutation, that segregated with elevated HDLc. Dyneins are microtubule-associated motor protein complexes composed of several heavy, light, and intermediate chains. They are found in cilia and flagella, where they facilitate ATP-driven movement (36). DNAH10 is an inner arm dynein heavy chain (37). Mutations in other dynein heavy chain genes cause primary ciliary dyskinesia (38). DNAH10 protein is expressed in platelets and the liver. However, its role in HDLc metabolism remains unknown.
In addition, we found a novel gene not previously implicated in the regulation of HDLc levels, RNASEL. The role of this gene in HDLc metabolism is unclear. We sequenced RNASEL because of its close proximity to a noncoding variant identified by a GWAS for HDLc (data not shown). It is widely expressed, with high levels in white blood cells, lung, liver, kidney, and adipocytes in humans (Illumina human body map). It functions in anti-viral and anti-proliferative roles of interferons through modulating RNA stability (39), and is part of the body’s innate immune defense. RNASEL is likely to mediate its antiviral effects through a combination of direct cleavage of single-stranded viral RNAs, inhibition of protein synthesis through the degradation of rRNA, induction of apoptosis, and induction of other antiviral genes. Mutations in this gene have been associated with predisposition to prostate cancer (40, 41).
We identified two different mutations in the extremely high HDLc probands in our study, E265X, which results in a premature truncation of the protein, and G179R, a missense mutation. The E265X mutation (rs74315364) has been annotated recently in the 1000 Genomes Project, the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project, and the Exome Chip Project. In the Exome Sequencing Project population, it was found at a minor allele frequency of 0.003. Despite the low minor allele frequency observed in the Exome Sequencing Project population for this variant, of the 120 unrelated high HDLc probands sequenced here, a surprising 5 (4.2%) carried the E265X mutation, indicating a significant enrichment of this variant in those with extremely high HDLc. No association with plasma lipids has previously been described for RNASEL. Our studies implicate new signaling pathways that may regulate HDL, which may provide a better understanding of both the biological functions of this complex particle.
Importantly, these discoveries would not have been readily possible without the use of Mendelian family-based resources, emphasizing the power of family data to supplement and interpret genome-wide population datasets. Indeed, a recent study used exome array screening of 56,000 individuals to identify a handful of novel mutations with significant effects on plasma HDLc, including mutations in PAFAH1B1, ANGPTL8, COL18A1, and PCSK7 (42). However, while this approach has clearly identified new genetic factors that underlie HDLc levels in humans, it is not sufficient to detect extremely rare variants (i.e., <0.1% in the general population) that may be unique to single families. In contrast, by restricting our study to probands with extreme HDLc in whom other confounding factors, such as elevated BMI, complicated medical history and medication use, excessive smoking, alcohol, drug use, and extremes in other lipid parameters are minimized, we have identified four new but separate causes of elevated plasma HDLc levels by screening only 200 unrelated probands and 59 families consisting of 685 family members. Thus both population- and family-based strategies will likely continue to contribute to discovery of new factors underlying plasma HDLc.
The following limitations to our study should be considered. First, as mentioned above, our study reflects an initial analysis of a very large dataset, and more mutations will likely be identified using additional analysis strategies. For example, additional analyses of putative mutations with low frequency (i.e., 0.1–2% in the general population) in dbSNP or 1000 Genomes are in progress. Second, we lack sufficient family members to confirm or rule out statistically many of the SNPs identified here. This limitation is likely to confound many independent researchers using next-generation sequencing approaches and argues for the need for researchers with family-based genetic resources to initiate broad collaborations, replicating the success of very large groups of individuals that identified new genetic susceptibility loci through GWAS of large populations. To this end, the segregation of mutations reported here should be replicated in independent groups. Third, although family members were genotyped for mutations in ABCA1, APOA1, LCAT, CETP, LIPG, and GALNT2 that were found in the original probands, these genes were not sequenced in their entirety in nonproband family members. The presence, in family members, of additional mutations not present in the probands may in part explain the presence of phenocopy or nonpenetrant genotypes in the families, thereby lowering our statistical power during segregation analyses. A recent study on Dutch individuals showed that variants in more than one putative HDL gene can be present per individual (43), reflecting the challenges of identifying mutations with direct Mendelian inheritance. Fourth, we limited our study to coding regions and adjacent sequence, so rare untranslated region, promoter, and other regulatory mutations are not considered here. In vitro functional studies, in addition to detailed phenotyping of HDL particles from individuals described here, are also desired in order to confirm the deleterious effects of some mutations described here, particularly in genes not previously implicated in HDLc regulation. Finally, while our study provides strong support for a genetic role for GCKR, RNASEL, LILRA3, and DNAH10 in HDLc regulation, the biological roles of these four genes in HDLc metabolism, in addition to their clinical significance to cardiovascular and other metabolic diseases, remain targets for future investigation.
In summary, we have performed sequencing of 456 genes in 200 individuals with extreme HDLc levels to identify novel monogenic causes of altered HDLc in humans. Using a family-based Mendelian approach, a total of 59 different families were genotyped to assess segregation of mutations found in probands. Overall, we identified four novel genes with mutations segregating in families with HHDL. We substantially increase the number of known monogenic susceptibility loci for elevated plasma HDLc levels and outline methods that help interpret and prioritize novel mutations associated with a phenotypic trait of interest from large next-generation sequencing datasets.
Supplementary Material
Footnotes
Abbreviations:
- CETP
- cholesteryl ester transfer protein
- dbSNP
- database of SNPs
- DNAH10
- dynein axonemal heavy chain 10
- GALNT2
- UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2
- GCKR
- glucokinase regulatory protein
- GK
- glucokinase
- GWAS
- genome-wide association study
- HDLc
- HDL cholesterol
- HHDL
- high HDL cholesterol or HDL cholesterol ≥90th percentile
- LHDL
- low HDL cholesterol or HDL cholesterol ≤10th percentile adjusted for age and gender
- LILRA3
- leukocyte immunoglobulin-like receptor 3 subfamily A member 3
- LIPG
- endothelial lipase
- RNASEL
- RNase L
- SCARB1
- scavenger receptor class B member 1
These studies were funded by Xenon Pharmaceuticals and Merck Research Laboratories. G.K.H. is the recipient of a Veni Grant (91612122) from the Netherlands Organization for Scientific Research (NWO), the CardioVascular Research Initiative (CVON GENIUS 2011-19; Genius), and the European Union (TransCard: FP7-603091-2). J.J.K. is the recipient of the Lifetime Achievement Award of the Dutch Heart Foundation Excellence (2010T082). R.R.S., I.T., P.L.F., C.R., I.M.S., M.W., M.M., A.L., and R.S. were employees at Xenon Pharmaceuticals at the time of these studies. K.W., M.vH., L.L., L.W., L.M., B.H., K.A., and A.P. were employees of Merck at the time of these studies. J.J.K. and M.R.H. are founding members of Xenon Pharmaceuticals Inc.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five tables.
REFERENCES
- 1.Goode E. L., Cherny S. S., Christian J. C., Jarvik G. P., de Andrade M. 2007. Heritability of longitudinal measures of body mass index and lipid and lipoprotein levels in aging twins. Twin Res. Hum. Genet. 10: 703–711. [DOI] [PubMed] [Google Scholar]
- 2.Qasim A., Rader D. J. 2006. Human genetics of variation in high-density lipoprotein cholesterol. Curr. Atheroscler. Rep. 8: 198–205. [DOI] [PubMed] [Google Scholar]
- 3.Morrison A. C., Voorman A., Johnson A. D., Liu X., Yu J., Li A., Muzny D., Yu F., Rice K., Zhu C., et al. ; Cohorts for Heart and Aging Research in Genetic Epidemiology (CHARGE) Consortium. 2013. Whole-genome sequence based analysis of high-density lipoprotein cholesterol. Nat. Genet. 45: 899–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., et al. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 466: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen J. C., Kiss R. S., Pertsemlidis Y. L., Marcel R., McPherson R., Hobbs H. H. 2004. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 305: 869–872. [DOI] [PubMed] [Google Scholar]
- 6.Tietjen I., Hovingh G. K., Singaraja R. R., Radomski C., McEwen J., Chan E., Mattice M., Legendre A., Kastelein J. J. P., Hayden M. R. 2012. Increased risk of coronary artery disease in Caucasians with extremely low HDL cholesterol due to mutations in ABCA1, APOA1, and LCAT. Biochim. Biophys. Acta. 1821: 416–424. [DOI] [PubMed] [Google Scholar]
- 7.Frikke-Schmidt R., Nordestgaard B. G., Jensen G. B., Tybjaerg-Hansen A. 2004. Genetic variation in ABC transporter A1 contributes to HDL cholesterol in the general population. J. Clin. Invest. 114: 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berge K. E., Leren T. P. 2010. Mutations in APOA-I and ABCA1 in Norwegians with low levels of HDL cholesterol. Clin. Chim. Acta. 411: 2019–2023. [DOI] [PubMed] [Google Scholar]
- 9.Tietjen I., Hovingh G. K., Singaraja R. R., Radomski C., Barhdadi A., McEwen J., Chan E., Mattice M., Legendre A., Franchini P. L., et al. 2012. Segregation of LIPG, CETP, and GALNT2 mutations in Caucasian families with extremely high HDL cholesterol. PLoS ONE. 7: e37437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asselbergs F. W., Guo Y., van Iperen E.P., Sivapalaratnam S., Tragante V., Lanktree M. B., Lange L. A., Almoguera B., Appelman Y. E., Barnard J., et al. 2012. LifeLines Cohort Study. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am. J. Hum. Genet. 91: 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pilia G., Chen W. M., Scuteri A., Orrú M., Albai G., Dei M., Lai S., Usala G., Lai M., Loi P., et al. 2006. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clee S. M., Kastelein J. J., van Dam M., Marcil M., Roomp K., Zwarts K. Y., Collins J. A., Roelants R., Tamasawa N., Stulc T., et al. 2000. Age and residual cholesterol efflux affect HDL cholesterol levels and coronary artery disease in ABCA1 heterozygotes. J. Clin. Invest. 106: 1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karathanasis S. K., Norum R. A., Zannis V. S., Breslow J. L. 1983. An inherited polymorphism in the human apolipoprotein A-I gene locus related to the development of atherosclerosis. Nature. 301: 718–720. [DOI] [PubMed] [Google Scholar]
- 14.Hovingh G. K., Brownlie A., Bisoendial R. J., Dube M. P., Levels J. H., Petersen W., Dullaart R. P., Stroes E. S., Zwinderman A. H., de Groot E., et al. 2004. A novel apoA-I mutation (L178P) leads to endothelial dysfunction, increased arterial wall thickness, and premature coronary artery disease. J. Am. Coll. Cardiol. 44: 1429–1435. [DOI] [PubMed] [Google Scholar]
- 15.von Eckardstein A. 2006. Differential diagnosis of familial high density lipoprotein deficiency syndromes. Atherosclerosis. 186: 231–239. [DOI] [PubMed] [Google Scholar]
- 16.Heiss G., Johnson N. J., Reiland S., Davis C. E., Tyroler H. A. 1980. The epidemiology of plasma high-density lipoprotein cholesterol levels. The Lipid Research Clinics Program Prevalence Study. Circulation. 62: IV116–IV136. [PubMed] [Google Scholar]
- 17.Rogler G., Trumbach B., Klima B., Lackner K. J., Schmitz G. 1995. HDL-mediated efflux of intracellular cholesterol is impaired in fibroblasts from Tangier disease patients. Arterioscler. Thromb. Vasc. Biol. 15: 683–690. [DOI] [PubMed] [Google Scholar]
- 18.Fullwood M. J., Wei C. L., Liu E. T., Ruan Y. 2009. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 19: 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chasman D. I., Paré G., Mora S., Hopewell J. C., Peloso G., Clarke R., Cupples L. A., Hamsten A., Kathiresan S., Mälarstig A., et al. 2009. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 5: e1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen X., Wang W., Wang L., Houde C., Wu W., Tudor M., Thompson J. R., Sisk C. M., Hubbard B., Li J. 2012. Identification of genes affecting apolipoprotein B secretion following siRNA-mediated gene knockdown in primary human hepatocytes. Atherosclerosis. 222: 154–157. [DOI] [PubMed] [Google Scholar]
- 22.Schadt E. E., Woo S., Hao K. 2012. Bayesian method to predict individual SNP genotypes from gene expression data. Nat. Genet. 44: 603–608. [DOI] [PubMed] [Google Scholar]
- 23.Fortin A., Diez E., Gros P., Tietjen I. 2010. Methods for the treatment, prevention and diagnosis of lipid metabolism associated diseases. WO patent 2,010,009,534.
- 24.Kozanitis C., Saunders C., Kruglyak S., Bafna V., Varghese G. 2011. Compressing genomic sequence fragments using SlimGene. J. Comput. Biol. 18: 401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng P. C., Henikoff S. 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramensky V., Bork P., Sunyaev S. 2002. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30: 3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bochem A. E., van Capelleveen J. C., Dallinga-Thie G. M., Schimmel A. W. M., Motazacker M. M., Tietjen I., Singaraja R. R., Hayden M. R., Kastelein J. J. P., Stroes E. S. G., et al. 2014. Two novel mutations in apolipoprotein C3 underlie atheroprotective lipid profiles in families. Clin. Genet. 85: 433–440. [DOI] [PubMed] [Google Scholar]
- 28.Brunham L. R., Tietjen I., Bochem A. E., Singaraja R. R., Franchini P. L., Radomski C., Mattice M., Legendre A., Hovingh G. K., Kastelein J. J., et al. 2011. Novel mutations in scavenger receptor BI associated with high HDL cholesterol in humans. Clin. Genet. 79: 575–581. [DOI] [PubMed] [Google Scholar]
- 29.Vergeer M., Korporaal S. J., Franssen R., Meurs I., Out R., Hovingh G. K., Hoekstra M., Sierts J. A., Dallinga-Thie G. M., Motazacker M. M., et al. 2011. Genetic variant of the scavenger receptor BI in humans. N. Engl. J. Med. 364: 136–145. [DOI] [PubMed] [Google Scholar]
- 30.Bi M., Kao W. H., Boerwinkle E., Hoogeveen R. C., Rasmussen-Torvik L. J., Astor B. C., North K. E., Coresh J., Köttgen A. 2010. Association of rs780094 in GCKR with metabolic traits and incident diabetes and cardiovascular disease: the ARIC Study. PLoS ONE. 5: e11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stančáková A., Paananen J., Soininen P., Kangas A. J., Bonnycastle L. L., Morken M. A., Collins F. S., Jackson A. U., Boehnke M. L., Kuusisto J., et al. 2011. Effects of 34 risk loci for type 2 diabetes or hyperglycemia on lipoprotein subclasses and their composition in 6,580 nondiabetic Finnish men. Diabetes. 60: 1608–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiota C., Coffey J., Grimsby J., Grippo J. F., Magnuson M. A. 1999. Nuclear import of hepatic glucokinase depends upon glucokinase regulatory protein, whereas export is due to a nuclear export signal sequence in glucokinase. J. Biol. Chem. 274: 37125–37130. [DOI] [PubMed] [Google Scholar]
- 33.Edmondson A. C., Braund P. S., Stylianou I. M., Khera A. V., Nelson C. P., Wolfe M. L., Derohannessian S. L., Keating B. J., Qu L., He J., et al. 2011. Dense genotyping of candidate gene loci identifies variants associated with high-density lipoprotein cholesterol. Circ Cardiovasc Genet. 4: 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryu M., Chen Y., Qi J., Liu J., Fan Z., Nam G., Shi Y., Cheng H., Gao G. F. 2011. LILRA3 binds both classical and non-classical HLA class I molecules but with reduced affinities compared to LILRB1/LILRB2: structural evidence. PLoS ONE. 6: e19245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An H., Chandra V., Piraino B., Borges L., Geczy C., McNeil H. P., Bryant K., Tedla N. 2010. Soluble LILRA3, a potential natural anti-inflammatory protein, is increased in patients with rhtumatoid arthritis and is tightly regulated by interleukin 10, tumor necrosis factor-alpha, and interferon-gamma. J. Rheumatol. 37: 1596–1606. [DOI] [PubMed] [Google Scholar]
- 36.Roberts A. J., Kon T., Knight P. J., Sutoh K., Burgess S. A. 2013. Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 14: 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maiti A. K., Mattéi M. G., Jorissen M., Volz A., Zeigler A., Bouvagnet P. 2000. Identification, tissue specific expression, and chromosomal localisation of several human dynein heavy chain genes. Eur. J. Hum. Genet. 8: 923–932. [DOI] [PubMed] [Google Scholar]
- 38.Pazour G. J., Agrin N., Walker B. L., Witman G. B. 2006. Identification of predicted human outer dynein arm genes: candidates for primary ciliary dyskinesia genes. J. Med. Genet. 43: 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou A., Paranjape J., Brown T. L., Nie H., Naik S., Dong B., Chang A., Trapp B., Fairchild R., Colmenares C., et al. 1997. Interferon action and apoptosis are defective in mice devoid of 2’,5′-oligoadenylate-dependent RNase L. EMBO J. 16: 6355–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carpten J., Nupponen N., Isaacs S., Sood R., Robbins C., Xu J., Faruque M., Moses T., Ewing C., Gillanders E., et al. 2002. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat. Genet. 30: 181–184. [DOI] [PubMed] [Google Scholar]
- 41.Rökman A., Ikonen T., Seppälä E. H., Nupponen N., Autio V., Mononen N., Bailey-Wilson J., Trent J., Carpten J., Matikainen M. P., et al. 2002. Germline alterations of the RNASEL gene, a candidate HPC1 gene at 1q25, in patients and families with prostate cancer. Am. J. Hum. Genet. 70: 1299–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peloso G. M., Auer P. L., Bis J. C., Voorman A., Morrison A. C., Stitziel N. O., Brody J. A., Khetarpal S. A., Crosby J. R., Fornage M., et al. 2014. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am. J. Hum. Genet. 94: 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motazacker M. M., Peter J., Treskes M., Shoulders C. C., Kuivenhoven J. A., Hovingh G. K. 2013. Evidence of a polygenic origin of extreme high-density lipoprotein cholesterol levels. Arterioscler. Thromb. Vasc. Biol. 33: 1521–1528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.