Abstract
Nonalcoholic fatty liver disease (NAFLD) is a highly prevalent complication of obesity, yet cellular mechanisms that lead to its development are not well defined. Previously, we have documented hepatic steatosis in mice carrying a mutation in the Sec61a1 gene. Here we examined the mechanism behind NAFLD in Sec61a1 mutant mice. Livers of mutant mice exhibited upregulation of Pparg and its target genes Cd36, Cidec, and Lpl, correlating with increased uptake of fatty acid. Interestingly, these mice also displayed activation of the heat shock response (HSR), with elevated levels of heat shock protein (Hsp) 70, Hsp90, and heat shock factor 1. In cell lines, inhibition of Hsp90 function reduced Pparγ signaling and protein levels. Conversely, overexpression of Hsp90 increased Pparγ signaling and protein levels by reducing degradation. This may occur via a physical interaction as Hsp90 and Pparγ coimmunoprecipitated in vivo. Furthermore, inhibition of Hsp90 in Sec61a1 mutant hepatocytes also reduced Pparγ protein levels and signaling. Finally, overexpression of Hsp90 in liver cell lines increased neutral lipid accumulation, and this accumulation was blocked by Hsp90 inhibition. Our results show that the HSR and Hsp90 play an important role in the development of NAFLD, opening new avenues for the prevention and treatment of this highly prevalent disease.
Keywords: steatosis, peroxisome proliferator-activated receptor γ, heat shock protein 90, heat shock response
NAFLD, which has the potential to advance to nonalcoholic steatohepatitis, and ultimately cirrhosis and hepatocellular carcinoma (HCC), is a significant problem in the Western world (1). The overall prevalence of NAFLD in the United States is ∼30% and is positively correlated with BMI (2). NAFLD is also a risk factor for cardiovascular disease and is thought to negatively regulate insulin signaling (3). Thus, determining the causes of fatty liver disease at the molecular and cellular levels can have a significant effect on treatment of this disease and its comorbidities.
Different cellular and molecular mechanisms are hypothesized to play a role in the development of steatosis. The liver plays a key role in lipid metabolism. It must balance secretion of VLDL, which requires uptake of dietary lipids or de novo synthesis, with oxidation of fatty acids to fuel its own anabolic programs, such as gluconeogenesis (1). A disturbance in this balance can lead to a deleterious accumulation of excess lipids. Increases in synthesis, via constitutively active sterol-regulatory element binding protein (SREBP) 1c, for example, or increases in uptake, via overexpression of the fatty acid transporter CD36, are sufficient to induce steatosis (4, 5). On the other hand, reductions in fatty acid oxidation or VLDL secretion increase fatty acid accumulation (6, 7). Furthermore, insulin resistance is thought to exacerbate steatosis, as higher levels of insulin can drive increased lipogenesis in the liver (8).
With respect to rodent models, both dietary manipulations (e.g., high-fat or methionine-and-choline-deficient diet) and genetic obesity (e.g., the ob/ob mouse) model aspects of NAFLD (9). These models suggest many possible mechanisms for steatosis, such as inflammation, endoplasmic reticulum (ER) stress, and lipotoxic stress. Investigation in mouse models that are independent of dietary manipulation, obesity, or insulin resistance can therefore be helpful in describing primary etiological factors involved in the pathogenesis of NAFLD.
Previously, we have shown that mice homozygous for an ethylnitrosourea-induced point mutation in the Sec61a1 gene (Sec61a1Y344H/Y344H) develop diabetes due to β-cell apoptosis (10). Sec61a1 encodes Sec61α, a 10-pass transmembrane protein that forms an aqueous pore in the ER membrane, which serves as the entry point for all newly synthesized proteins destined for the secretory pathway (11). The cause of apoptosis in these mice may be increased basal ER stress and reduced ER-associated degradation (ERAD), as has been demonstrated in yeast that carry this mutation, but others have documented disrupted calcium homeostasis and ER import of some substrates in cells carrying the Y344H mutation (12–14). These mice also exhibit hepatic steatosis. While β-cell-specific expression of a wild-type Sec61a1 under control of the rat insulin promoter in these mice (Sec61a1Y344H/Y344H, RIP-Sec61) was sufficient to rescue β-cell apoptosis and diabetes, it did not rescue steatosis in the liver (10). Therefore, the Sec61a1Y344H/Y344H, RIP-Sec61 mouse represents a novel, nondiabetic, nonobese model of hepatic steatosis without the requirement for dietary or chemical induction.
MATERIALS AND METHODS
Animals
All mice were bred and housed at the Scripps Research Institute in accordance with institutional animal care and use committee and National Institutes of Health guidelines. All experiments used age- and sex-matched male littermates aged 12 weeks and maintained on a chow diet. The generation of the Sec61a1Y344H/Y344H, RIP-Sec61 mouse has been described previously (10).
Antibodies and reagents
The following antibodies were used in this study: Pparγ and clones E-8 and B-5 (Santa Cruz Biotechnology, Santa Cruz, CA); CCAAT/enhancer binding protein (Cebp) α, Cebpβ, and heat shock factor 1 (Hsf1) (Cell Signaling Technology, Danvers, MA); heat shock protein (Hsp) 90 (Stressgen, Farmingdale, NY); Hsp70 (Abcam, Cambridge, England); FLAG (Clone M-2 Stratagene, Santa Clara, CA); and Sec24d antibody (kindly provided by W. E. Balch). The 17-dimethylaminoethylamino-17-demethoxygeldanamycin (17-DMAG) and radicicol were provided by W. E. Balch. Rosiglitazone was provided by E. Saez. Plasmids for expressing N-terminally tagged Hsf1 and Hsp90 were provided by W. E. Balch. Plasmids for expressing FLAG-tagged Pparγ and luciferase under the control of the peroxisome proliferator response element (PPRE) promoter were provided by E. Saez.
Cell culture and transfection.
HEK293 and Huh7 cells were cultured in DMEM with 10% FBS (Hyclone) and penicillin and streptomycin. All transfections were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA). F442A-Ap2Luc preadipocytes were maintained in DMEM-10% FBS and seeded in 96-well plates at 5 × 103 cells/well density. Two days after confluency, medium was replaced with DMEM-10% FBS and differentiation cocktail (5 μg/ml insulin, 2 μM dexamethasone, and 0.5 mM isobutyl-xanthine). Two days later, medium was changed, and cells were maintained in DMEM-10% FBS and insulin 5 μg/ml, changing every other day. Treatments were performed in DMEM-2% charcoal dextran-treated FBS overnight.
Liver lipid content
Lipids were extracted from liver homogenates using the method of Blighe and Dyer (15). Cholesterol levels were measured using the Amplex Red cholesterol assay kit (Invitrogen), and triglyceride using a triglyceride assay kit (Cayman Chemicals, Ann Arbor, MI).
Quantitative PCR
All real-time PCR was performed on an ABI 7900HT using the Quantitect Probe RT-PCR Kit (Qiagen, Valencia, CA). Taqman primers and probes were from IDT (San Jose, CA). Data were analyzed using the comparative Ct method.
Primary hepatocyte isolation
Primary hepatocytes were isolated using a two-step collagenase perfusion method. Livers were perfused with 50 ml of wash buffer (5.5 mM KCl, 0.1% glucose, 2.1 g NaHCO3, 700 μM EDTA, 10 mM Hepes, 150 mM NaCl) followed with 50 ml of collagenase buffer (5.5 mM KCl, 0.1% glucose, 2.1 g NaHCO3, 3.5 mM CaCl2, 10 mM Hepes, 150 mM NaCl, 1% BSA, and 150 μg/ml of Liberase TM; Roche, Basel, Switzerland). Livers were disrupted and washed 2× with serum-free DMEM. Viable hepatocytes were then isolated by spinning through 45% Percoll (GE Healthcare, Fairfield, CT) at 50 g for 10 min without brake. Cells were plated onto collagen-coated plates (Sigma, St. Louis, MO) in DMEM with 10% FBS; penicillin/streptomycin; insulin, transferrin, selenium supplement (Cellgro, Manassas, VA); and 50 μM dexamethasone (Sigma).
Lipid synthesis
Primary hepatocytes grown in 3.5 cm dishes were labeled with 150 nCi of 14C-acetate (Perkin Elmer, Waltham, MA) for 16 h and then washed 3× with PBS, dissolved first in 500 μl of 0.1 M NaOH, then with 500 μl of water, and combined. An aliquot was saved for protein determination. One milliliter of ethanol and 500 μl of 75% KOH were added, and the sample was incubated at 85°C for 2 h followed by addition of 1 ml of ethanol. Cholesterol was extracted 3× using 1.5 ml of petroleum ether (Sigma). Afterward, 1 ml of concentrated HCl was added, and fatty acids were extracted 3× using 1.5 ml of petroleum ether. Counts associated with the different lipid fractions were then determined using a liquid scintillation counter and Ultima Gold XR cocktail (Perkin Elmer). Samples were normalized to protein content.
Fatty acid uptake
Primary hepatocytes were cultured in 24-well plates. Cells were washed 3× with PBS and incubated in 37°C assay buffer (PBS, 50 μM fatty-acid-free BSA, 37.5 μM sodium oleate, 500 nCi of 1-14C oleic acid) for 5 min. Reactions were stopped with 1 ml of cold stop buffer (PBS, 0.1% fatty-acid-free BSA, and 200 μM phloretin), washed 3×, and lysed with 300 μl of 0.1 N NaOH. An aliquot was saved for protein determination, and the rest was added to 5 ml of Ultima Gold XR scintillation fluid and read on a liquid scintillation counter. Nonspecific counts from cells incubated with ice-cold assay buffer mixed with stop buffer were subtracted from all values.
Hsp90 immunoprecipitation
Cells or tissues were lysed in lysis buffer (20 mM Tris pH 7.5, 125 mM NaCl, 1 mM EDTA, 20 mM Na2MoO4, 5 mM ATP, 0.5% NP-40). Lysates were precleared, and 100 μg of protein for cells or 1 mg for tissue was incubated with 2 μg of anti-FLAG antibody or anti-PPARγ B-5 clone overnight at 4°C, followed by the addition of a 50% slurry of γ-bind for 1 h. Lysates were washed, eluted in SDS sample loading buffer, and fractionated by SDS-PAGE electrophoresis for Western blot.
Luciferase assay
F442A cells were incubated in 96-well plates and treated with the specified compounds for 16 h before addition of Bright-glo substrate (Promega, Madison, WI) and read on a Synergy Mx multimode plate reader (BioTek, Winooski, VT).
AdipoRed staining
After transfection, HuH7 cells were incubated with 200 μM oleate for 16 h, washed with PBS, and stained with AdipoRed (Lonza, Basel, Switzerland), diluted 1:200 in PBS, for 20 min. Cells were then washed and read on a Synergy Mx multimode plate reader (excitation: 485 nm; emission 572 nm).
RESULTS
Sec61a1Y344H/Y344H, RIP-Sec61 mice display hepatic steatosis in the absence of diabetes and obesity
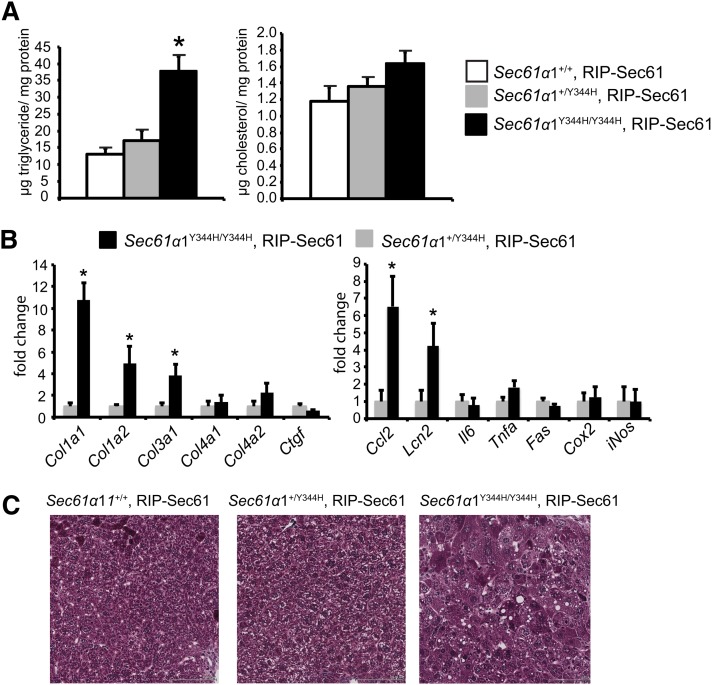
We have previously documented hepatic steatosis in Sec61a1Y344H/Y344H, RIP-Sec61 mice both on chow and high-fat diets (10). Here we analyzed lipids extracted from livers of young, chow-fed, wild-type Sec61a1+/+, RIP-Sec61; heterozygous Sec61a1+/Y344H, RIP-Sec61; and mutant Sec61a1Y344H/Y344H, RIP-Sec61 transgenic mice. Under these conditions, mutant mice showed a 3-fold increase in liver triglyceride content compared with wild-type or heterozygous mice (Fig. 1A). Cholesterol levels were not significantly different among the three genotypes (Fig. 1A).
Fig. 1.
Sec61a1Y344H/Y344H, RIP-Sec61 mice are steatotic. A: Biochemical assay for triglyceride and cholesterol from lipids extracted from livers of Sec61a1+/+, RIP-Sec61 (n = 5); Sec61a1+/Y344H, RIP-Sec61 (n = 5); and Sec61a1Y344H/Y344H, RIP-Sec61 (n = 6) mice. B: Abundance of the indicated mRNA as measured by quantitative PCR (qPCR) from livers of Sec61a1+/Y344H, RIP-Sec61 (n = 4) and Sec61a1Y344H/Y344H, RIP-Sec61 (n = 4) mice. C: Masson trichrome stained liver sections from mice of the indicated genotype. Bar = 100 μM. Error bars represent SEM, and * indicates P ≤ 0.05.
Examining expression levels of genes that encode proteins involved in inflammation and fibrosis indicated increased collagen synthesis and elevation of some markers of inflammation (Fig. 1B), including elevation of serum alanine aminotransferase and aspartate aminotransferase (supplementary Fig. IA, B). However, fibrosis was not yet widespread as indicated by staining of histological sections with Masson’s trichrome (Fig. 1C) and was mainly localized to the perisinusoidal space (data not shown). This is in contrast to these mice on a high-fat diet, which show a much more significant degree of fibrosis (10).
Because we have not previously seen differences between heterozygous and wild-type mice with respect to body mass, fat or lean mass distribution, fasting plasma lipid, and glucose or insulin levels, all subsequent experiments were performed with the unaffected heterozygous Sec61a1+/Y344H, RIP-Sec61 mice as controls (10). Sec61a1Y344H/Y344H, RIP-Sec61 mice weighed slightly less than heterozygotes (supplementary Fig. IC), consistent with reduced glucose and insulin levels, indicative of improved insulin sensitivity (supplementary Fig. ID, E).
Normal lipoprotein and lipid secretion in hepatocytes from Sec61a1Y344H/Y344H, RIP-Sec61 mice
A failure in lipoprotein secretion could account for increased triglyceride accumulation. Normal plasma levels of cholesterol, triglyceride, and nonesterified free fatty acid in Sec61a1Y344H/Y344H, RIP-Sec61 mice argue against a defect in lipid secretion (supplementary Fig. IF–H), but we nonetheless performed experiments in primary hepatocytes to further exclude such a defect. Hepatocytes from Sec61a1Y344H/Y344H, RIP-Sec61 mice exhibited no detectable decrease in albumin secretion compared with heterozygous mice (supplementary Fig. IIA). Furthermore, we saw no specific defect in secretion of ApoB, the main protein component of VLDL, by 35S pulse-chase analysis (supplementary Fig. IIB). Finally, triglyceride or cholesterol secretion into the media of primary hepatocytes from Sec61a1Y344H/Y344H, RIP-Sec61 or heterozygous mice appeared the same (supplementary Fig. IIC).
Upregulation of Pparg and its target genes in Sec61a1Y344H/Y344H, RIP-Sec61 mice
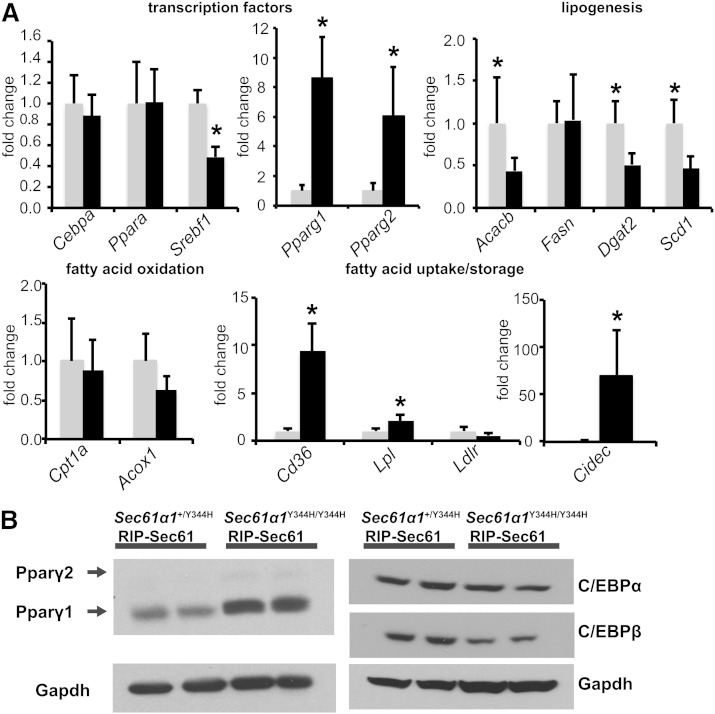
We next measured the expression of lipid metabolic genes in the livers of Sec61a1Y344H/Y344H, RIP-Sec61 or heterozygous control mice, to reveal transcriptional pathways that might affect the development of steatosis. Transcription factors that affect lipid metabolism include Srebpf1 and Cepba, which are associated with lipid synthesis; Pparg, which promotes lipid storage; and Ppara, which directs fatty acid oxidation.
We found a reduction in Srebf1 mRNA levels in Sec61a1Y344H/Y344H, RIP-Sec61 mice compared with heterozygous mice. This reduction coincided with decreased mRNA expression of Srebp-1c target genes Acacb, Scd1, and Dgat2 (Fig. 2A). Furthermore, we did not detect a change in gene expression levels of Ppara or its targets Cpt1a and Acox1 (Fig. 2A). Most notable, however, was an 8-fold increase in Pparg1 and a 6-fold increase in Pparg2 mRNA in livers of Sec61a1Y344H/Y344H, RIP-Sec61 mice and increased expression of their target genes: Cd36, Lpl, and Cidec (Fig. 2A).
Fig. 2.
Upregulation of Pparg and its targets in Sec61a1Y344H/Y344H, RIP-Sec61 mice. A: qPCR analysis of metabolically relevant genes from RNA isolated from livers of mice of the indicated genotype (n = 4 mice per genotype). B: Western blot analysis of Pparγ, C/EBPα, and C/EBPβ proteins from liver tissue homogenates. Error bars represent SEM, and * indicates P ≤ 0.05.
Increased expression of Pparg protein was confirmed by Western blot, which showed an elevation in protein levels of both the Pparγ1 and Pparγ2 isoform in Sec61a1Y344H/Y344H, RIP-Sec61 mice, although levels of Pparγ1 appeared to be the higher in absolute terms (Fig. 2B). It has been hypothesized that increased levels of Pparg mRNA and protein may be due to increased levels of C/EBPα/β proteins, which contribute to its expression in adipose tissue (16). However, we were unable to detect an increase in either C/EBPα or β levels by Western blot (Fig. 2B). Taken together, these results indicate that upregulation of Pparγ in Sec61a1Y344H/Y344H, RIP-Sec61 mice is through a C/EBPα/β-independent pathway.
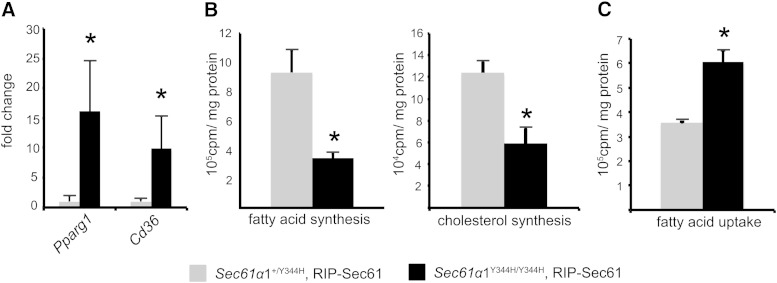
Reduced synthesis and increased uptake of lipid in hepatocytes from Sec61a1Y344H/Y344H, RIP-Sec61 mice
mRNA expression levels of lipid metabolic genes suggested that livers of Sec61a1Y344H/Y344H, RIP-Sec61 mice should exhibit lower rates of de novo lipogenesis and increased uptake of free fatty acids. We tested both of these hypotheses at the biochemical level in primary hepatocytes. qPCR on isolated hepatocytes verified that isolation did not alter gene expression with regard to Pparg and Cd36 mRNA levels (Fig. 3A). Using 14C-acetate, we observed a decrease in synthesis of both cholesterol and fatty acid (Fig. 3B) in Sec61a1Y344H/Y344H, RIP-Sec61 mice, in agreement with reduced levels of Srebf1, Acc2, Dgat2, and Scd1 mRNA. Using 14C-labeled oleic acid to monitor lipid uptake, we observed an ∼2-fold increase in fatty acid uptake (Fig. 3C), which agrees with elevated Cd36 mRNA. These data indicate that steatosis, despite reduced levels of de novo lipogenesis, in Sec61a1Y344H/Y344H, RIP-Sec61 mice is largely a result of increased uptake and storage of fatty acid.
Fig. 3.
Sec61a1Y344H/Y344H, RIP-Sec61 mice display increased uptake of free fatty acid. A: qPCR analysis on RNA isolated from hepatocytes from Sec61a1+/Y344H, RIP-Sec61 or Sec61a1Y344H/Y344H, RIP-Sec61 mice (n = 4 per genotype). Error bars represent SEM, and * indicates P ≤ 0.05. B: Primary hepatocytes were isolated from age- and sex-matched, fasted mice. Rates of sterol and triglyceride synthesis were determined by labeling with 14C-acetate for 16 h. Triplicate assays were performed from each of four (Sec61a1+/Y344H, RIP-Sec61) or three (Sec61a1Y344H/Y344H, RIP-Sec61) mice. C: Primary hepatocytes were isolated as in B. Rate of fatty acid uptake was determined in triplicate after cells were incubated with 14C-labeled oleate for 5 min, washed, and lysed. Cell-associated radioactivity was read by a scintillation counter (n = 2 per genotype).
Sec61a1Y344H/Y344H, RIP-Sec61 mice have normal adipose tissue
Hepatic steatosis as a result of elevated Pparg expression and increased uptake of fatty acid is reminiscent of the phenotype seen in lipoatrophic mice, in which defects in adipose tissue lead to an accumulation of lipid in the liver (17, 18). Therefore, we examined the adipose tissue of Sec61a1Y344H/Y344H, RIP-Sec61 mice for defects. In contrast to lipoatrophic mice, histology of white adipose tissue depots appeared normal (supplementary Fig. IIIA), as did fat mass generally (supplementary Fig. IIIB) and plasma adiponectin levels (supplementary Fig. IIIC). We even observed an increase in Cebpa and Pparg mRNA levels by qPCR in adipose tissue from Sec61a1Y344H/Y344H, RIP-Sec61 mice (supplementary Fig. IIID). Furthermore, we observed no defect in adipocyte differentiation of mouse embryonic fibroblasts from Sec61a1 mutant mice when compared with wild-type or heterozygous mice (supplementary Fig. IIIE).
Activation of ER stress and the heat shock responses in Sec61a1Y344H/Y344H, RIP-Sec61 mice
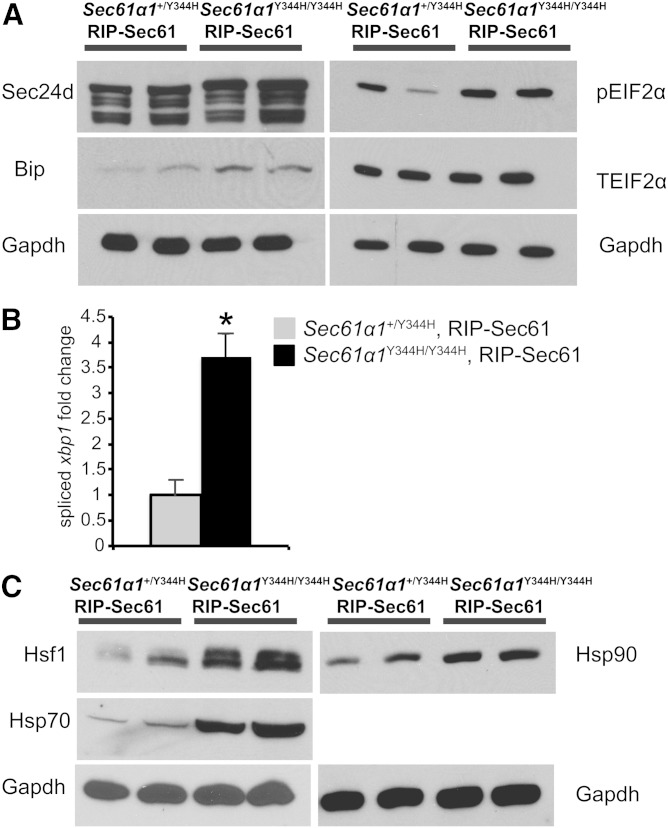
The Sec61α protein is involved in both translocation and the initial stages of protein processing in the ER (11). Schauble et al. (12) have reported excessive calcium leakage and a defective interaction between Bip and the Sec61αY344H mutant, which may impede protein translocation. Both our group and others have reported that the equivalent mutation in yeast leads to defective ERAD, which may lead to ER stress (13, 14). Taken together, these observations suggest a defect in protein processing in Sec61a1Y344H/Y344H cells, which may evoke a compensatory response that could contribute to the steatotic phenotype of mutant mice. We therefore surveyed expression of markers of the ER stress response and the heat shock response (HSR) in liver lysates from Sec61a1Y344H/Y344H, RIP-Sec61 mice.
We observed increases in markers of both the ER stress response and the HSR in Sec61a1Y344H/Y344H, RIP-Sec61 mice compared with heterozygous controls. Consistent with our earlier finding of ER stress [i.e., dilated ER and upregulation of both Bip and Chop mRNA (10)], we saw increased levels of spliced Xbp1 mRNA, Eif2α phosphorylated at serine 52, and Sec24d and Bip protein (Fig. 4A, B). In addition to elevation of ER stress markers, Sec61a1Y344H/Y344H, RIP-Sec61 mice exhibited increased protein levels of key cytosolic chaperones, Hsp90 and Hsp70, and Hsf1, the transcription factor responsible for orchestrating the HSR, compared with controls (Fig. 4C).
Fig. 4.
Increased ER stress and HSR in Sec61a1Y344H/Y344H, RIP-Sec61 mice. A: Western blot analysis for markers of ER stress activation of liver extracts from mice of the indicated genotype. B: qPCR analysis of spliced Xbp1 message on RNA isolated from livers of the indicated genotype (n = 4 per genotype). Error bars represent SEM, and * indicates P ≤ 0.05 by Student’s t-test. C: Western blot analysis of Hsf1, Hsp70, and Hsp90 from liver homogenates prepared from mice of the indicated genotype.
Inhibition of Hsp90 activity reduces Pparγ signaling and expression
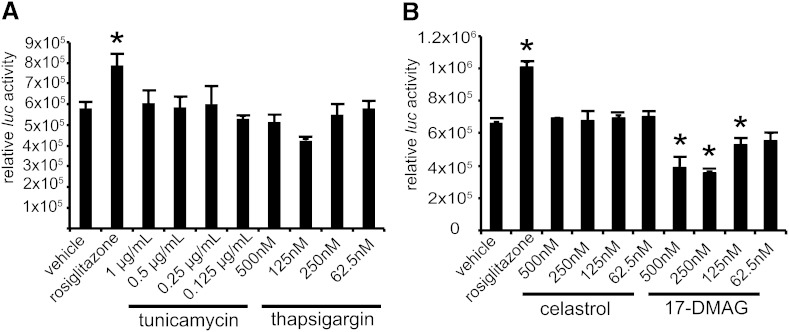
Because Sec61a1Y344H/Y344H, RIP-Sec61 mice exhibited signs of increased ER stress and an increased HSR along with elevations in Pparγ, we asked if perturbation of either pathway could affect Pparγ signaling. As an indicator of Pparγ signaling, we used the F442A-Ap2 reporter cell line, which carries an integrated luciferase gene under control of the Ap2 promoter, which is a Pparγ target gene (19). To induce the ER stress response, we used the small molecules thapsigargin and tunicamycin (20, 21); to induce the HSR, we used the small molecules celastrol, which inhibits cytosolic chaperone activity, and 17-DMAG, which inhibits Hsp90 (22, 23). Treatment with either thapsigargin or tunicamycin resulted in no detectable alteration in Pparγ signaling (Fig. 5A). Likewise, treatment with celastrol did not modify Pparγ signaling via the Ap2 promoter. However, treatment of cells with 17-DMAG did lead to a reduction in Ap2 luciferase activity (Fig. 5B), indicating that Hsp90 activity is necessary for Pparγ signaling.
Fig. 5.
Hsp90 inhibition reduces Pparγ signaling. A: F442A cells stably transfected with luciferase under the control of the AP2 promoter (F442A-Ap2Luc cells) were treated with rosiglitazone, tunicamycin, or thapsigargin at the indicated concentrations for 16 h, followed by lysis and homogeneous assay for luciferase activity. B: Cells were treated and assayed as in A, except that the HSR inducers celastrol and 17-DMAG were used. Error bars represent SD, and * indicates P ≤ 0.05 by Student’s t-test.
Hsp90 interacts with and stabilizes Pparγ facilitating signaling through the PPRE
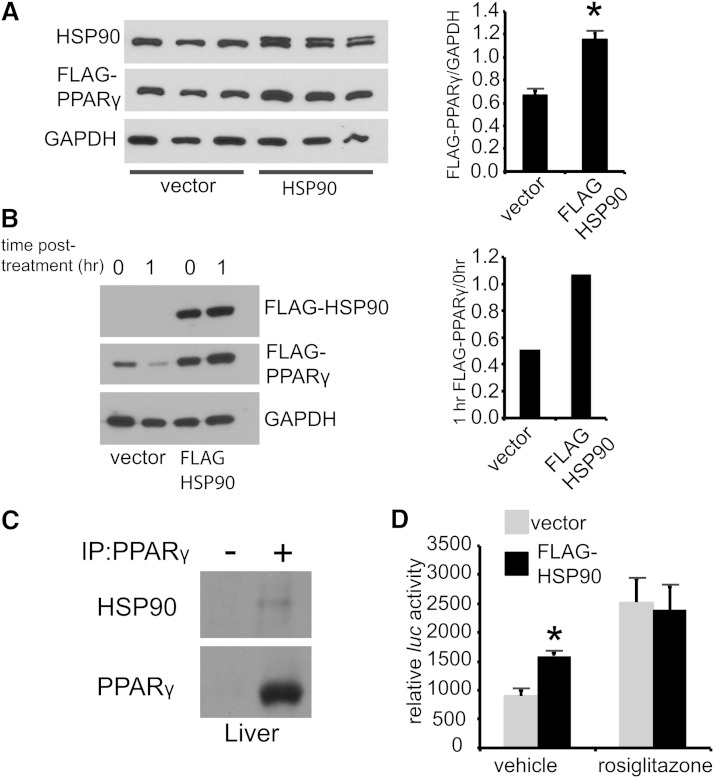
17-DMAG, which reduces Pparγ signaling (Fig. 5B), is specific for Hsp90, and Hsp90 affects stability of several nuclear hormone receptors (24). We therefore focused on Hsp90 as a modulator of Pparγ and asked if Hsp90 inhibition reduces Pparγ protein levels. In HEK293 cells transfected with a cytomegalovirus (CMV)-promoter driven, N-terminally FLAG-tagged version of Pparγ (CMV:FLAG-Pparγ), 17-DMAG dose dependently lowered Pparγ protein levels (supplementary Fig. IVA). Because Sec61a1Y344H/Y344H, RIP-Sec61 mice exhibited both increased Hsp90 and Pparγ protein levels; we next asked if overexpression of Hsp90 was sufficient to increase Pparγ protein levels. In this case, Hsp90 overexpression led to a nearly 60% increase in Pparγ protein levels (Fig. 6A). Furthermore, overexpression of Hsp90 reduced the degradation of Pparγ, following treatment with cycloheximide (Fig. 6B).
Fig. 6.
Hsp90 regulates Pparγ activity. A: Western blot analysis of HEK293 cells cotransfected with CMV:FLAG-Pparγ and either FLAG-Hsp90 or empty vector. Densitometry is shown to the right. B: Cycloheximide treatment (50 μg/ml) of HEK293 cells transfected with CMV:FLAG-Pparγ and either CMV:FLAG-Hsp90 or vector. C: Immunoprecipitation of Pparγ from liver of chow-fed wild-type mice; (-) indicates isotype control. D: HEK293 cells transfected with both a CMV:FLAG-Pparγ and a PPRE luciferase construct and either vector or a FLAG:Hsp90 for 24 h followed by 16 h of treatment with vehicle or rosiglitazone (10 μM) before luciferase assay. Error bars represent SD, and * indicates P ≤ 0.05.
These experiments suggest a mechanism whereby Hsp90 interacts with and stabilizes the Pparγ protein. To test this hypothesis, we asked if Pparγ and Hsp90 physically interact. Endogenous Pparγ coimmunoprecipitated with endogenous Hsp90 from livers of chow-fed C57Bl/6 mice (Fig. 6C), as has been shown previously in 3T3-L1 adipocytes (25). To determine whether Pparγ ligand modifies this interaction, we transfected HEK293 cells with CMV:FLAG-Pparγ. Here also anti-FLAG antibody coimmunoprecipitated Pparγ and endogenous Hsp90 (supplementary Fig. IVB), and treatment of cells with rosiglitazone increased the amount of Hsp90 coimmunoprecipitated with Pparγ. Finally, to see if Hsp90 had a functional effect on Pparγ signaling, we used a PPRE-luciferase reporter assay. Hsp90 overexpression had a positive effect on basal Pparγ signaling with a PPRE luciferase reporter in HEK293 cells, though this effect disappeared upon treatment with rosiglitazone (Fig. 6D).
Hsp90 inhibition blocks Pparγ signaling in primary hepatocytes
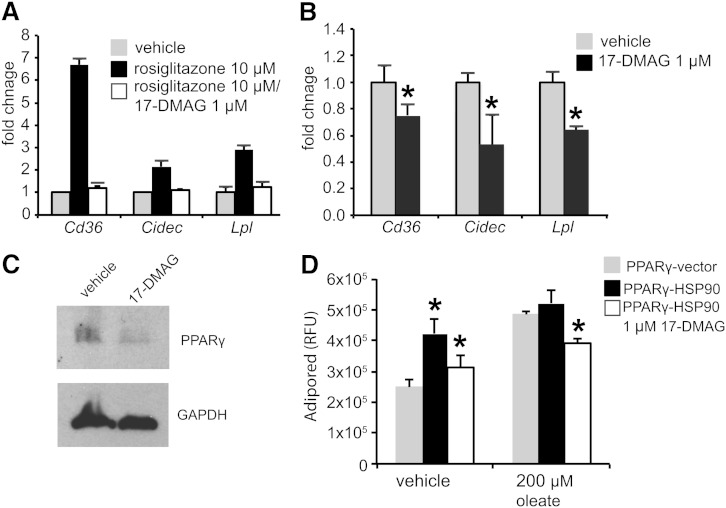
The previous experiments indicate that Hsp90 enhances Pparγ signaling. To place this in physiological context with respect to the development of NAFLD, we performed experiments in primary hepatocytes to see if Hsp90 inhibition could block ligand-induced expression of Pparγ target genes. In hepatocytes from unaffected mice, 17-DMAG completely blocked the upregulation of Cd36, Cidec, and Lpl mRNA in response to rosiglitazone, indicating that Hsp90 activity is required for the transcriptional activity of Pparγ (Fig. 7A).
Fig. 7.
Inhibition of Hsp90 reduces Pparγ target gene induction in Sec61a1Y344H/Y344H, RIP-Sec61 mice. A: qPCR analysis of RNA extracted from primary hepatocytes from Sec61a1+/Y344H, RIP-Sec mice (n = 2) after treatment with 10 μM rosiglitazone for 20 h following 4 h of pretreatment with or without 1 μM 17-DMAG. B: qPCR analysis on RNA extracted from primary hepatocytes from Sec61a1Y344H/Y344H, RIP-Sec mice (n = 2) after treatment with 1 μM 17-DMAG for 24 h. C: Western blot analysis of Pparγ in primary hepatocytes from Sec61a1+/Y344H, RIP-Sec mice treated with 1 μM 17-DMAG for 24 h. D: Huh7 cells, transfected with CMV:FLAG-Pparγ, and either vector or CMV:FLAG:Hsp90, were treated with 200 μM oleate before staining with AdipoRed. The 17-DMAG treatment was at 1 μM for the duration of oleate or vehicle treatment. Error bars represent SEM in A and B, and SD in D, and * indicates P ≤ 0.05 compared with cells transfected with only Pparγ.
We also performed this experiment using hepatocytes from Sec61a1Y344H/Y344H, RIP-Sec61 mice, to see if 17-DMAG treatment could lower the already elevated mRNA levels of these same Pparγ target genes. In this case, treatment of primary hepatocytes lowered levels of Cd36, Cidec, and Lpl (Fig. 7B). This was concomitant with reduced levels of Pparγ protein (Fig. 7C)
To determine whether Hsp90 overexpression has an effect on lipid storage in hepatocyte cell lines, we examined Huh7 cells treated with oleate, which promotes lipid accumulation (26), and stained with the fluorescent dye AdipoRed. Oleate increased lipid accumulation in Pparγ-transfected cells. Additional transfection of Hsp90 also increased lipid accumulation, with little additional effect of oleate (Fig. 7D). This effect required Hsp90 activity as treatment with 17-DMAG reversed lipid accumulation.
DISCUSSION
While NAFLD is a significant public health problem, little is known of its etiology at the cellular level. Sec61a1Y344H/Y344H, RIP-Sec61 mice develop steatosis in the absence of high-fat diet, obesity, hyperinsulinemia, or chemical induction, and we used them here to identify metabolic pathways relevant to the development of NAFLD. Fatty acid uptake was higher in primary hepatocytes from Sec61a1Y344H/Y344H, RIP-Sec61 mice compared with nonsteatotic heterozygous mice. This increase in uptake correlated with an increase, at the mRNA and protein level, of Pparg1 and 2 and mRNA expression of their target genes Cd36, Lpl, and Cidec, which encode proteins involved in the uptake and storage of fatty acids. Conversely, fatty acid and sterol synthesis were lower in primary hepatocytes from Sec61a1Y344H/Y344H, RIP-Sec61 mice compared with hepatocytes from heterozygous mice. This correlated with lower levels of Srebpf1 and its targets in livers of homozygous mutant mice compared with heterozygous controls.
Pparγ plays a well-established role in liver lipid accumulation, as loss of Pparγ function prevents steatosis in genetic or diet-induced obesity (27), and its activation leads to lipid accumulation in both adipocytes and liver (16, 17). Indeed, overexpression of Cd36 or CideC, both Pparγ targets, is sufficient to increase hepatic lipid accretion (5, 28). In this way, the Sec61a1 mutant mouse emulates steatosis as seen in both rodent models and in humans, which are marked by increased levels of Pparγ and its targets (29, 30).
To investigate activation of Pparγ, we looked at stress pathways likely to be activated in Sec61a1Y344H/Y344H, RIP-Sec61 mice, ER stress and heat shock. We have previously documented elevated ER stress in livers of Sec61a1Y344H/Y344H mice. Oyadomari et al. (31) have shown that ER stress in the liver can lead to greater translation of C/EBPα/β protein, which could in turn activate Pparg expression. However, we saw no increase in C/EBPα/β in the liver of Sec61a1Y344H/Y344H, RIP-Sec61 mice and sought other explanations for Pparγ activation. In addition to elevation of markers of the ER stress response, we also saw an increase in the HSR. Hsp70, Hsp90, and Hsf1 protein levels were elevated in the livers of Sec61a1Y344H/Y344H, RIP-Sec61 mice. Furthermore, inhibition of Hsp90 activity with 17-DMAG reduced Pparγ signaling and protein levels. Hsp90 overexpression, on the other hand, increased both Pparγ protein levels and signaling. 17-DMAG also had the effect of blocking the rosiglitazone-induced expression of Pparγ target genes and lowering the levels of those same target genes in primary hepatocyte culture from heterozygous and Sec61a1Y344H/Y344H, RIP-Sec61 mice, respectively. Finally, anti-Pparγ immunoprecipitated Hsp90 in vivo, suggesting that a physical interaction underlies Hsp90’s effects on Pparγ protein abundance and signaling.
Recently, two different labs have found that Hsp90 blockade reduces Pparγ signaling and inhibits adipocyte differentiation in vitro, which requires Pparγ. Nguyen et al. (25) have shown that Hsp90 chaperones Pparγ, while Desarzens et al. (32) have shown that Hsp90 inhibition blocks adipocyte lipid accumulation in vivo. Indeed, Hsp90 interacts with and chaperones many different transcription factors (33), most notably the nuclear hormone receptors, providing precedent for this observation (34). Notably, mice that lack Hsf1, which is the major transcription factor that orchestrates the HSR, display reduced levels of Hsp90 transcript and Pparγ protein and are resistant to steatosis associated with HCC (35).
We are unaware of any human genetic variant of SEC61A1 associated with NAFLD, but this is not surprising given the essential role that SEC61 plays in protein translocation and processing, and the pleiotropic effects of the mouse mutation. Our investigations, however, uncover a role for the HSR in the development of NAFLD, specifically that an interaction between Hsp90 and Pparγ positively regulates levels of Pparγ and thus leads to increased lipid accretion in the liver. This link between NAFLD and Hsp90 is supported by proteomic analysis that indicates that both Hsp90 isoforms are elevated in patients with NAFLD (36).
In conclusion, our studies indicate a role for the evolutionarily conserved HSR in the development of NAFLD. By interacting with a key transcription factor involved in lipid storage, Pparγ, Hsp90 facilitates Pparγ signaling in the liver. This is not the only pathological state that implicates Hsp90, as the HSR and Hsp90 play a role in oncogenic transformation, possibly linking the initiation of steatosis with downstream effects such as the development of HCC.
Supplementary Material
Acknowledgments
The authors thank William E. Balch and Enrique Saez for reagents and experimental advice; Daniela Martino Roth, Darren Hutt, Cristina Godio, and Eduardo Dominguez for experimental advice; and Andrea Galmozzi for differentiation of F442A-Ap2-Luc cells and experimental advice.
Footnotes
Abbreviations:
- 17-DMAG
- 17-dimethylaminoethylamino-17-demethoxygeldanamycin
- C/EBP
- CCAAT/enhancer binding protein
- CMV
- cytomegalovirus
- ER
- endoplasmic reticulum
- HCC
- hepatocellular carcinoma
- Hsf1
- heat shock factor 1
- Hsp
- heat shock protein
- HSR
- heat shock response
- NAFLD
- nonalcoholic fatty liver disease
- PPRE
- peroxisome proliferator response element
- qPCR
- quantitative PCR
- SREBP
- sterol-regulatory element binding protein
This work was supported by the National Institutes of Health Grants K01DK090185 (M.C.W.), R01DK079925 (N.G.), and R21HL113696 (subcontract to N.G.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures.
REFERENCES
- 1.Cohen J. C., Horton J. D., Hobbs H. H. 2011. Human fatty liver disease: old questions and new insights. Science. 332: 1519–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browning J. D., Szczepaniak L. S., Dobbins R., Nuremberg P., Horton J. D., Cohen J. C., Grundy S. M., Hobbs H. H. 2004. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 40: 1387–1395. [DOI] [PubMed] [Google Scholar]
- 3.Targher G., Day C. P., Bonora E. 2010. Risk of cardiovascular disease in patients with nonalcoholic fatty liver disease. N. Engl. J. Med. 363: 1341–1350. [DOI] [PubMed] [Google Scholar]
- 4.Knebel B., Haas J., Hartwig S., Jacob S., Kollmer C., Nitzgen U., Muller-Wieland D., Kotzka J. 2012. Liver-specific expression of transcriptionally active SREBP-1c is associated with fatty liver and increased visceral fat mass. PLoS ONE. 7: e31812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koonen D. P., Jacobs R. L., Febbraio M., Young M. E., Soltys C. L., Ong H., Vance D. E., Dyck J. R. 2007. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes. 56: 2863–2871. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto T., Cook W. S., Qi C., Yeldandi A. V., Reddy J. K., Rao M. S. 2000. Defect in peroxisome proliferator-activated receptor alpha-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J. Biol. Chem. 275: 28918–28928. [DOI] [PubMed] [Google Scholar]
- 7.Wang S., Chen Z., Lam V., Han J., Hassler J., Finck B. N., Davidson N. O., Kaufman R. J. 2012. IRE1alpha-XBP1s induces PDI expression to increase MTP activity for hepatic VLDL assembly and lipid homeostasis. Cell Metab. 16: 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimomura I., Matsuda M., Hammer R. E., Bashmakov Y., Brown M. S., Goldstein J. L. 2000. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol. Cell. 6: 77–86. [PubMed] [Google Scholar]
- 9.Anstee Q. M., Goldin R. D. 2006. Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int. J. Exp. Pathol. 87: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lloyd D. J., Wheeler M. C., Gekakis N. 2010. A point mutation in Sec61alpha1 leads to diabetes and hepatosteatosis in mice. Diabetes. 59: 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park E., Rapoport T. A. 2012. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41: 21–40. [DOI] [PubMed] [Google Scholar]
- 12.Schäuble N., Lang S., Jung M., Cappel S., Schorr S., Ulucan O., Linxweiler J., Dudek J., Blum R., Helms V., et al. 2012. BiP-mediated closing of the Sec61 channel limits Ca2+ leakage from the ER. EMBO J. 31: 3282–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler M. C., Gekakis N. 2012. Defective ER associated degradation of a model luminal substrate in yeast carrying a mutation in the 4th ER luminal loop of Sec61p. Biochem. Biophys. Res. Commun. 427: 768–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tretter T., Pereira F. P., Ulucan O., Helms V., Allan S., Kalies K. U., Romisch K. 2013. ERAD and protein import defects in a sec61 mutant lacking ER-lumenal loop 7. BMC Cell Biol. 14: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 16.Cristancho A. G., Lazar M. A. 2011. Forming functional fat: a growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 12: 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gavrilova O., Haluzik M., Matsusue K., Cutson J. J., Johnson L., Dietz K. R., Nicol C. J., Vinson C., Gonzalez F. J., Reitman M. L. 2003. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J. Biol. Chem. 278: 34268–34276. [DOI] [PubMed] [Google Scholar]
- 18.Shimomura I., Hammer R. E., Richardson J. A., Ikemoto S., Bashmakov Y., Goldstein J. L., Brown M. S. 1998. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 12: 3182–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waki H., Park K. W., Mitro N., Pei L., Damoiseaux R., Wilpitz D. C., Reue K., Saez E., Tontonoz P. 2007. The small molecule harmine is an antidiabetic cell-type-specific regulator of PPARgamma expression. Cell Metab. 5: 357–370. [DOI] [PubMed] [Google Scholar]
- 20.Duksin D., Mahoney W. C. 1982. Relationship of the structure and biological activity of the natural homologues of tunicamycin. J. Biol. Chem. 257: 3105–3109. [PubMed] [Google Scholar]
- 21.Kijima Y., Ogunbunmi E., Fleischer S. 1991. Drug action of thapsigargin on the Ca2+ pump protein of sarcoplasmic reticulum. J. Biol. Chem. 266: 22912–22918. [PubMed] [Google Scholar]
- 22.Jez J. M., Chen J. C., Rastelli G., Stroud R. M., Santi D. V. 2003. Crystal structure and molecular modeling of 17-DMAG in complex with human Hsp90. Chem. Biol. 10: 361–368. [DOI] [PubMed] [Google Scholar]
- 23.Westerheide S. D., Bosman J. D., Mbadugha B. N., Kawahara T. L., Matsumoto G., Kim S., Gu W., Devlin J. P., Silverman R. B., Morimoto R. I. 2004. Celastrols as inducers of the heat shock response and cytoprotection. J. Biol. Chem. 279: 56053–56060. [DOI] [PubMed] [Google Scholar]
- 24.Segnitz B., Gehring U. 1997. The function of steroid hormone receptors is inhibited by the hsp90-specific compound geldanamycin. J. Biol. Chem. 272: 18694–18701. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen M. T., Csermely P., Sőti C. 2013. Hsp90 chaperones PPARγ and regulates differentiation and survival of 3T3-L1 adipocytes. Cell Death Differ. 20: 1654–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Listenberger L. L., Han X., Lewis S. E., Cases S., Farese R. V., Jr, Ory D. S., Schaffer J. E. 2003. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA. 100: 3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsusue K., Haluzik M., Lambert G., Yim S. H., Gavrilova O., Ward J. M., Brewer B., Jr, Reitman M. L., Gonzalez F. J. 2003. Liver-specific disruption of PPARgamma in leptin-deficient mice improves fatty liver but aggravates diabetic phenotypes. J. Clin. Invest. 111: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsusue K., Kusakabe T., Noguchi T., Takiguchi S., Suzuki T., Yamano S., Gonzalez F. J. 2008. Hepatic steatosis in leptin-deficient mice is promoted by the PPARgamma target gene Fsp27. Cell Metab. 7: 302–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miquilena-Colina M. E., Lima-Cabello E., Sanchez-Campos S., Garcia-Mediavilla M. V., Fernandez-Bermejo M., Lozano-Rodriguez T., Vargas-Castrillon J., Buque X., Ochoa B., Aspichueta P., et al. 2011. Hepatic fatty acid translocase CD36 upregulation is associated with insulin resistance, hyperinsulinaemia and increased steatosis in non-alcoholic steatohepatitis and chronic hepatitis C. Gut. 60: 1394–1402. [DOI] [PubMed] [Google Scholar]
- 30.Pettinelli P., Videla L. A. 2011. Up-regulation of PPAR-gamma mRNA expression in the liver of obese patients: an additional reinforcing lipogenic mechanism to SREBP-1c induction. J. Clin. Endocrinol. Metab. 96: 1424–1430. [DOI] [PubMed] [Google Scholar]
- 31.Oyadomari S., Harding H. P., Zhang Y., Oyadomari M., Ron D. 2008. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 7: 520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desarzens S., Liao W. H., Mammi C., Caprio M., Faresse N. 2014. Hsp90 blockers inhibit adipocyte differentiation and fat mass accumulation. PLoS ONE. 9: e94127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taipale M., Krykbaeva I., Koeva M., Kayatekin C., Westover K. D., Karras G. I., Lindquist S. 2012. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 150: 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tata J. R. 2002. Signalling through nuclear receptors. Nat. Rev. Mol. Cell Biol. 3: 702–710. [DOI] [PubMed] [Google Scholar]
- 35.Jin X., Moskophidis D., Mivechi N. F. 2011. Heat shock transcription factor 1 is a key determinant of HCC development by regulating hepatic steatosis and metabolic syndrome. Cell Metab. 14: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodríguez-Suárez E., Duce A. M., Caballería J., Martínez Arrieta F., Fernández E., Gómara C., Alkorta N., Ariz U., Martínez-Chantar M. L., Lu S. C., et al. 2010. Non-alcoholic fatty liver disease proteomics. Proteomics Clin. Appl. 4: 362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.