Abstract
An ultraperformance LC (UPLC) method for the separation of different lipid molecular species and lipid isomers using a stationary phase incorporating charged surface hybrid (CSH) technology is described. The resulting enhanced separation possibilities of the method are demonstrated using standards and human plasma extracts. Lipids were extracted from human plasma samples with the Bligh and Dyer method. Separation of lipids was achieved on a 100 × 2.1 mm inner diameter CSH C18 column using gradient elution with aqueous-acetonitrile-isopropanol mobile phases containing 10 mM ammonium formate/0.1% formic acid buffers at a flow rate of 0.4 ml/min. A UPLC run time of 20 min was routinely used, and a shorter method with a 10 min run time is also described. The method shows extremely stable retention times when human plasma extracts and a variety of biofluids or tissues are analyzed [intra-assay relative standard deviation (RSD) <0.385% and <0.451% for 20 and 10 min gradients, respectively (n = 5); interassay RSD <0.673% and <0.763% for 20 and 10 min gradients, respectively (n = 30)]. The UPLC system was coupled to a hybrid quadrupole orthogonal acceleration time-of-flight mass spectrometer, equipped with a traveling wave ion-mobility cell. Besides demonstrating the separation for different lipids using the chromatographic method, we demonstrate the use of the ion-mobility MS platform for the structural elucidation of lipids. The method can now be used to elucidate structures of a wide variety of lipids in biological samples of different matrices.
Keywords: charge surface hybrid column, ultraperformance liquid chromatography, ion-mobility/high-resolution mass spectrometry, structural elucidation
Lipids are the building blocks of all cell membranes and are thus essential for various biological functions varying from membrane trafficking to signal transduction. Disorders in lipid metabolism play a key role in various diseases including cardiovascular disease, cancer, diabetes mellitus, and inflammation (1, 2). Lipids have been classified into eight categories (fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids, prenol lipids, saccharolipids, and polyketides) by the International Committee for the Classification and Nomenclature of Lipids in conjunction with the LIPID MAPS consortium, based on their chemically distinct functional backbones and hydrophobic nature (3, 4).
As the study of lipids attracts more and more attention, lipidomics, as a branch of metabolomics, is a quickly emerging field. It has been defined as “the full characterization of lipid molecular species and of their biological roles with respect to expression of proteins involved in lipid metabolism and function, including gene regulation” (5).
In order to achieve the goal of full characterization, different MS-based approaches have been described. Some groups describe shotgun lipidomics using direct infusion into the mass spectrometer (6–11). Direct infusion MS has the advantage of short analysis times and consuming very small amounts of sample. However, structural elucidation of lipid isomers is not possible. In order to elucidate structures of lipid isomers, separation needs to be executed. LC is a widely applied technique for separation of lipids, and additionally, it can be readily coupled to electrospray ionization MS. Focusing on only the triacylglycerols, nonaqueous reversed-phase (NARP) LC or silver-ion LC has been successfully applied (12–15). However, this technique cannot be used for analysis of all lipid classes. Different chromatographic systems have been described for analysis of multiple lipid classes on the same platform. Normal-phase LC or hydrophilic interaction LC (HILIC) have been described to separate different lipid classes (16–19). However, run times are often more than 30 min, which is not favorable for high-throughput analysis, and additionally, separation of one lipid class into different molecular species is not possible. A long run time is also a disadvantage for two-dimensional LC [HILIC combined with reversed-phase (RP) LC] (20). RP chromatography (RP-LC) has been widely applied for lipidomic analyses using different stationary phases [e.g., diphenyl (21), C8 (22–24), and C18 (23, 25–34)]. For several years, our group has used high strenght silica (HSS) T3 columns (C18 material) for our lipidomic analyses (33). Although we have extensive experience with this type of chromatography, and in separating both inter- and intralipid classes, optimal separation of different isomers has not been achieved.
Recently, Bird et al. (27) described the separation of cis/trans phospholipids, exploring different RP C18 columns. An Ascentis Express C18 (2.7 µm particles) column gave identical separations and peak shape for cis/trans isomers as a charged surface hybrid (CSH) C18 column (1.7 µm particles). However, due to the lack of an ultra-high-pressure LC system [ultraperformance LC (UPLC)], the CSH C18 column was run at suboptimum conditions; hence, it was suggested to increase the flow rate to optimum conditions and thus improve peak shape and separation and shorten analysis time. The CSH material contains a low-level positive surface charge, in acidic mobile phases, in order to enhance the separation, in addition to increasing the loading capacity.
Herein, we demonstrate the use of CSH C18 material for the separation of different lipid molecular species and different isomers within these classes in complex biological samples, in combination with ion-mobility spectrometry coupled with high-resolution MS.
Ion mobility is, besides chromatographic separation, adding an additional separation dimension located within the mass spectrometer and therefore enhancing structural elucidation of lipids. Using ion mobility, collisional cross-sections (CCSs) of ions can be calculated, and besides accurate mass, fragmentation information, and retention time (Rt), these CCS values can be added to a searchable library for a routine work flow to increase the identification confidence. It was recently demonstrated that the CCS values are highly reproducible even using different machines (35).
The chromatographic separation in combination with the ion-mobility separation is of pivotal importance for structural elucidation of these lipid isomers.
EXPERIMENTAL
Chemicals and materials
HPLC-grade methanol originated from Actu-All Chemicals BV (Oss, The Netherlands). HPLC-grade chloroform, ultra liquid chromatography (ULC)-MS-grade water, 2-propanol (IPA), acetonitrile (ACN), and 99% pure ULC-MS formic acid were purchased from Biosolve BV (Valkenswaard, The Netherlands). Ammonium formate, dichloromethane (DCM), leucine enkephalin (Leu-Enk), and poly-dl-alanine (product number P9003) were from Sigma Aldrich (St. Louis, MO). All lipid standards (as shown in supplementary Table I), and extracts of bovine heart, liver, and brain were purchased from Avanti Lipids (Alabaster, AL) and from Nu-Chek Prep (Elysian, MN). Human plasma with heparin as anticoagulant was obtained from healthy volunteers who had given their informed consent for use of their plasma for method development.
Lipid nomenclature
Throughout the entire paper and to follow a common standard lipid language, the lipid nomenclature described by LIPID MAPS (http://www.lipidmaps.org) was followed (3, 4). For example, 1,2-diheptadecanoyl-sn-glycero-3-phosphocholine is PC (17:0/17:0), 1-nonadecanoyl-2-hydroxy-sn-glycero-3-phosphocholine is LPC (19:0/0:0), 1,2-dipentadecanoyl-sn-glycero-3-phosphoethanolamine is PE (15:0/15:0), and so forth.
Preparation of lipid standards
A mixture of 66 lipids was prepared as shown in Table 1. Stock solutions of 1 mg/ml were prepared in chloroform-methanol (2:1, v/v) and stored at −20°C. Stock solutions were diluted prior to analysis in IPA-ACN-water (2:1:1, v/v/v).
TABLE 1.
List of mixture of 66 lipid standards with corresponding concentration and Rt
| Lipid Subclass | Lipid Molecular Species | Concentration (pmol/μl) | Rt (min) | Peak number (as marked in Fig. 1) |
| FA | C13:1 (12Z) | 4 | 1.24 | a |
| C17:1 (10Z) | 4 | 2.30 | a | |
| C23:1 (14Z) | 4 | 5.44 | a | |
| MG | 14:1 (9Z) | 4 | 1.11 | 5 |
| 17:1 (10Z) | 4 | 1.70 | 9 | |
| 19:2 (10Z, 13Z) | 4 | 1.87 | 10 | |
| D5-DG | 1,3-14:0/14:0 | 2 | 7.60 | 27 |
| 1,3-15:0/15:0 | 2 | 10.17 | 32 | |
| 1,3-16:0/16:0 | 2 | 12.98 | 36 | |
| 1,3-17:0/17:0 | 2 | 13.60 | 40 | |
| 1,3-19:0/19:0 | 2 | 14.55 | 46 | |
| 1,3-20:5 (5Z, 8Z, 11Z, 14Z, 17Z)/20:5 (5Z, 8Z, 11Z, 14Z, 17Z) | 2 | 5.41 | 16 | |
| 1,3-20:4 (5Z, 8Z, 11Z, 14Z)/20:4 (5Z, 8Z, 11Z, 14Z) | 2 | 7.88 | 28 | |
| 1,3-20:2 (11Z, 14Z)/20:2 (11Z, 14Z) | 2 | 13.23 | 38 | |
| 1,3-20:0/20:0 | 2 | 14.95 | 49 | |
| DG | 19:1/19:1 (10Z) | 4 | 13.65 | 42 |
| 19:1/19:1 (10Z) 1,3 isomer | 4 | 13.65 | 43 | |
| D5-TG | 14:0/16:1 (9Z)/14:0 | 2 | 14.99 | 50 |
| 15:0/18:1 (9Z)/15:0 | 2 | 15.65 | 53 | |
| 16:0/18:0/16:0 | 2 | 16.24 | 57 | |
| 19:0/12:0/19:0 | 2 | 16.24 | 58 | |
| 17:0/17:1 (10Z)/17:0 | 2 | 16.08 | 55 | |
| 20:4 (5Z, 8Z, 11Z, 14Z)/18:2 (9Z, 12Z)/20:4 (5Z, 8Z, 11Z, 14Z) | 2 | 14.90 | 48 | |
| 20:2 (11Z, 14Z)/18:3 (6Z, 9Z, 12Z)/20:2 (11Z, 14Z) | 2 | 15.46 | 51 | |
| 20:5 (5Z, 8Z, 11Z, 14Z, 17Z)/22:6 (4Z, 7Z, 10Z, 13Z, 16Z, 19Z)/20:5 (5Z, 8Z, 11Z, 14Z, 17Z) | 2 | 14.10 | 44 | |
| 20:0/20:1 (11Z)/20:0 | 2 | 17.13 | 62 | |
| TG | 19:2 (10Z, 13Z)/19:2 (10Z, 13Z)/19:2 (10Z, 13Z) | 4 | 15.57 | 52 |
| PC | 17:0/20:4 (5Z, 8Z, 11Z, 14Z) | 2 | 7.25 | 25 |
| 18:1 (9Z)/18:0 | 4 | 10.84 | 33 | |
| 19:0/19:0 | 4 | 13.60 | 41 | |
| 21:0/22:6 (4Z, 7Z, 10Z, 13Z, 16Z, 19Z) | 2 | 11.74 | 35 | |
| LPC | 17:0 LPC | 4 | 1.50 | 8 |
| PA | 16:0/18:1 | 4 | 7.92 | 29 |
| PE | 15:0/15:0 | 4 | 6.46 | 20 |
| 17:0/17:0 | 4 | 11.38 | 34 | |
| 18:0/18:0 | 4 | 13.20 | 37 | |
| LPE | 17:1 (10Z) LPE | 4 | 1.25 | 7 |
| PS | 16:0/18:1 (9Z) | 4 | 6.38 | 19 |
| 17:0/20:4 (5Z, 8Z, 11Z, 14Z) | 3 | 5.70 | 17 | |
| 18:1 (9Z)/18:1 (9Z) | 4 | 6.54 | 21 | |
| 21:0/22:6 (4Z, 7Z, 10Z, 13Z, 16Z, 19Z) | 2 | 9.20 | 31 | |
| LPS | 17:1 (10Z) LPS | 4 | 1.00 | 2 |
| PG | 14:0/14:0 | 4 | 3.81 | 13 |
| 17:0/17:0 | 4 | 8.36 | 30 | |
| 18:1 (9Z)/18:1 (9Z) | 4 | 6.71 | 22 | |
| 18:1 (9E)/18:1 (9E) | 4 | 7.36 | 26 | |
| 18:0/18:2 (9Z, 12Z) | 4 | 6.97 | 24 | |
| LPG | 17:1 (10Z) LPG | 4 | 1.00 | 3 |
| LPI | 17:1 (10Z) LPI | 25 | 1.00 | 1 |
| Cardiolipin | 14:1 (9Z)/14:1 (9Z)/14:1 (9Z)/15:1 (10Z) | 4 | 13.29 | 39 |
| 15:0/15:0/15:0/16:1 (9Z) | 4 | 14.82 | 47 | |
| 22:1 (13Z)/22:1 (13Z)/22:1 (13Z)/14:1 (9Z) | 4 | 16.32 | 59 | |
| 24:1 (15Z)/24:1 (15Z)/24:1 (15Z)/14:1 (9Z) | 4 | 16.89 | 61 | |
| Sphingolipid | d17:1 | 5 | 1.08 | 4 |
| d17:0 | 5 | 1.16 | 6 | |
| d18:1/12:0 SM | 5 | 3.49 | 11 | |
| d18:1/12:0 Cer | 5 | 4.72 | 15 | |
| d18:1/25:0 Cer | 5 | 14.49 | 45 | |
| d18:1/12:0 Gluc Cer | 5 | 3.84 | 14 | |
| d18:1/12:0 Lac Cer | 5 | 3.56 | 12 | |
| Sphingomyelin | d18:1/17:0 SM | 4 | 6.83 | 23 |
| Cho | Cho | 4 | 5.84 | 18 |
| CE | 17:0 | 4 | 16.33 | 60 |
| 18:2 (TT) | 4 | 15.86 | 54 | |
| 18:1 | 4 | 16.16 | 56 | |
| 23:0 | 4 | 17.19 | 63 |
Peak ID not shown in Fig. 1. Retention time value is obtained in negative ion mode only. CE, cholesterol ester; Cho, cholesterol; D5-DG, D5-diacylglycerol; LPE, lyso-phosphatidylethanolamide; LPG, lyso-phosphatidylglycerol; LPI, lyso-phosphatidylinositol; LPS, lyso-phosphatidylserine; MG, monoacylglycerol; PS, phosphatidylserine.
Lipid extraction from plasma
Lipids were extracted from human plasma with a slightly adapted protocol from Bligh and Dyer (36). In brief, to 25 µl of plasma 100 µl of cold (−20°C) chloroform-methanol (2:1, v/v) was added. The sample was vortex mixed for 30 s at ambient temperatures and allowed to stand for 5 min at ambient temperatures. After vortex mixing for 30 s, the sample was centrifuged at 12,000 g for 5 min. The lower organic phase was transferred to a clean tube and evaporated to dryness under a gentle stream of nitrogen. Immediately prior to analysis, the sample was reconstituted in 25 µl chloroform-methanol (1:1, v/v) and diluted to 20 and 10 times the original volume of plasma in IPA-ACN-water (2:1:1, v/v/v) for positive and negative ion mode, respectively. Five microliters was injected into the UPLC system for positive ion mode and 10 µl for negative ion mode.
UPLC analysis
Separation of lipids was carried out on an Acquity™ UPLC system (Waters Corporation, Milford, MA). Mobile phase A consisted of 10 mM ammonium formate with 0.1% formic acid in water-ACN (40:60, v/v), and mobile phase B was 10 mM ammonium formate with 0.1% formic acid in ACN-IPA (10:90, v/v). As weak wash, ACN-water-IPA (30:30:40, v/v/v) was used, and the strong wash was a mixture of IPA-water-formic acid-DCM (92:5:2:1, v/v/v/v). Gradient elution was applied at a flow rate of 0.4 ml/min through a CSH C18 column [100 × 2.1 mm inner diameter, particle size 1.7 µm (Waters Corporation, Milford, MA)] and thermostatted at 55°C. Initial conditions started with 40% B, and immediately a linear gradient (curve 6) was started from 40% to 43% B in 2 min. In the following 0.1 min, the percentage of mobile phase B was increased to 50%. Over the next 9.9 min, the gradient was further ramped up to 54% B, and the amount of mobile phase B was increased to 70% in 0.1 min. In the final part of the gradient, the % B was increased to 99% in 5.9 min. The eluent composition returned to the initial conditions in 0.1 min, and the column was equilibrated at the initial conditions for 1.9 min before the next injection, leading to a total run time of 20 min. Sample injections of 5 µl of both the lipid standard mixture and human plasma samples were carried out, and the autosampler temperature was set at 10°C. For both positive and negative ion mode, the same chromatographic conditions were used.
Lipidomics studies often involve hundreds of samples with multiple replicate injections, and therefore, a shorter chromatographic run of 10 min was explored as an alternative to reduce the analysis time for isomeric separation. For this short method, all conditions were the same except the gradient. Initial gradient conditions started with 40% B, and immediately a linear gradient (curve 6) was started from 40% to 43% B in 1 min. In 0.1 min, mobile phase B was increased to 50% B. Over the next 4.9 min, the gradient was further ramped to 54% B, and the amount of mobile phase B was increased to 70% in 0.1 min. In the final part of the gradient, the % B was increased to 99% in 2.9 min. The eluent composition returned to the initial conditions in 0.1 min, and the column was equilibrated at the initial conditions for 0.9 min before the next injection.
MS
The UPLC system was coupled to a traveling wave ion-mobility-enabled hybrid quadrupole orthogonal acceleration time-of-flight mass spectrometer (SYNAPT G2-S HDMS, Waters Corporation, Wilmslow, United Kingdom). Detailed descriptions of this mass spectrometer can be found elsewhere (37, 38). Electrospray positive and negative ionization modes were used. A capillary voltage and sampling cone voltage of (±) 0.6 kV and 30 V were used respectively for both polarities of electrospray ionization. The desolvation source conditions used nitrogen gas at 700 l/h with a constant desolvation temperature of 450°C. The source temperature was set at 120°C. Data were acquired over the m/z range of 50–1,200 Da. The mass spectrometer was operated in ion-mobility (HDMSE) mode for acquisition in both polarities. During this acquisition method, the first quadrupole Q1 was operated in a wide band radio frequency (RF) mode only, allowing all ions to enter the T-wave collision cell. The “trap” T-wave was operated at 4 V causing no fragmentation of the lipids. The intact lipid ions entered the helium cell region of the ion-mobility spectrometry (IMS) cell that was operated at 180 ml/min; the main function of the helium cell was to reduce the internal energy of ions and minimize fragmentation. The lipid ions then entered the IMS cell, held under 80 ml/min flow of nitrogen, to separate species according to their charge, mass, and CCS area. As the separated ions exited the IMS cell, they entered the “transfer” T-wave where two discrete and alternating acquisition functions were used. The first function, typically set at 2 eV, collects low-energy or unfragmented data, while the second function collects elevated-energy or fragment ion data, typically operated using a collision energy ramp from 30 to 55 eV. In both instances, argon gas is used for collision induced dissociation (CID).
The trap T-wave, IMS T-wave, and the transfer T-wave all carried different wave velocities; these were 314, 600, and 190 m/s, respectively. The Stepwave was operated at default settings with a wave velocity of 300 m/s and a wave height of 15.0 V.
Calibration of the ion-mobility cell for CCS calculations was performed using poly-dl-alanine at a concentration of 10 mg/L in water-ACN (50:50, v/v). The calibration procedure is performed automatically by the acquisition software using the fluidics system of the mass spectrometer and based on the principle as described in detail in the recent paper from Paglia et al. (35).
The system was equipped with an integral LockSpray unit with its own reference sprayer that was controlled automatically by the acquisition software to collect a reference scan every 10 s lasting 0.3 s. The LockSpray internal reference used for these experiments was Leu-Enk, and both were used as a lock mass as a lock CCS at a concentration of 0.1 mg/l in water-ACN (50:50, v/v) containing 0.1% formic acid. The reference calibrant was introduced into the lock-mass sprayer at a constant flow rate of 10 µl/min using the fluidics system of the mass spectrometer. A single point lock-mass calibration at m/z 556.2771 in positive ion mode and m/z 554.2615 in negative ion mode was used for the complete analysis.
Acquisition of the data were performed using MassLynx software version 4.1 SCN916 (Waters Corporation, Wilmslow, United Kingdom). The alignment of the low- and elevated-energy spectra and calculation of the CCS values using the mobility data were performed using UNIFI Research Edition, MSE dataviewer version 1.2 (both Waters Corporation, Wilmslow, United Kingdom) and Progenesis QI version 1.0 (Nonlinear Dynamics, Newcastle, United Kingdom). Lipid identification of these time-aligned data was performed using LIPID MAPS online searching tools (http://www.lipidmaps.org) and Progenis QI version 1.0 (Nonlinear Dynamics).
RESULTS AND DISCUSSION
Separation of different lipid subclasses
Our current method for routine lipidomic profiling using RP-UPLC on an HSS T3 column shows separation of different lipid molecular species (33). However, multiple lipid classes are coeluting, and in biological samples, different lipid classes have completely different concentrations. If different lipid classes are coeluting, this can lead to ion suppression effects that obscure the detection of the low abundant lipids. Therefore, enhanced separation of lipid classes is very much advantageous.
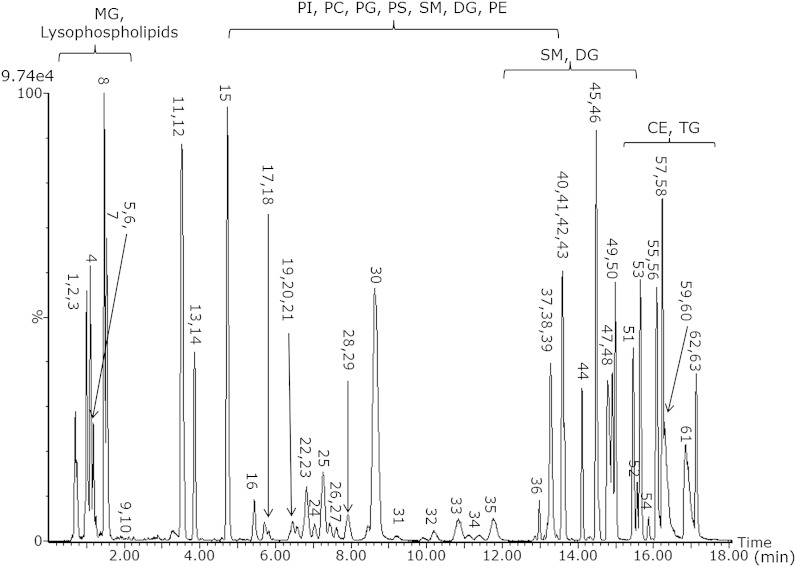
A mixture of 66 lipids as shown in Table 1 was injected on the CSH C18 UPLC column with the 20 min method. For the purpose of demonstrating the separation, all lipids, except the FFAs were analyzed in positive ion mode; however, it should be mentioned that multiple lipid classes [e.g., phosphatidylinositol (PI), phosphoethanolamine (PE), phosphatidic acid (PA), etc.] ionize much more efficiently in negative ion mode. Fig. 1 shows the separation of this mixture, demonstrating separation of different lipid classes. Within each class, an additional carbon atom increases the Rt, while an additional double bond reduces the Rt. This is demonstrated in more detail subsequently.
Fig. 1.
Base peak intensity chromatogram of 66 lipids (see Table 1) on CSH C18 acquired in positive ion mode in the 20 min gradient.
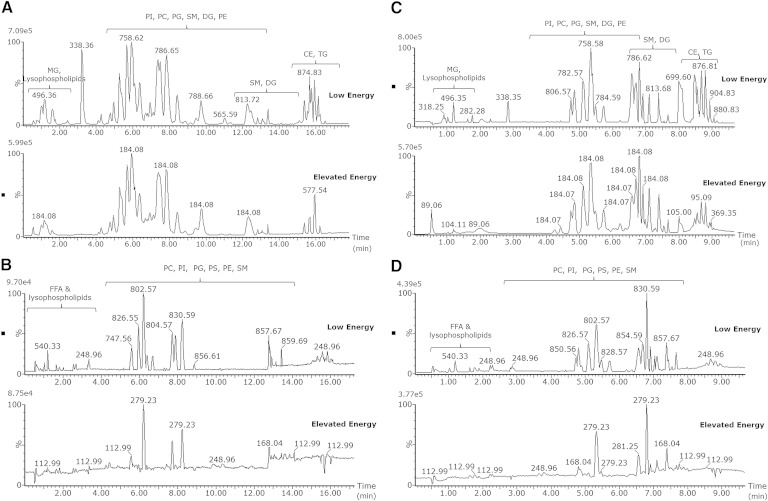
To demonstrate the applicability in biological material, extracts of human plasma were injected in both positive and negative ion mode using HDMSE acquisition as shown in Fig. 2A, B, respectively, for the 20 min gradient and in Fig. 2C, D both positive and negative ion mode for the 10 min gradient. Both low- and elevated-energy spectra are available from a single injection, and therefore, it is possible to focus on specific lipid classes by searching for the class-specific fragments in the elevated-energy chromatogram. In the elevated-energy trace for positive ion mode (Fig. 2A, C), for example, the most abundant fragment ion is in many cases m/z 184.08, which corresponds to the phosphatidylcholine (PC) head group. All peaks with this specific fragment belong to the same class. In negative ion mode, lipids fragment differently, not giving class-specific fragments but giving direct acyl chain fragments (e.g., m/z 279.23 corresponds to an 18:2 chain, while m/z 281.25 corresponds to an 18:1 chain). The elucidation of the lipid acyl chains using the chromatographic method in combination with the ion-mobility function to separate coeluting species will be demonstrated subsequently.
Fig. 2.
Base peak intensity chromatogram for human plasma for positive 20 min gradient (A), negative 20 min gradient (B), positive 10 min gradient (C), and negative 10 min gradient (D) polarity. All chromatograms are acquired in HDMSE mode and thus are from a single injection; both low- and elevated-energy data available.
Separation of isomers using CSH chromatography
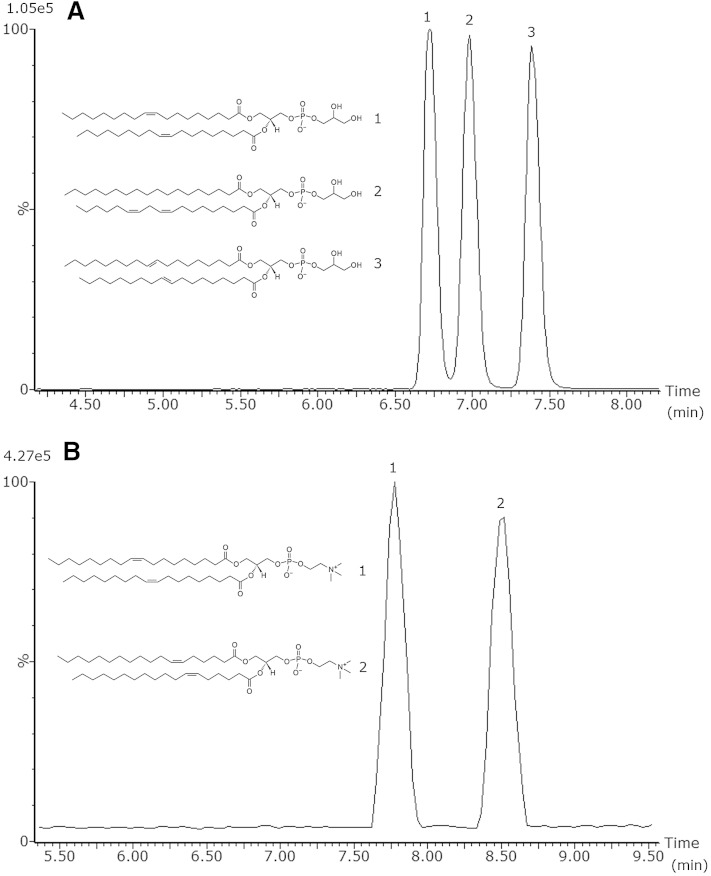
In order to elucidate the structures of lipid isomers in an untargeted platform, it is of importance to separate the individual isomers as much as possible. Humans only synthesize cis (Z) FAs endogenously; however, due to dietary intake, synthetic trans (E) FAs are present in the human body as well, and it is well known that they play an important role in various metabolic diseases (39–41). Therefore, it is of great interest to separate cis/trans isomers in the chromatographic method. To test the ability of the UPLC and CSH technology to separate cis/trans isobaric lipids and to separate structural isomers, lipid standards were injected either alone or as a mixture. Fig. 3A shows the separation of cis/trans isobaric phosphatidylglycerol (PG) species using the 20 min chromatographic method, PG 18:1 (9Z)/18:1 (9Z) and PG 18:1 (9E)/18:1 (9E), and additionally, the structural isomer PG 18:0/18:2 (9Z, 12Z) was added. In addition to PG, PC standards with a different double-bond position, PC 18:1 (6Z)/18:1 (6Z) and PC 18:1(9Z)/18:1(9Z), were also tested for their separation as shown in Fig. 3B. The CCSs of these lipids are quite similar and most likely not significantly different, 302.8 and 305.6 A2, respectively, demonstrating that chromatographic separation and ion-mobility separation are complementary. The mobility mobilogram of these two lipids is depicted in supplementary Fig. I. Biological samples are far more complex, and when an extracted ion chromatogram (XIC) for PC 36:2 ([M+H]+ m/z 786.6007) is generated from a porcine brain extract, three peaks are visible as shown in supplementary Fig. II. Using the elevated-energy trace, it appears that the first and third peaks are indeed PC 18:1/18:1. Besides the same Rt as in the standards (7.74 and 8.49 min, respectively), the species also have a quite similar CCSs (302.8 and 305.9 A2, respectively) as obtained with the standards. As our method is not suited for double-bond localization, we cannot claim with full certainty that the peaks are PC 18:1 (9Z)/18:1 (9Z) and PC 18:1 (6Z)/18:1 (6Z), respectively. However, besides an Rt, the additional value of the mobility cell is that CCS values are obtained that give an additional confirmation value. Besides the two PC 18:1/18:1 peaks, in the middle a third peak is visible (Rt 8.03 min). When using the elevated-energy trace, it appears that this peak is either PC 18:0/18:2 or PC 18:2/18:0, with CCS 301.8 A2.
Fig. 3.
Separation of PG 18:1 (9Z)/18:1 (9Z) (1) and PG 18:1 (9E)/18:1 (9E) (3), as well as PG 18:0/18:2 (9Z, 12Z) (2) (A), and separation of PC 18:1 (9Z)/18:1 (9Z) (1) and PC 18:1 (6Z)/18:1 (6Z) (2) (B) using the 20 min gradient.
As shown with the examples in Fig. 3, the cis/trans isobaric isomers are baseline separated in chromatography, while the PG structural isomer PG 18:0/18:2 (9Z, 12Z) is baseline separated from the trans isomer and almost baseline separated from the cis isomer. Additionally, with the PC example, it is demonstrated that also structural isomers with a different double-bond location are fully baseline separated. This information would typically not be available using an infusion or traditional HPLC method. In comparison, this separation was not achieved using the HSS T3 chromatographic method (33) as will be demonstrated with examples in human plasma later in this manuscript.
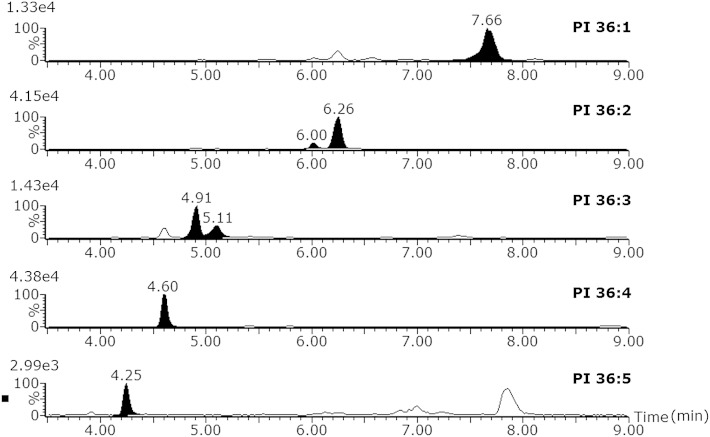
Fig. 4 demonstrates the separation of a lipid class with as example the PIs. XICs from a plasma extract injection in negative ion mode were created using accurate mass of the low-energy trace and additionally using the elevated-energy trace for additional confirmation of the lipid identity. The lipids are separated according to the acyl chain lengths and the number of double bonds. This chromatographic elution profile partially fits the well-known model used for TGs on NARP chromatography, where the ECN is defined as the total carbon number (CN) of fatty acyls minus two times the double-bond (DB) number (ECN = CN – 2DB). The Rt of lipids other than TGs in RP-HPLC also increases to their ECN as demonstrated by Lísa et al. (20) on a Kinetix C18 column and as shown in Fig. 4. Additionally, for TGs the separation of cis/trans isomers, double-bond positional isomers, and linear/branched isomers has also been described (13, 20) using NARP, and as demonstrated in Fig. 3, this is also possible for RP-LC for other lipid classes than TGs.
Fig. 4.
Separation of phosphatidylinositol lipids in human plasma according to their equivalent carbon number (ECN). XIC for every PI was made on [M-H] with 0.05 Da extraction window.
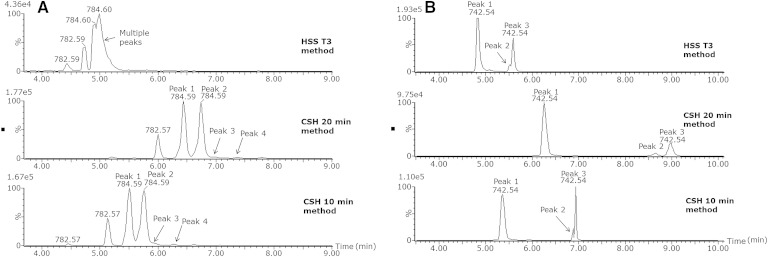
To demonstrate the applicability of this new chromatographic method in real biological samples and to compare it with our previous method using an HSS T3 column (33), we analyzed the same human plasma extracts on both platforms as shown in Fig. 5. The XIC for PC 36:3 ([M+H]+ m/z 784.5851) clearly shows that on the T3 column (Fig. 5A) there are at least two peaks visible that are not separated at all. Most likely, these peaks consist of multiple components as the peaks shape tends to indicate this. The same XIC on the CSH column in the 20 min method results in two near baseline separated peaks. The second peak has a shoulder that is also PC 36:3, and additionally, a fourth peak is visible. For the CSH column in the 10 min method, the separation and peak shapes are comparable with the 20 min method. This example demonstrates the superior separation of different isomers of the CSH method compared with the HSS T3 method.
Fig. 5.
XIC for a human plasma sample for PC 36:3 ([M+H]+ m/z 784.585 with 0.05 Da extraction window) on either HSS T3, CSH 20 min method, or CSH 10 min method (A) and XIC for PE 36:2 ([M-H] m/z 742.5392 with 0.05 Da extraction window) on either HSS T3, CSH 20 min method, or CSH 10 min method (B).
For PE 36:2 ([M-H]− m/z 742.5392) using the HSS T3 column (Fig. 5B), two separate peaks are visible; however, the peak shape of the second peak already indicates that it consists of two components. On the CSH column with the 20 min method, it is clearly visible that three different isomers are present as peaks 2 and 3 are completely baseline separated, demonstrating again the superior separation of isomers that can be achieved with the CSH method compared with the T3 method. With the 10 min method on the CSH column, the separation of peaks 2 and 3 is slightly better than on the HSS T3 column but is definitely not comparable with the 20 min method. In the 20 min method, the amount of mobile phase is increased from 50% to 54% from 2.1 to 12 min. On the 10 min method, this same increase in % B is from 1.1 to 6 min. Peaks that elute in the 20 min method relatively late in this part of the gradient appear not to elute in this part in the 10 min method. They elute in the next part (70–99% B from 6.1 to 9 min), and because of this steep gradient, a separation loss is observed. However, the 10 min method was developed as an alternative method for the analysis of hundreds of biological samples, and depending on the research question, it might be a useful method. We recommend using the 20 min method when an optimal separation of isomers is required. If, however, separation of isomers is not the major point of interest, but the separation of different lipid classes is of importance, then the 10 min method might be a good alternative for higher-throughput studies.
Robustness of the method
To test the robustness of the Rts on the CSH chromatography, human plasma extracts were injected in both positive and negative ion mode for both gradients. Samples were injected in each run and each condition for five times, and this was repeated in six separate runs. Between the runs, the UPLC-MS system was used for different application. From each lipid class, a single lipid was chosen as an example of the entire class. If the lipids of one class elute over a wide Rt window, then a lipid eluting approximately halfway through this elution window was chosen.
The intra-assay RSDs for Rt were ≤0.385% and ≤0.451% for the 20 and 10 min gradients, respectively. The interassay RSDs were ≤0.673% and ≤0.763% for the 20 and 10 min gradients, respectively. In absolute time between the longest and shortest Rt for a lipid, these numbers are 0.20 min for the 20 min gradient (diacylglycerol (DG) 36:3) and 0.10 min for the 10 min gradient (sterol lipids (STs) d18:1/20:0 and HexCer d18:1/16:0) demonstrating the excellent Rt stability for these methods.
Detailed results of this robustness experiment can be found in supplementary Tables I, II for the 20 and 10 min gradients, respectively.
To obtain these stable Rts from run to run, preparation of the mobile phase is of pivotal importance. We experienced that switching from 99% pure formic acid to 90% pure formic acid caused a shift in Rt of ∼3 min later (in the 20 min gradient). Additionally, flushing out the column with, for example, water-ACN (50:50, v/v) after a run and reconditioning of the column for the next run with mobile phase causes a slightly earlier elution time (maximum 0.15 min) for some lipids. Most likely, these effects are due to charge changes of the column material. However, this can be circumvented by storing the column in the mobile phase and not flushing with water-ACN (50:50, v/v). We have practical experience of leaving the column stored in the mobile phase for more than 5 months. After this period, the column can be used without any problem, back pressure and peak shape are identical to those obtained before, and additionally the Rts are also exactly the same as before. Besides using a high quality of formic acid and always leaving the column in the mobile phase, care should be taken with the preparation of especially mobile phase B. It is of critical importance that the ammonium formate is completely dissolved in the mixture of ACN-IPA (10:90, v/v), and this can be achieved by continuously stirring and heating the solvent during ∼1 h.
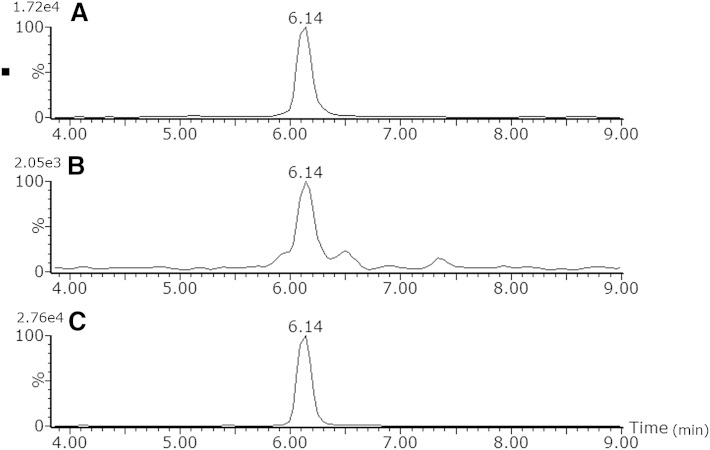
To test the applicability of the method in different matrices besides plasma, extracts of bovine liver, brain, and heart tissue were injected. Fig. 6 shows an extracted ion chromatogram of PC 34:2 using the 20 min chromatographic method. It can be clearly seen that the Rt of this lipid is the same in all three matrices demonstrating the applicability of the chromatographic method in various biological matrices.
Fig. 6.
Comparison of Rt of bovine liver (A), brain (B), and heart (C) extracts for PC 34:2 [M+H]+ m/z 758.5695 (with 0.05 Da extraction window) using the 20 min chromatographic method.
APPLICATION OF THE METHOD: ELUCIDATION OF LIPID ISOMERS
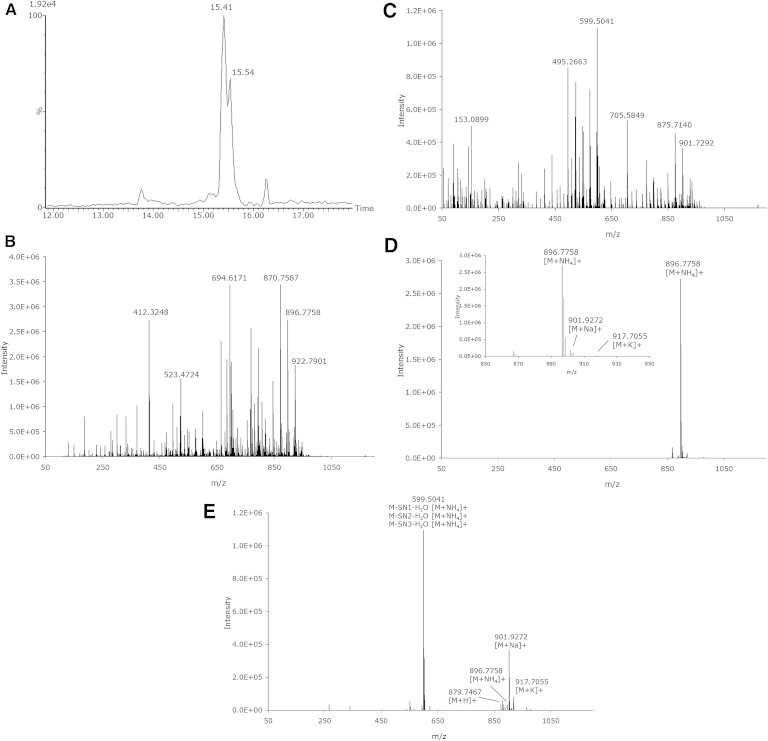
The goal of developing the described powerful method for separation of different lipid classes and different isomers within these classes is to elucidate the structures of the individual lipids. With the Synapt G2-S system, when operated in HDMSE mode, a spectrum obtained with low collision energy and a spectrum with elevated collision energy are obtained from a single injection of the sample. The fragmentation of lipids using MS has been extensively described (33, 42–45). From the fragment ion spectrum, the acyl chain information can be obtained. In positive ion mode, the most abundant fragment is often the head group. However, acyl chain-specific fragments are also visible and can be observed as M-SN1 and/or M-SN1-water, M-SN2 and/or M-SN2-water. In negative ion mode, the elucidation of the acyl chains is even more straightforward as the most abundant fragments are the direct tail fragments SN1 and SN2. For additional confirmation, the lower abundant fragments M-SN1 and/or M-SN1-water, M-SN2 and/or M-SN2-water can also be used. Fig. 7A shows an extracted ion chromatogram of a human plasma sample of TG 54:6 ([M+NH4]+ m/z 896.7702) containing two peaks at 15.41 and 15.54 min. Only the first peak at 15.41 min corresponds to the accurate mass of TG 54:6 (CCS 330.2 A2). In Fig. 7B, C, the associated low and elevated spectra are shown, respectively, when the mobility function is not used. It is clearly visible that at this Rt not only TG 54:6 elutes but also other highly abundant lipids. As all ions enter the “transfer” region of the mass spectrometer, all the lipids with the same Rt are fragmented. The obtained spectrum is thus a mixture of all fragments derived from the different precursor ions, thus making structural elucidation a challenge, especially with this class of compounds where fragments of various precursor ions have a high degree of similarity. However, due to the use of the ion-mobility cell and exploiting its additional orthogonal separation technique, intact lipid ions are separated on basis of the CCS, mass, and charge. The fragment ions derived in the transfer cell can therefore be time aligned with the precursor lipid ions, not just by Rt but also using the ion-mobility CCS, significantly “cleaning up” the spectrum, showing only the fragment ions of that specific drift time. Fig. 7D demonstrates for the peak at 15.41 min, when only the peaks with the same drift time are selected that the low-energy spectrum is easy to interpret. For TG 54:6 at an Rt of 15.41 min with CCS 330.2 A2, the most abundant ion is clearly [M+NH4]+; however, [M+Na]+ and [M+K]+ are also visible. Fig. 7E shows the time-aligned elevated-energy spectrum. Almost all [M+NH4]+ has been fragmented, but it is visible that almost no fragmentation has occurred for [M+Na]+ and [M+K]+. Most likely, these adducts require higher energies to induce fragmentation than applied in this experiment. The use of the ion-mobility function allows this lipid structure to be elucidated as TG 18:2/18:2/18:2.
Fig. 7.
XIC of a human plasma sample of TG 54:6 ([M+NH4]+ m/z 896.7702 with 0.05 Da extraction window) (A) with the associated low-energy (B) and elevated-energy (C) spectra without mobility data used for the peak at 15.41 min (± 0.08 min) and the same low-energy (D) and elevated-energy (E) spectra using the mobility data making structural elucidation possible.
To demonstrate an example from another lipid class and additionally in negative ion mode, PI 36:1 as shown in Fig. 4 is elucidated. This peak has an Rt of 7.66 min, and when a spectrum is made of the low-energy trace without using mobility (supplementary Fig. IIIA), it is visible that other lipids elute at the same Rt. The elevated spectrum of all these lipids leads to a mixture of many different fragments (supplementary Fig. IIIB) making structural elucidation. However, if at this Rt only the peak with CCS 303.0 A2 is selected by using the mobility information in the low-energy trace, only PI 36:1 is visible (supplementary Fig. IIIC), and as the elevated-energy spectrum (supplementary Fig. IIID) is time aligned, all obtained fragments are derived only from this parent making the elucidation of PI 18:0/18:1 or PI 18:1/18:0 possible. For PI lipids, the elevated-energy ramp of 30–55 V appears to be on the low side as quite some parent ion is left in the elevated-energy trace. However, all lipid-specific fragments are obtained, and thus acyl chain elucidation is possible.
With this method, it cannot be concluded which tail is actually SN1 and which one is SN2, and therefore, both PI 18:0/18:1 and PI 18:1/18:0 are possible.
Next to these two examples for both positive and negative ion mode, this feature is as well exploited for all other lipid subclasses and allows us to assign an identity to a large number of the lipid structures.
CONCLUSION
In this paper, the development and application of an UPLC method for the separation of lipid molecular species and lipid isomers using a CSH C18 column has been described. We have coupled the chromatographic system to a traveling wave ion-mobility-enabled hybrid quadrupole orthogonal acceleration time-of-flight mass spectrometer. As lipid isomers are chromatographically separated, structural elucidation of the lipids can be obtained. The additional separation in the ion-mobility part of the mass spectrometer is of pivotal importance for this process as was demonstrated by the structural elucidation of the TG and PI species. Additionally, a CCS value is available that gives an additional confirmation for the compound. The improved peak capacity and separation efficiency afforded by the chromatography and ion-mobility are complementary, and without one or the other, structural elucidation is a complex process. The method described here opens up future opportunities for the structural elucidation of many different lipids in biological samples. Future work will focus on the creation of a lipid database including Rts, acyl chain information, and CCS to be implemented for routine lipidomic screening.
Supplementary Material
Acknowledgments
The authors thank Unilever R&D (Vlaardingen, The Netherlands) for their hospitality regarding the use of their equipment.
Footnotes
Abbreviations:
- ACN
- acetonitrile
- CCS
- collisional cross-section
- CSH
- charged surface hybrid
- ECN
- equivalent carbon number
- IMS
- ion-mobility spectrometry
- IPA
- 2-propanol
- NARP
- nonaqueous reversed-phase
- PC
- phosphatidylcholine
- PE
- phosphoethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- RP
- reversed-phase
- Rt
- retention time
- UPLC
- ultraperformance LC
- XIC
- extracted ion chromatogram
This work was supported by the Netherlands Metabolomics Centre, which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables and three figures.
REFERENCES
- 1.Maxfield F. R., Tabas I. 2005. Role of cholesterol and lipid organization in disease. Nature. 438: 612–621. [DOI] [PubMed] [Google Scholar]
- 2.van Meer G., Voelker D. R., Feigenson G. W. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fahy E., Subramaniam S., Brown H. A., Glass C. K., Merrill A. H., Murphy R. C., Raetz C. R. H., Russell D. W., Seyama Y., Shaw W., et al. 2005. A comprehensive classification system for lipids. J. Lipid Res. 46: 839–861. [DOI] [PubMed] [Google Scholar]
- 4.Fahy E., Subramaniam S., Murphy R. C., Nishijima M., Raetz C. R., Shimizu T., Spener F., van Meer G., Wakelam M. J., Dennis E. A. 2009. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 50 (Suppl.): S9–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spener F., Lagarde M., Géloên A., Record M. 2003. Editorial: what is lipidomics? Eur. J. Lipid Sci. Technol. 105: 481–482. [Google Scholar]
- 6.Han X., Yang K., Gross R. W. 2012. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom. Rev. 31: 134–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han X., Gross R. W. 2003. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 44: 1071–1079. [DOI] [PubMed] [Google Scholar]
- 8.Basconcillo L. S., Zaheer R., Finan T. M., McCarry B. E. 2009. A shotgun lipidomics study of a putative lysophosphatidic acid acyl transferase (PlsC) in Sinorhizobium meliloti. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2873–2882. [DOI] [PubMed] [Google Scholar]
- 9.Basconcillo L. S., Zaheer R., Finan T. M., McCarry B. E. 2009. A shotgun lipidomics approach in Sinorhizobium meliloti as a tool in functional genomics. J. Lipid Res. 50: 1120–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ejsing C. S., Sampaio J. L., Surendranath V., Duchoslav E., Ekroos K., Klemm R. W., Simons K., Shevchenko A. 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 106: 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy R. C., Leiker T. J., Barkley R. M. 2011. Glycerolipid and cholesterol ester analyses in biological samples by mass spectrometry. Biochim. Biophys. Acta. 1811: 776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holčapek M., Dvořáková H., Lísa M., Girón A. J., Sandra P., Cvačka J. 2010. Regioisomeric analysis of triacylglycerols using silver-ion liquid chromatography-atmospheric pressure chemical ionization mass spectrometry: comparison of five different mass analyzers. J. Chromatogr. A. 1217: 8186–8194. [DOI] [PubMed] [Google Scholar]
- 13.Lísa M., Holčapek M. 2013. Characterization of triacylglycerol enantiomers using chiral HPLC/APCI-MS and synthesis of enantiomeric triacylglycerols. Anal. Chem. 85: 1852–1859. [DOI] [PubMed] [Google Scholar]
- 14.Lísa M., Holcapek M., Sovová H. 2009. Comparison of various types of stationary phases in non-aqueous reversed-phase high-performance liquid chromatography-mass spectrometry of glycerolipids in blackcurrant oil and its enzymatic hydrolysis mixture. J. Chromatogr. A. 1216: 8371–8378. [DOI] [PubMed] [Google Scholar]
- 15.Lísa M., Netušilová K., Franěk L., Dvořáková H., Vrkoslav V., Holčapek M. 2011. Characterization of fatty acid and triacylglycerol composition in animal fats using silver-ion and non-aqueous reversed-phase high-performance liquid chromatography/mass spectrometry and gas chromatography/flame ionization detection. J. Chromatogr. A. 1218: 7499–7510. [DOI] [PubMed] [Google Scholar]
- 16.Hutchins P. M., Barkley R. M., Murphy R. C. 2008. Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J. Lipid Res. 49: 804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu C., Dane A., Spijksma G., Wang M., van der Greef J., Luo G., Hankemeier T., Vreeken R. J. 2012. An efficient hydrophilic interaction liquid chromatography separation of 7 phospholipid classes based on a diol column. J. Chromatogr. A. 1220: 26–34. [DOI] [PubMed] [Google Scholar]
- 18.Kim J., Hoppel C. L. 2013. Comprehensive approach to the quantitative analysis of mitochondrial phospholipids by HPLC-MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 912: 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cífková E., Holčapek M., Lísa M., Ovčačiková M., Lyčka A., Lynen F., Sandra P. 2012. Nontargeted quantitation of lipid classes using hydrophilic interaction liquid chromatography-electrospray ionization mass spectrometry with single internal standard and response factor approach. Anal. Chem. 84: 10064–10070. [DOI] [PubMed] [Google Scholar]
- 20.Lísa M., Cífková E., Holčapek M. 2011. Lipidomic profiling of biological tissues using off-line two-dimensional high-performance liquid chromatography-mass spectrometry. J. Chromatogr. A. 1218: 5146–5156. [DOI] [PubMed] [Google Scholar]
- 21.Hellmuth C., Weber M., Koletzko B., Peissner W. 2012. Nonesterified fatty acid determination for functional lipidomics: comprehensive ultrahigh performance liquid chromatography-tandem mass spectrometry quantitation, qualification, and parameter prediction. Anal. Chem. 84: 1483–1490. [DOI] [PubMed] [Google Scholar]
- 22.Chen S., Hoene M., Li J., Li Y., Zhao X., Häring H-U., Schleicher E. D., Weigert C., Xu G., Lehmann R. 2013. Simultaneous extraction of metabolome and lipidome with methyl tert-butyl ether from a single small tissue sample for ultra-high performance liquid chromatography/mass spectrometry. J. Chromatogr. A. 1298: 9–16. [DOI] [PubMed] [Google Scholar]
- 23.Gao X., Zhang Q., Meng D., Isaac G., Zhao R., Fillmore T. L., Chu R. K., Zhou J., Tang K., Hu Z., et al. 2012. A reversed-phase capillary ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) method for comprehensive top-down/bottom-up lipid profiling. Anal. Bioanal. Chem. 402: 2923–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu C., van Dommelen J., van der Heijden R., Spijksma G., Reijmers T. H., Wang M., Slee E., Lu X., Xu G., van der Greef J., et al. 2008. RPLC-ion-trap-FTMS method for lipid profiling of plasma: method validation and application to p53 mutant mouse model. J. Proteome Res. 7: 4982–4991. [DOI] [PubMed] [Google Scholar]
- 25.Bird S. S., Marur V. R., Sniatynski M. J., Greenberg H. K., Kristal B. S. 2011. Serum lipidomics profiling using LC-MS and high-energy collisional dissociation fragmentation: focus on triglyceride detection and characterization. Anal. Chem. 83: 6648–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bird S. S., Marur V. R., Sniatynski M. J., Greenberg H. K., Kristal B. S. 2011. Lipidomics profiling by high-resolution LC-MS and high-energy collisional dissociation fragmentation: focus on characterization of mitochondrial cardiolipins and monolysocardiolipins. Anal. Chem. 83: 940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bird S. S., Marur V. R., Stavrovskaya I. G., Kristal B. S. 2012. Separation of cis-trans phospholipid isomers using reversed phase LC with high resolution MS detection. Anal. Chem. 84: 5509–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bird S. S., Marur V. R., Stavrovskaya I. G., Kristal B. S. 2013. Qualitative characterization of the rat liver mitochondrial lipidome using LC-MS profiling and high energy collisional dissociation (HCD) all ion fragmentation. Metabolomics. 9 (1 Suppl.): 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada T., Uchikata T., Sakamoto S., Yokoi Y., Fukusaki E., Bamba T. 2013. Development of a lipid profiling system using reverse-phase liquid chromatography coupled to high-resolution mass spectrometry with rapid polarity switching and an automated lipid identification software. J. Chromatogr. A. 1292: 211–218. [DOI] [PubMed] [Google Scholar]
- 30.Laaksonen R., Katajamaa M., Päivä H., Sysi-Aho M., Saarinen L., Junni P., Lütjohann D., Smet J., Van Coster R., Seppänen-Laakso T., et al. 2006. A systems biology strategy reveals biological pathways and plasma biomarker candidates for potentially toxic statin-induced changes in muscle. PLoS ONE. 1: e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietiläinen K. H., Sysi-Aho M., Rissanen A., Seppänen-Laakso T., Yki-Järvinen H., Kaprio J., Oresic M. 2007. Acquired obesity is associated with changes in the serum lipidomic profile independent of genetic effects – a monozygotic twin study. PLoS ONE. 2: e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nygren H., Poho P., Seppanen-Laakso T., Lahtinen U., Oreaic M., Hyotylainen T. 2013. Ultrahigh-performance liquid chromatography-mass spectrometry in lipidomics. LC GC Eur. 26: 142. [Google Scholar]
- 33.Castro-Perez J. M., Kamphorst J., DeGroot J., Lafeber F., Goshawk J., Yu K., Shockcor J. P., Vreeken R. J., Hankemeier T. 2010. Comprehensive LC-MS E lipidomic analysis using a shotgun approach and its application to biomarker detection and identification in osteoarthritis patients. J. Proteome Res. 9: 2377–2389. [DOI] [PubMed] [Google Scholar]
- 34.Knittelfelder O. L., Weberhofer B. P., Eichmann T. O., Kohlwein S. D., Rechberger G. N. 2014. A versatile ultra-high performance LC-MS method for lipid profiling. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 951–952: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paglia G., Williams J. P., Menikarachchi L., Thompson J. W., Tyldesley-Worster R., Halldórsson S., Rolfsson O., Moseley A., Grant D., Langridge J., et al. 2014. Ion mobility derived collision cross sections to support metabolomics applications. Anal. Chem. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 37.Konijnenberg A., Butterer A., Sobott F. 2013. Native ion mobility-mass spectrometry and related methods in structural biology. Biochim. Biophys. Acta. 1834: 1239–1256. [DOI] [PubMed] [Google Scholar]
- 38.Pringle S. D., Giles K., Wildgoose J. L., Williams J. P., Slade S. E., Thalassinos K., Bateman R. H., Bowers M. T., Scrivens J. H. 2007. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument. Int. J. Mass Spectrom. 261: 1–12. [Google Scholar]
- 39.Mozaffarian D., Aro A., Willett W. C. 2009. Health effects of trans-fatty acids: experimental and observational evidence. Eur. J. Clin. Nutr. 63 (Suppl. 2): S5–S21. [DOI] [PubMed] [Google Scholar]
- 40.Teegala S. M., Willett W. C., Mozaffarian D. 2009. Consumption and health effects of trans fatty acids: a review. J. AOAC Int. 92: 1250–1257. [PubMed] [Google Scholar]
- 41.Menaa F., Menaa A., Menaa B., Treton J. 2013. Trans-fatty acids, dangerous bonds for health? A background review paper of their use, consumption, health implications and regulation in France. Eur. J. Nutr. 52: 1289–1302. [DOI] [PubMed] [Google Scholar]
- 42.Hsu F-F., Turk J. 2003. Electrospray ionization/tandem quadrupole mass spectrometric studies on phosphatidylcholines: the fragmentation processes. J. Am. Soc. Mass Spectrom. 14: 352–363. [DOI] [PubMed] [Google Scholar]
- 43.Hsu F. F., Bohrer A., Turk J. 1998. Formation of lithiated adducts of glycerophosphocholine lipids facilitates their identification by electrospray ionization tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 9: 516–526. [DOI] [PubMed] [Google Scholar]
- 44.Shah V., Castro-Perez J. M., McLaren D. G., Herath K. B., Previs S. F., Roddy T. P. 2013. Enhanced data-independent analysis of lipids using ion mobility-TOFMSE to unravel quantitative and qualitative information in human plasma. Rapid Commun. Mass Spectrom. 27: 2195–2200. [DOI] [PubMed] [Google Scholar]
- 45.Kliman M., May J. C., McLean J. A. 2011. Lipid analysis and lipidomics by structurally selective ion mobility-mass spectrometry. Biochim. Biophys. Acta. 1811: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.