SUMMARY
Glioblastoma multiforme (GBM) displays cellular hierarchies harboring a subpopulation of stem-like cells (GSCs). Enhancer of Zeste Homolog 2 (EZH2), the lysine methyl transferase of Polycomb repressive complex 2, mediates transcriptional repression of pro-differentiation genes in both normal and neoplastic stem cells. An oncogenic role of EZH2 as a transcriptional silencer is well established; however, additional functions of EZH2 are incompletely understood. Here we show that EZH2 binds to and methylates STAT3, leading to enhanced STAT3 activity by increased tyrosine phosphorylation of STAT3. The EZH2-STAT3 interaction preferentially occurs in GSCs relative to non-stem bulk tumor cells, and it requires a specific phosphorylation of EZH2. Inhibition of EZH2 reverses the silencing of Polycomb target genes and diminishes STAT3 activity, suggesting therapeutic strategies.
INTRODUCTION
Glioblastoma multiforme (GBM) is the most common and the most lethal primary brain cancer (Louis et al., 2007). Current standard-of-care for GBM patients provide only palliation with the median survival of about 15 months (Furnari et al., 2007; Stupp et al., 2005).
Cancer stem/propagating cells (CSCs) are functionally defined by the enriched capacity to propagate tumors in vivo, and have characteristics of normal stem cells such as self-renewal capacity and differentiation potential to establish cellular hierarchy and heterogeneity (Dirks, 2010; Reya et al., 2001). While some cancers may not follow CSC model, numerous studies support that GBMs harbor a subpopulation of highly tumorigenic, stem-like cells (GSCs) (Dirks, 2010; Hemmati et al., 2003; Singh et al., 2004), and that GSCs are responsible for glioma propagation, resistance to conventional therapy, and tumor recurrence (Bao et al., 2006; Chen et al., 2012; Gilbert and Ross, 2009; Zhou et al., 2009). Therefore, it may be crucial to identify the mechanisms involved in GSC maintenance.
Polycomb group (PcG) proteins are important epigenetic regulators of embryonic development and cell fate decision (Margueron and Reinberg, 2011; Richly et al., 2011; Sparmann and van Lohuizen, 2006). They execute transcriptional repression in two multi-protein complexes named Polycomb repressive complexes 1 and 2 (PRC1 and PRC2). Core components of PRC2 include EZH2 (Enhancer of Zeste Homolog 2), Suz12 (Suppressor of Zeste 12), and EED (Embryonic Ectoderm Development) (Sparmann and van Lohuizen, 2006). EZH2 functions as a lysine methyl transferase and EZH2-containing PRC2 catalyzes trimethylation of Histone 3 at lysine 27 (H3K27me3) (Cao et al., 2002). PRC1 in turn recognizes the H3K27me3 mark and maintains gene silencing (Shao et al., 1999; Sparmann and van Lohuizen, 2006).
Many of the PRC2 target genes in embryonic and tissue-specific stem cells are lineage-committed pro-differentiation genes, supporting Polycomb-mediated maintenance of stem cells (Boyer et al., 2006; Lee et al., 2006b; Mikkelsen et al., 2007). Several genome-wide integrative studies have revealed that a significant subset of PRC2 target genes is repressed in various tumors, some of which are further silenced by promoter hypermethylation, implying crucial roles of the Polycomb pathway in cancer initiation and progression (Schlesinger et al., 2007; Vire et al., 2006; Widschwendter et al., 2007). In a wide range of cancers including GBM, EZH2 is highly expressed and its expression positively correlates with tumor malignancy and invasiveness (Crea et al., 2010; Kleer et al., 2003; Varambally et al., 2002). We and others have previously shown that EZH2 is a critical regulator for GSC maintenance and GBM propagation (Abdouh et al., 2009; Lee et al., 2008; Suva et al., 2009).
The reported roles of EZH2 have been attributed to its ability to drive transcriptional repression via a repressive histone mark, especially H3K27 trimethylation (Esteller, 2008; Morey and Helin, 2010; Simon and Kingston, 2009; Simon and Lange, 2008). However, emerging evidence suggests the presence of additional downstream effectors of EZH2 signaling (Cha et al., 2005; He et al., 2012; Lee et al., 2011; Wei et al., 2008; Xu et al., 2012). Consistent with this hypothesis, recent studies reported that EZH2 interacts with various transcription factors including androgen receptor (AR), GATA4, and RORα (He et al., 2012; Lee et al., 2012; Xu et al., 2012). A series of reports showed that histone methyl transferases such as SET7/9 can regulate signaling pathways through direct methylation of p53, NF-κB, and STAT3 (Huang et al., 2006; Lu et al., 2010; Stark et al., 2011; Yang et al., 2010), raising the possibility that EZH2 might have such a property. Based on this background, we investigated the histone methylation-independent role of EZH2 in GSC self-renewal and GBM propagation.
RESULTS
EZH2 interacts with STAT3 in GSCs
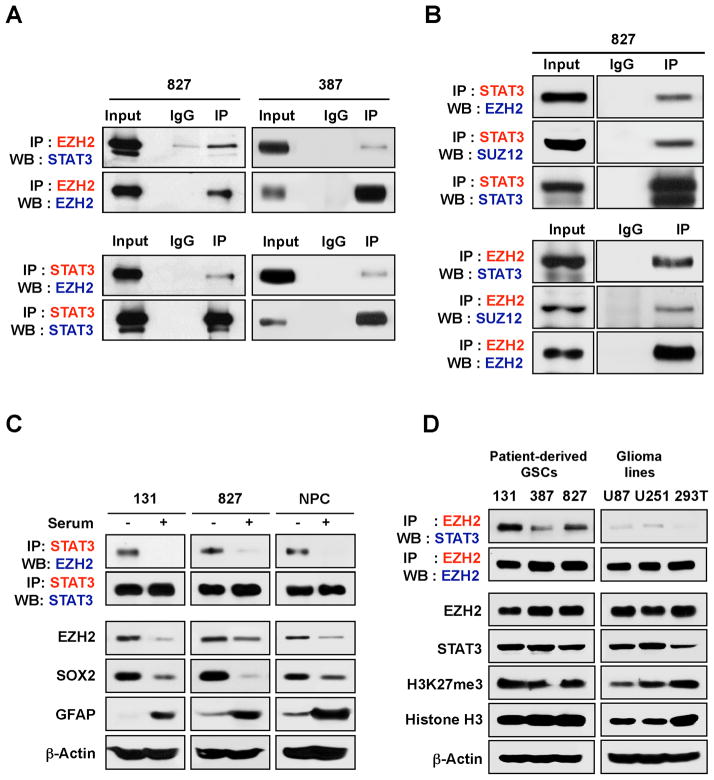
To identify proteins that interact with EZH2, we performed co-immunoprecipitation (IP) experiments using an anti-EZH2 antibody and characterized proteins that co-precipitate with EZH2 by mass spectrometry (data not shown). GBM cells prospectively enriched by the putative GSC enrichment markers CD133 and/or CD15 (Singh et al., 2004; Son et al., 2009) were isolated from surgical specimens of GBM patients or their derivative xenograft tumors, and functionally validated in assays of self-renewal and tumor propagation (Joo et al., 2013; Lee et al., 2008; Pastrana et al., 2011; Pollard et al., 2009). We found that EZH2 co-precipitated with STAT3 in protein lysates isolated from CD133 and/or CD15-enriched GSCs (Figure 1A). Co-IP experiments using an anti-STAT3 antibody in the same lysates revealed that STAT3 co-precipitated with EZH2 as well as SUZ12 and EED, other core components of PRC2, suggesting that STAT3 and PRC2 may interact in GSCs (Figure 1B).
Figure 1. EZH2 protein interacts with STAT3 in GSCs.
A and B) Co-immunoprecipitation (Co-IP) of EZH2 and STAT3 in GSCs (827 and 387). IgG represents a control antibody used for IPs. For IP-immunoblotting data, antibodies used for IP and Western blotting (WB) were labeled as red and blue, respectively. Two hundred μg of lysates were used for each IP reaction and total lysates (20 μg) were used as input controls. (C) Co-IP and immunoblot analysis of EZH2 and STAT3 in GSCs and NPCs vs. differentiated progenies. Differentiation was induced by culturing these cells in serum (10%)-containing media for three days. The EZH2-STAT3 complex was analyzed by co-IP, followed by immunoblot analysis. Protein levels of EZH2, SOX2 (a GSC-specific transcription factor), and GFAP (an astroglial differentiation marker) were examined. β-actin was used as a loading control. (D) Co-IP and immunoblots of the EZH2-STAT3 complexes in various cells. A 293T cell line (derived from human embryonic kidney cells) was used as a reference. See also Figure S1.
As EZH2 is implicated in the maintenance of normal and neoplastic stem cells, we assessed whether the EZH2-STAT3 interaction is affected by the differentiation states of these cells. Addition of serum or the removal of epidermal growth factor (EGF) and fibroblast growth factor 2 (FGF2) in culture media decreased expression of stem cell associated markers such as SOX2 and concurrently increased an astroglial marker expression (e.g. GFAP), consistent with “differentiated” phenotype (Galli et al., 2004; Lee et al., 2006a; Piccirillo et al., 2006; Zheng et al., 2010). We determined the relative abundance of the EZH2-STAT3 complex in two pairs of GSCs and their differentiated progenies by co-IP assays. An association between EZH2 and STAT3 was prominent in GSCs, but lost in the cells cultured in the presence of serum or the absence of growth factors (Figure 1C and Figure S1), providing an initial link between the EZH2-STAT3 interaction and GSC maintenance. As GSCs share many characteristics with normal human neural stem/progenitor cells (NPCs), we assessed whether the EZH2-STAT3 association is present in NPCs derived from fetal brain tissues. Similar to GSCs, EZH2 and STAT3 were co-immunoprecipitated in NPCs but not in differentiated cells (Figure 1C). Collectively, these data demonstrate an interaction between EZH2 and STAT3 in GSCs.
We performed additional co-IP experiments to determine the EZH2-STAT3 interaction in GSC-derived GBM xenografts or other glioma cell lines. U87 and U251 are two most widely used, established glioma cell lines. EZH2 and STAT3 proteins were expressed in these cells at comparable levels. While the EZH2-STAT3 co-precipitation was abundant in GSCs and GBM xenografts, it was barely detectable in lysates isolated from U87 and U251 (Figure 1D).
EZH2 targeting decreases STAT3 activation in GSCs
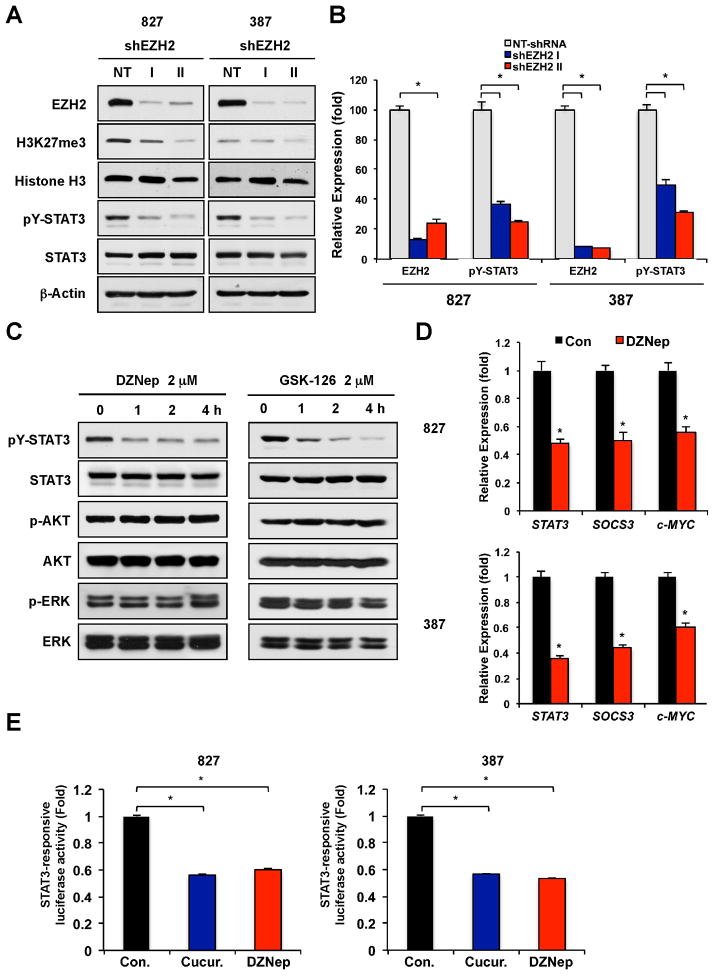
As an association between EZH2 and STAT3 is abundant in GSCs, and STAT3 is a key signaling node in GSCs (Cao et al., 2010; Sherry et al., 2009; Yang et al., 2012), we hypothesized that EZH2 may positively regulate STAT3 signaling in GSCs. To investigate the role of EZH2 in STAT3 activation, we targeted EZH2 expression by lentiviral-mediated short hairpin RNA (shRNA) directed against EZH2 (shEZH2) and measured STAT3 activation in GSCs (Figure 2A). Some GSCs appear to have a constitutively active STAT3 signaling, evidenced by a high basal level of tyrosine phosphorylated STAT3 at 705 residue (pY-STAT3) (Cao et al., 2010). As expected, EZH2 knockdown greatly decreased global levels of H3K27 trimethylation in GSCs. EZH2 knockdown in two different GSCs also reduced the levels of pY-STAT3 to ~30% of those in non-targeting shRNA-transduced GSCs (Figure 2A and 2B). We complemented these findings using a prototype EZH2 inhibitor (3-deazaneplanocin A, DZNep) (Tan et al., 2007) and a highly selective EZH2 inhibitor GSK126 (McCabe et al., 2012). pY-STAT3 in GSCs was rapidly decreased by treatment with either DZNep or GSK126 (Figure 2C). In contrast, the levels of phosphorylated AKT and ERK in GSCs were not changed within 4 hours of drug treatment (Figure 2C).
Figure 2. EZH2 targeting decreases STAT3 activation in GSCs.
(A) Immunoblots of EZH2, H3K27me3, pY-STAT3 (Y705) and total STAT3 in GSCs transduced with shEZH2-I, shEZH2-II, or non-targeting (NT) control shRNA. (B) Quantification of protein intensities in immunoblots including the one shown in (A) was performed by densitometry. (C) Immunoblots of pY-STAT3 and total STAT3 in GSCs treated with EZH2 inhibitors DZNep (2 μM) or GSK-126 (2 μM) for the indicated times (0 hr to 4 hr). Representative blots using 827 cells were shown. (D) Semi-quantitative real time RT-PCR analysis to determine mRNA expression of STAT3 target genes in GSCs (827 and 387) treated with DZNep for 1 day. Expression levels of these genes were normalized and compared to those of the untreated control cells. Data are means ± SD (n = 3). (E) Determination of STAT3 transcription activity by STAT3-responsive luciferase reporter assays. Luciferase activities in GSCs treated with DZNep were compared to those of the untreated control. Cells treated with cucurbitacin (a STAT3 inhibitor) were used as a positive control. Error bar represents SD. *p < 0.01.
As STAT3 functions as a transcription factor, we determined the functional effects of decreased STAT3 activity in GSCs after EZH2 inhibition. We performed real-time RT-PCR analysis to measure mRNA expression of validated STAT3 transcriptional target genes including STAT3, SOCS3 and c-MYC (Figure 2D) in EZH2-inhibited cells (Frank, 2007). Messenger RNA levels of these genes were significantly decreased in GSCs treated with DZNep or EZH2 knockdown cells compared to the control (Figure 2D and data not shown). In addition, we determined STAT3 transcriptional activity by STAT3 responsive promoter reporter assays in which a luciferase reporter was driven by a STAT3 DNA-binding sequence-containing promoter. STAT3 transcriptional activity was substantially lower in cells treated with DZNep or EZH2 knockdown cells than the control (Figure 2E and data not shown). Together, these data supporting that EZH2 may directly regulate STAT3 activity in GSCs.
STAT3 is methylated by EZH2 in GSCs
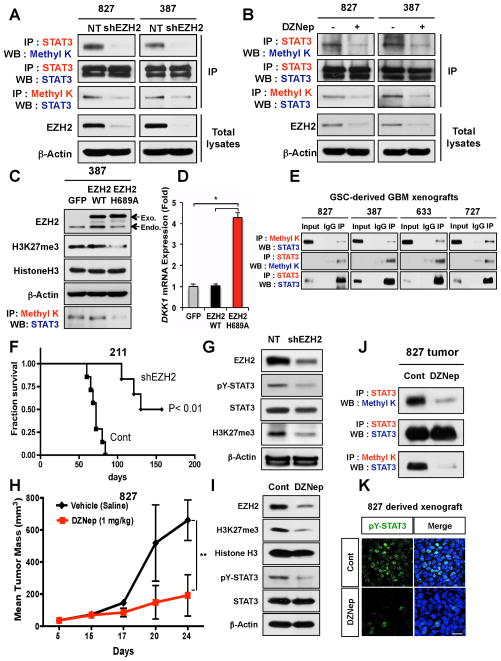
Since EZH2 methylates specific lysine residues of histone proteins and EZH2 interacts with STAT3, we hypothesized that EZH2 methylates STAT3. By immnuoprecipitation with an anti-STAT3 antibody followed by immunoblotting using a pan-methyl lysine antibody, we found that STAT3 is methylated in GSCs (Figure 3A). To interrogate the role of EZH2 in STAT3 methylation, we targeted EZH2 genetically (shRNA-mediated knockdown) or pharmacologically (with DZNep), and then assessed the methylation status of STAT3 protein in GSCs. In both cases, STAT3 methylation was markedly decreased (Figure 3A and 3B). To determine whether a methyl transferase activity of EZH2 is required for STAT3 methylation, we overexpressed a methyl transferase-inactive EZH2 H689A mutant (Kuzmichev et al., 2002) in GSCs via lentiviral transduction and assessed the level of STAT3 methylation (Figure 3C). Ectopic expression of EZH2 H689A mutant decreased H3K27 trimethylation and reversed transcriptional repression of Dickkopf-1 (Dkk1), a Wnt antagonist and Polycomb target gene (Gotze et al., 2010; Hussain et al., 2009), suggesting that this mutant functions as a dominant negative form (Figure 3C and 3D). Compared to the GFP-transduced cells or the wild-type EZH2-transduced cells, EZH2 H689A mutant-transduced cells showed decreased STAT3 methylation (Figure 3C). Collectively, these data support that STAT3 methylation in GSCs is mediated by EZH2.
Figure 3. EZH2 methylates STAT3 protein in GSCs.
(A and B) Co-IP and immunoblots of the methylated STAT3 in GSCs. Either EZH2 knockdown (A) or treatment with DZNep (B) decreased methylated STAT3 in GSCs. Methyl K represents an antibody recognizing the methylated lysine. (C) Co-IP and immunoblots of the methylated STAT3 in GSCs that express GFP, wild-type EZH2, and EZH2 H689A. Arrows indicate protein bands of endogenous EZH2 (endo) and exogenous EZH2 transgenes (exo). (D) Expression levels of DKK1, a PRC2 target gene, in GSCs expressing EZH2 transgenes were determined by real time RT-PCR analysis. Data are means ± SD (n = 3). * p < 0.01. (E) IP and immunoblots of methylated STAT3 in various GSC-derived GBM tumors. (F) Kaplan-Meier survival curves of mice orthotopically implanted with 211 GSCs transduced with either control or EZH2 shRNA expressing lentivirus (n = 7). p < 0.01. (G) Immunoblots of pY-STAT3 and H3K27me3 in xenograft tumors (F). Human tumor cells were harvested three months after intracranial implantation and processed for immunoblot analysis. (H) Measurement of subcutaneous xenograft tumor size after treatment with DZNep. Error bar represents SD (n = 5). ** p <0.01. GSC-derived xenograft tumors were treated with DZNep (1 mg/kg body weight twice a week via intraperitoneal injection) for two weeks, harvested, and processed for immunoblots and co-IP analysis (I and J) and IF staining (K). (K) IF staining of pY-STAT3 (green) on the frozen sections of GBM xenografts treated with vehicle or DZNep. Nuclei were stained with DAPI. Bar represents 10 microns.
EZH2 inhibition decreases STAT3 methylation and activity in GBM xenografts
As EZH2-STAT3 interaction is prominent in GSCs in vitro, we determined STAT3 methylation status in GBM xenografts. Protein lysates of GBM xenograft tumors derived from four different GSCs were interrogated by reciprocal co-IP experiments using anti-STAT3 and methylated lysine-specific antibodies. Methylated STAT3 was present in lysates from all tumors tested (Figure 3E). Next, we determined whether EZH2 inhibition could affect STAT3 methylation and STAT3 activity in GBM in vivo by using two complementary approaches. First, we implanted GSCs expressing either EZH2 shRNA or control shRNA into the brains of immunocompromised mice. Second, we generated GSC-derived xenograft tumors and treated the animals with DZNep after tumors were established. EZH2 knockdown (Figure 3F and 3G) or DZNep treatment (Figure 3H and 3I) decreased the growth of xenograft tumors. Tumors obtained from both experiments were dissected and processed for Western blot analysis and co-IP experiments. Compared to the controls, tumors treated with DZNep or tumors expressing EZH2 shRNA showed low levels of STAT3 methylation and pY-STAT3 (Figure 3F to 3J). Furthermore, immunohistochemcial analyses on tumor sections showed the decreased number of pY-STAT3 positive cells in tumors treated with DZNep compared to the control (Figure 3K). Taken together, these data demonstrate that targeting EZH2 effectively inhibits H3K27 methylation, STAT3 methylation and STAT3 activation in GBM xenografts.
STAT3 methylation positively regulates STAT3 activity in GSCs
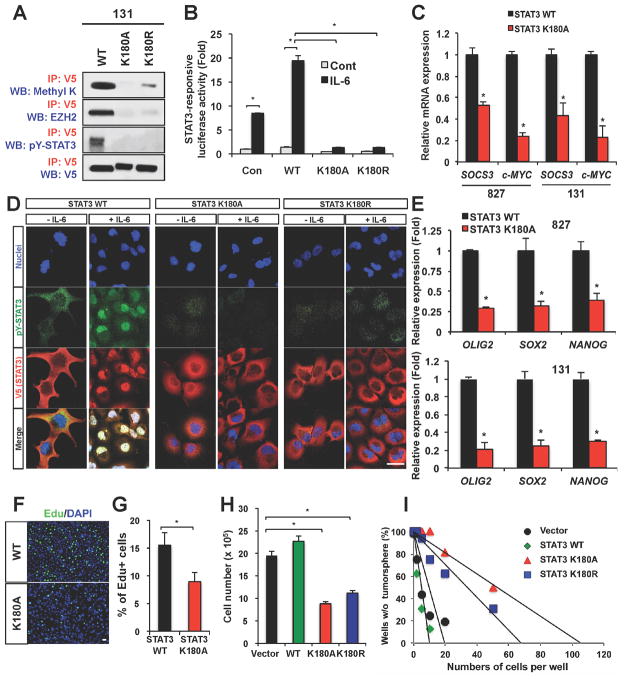
To analyze the methylation status of STAT3 protein, we overexpressed Flag-tagged STAT3 (Flag-STAT3) in GSCs and generated xenograft tumors using these cells. The tagged STAT3 was phosphorylated normally in response to EGF and Interleukin-6 (IL-6), potent inducers for pY-STAT3 (Zhong et al., 1994). Flag-STAT3 protein was purified from GSC-derived xenograft tumors and analyzed by mass spectrometry (MS). Mass shift of +42 was observed for a STAT3 peptide, consistent with trimethyl modifications (Figure S2A). The previously reported Lysine 140 methylation of STAT3 was not detected in these tumors by MS analysis (Yang et al., 2010) (data not shown). As the methylation was in a peptide that includes residues 178 to 197, and tandem MS analysis localized the site to Lysine 180 (K180) (Figure S2A), we generated lentiviruses expressing STAT3 mutants in which a K180 residue of STAT3 protein was replaced by Alanine (A) or Arginine (R). We overexpressed these STAT3 mutants in GSCs and performed co-IP experiments to assess the methylation status of these proteins and the interaction with EZH2 (Figure 4). While exogenous wild-type STAT3 was methylated, both K180A and K180R mutants showed little or no methylation (Figure 4A). Similarly, STAT3 K180 mutants were unable to immunoprecipitate with EZH2 in contrast to the wild-type STAT3 (Figure 4A). These data support K180 of STAT3 as a key residue required for the EZH2-STAT3 interaction and EZH2-mediated STAT3 methylation.
Figure 4. Effects of STAT3 methylation on STAT3 activity and GSC self-renewal.
(A) Co-IP analyses of methylated STAT3, pY-STAT3 and the EZH2-STAT3 complex in 131 GSCs expressing STAT3 K180 mutants. V5 is a protein tag fused with STAT3. Methyl K denotes an antibody recognizing either di- or tri-methylated lysine. (B) STAT3 transcriptional activity in response to IL-6 in 131 GSCs expressing the empty vector control, wild-type STAT3, or the STAT3 K180 mutants. * p < 0.01. (C) Real time RT-PCR analysis to determine mRNA levels of SOCS3 and c-MYC in GSCs (827 and 131) expressing either wild-type or STAT3 K180A mutant. Messenger RNA levels of SOCS3 and c-MYC in STAT3 K180 expressing cells were compared to those in wild-type STAT3 expressing cells (set to 1). Data are means ± SD (n = 3). * p < 0.01. (D) IF staining of pY-STAT3 (green) on GSCs expressing the wild-type STAT3 and STAT3 K180 mutants in response to IL-6 (50ng/ml). V5 (red) was used to stain exogenous STAT3 and DAPI (blue) was used to stain nuclei. (E) Real time RT-PCR analysis to determine mRNA expression of stem cell associated transcription factors (OLIG2, SOX2, and NANOG) in GSCs (827 and 131) expressing STAT3 mutants. Data are means ± SD (n = 3). * p < 0.01. (F to I) Effects of STAT3 K180 mutant overexpression on proliferation and clonogenic growth of GSCs. 5′-ethynyl-2′-deoxyuridine (EdU)-positive cells (stained green) were counted in three random fields and plotted (in F and G). *p < 0.01. Cell counts of STAT3 mutant-expressing GSCs after 6 days of culture (H) and limiting dilution assay results (I) were shown. Error bars represent SD. Bars (in D and F) represent 10 microns. See also Figure S2.
As pY-STAT3 is an important indicator of STAT3 activity, we performed IP experiments to determine the phosphorylation status of exogenous STAT3 proteins. While wild-type STAT3 was phosphorylated at tyrosine 705, both K180A and K180R mutants showed little or no phosphorylation (Figure 4A). To further investigate the effects of K180 mutation on STAT3 function, we overexpressed wild-type STAT3 and STAT3 K180 mutants in PC3, a STAT3-null prostate cancer cell line. After treatment with IL-6, activating tyrosine phosphorylation of STAT3 was evident in the wild-type STAT3-PC3 cells but not in STAT3 K180 mutant-PC3 cells (Figure S2 B and C). Consistent with this, overexpression of K180 STAT3 mutant in PC3 failed to increase STAT3 transcriptional activity responding to IL6. To interrogate the role of these STAT3 mutants in GSCs, we performed similar experiments in GSCs in which endogenous STAT3 signaling is active. Luciferase reporter assays to monitor STAT3 transcriptional activation revealed that STAT3 K180 mutant-expressing cells were impaired in the ability to activate STAT3, in contrast to the wild-type STAT3-expressing GSCs (Figure 4B). Unlike wild-type STAT3, overexpression of STAT3 K180A in GSCs did not increase transcription of STAT3 target genes, such as SOCS3 and c-MYC (Figure 4C). As nuclear accumulation of pY-STAT3 is another indicator of STAT3 activation, we assessed the levels of nuclear pY-STAT3 in STAT3 mutant-expressing GSCs. pY-STAT3 was highly accumulated after IL6 challenge in empty vector control GSCs or the wild-type STAT3-expressing GSCs but not in STAT3 K180 mutant-expressing cells (Figure 4D and data not shown). In GSCs, STAT3 signaling promotes activation of stem cell associated transcriptional factors including Olig2, Sox2, and Nanog (Guryanova et al., 2011). Overexpression of wild-type STAT3 but not STAT3 K180A mutant induced transcription of these genes (Figure 4E). We then assessed the effect of STAT3 mutants on the growth of GSCs. Compared to the wild-type STAT3-expressing cells, STAT3 K180 mutant-expressing cells were less proliferative (Figure 4F to 4H). Clonogenic growth as a form of sphere is an in vitro indicator of GSC self-renewal. To determine the role of STAT3 K180 mutants on clonogenic growth of GSCs, we performed neurosphere formation limiting dilution assays. Tumor cells were plated into 96-well plates with various seeding densities and allowed to grow. Compared to the empty vector control and the wild-type STAT3, ectopic expression of STAT3 K180 mutants reduced the efficiency of sphere formation (Figure 4I). Together, these data are consistent with the possibility that EZH2 methylates STAT3 at K180 and this methylation enhances STAT3 activation and promotes clonogenic growth of GSCs.
AKT signaling is essential for the EZH2-STAT3 interaction in GSCs
As our data indicate that the EZH2-STAT3 interaction leads to STAT3 activation, we investigated a potential upstream effector(s) to facilitate this interaction. An initial clue came from a previous study in which the authors reported that EZH2 is phosphorylated at serine residue 21 (pS21 EZH2) by AKT (Cha et al., 2005). Cha et al. showed that phosphorylation of EZH2 at S21 significantly decreased H3K27 trimethylation leading to the derepression of PRC2 target genes (Cha et al., 2005). However, breast cancer cells overexpressing EZH2 S21D (phosphorylation mimetic mutant of pS21 EZH2) showed increased proliferation and invasiveness. H3K27 trimethylation and PRC2-mediated transcriptional repression is generally associated with tumor malignancy, thus an oncogenic role of EZH2 phosphorylation appears to be counter-intuitive. A potential explanation for this discrepancy is that S21 phosphorylation of EZH2 may exert pro-tumorigenic functions in H3K27 trimethylation-independent manner, as suggested in a recent report (Xu et al., 2012).
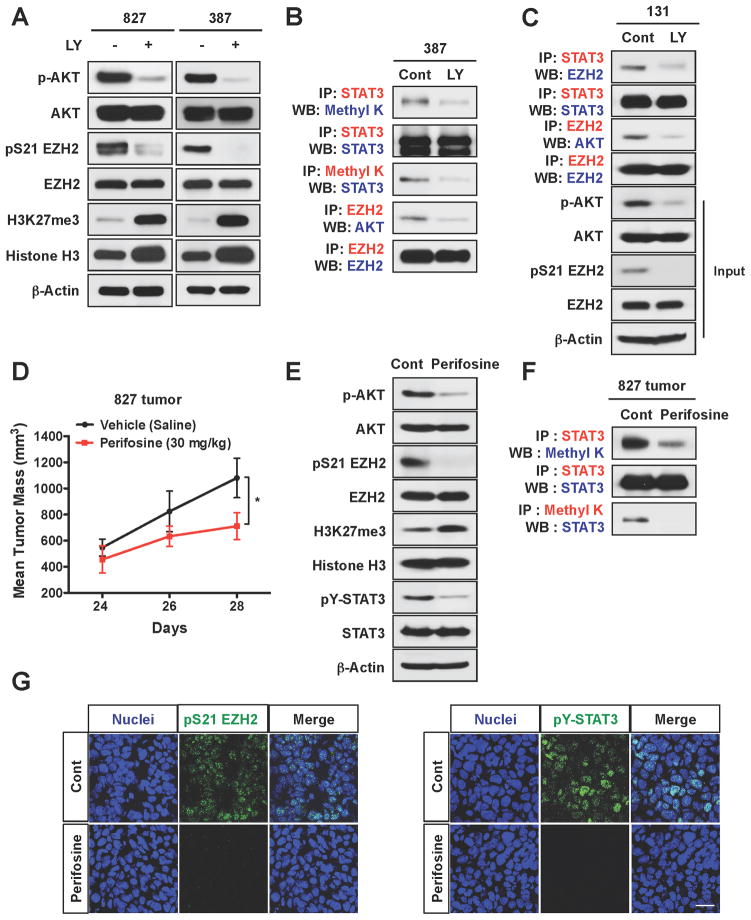
As EZH2 interacts with and methylates STAT3, we hypothesized that STAT3 is one such molecule to exert pro-tumorigenic functions in GSCs. PI3K/AKT signaling is highly active in about 90% of GBM specimens (Fan et al., 2006; Fan and Weiss, 2010; TCGA, 2008; Verhaak et al., 2010) and all of GSCs we tested have a high level of activating phosphorylation of AKT (Figure 2C and data not shown). In agreement with the previous reports (Cha et al., 2005; Costa et al., 2010), treatment with LY294002 (a PI3K-AKT inhibitor) or perifosine (an AKT inhibitor) blocked S21 phosphorylation of EZH2 but increased global levels of H3K27 trimethylation in GSCs (Figure 5A). In GSCs treated with these inhibitors, levels of EZH2-STAT3 complexes and methylated STAT3 were markedly reduced compared to controls (Figure 5B and 5C). Overexpression of a dominant-negative AKT in GSCs showed the similar results (data not shown). Together, these data suggest that AKT signaling is required for the EZH2-STAT3 interaction and STAT3 methylation in GSCs.
Figure 5. AKT signaling is essential for the EZH2-STAT3 interaction in GSCs and AKT inhibition decreases STAT3 activity in GBM xenografts.
(A) Immunoblot analysis of AKT, pS473 AKT, EZH2, pS21 EZH2, and trimethylated H3K27 in GSCs treated with a PI3K/AKT pathway inhibitor LY294002. (B and C) Co-IP analysis of methylated STAT3 and the EZH2-STAT3 complex in GSCs treated with LY294002 for one day. (D) Tumor growth of 827 GSC-derived xenografts treated with an AKT inhibitor perifosine (30 mg/kg body weight). Twenty-three days after tumor implantation in mice, perifosine was administered by intraperitoneal injection (daily for 5 days). Error bars represent SD. Five mice per group, * p < 0.01. (E) Immunoblot analysis of pY-STAT3, EZH2, AKT, and trimethylated H3K27 in lysates from xenograft tumors in (D). (F) Co-IP analysis of methylated STAT3 in GBM xenograft tumors treated with perifosine. (G) IF staining of pS21 EZH2 and pY-STAT3 on the frozen sections of GBM xenografts treated with vehicle or perifosine. Nuclei were stained with DAPI. Bar represents 10 microns.
AKT inhibition decreases methylation and activation of STAT3 in GBM xenografts
To address the relevance of the above findings in vivo, we established GSC-derived xenograft tumors and evaluated the effects of AKT inhibition on STAT3 activity and tumor growth (Figure 5). Animals treated with perifosine for 5 days displayed reduced tumor size relative to those treated with vehicle control (Figure 5D). One day after the final injection of perifosine, we dissected tumors for immunoblot analysis and co-IP experiments. Compared to the control, tumors treated with perifosine showed low levels of phosphorylated AKT and EZH2 phosphorylation (Figure 5E). Consistent with in vitro results, AKT inhibition in vivo decreased STAT3 methylation and pY-STAT3, but increased global levels of H3K27 trimethylation (Figure 5E and 5F). Immunofluorescence analyses on tumor sections further confirmed that perifosine treatment decreased pS21 EZH2 and nuclear PY-STAT3 (Figure 5G). Together, these data suggest that AKT regulates STAT3 signaling in part via EZH2 phosphorylation and STAT3 methylation.
GSCs have high level of EZH2 phosphorylation at Serine 21
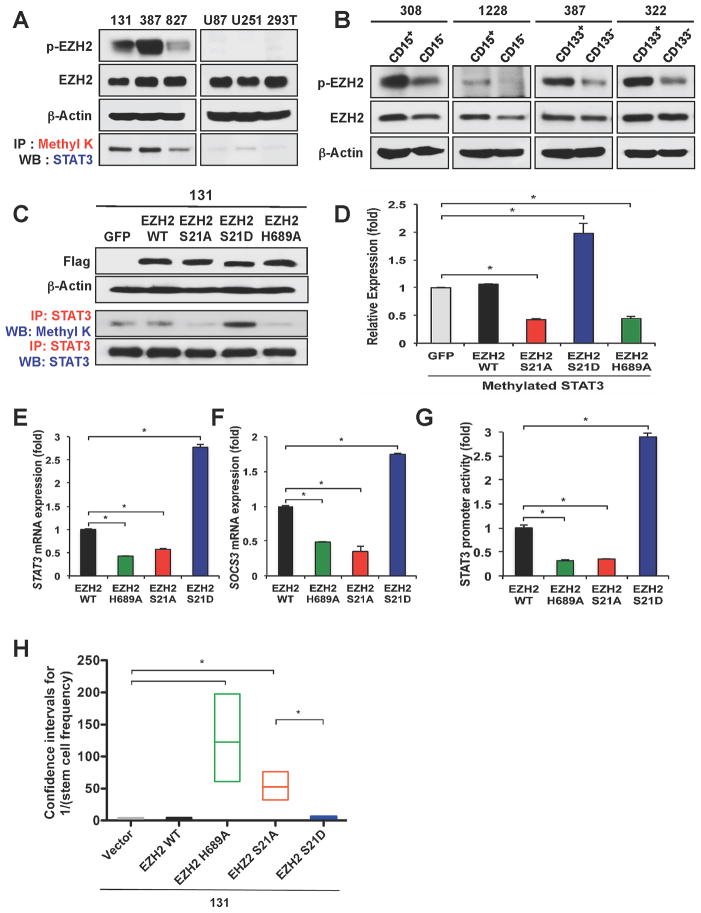
As phosphorylation of EZH2 at S21 appears to be an important signal node between AKT signaling and STAT3 methylation in GSCs, we determined the levels of pS21 EZH2 and STAT3 methylation in various GBM cells. Immunoblot analysis showed that pS21 EZH2 is highly expressed in GSCs but not in U87 and U251 glioma cell lines (Figure 6A). Similarly, STAT3 methylation was abundant in GSCs but much lower in glioma cell lines (Figure 6A). This trend is entirely consistent with the level of the EZH2-STAT3 complex in these cells (Figure 1D). To further examine preferential expression of pS21 EZH2 in GSCs, we used four additional GBM cells with prospective sorting for CD133 and/or CD15. CD133 and/or CD15 positive GSCs have higher level of pS21 EZH2 compared to CD133 and/or CD15 negative cells (Figure 6B). Together, these data show that pS21 EZH2 is preferentially expressed in GSCs and suggest that pS21 EZH2 is required for the EZH2-STAT3 interaction and STAT3 methylation in GSCs.
Figure 6. Phosphorylation of EZH2 at Serine 21 is critical for STAT3 methylation and STAT3 activity in GSCs.
(A) Co-IP and Immunoblot analysis of pS21 EZH2 and methylated STAT3. (B) Immunoblot analysis of pS21 EZH2 in GSC-enriched or GSC-depleted GBM cells (CD133+ or − and CD15+ or −). (C) Co-IP and immunoblot analyses of methylated STAT3 in GSCs transduced with various EZH2 mutants. EZH2 transgenes were detected by Flag tag. (D) Quantification of methylated STAT3 shown in (C). Protein amounts were estimated by densitometry of immunoblots. Error bars represent SD. * p < 0.01. (E and F) Real time RT-PCR analysis to determine mRNA expression of STAT3 and SOCS3 in 131 GSCs expressing various EZH2 mutants. Data are means ± SD (n = 3). *p < 0.01. (G) Determination of STAT3 transcription activity by reporter promoter assays. Luciferase activity in GSCs expressing various EZH2 constructs was compared to that of the GFP-expressing control cells. Error bars represent SD. *p < 0.01. (H) Effects of ectopic expression of various EZH2 constructs on GSC tumorsphere formation. GSC cells (131) transduced with lentivirus expressing various EZH2 transgenes were cultured for limiting dilution assays. Frequency of stem-like clonogenic cells was determined by a web-based tool “ELDA” (extreme limiting dilution analysis, available on http://bioinf.wehi.edu.au/software/elda/) and represented as a box plot. Boxes indicate the upper and lower confidence intervals, and the lines denote the estimated values. *p < 0.01. See also Figure S3.
EZH2 phosphorylation at Serine 21 enhances STAT3 methylation and activation
To interrogate the role of pS21 EZH2 in regulating STAT3, we overexpressed a phosphorylation-defective or phosphorylation-mimetic EZH2 mutant in GSCs and examined the effects on STAT3 methylation and STAT3 activity (Figure 6C to 6G). Expression of EZH2 S21A mutant markedly decreased STAT3 methylation similar to a catalytic activity-dead EZH2 H689A mutant. In sharp contrast, expression of EZH2 S21D mutant significantly increased STAT3 methylation (Figure 6C and 6D). These data prompted us to test whether low level of STAT3 methylation in U87 glioma cells is due to the low expression of pS21 EZH2. Exogenous expression of EZH2 S21D mutant significantly increased STAT3 methylation and STAT3 activity in U87 cells and NPCs, similar to GSCs (Figure S3 and data not shown). Furthermore, STAT3 target gene expression (e.g. STAT3 and SOCS3) and luciferase-based STAT3 transcriptional activity were significantly high in EZH2 S21D mutant-expressing cells but low in EZH2 S21A mutant-expressing cells compared to the control (Figure 6E to 6G). Together, these data support the notion that STAT3 methylation and activation are mediated by pS21 EZH2.
To interrogate the role of pS21 EZH2 in GSC growth, we overexpressed various EZH2 constructs in GSCs via lentiviral transduction and determined the effects on clonogenic growth of GSCs (Figure 6H). Limiting dilution assays were performed and the frequencies of sphere-forming clonogenic cells were estimated. The frequencies of sphere-forming clonogenic cells were about 1 out of 5 cells in the control or wild-type EZH2 expressing cells. In contrast, most of EZH2 H689A expressing cells failed to generate colonies. Ectopic expression of EZH2 S21A also significantly reduced the number of clonogenic cells to less than 10% of the controls (Figure 6H).
STAT3 signaling is a crucial downstream effector of EZH2 signaling in GBM
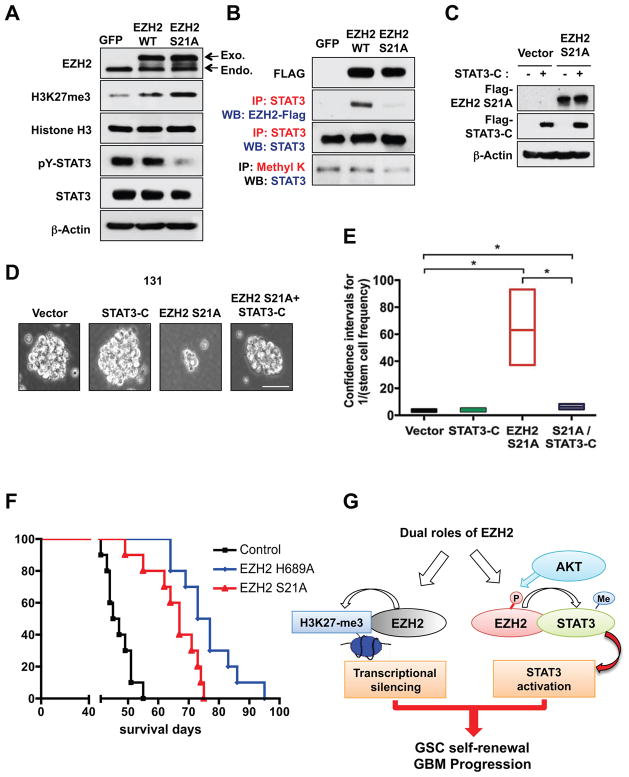
To interrogate the role of pS21 EZH2, we further analyzed the effects of EZH2 S21A overexpression in GSCs. Ectopic expression of a catalytic activity-dead EZH2 H689A mutant in GSCs decreased methylation of both H3K27 and STAT3 (Figure 3C). In contrast, expression of EZH2 S21A mutant significantly decreased STAT3 methylation but increased global levels of H3K27 trimethylation in GSCs (Figure 7A). Overexpression of EZH2 S21A mutant in GSCs did not change the level of Dkk1 mRNA expression, suggesting that PRC2-mediated silencing is unaffected by EZH2 S21A (data not shown). However, EZH2 S21A mutant was unable to immunoprecipitate with STAT3 in contrast to the wild-type EZH2 (Figure 7B) and PY-STAT3 was significantly low in EZH2 S21A expressing cells compared to the controls (Figure 7A). As EZH2 S21A mutant-expressing cells show the decreased STAT3 activity, we then determined whether forced activation of STAT3 could rescue the reduced clonogenic growth in these cells (Figure 7C to 7E). We overexpressed a constitutively active STAT3 mutant (STAT3C) in EZH2 S21A-expressing GSCs and determined their clonogenic growth by limiting dilution assays. In the control and STAT3C-expressing cells, the numbers of clonogenic cells are similar. Expression of STAT3C in EZH2 S21A mutant cells significantly increased the number of clonogenic cells (Figure 7E), suggesting that STAT3 signaling is a crucial downstream effector of EZH2 signaling in GBM.
Figure 7. EZH2 contributes to GBM tumor growth by both H3K27 trimethylation-dependent and STAT3 dependent pathways.
(A and B) Immunoblot and Co-IP analysis of methylated STAT3 and H3K27me3 in GSCs that express GFP, wild-type EZH2, or EZH2 S21A. (C) Immunoblots of exogenous EZH2 S21A and STAT3-C proteins in 131 GSCs. (D) Representative images of 131 GSCs expressing EZH2 S21A and STAT3-C protein. Bar represent 50 microns. (E) Effects of ectopic expression of EZH2 S21A and STAT3-C protein on GSC tumorsphere formation. From limiting dilution assays, frequencies of stem-like clonogenic cells were calculated using ELDA and presented as a box plot. *p < 0.01. (F) Kaplan-Meier survival curves of mice (n = 10 for each group) intracranially implanted with 131 GSCs expressing control, EZH2 689A or EZH2 S21A proteins (control vs. EZH2 H689A, p <0.0001; control vs. EZH2 S21A, p <0.0001; EZH2 H689A vs. EZH2 S21A, p = 0.015 by log ranked analysis). (G) A schematic model that illustrates two functional arms of EZH2. P and Me represent phosphorylation and methylation, respectively. See also Figure S4.
To interrogate the role of EZH2 in tumor growth in vivo, we overexpressed EZH2 H689A or EZH2 S21A proteins in GSCs and implanted these cells into the brains of SCID mice (50,000 cells per mouse, n = 10 per each group). While animals bearing control cells died within 2 months (median survival: 47 days), animals injected with EZH2 H689A expressing cells survived significantly longer (median survival: 75 days; p <0.001). Median survival of the mice bearing EZH2 S21A expressing cells was 67 days (p <0.001), which is significantly longer than that of the control (Figure 7F). Kaplan-Meier curves showed the increase in survival of mice bearing tumor expressing EZH2 S21A- and EZH2 H689A-expressing tumor (p < 0.001 compared to the control). These in vivo data demonstrate that the selective blocking of a STAT3-related arm of EZH2 function can increase the survival of tumor-bearing mice. Our findings, therefore, support that EZH2 contributes to GBM tumor growth by both H3K27 trimethylation-dependent and STAT3 dependent pathways.
Our data obtained in the models of various patient-derived GSCs and GBM xenografts in vitro and in vivo suggest that EZH2 phosphorylation has pro-tumorigenic roles in part via STAT3 activation. To assess the in vivo relevance of the EZH2-STAT3 axis, we determined the expression of pS21 EZH2 and pY-STAT3 in protein lysates from the patient-derived GBM specimens (Figure S4). The correlation between the expression of EZH2 and pS21 EZH2 is not always linear; however, there appears to be a potentially positive correlation between pS21 EZH2 and pY-STAT3 (Figure S4). To determine whether pS21 EZH2 is associated with GBM patient survival, we performed immunohistochemical analysis using the tissue microarray (TMA) sections containing 87 human GBM patient specimens (Figure S4). Expression of pS21 EZH2 displayed inter- and intratumoral heterogeneity. High frequency of cells positive for pS21 EZH2 in GBM specimens was positively correlated with the shorter overall survival of the patients (p < 0.001 by log ranked analysis).
Collectively, these data support a model in which EZH2 not only mediates transcriptional silencing, but also contributes to the activation of STAT3 signaling in GBM (Figure 7G). We propose that the molecular mechanism through which EZH2 activates STAT3 involves a specific EZH2 phosphorylation and STAT3 methylation.
DISCUSSION
Altered profile of histone modification is a major epigenetic mechanism, deregulation of which leads to tumor initiation and propagation (Margueron and Reinberg, 2011; Schlesinger et al., 2007; Widschwendter et al., 2007). Polycomb-mediated transcriptional silencing plays a critical role in the maintenance of the stem cell status in both normal and malignant stem cells. In this study, we demonstrate that EZH2 methylates STAT3 leading to the enhanced STAT3 activity.
Our data indicate that EZH2 not only mediates transcriptional silencing, but also contributes to the activation of STAT3 signaling in stem-like GBM cells. As supporting evidence, we found that the EZH2-STAT3 interaction and EZH2 S21 phosphorylation preferentially occur in GSCs relative to non-stem tumor cells. Overexpression of EZH2 S21D induced STAT3 methylation and enhanced STAT3 activity, suggesting that EZH2 S21 phosphorylation is a molecular switch that facilitates STAT3 methylation. We do not know the underlying molecular mechanism by which the EZH2 S21 phosphorylation is more prominent in stem-like cells. One possibility is that a cofactor of PRC2 component preferentially expressed in stem cell population (Simon and Kingston, 2009) might facilitate the AKT-EZH2 interaction and subsequent interaction with STAT3. It is also possible that EZH2 S21 phosphorylation is regulated by differential AKT or AKT-related downstream effects which may not be directly associated with stem-like characteristics. Further studies to decipher underlying molecular mechanisms are warranted as a recent study reported EZH2 S21 phosphorylation as a critical modification for EZH2 to function as a transcriptional co-activator with androgen receptor (Xu et al., 2012).
Our data support that EZH2-mediated STAT3 methylation is a critical modification that leads to STAT3 activation, although the precise molecular events that link STAT3 methylation to STAT3 activation are unknown. STAT3 activation involves a series of sequential events including tyrosine phosphorylation, dimerization, nuclear import, DNA binding, serine phosphorylation, nuclear retention, and binding to protein partners (Akira et al., 1994; Yang and Stark, 2008; Zhong et al., 1994). It is possible that K180 methylation of STAT3 enhances its tyrosine phosphorylation by protecting from dephosphorylation, as supported by K180 STAT3 mutant data. It has been proposed that K140 methylation of STAT3 destabilizes STAT3 tyrosine phosphorylation, thereby negatively affecting STAT3-dependent transcription (Yang et al., 2010). In contrast to K180 STAT3 methylation by EZH2, K140 methylation can be viewed as a negative event for STAT3 signaling cascade. By analogy with the cases in P53 and NFκB, specific lysine site and the degree of methylation will likely determine STAT3 activity. Although K140 methylation of STAT3 was not detected in our studies, these findings may suggest a critical role of methylation in STAT3 signaling cascade in various cancers. In addition to K180 methylation, we also found K709 dimethylation of STAT3 by MS analysis (data not shown), suggesting that there might be additional lysine residues of STAT3 that could be methylated by EZH2. Further studies are required to precisely define when lysine methylation of STAT3 occurs during the STAT3 signaling cascade and interrogate the role of STAT3 methylation in STAT3 activation.
Despite these caveats, there are important biological and clinical implications derived from our findings. First, the EZH2 and STAT3 signaling pathways are critical therapeutic targets for GBMs, and our findings suggest that EZH2 targeting may effectively inhibit oncogenic activities of both pathways. Significant research efforts have been aimed at developing therapeutic agents targeted for EZH2, as highly specific EZH2 inhibitors have been reported recently (Knutson et al., 2012; McCabe et al., 2012; Qi et al., 2012). Our findings may lead to accelerated development of Polycomb/EZH2 targeting agents and provide a potential biomarker for evaluating therapeutic efficacy.
Second, STAT3 signaling is hyperactive in various cancers including GBMs and an important therapeutic target, however, the specific inactivation of STAT3 has been challenging (Guryanova et al., 2011; Yue and Turkson, 2009). Our finding adds EZH2 to a growing list of upstream pathways leading to the STAT3 activation in GSCs, including IL-6, Notch, PI3K, LIF, BMX, and various RTKs (EGFR, PDGF, and MET) (Bromberg et al., 1999; Fan et al., 2010; Guryanova et al., 2011). It will be highly desirable if EZH2 targeting achieves the effective inhibition of both transcriptional silencing and STAT3 activity in a clinical setting.
A recent study reported that STAT3 signaling is a master regulator of mesenchymal transformation of gliomas, and that STAT3 downstream genes are highly expressed in the mesenchymal GBM subtype (Carro et al., 2010; Verhaak et al., 2010). Further studies are aimed to identify the subset of tumors in which the EZH2-STAT3 axis is particularly active, and to demonstrate the mechanistic correlations between these pathways. It is enticing to propose that EZH2 targeting agents may be particularly useful for patients harboring high level of phosphorylated EZH2.
Inhibition of AKT signaling decreases STAT3 activity via EZH2 phosphorylation, implicating that PI3K/AKT signaling is an upstream mediator of the EZH2-STAT3 interaction in GSCs. Activation of the PI3K/AKT pathway in glioma is associated with adverse clinical outcome and various PI3K/AKT inhibitors are being tested in clinical trials (Fan et al., 2006; Fan and Weiss, 2010; TCGA, 2008; Verhaak et al., 2010). Our data indicate that EZH2 S21 phosphorylation is required for the EZH2-STAT3 interaction and the enhanced STAT3 activity, and AKT inhibition in vivo effectively abolished pS21 EZH2. Therefore, it would be interesting to examine pS21 EZH2 expression as a biomarker to predict therapeutic responses to PI3K/AKT targeted therapies.
In conclusion, we demonstrate that EZH2 binds to and methylates STAT3, thereby enhancing STAT3 activity in GSCs. EZH2 phosphorylation by AKT is critical for the EZH2-STAT3 interaction. By utilizing EZH2 constructs that allow the dissection of two distinct functions of EZH2, we showed the role of STAT3-dependent EZH2 function in GBMs. We propose that two arms of EZH2 function, namely a histone methylation- dependent transcriptional silencing and STAT3 activation by direct protein-protein interaction, can cooperatively contribute to GSC self-renewal and GBM malignancy. These data suggest that functional disruption of EZH2 may attenuate multiple key signals involved in GSC self-renewal and survival, and further support that EZH2 is a promising therapeutic target for GBM.
EXPERIMENTAL PROCEDURES
Human GBM specimens and derivative GSCs
Following informed consent, glioblastoma specimens were obtained from patients undergoing surgery at the Samsung Medical Center in accordance with the Institutional Review Boards. Within hours after surgical removal, tumor specimens were enzymatically dissociated into single cells, following the procedures previously reported (Lee et al., 2006a; Son et al., 2009). For short-term in vitro expansion of GSCs, tumor cells were cultured in Neurobasal media with N2 and B27 supplements (0.5 x each; Invitrogen), human recombinant bFGF and EGF (25 ng/ml each; R&D systems). Human neural stem/progenitor cells (NPCs) were purchased from Lonza and cultured similar to GSCs. For differentiation of GSCs and NPCs, cells were cultured in the absence of growth factors or in the presence of 10 % fetal bovine serum (Cellgro).
Intracranial tumor cell injection
All mice experiments were performed according to the guidelines of the Animal Use and Care Committees at Samsung Medical Center. GBM cells were dissociated, resuspended in 2 μl of HBSS, and injected intracranially into the striatum of adult nude mice by using a stereotactic device (Kopf instruments) (coordinates: 2 mm anterior, 2 mm lateral, 2.5 mm depth from the dura). Mice with neurological signs were killed for the analysis of tumor histology and immunohistochemistry. Brains were perfused with 4% paraformaldehyde by cardiac perfusion and further fixed at 4°C overnight.
Chemicals and Antibodies
3-Deazaneplanocin A (DZNep) was kindly provided from Dr. Victor Marquez (National Cancer Institute). GSK126 was purchased from Xcessbio. Cucurbitacin and perifosine were purchased from Tocris and Selleckchem, respectively. The following antibodies were used as primary antibodies: EZH2 (BD Transduction Laboratories; 1:4000); phospho-EZH2 (Bethyl Laboratories; 1:500); Methylated Lysine (Enzo Life Science and Abcam; 1:500); STAT3, phospho-STAT3 (Y705), Histone H3, AKT, and phospho-AKT (S473) (Cell Signaling; 1:2000); SOX2 and OLIG2 (R &D Systems; 1:2000 for WB and 1:400 for IF); Tri-Methylated Histone H3K27 and SUZ12 (Millipore; 1:1000); GFAP (Dako; 1:1000); β-actin (Sigma-Aldrich; 1:5000).
Immunoprecipitation
Cells were collected and lysed in lysis buffer supplemented with protease inhibitors, incubated on ice for 15 min and cleared by centrifugation at 13,200 rpm at 4°C for 15 min. Total protein lysate (500 μg) was subjected to immunoprecipitation with the Agarose-immobilized Antibody (1 μg of anti-STAT3, EZH2, methylated Lysine antibody or isotype control antibodies) for overnight at 4 °C.
Supplementary Material
SIGNIFICANCE.
Elucidation of the regulatory pathways operating in stem-like cancer cells will be critical to understand the pathogenesis of primary human tumors and may help lead to better therapies. We demonstrate that EZH2 not only mediates transcriptional silencing but also contributes to the activation of STAT3, a key GSC signaling node. Phosphorylation of EZH2 at serine 21 (pS21 EZH2) by AKT signaling facilitates STAT3 methylation by EZH2 and enhances STAT3 activity. Selective blocking of histone methylation- independent EZH2-STAT3 signaling decreases tumor growth in an orthotopic animal model. Together, these data suggest that the AKT-EZH2-STAT3 axis is a positive regulator of GSC self-renewal and tumor malignancy, and a promising therapeutic target for GBM.
HIGHLIGHTS.
EZH2 activates STAT3 signaling via lysine methylation of STAT3.
EZH2 and STAT3 preferentially interact in stem-like tumor cells.
AKT serves as an upstream inducer of EZH2 to promote activation of STAT3.
EZH2 inhibition reverses Polycomb-mediated silencing and decreases STAT3 activity.
Acknowledgments
We thank Drs. George Stark and Bingcheng Wang for critical reading of the manuscript. This work was supported by R01 NS082312 (JL), the Case Comprehensive Cancer Center pilot grant P30CA043703 (JL), and Samsung Medical Center grant GFO1130011 (D-H.N).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdouh M, Facchino S, Chatoo W, Balasingam V, Ferreira J, Bernier G. BMI1 sustains human glioblastoma multiforme stem cell renewal. J Neurosci. 2009;29:8884–8896. doi: 10.1523/JNEUROSCI.0968-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cao Y, Lathia JD, Eyler CE, Wu Q, Li Z, Wang H, McLendon RE, Hjelmeland AB, Rich JN. Erythropoietin Receptor Signaling Through STAT3 Is Required For Glioma Stem Cell Maintenance. Genes Cancer. 2010;1:50–61. doi: 10.1177/1947601909356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa BM, Smith JS, Chen Y, Chen J, Phillips HS, Aldape KD, Zardo G, Nigro J, James CD, Fridlyand J, et al. Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res. 2010;70:453–462. doi: 10.1158/0008-5472.CAN-09-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea F, Hurt EM, Farrar WL. Clinical significance of Polycomb gene expression in brain tumors. Molecular cancer. 2010;9:265. doi: 10.1186/1476-4598-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks PB. Brain tumor stem cells: the cancer stem cell hypothesis writ large. Mol Oncol. 2010;4:420–430. doi: 10.1016/j.molonc.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Fan QW, Knight ZA, Goldenberg DD, Yu W, Mostov KE, Stokoe D, Shokat KM, Weiss WA. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 2006;9:341–349. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QW, Weiss WA. Targeting the RTK-PI3K-mTOR axis in malignant glioma: overcoming resistance. Curr Top Microbiol Immunol. 2010;347:279–296. doi: 10.1007/82_2010_67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, Koh C, Zhang J, Li YM, Maciaczyk J, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer letters. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- Gilbert CA, Ross AH. Cancer stem cells: cell culture, markers, and targets for new therapies. J Cell Biochem. 2009;108:1031–1038. doi: 10.1002/jcb.22350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotze S, Wolter M, Reifenberger G, Muller O, Sievers S. Frequent promoter hypermethylation of Wnt pathway inhibitor genes in malignant astrocytic gliomas. Int J Cancer. 2010;126:2584–2593. doi: 10.1002/ijc.24981. [DOI] [PubMed] [Google Scholar]
- Guryanova OA, Wu Q, Cheng L, Lathia JD, Huang Z, Yang J, MacSwords J, Eyler CE, McLendon RE, Heddleston JM, et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell. 2011;19:498–511. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P, Wang G, Marquez VE, Orkin SH, Pu WT. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26:37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- Hussain M, Rao M, Humphries AE, Hong JA, Liu F, Yang M, Caragacianu D, Schrump DS. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 2009;69:3570–3578. doi: 10.1158/0008-5472.CAN-08-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo KM, Kim J, Jin J, Kim M, Seol HJ, Muradov J, Yang H, Choi YL, Park WY, Kong DS, et al. Patient-Specific Orthotopic Glioblastoma Xenograft Models Recapitulate the Histopathology and Biology of Human Glioblastomas In Situ. Cell Rep. 2013 doi: 10.1016/j.celrep.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006a;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Lee J, Son MJ, Woolard K, Donin NM, Li A, Cheng CH, Kotliarova S, Kotliarov Y, Walling J, Ahn S, et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Lee JS, Kim H, Kim K, Park H, Kim JY, Lee SH, Kim IS, Kim J, Lee M, et al. EZH2 Generates a Methyl Degron that Is Recognized by the DCAF1/DDB1/CUL4 E3 Ubiquitin Ligase Complex. Mol Cell. 2012;48:572–586. doi: 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, Liou YC, Yu Q. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006b;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, Gudkov AV, Stark GR. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci U S A. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35:323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8:486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, Vescovi AL. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, Bayani J, Head R, Lee M, Bernstein M, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, Zeng J, Li M, Fan H, Lin Y, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Richly H, Aloia L, Di Croce L. Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis. 2011;2:e204. doi: 10.1038/cddis.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nature reviews Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Stark GR, Wang Y, Lu T. Lysine methylation of promoter-bound transcription factors and relevance to cancer. Cell Res. 2011;21:375–380. doi: 10.1038/cr.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, Baumer K, Le Bitoux MA, Marino D, Cironi L, et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Wei Y, Xia W, Zhang Z, Liu J, Wang H, Adsay NV, Albarracin C, Yu D, Abbruzzese JL, Mills GB, et al. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Molecular carcinogenesis. 2008;47:701–706. doi: 10.1002/mc.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, Chance MR, Chen X, Du Y, Wang Y, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci U S A. 2010;107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. Cell Res. 2008;18:443–451. doi: 10.1038/cr.2008.41. [DOI] [PubMed] [Google Scholar]
- Yang YP, Chang YL, Huang PI, Chiou GY, Tseng LM, Chiou SH, Chen MH, Chen MT, Shih YH, Chang CH, et al. Resveratrol suppresses tumorigenicity and enhances radiosensitivity in primary glioblastoma tumor initiating cells by inhibiting the STAT3 axis. J Cell Physiol. 2012;227:976–993. doi: 10.1002/jcp.22806. [DOI] [PubMed] [Google Scholar]
- Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert opinion on investigational drugs. 2009;18:45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Wiedemeyer R, Yan H, Quayle SN, Ivanova EV, Paik JH, Zhang H, Xiao Y, Perry SR, et al. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell. 2010;17:497–509. doi: 10.1016/j.ccr.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Wen Z, Darnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nature reviews Drug discovery. 2009;8:806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.