Abstract
Preimplantation embryo metabolism demonstrates distinctive characteristics associated with the developmental potential of embryos. On this basis, metabolite content of culture media was hypothesized to reflect the implantation potential of individual embryos. This hypothesis was tested in consecutive studies reporting a significant association between culture media metabolites and embryo development or clinical pregnancy. The need for a noninvasive, reliable, and rapid embryo assessment strategy promoted metabolomics studies in vitro fertilization (IVF) in an effort to increase success rates of single embryo transfers. With the advance of analytical techniques and bioinformatics, commercial instruments were developed to predict embryo viability using spectroscopic analysis of surplus culture media. However, despite the initial promising results from proof-of-principal studies, recent randomized controlled trials using commercial instruments failed to show a consistent benefit in improving pregnancy rates when metabolomics is used as an adjunct to morphology. At present, the application of metabolomics technology in clinical IVF laboratory requires the elimination of factors underlying inconsistent findings, when possible, and development of reliable predictive models accounting for all possible sources of bias throughout the embryo selection process.
Keywords: embryo assessment, metabolomics
Within the past decade, utilization of in vitro fertilization (IVF) for infertility treatment increased significantly in the United States, from 107,587 cycles in 2001 to 147,260 in 2010, accounting for 1% of all live births.1 In Europe, where IVF accounts for 3% of live births, an estimated 1,000,000 IVF cycles is expected to occur yearly by 2015.2
While IVF offers the highest success rates in achieving pregnancy for couples undergoing infertility treatment, multiple pregnancies resulting from multiple embryo transfer continue to have serious medical, social, and financial implications.3 Consequently, regulatory legislations came into effect in many countries, restricting the number of embryos transferred.3 Corresponding transition effort in clinical practice stressed out the problem of embryo selection for IVF specialists. Accordingly, investigations focused on the development of new methods for the selection of the most viable embryo(s) to be transferred with the aim of reducing multiple gestations without decreasing overall success rates.
At present, embryo viability is mainly assessed by means of morphological evaluation throughout the embryo culture period. Observation of embryo morphology at certain time points using a light microscope has practical advantages in embryo selection process, and resulted in significant improvement in pregnancy rates.4 The efficiency of morphological observation is likely to be further improved as a result of the new instruments that provide continuous monitoring of the embryo development in vitro and selection of the embryo(s) based on additional dynamic morphological parameters.5,6 However, the precision of morphological criteria remains less than desired and novel invasive and non-invasive embryo assessment strategies have been proposed with the advance of recent genomics, transcriptomics, proteomics, and metabolomics (OMICs) technologies (reviewed by Uyar and Seli7).
The invasive genomic approach to embryo assessment, referred to as preimplantation genetic screening (PGS) allows the exclusion of aneuploid embryos through genetic profiling of biopsied polar body(ies), blastomere(s), or trophoectodermal cells. Available technologies for PGS include fluorescent in situ hybridization (FISH), comparative genomic hybridization (CGH)-array, or single nucleotide polymorphism (SNP)-array (reviewed by Seli et al8). Among these methods, FISH has been used widely in the past and the studies applying FISH for PGS have reported conflicting results. More recently, a meta-analysis of 10 randomized controlled trials (RCTs) assessing FISH method concluded that PGS with FISH is associated with lower pregnancy and live birth rates,9 leading to a statement from the American Society of Reproductive Medicine (ASRM) and the European Society for Human Reproduction and Embryology (ESHRE) advising against its use in clinical practice.10,11 It is noteworthy that, chromosomal mosaicism and the invasiveness of biopsy have been considered as major drawbacks of PGS, potentially underlying the associated decrease in clinical success rates. On the other hand, PGS using more recent CGH-array or SNP-array technologies has shown promising results.12,13
Beyond the genomic constitution of embryos, transcriptomic analysis of cumulus/granulosa cells has been proposed as a noninvasive tool to assess oocyte quality as a means of embryo viability (reviewed by Uyar et al14). As the genes are expressed by transcription of the genetic code into messenger RNAs (mRNAs) and subsequent translation of mRNAs into proteins, relative quantities of individual mRNAs can be evaluated as an approximation to the expression levels of the corresponding genes. Using quantitative reverse transcriptase polymerase chain reaction or microarrays for quantification of mRNA transcripts in follicular somatic cells, the expression levels of the candidate genes in cumulus/granulosa cells were shown to be associated with oocyte maturation, fertilization, embryo competence or pregnancy (reviewed by Uyar et al14). Although, several genes have been identified in these studies as potential biomarkers of oocyte/embryo competence, a clinical benefit from transcriptomic analysis of cumulus/granulosa cells remains to be demonstrated in RCTs.
Alternative noninvasive approaches to embryo assessment are currently based on the analysis of embryo culture media. Preliminary proteomics studies in this direction hypothesized that secretome profiles of the culture media could potentially correlate with embryo viability.15–17 Using mass spectrometry or protein-array technologies, these studies reported altered expression of specific proteins associated with blastocyst development or implantation. On the other hand, proteomics is still considered a limited source of information for assessing reproductive potential of embryos possibly due to lack of sensitivity of proteomics platforms in this domain.8
Metabolite content of the spent embryo culture media has also been proposed as a noninvasive diagnostic platform to assess embryo viability. Because certain key nutrients are required for normal preimplantation embryo development, the change in the levels of these nutrients and their metabolites were suggested as indicators of metabolic activity during in vitro embryo culture. Conveniently, metabolomic profiling of culture media was analyzed by taking advantage of the Raman, near infrared (NIR) or Fourier transform infrared (FTIR) spectroscopic technologies. Although, a multitude of retrospective cohort studies reported significant associations between culture media metabolome and assessed clinical outcomes,18–21 recent RCTs evaluating the efficiency of a commercial metabolomics device showed no clinical benefit.22,23
To date, metabolomics has been the most widely investigated noninvasive OMICs approach in embryo assessment where its efficiency is still under debate. In this review, we first present basic aspects of preimplantation embryo metabolism and provide a summary of the studies demonstrating correlation between culture media metabolome and clinical outcomes. Current metabolomics technologies and related data analysis procedures are then outlined followed by an overview of literature and commercialization efforts on metabolomic assessment of embryo viability. Lastly, advantages and limitations of metabolomics research in IVF have been explored with an emphasis on potential future applications.
Preimplantation Embryo Metabolism as an Indicator of Viability
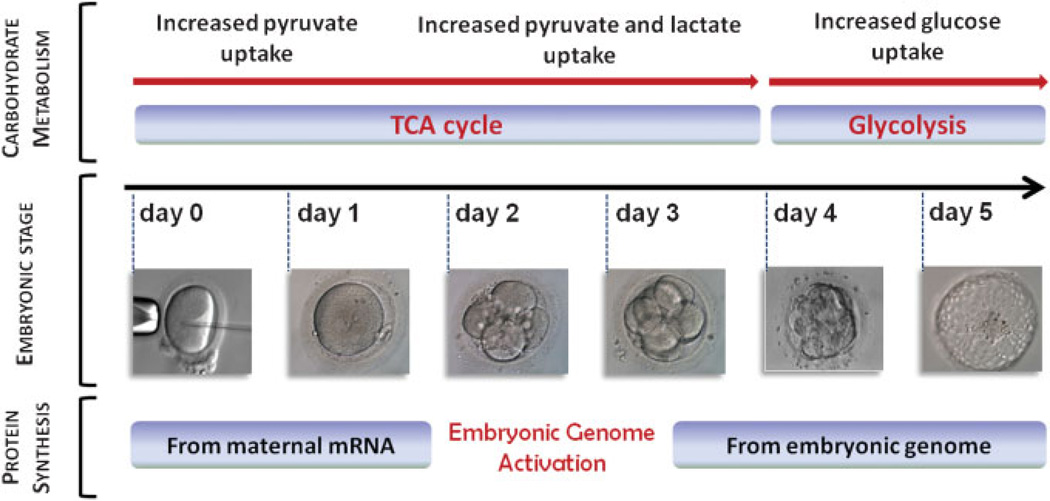
Oocyte and preimplantation embryo development shows unique characteristics in terms of regulation of gene expression and metabolism. Transcription (RNA synthesis) becomes suppressed upon stimulation of oocyte maturation and remains suppressed until zygotic genome activation (ZGA) (Fig. 1). Associated with transcriptional silencing is the translational regulation of gene expression through activation and silencing of maternally derived messenger mRNAs. Key regulatory proteins such as embryonic poly(A) binding protein, cytoplasmic polyadenylation element binding protein, and deleted in azoospermia-like mediate translational activation of maternal mRNAs and protein synthesis, and are required for female fertility.24–29 ZGA occurs at two-cell stage in the mouse and four- to eight-cell stage in human,30–32 and is associated with degradation of maternal mRNAs and initiation of zygotic transcription (Fig. 1).
Figure 1.
Alteration of carbohydrate metabolism and protein synthesis during preimplantation embryo development. In the early stages of development, TCA cycle predominate using pyruvate and lactate as the main sources of energy. From the zygote stage to compaction, glycolysis becomes increasingly more utilized for energy production and at last glucose is consumed preferentially at the blastocyst stage. Protein synthesis, on the other hand, is regulated through embryonic genome activation that occurs at 4- to 8-cell stage in human and is associated with degradation of maternal mRNAs and initiation of embryonic transcription. mRNA, messenger RNA; TCA, tricarboxylic acid.
Importantly, the preferred energy source for embryonic cellular metabolism seems to change during preimplantation development, soon after ZGA. In the early stages of development, carboxylic acid-based metabolism (aerobic glycolysis also called the Krebs cycle) predominates, using pyruvate and lactate as the main sources of energy, while glucose uptake is minimal.33–38 As development progresses, glycolysis becomes increasingly more utilized for energy production, and glucose uptake steadily increases from the zygote stage to compaction. Finally, glucose metabolism predominates as the energy source at the blastocyst stage. This increase in glucose consumption in late-stage preimplantation embryos has been observed across several species, including mouse, rat, human, cow, sheep, and pig embryos.39,40
Over the last two decades, preimplantation embryo metabolism has been studied extensively with the objective of correlating metabolic activity with the viability of an embryo. Within the context of IVF, several studies identified metabolites associated with carbohydrate metabolism or amino acid turnover during the embryo culture period as potential biomarkers of embryo viability (reviewed by Botros et al41). These studies are summarized in Table 1 emphasizing the major findings, embryo stages examined, assessed clinical outcomes, and study populations.
Table 1.
Summary of the studies evaluating preimplantation embryo metabolism as an indicator of reproductive viability
| Study | Embryonic stage examined |
Analyzed metabolite |
Assessed outcome |
Study population |
Technology | Findings |
|---|---|---|---|---|---|---|
| Hardy etal (1989)42 | Days 1–6 | Pyruvate, glucose | Embryo and blastocyst development |
25/43/73 (Nb/Ne/No) |
Ultramicrofluorescence assay | ↑Pyruvate uptake on days 1–4 ∝ embryo development. ↔ Glucose uptake on days 1–4. ↑ Pyruvate and glucose uptake on day 5 ∝ blastocyst development. |
| Gottetal (1990)35 | Days 2–6 | Pyruvate, glucose, lactate |
Blastocyst development |
9/40 (Nb/Ne) | Ultramicrofluorescence assay | ↑ Pyruvate uptake on days 2–4 oc embryo development. ↔ Glucose uptake on days 2–4. ↑ Pyruvate and glucose uptake on day 5 ∝ blastocyst development. ↑ Lactate production on days 3–5 ∝ blastocyst development. |
| Conaghan et al (1993)36 |
Days 2–3 | Pyruvate | Implantation | 42/173 (Ni/Net) | Ultramicrofluorescence assay | ↓ Pyruvate uptake oc implantation. |
| Turner et al (1994)43 |
Day 2 | Pyruvate | Pregnancy | 1 8/80 (Ni/Net) | Ultramicrofluorescence assay | Morphologically good embryos were more likely to implant if they demonstrated an intermediate pyruvate uptake (10–30 pmol/embryo/h). |
| Gardner et al (2001 )38 |
Days 4–6 | Pyruvate, glucose | Blastocyst development and quality |
25/43/60 (Nb/Ne/Npe) |
Ultramicrofluorescence assay | ↑ Pyruvate and glucose uptake on day 4 ∝ blastocyst development. ↑ Glucose uptake on days 5–6 oc blastocyst quality. |
| Houghton et al (2002)46 |
Days 2–3 | 18 Amino acids including glutamine, arginine, methionine, alanine, asparagine |
Blastocyst development |
14/42 (Nb/Ne) | High-performance liquid chromatography |
↓ Glutamine, arginine, methionine uptake ∝ blastocyst development. ↓ Alanine and asparagine production ∝ blastocyst development. |
| Compacting 8-cell |
18 Amino acids including serine, alanine, glycine |
Blastocyst development |
23/55 (Nb/Ne) | High-performance liquid chromatography |
↓ Serine uptake oc blastocyst development. ↓ Alanine and glycine release ∝ blastocyst development. |
|
| Brison et al (2004)48 | Day 2 | Glycine, leucine, asparagine |
Pregnancy, live birth |
22/113/221 (Ni/Net/Ne) |
High-performance liquid chromatography |
↑ Asparagine, ↓ Glycine and leucine ∝ pregnancy and live birth. |
| Sell et al (2008)49 | Day 3 | Glutamate | Pregnancy, live birth |
17/34 (Ni/Net) | Proton nuclear magnetic resonance |
↑ Glutamate ∝ pregnancy and live birth. |
Abbreviations: Nb, numberof blastocysts; Nc, number of cycles; Ne, numberof embryos; Net, number of embryos transferred; Ni, numberof embryos implanted; N0, number of oocytes; Np, number of patients; Npe, number of pronucleate embryos.
Source: Adapted from Botros et al.41
Note: Symbols ↑, ↓,↔, and ∝ stands for high level, low level, no difference in levels, and associated with, respectively.
Carbohydrate Metabolism
Carbohydrate metabolism of preimplantation human embryos was mainly investigated in terms of pyruvate and glucose uptake and lactate production. Initially in 1989, Hardy et al reported higher pyruvate uptake in embryos that develop to blastocyst stage.42 Their findings were confirmed by Gott et al.35 Subsequently, Conaghan et al measured pyruvate uptake in relation to implantation and clinical pregnancy.36 This study demonstrated that embryo implantation after days 2 and 3 transfers was inversely correlated with pyruvate up take by two to eight cell embryos. Although, these findings were considered contradictory to initial reports, it should be noted that the assessed outcomes were different; blastocyst development in the former studies and implantation in the latter one. Another study by Turner et al showed that embryos had a wide range of pyruvate uptake values, but this variation was reduced to an intermediate range in the embryos that implanted.43
More recently, Gardner et al investigated the pyruvate metabolism again in relation to blastocyst development and suggested that pyruvate uptake on day 4 was significantly higher by embryos that formed blastocysts compared with ones that failed to develop.38 These results were consistent with the initial studies showing an association between pyruvate uptake and blastocyst development. On the basis of the findings summarized above, pyruvate uptake seems to be correlated with blastocyst development where its relation to implantation or pregnancy outcome remains inconclusive.
As another major aspect of carbohydrate metabolism, glucose uptake was also hypothesized to reflect the embryo’s developmental potential, and most studies investigating pyruvate levels in culture media also investigated glucose consumption of embryos simultaneously. According to initial findings, from days 2 to 4, glucose uptake by human embryos developing to blastocyst stage was similar to those which were arrested during cleavage.35,42 In contrast, Gardner et al reported a significant association between increased levels of glucose uptake on day 4 and subsequent blastocyst formation and quality.38
It is noteworthy that the composition of embryo culture media used in different studies display variations that could affect nutrient uptake or metabolite secretion by embryos. This factor, together with other potential confounding variables related to embryo culture, conditions, and measurement techniques, possibly underlies the contradictory results and complicates interpretation of the findings. Overall, analysis of carbohydrate metabolism during the embryo culture period may provide useful information about developmental potential of embryos but cannot be suggested alone as a reliable biomarker for embryo selection.
Amino Acid Turnover
The improvement in the rates of blastocyst formation was mainly achieved by inclusion of mixtures of essential and nonessential amino acids in the culture medium.44,45 Consequently, depletion and appearance of certain amino acids were profiled to explore specific patterns during preimplantation embryo development in correlation to blastocyst formation.
Initially, using high performance liquid chromatography, Houghton et al examined 18 amino acids at different stages of embryo development and showed that embryos that form a blastocyst displayed a different profile of amino acid metabolism than those that were arrested.46 Specifically, lower uptake of glutamine, arginine, and methionine and lower release of alanine and asparagine by embryos on days 2 and 3 was found to be associated with blastocyst development. In the same study, analysis of amino acid metabolism at eight cell- and morula stage embryos revealed correlation of blastocyst development with lower uptake of serine and lower release of alanine and glycine. Moreover, sum of depletion and appearance of the amino acids examined, that is, amino acid turnover, was shown to be lower in developing embryos compared with the arrested ones consistent with the “quite embryo hypothesis.”47
In a follow-up study by the same group, Brison et al investigated the correlation of amino acid turnover with implantation and pregnancy outcomes when the embryos were selected according to routine morphological criteria and transferred on day 2.48 They reported that decreased glycine and leucine, and increased asparagine levels in the culture media were associated with increased clinical pregnancy and live birth rates. More recently, Seli et al used proton nuclear magnetic resonance (1H NMR) and demonstrated an association between increased glutamate levels in the culture media and clinical pregnancy and live birth.49
Metabolomics: Technology and Data Analysis
Preliminary studies examining specific nutrients or amino acids in embryo culture media demonstrated metabolic differences related to embryo viability and hence motivated further analysis of metabolome for embryo selection. Metabolome is the complete set of small molecules (< 1 kDa) including metabolic intermediates (amino acids, lipids, and nucleotides), hormones, signaling molecules, and secondary metabolites within a biological sample. These molecules are the final downstream products of gene expression and their inventory provides a valuable database to explore genotype-phenotype relationships as well as genotype-environment interactions.
The metabolome of a cell is inherently dynamic as a result of the changing metabolic activity depending on the stage of the cell cycle. Metabolomic profile also changes markedly under disease conditions or in response to a treatment. Comparative metabolomic profiling of biological samples under different conditions allows the determination of phenotypic differences among the samples under investigation.
Metabolomics refers to rapid, high-throughput characterization of the metabolome and enables systematic analysis of the condition-dependent metabolic regulations. The human metabolome is estimated to include about 3,000 metabolites, a number significantly lower compared with approximately 30,000 genes, 200,000 transcripts, and 1 million proteins, included in human genome, transcriptome, and proteome, respectively. This relatively small set of metabolites can be efficiently profiled by analytical techniques favoring metabolomics as a powerful tool in biomedical research. Particularly in IVF, using the culture media metabolomics data, predictive models have been developed for selection of the most viable embryos. In this section, we provide a brief overview of the analytical platforms, which are commonly used for metabolomics research in IVF and related data analysis methods.
Spectroscopic Techniques
The analytical approaches applied for metabolomic analysis of embryo culture media can be categorized as NMR spectroscopy, mass spectrometry (MS), or vibrational spectroscopy.
Nuclear Magnetic Resonance Spectroscopy
NMR techniques exploit the interaction of the magnetic moment of certain atomic nuclei with external magnetic field and provide information about metabolites that contain elements with nonzero magnetic moments. NMR-based metabolomic analysis was pioneered and mostly driven by Nicholson et al.50,51 As a nondestructive analytical tool, the technique is efficiently utilized for biomarker analysis by enabling detection and quantification of specific metabolites within a biological fluid or tissue. On the other hand, limitations of NMR were attributed to requirement for large amounts of sample, higher costs, and lack of sensitivity for low abundance targets favoring the method for analysis of high abundance metabolites.
Mass Spectrometry
MS processes through three steps: ion formation, separation of ions according to their mass-to-charge ratios (m/z), and detection of the separated ions.52 MS is the most widely used analytical platform in metabolomics studies enabling simultaneous characterization of several hundreds of metabolites with higher sensitivity compared with NMR approaches. The sensitivity and specificity of MS is further enhanced when coupled with chromatography or electrophoresis-based separation techniques.
Specifically, gas chromatography (GC) utilizes a capillary column to separate volatile compounds or compounds that can become volatile after chemical derivatization. GC–MS is highly efficient for detection and quantification of low molecular weight metabolites.53 The technique, however, exhibit drawbacks associated with extra derivatization step and long chromatographic analysis.
The other chromatography-based separation method is liquid chromatography (LC). Both GC and LC techniques are based on the distribution of the sample components between mobile and stationary phases. The mobile phase is liquid in LC increasing its coverage compared with GC, since all compounds may not reach to a volatility level required by GC. LC– MS approaches are commonly used and favored for metabolomics analysis due to its high-throughput, soft ionization, and good coverage of metabolites.54
As an alternative separation technique, capillary electrophoresis (CE) separates metabolites based on their electrophoretic mobility. CE–MS based metabolomics approaches enable simultaneous determination of over 1,000 charged species in a wide range of m/z values that can be readily applied to various types of biological samples.55 As only charged ions are affected by the electric field, the efficiency of CE is limited due to lack of separation of neutral analytes. Nevertheless, a major fraction of metabolites are polar and ionic. Therefore, CE-MS can be reserved as an efficient separation tool which does not require rigorous sample pretreatment and can operate on small amounts of material.56
Overall, Buscher et al provided a systematic cross-platform analysis of GC-, LC- and CE-MS techniques and suggested GC or LC platforms for metabolomic analysis since the separation power and sensitivity of CE are equivalent to both LC and GC, but CE suffers from poor reproducibility.57
Vibrational Spectroscopy
The other group of analytical techniques is the vibrational spectroscopy which includes Raman and IR approaches.58 The main principle behind this kind of techniques is that, when a sample is exposed to an electromagnetic radiation, the chemical bonds within the molecules will absorb the energy and vibrate to a greater extent. Raman spectroscopy measures the scattering of electromagnetic radiation by the vibrating molecules under exposure to a particular wavelength light (usually in the form of a laser). In contrast, IR spectrum is the result of absorption of electromagnetic radiation by vibrating molecules when the sample is interrogated with an IR beam.
IR spectroscopy is categorized as NIR and FTIR according to the wavelength of the light used. NIR measures the spectra in the 14,000 to 4,000 cm−1 region whereas FTIR looks at the mid-IR part of the spectrum (4,000–600 cm−1). Although, FTIR is a faster and higher throughput method, NIR has the advantage of quantitation with higher sensitivity in metabolomics research. On the other hand, at a higher cost of instrumentation, Raman spectroscopy also has certain advantages since it does not suffer from water interference and measurements can be made directly from biofluids.
To summarize, there is no single best analytical technique for all kinds of metabolomics investigations. The choice of the technique depends on metabolite class of interest, required sensitivity, dynamic range of measurements, sample size, sample specific pretreatment requirements, and the relative time and cost-efficiency of the method.
Analysis of Metabolomics Data and Predictive Model Development
Metabolomics studies may consider three main approaches: targeted metabolite profiling, nontargeted profiling, and metabolic fingerprinting.59,60 Targeted analysis is based on measurement of preidentified metabolites for which chemical structures are known. In contrast, nontargeted profiling aims at quantitation of all the peaks in the spectrum without associating the peaks to chemical structure of certain compounds. Metabolomic fingerprinting is also a nontargeted approach considering the whole metabolic profile as a pattern that can be used for sample classification.
Analysis of targeted metabolite profiling is relatively simple since spectral regions corresponding to specific compounds have been identified previously and only these regions are analyzed. On the other hand, nontargeted profiling and metabolic fingerprinting requires bioinformatics support for efficient and accurate analysis of high-dimensional complex metabolomics data.60
A critical preprocessing step in the analysis of metabolomics data are normalization. The multistage experimental setting underlying metabolomics studies is likely to introduce systematic variations in the resulting spectral data. Main source of nonbiological variations in embryo assessment is attributed to the culture environment. Spectral profiles of spent culture media (where individual embryo has been cultured) may be normalized to that of blank samples (culture media incubated under the same conditions but without an embryo) to eliminate possible impact of variations in culture conditions.
After normalization, metabolomics data can be analyzed using a wide range of statistical methods and predictive models can be developed using machine learning algorithms to predict implantation potentials of individual embryos. In construction of such models, it is crucial to deal with possible sources of bias to ensure reliability and consistency of the predictions. The most common sources of bias relate to size of the study population, model selection, sampling procedure utilized in training and testing stages, and performance evaluation.
Metabolomics data are considered a typical example of high dimension low sample size data with thousands of measurements coming from the spectra and only tens or hundreds of samples to examine the relevant metabolome content. Increasing the number of samples would improve the power of analysis; however biological samples are usually limited, especially in case of human subjects. On the other hand, using dimensionality reduction methods such as principal component analysis or filtering the predictive regions in the spectrum would eliminate the redundancy in the data and reduce the computational cost of the analysis.
Machine learning literature provides a variety of classification methods with diverse mathematical backgrounds, such as decision trees, artificial neural networks, support vector machines, nearest neighbors, genetic algorithms, and so on.61 Selection of the best-fitting classification method for prediction of embryo viability requires comprehensive benchmarking analysis. Predictive model development consists of training and testing stages, where the model is constructed on the training samples and the performance of predictions is assessed on blinded test samples. During this process, it is necessary to partition the original dataset into training and test groups in a structured and unbiased manner. The most common approaches for data partitioning include k-fold cross-validation, leave-one-out, and bootstrap methods.62 Finally, the performance of the model should be evaluated considering both the sensitivity and specificity of the predictions, preferably using receiver operating characteristics (ROC) curves.
Most importantly, metabolomics-based embryo selection models should be either developed using pooled multicenter data or validated in different centers to ensure the robustness of the model against center-specific variations. Once the success and robustness of a prediction model is guaranteed, it can be utilized as an embryo selection tool alone or in combination with other available criteria in clinical practice. Furthermore, culture media metabolomics can be integrated to data from other OMICs platforms to provide an extensive knowledge base for functional genomics research in reproductive sciences.63
Clinical Application: Metabolomic Assessment of Embryo Viability
Efficiency of metabolomics-based embryo selection strategies was first evaluated in retrospective studies and then in subsequent RCTs, as summarized in ?Tables 2 and 3, respectively.
Table 2.
Summary of the proof-of-principle studies evaluating culture media metabolomics as a potential biomarker of embryo viability
| Study | Outcome measure |
Study population (number of embryos) |
Day of transfer | Technology | Statistical method/ performance criteria |
Result |
|---|---|---|---|---|---|---|
| Seli et al (2007)18 | Pregnancy | 69 | 3 | NIR Raman | Genetic algorithm (model development)/ sensitivity, specificity |
Metabolomic profiling of embryo culture media correlates with pregnancy. |
| Scott etal(2008)19 | Implantation | 41 | 3 and 5 | Raman | ROC analysis | Metabolomic profiling of embryo culture media correlates with implantation. |
| Vergouw et al (2008)20 | Pregnancy | 333 | 3 | NIR | Accuracy, PPV, NPV | Metabolomic profiling of embryo culture media correlates with pregnancy. |
| Seli etal (2010)21 | Implantation | 485 | 2 and 3 | NIR | ROC analysis | Metabolomic profiling of embryo culture media is independent of morphology and correlates with implantation. |
| Ahlstrom et al (2011)64 | Implantation | 137 | 5 | NIR | ROC analysis | Metabolomic profiling of embryo culture media can predict the implantation potential of blastocysts. |
| Seli etal (2011)65 | Pregnancy | 198 | 5 | NIR | ROC analysis | Metabolomic profiling alone or in combination with morphologic grading has the potential to be a better classifier for pregnancy outcome. |
Table 3.
RCTs assessing culture media metabolomics for embryo selection
| Study | Outcome measure |
Study population (number of embryos) |
Day of transfer | Technology | Result |
|---|---|---|---|---|---|
| Hardarson et al (2012)22 | Pregnancy rate | 327 | 2 and 5 | NIR | No significant improvement of pregnancy rates. |
| Vergouw et al (2012)23 | Live birth rate | 417 | 3 | NIR | No significant improvement of live birth rates. |
| Sfontouris et al (2011)67 | Implantation rate | 125 | 2, 3, and 5 | NIR | Increased implantation rates following embryo selection based on noninvasive metabolomic analysis. |
Abbreviations: NIR, near infrared; RCT, randomized controlled trials.
Proof-of-Principal Studies
A correlation between embryo viability and culture media metabolome was first reported in 2007.18 In this initial proof-of-principal study, Seli et al analyzed day 3 spent embryo culture media samples using Raman and/or NIR spectroscopy. The mean spectrum obtained from embryos that failed to implant was compared with that obtained from embryos that resulted in a live birth. Algorithms that help predict embryo viability was developed using both Raman and NIR spectroscopy. The algorithm established for Raman spectroscopy in the initial study was then successfully tested by Scott et al, analyzing spent culture media collected at a different center, where embryos were cultured in a different type and volume of culture medium.19
In a following study by Vergouw et al20 embryo culture media samples from single embryo transfer (SET) cycles were analyzed using NIR spectroscopy. A new algorithm predictive of embryo implantation potential was developed. A “Viability Score” was calculated for each embryo based on the NIR-metabolomic profiling of culture medium and increasing Viability Scores were correlated with an increasing ability of the embryo to implant. Following studies with large number of samples collected in SET) cycles reported similar findings.21,64,65
Importantly, when culture media from human embryos of similar morphology were examined, their metabolomic profile varied remarkably indicating that the metabolome of embryos that looked similar could be significantly different. This observation was in agreement with the studies of Katz-Jaffe et al,15,66 which suggested that the proteome of individual human blastocysts of the same grade differed between embryos, again indicating that embryo morphology is not completely reflective of its physiology.
Commercialization Efforts and Randomized Controlled Trials
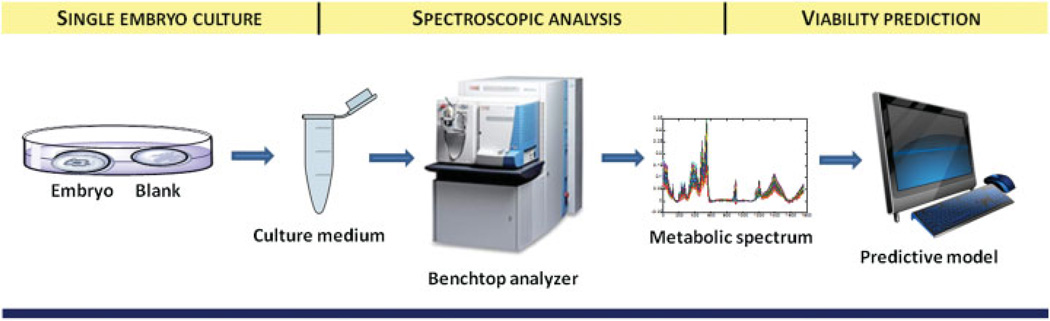
The studies summarized above suggested a potential benefit from metabolomics technology in embryo viability assessment either alone or as an adjunct to morphology. However, these studies were retrospective, and were performed in a single research laboratory using frozen and transported culture media samples. As the promise of metabolomics approach is its potential use as a rapid on-site technology in the IVF laboratory, it was further developed by a private company (Molecular Biometrics Inc., Norwood, MA). A series of prototypes and commercial instruments were built and tested by using an NIR-based system. The typical workflow of metabolomics based embryo assessment is shown in Fig. 2.
Figure 2.
Metabolomic assessment of embryo viability through spectroscopic analysis of spent culture media and computerized predictive models.
Using the prototype instruments, two RCTs were conducted in centers performing SET.22,23 In these studies, women in the control arm underwent SET with embryos selected using standard morphological embryo assessment protocols. In the treatment arm, a combination of metabolomic profiling and morphological assessment was used by first identifying embryos with a good morphology, and then selecting the embryo to be transferred based on the metabolomic Viability Score generated by the commercial instrument.
In one of these studies, Hardarson et al22 analyzed spent culture media from SETs on days 2 and 5. While not statistically significant, their findings suggested a potential benefit of NIR for the assessment of embryos transferred on day 2. Of 87 and 83 SETs in the NIR/morphology and morphology alone groups the pregnancy rates were 31 and 26.5%, respectively. In the same study no benefit for selection of day 5 SET was observed. Of 77 and 80 SETs in the NIR/morphology and morphology alone groups (day 5) the pregnancy rates were 39 and 45%, respectively.
In another RCT performed by Vergouw et al23 women undergoing SETon day 3 were similarly studied. No difference in live birth rates was found between the groups. Live birth rate was 29.5% in the NIR/morphology group (n = 146) compared with the 31.3% in morphology alone group (n = 163 patients).
Conversely, Sfontouris et al67 performed a RCT using the commercial platform where two or three embryos were selected for transfer on the basis of NIR/morphology or morphology alone, and showed an improvement in pregnancy rates in the NIR-assisted group. In this study, for every two patients, where embryos were selected based on morphology, one patient hada transfer based on NIR-assisted embryo selection (randomly allocated with a 2:1 ratio). Implantation rates (IRs) were significantly improved in the NIR/morphology group (n = 39 patients, IR = 33/102 [32.4%]) compared with the morphology alone group (n = 86 patients, 55/257 [21.4%]).67
An important problem encountered in the commercial NIR system was that the different instruments were variable in performance, which in turn masked the low intensity of the signal generated from the embryo within the culture media. This meant that any algorithm created on one group of instruments suffered when translated on other instruments as their noise thresholds and behavior may have differed significantly. Therefore, the commercial version of the NIR instrument was withdrawn due to the wide variability in performance between clinics and inconsistent results in clinical trials. The technology will hopefully be improved and be tested again in the near future.
Conclusion
Levels of uptake/secretion of specific metabolites by preimplantation human embryos display changes in a timely manner during preimplantation development. Measured metabolite levels in the culture media from cleavage stage the embryos demonstrated certain patterns correlating to the blastocyst formation and/or pregnancy. On the basis of these observations, the entire metabolome content of culture media has been profiled to gain insight into the embryo viability.
With the advance of analytical and bioinformatics resources, “metabolomics” investigations emerged as a promising noninvasive tool for embryo assessment. Proof-of-principal studies represented significant associations between culture media metabolomics and pregnancy outcomes. These studies led to the commercialization of benchtop instruments that provided prediction of embryo viability by spectroscopic analysis. However, subsequent RCTs failed to confirm the efficiency of metabolomics based commercial devices for embryo selection.
Inconsistencies observed in these studies may be attributed to the omitted confounding factors related to patient demographics, cycle characteristics, stimulation protocols, embryo culture, instrumentation, and so on. Metabolomic assessment of embryo viability is a multistage complex procedure where it is extremely challenging to account for all intra- and intercenter variations during predictive model development. With meticulous care to overcome all possible sources of biases in embryo selection and development of robust models, metabolomics approach reserves the potential to be a useful adjunct to conventional morphological criteria for identification of most viable embryos.
Acknowledgement
E.S. is supported by award no. R01HD059909 from the National Institutes of Health (NIH).
References
- 1.Center for Disease Control and Prevention Web site. [Accessed April 10, 2013];Assisted Reproductive Technology (ART) Available at: http://www.cdc.gov/art/index.htm.
- 2.Ata B, Seli E. Economics of assisted reproductive technologies. Curr Opin Obstet Gynecol. 2010;22(3):183–188. doi: 10.1097/GCO.0b013e3283373c13. [DOI] [PubMed] [Google Scholar]
- 3.Bromer JG, Seli E. Assessment of embryo viability in assisted reproductive technology: shortcomings of current approaches and the emerging role of metabolomics. Curr Opin Obstet Gynecol. 2008;20(3):234–241. doi: 10.1097/GCO.0b013e3282fe723d. [DOI] [PubMed] [Google Scholar]
- 4.Sakkas D, Gardner DK. Noninvasive methods to assess embryo quality. Curr Opin Obstet Gynecol. 2005;17(3):283–288. doi: 10.1097/01.gco.0000169106.69881.3e. [DOI] [PubMed] [Google Scholar]
- 5.Wong CC, Loewke KE, Bossert NL, et al. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol. 2010;28(10):1115–1121. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- 6.Meseguer M, Rubio I, Cruz M, Basile N, Marcos J, Requena A. Embryo incubation and selection in a time-lapse monitoring system improves pregnancy outcome compared with a standard incubator: a retrospective cohort study. Fertil Steril. 2012;98(6):1481–1489. doi: 10.1016/j.fertnstert.2012.08.016. e10. [DOI] [PubMed] [Google Scholar]
- 7.Uyar A, Seli E. Embryo assessment strategies and their validation for clinical use: a critical analysis of methodology. Curr OpinObstet Gynecol. 2012;24(3):141–150. doi: 10.1097/GCO.0b013e328352cd17. [DOI] [PubMed] [Google Scholar]
- 8.Seli E, Robert C, Sirard MA. OMICS in assisted reproduction: possibilities and pitfalls. Mol Hum Reprod. 2010;16(8):513–530. doi: 10.1093/molehr/gaq041. [DOI] [PubMed] [Google Scholar]
- 9.Checa MA, Alonso-Coello P, Solà I, Robles A, Carreras R, Balasch J. IVF/ICSI with or without preimplantation genetic screening for aneuploidy in couples without genetic disorders: a systematic review and meta-analysis. J Assist Reprod Genet. 2009;26(5):273–283. doi: 10.1007/s10815-009-9328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Practice Committee of Society for Assisted Reproductive Technology; Practice Committee of American Society for Reproductive Medicine. Preimplantation genetic testing: a Practice Committee opinion. Fertil Steril. 2008;90(5, Suppl):S136–S143. doi: 10.1016/j.fertnstert.2008.08.062. [DOI] [PubMed] [Google Scholar]
- 11.Harper J, Coonen E, De Rycke M, et al. What next for preimplantation genetic screening (PGS)? A position statement from the ESHRE PGD Consortium Steering Committee. Hum Reprod. 2010;25(4):821–823. doi: 10.1093/humrep/dep476. [DOI] [PubMed] [Google Scholar]
- 12.Sher G, Keskintepe L, Keskintepe M, Maassarani G, Tortoriello D, Brody S. Genetic analysis of human embryos by metaphase comparative genomic hybridization (mCGH) improves efficiency of IVF by increasing embryo implantation rate and reducing multiple pregnancies and spontaneous miscarriages. Fertil Steril. 2009;92(6):1886–1894. doi: 10.1016/j.fertnstert.2008.11.029. [DOI] [PubMed] [Google Scholar]
- 13.Schoolcraft WB, Treff NR, Stevens JM, Ferry K, Katz-Jaffe M, Scott RT., Jr Live birth outcome with trophectoderm biopsy, blastocyst vitrification, and single-nucleotide polymorphism microarray-based comprehensive chromosome screening in infertile patients. Fertil Steril. 2011;96(3):638–640. doi: 10.1016/j.fertnstert.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 14.Uyar A, Torrealday S, Seli E. Cumulus and granulosa cell markers of oocyte and embryo quality. Fertil Steril. 2013;99(4):979–997. doi: 10.1016/j.fertnstert.2013.01.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz-Jaffe MG, Schoolcraft WB, Gardner DK. Analysis of protein expression (secretome) by human and mouse preimplantation embryos. Fertil Steril. 2006;86(3):678–685. doi: 10.1016/j.fertnstert.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 16.Domínguez F, Gadea B, Esteban FJ, Horcajadas JA, Pellicer A, Simón C. Comparative protein-profile analysis of implanted versus non-implanted human blastocysts. Hum Reprod. 2008;23(9):1993–2000. doi: 10.1093/humrep/den205. [DOI] [PubMed] [Google Scholar]
- 17.Dominguez F, Gadea B, Mercader A, Esteban FJ, Pellicer A, Simón C. Embryologic outcome and secretome profile of implanted blastocysts obtained after coculture in human endometrial epithelial cells versus the sequential system. Fertil Steril. 2010;93(3):774–782. doi: 10.1016/j.fertnstert.2008.10.019. e1. [DOI] [PubMed] [Google Scholar]
- 18.Seli E, Sakkas D, Scott R, Kwok SC, Rosendahl SM, Burns DH. Noninvasive metabolomic profiling of embryo culture media using Raman and near-infrared spectroscopy correlates with reproductive potential of embryos in women undergoing in vitro fertilization. Fertil Steril. 2007;88(5):1350–1357. doi: 10.1016/j.fertnstert.2007.07.1390. [DOI] [PubMed] [Google Scholar]
- 19.Scott R, Seli E, Miller K, Sakkas D, Scott K, Burns DH. Noninvasive metabolomic profiling of human embryo culture media using Raman spectroscopy predicts embryonic reproductive potential: a prospective blinded pilot study. Fertil Steril. 2008;90(1):77–83. doi: 10.1016/j.fertnstert.2007.11.058. [DOI] [PubMed] [Google Scholar]
- 20.Vergouw CG, Botros LL, Roos P, et al. Metabolomic profiling by near-infrared spectroscopy as a tool to assess embryo viability: a novel, non-invasive method for embryo selection. Hum Reprod. 2008;23(7):1499–1504. doi: 10.1093/humrep/den111. [DOI] [PubMed] [Google Scholar]
- 21.Seli E, Vergouw CG, Morita H, et al. Noninvasive metabolomic profiling as an adjunct to morphology for noninvasive embryo assessment in women undergoing single embryo transfer. Fertil Steril. 2010;94(2):535–542. doi: 10.1016/j.fertnstert.2009.03.078. [DOI] [PubMed] [Google Scholar]
- 22.Hardarson T, Ahlström A, Rogberg L, et al. Non-invasive metabolomic profiling of Day 2 and 5 embryo culture medium: a prospective randomized trial. Hum Reprod. 2012;27(1):89–96. doi: 10.1093/humrep/der373. [DOI] [PubMed] [Google Scholar]
- 23.Vergouw CG, Kieslinger DC, Kostelijk EH, et al. Day 3 embryo selection by metabolomic profiling of culture medium with near-infrared spectroscopy as an adjunct to morphology: a randomized controlled trial. Hum Reprod. 2012;27(8):2304–2311. doi: 10.1093/humrep/des175. [DOI] [PubMed] [Google Scholar]
- 24.Guzeloglu-Kayisli O, Lalioti MD, Aydiner F, et al. Embryonic poly (A)-binding protein (EPAB) is required for oocyte maturation and female fertility in mice. Biochem J. 2012;446(1):47–58. doi: 10.1042/BJ20120467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tay J, Richter JD. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell. 2001;1(2):201–213. doi: 10.1016/s1534-5807(01)00025-9. [DOI] [PubMed] [Google Scholar]
- 26.Ruggiu M, Speed R, Taggart M, et al. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389(6646):73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 27.Lin Y, Page DC. Dazl deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. Dev Biol. 2005;288(2):309–316. doi: 10.1016/j.ydbio.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 28.Racki WJ, Richter JD. CPEB controls oocyte growth and follicle development in the mouse. Development. 2006;133(22):4527–4537. doi: 10.1242/dev.02651. [DOI] [PubMed] [Google Scholar]
- 29.Chen J, Melton C, Suh N, et al. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011;25(7):755–766. doi: 10.1101/gad.2028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clegg KB, Pikó L. RNA synthesis and cytoplasmic polyadenylation in the one-cell mouse embryo. Nature. 1982;295(5847):343–344. doi: 10.1038/295342a0. [DOI] [PubMed] [Google Scholar]
- 31.Flach G, Johnson MH, Braude PR, Taylor RA, Bolton VN. The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J. 1982;1(6):681–686. doi: 10.1002/j.1460-2075.1982.tb01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332(6163):459–461. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- 33.Biggers JD, Whittingham DG, Donahue RP. The pattern of energy metabolism in the mouse oöcyte and zygote. Proc Natl Acad Sci U S A. 1967;58(2):560–567. doi: 10.1073/pnas.58.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardner DK, Leese HJ. The role of glucose and pyruvate transport in regulating nutrient utilization by preimplantation mouse embryos. Development. 1988;104(3):423–429. doi: 10.1242/dev.104.3.423. [DOI] [PubMed] [Google Scholar]
- 35.Gott AL, Hardy K, Winston RM, Leese HJ. Non-invasive measurement of pyruvate and glucose uptake and lactate production by single human preimplantation embryos. Hum Reprod. 1990;5(1):104–108. doi: 10.1093/oxfordjournals.humrep.a137028. [DOI] [PubMed] [Google Scholar]
- 36.Conaghan J, Hardy K, Handyside AH, Winston RM, Leese HJ. Selection criteria for human embryo transfer: a comparison of pyruvate uptake and morphology. J Assist Reprod Genet. 1993;10(1):21–30. doi: 10.1007/BF01204436. [DOI] [PubMed] [Google Scholar]
- 37.Conaghan J, Handyside AH, Winston RM, Leese HJ. Effects of pyruvate and glucose on the development of human preimplantation embryos in vitro. J Reprod Fertil. 1993;99(1):87–95. doi: 10.1530/jrf.0.0990087. [DOI] [PubMed] [Google Scholar]
- 38.Gardner DK, Lane M, Stevens J, Schoolcraft WB. Noninvasive assessment of human embryo nutrient consumption as a measure of developmental potential. Fertil Steril. 2001;76(6):1175–1180. doi: 10.1016/s0015-0282(01)02888-6. [DOI] [PubMed] [Google Scholar]
- 39.Leese HJ, Barton AM. Pyruvate and glucose uptake by mouse ova and preimplantation embryos. J Reprod Fertil. 1984;72(1):9–13. doi: 10.1530/jrf.0.0720009. [DOI] [PubMed] [Google Scholar]
- 40.Brison DR, Leese HJ. Energy metabolism in late preimplantation rat embryos. J Reprod Fertil. 1991;93(1):245–251. doi: 10.1530/jrf.0.0930245. [DOI] [PubMed] [Google Scholar]
- 41.Botros L, Sakkas D, Seli E. Metabolomics and its application for non-invasive embryo assessment in IVF. Mol Hum Reprod. 2008;14(12):679–690. doi: 10.1093/molehr/gan066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hardy K, Hooper MA, Handyside AH, Rutherford AJ, Winston RM, Leese HJ. Non-invasive measurement of glucose and pyruvate uptake by individual human oocytes and preimplantation embryos. Hum Reprod. 1989;4(2):188–191. doi: 10.1093/oxfordjournals.humrep.a136869. [DOI] [PubMed] [Google Scholar]
- 43.Turner K, Martin KL, Woodward BJ, Lenton EA, Leese HJ. Comparison of pyruvate uptake by embryos derived from conception and non-conception natural cycles. Hum Reprod. 1994;9(12):2362–2366. doi: 10.1093/oxfordjournals.humrep.a138453. [DOI] [PubMed] [Google Scholar]
- 44.Devreker F, Hardy K, Van den Bergh M, Vannin AS, Emiliani S, Englert Y. Amino acids promote human blastocyst development in vitro. Hum Reprod. 2001;16(4):749–756. doi: 10.1093/humrep/16.4.749. [DOI] [PubMed] [Google Scholar]
- 45.Lane M, Gardner DK. Nonessential amino acids and glutamine decrease the time of the first three cleavage divisions and increase compaction of mouse zygotes in vitro. J Assist Reprod Genet. 1997;14(7):398–403. doi: 10.1007/BF02766148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houghton FD, Hawkhead JA, Humpherson PG, et al. Non-invasive amino acid turnover predicts human embryo developmental capacity. Hum Reprod. 2002;17(4):999–1005. doi: 10.1093/humrep/17.4.999. [DOI] [PubMed] [Google Scholar]
- 47.Leese HJ, Baumann CG, Brison DR, McEvoy TG, Sturmey RG. Metabolism of the viable mammalian embryo: quietness revisited. Mol Hum Reprod. 2008;14(12):667–672. doi: 10.1093/molehr/gan065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brison DR, Houghton FD, Falconer D, et al. Identification of viable embryos in IVF by non-invasive measurement of amino acid turnover. Hum Reprod. 2004;19(10):2319–2324. doi: 10.1093/humrep/deh409. [DOI] [PubMed] [Google Scholar]
- 49.Seli E, Botros L, Sakkas D, Burns DH. Noninvasive metabolomic profiling of embryo culture media using proton nuclear magnetic resonance correlates with reproductive potential of embryos in women undergoing in vitro fertilization. Fertil Steril. 2008;90(6):2183–2189. doi: 10.1016/j.fertnstert.2008.07.1739. [DOI] [PubMed] [Google Scholar]
- 50.Nicholson JK, O’Flynn MP, Sadler PJ, Macleod AF, Juul SM, Sönksen PH. Proton-nuclear-magnetic-resonance studies of serum, plasma and urine from fasting normal and diabetic subjects. Biochem J. 1984;217(2):365–375. doi: 10.1042/bj2170365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1(2):153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 52.Dunn WB, Ellis DI. Metabolomics: Current analytical platforms and methodologies. Trends Analyt Chem. 2005;24(4):285–294. [Google Scholar]
- 53.Garcia A, Barbas C. Gas chromatography-mass spectrometry (GC-MS)-based metabolomics. Methods Mol Biol. 2011;708:191–204. doi: 10.1007/978-1-61737-985-7_11. [DOI] [PubMed] [Google Scholar]
- 54.Zhou B, Xiao JF, Tuli L, Ressom HW. LC-MS-based metabolomics. Mol Biosyst. 2012;8(2):470–481. doi: 10.1039/c1mb05350g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soga T. Capillary electrophoresis-mass spectrometry for metabolomics. Methods Mol Biol. 2007;358:129–137. doi: 10.1007/978-1-59745-244-1_8. [DOI] [PubMed] [Google Scholar]
- 56.Monton MR, Soga T. Metabolome analysis by capillary electrophoresis-mass spectrometry. J Chromatogr A. 2007;1168(1–2):237–246. doi: 10.1016/j.chroma.2007.02.065. discussion 236. [DOI] [PubMed] [Google Scholar]
- 57.Büscher JM, Czernik D, Ewald JC, Sauer U, Zamboni N. Cross-platform comparison of methods for quantitative metabolomics of primary metabolism. Anal Chem. 2009;81(6):2135–2143. doi: 10.1021/ac8022857. [DOI] [PubMed] [Google Scholar]
- 58.Ellis DI, Goodacre R. Metabolic fingerprinting in disease diagnosis: biomedical applications of infrared and Raman spectroscopy. Analyst (Lond) 2006;131(8):875–885. doi: 10.1039/b602376m. [DOI] [PubMed] [Google Scholar]
- 59.Bajad S, Shulaev V. LC-MS-based metabolomics. Methods Mol Biol. 2011;708:213–228. doi: 10.1007/978-1-61737-985-7_13. [DOI] [PubMed] [Google Scholar]
- 60.Shulaev V. Metabolomics technology and bioinformatics. Brief Bioinform. 2006;7(2):128–139. doi: 10.1093/bib/bbl012. [DOI] [PubMed] [Google Scholar]
- 61.Alpaydin E. Introduction to Machine Learning. 2nd ed. Cambridge, MA: MIT Press; 2010. [Google Scholar]
- 62.Kim JH. Estimating classification error rate: Repeated cross-validation, repeated hold-out and bootstrap. Comput Stat Data Anal. 2009;53(11):3735–3745. [Google Scholar]
- 63.Mehrotra B, Mendes P. Bioinformatics approaches to integrate metabolomics and other systems biology data. In: Saito K, Dixon RA, Willmitzer L, editors. Plant Metabolomics. Vol 57. Springer-Verlag: Berlin, Heidelberg; 2006. pp. 105–115. [Google Scholar]
- 64.Ahlström A, Wikland M, Rogberg L, Barnett JS, Tucker M, Hardar-son T. Cross-validation and predictive value of near-infrared spectroscopy algorithms for day-5 blastocyst transfer. Reprod Biomed Online. 2011 Jan 24; doi: 10.1016/j.rbmo.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 65.Seli E, Bruce C, Botros L, et al. Receiver operating characteristic (ROC) analysis of day 5 morphology grading and metabolomic Viability Score on predicting implantation outcome. J Assist Reprod Genet. 2011;28(2):137–144. doi: 10.1007/s10815-010-9501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katz-Jaffe MG, Gardner DK, Schoolcraft WB. Proteomic analysis of individual human embryos to identify novel biomarkers of development and viability. Fertil Steril. 2006;85(1):101–107. doi: 10.1016/j.fertnstert.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 67.Sfontouris IA, Lainas GT, Sakkas D, et al. Assessment of embryo selection using non-invasive metabolomic analysis as an adjunct to morphology indicates improvement in implantation and fetal cardiac activity rates. Hum Reprod. 2011;26(Suppl 1):86. [Google Scholar]