Abstract
To track the activity of cellular signaling molecules within the endogenous cellular environment, researchers have developed a diverse set of genetically encodable fluorescent biosensors. These sensors, which can be targeted to specific subcellular regions to monitor specific pools of a given signaling molecule in real time, rely upon conformational changes in a sensor domain to alter the photophysical properties of green fluorescent protein (GFP) family members. In this introductory chapter, we first discuss the properties of GFP family members before turning our attention to the design and application of genetically encodable fluorescent biosensors to live cell imaging.
Keywords: Fluorescent proteins, Fluorescent biosensor, Biosensor design, FRET, Cell signaling, Live cell imaging
1 Introduction
Cells do not exist in a static environment. Instead, they must simultaneously sense multiple extracellular cues and respond accordingly. This task is accomplished by a host of cellular signaling molecules whose activities are precisely coordinated by endogenous regulatory factors. Together, signaling molecules and their cofactors form tightly regulated signaling networks that impact nearly every aspect of cellular physiology. Therefore, to better understand how individual signaling molecules are controlled inside cells, it is necessary to develop tools that are able to monitor their activities within the context of intact, fully functional signaling networks. To this end, researchers have recently developed a diverse set of genetically encodable fluorescent biosensors designed to probe dynamic cellular events in living cells with high spatial and temporal resolution. These sensors, which typically involve the fusion of green fluorescent protein (GFP) or a related fluorescent protein (FP) color variant(s) into the primary amino acid sequence of a protein or a selected protein domain, have enabled researchers to track various components of intracellular signaling networks in real time within the native cellular environment.
In this chapter, we provide a general overview of genetically encodable fluorescent biosensors. To this end, we will briefly examine the properties of GFP and several FP family members commonly used to construct fluorescent biosensors before turning our attention to the development and application of fluorescent biosensors for studying dynamic signaling processes in living cells.
2 Fluorescent Proteins for Biosensor Development
Aequorea victoria GFP (avGFP) is the founding member of a large family of autofluorescent proteins isolated from bioluminescent marine organisms such as hydrozoa and reef-building corals [1, 2]. FP family members, which require only molecular oxygen to generate their intrinsically derived fluorophores, form a highly stable 11-stranded β-barrel structure whose architecture aids in both the formation and stabilization of the conjugated ring systems that account for their spectral properties (Fig. 1a) [1, 2]. Importantly, in addition to the extent of conjugation within the ring system, the fluorescent properties of FP fluorophores can also be substantially influenced by their local protein microenvironment [1, 3]. For instance, the p-hydroxylbenzylideneimidazolinone (p-HBI) fluorophore generated by wild-type avGFP is formed through a three- step autocatalytic cyclization reaction involving a triplet of amino acids, Ser65-Tyr66-Gly67, located along a central α-helix that runs through the interior of the β-barrel (Fig. 1a) [1–3]. In the context of the popular avGFP derivative, enhanced GFP (EGFP), the p-HBI chromophore emits green light (507 nm) following excitation with 484 nm light. Meanwhile, the excitation and emission spectra of this same p-HBI species is red-shifted by ~20 nm in the closely related color variant, enhanced yellow FP (EYFP) [4, 5]. The dramatic changes observed in the spectral properties of the EYFP fluorophore are attributed to π-stacking interactions caused by the introduction of a Tyr residue (T203Y) in the vicinity of the chromophore (Fig. 1a). In fact, together with the surprising tolerance of the avGFP fluorophore to mutation (only Gly67 appears to be essential for its formation), modification and refinement of the protein microenvironment surrounding the fluorophore is a primary focus in the development of many of the avGFP color variants used to construct fluorescent biosensors [6, 7].
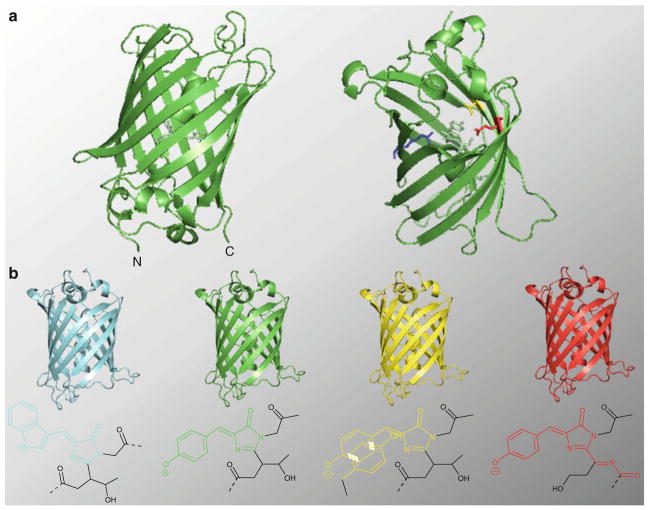
Fig. 1.
Structure and function of GFP. (a) Structure of A. victoria GFP, highlighting the protein’s β-barrel architecture. In the structure on the right, which is rotated approximately 180° along both the X and Y axes relative to the structure on the left, four residues (V68 through F71) have been removed in order to highlight the location of the fluorophore (pale green) on the alpha helix that runs through the core of the protein. Two residues, E222 (red) and R96 (blue), involved both in fluorophore maturation and the formation of the hydrogen bonding network are shown. In addition, the position of T203 (yellow), which is mutated to Tyr in YFP variants, is shown. (b) FP color variants frequently used in biosensor development, along with the structure of their fluorophores, are shown. The conjugated ring structure responsible for the spectral properties of each fluorophore is colored accordingly. The fluorophore structures correspond to those found in ECFP (Ex: 439 nm; Em: 476 nm), EGFP (Ex: 484 nm; Em: 507 nm), EYFP (Ex: 514 nm; Em: 527 nm), and mCherry (Ex: 587 nm; Em: 610 nm). All protein structures were generated using Pymol using coordinates for avGFP (PDB ID: 1gfl) originally published by Yang et al. [57]
The FP toolbox contains a broad range of color variants whose emission profiles span most of the visible spectrum. Though in reality there is no clear line of demarcation between them, FP family members are often divided into seven spectral classes according to their emission maxima. These include blue FPs (BFPs), which emit between 440–470 nm, cyan FPs (CFPs; 471–500 nm), GFPs (501–520 nm), YFPs (521–550 nm), orange FPs (OFPs; 551–575 nm), red FPs (RFPs: 576–610 nm) and far-red FPs (FRFPs; 611–660 nm). In this chapter, we will examine the molecular characteristics of representative members from each of the spectral classes most commonly used in biosensor development, namely CFP, GFP, YFP, and RFP family members (Fig. 1a). Though an in-depth discussion of the many FP color variants currently available for live cell imaging is beyond the scope of this chapter, the interested reader is referred to our recent reviews on the subject, [8, 9] as well as excellent reviews by Day and Davidson [10] and Pakhomov and Martynov [11].
2.1 Green Fluorescent Protein
As alluded to above, avGFP contains an intrinsically derived p-HBI fluorophore that emits green light when excited with 396 nm light. Early studies of avGFP revealed that, in the context of the wild-type protein, the p-HBI fluorophore alternates between two ion-ization states—a weakly fluorescent, neutral species dubbed the A state (Ex: 396 nm, Em: 508 nm) and a highly fluorescent phenolate species termed the B state (Ex: 475 nm; Em: 503 nm) [12]. Due to the stabilizing effects of an extensive hydrogen bonding network within the protein core, the A state is nearly six times more highly populated than the B state [1, 2]. However, upon excitation, the A state is converted to the highly fluorescent anionic species (i.e., the B state) through a phenomenon known as excited state proton transfer (ESPT) [12]. During ESPT, a nearby glutamate residue, E222, abstracts a high energy proton from the phenol moiety of Tyr66 [13]. The conversion of E222 to a neutral acid not only relieves the electrostatic repulsion which destabilizes the phenolate oxyanion of B prior to excitation, but it also triggers structural rearrangements in the protein core which further stabilize the B state after excitation [13].
Though the photoisomerization of the avGFP fluorophore is intriguing from both a biological and a photochemical perspective, this behavior is not ideal for biosensor development. For instance, the existence of two isomerization states with distinct excitation/emission profiles can complicate the interpretation of experimental data. Moreover, the ultraviolet (UV) light required to excite the A state not only induces cellular autofluorescence, but it can also adversely affect the cellular system under study. Therefore, several mutations have been introduced into the wild-type protein that simplify its excitation/emission spectra and improve its spectral properties for live cell imaging. One of these mutations, the substitution of a Thr residue for Ser at position 65 (S65T), introduces a single methyl group into the fluorophore [14]. Though this mutation does not dramatically affect the structure of the fluorophore itself, it has a profound impact on the spectral properties of the resultant GFP variant. This is because the methyl group causes local conformational changes in the protein core that disrupt the ground state hydrogen bonding network originally involved in stabilization of the A state [13, 14]. As a consequence, the resulting fluorophore is converted almost exclusively to the phenolate species in the unexcited state, simplifying its excitation/emission spectra and making it better suited for live cell imaging. For this reason, the S65T mutation, along with a series of mutations that improve the expression and maturation of avGFP in mammalian cells, has been incorporated into EGFP (avGFP/F64L/S65T). For both practical and historical reasons, EGFP is the most widely used FP in the biological sciences and the starting point for the development of many FP-based biosensors.
2.2 Yellow Fluorescent Proteins
Due to their relatively high extinction coefficients (ε) and quantum yields (ϕ), YFPs represent some of the brightest monomeric FPs reported to date [10, 14]. As alluded to above, FPs from this class are generated by the introduction of an aromatic Tyr residue near the phenolate ion of the p-HBI fluorophore (T203Y) [1, 14]. This mutation promotes π–π stacking interactions that stabilize the excited state dipole moment of the fluorophore, shifting the excitation and emission spectra of YFP variants by ~20 nm toward the red end of the spectrum.
The introduction of a bulky Tyr residue within the protein core is believed to disrupt internal hydrogen bonding networks and reduce steric packing interactions around the fluorophore [5]. As a consequence, YFP family members often exhibit acute sensitivities to several cellular parameters. Though not ideal for general biosensor development, the environmental sensitivities of YFP family members have been exploited to probe changes in the cellular environment. For example, EYFP, a first generation YFP derived from EGFP, exhibits a relatively high pKa (pKa = 6.2) and is highly sensitive to halide ions [5, 15]. The acute sensitivity of EYFP to these cellular parameters appears to be related to the stabilization of the weakly fluorescent neutral form of p-HBI at low pH or in the presence of a bound halide ion in the vicinity of the fluorophore. By exploiting these sensitivities, researchers have designed a series of EYFP-based biosensors capable of tracking dynamic changes in the concentration of halide ions and/or fluctuations in pH within different subcellular compartments [16–18].
To produce a YFP variant that is useful for general biosensor development, Griesbeck et al. employed a molecular evolution strategy to produce mCitrine (EYFP-V68L, Q69M) [19]. Compared to the parent EYFP species, mCitrine exhibits a markedly reduced sensitivity to halide ions, better photostability, and a lower pKa (pKa = 5.7). Structural analysis revealed that the reduced halide sensitivity exhibited by mCitrine is likely conferred by a M69Q point mutation that plugs a large halide-binding pocket next to the fluorophore [19]. By preventing the binding of halide ions in the vicinity of the fluorophore, this substitution relieves repulsive forces caused by the binding of a negatively charged halide ion in close proximity to the fluorophore which destabilizes the highly fluorescent phenolate anion in the parent species. As a consequence, the phenolate form of p-HBI is more highly populated than it is in EYFP. Moreover, the tight packing interactions caused by the introduction of the neutral methionine side chain within the core of the protein are believed to further stabilize the phenolate species. Together, these modifications help to shift the equilibrium away from the neutral form of the p-HBI chromophore and toward the phenolate anion, leading to the enhanced fluorescence exhibited by this variant. In addition to mCitrine, another popular EYFP variant, Venus, also exhibits enhanced photostability and reduced sensitivity to both pH and fluctuations in halide ion concentrations [20]. As a consequence, mCitrine and Venus have been incorporated into a large number of biosensors.
2.3 Cyan Fluorescent Proteins
Perhaps surprisingly, the SYG tripeptide used to generate the p-HBI fluorophore found in wild-type avGFP is quite tolerant of substitutions. This conformational flexibility permits the spontaneous formation of several different fluorophore structures, each of which exhibits unique photophysical properties. For instance, in the case of CFP family members derived from avGFP, substitution of Trp for Y66 (Y66W) results in the formation of a fluorophore with an indole moiety in place of the phenol ring [21]. As a consequence, the excitation and emission wavelengths of CFPs are blue- shifted relative to the parent protein, giving members of this family a bluish-green appearance following excitation with ~450 nm light. However, because the protein core of the parent avGFP species is designed to accommodate the phenol-containing p-HBI species, modification of the fluorophore structure often comes at a price. For instance, the indole-containing fluorophore present in the enhanced CFP (ECFP) variant behaves as two distinct species, each characterized by a unique fluorescence lifetime decay curve and exhibiting distinct excitation and emission spectra. Using the crystal structure of ECFP as a guide, Rizzo and colleagues hypothesized that the distinct fluorogenic species observed in ECFP arise from isostable conformations of the protein caused by the dynamic exchange of two bulky residues, Tyr145 and His148, in the region surrounding the rigid fluorophore [22]. Therefore, to eliminate the biphasic character of ECFP, a combination of site-directed and random mutagenesis was employed to produce mCerulean (ECFP-S72A/H148D/Y154A), a 2.5-fold brighter ECFP variant that exhibits a monophasic decay curve [22]. The key mutation in mCerulean is a H148D substitution that eliminates the exchange between Tyr145 and His148, thus stabilizing the first conformational state and improving the fluorescence lifetime characteristics of the protein. Recently, the photophysical properties of mCeru-lean were further improved by (1) sealing a gap between β-strands 7 and 8 in the β-barrel and (2) converting T65 in ECFP back to the Ser residue found in wild-type avGFP [22]. By improving the stability and ϕ of the fluorophore, these mutations increase the relative brightness of the resulting CFP variant, mCerulean3, by approximately 65 % compared to mCerulean. Likewise, the recently described CFP variant, mTurquoise, which contains the same T65S mutation as mCerulean3, is 1.5 times brighter than mCerulean and exhibits a monoexponential decay curve [23]. Together, their photophysical properties suggest that these new CFP variants will be broadly useful in biosensor development (e.g., see Chapter 4 in this textbook and ref. 24).
2.4 Red Fluorescent Proteins
In the same way that the spectral properties of CFP color variants can be altered by direct chemical modification of their fluorophores, the fluorophores generated by RFP family members also exhibit altered chemical structures whose excitation and emission spectra are shifted relative to the p-HBI species utilized by avGFP. However, unlike CFP variants and other avGFP derivatives, red-emitting FPs undergo a second oxidation step along the Cα-N bond of residue 65 (according to the GFP numbering system) to form an acylimine linkage with the polypeptide backbone [25, 26]. As a consequence, the conjugated π-system of RFP-derived fluorophores are extended over a greater distance, thereby lowering the energy barrier separating the ground (S0) and excited states (S1) of the fluorophore and leading to red-shifted excitation and emission spectra [6, 27, 28].
Importantly, the longer wavelength light used to excite RFPs (>550 nm) substantially reduces the effects of cellular autofluorescence caused by the excitation of abundant fluorescent biomolecules inside the cell. Moreover, the light emitted by RFPs (>580 nm) is less susceptible to interference, such as light scattering, that can reduce the signal-to-noise ratio in an imaging experiment. Together, these spectral properties make RFPs particularly attractive for deep tissue and whole-body imaging, where the excitation and emission light must traverse several layers of cells. As a consequence, much effort has been devoted to the development of monomeric RFP variants suitable for biosensor development. These efforts have culminated in the evolution of two major families of mRFPs: the mFruits and a series of eqFP578 derivatives consisting of TagRFP [29], TagRFP-T [30], mKate [31], mKate2 [32], and Neptune [33]. Together, these proteins provide researchers with several monomeric FPs that are excited at wavelengths >550 nm. Though no single variant is optimal for all applications, the photophysical properties of mCherry, mRuby2 [34], and TagRFP-T make them the best general-purpose mRFPs for biosensor development.
3 Design and Application of Fluorescent Protein-Based Biosensors
Aside from allowing multiple cellular parameters, such as protein dynamics or gene expression profiles, to be probed simultaneously, the availability of spectrally distinct FP variants has also prompted the development of novel imaging techniques, such as fluorescence resonance energy transfer (FRET)-based approaches. Such techniques extend the application of FP technologies, enabling researchers to visualize other dynamic cellular events, such as the turnover of small molecule second messengers like Ca2+ and cyclic AMP (cAMP) or the activity profiles of cellular signaling enzymes. In some cases, two or more cellular parameters can even be measured simultaneously in the same cell using biosensors that employ three spectrally distinct FPs, such as CFP, YFP, and RFP (e.g., see Chapter 16 in this textbook). Below, we explore several of the design principles used to construct FP-based biosensors and briefly discuss some of the ways in which these sensors have been used to gain unique insights into the activation and regulation of cellular signaling molecules within their native cellular environment. During the course of this discussion, we will also highlight several of the key parameters that must be considered when utilizing fluorescent biosensors for live cell imaging. These include parameters that are intrinsic to the sensors themselves, such as sensitivity, reversibility, response kinetics and dynamic range, as well as cellular factors, such as interference from endogenous proteins or small molecules, which can impact sensor performance.
Under certain circumstances, biosensors can be constructed based on the intrinsic environmental sensitivities of FP color variants, as illustrated by the EYFP-based probes discussed above. However, in addition to environmental parameters, such as pH and halide ion concentration, FP-based biosensors can also be engineered to directly sense other important cellular parameters. In general, engineered fluorescent biosensors contain two basic components: (1) a “sensor unit”, which undergoes a conformational change in response to a given cellular parameter and (2) a “reporter unit”, which converts the induced conformational change into a fluorescent readout (Fig. 2) [8]. In the case of FP-based sensors, the reporter unit usually consists of a pair of FPs that undergo FRET or a single FP whose fluorescent properties are altered in response to a conformational change in the sensor unit. Meanwhile, the molecular switch utilized by the sensor unit can be generated in many different ways, provided that it promotes a conformational change in response to the cellular parameter under study. For instance, a relatively simple molecular switch is employed by two related classes of redox-sensitive FP indicators known as roGFPs and rxYFPs [35]. These biosensors, which change either their spectral properties (roGFPs) or fluorescence intensity (rxY-FPs) in response to changes in the cellular redox potential, use disulfide bond formation between pairs of carefully positioned cysteine residues on the surface of their respective FPs to induce conformational changes in the FP [35]. In the case of rxYFP, structural analysis suggests that disulfide bond formation leads to the reorganization of residues in close proximity to the fluorophore. Reorganization of the fluorophore microenvironment, particularly the hydrogen bonding network, shifts the equilibrium between the neutral (A) state and the phenolic (B) state, leading to weak fluorescence in oxidizing environments and increased fluorescence under reducing conditions [36].
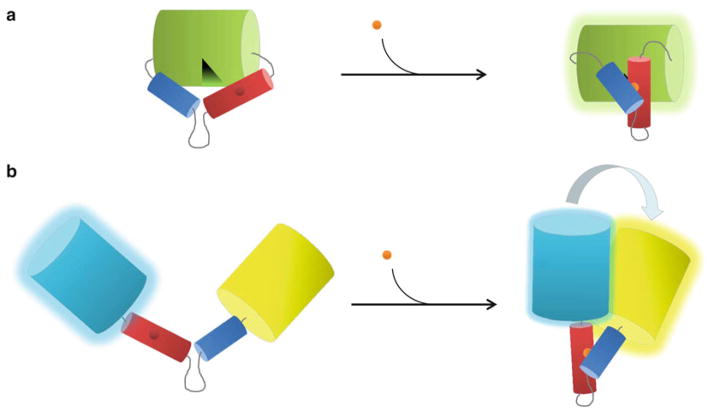
Fig. 2.
FP-based biosensor designs. (a) Cartoon depicting a single FP biosensor based on an engineered molecular switch. In this design, a sensor domain composed of a receiver unit (red) and a switching unit (blue) is grafted into a circularly permuted version of GFP (green cylinder). In the unbound “open” state, a hole in the β-barrel created by circular permutation allows bulk solvent access to the fluorophore, quenching its fluorescence. In the presence of ligand (orange sphere), the sensor domain undergoes a conformational change that plugs the hole and increases fluorescence (green glow). (b) Cartoon depicting a FRET-based FP biosensor using the same engineered molecular switch described in (a). In this case, conformational changes induced by ligand binding alter the relative distance and orientation of the donor CFP (cyan cylinder) and the acceptor YFP (yellow cylinder), increasing FRET between them
Though roGFPs and rxYFPs employ a relatively simple molecular switch to convert changes in the cellular environment into a fluorescence output, in general, more complex switches are necessary to detect changes in other cellular parameters. Under these circumstances, the molecular switch can be derived from a conformational change intrinsic to an endogenous protein or it can be generated via an engineered switch [8]. By combining the molecular switch with an appropriate reporter unit—either by flanking the switch region with complementary FRET pairs or by grafting it into the FP itself—conformational changes in the sensor unit can be translated into a fluorescence readout from the reporter unit (Fig. 2). For instance, to construct a biosensor that monitors the NADH/NAD+ redox state, Hung et al. employed an intrinsic molecular switch based on the bacterial NADH binding protein, Rex [37]. In this sensor design, a circularly permuted version of the GFP variant, T-Sapphire (cpT-Sapphire), is grafted into the Rex protein between two NADH binding domains. In the absence of NADH, the two domains do not interact with one another, straining the structure of cpT-Sapphire and causing a decrease in GFP fluorescence. Upon binding of NADH (but not NAD+), Rex undergoes a conformational change that restores the native structure of cpT-Sapphire and increases its fluorescence intensity. As a consequence, the resulting sensor, termed Paradox, is able to sense the NADH/NAD+ redox state in different cytosolic regions or in response to different environmental stimuli (for more information, please see the Paradox imaging protocol described in Chapter 7 in this textbook) [37].
Similarly, the Palmer lab recently developed a series of FRET- based biosensors based on an intrinsic molecular switch to measure intracellular Zn2+ concentrations ([Zn2+]i) in multiple subcellular regions [38–40]. These Zn-sensors, which can be calibrated either in vitro or in cells to measure absolute [Zn2+]i (see Chapter 3 in this textbook), exploit a structural rearrangement in the canonical Cys2His2 Zn2+ finger domain of the transcription factor, Zif268, to change the relative distance/orientation of flanking FP color variants [38]. In the absence of Zn2+, the Zn2+-finger domain is largely unstructured, leading to large degree of separation between the FPs in space. Zn2+ binding causes the domain to fold into a compact structure that brings the FPs into close proximity with one another, facilitating FRET between them. Importantly, the Zn2+ binding affinity of Zn-sensor can be reduced ~100-fold by substituting His for Cys in the Cys2His2 motif [38]. Due to its reduced sensitivity for Zn2+, the resulting His4 Zn-sensor is able to detect changes in intracellular Zn2+ at concentrations that would otherwise saturate biosensors based on the canonical Cys2His2 motif. Thus, by tuning the sensitivity of the sensor, researchers have effectively expanded the range of [Zn2+]i that can be reliably measured in cells [38]. Together, this family of Zn-sensors has been used to gain a better understanding of [Zn2+]i regulation in several organelles, including the cytosol [38], ER [40], Golgi [40], and mitochondria [39].
In contrast to the intrinsic molecular switches utilized by Paradox and Zn-sensor, which each rely upon a naturally occuring conformational change to alter the fluorescent properties of the reporter unit, engineered switches can be constructed based on a modular design. Engineered molecular switches typically consist of a “receiver” module and a “switching” module. The receiver module “senses” the cellular parameter under study (be it through the binding of a small molecule or via posttranslational modification by a signaling enzyme) while the switching module converts changes in the receiver module into a conformational change that alters the fluorescent properties of the FPs in the reporter unit (Fig. 2). For instance, two of the most popular families of genetically targetable Ca2+ sensors, the GCaMPs and yellow cameleons (YC’s), both rely upon an engineered molecular switch composed of the Ca2+ binding protein, calmodulin (CaM), and the CaM-binding peptide, M13 [5, 41–46]. In the case of the GCaMPs, the CaM-M13 switch is fused to a circularly permuted version of EGFP. In the presence of Ca2+, CaM binds M13 and induces a conformational change in the reporter that leads to an increase in its fluorescence intensity. In the context of the FRET-based YC’s, in which the same CaM-M13 switch is sandwiched between a CFP-YFP FRET pair, the Ca2+-dependent conformational change in CaM-M13 alters the distance and orientation of the CFP donor relative to the YFP acceptor, altering their FRET emission ratio. Importantly, because the CaM-M13 interaction is readily reversible, both YC’s and GCaMPs can report transient changes in intracellular Ca2+ levels ([Ca2+]i) in real-time. Moreover, since the CaM-based receiver module is tethered directly to the M13-based switching module, the response kinetics of the sensors is largely determined by the intrinsic rates of association and dissociation of Ca2+-CaM for M13.
Aside from simplifying the response kinetics, the modular design of the sensor domain also ensures that the effective concentrations of the two halves of the molecular switch are relatively high in the context of the sensor. As a consequence, in most cases, the performance of GCaMP and YC family members is not dramatically influenced by nonproductive interactions with endogenous binding partners. Nonetheless, there are circumstances in which interference becomes a concern. For instance, in cellular environments where the concentration of endogenous CaM is extremely high, such as at the plasma membranes of hippocampal neurons, the ability of YC biosensors to report changes in [Ca2+]i is hindered by interactions with endogenous CaM. Thus, to detect changes in [Ca2+]i in regions of high endogenous CaM, the Tsien lab used a “bump and hole” strategy to create CaM and M13 variants that exhibit a very low affinity for wild-type CaM but were still able to interact with one another efficiently [45].
A modular design similar to that described above has been used to construct engineered molecular switches for a wide range of cellular parameters. For instance, in order to monitor changes in the activity profiles of different protein kinases inside the cell, a series of FRET-based kinase activity reporters have been developed based on an engineered molecular switch [47]. In this type of sensor, a short peptide sequence that is specifically phosphorylated by the kinase-of-interest serves as the receiver module while a phospho-amino acid binding domain (PAABD) that reversibly associates with the phosphorylated form of the substrate region serves as the switching module. In order to detect both activation and attenuation of kinase activity, the PAABD must be carefully selected when designing kinase activity reporters. Ideally, the PAABD would associate with the phosphorylated form of the substrate region rapidly but not bind so tightly as to prevent dephosphorylation by cellular phosphatases. For instance, though the first protein kinase A (PKA) activity sensor, AKAR1, accurately reported increases in PKA activity, the high affinity of the 14-3-3τ PAABD employed by this sensor blocked access of cellular phosphosphatases to the substrate domain, preventing its dephosphorylation once the activity of PKA decreased [48]. Therefore, to improve the reversibility of subsequent AKAR variants, 14-3-3τ was replaced with the forkhead associated 1 (FHA1) domain from Rad53p [49–51]. The FHA1 domain exhibits a lower Kd for the phosphorylated substrate domain than 14-3-3, allowing both the activation and attentuation of PKA to be visualzed in cells. As described in Chapter 10, kinase activity reporters can be targeted to specific subcellular compartments through the incorporation of short targeting sequences, allowing researchers to gain important insights into the regulation of specific pools of a given kinase under various cellular conditions.
It is important to note that, due to the modular design of their sensor unit, unimolecular FRET-based probes that utilize an engineered molecular switch can be converted into bimolecular probes simply by removal of the flexible linker between the receiver domain and the switching domain (Fig. 3). One of the primary advantages of a bimolecular design is that the reporter typically exhibits a larger dynamic range than its unimolecular counterpart [52]. Presumably, this is due to lower basal FRET caused by a large degree of separation between the donor and acceptor fluorophores in the uninduced state [41, 53, 54]. However, despite this potential advantage, bimolecular FRET-based reporter systems also present researchers with unique challenges not typically encountered when using unimolecular sensors. For instance, because both halves of the sensor are required to generate a measurable FRET response, the stoichiometric ratio between them must be strictly regulated when using bimolecular reporter systems. This task is nontrivial considering the variability that often exists in parameters such as DNA transfection efficiency, transcriptional regulation, and protein translation, to name a few. As a consequence, changes in FRET are often measured differently when utilizing unimolecular versus bimolecular biosensor designs. For instance, when the stoichiometry between the donor and acceptor fluorophores is fixed, as it is in the case of unimolecular probes, the donor-to-acceptor emission ratio is generally the easiest and most convenient means of measuring changes in FRET [55]. On the other hand, if the stoichiometries between the donor and acceptor are variable, as is often the case when using bimolecular reporter systems, more sophisticated measures of FRET efficiency, such as donor fluorescence recovery after acceptor photobleaching and fluorescence lifetime imaging (FLIM), need to be used (for a more detailed discussion about commonly used FRET imaging techniques, please see Chapter 2 in this textbook) [55]. Moreover, because the two sensor halves exist independently of one another inside the cell, they are more susceptible to artifacts such as interactions with endogenous, untagged binding partners. As alluded to above, such nonproductive interactions can both reduce the sensitivity and slow the response kinetics of the sensor. With regard to response kinetics, it is also important to note that when measuring cellular processes that occur on a rapid time scale, even in the absence of interference, a bimolecular probe may exhibit slower response times compared to its unimolecular counterpart due to the fact that the two sensor halves must rely upon random diffusion to “find” one another inside the cell.
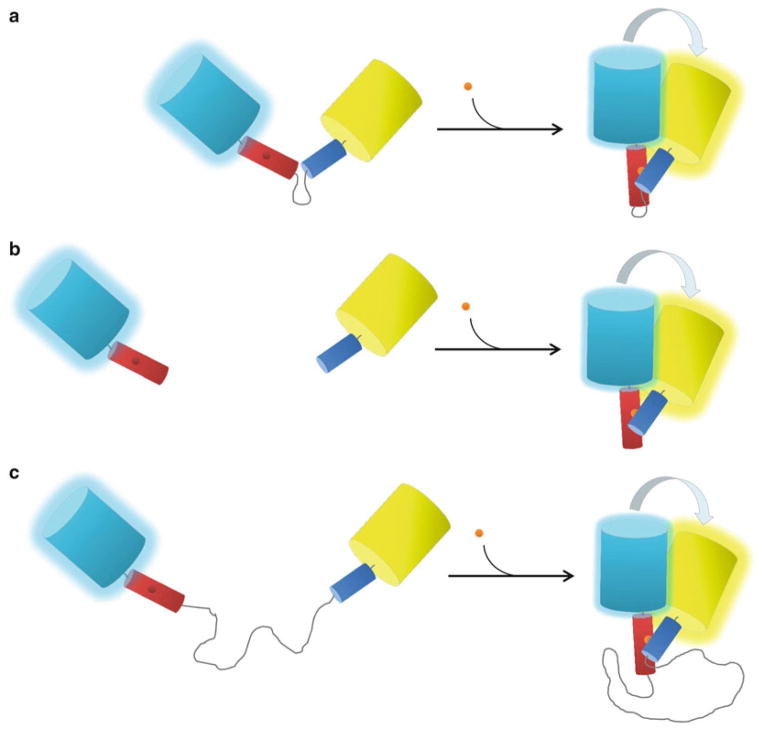
Fig. 3.
Variations on an engineered molecular switch. (a) The “classic” unimolecular biosensor design. According to this design, the receiver unit (red) is tethered to the switching unit (blue) by a short flexible linker that is typically between 5 and 15 residues in length. In the presence of ligand (orange sphere), the sensor domain undergoes a conformational change that alters the relative distance and orientation of the donor CFP (cyan cylinde r) and the acceptor YFP (yellow cylinder), increasing FRET between them. Most current FRET-based FP biosensors that employ an engineered molecular switch are constructed according to this design. (b) A bimolecular reporter design. By removing the flexible linker between the receiver and switching units, the unimolecular biosensor shown in (a) is converted to a bimolecular reporter. In the absence of ligand, each half of the sensor behaves independently of the other, leading to lower basal FRET and a higher dynamic range for the sensor (see text for details). (c) A pseudo-bimolecular biosensor design. In this design, the receiver unit and the sensor unit are separated by a very long flexible linker that is between 52 and 244 amino acids in length. As a consequence, pseudo-bimolecular reporters exhibit features of both unimolecular and bimolecular probes (see text for details)
One interesting way to improve the dynamic range of unimolecular FRET-based biosensors while retaining many of their beneficial properties was recently described by Komatsu et al., who developed a generalizable strategy to convert unimolecular FRET- based reporters into pseudo-bimolecular probes [56]. Their strategy, which involved the incorporation of an extremely long flexible linker between the receiver and switching domains, rendered the two halves of the biosensor virtually independent of one another while, at the same time, maintaining a one-to-one stoichiometry between the attached FP fluorophores (Fig. 3c). As a consequence, biosensors using the extended “EV” linker design generally exhibited an improved dynamic range relative to their predecessors while maintaining similar sensitivities and response kinetics [56].
4 Conclusions
Whether they utilize an intrinsic or an engineered molecular switch or rely on FRET or changes in FP fluorescence intensity, the FP-based biosensors outlined above have equipped researchers with the molecular tools necessary to monitor the activities of a variety of cellular signaling molecules within the native cellular environment with high spatial and temporal resolution. Due to their versatility and relative ease of use, these sensors have found applications in a variety of areas, including in situ and in vivo imaging (e.g., see Chapters 14 and 15), compound screening (e.g., see Chapter 17), and computational modeling (e.g., see Chapter 18). In the following chapters, we provide in-depth protocols describing not only how to conduct imaging experiments using these sensors, but also how the quantitative data generated from these experiments can be used to better understand the inner-workings of the cell.
References
- 1.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Zimmer M. Green fluorescent protein (GFP): applications, structure, and related photophysical behavior. Chem Rev. 2002;102:759–781. doi: 10.1021/cr010142r. [DOI] [PubMed] [Google Scholar]
- 3.Remington SJ. Fluorescent proteins: maturation, photochemistry and photophysics. Curr Opin Struct Biol. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 5.Miyawaki A, Griesbeck O, Heim R, Tsien RY. Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc Natl Acad Sci U S A. 1999;96:2135–2140. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson MW, Campbell RE. Engineered fluorescent proteins: innovations and applications. Nat Methods. 2009;6:713–717. doi: 10.1038/nmeth1009-713. [DOI] [PubMed] [Google Scholar]
- 7.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 8.Newman RH, Fosbrink MD, Zhang J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem Rev. 2011;111:3614–3666. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sample V, Newman RH, Zhang J. The structure and function of fluorescent proteins. Chem Soc Rev. 2009;38:2852–2864. doi: 10.1039/b913033k. [DOI] [PubMed] [Google Scholar]
- 10.Day RN, Davidson MW. The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev. 2009;38:2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pakhomov AA, Martynov VI. GFP family: structural insights into spectral tuning. Chem Biol. 2008;15:755–764. doi: 10.1016/j.chembiol.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Chattoraj M, King BA, Bublitz GU, Boxer SG. Ultra-fast excited state dynamics in green fluorescent protein: multiple states and proton transfer. Proc Natl Acad Sci U S A. 1996;93:8362–8367. doi: 10.1073/pnas.93.16.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brejc K, Sixma TK, Kitts PA, Kain SR, et al. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proc Natl Acad Sci U S A. 1997;94:2306–2311. doi: 10.1073/pnas.94.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ormo M, Cubitt AB, Kallio K, Gross LA, et al. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 15.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 16.Abad MF, Di Benedetto G, Magalhaes PJ, Filippin L, et al. Mitochondrial pH monitored by a new engineered green fluorescent protein mutant. J Biol Chem. 2004;279:11521–11529. doi: 10.1074/jbc.M306766200. [DOI] [PubMed] [Google Scholar]
- 17.Kneen M, Farinas J, Li Y, Verkman AS. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys J. 1998;74:1591–1599. doi: 10.1016/S0006-3495(98)77870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, et al. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci U S A. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griesbeck O, Baird GS, Campbell RE, Zacharias DA, et al. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 20.Nagai T, Ibata K, Park ES, Kubota M, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 21.Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 23.Goedhart J, van Weeren L, Hink MA, Vischer NO, et al. Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat Methods. 2010;7:137–139. doi: 10.1038/nmeth.1415. [DOI] [PubMed] [Google Scholar]
- 24.Klarenbeek JB, Goedhart J, Hink MA, Gadella TW, et al. A mTurquoise-based cAMP sensor for both FLIM and ratiometric read-out has improved dynamic range. PLoS One. 2011;6:e19170. doi: 10.1371/journal.pone.0019170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verkhusha VV, Lukyanov KA. The molecular properties and applications of Anthozoa fluorescent proteins and chromo-proteins. Nat Biotechnol. 2004;22:289–296. doi: 10.1038/nbt943. [DOI] [PubMed] [Google Scholar]
- 26.Shu X, Shaner NC, Yarbrough CA, Tsien RY, et al. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 27.Chudakov DM, Lukyanov S, Lukyanov KA. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005;23:605–613. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Wachter RM, Watkins JL, Kim H. Mechanistic diversity of red fluorescence acquisition by GFP-like proteins. Biochemistry. 2010;49:7417–7427. doi: 10.1021/bi100901h. [DOI] [PubMed] [Google Scholar]
- 29.Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 30.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, et al. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 32.Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin MZ, McKeown MR, Ng HL, Aguilera TA, et al. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol. 2009;16:1169–1179. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam AJ, St Pierre F, Gong Y, Marshall JD, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer AJ, Dick TP. Fluorescent protein-based redox probes. Antioxid Redox Signal. 2010;13:621–650. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- 36.Hanson GT, Aggeler R, Oglesbee D, Cannon M, et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 37.Hung YP, Albeck JG, Tantama M, Yellen G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dittmer PJ, Miranda JG, Gorski JA, Palmer AE. Genetically encoded sensors to elucidate spatial distribution of cellular zinc. J Biol Chem. 2009;284:16289–16297. doi: 10.1074/jbc.M900501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park JG, Qin Y, Galati DF, Palmer AE. New sensors for quantitative measurement of mitochondrial Zn(2+) ACS Chem Biol. 2012;7:1636–1640. doi: 10.1021/cb300171p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin Y, Dittmer PJ, Park JG, Jansen KB, et al. Measuring steady-state and dynamic endoplasmic reticulum and Golgi Zn2+ with genetically encoded sensors. Proc Natl Acad Sci U S A. 2011;108:7351–7356. doi: 10.1073/pnas.1015686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyawaki A, Llopis J, Heim R, McCaffery JM, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 42.Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci U S A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagai T, Yamada S, Tominaga T, Ichikawa M, et al. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci U S A. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohkura M, Matsuzaki M, Kasai H, Imoto K, et al. Genetically encoded bright Ca2+ probe applicable for dynamic Ca2+ imaging of dendritic spines. Anal Chem. 2005;77:5861–5869. doi: 10.1021/ac0506837. [DOI] [PubMed] [Google Scholar]
- 45.Palmer AE, Giacomello M, Kortemme T, Hires SA, et al. Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem Biol. 2006;13:521–530. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 46.Tallini YN, Ohkura M, Choi BR, Ji G, et al. Imaging cellular signals in the heart in vivo: cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc Natl Acad Sci U S A. 2006;103:4753–4758. doi: 10.1073/pnas.0509378103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Allen MD. FRET-based biosensors for protein kinases: illuminating the kinome. Mol Biosyst. 2007;3:759–765. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J, Ma Y, Taylor SS, Tsien RY. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Natl Acad Sci U S A. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen MD, Zhang J. Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem Biophys Res Commun. 2006;348:716–721. doi: 10.1016/j.bbrc.2006.07.136. [DOI] [PubMed] [Google Scholar]
- 50.Depry C, Allen MD, Zhang J. Visualization of PKA activity in plasma membrane microdomains. Mol Biosyst. 2011;7:52–58. doi: 10.1039/c0mb00079e. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, et al. Insulin disrupts beta-adrenergic signalling to protein kinase A in adipocytes. Nature. 2005;437:569–573. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]
- 52.Zhou X, Herbst-Robinson KJ, Zhang J. Visualizing dynamic activities of signaling enzymes using genetically encodable FRET- based biosensors from designs to applications. Methods Enzymol. 2012;504:317–340. doi: 10.1016/B978-0-12-391857-4.00016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Knopfel T, Tomita K, Shimazaki R, Sakai R. Optical recordings of membrane potential using genetically targeted voltage- sensitive fluorescent proteins. Methods. 2003;30:42–48. doi: 10.1016/s1046-2023(03)00006-9. [DOI] [PubMed] [Google Scholar]
- 54.Lundby A, Mutoh H, Dimitrov D, Akemann W, et al. Engineering of a genetically encodable fluorescent voltage sensor exploiting fast Ci-VSP voltage-sensing movements. PLoS One. 2008;3:e2514. doi: 10.1371/journal.pone.0002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 56.Komatsu N, Aoki K, Yamada M, Yukinaga H, et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang F, Moss LG, Phillips GN., Jr The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]