Abstract
Although regular exercise improves submaximal aerobic capacity, there is large variability in its response to exercise training. While this variation is thought to be partly due to genetic differences, relatively little is known about the causal genes. Submaximal aerobic capacity traits in the current report include the responses of oxygen consumption (ΔVO260), power output (ΔWORK60), and cardiac output (ΔQ60) at 60% of VO2max to a standardized 20-week endurance exercise training program. Genome-wide linkage analysis in 475 HERITAGE Family Study Caucasians identified a locus on chromosome 13q for ΔVO260 (LOD = 3.11). Follow-up fine mapping involved a dense marker panel of over 1,800 single-nucleotide polymorphisms (SNPs) in a 7.9-Mb region (21.1–29.1 Mb from p-terminus). Single-SNP analyses found 14 SNPs moderately associated with both ΔVO260 at P ≤ 0.005 and the correlated traits of ΔWORK60 and ΔQ60 at P < 0.05. Haplotype analyses provided several strong signals (P<1.0 × 10−5) for ΔVO260. Overall, association analyses narrowed the target region and included potential biological candidate genes (MIPEP and SGCG). Consistent with maximal heritability estimates of 23%, up to 20% of the phenotypic variance in ΔVO260 was accounted for by these SNPs. These results implicate candidate genes on chromosome 13q12 for the ability to improve submaximal exercise capacity in response to regular exercise. Submaximal exercise at 60% of maximal capacity is an exercise intensity that falls well within the range recommended in the Physical Activity Guidelines for Americans and thus has potential public health relevance.
Keywords: SNP, Fitness, MIPEP, SGCG, Genetics
Introduction
Numerous studies have established a strong association between low cardiorespiratory fitness and increased cardiovascular disease morbidity and mortality (Blair et al. 1996; Physical Activity Guidelines Advisory Committee 2008; US Department of Health and Human Services 1996). Maximal oxygen uptake (VO2max), the maximal amount of oxygen per unit of time that can be delivered to peripheral organs, is considered the gold standard measure of cardiorespiratory fitness. Regular exercise results in increased VO2max due to peripheral changes in muscle and to cardiovascular changes with increased stroke volume and cardiac output in normal persons.
Endurance exercise training enhances submaximal exercise capacity, which is also associated with cardiovascular health benefits (e.g. Cornelissen et al. 2009; Mendoza et al. 1991). It is believed that many of the benefits of endurance training occur at submaximal exercise intensities that are close to everyday activity levels (Larson and Bruce 1987). However, there is considerable inter-individual variation in submaximal aerobic capacity and its response to exercise training. For example, in the HERITAGE Family Study (HERITAGE) 475 adult Caucasians from 99 nuclear families completed a fully standardized 20-week endurance training program. Submaximal VO2 was measured at the power output associated with 60% of VO2max (VO260). The training program induced an average VO260 increase of about 198 mL/min with a standard deviation (SD) of 170 mL/min and a range from −231 to +780 mL/min (Perusse et al. 2001).
Results from twin and family studies suggest that genetic factors are important in determining the inter-individual variability in the response of submaximal aerobic capacity traits to exercise training (Bouchard et al. 1992; Perusse et al. 2001). Studies with monozygotic (MZ) twins trained under standardized endurance cycle exercise programs for periods of 15 or 20 weeks revealed that changes in O2 consumption measured at a given submaximal power output were characterized by a significant within MZ twin pair resemblance (Bouchard et al. 1992), suggesting that genetic factors are involved in the trainability of these phenotypes. The response of VO260 to endurance training (ΔVO260) in HERITAGE Family Study was characterized by a significant heritable component, with genetic factors accounting for 23% of the phenotypic variance (Perusse et al. 2001). However, little is known about the genes and DNA sequence variants that account for this genetic effect (Hagberg et al. 2011).
Here we present the identification of a putative quantitative trait locus (QTL) for ΔVO260 using a genome-wide linkage scan approach, followed by fine mapping of the QTL via genotyping of a dense set of over 1,800 single-nucleotide polymorphisms (SNPs) within a ∼7.9 Mb region on chromosome 13q12. The purpose of the current report is to document the association results from this dense SNP panel on chromosome 13q12.
Materials and methods
The HERITAGE Family Study was designed to investigate the role of the genotype in cardiovascular and metabolic responses to aerobic exercise training. The sample, study design, and exercise training protocol have been described elsewhere (Bouchard et al. 1995) and will be briefly summarized below.
Study design
Families having both parents and at least two offspring who were healthy (i.e., no cardiovascular, metabolic, or other chronic diseases) and able to complete a 20-week exercise training program were targeted for recruitment. Parents were 65 years of age or younger and offspring at least 17 years of age. Subjects were required to be sedentary at baseline (i.e. not engaged in regular strenuous exercise lasting more than 30 min per week for at least 6 months prior to entry into the study). Additionally, individuals with a body mass index ≥40 kg/m2 were excluded unless they were approved by a physician to be able to complete the exercise test and training requirements. Individuals were also excluded if they had had a resting blood pressure ≥159/99 mmHg or were on antihypertensive or lipid lowering or hypoglycemic medications. Several other exclusion criteria pertaining to the ability to tolerate the exercise program and the exercise tests were used also as defined elsewhere (Bouchard et al. 1995). Informed written consent was obtained from all participants and the study was approved by the Institutional Review Board of each of the clinical and data coordinating center institutions.
For the current report, a total of 475 individuals (228 males, 247 females) from 99 two-generation Caucasian families had genotype and training response phenotype data. Sample sizes and family structures are described in more detail in Online Resource 1, which outlines the covariate adjustments, the sample statistics, and the frequency distributions.
Exercise tests
Maximal, submaximal, and submaximal–maximal exercise tests were each performed on separate days before and after the 20-week training program. Details of the exercise test protocols are found elsewhere (Skinner et al. 2000). The tests were performed at about the same time each day (about 48 h apart) on a SensorMedics Ergometrics 800S cycle ergometer (Yorba Linda, CA, USA) connected to a SensorMedics 2900 metabolic measurement cart. Before, during, and after each exercise test, heart rate (HR) was monitored using an electrocardiogram. Gas exchange variables were recorded during each exercise stage (O2 and CO2, among others) as rolling averages of three 20-s intervals. For the maximal exercise test, subjects exercised at a power output of 50 W for 3 min, followed by increases of 25 W each 2 min until volitional exhaustion. For older, smaller, or less fit individuals, the test started at 40 W, with increases of 10–20 W each 2 min thereafter. The criteria for attaining VO2max (during the maximum test) were defined as the respiratory exchange ratio being >1.1, a plateau in O2 uptake (change of <100 mL/min in the last three 20-s intervals), or a HR within 10 beats/min of the maximal HR predicted for the individual's age. Each subject achieved VO2max using one or more of the criteria.
During the submax test, subjects performed at submaximal levels at an absolute power output of 50 W for 8–12 min and then 8–12 min at a relative power output equivalent to 60% of the associated VO2max (baseline or post-training), based on whether the test was performed pre- or post-training. During the submaximal/maximal test, subjects completed the submax protocol described above and then exercised for 3 min at 80% of VO2max. Resistance was then increased to the highest power output attained in the maximal exercise test. If subjects were able to pedal after 2 min, power output was increased each 2 min thereafter until the subject reached volitional exhaustion.
During each of the submax and submaximal/maximal tests the same measurements were made as during the maximal test. Other measurements included cardiac output (Q), which was determined with a Collier CO2 rebreathing procedure (Collier 1956; Wilmore et al. 1982). The power output level or work level (WORK) indexed the amount of exertion in Watts on the SensorMedics Ergometrics 800S cycle ergometer. WORK at 60% of maximum (WORK60) corresponded to the power output in Watts when the subject was exercising at 60% of their VO2max. Thus, during submaximal testing all subjects performed at the same relative work load (60% of maximum), although the absolute work load varied depending on maximal fitness levels, which were based on baseline or post-training VO2max values depending on whether the tests were performed pre- or post-training.
Submaximal aerobic capacity traits in the current report include oxygen consumption (VO260 in mL/min), power output (WORK60 in Watts), and cardiac output (Q60 in L/min) at the power output associated with 60% of VO2max (baseline or post-training). The Q60 was based on the average of two measurements as derived from the separate submaximal, and submaximal/maximal exercise tests, while VO260 and WORK60 were continuously monitored. The submaximal aerobic capacity traits in HERITAGE were reliably measured (see Online Resource 1). For baseline VO260, the coefficients of variation (CVs) and intraclass correlations (ICCs) were 3.6% and 0.99 for reliability. Similarly, for baseline Q60 the CVs and ICCs for reliability were 5.9% and 0.93 (Wilmore et al. 1998). Thus, both within subject variation and measurement unreliabilities for day-to-day and across-center are generally small, particularly as compared with the between-subject variance in the responses to the submaximal exercise.
20-week exercise training protocol
During the interval between baseline and post-training assessments, each subject trained by exercising three times per week for 20 weeks on cycle ergometers. The intensity of the training was customized for each individual based on their HR and VO2 measurements taken at baseline. Subjects trained at the HR associated with 55% of baseline VO2max for 30 min for the first 2 weeks. The duration and intensity were gradually increased every 2 weeks, until reaching 50 min and the HR associated with 75% of baseline VO2max. This level was maintained for the final 6 weeks of training. All training was performed on Universal Aerobicycles (Cedar Rapids, IA, USA) and power output was controlled by direct HR monitoring using the Universal Gym Mednet (Cedar Rapids) computerized system. The protocol was standardized across all clinical centers and supervised by trained exercise specialists who ensured that the equipment was working properly and that the participants were compliant.
Genotyping
Genomic DNA was prepared from immortalized lymphoblastoid cell lines by commercial DNA extraction kit (Gentra Systems, Inc., Minneapolis, MN, USA). For the genome-wide linkage scan, a total of 701 markers covering the 22 autosomes with a mean spacing of 4.1 Mb on the physical map were used. PCR conditions and genotyping methods have been reported previously (Chagnon et al. 2000). DNA sequencers from LI-COR were used to detect the PCR products, and genotypes were scored semiautomatically using the software SAGA.
The SNPs for fine mapping studies were selected from the Caucasian (CEU) data set of the International HapMap consortium (data release 20 January 2006) using the pair-wise algorithm of the Tagger program (de Bakker et al. 2005). The pairwise linkage disequilibrium (LD) threshold for the LD clusters was set to r2 ≥ 0.80 and minimum minor allele frequency (MAF) to 5%. The HapMap data set contained 7,256 SNPs with MAF of at least 5% within the target region (Chromosome 13 from 21 to 29 Mb) and Tagger identified 1,902 tagSNPs. In addition, confirmed coding region SNPs were checked from the NCBI dbSNP data base for each known gene within the region. Additional 18 SNPs were uncovered from dbSNP, bringing the total number of SNPs to 1,920.
Genotyping of the SNPs was done using the Illumina (San Diego, CA, USA) GoldenGate chemistry and Sentrix Array Matrix technology on the BeadStation 500GX. The 1,920 SNPs were divided on one 1536-plex and one 384-plex Array Matrix. Genotype calling was done with the Illumina BeadStudio software, with calls confirmed manually. Of the 1,920 SNPs, 1,815 (>94%) were successfully genotyped.
For quality control purposes, one sample on each 96-sample Sentrix Array Matrix was included in duplicate. In addition, five CEPH control DNA samples (NA10851, NA10854, NA10857, NA10860, NA10861; all samples included in the HapMap CEU panel) were genotyped. Concordance between the replicates as well as with the genotypes from the HapMap database was 100%. Finally, two gender-specific control markers included in each Illumina GoldenGate assay agreed 100% with the gender of the subjects.
A series of quality control tests were performed on the 1,815 SNPs including tests for Mendelian consistency and for deviations from Hardy–Weinberg Equilibrium (HWE). About 5% of the markers had a MAF of ≤5% (1 SNP = 0%, 4 SNPs = 2%, 16 SNPs = 3% and 37 SNPs = 5%). Only the monomorphic SNP was excluded from further analyses. Deviations from HWE were minimal, with a P value of <0.05 for 5.2% of the SNPs and the smallest P value being 0.0021. Plots comparing the HWE and the MAF statistics with the association tests for ΔVO260 demonstrate that there are no violations of HWE or MAF for SNPs that are associated with ΔVO260 (see Online Resource 2, which provides plots for HWE and MAF). After QC of the marker data, a total of 1,814 SNPs were available for association analyses.
Statistical analyses
The computer program SAS was used to manage the data, conduct SNP quality control analysis (e.g. allele frequencies and Hardy–Weinberg equilibrium), perform phenotype adjustments, and compute sample statistics (means, variances, kurtosis). The response to exercise training (Δ) was computed as the difference between the average post-training and the average baseline measures. Responses were corrected for the effects of confounding variables in both the mean and variance using a stepwise regression procedure (see Online Resource 1, which outlines the covariate adjustments, sample statistics and frequency distributions). The final analysis variables consisted of the corrected and standardized responses to exercise for VO260 (ΔVO260), WORK60 (ΔWORK60), and Q60 (ΔQ60).
Multipoint linkage analysis was performed using a regression-based model as implemented in MERLIN (Sham et al. 2002; Abecasis et al. 2002). Briefly, siblings who share a greater proportion of alleles identical-by-descent (IBD) at the marker locus will also show a greater resemblance in the phenotype. The phenotypic resemblance of the siblings is linearly regressed on the estimated proportion of alleles that the sibling pair shares IBD at each marker locus.
The computer programs QTDT version 2.6.1 (Abecasis et al. 2000) and PLINK version 1.04 (Purcell et al. 2007) were used to perform family-based SNP associations for the covariate-adjusted phenotypes. Haploview version 4.1 (Barrett et al. 2005) was used to compute pair-wise LD among all known SNPs in specific regions using the comparative (CEU) HapMap population. PLINK was used to compute LD and to compute and analyze haplotypes in the HERITAGE sample. Family-based association tests were performed using the computer program QTDT. The total association model with additional sources of variance due to a family component and a residual (-at -wega) was used. This model adjusts for family membership using a between/within maximum likelihood variance components model as described by Fulker et al. (1999), and the non-independence of family members is modeled using kinship coefficients in the total association model.
P values were adjusted for the effects of multiple tests using the SAS procedure multtest. The adjusted P value is the significance level at which a hypothesis is rejected when all tests are considered simultaneously. The adjusted P value is the proportion of P values from the n samples that were smaller than the raw P value based on the original data.
To explore multi-marker hypotheses among SNPs, haplotype analysis for association with ΔVO260 was performed using PLINK. Haplotypes were imputed by first phasing founders using the E-M algorithm and then phasing all descendents of the founders given the set of possible parental phases and assuming random-mating. In this application, haplotypes across the entire region were imputed using a sliding window procedure by walking along the chromosome one SNP at a time. Different window sizes were explored (3 through 9 SNPs). When a larger window size was significant, we explored alternative (shorter) haplotypes using only SNPs from each end of the longer haplotype to construct shorter ones.
Results
The submaximal fitness traits in HERITAGE were responsive to the training program. There were significant increases in mean levels of VO260 (14%), Q60 (8%), and in WORK60 (30%) in HERITAGE Caucasians, with considerable between-subject variability for each trait (Wilmore et al. 2001a, b).
Genome-wide linkage scan for ΔVO260
A genome-wide linkage scan was performed in Caucasian HERITAGE families to identify genomic regions that may harbor gene(s) affecting ΔVO260. Results of the complete scan for each autosome can be found in Figure 2 of the Online Resource 3. A total of five regions (chromosomes 2p11.2, 2q13, 4p16.1, 13q12, 15q26.3) yielded suggestive linkage evidence (logarithm of odds [LOD] score >1.1; Table 1). However, only linkage on chromosome 13q12 reached genome-wide significance (LOD > 3.0); the maximum linkage of LOD = 3.11 was detected with marker D13S787, located in intron 15 of the MIPEP gene, at ∼23.8 Mb from pter of chromosome 13. The 2-LOD drop interval around the maximum LOD score of 3.11 ranged from ∼21.1 to ∼29.1 Mb from the p-terminus (see Figure 3 of Online Resource 3). The 2-LOD drop interval represents a confidence interval for locating the QTL (Lander and Botstein 1989).
Table 1. Summary of the LOD-scores > 1.1 observed in the genome-wide linkage scan for VO260 training response.
| Chr | Map | MARKER | LOD | P value |
|---|---|---|---|---|
| 2 | 84.987 | D2S1790 | 1.102 | 0.012 |
| 2 | 114.26 | D2S121 | 1.156 | 0.011 |
| 4 | 13.427 | D4S403 | 1.485 | 0.004 |
| 13 | 21.74 | D13S115 | 1.442 | 0.005 |
| 13 | 21.849 | D13S1275 | 1.321 | 0.007 |
| 13 | 23.279 | D13S787 | 3.112 | 0.00008 |
| 13 | 23.706 | D13S1243 | 2.094 | 0.001 |
| 13 | 25.375 | D13S1294 | 1.368 | 0.006 |
| 13 | 27.186 | D13S243 | 1.328 | 0.007 |
| 13 | 28.043 | D13S1242 | 1.409 | 0.005 |
| 15 | 97.41 | D15S120 | 1.493 | 0.004 |
See Figure 2 of the Online Resource 3 for complete linkage scan results
Associations for single-SNP analyses
The ∼7.9 Mb linkage region was saturated with a dense set of about 1,800 (primarily tag) SNPs using the Illumina platform in order to identify genetic variants that underlie the ΔVO260 QTL. The average density of the marker panel is 227 SNPs/Mb. To assist in the visualization and interpretation of results, the ∼7.9 Mb region was arbitrarily divided into six general regions ranging from 0.2 to 1.0 Mb, shown in Fig. 1.
Fig. 1.
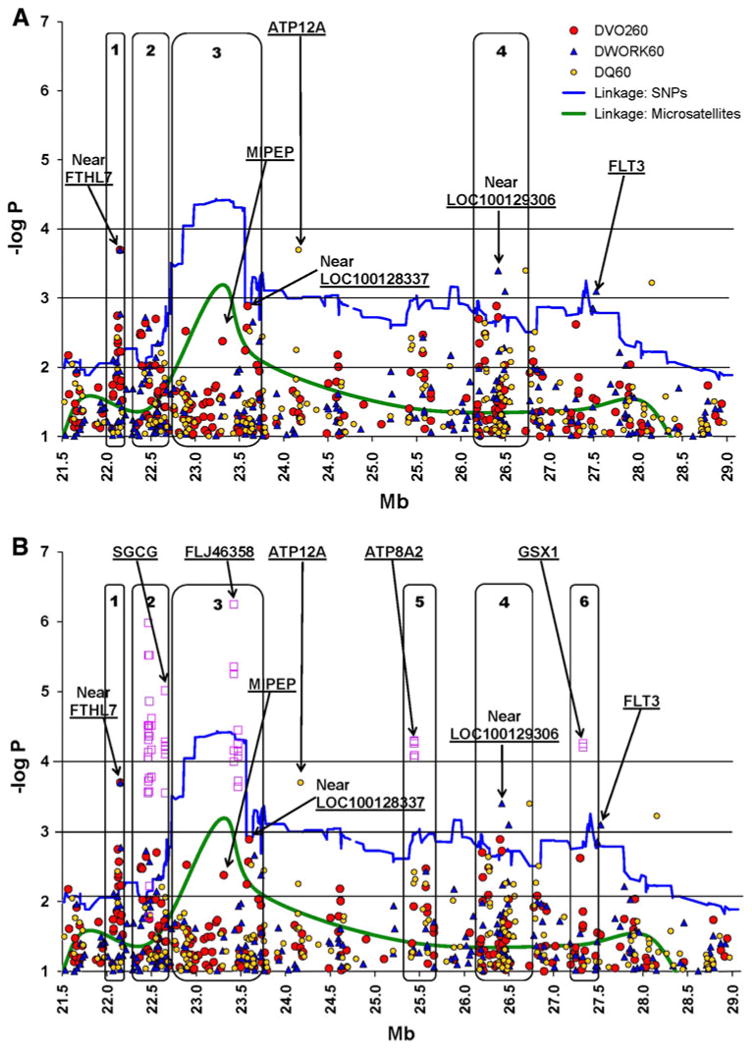
Family-based associations for ΔVO260 (labeled as DVO260) and correlated traits (ΔWORK60 and ΔQ60, labeled as DWORK60 and DQ60), with –log P values adjusted for multiple comparisons for chromosome 13q12.11-q12.3. The X-axis represents the SNP locations (in Mb units) and the Y-axis represents the –log P values for the association models (circle) or the LOD scores for the linkage models (solid lines). Noise was reduced by plotting only for –log P values that are at least 1.0. Linkage plots are presented for both micro satellites (original linkage scan) and SNPs (fine mapping). Blocks 1–6 represent arbitrary regions where the signals are large or concentrated. The top panel (a) represents the results for the single-SNP analyses. In the bottom panel (b), the superimposed square symbols represent the haplotype analysis results. SNP (or haplotype) signals that are located within genes are shown at the top of the bottom panel (SGCG, FLJ46358, MIPEP, ATP12A, ATP8A2, GSX1, FLT3, FLT1, PAN3). For the four best single-SNP results (–log P > 3.0), the closest genes are defined as Near FTHL7 (rs9506903 138,483 bp), Near LOC646208 (rs971206 6,460 bp), Near LOC100128337 (rs2104257 1,421 bp), Near LOC100129306 (rs7326591 69,878). See text for details
The association results for the dense SNP panel in this region of Chromosome 13 are presented in Table 2 for all single-SNP results with adjusted P values ≤0.05, and graphically in Fig. 1a. The figure also shows the linkage results (using the original linkage scan microsatellites and using the SNPs) superimposed.
Table 2. Single SNP association results for top SNPs (adjusted P values).
| Region | SNP name | Mb location | ΔVO260 | ΔWORK60 | ΔQ60 |
|---|---|---|---|---|---|
| rs7332465 | 21.567707 | 0.0068 | 0.2818 | 0.4898 | |
| rs1408051 | 21.725752 | 0.0427 | 0.0617 | 0.1230 | |
| 1 | rs9578482 | 22.068536 | 0.0182 | 0.0977 | 0.8710 |
| 1 | rs7342452 | 22.109944 | 0.0138 | 0.0120 | 0.3311 |
| 1 | rs4436648 | 22.118204 | 0.0148 | 0.0083 | 0.0037 |
| 1 | rs9578484 | 22.121090 | 0.0018 | 0.0617 | 0.0105 |
| 1 | rs2334946 | 22.126060 | 0.0045 | 0.1096 | 0.3802 |
| 1 | rs1034200 | 22.126691 | 0.0041 | 0.1000 | 0.3715 |
| 1 | rs2151499 | 22.127153 | 0.0062 | 0.4571 | 0.0871 |
| 1 | rs9550855 | 22.134792 | 0.0027 | 0.0603 | 0.3715 |
| 1 | rs9510292 | 22.149737 | 0.0191 | 0.0115 | 0.1585 |
| 1 | rs9506903 | 22.150146 | 0.0002 | 0.0002 | 0.2042 |
| 1 | rs17266994 | 22.155044 | 0.0059 | 0.0017 | 0.0741 |
| 2 | rs4770336 | 22.378267 | 0.0034 | 0.0219 | 0.7586 |
| 2 | rs971206 | 22.381330 | 0.0032 | 0.1445 | 0.4074 |
| 2 | rs9506969 | 22.394997 | 0.0034 | 0.0174 | 0.7586 |
| 2 | rs9510428 | 22.397333 | 0.0095 | 0.1862 | 0.6761 |
| 2 | rs2019091 | 22.437060 | 0.0023 | 0.0019 | 0.8913 |
| 2 | rs9510516 | 22.476548 | 0.0178 | 0.0027 | 0.0676 |
| 2 | rs9580513 | 22.488702 | 0.0095 | 0.0095 | 0.1862 |
| 2 | rs17078161 | 22.548298 | 0.0093 | 0.1413 | 0.0324 |
| 2 | rs7335200 | 22.555518 | 0.0020 | 0.0072 | 0.0617 |
| 2 | rs7319068 | 22.579661 | 0.0234 | 0.0045 | 0.8913 |
| 3 | rs9510730 | 22.892263 | 0.0030 | 0.0832 | 0.0398 |
| 3 | rs17330522 | 22.962269 | 0.0347 | 0.3311 | 0.9120 |
| 3 | rs9551012 | 23.308057 | 0.0042 | 0.0324 | 0.3162 |
| 3 | rs9553166 | 23.565701 | 0.0058 | 0.0933 | 0.3981 |
| 3 | rs17080058 | 23.582320 | 0.0027 | 0.0661 | 0.2455 |
| 3 | rs2104257 | 23.591742 | 0.0013 | 0.1288 | 0.0347 |
| rs9511156 | 23.695786 | 0.0093 | 0.1738 | 0.3981 | |
| rs9511655 | 24.609337 | 0.0066 | 0.0245 | 0.8710 | |
| rs9551180 | 24.615548 | 0.0098 | 0.1122 | 0.6026 | |
| 5 | rs1960061 | 25.568374 | 0.0062 | 0.0245 | 0.0204 |
| 5 | rs2038798 | 25.570294 | 0.0034 | 0.0037 | 0.9333 |
| 4 | rs2441075 | 26.199854 | 0.0020 | 0.0105 | 0.1622 |
| 4 | rs9512361 | 26.206857 | 0.0045 | 0.0219 | 0.1820 |
| 4 | rs2182880 | 26.268669 | 0.0081 | 0.1820 | 0.0023 |
| 4 | rs7332384 | 26.271154 | 0.0033 | 0.1023 | 0.0032 |
| 4 | rs17566649 | 26.327202 | 0.0562 | 0.1778 | 0.2951 |
| 4 | rs1576168 | 26.400279 | 0.0013 | 0.0155 | 0.5495 |
| 4 | rs7326591 | 26.419810 | 0.0019 | 0.0004 | 0.6607 |
| 4 | rs2479561 | 26.497574 | 0.0093 | 0.0008 | 0.0724 |
| 4 | rs17523405 | 26.777607 | 0.0120 | 0.0302 | 0.0115 |
| 4 | rs9512630 | 26.816639 | 0.0083 | 0.0102 | 0.0537 |
| 4 | rs1040988 | 26.875300 | 0.0117 | 0.0195 | 0.1380 |
| 4 | rs17085633 | 26.920617 | 0.0135 | 0.0117 | 0.0741 |
| 6 | rs9554166 | 27.295996 | 0.0024 | 0.0288 | 0.1072 |
| rs2281827 | 27.899721 | 0.0195 | 0.0525 | 0.0309 | |
| rs4771249 | 27.911414 | 0.0091 | 0.0263 | 0.0575 | |
| rs11840712 | 28.787442 | 0.0182 | 0.0309 | 0.0794 |
Mb megabase location from p-terminal
See Fig. 1 for location of Regions 1-6. SNPs listed if adjusted P value ≤ 0.05 for ΔVO260; represents 3% of the tested markers. Bold entries reflect: (1) primary signal (ΔVO260) significant (P value < 0.005) and (2) supporting evidence from correlated traits (ΔQ60 or ΔWORK60 P value ≤ 0.05) either at or within 1 kb of the ΔVO260 signal. None of these single-SNP results are located within a gene
The best results from the single-SNP analyses (highlighted rows in Table 2) are defined as having a ΔVO260 association adjusted P value of ≤0.005 and corroborating evidence (adjusted P value of ≤0.05) from either of the correlated traits of ΔWORK60 or ΔQ60. The best single-SNP signal for ΔVO260 in Fig. 1 is rs9506903 (at 22.15 Mb, adjusted P = 0.0002), with correlated support from ΔWORK60 (P = 0.0002). The closest gene to this SNP is FTHL7 (ferritin, heavy polypeptide-like 7) located about 17.9 kb downstream (see Online Resource 4, which provides a list of genes and SNPs in the region). FTHL7 is a pseudogene with no known function. Note that this general area (21.982–22.228 Mb) is labeled Region 1 for convenience.
The general area labeled Region 2 (22.279–22.712 Mb) has several SNPs meeting the criteria as shown in Table 2 (rs4770336, rs9506969, rs2019091 and rs7335200). While the first marker (rs4770336) is within 20.9 kb of FTHL7, there are several other undefined loci that are closer (LOC100131224, LOC401730, LOC100129167, LOC64 6201 and LOC646208). In fact, the rs4770336 is only 76 bp from LOC646201.
Region 3 (22.735–23.767 Mb) maps underneath the original maximal linkage signal peak (microsatellite D13S787). It is the largest region with at least eight known genes in the area. The best single-SNP associations in this region for ΔVO260 are with rs9510730 (adjusted P = 0.003) and rs9551012 (adjusted P = 0.004) with corroborating evidence from ΔQ60 (P = 0.04) and ΔWORK60 (P = 0.03), respectively. SNP rs9510730 is in an intron of the SACS gene (spastic ataxia of Charlevoix-Saguenay (sacsin)), while rs9551012 is located in the intron of the MIPEP gene (mitochondrial intermediate peptidase). Another SNP associated with ΔVO260 (P = 0.009) in this region was also located in a gene, as rs9511156 is in an intron of SPATA13 (spermatogenesis associated 13).
Region 4 (26.180–26.780 Mb) contains several SNPs (rs 2441075, rs9512361, rs1576168, rs7326591) that are associated with ΔVO260 and ΔWORK60. The closest gene to the first two SNPs is GPR12 (G-protein-coupled receptor 12). The closest gene to the last two markers is LOC100129306 (a pseudogene).
Associations for haplotype analyses
The single-SNP results were inconclusive in terms of strongly implicating any particular region or gene associated with ΔVO260. That is, signals that were within known genes are only moderate in magnitude. Consequently, additional analyses for ΔVO260 were performed using haplotypes, as shown in Fig. 1b and in Table 3. In the figure, open squares represent association results for the haplotypes with adjusted –log P values of 1.3 or greater. Note that multiple sets of analyses were performed using various window sizes ranging from three to nine SNPs and using a sliding window approach. In contrast to most of the single-SNP analysis, several haplotypes have strong signals (adjusted P value <0.001) that are located within known genes.
Table 3. Haplotypes with experiment-wide P values <0.0001 for ΔVO260.
| Region | Location | Maximum P value | SNPs (rs) | Allele values | Gene |
|---|---|---|---|---|---|
| 2 | 22.46 | 1.0 × 10−6 | rs9552790 | 3 | |
| rs9578521 | 3 | ||||
| rs9506995 | 3 | ||||
| rs9510516 | 3 | ||||
| rs17078046 | 3 | ||||
| 2 | 22.65 | 9.8 × 10−6 | rs1415130 | 3 | |
| rs4770402 | 2 | ||||
| rs4770403 | 1 | SGCG | |||
| rs3794370 | 1 | SGCG | |||
| s512444 | 3 | SGCG | |||
| rs7322327 | 3 | SGCG | |||
| rs3794368 | 3 | SGCG | |||
| 3 | 23.41 | 5.6 × 10−7 | rs12876596 | 3 | FLJ46358 |
| rs2147995 | 1 | ||||
| rs2765114 | 1 | ||||
| rs4769296 | 1 | ||||
| rs7331024 | 1 | ||||
| 5 | 25.43 | 9.3 × 10−5 | rs3783139 | 1 | ATP8A2 |
| rs912514 | 3 | ATP8A2 | |||
| rs11616429 | 2 | ATP8A2 | |||
| rs975508 | 3 | ATP8A2 | |||
| rs975506 | 1 | ATP8A2 | |||
| 6 | 27.25 | 4.0 × 10−5 | rs1326383 | 1 | |
| rs1326384 | 3 | ||||
| rs1231051 | 2 | ||||
| rs1231054 | 1 | ||||
| rs3742112 | 3 | GSX1 | |||
| rs1231064 | 3 | ||||
| rs10162043 | 1 | ||||
| rs9554166 | 3 |
Haplotype from among alternatives with different window sizes and different start SNPs
The strongest haplotype results (P values <0.0001) are in Regions 2 and 3. In Region 2, three unique haplotypes occur between 22.464 and 22.652 Mb. The haplotype with the strongest signal (P = 1.0 × 10−6) includes several SNPs (rs9552790, rs9578521, rs9506995 rs9510516, and rs17078046, spanning 22.464 to 22.484 Mb), one of which was also detected in the single-SNP analysis (rs9510516 at 22.476 Mb) for ΔVO260 (P = 0.018) and ΔWORK60 (P = 0.003). The third haplotype in this region (rs1415130, rs47700402, rs4770403, rs3794370, rs512444, rs7322327 and rs3794368 between 22.651 and 22.659 Mb) is highly significant (P = 9.7 × 10−6) and implicates the SGCG gene (sarcoglycan, gamma (35 kDa dystrophin-associated glycoprotein). The SNP rs4770403 is located in the first (untranslated) exon of SGCG, and the other SNPs are in intronic regions.
There were three unique haplotypes in Region 3 which corresponded to the original maximal linkage peak region. One haplotype in this region (rs12876596, rs2147995, rs2765114, rs4769296, and rs7330124 from 23.419 to 23.436 Mb) was significantly associated with ΔVO260 (P = 5.6 × 10−7). The rs12876596 marker is located in an intron of FLJ46358 which is a putative protein-coding gene, although no functional significance has been attributed to it.
Haplotype analysis also implicates two additional genes. First, an additional region upstream, noted Region 5 in Fig. 1, spans 25.377–25.660 Mb, and includes a haplotype (rs3783139, rs912514, rs11616429, rs975508, and rs975506) associated with ΔVO260 (P = 9.3 × 10−5) and located near the ATPase aminophospholipid transporter-like, class 1, type 8A, member 2 gene (ATP8A2). Another downstream area labeled Region 6 (27.134–27.540 Mb) included a haplotype associated with ΔVO260 (P = 4.0 × 10−5) near the GS homeobox protein 1 gene (GXS1).
Online Resource 5 provides a short summary of the association results for the 6 regions labeled in the previous figures and tables. Summarized are the density of genotyped and significant SNPs and characteristics concerning the haplotypes and genes in each region.
Post hoc regressions
In order to determine the magnitude of the effect of these SNPs on the ΔVO260 phenotype, stepwise regression analysis was performed using the SNPs that were significant in single-SNP (N = 16) and in haplotype analyses (N = 27) (see Online Resource 6). For the model including the 16 SNPs that were significant in the single-SNP analyses, only ten SNPs were retained in the model and accounted for 18.73% of the adjusted phenotypic variance. For the model including only the SNPs from significant haplotypes, the five SNPs remaining in the model accounted for 6.46% of the variance. Finally, for the model that included both the single SNPs and the haplotype SNPs, a total of 20.10% of the variance was accounted for by 13 SNPs (see Online Resource 6).
Discussion
The QTL for VO260 response to exercise training on chromosome 13q12, identified by genome-wide linkage scan, was followed-up with a fine mapping association study using a dense set of 1,814 SNPs covering the 7.9-Mb linkage region. While the results from this study do not unequivocally implicate a single causal variant, they contribute valuable information for further follow up. This is not unexpected for a complex trait such as ΔVO260 since by definition the etiology of complex traits includes a combination of multiple genetic, environmental and epigenetic factors.
An interesting and significant feature of these results is that the combination of SNPs in this general linkage region accounts for a significant portion of the phenotypic variance in ΔVO260. That is, post hoc regressions show that the 16 single-SNPs and 5 haplotypes (incorporating 30 SNPs) account for up to 20% of the total phenotypic variance. This estimate of the magnitude of the effect is consistent with previously reported heritability estimates (Perusse et al. 2001) which suggested that 23% of the variance in ΔVO260 is attributed to genetic factors. Since the single SNPs (regions 1, 2, 3 and 4) and haplotypes (regions 2, 3, 5 and 6) are spread across different regions, it is possible there are multiple signals with additive or interactive effects. The genes that are implicated in the present study include SGCG, MIPEP, ATP8A2, and GSX11, as well as several pseudogenes (FTHL7, LOC646208, LOC1001 28337, LOC100129306 and FLJ46358).
Mitochondrial intermediate peptidase (MIPEP) is the most likely candidate gene and is located within the peak linkage area in region 3 (microsatellite D13S787 is located within intron 15 of the MIPEP gene). MIPEP targets the mitochondrial matrix or inner membrane and is most highly expressed in the heart, skeletal muscle, and pancreas. The protein coded by MIPEP is primarily involved in the maturation of oxidative phosphorylation proteins by cleaving pro-peptides that are imported to the mitochondrion to their mature size (Chew et al. 1997). It is also related to the clinical manifestations of Freidreich ataxia which is characterized by defective coordination of limb movement as well as a host of other disorders (Chew et al. 2000). This disease involves an iron build-up in the mitochondria which results in secondary oxidative damage.
The SGCG gene is located upstream from the MIPEP gene and it is a plausible candidate due to location and function. The gene is located on the hip of the original peak linkage region (<500 kb from D13S787) and in the boundary between regions 2 and 3 in this dense association scan. The primary phenotype associated with SGCG is limb girdle muscular dystrophy, an autosomal recessive form of myopathy (Crosbie et al. 2000; Trabelsi et al. 2008). SGCG encodes a protein that, together with other isoforms of sarcoglycan (α and β), participates in binding actin to the extracellular matrix of skeletal and cardiac muscle cells and is involved in contractility. While the protein expression of the γ-form of sarcoglycan is traditionally confined to striated muscle cells (Noguchi et al. 1995) its presence in smooth muscle cells of the lung (and other organs) also has been demonstrated (Durbeej and Campbell 1999; Barresi et al. 2000). Our report of an association of SGCG with a cardiorespiratory trait (ΔVO260) appears to be novel. We noted at least two unique haplotypes in the SGCG gene that were associated with ΔVO260. One is located in the 5' untranslated exon region (P = 9.8 × 10−6). Another (rs1800354) was also located in an exon but was not reported in Table 1 since the signal (unadjusted P = 0.0009) did not remain significant after adjusting for multiple tests. This exon variant leads to a missense mutation by substituting Asn for Ser at amino acid position 287 (N287S). This variant also occurs in the same exon as the C283Y variant that was recently identified as a variant for limb girdle muscular dystrophy in Bulgarian gypsies (Todorova et al. 2002).
Other genes were statistically implicated in the current study, although we are unable to define a potential reference to cardiorespiratory fitness for any of them. In region 1, FTHL7 (ferritin, heavy polypeptide-like 7), a pseudogene, is near the largest single-SNP signal. Another gene (FLJ46358) is in the linkage region (region 4). While the location of this gene is relevant with a strong association signal (adjusted P = 5.6 × 10−7), it is unclassified with unknown function. The remaining two signals are further away from the linkage region (2–4 Mb upstream). The ATP8A2 gene drives the uphill transport of ions across membranes. Although the association signal with ΔVO260 was robust (adjusted P = 9.3 × 10−5), this haplotype is distant from the linkage region (over 2 Mb), with no primary phenotype associated with this gene in the Online Mendelian Inheritance in Man (OMIM 605870) and no associations with cardiorespiratory function found in the literature. Similarly, the GSX1 gene is distant from the linkage region (over 4 Mb) and the association signal with ΔVO260 is quite strong (P = 4.0 × 10−5). The only associations previously reported with this gene have been with conduct disorder and attention deficit hyperactivity disorder (Anney et al. 2008).
In conclusion, our genome-wide linkage scan followed by a fine mapping study narrowed the most promising signal close to the original linkage peak on chromosome 13. The genomic area includes two candidates with possible biological relevance (MIPEP and SGCG). However, these findings need to be confirmed, replicated, and potentially extended to other types of exercise programs in future studies. Identifying the exact causes of human variation in the response to regular exercise of submaximal exercise traits may have public health implications. Submaximal exercise capacity at 60% of maximum falls within the Physical Activity Guidelines for Americans (Physical Activity Guidelines Advisory Committee 2008). Identification of the true genetic causes of human variation in responsiveness of submaximal exercise capacity to exercise training would potentially open possibilities for more targeted exercise recommendations, perhaps in the context of personalized preventive medicine.
Supplementary Material
Acknowledgments
The HERITAGE Family Study is currently supported by NIH/NHLBI grant R01 HL045670. C. Bouchard is partially supported by the John W. Barton Sr. Chair in Genetics and Nutrition. T. Rice and D.C. Rao are also partly supported by NIH/NIGMS grant R01 GM28719. We wish to thank all PIs who contributed to the HERITAGE Family Study in the past via support from the National Heart, Lung, and Blood Institute through the following grants: C Bouchard (HL-45670); AS Leon (HL-47323); DC Rao (HL-47317); JS Skinner (HL-47327); JH Wilmore (HL-47321). The present address for author George Argyropoulos is Weis Center for Research, Geisinger Clinic, Danville, PA 17822-2615, for author Adrian M. Stütz is European Molecular Biology Laboratory, 69117 Heidelberg, Germany, and for author Margarita Teran-Garcia is Division of Nutritional Sciences, University of Illinois at Urbana-Champaign, Urbana, IL 61801.
Footnotes
Conflict of interest The authors declare there are no conflicts of interest.
Electronic supplementary material The online version of this article (doi:10.1007/s00421-011-2274-8) contains supplementary material, which is available to authorized users.
Contributor Information
Treva K. Rice, Division of Biostatistics, Washington University, St. Louis, MO 63110, USA
Mark A. Sarzynski, Human Genomics Laboratory, Pennington Biomedical Research Center, 6400 Perkins Road, Baton Rouge, LA 70808-4124, USA
Yun Ju Sung, Division of Biostatistics, Washington University, St. Louis, MO 63110, USA.
George Argyropoulos, Human Genomics Laboratory, Pennington Biomedical Research Center, 6400 Perkins Road, Baton Rouge, LA 70808-4124, USA.
Adrian M. Stütz, Human Genomics Laboratory, Pennington Biomedical Research Center, 6400 Perkins Road, Baton Rouge, LA 70808-4124, USA
Margarita Teran-Garcia, Human Genomics Laboratory, Pennington Biomedical Research Center, 6400 Perkins Road, Baton Rouge, LA 70808-4124, USA.
D. C. Rao, Division of Biostatistics, Washington University, St. Louis, MO 63110, USA
Claude Bouchard, Human Genomics Laboratory, Pennington Biomedical Research Center, 6400 Perkins Road, Baton Rouge, LA 70808-4124, USA.
Tuomo Rankinen, Email: rankint@pbrc.edu, Human Genomics Laboratory, Pennington Biomedical Research Center, 6400 Perkins Road, Baton Rouge, LA 70808-4124, USA.
References
- Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Anney RJ, Lasky-Su J, O'Dushlaine C, Kenny E, Neale BM, Mulligan A, Franke B, Zhou K, Chen W, Christiansen H, Arias-Vasquez A, Banaschewski T, Buitelaar J, Ebstein R, Miranda A, Mulas F, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Sonuga-Barke E, Steinhausen H, Asherson P, Faraone SV, Gill M. Conduct disorder and ADHD: evaluation of conduct problems as a categorical and quantitative trait in the international multicentre ADHD genetics study. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1369–1378. doi: 10.1002/ajmg.b.30871. [DOI] [PubMed] [Google Scholar]
- Barresi R, Moore SA, Stolle CA, Mendell JR, Campbell KP. Expression of gamma–sarcoglycan in smooth muscle and its interaction with the smooth muscle sarcoglycan-sarcospan complex. J Biol Chem. 2000;275:38554–38560. doi: 10.1074/jbc.M007799200. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Blair SN, Kampert JB, Kohl HW, Barlow CE, Macera CA, Paffenbarger RS, Gibbons LW. Influences of cardiorespiratory fitness and other precursors on cardiovascular disease and all-cause mortality in men and women. JAMA. 1996;276:205–210. [PubMed] [Google Scholar]
- Bouchard C, Dionne FT, Simoneau JA, Boulay MR. Genetics of aerobic and anaerobic performances. Exerc Sport Sci Rev. 1992;20:27–58. [PubMed] [Google Scholar]
- Bouchard C, Leon AS, Rao DC, Skinner JS, Wilmore JH, Gagnon J. The HERITAGE family study. Aims, design, and measurement protocol. Med Sci Sports Exerc. 1995;27:721–729. [PubMed] [Google Scholar]
- Chagnon YC, Borecki IB, Perusse L, Roy S, Lacaille M, Chagnon M, Ho-Kim MA, Rice T, Province MA, Rao DC, Bouchard C. Genome-wide search for genes related to the fat-free body mass in the Quebec family study. Metabolism. 2000;49:203–207. doi: 10.1016/s0026-0495(00)91299-x. [DOI] [PubMed] [Google Scholar]
- Chew A, Buck EA, Peretz S, Sirugo G, Rinaldo P, Isaya G. Cloning, expression, and chromosomal assignment of the human mitochondrial intermediate peptidase gene (MIPEP) Genomics. 1997;40:493–496. doi: 10.1006/geno.1996.4586. [DOI] [PubMed] [Google Scholar]
- Chew A, Sirugo G, Alsobrook JP, II, Isaya G. Functional and genomic analysis of the human mitochondrial intermediate peptidase, a putative protein partner of frataxin. Genomics. 2000;65:104–112. doi: 10.1006/geno.2000.6162. [DOI] [PubMed] [Google Scholar]
- Collier CR. Determination of mixed venous CO2 tensions by rebreathing. J Appl Physiol. 1956;9:25–29. doi: 10.1152/jappl.1956.9.1.25. [DOI] [PubMed] [Google Scholar]
- Cornelissen VA, Arnout J, Holvoet P, Fagard RH. Influence of exercise at lower and higher intensity on blood pressure and cardiovascular risk factors at older age. J Hypertens. 2009;27:753–762. doi: 10.1097/HJH.0b013e328322cf60. [DOI] [PubMed] [Google Scholar]
- Crosbie RH, Lim LE, Moore SA, Hirano M, Hays AP, Maybaum SW, Collin H, Dovico SA, Stolle CA, Fardeau M, Tome FM, Campbell KP. Molecular and genetic characterization of sarcospan: insights into sarcoglycan–sarcospan interactions. Hum Mol Genet. 2000;9:2019–2027. doi: 10.1093/hmg/9.13.2019. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37(11):1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Durbeej M, Campbell KP. Biochemical characterization of the epithelial dystroglycan complex. J Biol Chem. 1999;274:26609–26616. doi: 10.1074/jbc.274.37.26609. [DOI] [PubMed] [Google Scholar]
- Fulker DW, Cherny SS, Sham PC, Hewitt JK. Combined linkage and association sib-pair analysis for quantitative traits. Am J Hum Genet. 1999;64:259–267. doi: 10.1086/302193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg JM, Rankinen T, Loos RJ, Perusse L, Roth SM, Wolfarth B, Bouchard C. Advances in exercise, fitness, and performance genomics in 2010. Med Sci Sports Exerc. 2011;43:743–752. doi: 10.1249/MSS.0b013e3182155d21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EB, Bruce RA. Health benefits of exercise in an aging society. Arch Intern Med. 1987;147:353–356. [PubMed] [Google Scholar]
- Mendoza SG, Carrasco H, Zerpa A, Briceno Y, Rodriguez F, Speirs J, Glueck CJ. Effect of physical training on lipids, lipoproteins, apolipoproteins, lipases, and endogenous sex hormones in men with premature myocardial infarction. Metabolism. 1991;40:368–377. doi: 10.1016/0026-0495(91)90147-o. [DOI] [PubMed] [Google Scholar]
- Noguchi S, McNally EM, Ben Othmane K, Hagiwara Y, Mizuno Y, Yoshida M, Yamamoto H, Bonnemann CG, Gussoni E, Denton PH, Kyriakides T, Middleton L, Hentati F, Ben Hamida M, Nonaka I, Vance JM, Kunkel LM, Ozawa E. Mutations in the dystrophin-associated protein gamma–sarcoglycan in chromosome 13 muscular dystrophy. Science. 1995;270:819–822. doi: 10.1126/science.270.5237.819. [DOI] [PubMed] [Google Scholar]
- Perusse L, Gagnon J, Province MA, Rao DC, Wilmore JH, Leon AS, Bouchard C, Skinner JS. Familial aggregation of submaximal aerobic performance in the HERITAGE Family study. Med Sci Sports Exerc. 2001;33:597–604. doi: 10.1097/00005768-200104000-00014. [DOI] [PubMed] [Google Scholar]
- Physical Activity Guidelines Advisory Committee. Physical activity guidelines advisory committee report. Department of Health and Human Services; Washington, DC: 2008. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Purcell S, Cherny SS, Abecasis GR. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet. 2002;71:238–253. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner JS, Wilmore KM, Krasnoff JB, Jaskolski A, Jaskolska A, Gagnon J, Province MA, Leon AS, Rao DC, Wilmore JH, Bouchard C. Adaptation to a standardized training program and changes in fitness in a large, heterogeneous population: the HERITAGE Family Study. Med Sci Sports Exerc. 2000;32:157–161. doi: 10.1097/00005768-200001000-00023. [DOI] [PubMed] [Google Scholar]
- Todorova A, Tournev I, Ninova N, Georgieva V, Kremensky I. Screening for C283Y gamma–sarcoglycan mutation in a high-risk group of Bulgarian Gypsies: evidence for a geographical localization and a non-random distribution among Gypsy subgroups. Community Genet. 2002;5:217–221. doi: 10.1159/000066687. [DOI] [PubMed] [Google Scholar]
- Trabelsi M, Kavian N, Daoud F, Commere V, Deburgrave N, Beugnet C, Llense S, Barbot JC, Vasson A, Kaplan JC, Leturcq F, Chelly J. Revised spectrum of mutations in sarcoglycanopathies. Eur J Hum Genet. 2008;16:793–803. doi: 10.1038/ejhg.2008.9. [DOI] [PubMed] [Google Scholar]
- US Department of Health and Human Services. Physical activity and health: a report of the Surgeon General US Dept of Health and Human Services, Centers for Disease Control and Prevention. National Center for Chronic Disease Prevention and Health Promotion; Atlanta, GA: 1996. [Google Scholar]
- Wilmore JH, Farrell PA, Norton AC, RWd Cote, Coyle EF, Ewy GA, Temkin LP, Billing JE. An automated, indirect assessment of cardiac output during rest and exercise. J Appl Physiol. 1982;52:1493–1497. doi: 10.1152/jappl.1982.52.6.1493. [DOI] [PubMed] [Google Scholar]
- Wilmore JH, Stanforth PR, Turley KR, Gagnon J, Daw EW, Leon AS, Rao DC, Skinner JS, Bouchard C. Reproducibility of cardiovascular, respiratory, and metabolic responses to submaximal exercise: the HERITAGE Family Study. Med Sci Sports Exerc. 1998;30:259–265. doi: 10.1097/00005768-199802000-00014. [DOI] [PubMed] [Google Scholar]
- Wilmore JH, Stanforth PR, Gagnon J, Rice T, Mandel S, Leon AS, Rao DC, Skinner JS, Bouchard C. Cardiac output and stroke volume changes with endurance training: the HERITAGE Family Study. Med Sci Sports Exerc. 2001a;33:99–106. doi: 10.1097/00005768-200101000-00016. [DOI] [PubMed] [Google Scholar]
- Wilmore JH, Stanforth PR, Gagnon J, Rice T, Mandel S, Leon AS, Rao DC, Skinner JS, Bouchard C. Heart rate and blood pressure changes with endurance training: the HERITAGE Family Study. Med Sci Sports Exerc. 2001b;33:107–116. doi: 10.1097/00005768-200101000-00017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


