Abstract
What gives an organism the ability to regrow tissues and to recover function where another organism fails is the central problem of regenerative biology. The challenge is to describe the mechanisms of regeneration at the molecular level, delivering detailed insights into the many components that are cross-regulated. In other words, a broad, yet deep dissection of the system-wide network of molecular interactions is needed. Functional genomics has been used to elucidate gene regulatory networks (GRNs) in developing tissues, which, like regeneration, are complex systems. Therefore, we reason that the GRN approach, aided by next generation technologies, can also be applied to study the molecular mechanisms underlying the complex functions of regeneration. We ask what characteristics a model system must have to support a GRN analysis. Our discussion focuses on regeneration in the central nervous system, where loss of function has particularly devastating consequences for an organism. We examine a cohort of cells conserved across all vertebrates, the reticulospinal (RS) neurons, which lend themselves well to experimental manipulations. In the lamprey, a jawless vertebrate, there are giant RS neurons whose large size and ability to regenerate make them particularly suited for a GRN analysis. Adding to their value, a distinct subset of lamprey RS neurons reproducibly fail to regenerate, presenting an opportunity for side-by-side comparison of gene networks that promote or inhibit regeneration. Thus, determining the GRN for regeneration in RS neurons will provide a mechanistic understanding of the fundamental cues that lead to success or failure to regenerate.
Introduction
What are the molecular factors that are necessary and sufficient to accomplish successful regeneration, as defined as regrowth after injury, leading to functional recovery? Many studies have beautifully illustrated the cellular complexity of regeneration, including the disruption of tissue boundaries, the simultaneous induction of multiple cell types, and the induction of wound healing and inflammatory processes (for review, see Sanchez Alvarado and Tsonis, 2006; Ingber and Levin, 2007; Zhou and Melton, 2008; Tanaka and Ferretti, 2009). However, a comprehensive understanding of regeneration at the molecular level remains elusive. To reveal how such simultaneous cellular processes are regulated at the molecular level requires a method that can take into account the complexity of parallel biological processes. The use of functional genomics for the study of developmental biology illustrates an approach that balances both breadth and depth of functional analysis across the components of a complex system. Indeed functional genomics has been used successfully to provide insights into the gene regulatory networks (GRNs) controlling the early stages of development, such as the specification and growth of new embryonic tissues. Understanding the mechanisms that promote tissue regrowth after injury, or regeneration, demands a similar approach.
Parallels between the challenges of understanding the processes regulating regeneration and development have long been appreciated. In fact, a major question in regenerative biology is the degree to which mechanisms that promote regeneration resemble the developmental programs by which the relevant cell types and tissues were originally specified and organized (Harel and Strittmatter, 2006; Ingber and Levin, 2007). Historically, progress in both regenerative and developmental biology research relied heavily on a descriptive foundation, establishing the cell types involved and the temporal sequence of events that build tissues in the first place or that repair them after injury. Recently, developmental biologists have gained novel insights into the mechanisms by which new tissues and cell types are determined, or “specified,” by elucidating and analyzing the GRNs that govern these processes. A GRN describes the set of genes or gene products that functionally interact during any biological process. As an example, GRN analyses have been successfully applied toward understanding early developmental processes, such as the specification of endomesoderm in the sea urchin embryo (Davidson et al., 2002; Smith and Davidson, 2008). Other examples include developmental questions in insects and ascidians, specification of the chordate central nervous system (CNS), and vertebrate thymopoesis (Brent et al., 2007; Goltsev et al., 2007; Bajoghli et al., 2009; Imai et al., 2009; Christiaen et al., 2010). In these instances, the ability to determine regulatory gene networks benefited from several practical features, such as the spatiotemporal reproducibility of the process being analyzed, accessibility to observation and manipulation, and reliability of harvesting sufficient genetic material. These same experimental requirements will also be necessary for using the GRN approach to address open questions in regeneration.
In this position paper, we discuss the specific characteristics within regenerating organisms that are needed to conduct a GRN analysis. We focus our discussion on the application of GRN analysis to understand regeneration of the vertebrate central nervous system neurons, whose loss is devastating to the overall functionality of an organism. As we move away from the paradigm that the mammalian CNS is unable to regenerate and toward a better appreciation of its adaptive plasticity (e.g., Blesch and Tuszynski, 2009; Bradbury and Carter, 2011), this strategy will be particularly valuable in understanding how regeneration is differentially regulated across different vertebrate species. We postulate that using the GRN approach in the lamprey, as well as in other non-mammalian vertebrates, will allow us to gain a better evolutionary perspective on the mechanisms used by these organisms to regenerate their CNS, providing us with reference mechanisms that can be investigated in other species, such as mammals (Blau and Pomerantz, 2010).
The Gene Regulatory Network Approach
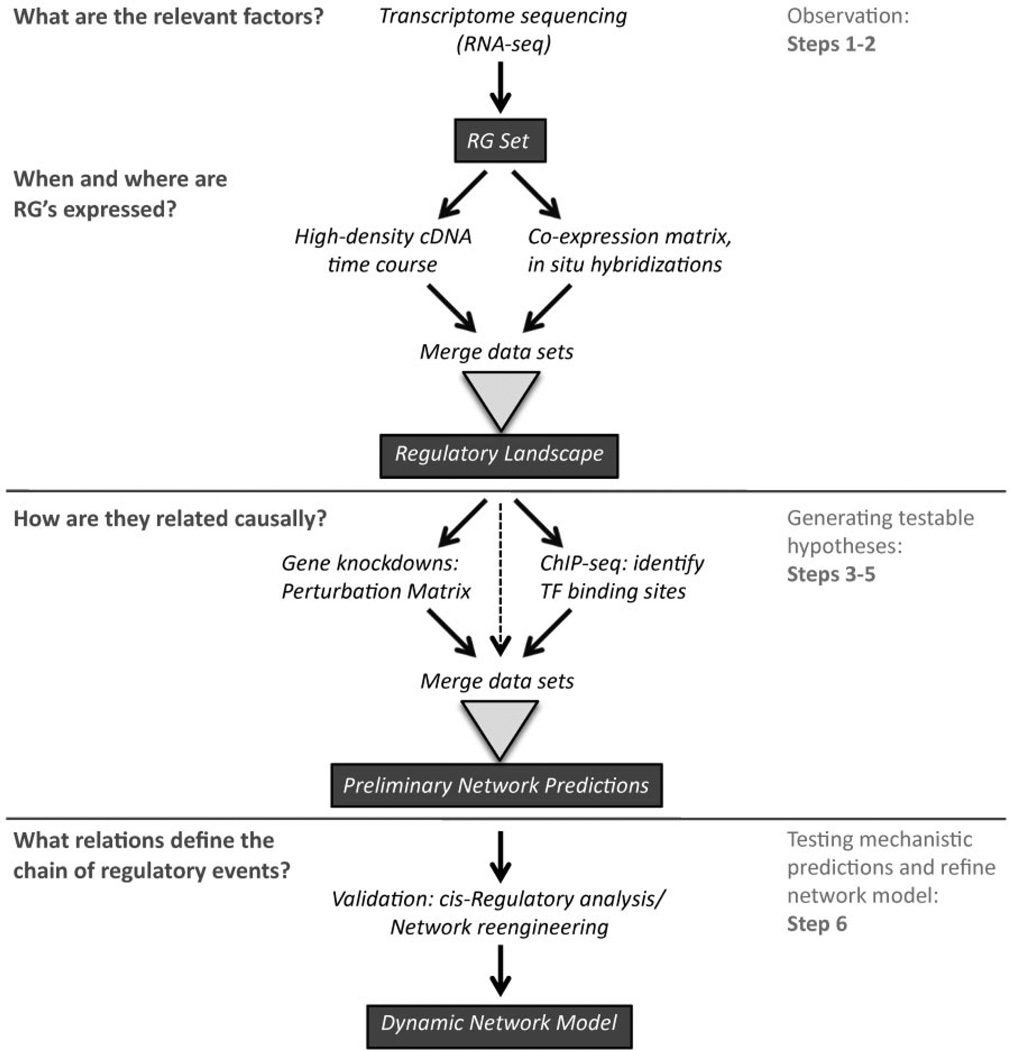
The dissection of GRNs couples large-scale gene expression data with methodical perturbation of potentially critical pathways. Network models derived from such experiments can then be used to generate targeted and testable hypotheses for further refinement (Geschwind and Konopka, 2009). One way to understand the GRN approach is, therefore, as a progressive focus on the classic parameters of observation, perturbation, and hypothesis testing (Fig. 1), distinct from the candidate-gene approach in that it has an integral discovery component. From this viewpoint, we identify six components that define a general framework for GRN analysis: (1) identification of all potentially relevant regulatory genes; (2) determination of the differential spatiotemporal expression of these genes, and if possible, their gene product distribution and post-translational modification state; (3) methodical perturbation of each gene within the regulatory gene set, followed by quantitative measurement of transcript levels of all other genes; (4) factor-factor and factor-DNA binding analysis, as a complementary dimension to elucidate the mechanisms underlying in vivo functional perturbations; (5) integration of data sets in (1– 4) to generate a preliminary network model of direct functional interactions across all system components that can be tested experimentally; and (6) verification of the predicted gene transcription mechanisms. Each of these steps of the GRN approach will be described more fully below and in the Appendix. Of note, both the cost and time requirements for this approach are decreasing as the technology advances, making them accessible to single laboratories or small collaborative groups on a timescale commensurate with funding cycle requirements.
Figure 1.
Overview of gene regulatory network (GRN) approach. The middle column depicts the general work flow for the GRN approach with landmark data sets shown in the Appendix. The left column highlights the scientific questions addressed by each component; the right column identifies the corresponding steps as labeled in text. To sum up, the GRN approach follows the classic parameters of observation, controlled experimentation, and hypothesis testing. During the observation phase, expressed regulatory factors are identified by deep transcriptome sequencing, and their spatial and temporal expression patterns are determined at high resolution (Appendix Part A). Next, an individual regulatory gene is perturbed and transcript levels for all other genes quantitatively assessed by high-throughput methods (see text and Appendix Part B); this process is then repeated for all expressed genes, while parallel ChIP-seq assays provide a complementary data set to identify candidate cis-acting genomic elements. All data sets are then cross-referenced to generate predictions of the network of interactions. These predictions are validated in mechanistic detail by reporter assays; the dynamic series of regulatory steps is then tested by network re-engineering (Appendix Part C).
Identification of the regulatory gene set and determination of the spatiotemporal expression map (Steps 1 and 2)
The initial step in the GRN approach is to cast a maximally broad net to identify the complete set of potential regulatory genes expressed during regeneration. Deep-sequencing of the entire transcriptome (using RNA-seq) provides the essential discovery platform to determine in an inclusive and unbiased manner the set of all expressed genes, which will contain all relevant regulatory genes: the regulatory gene (RG) set. Next, a high-resolution map of the spatial and temporal expression patterns of all genes in the RG set is determined by a combination of standard methods and high-throughput transcriptome and genetic analyses (See Appendix Part A). This provides the spatiotemporal expression map, also referred to as the regulatory landscape. Precisely when and where all potentially relevant regulatory genes are first transcribed is critical for determining the regulatory events driving differential gene expression. In the next step of the GRN approach, this information will be used to guide the functional perturbation of potentially relevant genes.
Generating testable hypotheses of regulatory mechanisms: systematic perturbation of individual genes and quantitative analysis of molecular consequences (Steps 3–5)
The next steps of the GRN approach are to systematically perturb individual genes within the RG set and measure the effects on the other genes within the set. As individual genes in the putative RG set are altered by knockdown or overexpression approaches, including drug, RNAi, or morpholino (MO) antisense oligonucleotide treatments, expression levels of all other genes in the RG set are measured by high-throughput transcriptome analysis. When repeated for each gene in the putative RG set, this produces the perturbation matrix, an experimentally defined set of potentially direct and indirect functional relationships. These experiments represent the central step of functional GRN analysis. Several decisions on methodology are required; for instance, genetic knockdowns versus gain of function perturbations. An in-depth discussion of this step is found in Appendix Part B.
Next, to determine the mechanisms underlying the observed functional interactions, the perturbation matrix data are combined with in vitro methods to identify direct binding of transcription factors or other DNA-associated proteins to chromatin, which can be achieved using chromatin- immunoprecipitation followed by next-generation sequencing (i.e., ChIP-seq) (Mortazavi et al., 2006; Park, 2009). This method determines the set of direct interactions by transcriptional regulatory genes throughout an entire genome, which can be derived from samples at different locations and stages of the regenerative process. In practice, however, ChIP-seq cannot be performed for all transcriptional regulators in the RG set, short of an epic effort, so additional strategies are necessary (discussed in Appendix Part B). Perturbation matrix data are then cross-referenced with ChIP-seq interaction data, to examine whether interactions predicted by the matrix data are confirmed by direct binding interactions in the ChIP-Seq data. Together with the spatiotemporal expression map, these data are used to predict the set of regulatory interactions relevant throughout stages of the regenerative process: the preliminary network model.
Verification: testing predictions of gene regulatory mechanisms (Step 6)
At this step of the process, the regulatory events predicted by the preliminary gene regulatory network model are rigorously tested. All genomic regulatory sequences are mutated or disrupted to affect particular transcription-factor binding sites. Individual regulatory mechanisms predicted to be central to network function are thereby tested at a detailed molecular level by this cis-regulatory analysis, which can be achieved using reporter gene assays. Identifying the relevant cis-regulatory modules is technically intensive, as binding elements may be anywhere within hundreds of kilobases of genomic sequence (Lettice et al., 2003; Vokes et al., 2008; Visel et al., 2009). However, ChIP-seq analysis from the previous step may reduce the potentially relevant areas of sequence, since ChIP-seq typically measures binding of regulatory elements within a 100-bp stretch of DNA (Mortazavi et al., 2006). Bacterial artificial chromosome (BAC) reporter constructs supply the required capacity to test binding on relatively large stretches of DNA sequence. Once regulatory sites have been determined, expression constructs with disrupted or altered cisregulatory binding sequences are designed to test select sub-network functions driving differential gene expression, also known as network re-engineering (Smith et al., 2008; Smith and Davidson, 2009). Overall, this component of the GRN approach represents the synthesis of all other data sets (for further discussion, see Appendix Part C). In practice, these results often inspire further iterations of perturbation experimentation and refinement of the regulatory gene expression map at higher spatial and temporal resolution.
What Makes a Model System Tractable for Gene Regulatory Network Analysis?
To be an appropriate subject for GRN analysis, the biological process or organism under investigation must have a certain degree of reproducibility and accessibility to observation and manipulation, as is the case for virtually any experimental study. In particular, reproducible spatiotemporal parameters to facilitate controlled and quantitative experimentation are essential. For example, the success of the GRN approach in embryological systems derived from the fact that developmental stages, tissue boundaries, and cell lineages were well defined in terms of time and space. As will be described below, application of the GRN approach to studying regeneration requires the system to be well characterized in similar ways. The amount of genetic material that can be harvested is a second consideration and related to the ability to define reproducible stages and cell lineages. In general, this should not be a problem, as quantitative assessment of transcript levels can now be achieved with relatively small quantities of RNA. In special cases, it may be possible and desirable to extend gene transcript analysis to the level of the single cell. The third general consideration is how amenable the system is to experimental manipulation, primarily by genetic approaches to alter expression of specific gene targets. One straightforward strategy for genetic manipulation, from a technical perspective, is achieved by delivery of MOs or RNA constructs to inhibit translation of specific genes. Efficient methods of gene delivery are equally important for gene-specific reporter assays in order to perform cis-regulatory analysis. Use of gene-specific reporter constructs may also enhance determination of the regulatory landscape, as discussed in Appendix Part A. In sum, to be amenable to the GRN approach a system should be as follows:
Temporally defined, with not just a beginning and end, but also discrete identifiable stages; and spatially defined, particularly with specific cell lineages;
Large or available in abundance in order to provide adequate genetic material;
Open to genetic manipulation, such as gene delivery for genetic perturbation and gene reporter assays.
Choosing a System for Study of CNS Regeneration Using the Gene Regulatory Network Approach
While the regenerative ability of non-mammalian vertebrates has long been appreciated, identification of the molecular pathways underlying this remarkable capacity has often been hampered by a paucity of molecular resources to study these organisms. Recent advances in high throughput sequencing, coupled with additional features of non-mammalian vertebrates that can regenerate, make the GRN approach now feasible in these species. In addition to access to these technological advances, we identify six biological criteria for evaluating the suitability of an organism to study CNS regeneration by the GRN approach:
-
(1)
Damaged neuron exhibits a robust capacity to regrow.
-
(2)
Regrowth of damaged tissue leads to functional recovery.
-
(3)
The relevant cell type or tissue undergoing regrowth is large enough and in sufficient quantity to achieve high-quality transcriptome analysis.
In short, these first three characteristics define system reproducibility and observability. In addition:
-
(4)
A sequenced genome, though not strictly essential, provides an established template for molecular analyses.
-
(5)
A means of gene delivery is critical.
-
(6)
Relevance to the CNS in other vertebrates is indicated by conservation.
There are several non-mammalian vertebrates that achieve CNS regeneration and have some or all of these features (Tanaka and Ferretti, 2009), such as the zebrafish (Becker and Becker, 2008), the African clawed frog (Tseng and Levin, 2008), and the main focus of our discussion, the lamprey species Petromyzon marinus. The lamprey achieves robust anatomical and functional recovery after complete spinal cord injury and as yet is the only vertebrate organism to satisfy the criteria of the National Institutes of Health (NIH) for full functional recovery after spinal cord injury (Fig. 2) (Guth et al., 1980, Cohen et al., 1988). As will be discussed below, injured neurons, some of which are exceptionally large, can be reliably followed through the stages of regeneration. Gene expression in lamprey can be evaluated by transcriptome analysis (O. Bloom, C. Brown, J. Buxbaum, J. Morgan, J. Smith, W. Li, and colleagues, unpubl. data) and functionally manipulated by gene delivery of MOs (McCauley and Bronner-Fraser, 2006). Moreover, there is an effort sponsored by the NIH and the National Human Genome Research Institute (NHGRI) to sequence the lamprey genome, which is publicly available with 5.9X coverage (Genome Institute at Washington University, 2007), as well as ongoing efforts to improve the genome data, assembly, and annotation (Li, W., and Lamprey Genome Project Consortium, 2011).
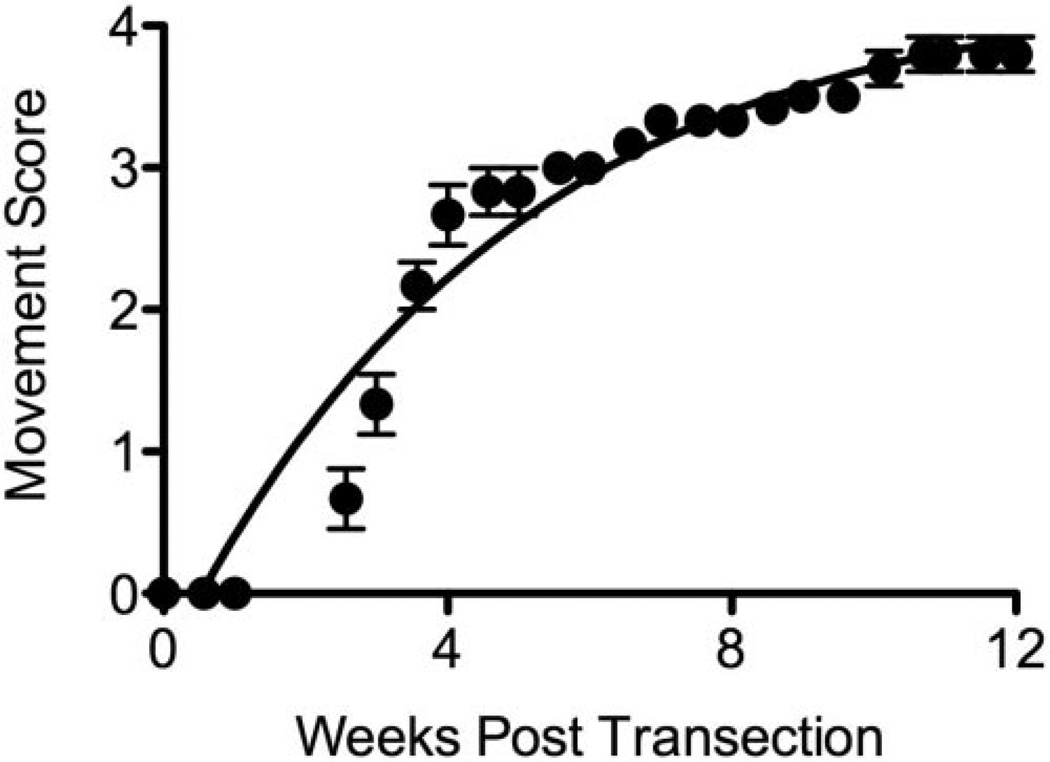
Figure 2.
Time course of behavioral recovery in lamprey after spinal cord transection. The lamprey spontaneously recovers swimming movements after complete spinal cord transection in a stereotypic manner over the course of 12 weeks. The numbers on the y-axis correspond to reproducible stages of recovery and are adapted from Oliphint et al. (2010). Briefly, a score of 0 indicates that the animal can move its head rostral to the transection site and is paralyzed below it. A score of 1 indicates that the animal can curl into a C- or S-shape. A score of 2 indicates the ability to accomplish abnormal, brief bouts of self-initiated swimming. A 3 indicates the ability to sustain more persistent, but still abnormal, bouts of swimming, including abnormal body contractions and difficulty righting. Finally, a score of 4 indicates that the animal can swim consistently, persistently, in a manner that appears as if uninjured. Data are derived from the mean ± sem of 5–6 animals (Bloom, O, unpubl. data).
Therefore, in light of the above criteria, 1–4 in particular, we propose to study CNS regeneration in the lamprey by using the GRN approach, with the specific goal of determining the molecular pathways that define successful axonal regrowth and functional recovery. Within the lamprey CNS, a subset of giant reticulospinal (RS) neurons fail to regenerate after injury, while others in their cohort reproducibly do, offering an additional opportunity: to compare the GRNs of “good” and “bad” regenerators (Jacobs et al., 1997). As these neurons are large and readily identifiable, determining the different GRNs in both regenerating and non-regenerating neurons, at the single-cell level, becomes feasible.
After describing how we propose to apply a GRN analysis to regenerating and non-regenerating giant RS neurons of lampreys, we return to a discussion of RS neuron regeneration in other non-mammalian vertebrates—zebrafish and African clawed frog—and how the GRN approach can be used to elucidate additional molecular features underlying successful CNS regeneration. Broadly, we suggest that applying the GRN approach in non-mammalian vertebrates that spontaneously achieve robust regeneration takes advantage of what the lay press has referred to as “the free lesson that nature is offering” on the regulation of CNS regeneration (Wade, 2010).
Conserved Features of the Lamprey Central Nervous System
The lamprey CNS, both brain and spinal cord, shares its basic organization and structure with other vertebrates (Hardisty and Potter, 1971; Rovainen, 1979; Grillner and Jessell, 2009). As is characteristic of vertebrates, the lamprey brain is divided into the morphologically distinct regions of hindbrain, midbrain, and forebrain (Watanabe et al., 2008; Murakami and Watanabe, 2009). The lamprey spinal cord is about 1 mm wide and 250 µm thick, with a flat ribbon-like appearance and a central canal (Fig. 3) (Rovainen, 1979). It contains axons of ascending and descending neurons, motor neurons, several types of sensory neurons, and interneurons, many of which are large and identifiable (Parker and Grillner, 200; Buchanan, 2001; Bevan et al., 2008; Cooke and Parker, 2009).
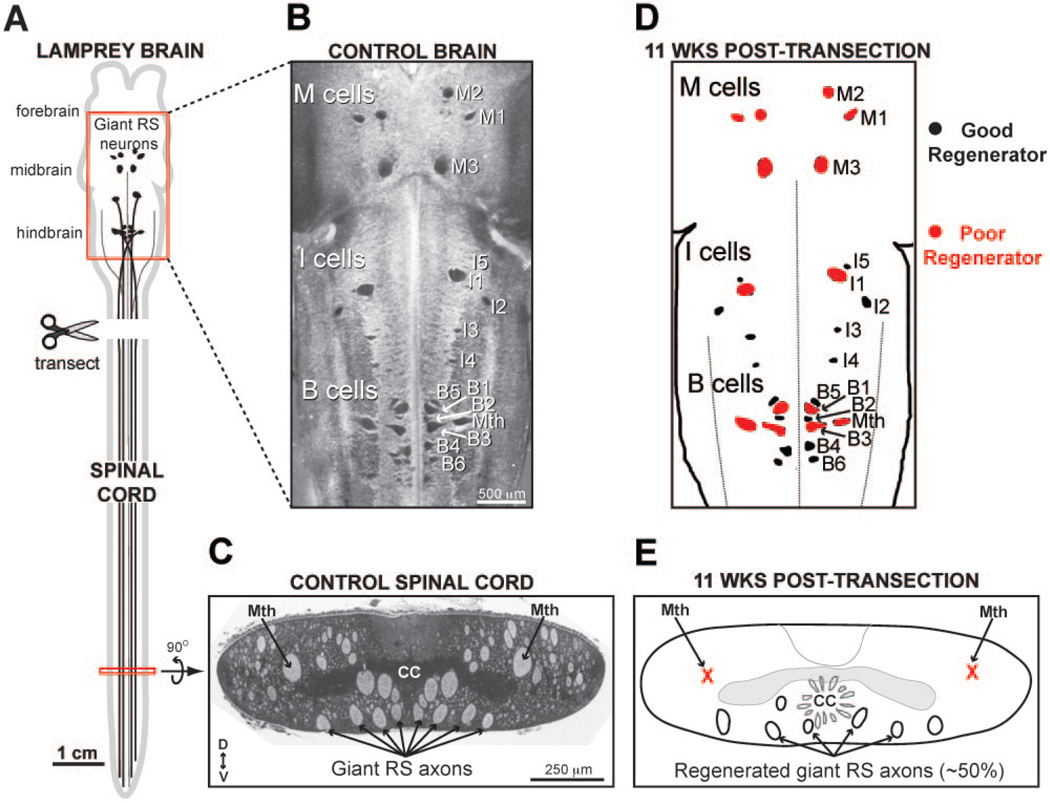
Figure 3.
Regeneration in the lamprey nervous system. (A) Diagram of lamprey CNS. Giant reticulospinal (RS) neurons in midbrain and hindbrain project directly to spinal cord. The typical spinal cord injury paradigm is a complete spinal cord transection, which axotomizes all descending neurons, including giant RS neurons. (B) A Nissl-stained lamprey brain showing the locations of all identified giant RS neurons. Giant RS neurons include the large Müller cells in the mesencephalic (M), isthmic (I), and bulbar (B) brain regions, as well as the Mauthner (Mth) cell. (C) A Nissl-stained cross-section of the lamprey spinal cord showing locations of giant RS axons in the ventromedial tract. The axons of the Mauthner (Mth) neurons are located more dorsolaterally. Dorsal (D) and ventral (V) orientations are indicated. CC = central canal. (D) After transection, some giant RS neurons regenerate reliably, while others do not. Here, “poor regenerators” (red) are defined as those neurons that regenerate <50% of the time (see Jacobs et al., 1997). (E) A schematic of the distal lamprey spinal cord at 11 weeks post-transection. Mauthner axons are rarely observed because they are poor regenerators. Only about 50% of the giant RS neurons regenerated.
Within this conserved structure are conserved nerve cells and functionally equivalent sets of neurons (Bullock, 1984, 2000; Buchanan, 2001). Notable examples are the reticulospinal (RS) neurons, which compose one of the most conserved descending pathways in vertebrates (Murakami et al., 2005). RS neuron homologs are identifiable across all vertebrate orders (Rovainen, 1967a, b; Newman et al., 1983; Cruce et al., 1988; Ronan, 1989; Fetcho, 1992; Buchanan, 2001). These neurons function as integration centers for sensory inputs and motor outputs, and they ultimately initiate, coordinate, and regulate motor functions in the spinal cord (e.g., Rovainen, 1967a, b; Nissanov and Eaton, 1989; Eaton et al., 1991, 2001; Lingenhohl and Friauf, 1994; Yeomans et al., 2002; Grillner and Jessell, 2009). The utilization of various tract-tracing methods has allowed the description of an expanded number of identifiable RS neurons that can be found in a segmental pattern in many vertebrates, including lamprey (Ronan, 1989; Murakami and Kuratani, 2008; Watanabe et al., 2008), larval and adult zebrafish (Kimmel, 1982; Kimmel et al., 1982; Metcalfe et al., 1986; Lee and Eaton, 1991; Kinkhabwala et al., 2011), juvenile goldfish (Lee et al., 1993), and larval and adult frogs (Straka et al., 1998, 2001, 2006). Also see Murakami and Kuratani (2008) for an illustration of their conservation across vertebrates. Furthermore, as large RS neurons can be found in vertebrates as diverse as lamprey and mouse (Rovainen, 1967a, b; Nissanov and Eaton, 1989; Eaton et al., 1991, 2001; Lingenhohl and Friauf, 1994; Yeomans et al., 2002), we expect that this approach in the lamprey will help to identify mechanisms that promote and, when compared to GRNs in non-regenerating species, also those that inhibit regeneration.
In lamprey, there are 1200 RS neurons in the midbrain and hindbrain (Dubuc et al., 2008). Of these, there are about 30 identified giant RS neurons: the Mauthner cells (Mth), and the Müller cells, in the mesencephalic (M), isthmic (I), and bulbar (B) regions of the brain (Ronan, 1989; Swain et al., 1993; Jacobs et al., 1997; Oliphint et al., 2010). Giant RS neurons are present in stereotypical locations in the brain (Fig. 3A–B), and they project large unbranched axons (20–60 µm in diameter in larvae and 40–80 µm in adults), that run the length of the spinal cord and form en passant synapses with spinal moto- and interneurons (Rovainen, 1967a, b). The large size of both the somata and axons of lamprey giant RS neurons renders them highly accessible for experimental manipulation, including microinjection in either the cell body or axon with tracers, antibodies, recombinant proteins, or MOs. Critically, the size and stereotypical position of giant RS neurons in lamprey make them identifiable under a variety of conditions, allowing a detailed examination of their responses to injury and during regeneration, as discussed in the next section.
The Lamprey Spinal Cord Regenerates: An Experimental Opportunity
In contrast to mammals, the lamprey spinal cord regenerates spontaneously and recovers nearly full function after a complete spinal cord transection (Fig. 2) (Maron, 1959; Hibbard, 1963; Rovainen, 1976; Selzer, 1978; Ayers et al., 1983; Currie and Ayers, 1983; Cohen et al., 1986, 1988, 1989; Lurie and Selzer, 1991a; Oliphint et al., 2010). As mentioned above, the lamprey is the only vertebrate for which sufficient experimental data exist to satisfy what the National Institute of Neurological Disorders and Stroke (NINDS) has specified as the definition of functional regeneration in the spinal cord after injury (Guth et al., 1980; Cohen et al., 1988). This observation drives our current discussion of GRN analysis in the lamprey CNS. As we will see, other criteria relating to staging, reproducibility, and accessibility are satisfied as well.
Complete spinal cord transection, the most common paradigm used for these studies, is followed by discrete stages of recovery that can be marked by changes at the lesion site. Immediately after injury, ependymal cells from the central canal proliferate across the lesion (Hibbard, 1963; Rovainen, 1976; Selzer, 1978; Lurie and Selzer, 1991a, b, c; Lurie et al., 1994). Within the first 1–2 weeks after injury, the proximal portions of giant RS axons retract an average of 1–2 mm while the distal portion undergoes Wallerian degeneration (Roederer et al., 1983; Yin and Selzer, 1983; Zhang et al., 2005). At this time, activated microglia and macrophages accumulate, presumably via proliferation, as in other species (Shifman and Selzer, 2007; Shifman et al., 2009). By 3–4 weeks post-transection, giant RS axons begin to regenerate (Rovainen, 1976; Lurie et al., 1994). By 10–12 weeks post-transection, in both larvae and adult animals, about 50% of neurons cross the lesion site (Rovainen et al., 1973; Rovainen, 1976, 1979; Lurie and Selzer, 1991a). As mentioned in the description of the GRN approach, such reproducible responses are an important feature for controlled experimentation.
Of considerable interest in the present discussion is that regenerative capacity varies between individual giant RS neurons, but is remarkably reproducible (Fig. 3D). The most robust regeneration is consistently observed in mesencephalic cell M1, isthmic cell I2, and the auxiliary Mauthner; whereas the least regeneration is observed in isthmic cell I1, bulbar cells B1 and B3, and the Mauthner cell (Fig, 3E) (Yin and Selzer, 1983; Davis and McClellan, 1994a, b; Jacobs et al., 1997; Shifman et al., 2008). A series of elegant studies characterized the response of giant RS neurons after spinal cord transection and established that the “good regenerators” undergo robust axon regeneration through the injury scar and form synapses with downstream targets (Rovainen, 1976; Selzer, 1978; Wood and Cohen, 1979; Mackler and Selzer, 1987; Lurie and Selzer, 1991a, b, c; Oliphint et al., 2010). The extensive characterization of RS neuron populations that are either predictably good or bad regenerators presents a significant opportunity to identify the functional GRNs supporting neuronal regeneration within the same animal.
The time course of functional recovery correlates with regeneration of descending reticulospinal axons and reestablishment of appropriate synaptic connections (Fig. 2) (Rovainen, 1967a, b, 1976; Selzer, 1978; Wood and Cohen, 1979, 1981; Yin et al., 1984; Cohen et al., 1986, 1988, 1989; McClellan, 1990, 1994; Davis et al., 1993; Davis and McClellan, 1994a, Lurie et al., 1994; Parker and Grillner, 2000; Oliphint et al., 2010). While most studies on spinal cord regeneration in lampreys have been conducted in the larval animals (~5–7 years old), some adult lamprey reticulospinal axons regrow across the lesion site with directional specificity (Cohen et al., 1989; Lurie et al., 1994), and a percentage of adult animals accomplish functional recovery in the form of intersegmental coordination (Cohen et al., 1989). Thus, although less well-studied and apparently less complete than in larval animals, both axonal regrowth and functional recovery can be achieved to some extent in the adult lamprey, providing an opportunity to compare the GRNs associated with regeneration throughout different developmental stages.
In sum, regeneration in the lamprey spinal cord shares key characteristics with developmental systems: defined stages, and traceable cell lineages and fates. Most regenerating reticulospinal axons grow in the appropriate direction (i.e., caudally), indicating some maintenance or restoration of the body axis during regeneration (Rovainen, 1976; Borgens et al., 1981; Roederer et al., 1983; Yin et al., 1984; Lurie and Selzer, 1991b). Ascending axons also regenerate in the proper direction (Yin and Selzer, 1983; Armstrong et al., 2003). Moreover, injury induces changes in the expression levels of several axon guidance molecules (Shifman and Selzer, 2000a, b, 2006, 2007; Shifman et al., 2009). Some of these guidance molecules, such as UNC5 and neogenin, are differentially expressed between good and bad regenerators, providing proof-of-concept for the identification of differences in gene expression between the two populations. Thus, there is already evidence from the candidate- gene approach that regeneration in the lamprey shares some elements with the normal developmental program and that there are different transcriptional responses post-injury. Interestingly, the regenerative process in lamprey spinal cord, as in mammals, is influenced by age, temperature, conditioning lesion, and cAMP, as well as by electric fields; indicating that at least some of the networks governing regeneration are shared across species—a point we will return to later in the article (Borgens et al., 1981; Cohen et al., 1989, 1999; Zhang et al., 2004; Jin et al., 2009). In the next section, we look at the specific components of a GRN analysis in the lamprey RS neurons, which would go beyond the candidate-gene approach to allow us to define in a comprehensive manner the molecular mechanisms underlying CNS regeneration.
Applying Gene Regulatory Network Analysis to Lamprey CNS Regeneration
Defining the regulatory gene set and the regulatory landscape in the regenerating lamprey spinal cord
The GRN approach has already been used successfully in lamprey in developmental contexts—specifically, to identify networks relevant to neural crest specification (Nikitina et al., 2008; Sauka-Spengler and Bronner-Fraser, 2008; Nikitina and Bronner-Fraser, 2009) and thymopoesis (Bajoghli et al., 2009), indicating that lamprey is a suitable model for the proposed experiments on regeneration. The first step is to define the RG set relevant to neuronal regeneration in the lamprey spinal cord after injury. To do so will require an analysis of the complete transcriptome from injured and uninjured tissue or cells, which will contain all potentially relevant regulatory genes. With the use of RNA-Seq methods, both brain and spinal cord tissue can yield high-quality transciptome libraries for this purpose (O. Bloom, J. Morgan, J. D. Buxbaum, C. Brown, W. Li, and colleagues, unpubl. data). In addition, cell bodies of giant RS neurons can be selectively sampled, providing an opportunity to pursue single-cell transcriptome analyses. Once the set of relevant regulatory genes has been identified, individual genes will be examined at higher temporal resolution to determine their time course of expression in order to resolve the spatiotemporal expression map, as described in Appendix Part A. Ideally, data would be sampled at high frequency over time, to best include transient expression of transcripts relevant to different aspects or stages of regeneration. In the lamprey, identification of the relevant time points for sampling transcriptome data will be greatly assisted by the well-defined time course of functional recovery (Fig. 2) and the accompanying anatomical changes, which serve as appropriate downstream indicators of presumed upstream key gene regulatory events. By using next-generation sequencing platforms to determine the transcriptome of tissues or cells at different stages of regeneration, it will be possible to achieve an optimal balance between inclusiveness and resolution (Wang et al., 2009; Metzker, 2010). In terms of spatial resolution, lamprey giant RS axons and cell bodies can be sampled by microdissection, potentially as single cells or as aggregates (McClellan et al., 2008; Tang et al., 2011) These experimental strategies allow for added depth of coverage within regenerating neurons, as some regulatory genes may be relevant at low mRNA copy numbers. By contrast, sampling all tissue around the site of spinal cord injury detects gene expression in non-neuronal as well as neuronal cells, such as invading immune cells or resident microglia that may be part of the wound healing response. By pursuing molecular analyses of both the injury site and of individual neurons that do or do not regenerate, we gain resolution into active networks intrinsic to distinct cell types and the same cell types that achieve different functional fates.
Perturbation analysis in reticulospinal neurons and surrounding tissue
The next step of the GRN approach, perturbation analysis, rests on the ability to alter cells genetically. Successfully employed methods to alter gene expression in lamprey include MOs or RNA constructs to inhibit translation or splicing of specific expressed target genes (McCauley and Bronner-Fraser, 2006; Sauka-Spengler et al., 2007; Nikitina and Bronner-Fraser, 2009). Efficient gene delivery methods, including microinjection, gene gun, and electroporation, have also been used in lamprey and in many cases, the giant RS neurons (Hall et al., 1997; Zhang and Selzer, 2001; Kim et al., 2009; Nikitina and Bronner-Fraser, 2009) (and J. R. Morgan, unpubl. data). Ultimately, there are many possible strategies for delivering expression plasmids and MOs to giant RS neurons, and the choice of experimental strategy will depend on the desired selectivity of the targeting.
The second critical experimental parameter for perturbation analysis involves staging of the regeneration process. While the recovery of spinal cord function after injury is quite reproducible across a population of lampreys (See Fig. 2 and e.g., Davis et al., 1993; Davis and McClellan, 1994a), the limited variations between animals can be monitored by internal controls within the same animal. For example, a MO, or other gene-altering strategy, can be delivered in a targeted manner to one-half of the brain or one cell type, while comparable cells or tissues in the same animal can be treated with a control MO or other reagent. An example of how this method has been used extensively is in the developing neural plate of chick, where one lateral half is perturbed by delivery of a gene-specific MO by electroporation, while the opposite side serves as an internal control (Basch et al., 2006).
Toward a Comparative Gene Regulatory Network Approach: Central Nervous System Regeneration in Other Species
What other organisms satisfy the criteria we set out for studying CNS regeneration with the GRN approach? While axonal regrowth and behavioral recovery after spinal cord injury is best studied in the larval and adult lamprey, there are several other vertebrate species known to be capable of regenerating various CNS components (Bhatt et al., 2007; Tanaka and Ferretti, 2009). In teleost fishes, as in lampreys, some RS neurons have the capacity to regenerate and recover functionality, while others do not. For example, in the adult goldfish, the largest of these RS neurons are identifiable, the Mauthner cells, which are capable of regrowth after spinal cord crush, but they contribute minimally to the return of function (Zottoli et al., 1994; and Zottoli et al., unpubl. obs.). Morphological homologs are nevertheless hypothesized to underlie the functional recovery of a fast startle response (Zottoli and Freemer, 2003). Although the regenerative processes are often not as well defined in the teleosts and other non-mammalian vertebrates as in lamprey, some of these models do offer other alternate advantages and therefore serve a valuable comparative role in dissecting gene networks controlling CNS regeneration.
One example of a well-studied genetic model is the zebrafish, Danio rerio. The larval zebrafish has many of the critical features for applying the GRN approach to understanding regeneration. Because of its transparency, for example, one can combine imaging with genetics to study neural circuits (McLean and Fetcho, 2008, 2010). RS neurons in larval zebrafish are optically accessible, allowing studies on the role of Mauthner neurons and other RS neurons in startle behavior (O’Malley et al., 1996; Fetcho et al., 1998; Liu and Fetcho, 1999; Gahtan et al., 2002; Nakayama and Oda, 2004). In addition, the functional consequences of neuronal regrowth can be assayed in a living zebrafish (Bhatt et al., 2007). CNS regeneration also occurs in the adult zebrafish (Becker et al., 1997; Becker and Becker, 2008), ultimately allowing a direct comparison with CNS regeneration in the larval zebrafish. As in lamprey, regenerative capacity can also be modulated in the larval zebrafish: Mauthner axons demonstrate minimal regeneration after spinal cord damage (Bhatt et al., 2004), but when cAMP is electroporated into the soma, axons sprout and functional regeneration occurs (Bhatt et al., 2004, 2007). Indeed, the ability to enhance regeneration or “convert” a non-regenerating neuron to a regenerating one by increasing intracellular cAMP has been observed in many species (Bhatt et al., 2004; Hannila and Filbin, 2008; Jin et al., 2009; Ghosh-Roy et al., 2010), and challenges the hypothesis that certain central neurons have an intrinsic inability to regenerate (Kiernan, 1979). This conversion validates the search for GRNs in the context of regeneration, suggesting that once identified, key nodes (perhaps those that contain cAMP responsive elements) may be evolutionarily conserved and can be turned on or off to promote regeneration.
Another example of an appropriate organism for GRN analysis of regeneration is one classically used to study development, the tadpole of the African clawed frog Xenopus laevis. In the tadpole, RA and raphe neurons reliably regrow after spinal cord injury (Gibbs and Szaro, 2006). Although the larval form undergoes functional recovery after spinal cord transection, this regenerative capacity is lost at metamorphosis (Sims, 1962; Forehand and Farel, 1982; Beattie et al., 1990; ten Donkelaar, 2000). This switch offers the possibility of elucidating the GRNs that convert regenerating neurons into non-regenerating neurons as it applies to the developing vertebrate, including mammalian, CNS (Mladinic et al., 2009). Another example of how Xenopus can be used to complement a comparative understanding of GRNs during regeneration relates to Mauthner neuron regeneration. Regrowth of Xenopus Mauthner axon results in its reconnection to motoneurons, as shown by ventral root activity after stimulation of a regenerated Mauthner cell (Lee, 1982). The optical accessibility of Mauthner axons in Xenopus tadpoles was used to document their anatomical regrowth with Lucifer yellow tracing after spinal cord transection. In 60% of the X. laevis tadpoles studied, the Mauthner axon had regenerated across the lesion site appropriately; in the other 40%, the Mauthner axons had not regenerated. In these cases, the Mauthner axons had either grown within the wound or grown in the wrong direction— rostrally (Lee, 1982). The fact that functional regeneration occurred in only about half of the damaged Mauthner axons provides yet another an opportunity to compare the GRNs in regenerating and non-regenerating RS neurons, perhaps in the same animal. Recently, it was shown in the tadpole that conditions that induced metamorphosis, such as the application of thyroid hormone, blocked reticular axon regeneration, while conditions that blocked metamorphosis permitted regeneration. Microarray and real-time quantitative polymerase chain reaction analyses of genetic material from tadpoles injured and allowed to recover in the presence or absence of thyroid hormone identified distinct changes in gene expression associated with regenerative and non-regenerative conditions (Gibbs et al., 2011). These experiments provide an exciting opportunity to focus on changes in gene regulatory networks of RS neurons after perturbations that impact regeneration.
Conclusions
The significance of the GRN approach is that it will allow us to determine the underlying molecular interactions that result in successful functional regeneration of the vertebrate CNS. Differences and similarities between species will provide insights into how animals have evolved alternate strategies to respond to similar perturbations. Across some vertebrate species, responses are distinct and opposed, as in the ability or inability to achieve functional recovery. In the lamprey, as in the zebrafish and the African clawed frog, different regenerative capacities exist even among cells in the same tissue. As described above, environmental conditions or perturbants, such as cAMP and thyroid hormone, can also affect the regenerative capacity of vertebrate CNS neurons. What are the fundamental differences in the regulatory systems that mediate this divergence? What are the similarities in the regulatory systems that allow us to converge upon the underlying molecular interactions that define successful CNS regeneration? A dissection of the underlying GRNs will be able to address these questions.
The strength of functional genomics derives in no small part from its capacity to act as a tool for discovery, displacing candidate-gene methods. The application of large-scale molecular analyses, the first components in the GRN approach, are now being applied in understanding various aspects of the nervous system, leading to insights that were impossible using standard approaches (Geschwind and Konopka, 2009; Jain and Heutink, 2010). For example, making use of genome-scale gene expression data, including spatial information derived from in situ hybridization, the CA3 region of the hippocampus can be unambiguously divided into nine distinct regions, with clearly defined boundaries and evidence for distinct connectivity (Thompson et al., 2008). This level of complexity was not fully appreciated previously, in spite of extensive molecular and functional analyses in this region. Other studies using the network approach in the CNS include those concerning synapse complexity across species (Emes et al., 2008), axonal sprouting after stroke (Li et al., 2010), Alzheimer’s pathways in mice and humans (Miller et al., 2010), motor neuron responses after spinal cord injury (Ryge et al., 2010), and differences between individual classes of neurons (Winden et al., 2009). Pioneering work on the network of cellular and molecular interactions governing peripheral nerve injury responses have also been described (Michaelevski et al., 2010). On a larger scale, analysis of weighted gene co-expression profiles in genome-scale expression data has begun to deepen our understanding of gene expression, moving away from simple lists of differential expression toward coherent, robust, and reproducible networks that are biologically plausible. These methods are revealing key nodal molecules that can then be used in confirmatory experiments. Systems-level analysis in whole-brain gene expression data identified modules that captured the transcriptional networks of major cell classes in the human brain on three levels—on a gross cellular level (e.g., neurons, oligodendrocytes, astrocytes), on the finer cellular level (e.g., parvalbumin-positive interneurons, Purkinje neurons, meningeal cells), and on the subcellular levels (e.g., glutamatergic synaptic function, mitochondrial) (Oldham et al., 2008).
The studies cited above go beyond providing proof-of-concept for the GRN approach by providing novel information on the function of known and heretofore unannotated genes that could not have been predicted. In addition, new insights have been provided to explain the molecular differences between and within cells over the time course of development (Goltsev et al., 2007; Smith and Davidson, 2008, 2009; Smith et al., 2008; Imai et al., 2009; Christiaen et al., 2010). Of course, perturbation of GRNs in genetically or otherwise experimentally modified systems can elucidate gene function and lead to a systems-level understanding of biological processes and disease pathogenesis (e.g., Michaelevski et al., 2010; Miller et al., 2010). In the particular setting of CNS regeneration, features of non-mammalian vertebrates such as the lamprey, whose nervous system is complex, experimentally accessible, and capable of robust functional recovery after injury, coupled with the GRN approach, will lead to important insights into relevant molecules and identify key regulatory nodes that can be targeted to modulate regenerative capability. The pursuit of such a strategy and comparison of data in multiple vertebrate species will identify nodes that are conserved throughout evolution and will also provide added perspective on the molecular programs needed for regeneration in mammals, which has obvious clinical relevance.
Acknowledgments
We thank all of our colleagues in functional genomics and in regeneration and apologize to those whose work was not discussed due to space limitations. In addition, the authors gratefully acknowledge support from The Marine Biological Laboratory, The Charles Evans Foundation (OB, JDB, JRM), AG005138 (JDB), and G. Harold and Leila Y. Mathers Research Professorship of Geriatrics and Adult Development (JDB); University of Texas, Austin start-up funds (JM), the Paralyzed Veterans of America Research Grant #2586 (JM) and the Morton Cure Paralysis Fund (JM); The Feinstein Institute for Medical Research (OB); The Essel Foundation (SJZ) and The Howard Hughes Medical Institute (Williams College).
Abbreviations
- CNS
central nervous system
- GRN
gene regulatory network
- MO
morpholino
- RG
regulatory gene
- RS
reticulospinal
Appendix
A. High-throughput spatiotemporal expression analysis
Determining the timing of gene expression for all genes in the regulatory gene (RG) set with high precision is an important element in network analysis. This requires a high density of sampling to capture what are often transient but critical changes in gene transcription, including the onset of expression of key regulatory genes and, often, cohorts of co-regulated genes. As relevant changes in gene expression are often rapid, it may be necessary to sample gene expression as often as at 30-min intervals. Profiling expression patterns at this level of resolution may define previously unknown stages of a biological process. While RNA-Seq is a sensitive method, it is not always practical to use for large numbers of samples. More sensitive and cost-effective methods, such as high-throughput quantitative PCR and other solution-phase hybridization methods (i.e., Nanostring) that have succeeded microarray technology can be used for quantitative analysis of gene expression over time.
How to determine spatial expression patterns for many genes? High-throughput in situ hybridization is not yet available at the level of throughput needed. During regeneration, moreover, the degree of stereotypic consistency that is found in embryological development may not be achievable. Two methods present alternatives. The first is fluorescence- activated cell-sorting to separate cell populations according to surface markers to add an additional qualitative step to quantitative RNA-seq. Another possibility, previously used on smaller scales, is to use translating-ribosome affinity purification to isolate in a single step the mRNA from highly specific cell populations (Heiman et al., 2008; Dougherty et al., 2010; Jiao and Meyerowitz, 2010). In short, a bacterial artificial chromosome (BAC) clone containing a gene of interest (GOI) is recombined to express a ribosomal protein in frame with a fluorescent protein; as BAC clones contain about 150 kb of surrounding noncoding sequence including regulatory elements, this serves as a faithful reporter for expression of the GOI and has proved to be a highly reliable method to achieve specific gene expression. Antibodies against the fluorescent protein conjugated to magnetic beads are then used to purify all ribosomes and mRNA that is being translated from the cells expressing the GOI. Next, mRNAs are amplified and sequenced by next-generation sequencing or directly analyzed by alternate transcriptomics platforms (i.e., Nanostring and HT-QPCR) for expression levels of all genes in the RG set. If these analyses are performed systematically for each gene in the RG set, they provide a multi-dimensional and quantitative map of co-expression covering every gene in the system, or a co-expression matrix. These analyses could be complemented by assays for histone modifications and other markers of active chromatin such as DNAse I hypersensitivity, within specific cell populations.
B. Generating network predictions by perturbation analysis: caveats and controls
Perturbation analysis is a key step in defining the essential nodes—that is, genes—within a regulatory network (Davidson et al., 2002). One potential issue with perturbation analysis is distinguishing between direct and indirect effects. By perturbing every gene in the network individually, one could be able to discriminate between effects that are direct and those that may be, at least in part, acting through other genes–that is, indirect (this method relies upon having a complete or near complete account of all genes in the network). For example, in developmental gene regulatory networks (GRNs), there may be several highly connected nodes. Perturbation of one of these would produce many downstream effects, a number of which may be several steps removed from the immediate action of the targeted gene. These downstream effects would therefore not be predictive of the direct function of the gene that has been knocked down. Thus, by perturbing any one of these genes, one might derive many false-positives within the GRN. Even under ideal circumstances the perturbation matrix produces these and other systemic errors, as is the case for all prospective analyses. In addition, these errors can be characterized as either false-positives (effects from the gene knockdown that are indirect only) or false-negatives (direct functions that are undetected due to low signal or other causes). Using an algorithm to minimize false-positive predictions necessarily increases the frequency of false negative interactions. For example, this occurs when a gene has both direct and indirect effects on a downstream gene or genes. Strict application of the perturbation matrix algorithm would predict only the direct interactions when, in fact, both direct and indirect interactions exist. Proper analysis requires consideration of this alternate possibility. Experience with developmental GRNs suggests that this network architecture is indeed common, perhaps because it allows information to flow through a low-pass filter. In other cases, gene perturbation effects are below the limits of detection of an assay, as when an activator/repressor gene product affects expression of a target gene in only a small subset of the cells expressing that target gene. Many examples of observations fall within this category. Overexpression of an effector gene may produce a detectable and interpretable signal in this scenario; however, gene overexpression itself produces artifacts, as it places the overexpressed gene in entirely novel contexts, both in time and space. Systematic gene knockdowns are therefore the preferred method of perturbation, despite the drawbacks stated above.
Several experimental variables can be optimized to reduce systemic errors and thereby improve the predictive power of perturbation analysis:
Time of incubation.One way to reduce systematic errors is to limit the time between induction of the genetic perturbation and harvest of the sample for quantitative analysis. In practice, this depends on the method of delivery of the agent for genetic perturbation— for example, morpholinos or DNA/RNA constructs. In many cases, as in the lamprey spinal cord, gene delivery into complex tissues at various time points can be achieved by electroporation (Hall et al., 1997; Zhang and Selzer, 2001; Kim et al., 2009; Nikitina and Bronner-Fraser, 2009; and J. R. Morgan, unpubl. data). By restricting sampling to about 2–8 h after perturbation, depending on transcription kinetics in an organism, detected effects should reflect direct effects on transcription. Increasing the sampling period should increase effects due to correspondingly more indirect interactions. This can and should be tested.
Dose responses. In the case of signaling pathways where small molecule inhibitors are available, doseresponse effects can bolster support for specific functional interactions, whether direct or indirect.
Local delivery. Delivery of perturbants to defined regions further allows direct comparison between control and experimental tissue within a single animal. This reduces the need for highly accurate staging, which could be particularly problematic outside of strictly defined developmental contexts.
ChIP-seq (and other complementary binding assays). This method uses antibodies against transcription factors or cofactors to identify direct binding sites following immunoprecipitation and next-generation sequencing (Mortazavi et al., 2006; Park, 2009). The simplest form of this assay requires a sequenced genome, which is available for many (or all) of the species that can (or will) be used for GRN analyses of successful CNS regeneration, including lampreys, zebrafish, and Xenopus. This produces a large number of signals indicating where in the genomic DNA regulatory element a given transcription-factor binds, at 20–100 bp resolution. Though there are many instances in which the transcription-factor interaction may not be functionally active, binding data can be cross-referenced with the perturbation matrix data to assess whether a functional interaction is implied at a given locus. In essence, ChIP-seq (measuring binding activity) and perturbation data (measuring functional consequences) produce complementary data sets and are optimally deployed where the two sets can be compared.
Unfortunately, the large-scale use of ChIP-seq is not often feasible, as it requires generating specific antibodies to 50–100 transcription factors. An alternative is to use antibodies against the relatively few co-activators and co-repressors involved with most transcriptional regulatory responses (Mortazavi et al., 2006). In theory, co-activator/corepressor (CoA/CoR) ChIP-seq should identify functional target sites with higher fidelity, in contrast to the unknown predictive capacity of transcription factor ChIP-seq (TF ChIP-seq), which may signal many non-functional interactions. CoA/CoR ChIP-seq may thus define the set of genomic regulatory elements for the regulatory genes of interest, the RG cis-regulatory module set (RG CRM set). Furthermore, consensus-binding sites are known for most transcription-factor classes and are highly conserved across species. These data indicate that transcription factors typically bind to eight nucleotide stretches, often with five to seven invariant or nearly invariant nucleotides—that is, sequences that would be found randomly once per 500– 2000 bp. At about 50 bp, the CoA/CoR ChIP-seq signal is an order of magnitude smaller, representing manageable search parameters for identifying candidate transcriptionfactor binding sites.
Sequence conservation may be a concern with this kind of approach. However, many transcription factors and regulatory proteins are highly conserved among vertebrate lineages. As an example, HMGb1, a DNA-binding protein, is highly conserved among vertebrates; HMG1 from Lampetra fluviatilis is 72% identical at the AA level to human HMG1 (Sharman et al., 1997). The Groucho/Tup1 family of corepressors is also well conserved among vertebrates (Buscarlet and Stifani, 2007). Although some DNA-binding proteins, co-repressors, and co-activators appear to be conserved across species, the possibility remains that this approach may require either raising of species-specific antibodies or use of exogenously tagged proteins, which would have to be expressed at sufficiently low levels and in a specific fashion so as to not alter normal development.
The result of the combined perturbation and CoA/CoR ChIP-seq analysis is a set of possible gene regulatory network interactions. Aligned with the spatiotemporal expression map, these data generate predictions of specific mechanisms or hypotheses describing the network of regulatory interactions driving differential gene expression to be validated. Crucially, in addition to its role in refining the set of predicted interactions, CoA/CoR ChIP-seq greatly reduces the search space for validation experiments at the level of cis-regulatory sequences. As limiting the analysis for putative target elements to less than 100 bp is the rate-limiting step of cis-regulatory analysis, and cis-regulatory analysis is itself the rate-limiting step for the overall network analysis process, achievement of this level of resolution has a high impact on the overall rate of the approach.
C. Cis-regulatory and cis-reengineering analysis
Detailed dissection of the functional transcription factor- DNA target site interactions represents the overall ratelimiting step in gene regulatory network (GRN) analysis. By testing specific mechanistic predictions, a solitary cisregulatory analysis accomplishes the immediate reductionist aim of describing the functional properties for an individual component of the system. Across a network, many regulatory analyses define the network architecture—specific, validated assessments of the transcription regulatory functions exerted by particular transcription factors on genes in the network. Accumulated mechanistic descriptions of individual regulatory interactions define the specific chain of events driving biological outcomes. In more directed experiments, altering specific transcription-factor target sequences in dominant acting transgenes, a process of network reengineering, tests the overarching network structure and critical biological information processing functions of network subcircuits.
Essential for both cis-analysis and reengineering, and beyond efficient gene delivery, is generation of reporter and expression constructs that faithfully reproduce the expression patterns of genes in the RG set. Bacterial artificial chromosome (BAC) clones containing 150 kb or more of sequence surrounding the transcription start site of genes of interest (GOIs) provide reliable starting material to capture all relevant cis-regulatory elements. BAC recombineering can be used for the targeted insertion coding sequence for fluorescent proteins in place of GOI coding sequence (these are the same recombinant BACs as described in Part A). By straightforward analysis, all necessary and sufficient regulatory elements can be identified employing the BAC-FP reporter and derivatives (Smith, 2008). Once BAC-FP reporters have been verified to reproduce expression faithfully, BAC clones without fluorescent protein knock-in provide the foundational material for creating expression constructs with reengineered transcriptional regulatory sequences to test specific network functions. This allows specific transcription- factor target sequences, often just 4 – 8 bp, to be tested for network function within what approximates a genomic context (e.g., Smith and Davidson, 2009).
Literature Cited
- Armstrong J, Zhang L, McClellan AD. Axonal regeneration of descending and ascending spinal projection neurons in spinal cord-transected larval lamprey. Exp. Neurol. 2003;180:156–166. doi: 10.1016/s0014-4886(02)00057-2. [DOI] [PubMed] [Google Scholar]
- Ayers J, Carpenter GA, Currie S, Kinch J. Which behavior does the lamprey central motor program mediate? Science. 1983;221:1312–1314. doi: 10.1126/science.6137060. [DOI] [PubMed] [Google Scholar]
- Bajoghli B, Aghaallaei N, Hess I, Rode I, Netuschil N, Tay BH, Venkatesh B, Yu JK, Kaltenbach SL, Holland ND, et al. Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell. 2009;138:186–197. doi: 10.1016/j.cell.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Beattie MS, Bresnahan JC, Lopate G. Metamorphosis alters the response to spinal cord transection in Xenopus laevis frogs. J. Neurobiol. 1990;21:1108–1122. doi: 10.1002/neu.480210714. [DOI] [PubMed] [Google Scholar]
- Becker CG, Becker T. Adult zebrafish as a model for successful central nervous system regeneration. Restor. Neurol. Neurosci. 2008;26:71–80. [PubMed] [Google Scholar]
- Becker T, Wullimann MF, Becker CG, Bernhardt RR, Schachner M. Axonal regrowth after spinal cord transection in adult zebrafish. J. Comp. Neurol. 1997;377:577–595. doi: 10.1002/(sici)1096-9861(19970127)377:4<577::aid-cne8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Bevan S, Vakharia V, Parker D. Changes in gene expression and integrin-mediated structural changes are associated with long-term plasticity of a spinal cord locomotor network. Neuroscience. 2008;152:160–168. doi: 10.1016/j.neuroscience.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Bhatt DH, Otto SJ, Depoister B, Fetcho JR. Cyclic AMP-induced repair of zebrafish spinal circuits. Science. 2004;305:254–258. doi: 10.1126/science.1098439. [DOI] [PubMed] [Google Scholar]
- Bhatt DH, Patzelova H, McLean DL, Fetcho JR, Zottoli SJ. Functional regeneration in the larval zebrafish spinal cord. In: Becker CG, Becker T, editors. Model Organisms in Spinal Cord Regeneration. Weinheim, Germany: Wiley VCH; 2007. pp. 263–288. [Google Scholar]
- Blau HM, Pomerantz JH. Re “evolutionary” regenerative medicine. JAMA Online First. 2010;305:87–88. doi: 10.1001/jama.2010.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blesch A, Tuszynski MH. Spinal cord injury: plasticity, regeneration and the challenge of translational drug development. Trends Neurosci. 2009;32:41–47. doi: 10.1016/j.tins.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Borgens RB, Roederer E, Cohen MJ. Enhanced spinal cord regeneration in lamprey by applied electric fields. Science. 1981;213:611–617. doi: 10.1126/science.7256258. [DOI] [PubMed] [Google Scholar]
- Bradbury EJ, Carter LM. Manipulating the glial scar: chondroitinase ABC as a therapy for spinal cord injury. Brain Res. Bull. 2011;84:306–316. doi: 10.1016/j.brainresbull.2010.06.015. [DOI] [PubMed] [Google Scholar]
- Brent AE, Yucel G, Small S, Desplan C. Permissive and instructive anterior patterning rely on mRNA localization in the wasp embryo. Science. 2007;315:1841–1843. doi: 10.1126/science.1137528. [DOI] [PubMed] [Google Scholar]
- Buchanan JT. Contributions of identifiable neurons and neuron classes to lamprey vertebrate neurobiology. Prog. Neurobiol. 2001;63:441–466. doi: 10.1016/s0301-0082(00)00050-2. [DOI] [PubMed] [Google Scholar]
- Bullock T. Identifiable and addressed neurons in the vertebrates. In: Faber DS, Korn H, editors. Neurobiology of the Mauthner Cell. New York: Raven Press; 1978. pp. 1–12. [Google Scholar]
- Bullock TH. Comparative neuroscience holds promise for quiet revolutions. Science. 1984;225:473–478. doi: 10.1126/science.6740319. [DOI] [PubMed] [Google Scholar]
- Bullock TH. Revisiting the concept of identifiable neurons. Brain Behav. Evol. 2000;55:236–240. doi: 10.1159/000006657. [DOI] [PubMed] [Google Scholar]
- Buscarlet M, Stifani S. The ‘Marx’ of Groucho on development and disease. Trends Cell Biol. 2007;17:353–361. doi: 10.1016/j.tcb.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Christiaen L, Stolfi A, Levine M. BMP signaling coordinates gene expression and cell migration during precardiac mesoderm development. Dev. Biol. 2010;340:179–187. doi: 10.1016/j.ydbio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Cohen AH, Mackler SA, Selzer ME. Functional regeneration following spinal transection demonstrated in the isolated spinal cord of the larval sea lamprey. Proc. Natl. Acad. Sci. USA. 1986;83:2763–2766. doi: 10.1073/pnas.83.8.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AH, Mackler SA, Selzer ME. Behavioral recovery following spinal transection: functional regeneration in the lamprey CNS. Trends Neurosci. 1988;11:227–231. doi: 10.1016/0166-2236(88)90131-2. [DOI] [PubMed] [Google Scholar]
- Cohen AH, Baker MT, Dobrov TA. Evidence for functional regeneration in the adult lamprey spinal cord following transection. Brain Res. 1989;496:368–372. doi: 10.1016/0006-8993(89)91090-1. [DOI] [PubMed] [Google Scholar]
- Cohen AH, Kiemel T, Pate V, Blinder J, Guan L. Temperature can alter the function outcome of spinal cord regeneration in larval lampreys. Neuroscience. 1999;90:957–965. doi: 10.1016/s0306-4522(98)00502-8. [DOI] [PubMed] [Google Scholar]
- Cooke RM, Parker D. Locomotor recovery after spinal cord lesions in the lamprey is associated with functional and ultrastructural changes below lesion sites. J. Neurotrauma. 2009;26:597–612. doi: 10.1089/neu.2008.0660. [DOI] [PubMed] [Google Scholar]
- Cruce WL, Stuesse SL, Newman DB. Evolution of the reticular formation. Acta Biol. Hung. 1988;39:327–333. [PubMed] [Google Scholar]
- Currie SN, Ayers J. Regeneration of locomotor command systems in the sea lamprey. Brain Res. 1983;279:238–240. doi: 10.1016/0006-8993(83)90183-x. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Davis GR, Jr, McClellan AD. Extent and time course of restoration of descending brainstem projections in spinal cord-transected lamprey. J. Comp. Neurol. 1994a;344:65–82. doi: 10.1002/cne.903440106. [DOI] [PubMed] [Google Scholar]
- Davis GR, Jr, McClellan AD. Long distance axonal regeneration of identified lamprey reticulospinal neurons. Exp. Neurol. 1994b;127:94–105. doi: 10.1006/exnr.1994.1083. [DOI] [PubMed] [Google Scholar]
- Davis GR, Jr, Troxel MT, Kohler VJ, Grossmann EM, McClellan AD. Time course of locomotor recovery and functional regeneration in spinal-transected lamprey: kinematics and electromyography. Exp. Brain Res. 1993;97:83–95. doi: 10.1007/BF00228819. [DOI] [PubMed] [Google Scholar]
- Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 2010;38:4218–4230. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuc R, Brocard F, Antri M, Fenelon K, Gariepy JF, Smetana R, Menard A, Le Ray D, Viana Di Prisco G, Pearlstein E, et al. Initiation of locomotion in lampreys. Brain Res. Rev. 2008;57:172–182. doi: 10.1016/j.brainresrev.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Eaton RC, DiDomenico R, Nissanov J. Role of the Mauthner cell in sensorimotor integration by the brain stem escape network. Brain Behav. Evol. 1991;37:272–285. doi: 10.1159/000114365. [DOI] [PubMed] [Google Scholar]
- Eaton RC, Lee RK, Foreman MB. The Mauthner cell and other identified neurons of the brainstem escape network of fish. Prog. Neurobiol. 2001;63:467–485. doi: 10.1016/s0301-0082(00)00047-2. [DOI] [PubMed] [Google Scholar]
- Emes RD, Pocklington AJ, Anderson CN, Bayes A, Collins MO, Vickers CA, Croning MD, Malik BR, Choudhary JS, Armstrong JD, Grant SG. Evolutionary expansion and anatomical specialization of synapse proteome complexity. Nat. Neurosci. 2008;11:799–806. doi: 10.1038/nn.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetcho JR. The spinal motor system in early vertebrates and some of its evolutionary changes. Brain Behav. Evol. 1992;40:82–97. doi: 10.1159/000113905. [DOI] [PubMed] [Google Scholar]
- Fetcho JR, Cox KJ, O’Malley DM. Monitoring activity in neuronal populations with single-cell resolution in a behaving vertebrate. Histochem. J. 1998;30:153–167. doi: 10.1023/a:1003243302777. [DOI] [PubMed] [Google Scholar]
- Forehand CJ, Farel PB. Anatomical and behavioral recovery from the effects of spinal cord transection: dependence on metamorphosis in anuran larvae. J. Neurosci. 1982;2:654–652. doi: 10.1523/JNEUROSCI.02-05-00654.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahtan E, Sankrithi N, Campos JB, O’Malley DM. Evidence for a widespread brain stem escape network in larval zebra-fish. J. Neurophysiol. 2002;87:608–614. doi: 10.1152/jn.00596.2001. [DOI] [PubMed] [Google Scholar]
- Genome Institute at Washington University. Petromyzon marinus. 2007 Sea Lamprey. [Online] Genome provided by Marianne Bonner-Fraser. Available: http://genome.wustl.edu/genomes/view/petromyzon_marinus/ (2011, July 15). [Google Scholar]
- Geschwind DH, Konopka G. Neuroscience in the era of functional genomics and systems biology. Nature. 2009;461:908–915. doi: 10.1038/nature08537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Roy A, Wu Z, Goncharov A, Jim Y, Chisholm AD. Calcium and cyclic AMP promote axonal regeneration in Caenorhabditis elegans and require DLK-1 kinase. J. Neurosci. 2010;30:3175–3183. doi: 10.1523/JNEUROSCI.5464-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs KM, Szaro BG. Regeneration of descending projections in Xenopus laevis tadpole spinal cord demonstrated by retrograde double labeling. Brain Res. 2006;1088:68–72. doi: 10.1016/j.brainres.2006.02.126. [DOI] [PubMed] [Google Scholar]
- Gibbs KM, Chittur SV, Szaro BG. Metamorphosis and the regenerative capacity of spinal cord axons in Xenopus laevis. Eur. J. Neurosci. 2011;33:9–25. doi: 10.1111/j.1460-9568.2010.07477.x. [DOI] [PubMed] [Google Scholar]
- Goltsev Y, Fuse N, Frasch M, Zinzen RP, Lanzaro G, Levine M. Evolution of the dorsal-ventral patterning network in the mosquito. Anopheles gambiae. Development. 2007;134:2415–2424. doi: 10.1242/dev.02863. [DOI] [PubMed] [Google Scholar]
- Grillner S, Jessell TM. Measured motion: searching for simplicity in spinal locomotor networks. Curr. Opin. Neurobiol. 2009;19:572–586. doi: 10.1016/j.conb.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth L, Brewer CR, Collins WF, Jr, Goldberger ME, Perl ER. Criteria for evaluating spinal cord regeneration experiments. Exp. Neurol. 1980;69:1–3. doi: 10.1016/0014-4886(80)90138-7. [DOI] [PubMed] [Google Scholar]
- Hall GF, Yao J, Lee G. Human tau becomes phosphorylated and forms filamentous deposits when overexpressed in lamprey central neurons in situ. Proc. Natl. Acad. Sci. USA. 1997;94:4733–4738. doi: 10.1073/pnas.94.9.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannila SS, Filbin MT. The role of cyclic AMP signaling in promoting axonal regeneration after spinal cord injury. Exp. Neurol. 2008;209:321–332. doi: 10.1016/j.expneurol.2007.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardisty M, Potter I. The Biology of Lampreys. London: Academic Press; 1971. [Google Scholar]
- Harel NY, Strittmatter SM. Can regenerating axons recapitulate developmental guidance during recovery from spinal cord injury? Nat. Rev. Neurosci. 2006;7:603–616. doi: 10.1038/nrn1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbard E. Regeneration in the severed spinal cord of chordate larvae of Petromyzon marinus. Exp. Neurol. 1963;7:175–185. [Google Scholar]
- Imai KS, Stolfi A, Levine M, Satou Y. Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development. 2009;136:285–293. doi: 10.1242/dev.026419. [DOI] [PubMed] [Google Scholar]
- Ingber DE, Levin M. What lies at the interface of regenerative medicine and developmental biology? Development. 2007;134:2541–2547. doi: 10.1242/dev.003707. [DOI] [PubMed] [Google Scholar]
- Jacobs AJ, Swain GP, Snedeker JA, Pijak DS, Gladstone LJ, Selzer ME. Recovery of neurofilament expression selectively in regenerating reticulospinal neurons. J. Neurosci. 1997;17:5206–5220. doi: 10.1523/JNEUROSCI.17-13-05206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Heutink P. From single genes to gene networks: high-throughput-high-content screening for neurological disease. Neuron. 2010;68:207–217. doi: 10.1016/j.neuron.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Meyerowitz EM. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 2010;6:419. doi: 10.1038/msb.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin LQ, Zhang G, Jamison C, Jr, Takano H, Haydon PG, Selzer ME. Axon regeneration in the absence of growth cones: acceleration by cyclic AMP. J. Comp. Neurol. 2009;515:295–312. doi: 10.1002/cne.22057. [DOI] [PubMed] [Google Scholar]
- Kiernan JA. Hypotheses concerned with axonal regeneration in the mammalian nervous system. Biol. Rev. Camb. Philos. Soc. 1979;54:155–197. doi: 10.1111/j.1469-185x.1979.tb00871.x. [DOI] [PubMed] [Google Scholar]
- Kim W, Lee S, Jung C, Ahmed A, Lee G, Hall GF. Interneuronal transfer of human tau between lamprey central neurons in situ. J. Alzheimers Dis. 2009;19:647–664. doi: 10.3233/JAD-2010-1273. [DOI] [PubMed] [Google Scholar]
- Kimmel CB. Reticulospinal and vestibulospinal neurons in the young larva of a teleost fish, Brachydanio rerio. Prog. Brain Res. 1982;57:1–23. doi: 10.1016/S0079-6123(08)64122-9. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Powell SL, Metcalfe WK. Brain neurons which project to the spinal cord in young larvae of the zebrafish. J. Comp. Neurol. 1982;205:112–127. doi: 10.1002/cne.902050203. [DOI] [PubMed] [Google Scholar]
- Kinkhabwala A, Riley M, Koyama M, Monen J, Satou C, Kimura Y, Higashijima SI, Fetcho J. A structural and functional ground plan for neurons in the hindbrain of zebrafish. Proc. Natl. Acad. Sci. USA. 2011;108:1164–1169. doi: 10.1073/pnas.1012185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT. Regeneration and functional reconnection of an identified vertebrate central neuron. J. Neurosci. 1982;2:1793–1811. doi: 10.1523/JNEUROSCI.02-12-01793.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RK, Eaton RC. Identifiable reticulospinal neurons of the adult zebrafish, Brachydanio rerio. J. Comp. Neurol. 1991;304:34–52. doi: 10.1002/cne.903040104. [DOI] [PubMed] [Google Scholar]
- Lee RK, Eaton RC, Zottoli SJ. Segmental arrangement of reticulospinal neurons in the goldfish hindbrain. J. Comp. Neurol. 1993;329:539–556. doi: 10.1002/cne.903290409. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Li S, Overman JJ, Katsman D, Kozlov SV, Donnelly CJ, Twiss JL, Giger RJ, Coppola G, Geschwind DH, Carmichael ST. An age-related sprouting transcriptome provides molecular control of axonal sprouting after stroke. Nat. Neurosci. 2010;13:1496–1504. doi: 10.1038/nn.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W Lamprey Genome Project. Consortium. Seattle, Washington: Unpublished Data, Michigan State University, Lansing, Michigan and WUSTL Genome Center; 2011. [Google Scholar]
- Lingenhohl K, Friauf E. Giant neurons in the rat reticular formation: a sensorimotor interface in the elementary acoustic startle circuit? J. Neurosci. 1994;14:1176–1194. doi: 10.1523/JNEUROSCI.14-03-01176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KS, Fetcho JR. Laser ablations reveal functional relationships of segmental hindbrain neurons in zebrafish. Neuron. 1999;23:325–335. doi: 10.1016/s0896-6273(00)80783-7. [DOI] [PubMed] [Google Scholar]
- Lurie DI, Selzer ME. Axonal regeneration in the adult lamprey spinal cord. J. Comp. Neurol. 1991a;306:409–416. doi: 10.1002/cne.903060305. [DOI] [PubMed] [Google Scholar]
- Lurie DI, Selzer ME. Preferential regeneration of spinal axons through the scar in hemisected lamprey spinal cord. J. Comp. Neurol. 1991b;313:669–679. doi: 10.1002/cne.903130410. [DOI] [PubMed] [Google Scholar]
- Lurie DI, Selzer ME. The need for cellular elements during axonal regeneration in the sea lamprey spinal cord. Exp. Neurol. 1991c;112:64–71. doi: 10.1016/0014-4886(91)90114-r. [DOI] [PubMed] [Google Scholar]
- Lurie DI, Pijak DS, Selzer ME. Structure of reticulospinal axon growth cones and their cellular environment during regeneration in the lamprey spinal cord. J. Comp. Neurol. 1994;344:559–580. doi: 10.1002/cne.903440406. [DOI] [PubMed] [Google Scholar]
- Mackler SA, Selzer ME. Specificity of synaptic regeneration in the spinal cord of the larval sea lamprey. J. Physiol. 1987;388:183–198. doi: 10.1113/jphysiol.1987.sp016609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron K. Regeneration capacity of the spinal cord in Lampetra fluviatilis larvae. Folia Biol. 1959;7:179–189. [Google Scholar]
- McCauley DW, Bronner-Fraser M. Importance of SoxE in neural crest development and the evolution of the pharynx. Nature. 2006;441:750–752. doi: 10.1038/nature04691. [DOI] [PubMed] [Google Scholar]
- McClellan AD. Locomotor recovery in spinal-transected lamprey: regenerated spinal coordinating neurons and mechanosensory inputs couple locomotor activity across a spinal lesion. Neuroscience. 1990;35:675–685. doi: 10.1016/0306-4522(90)90338-5. [DOI] [PubMed] [Google Scholar]
- McClellan AD. Functional regeneration and restoration of locomotor activity following spinal cord transection in the lamprey. Prog. Brain Res. 1994;103:203–217. doi: 10.1016/s0079-6123(08)61137-1. [DOI] [PubMed] [Google Scholar]
- McClellan AD, Kovalenko MO, Benes JA, Schulz DJ. Spinal cord injury induces changes in electrophysiological properties and ion channel expression of reticulospinal neurons in larval lamprey. J. Neurosci. 2008;28:650–659. doi: 10.1523/JNEUROSCI.3840-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean DL, Fetcho JR. Using imaging and genetics in zebrafish to study developing spinal circuits in vivo. Dev. Neurobiol. 2008;68:817–834. doi: 10.1002/dneu.20617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean DL, Fetcho JR. Movement, technology and discovery in the zebrafish. Curr. Opin. Neurobiol. 2010;21:105–110. doi: 10.1016/j.conb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe WK, Mendelson B, Kimmel CB. Segmental homologies among reticulospinal neurons in the hindbrain of the zebrafish larva. J. Comp. Neurol. 1986;251:147–159. doi: 10.1002/cne.902510202. [DOI] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Michaelevski I, Segal-Ruder Y, Rozenbaum M, Medzihradszky KF, Shalem O, Coppola G, Horn-Saban S, Ben-Yaakov K, Dagan SY, Rishal I, et al. Signaling to transcription networks in the neuronal retrograde injury response. Sci. Signal. 2010;3:ra53. doi: 10.1126/scisignal.2000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc. Natl. Acad. Sci. USA. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladinic M, Muller KJ, Nicholls JG. Central nervous system regeneration: from leech to opossum. J. Physiol. 2009;587:2775–2782. doi: 10.1113/jphysiol.2009.169938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Leeper Thompson EC, Garcia ST, Myers RM, Wold B. Comparative genomics modeling of the NRSF/REST repressor network: from single conserved sites to genome-wide repertoire. Genome Res. 2006;16:1208–1221. doi: 10.1101/gr.4997306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Uchida K, Rijli FM, Kuratani S. Evolution of the brain developmental plan: insights from agnathans. Dev. Biol. 2005;280:249–259. doi: 10.1016/j.ydbio.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Kuratani S. Brain segmentation and trigeminal projections in the lamprey; with reference to vertebrate brain evolution. Brain Res. Bull. 2008;75:218–224. doi: 10.1016/j.brainresbull.2007.10.057. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Watanabe A. Development of the central and peripheral nervous systems in the lamprey. Dev. Growth Differ. 2009;51:197–205. doi: 10.1111/j.1440-169X.2009.01087.x. [DOI] [PubMed] [Google Scholar]
- Nakayama H, Oda Y. Common sensory inputs and differential excitability of segmentally homologous reticulospinal neurons in the hindbrain. J. Neurosci. 2004;24:3199–3209. doi: 10.1523/JNEUROSCI.4419-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DB, Cruce WL, Bruce LL. The sources of supraspinal afferents to the spinal cord in a variety of limbed reptiles. I. Reticulospinal systems. J. Comp. Neurol. 1983;215:17–32. doi: 10.1002/cne.902150103. [DOI] [PubMed] [Google Scholar]
- Nikitina NV, Bronner-Fraser M. Gene regulatory networks that control the specification of neural-crest cells in the lamprey. Biochim. Biophys. Acta. 2009;1789:274–278. doi: 10.1016/j.bbagrm.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Nikitina N, Sauka-Spengler T, Bronner-Fraser M. Dissecting early regulatory relationships in the lamprey neural crest gene network. Proc. Natl. Acad. Sci. USA. 2008;105:20083–20088. doi: 10.1073/pnas.0806009105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissanov J, Eaton RC. Reticulospinal control of rapid escape turning maneuvers in fishes. Am. Zool. 1989;29:103–121. [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat. Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphint PA, Alieva N, Foldes AE, Tytell ED, Lau BY, Pariseau JS, Cohen AH, Morgan JR. Regenerated synapses in lamprey spinal cord are sparse and small even after functional recovery from injury. J. Comp. Neurol. 2010;518:2854–2872. doi: 10.1002/cne.22368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley DM, Kao YH, Fetcho JR. Imaging the functional organization of zebrafish hindbrain segments during escape behaviors. Neuron. 1996;17:1145–1155. doi: 10.1016/s0896-6273(00)80246-9. [DOI] [PubMed] [Google Scholar]
- Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D, Grillner S. Neuronal mechanisms of synaptic and network plasticity in the lamprey spinal cord. Prog. Brain Res. 2000;125:381–398. doi: 10.1016/S0079-6123(00)25027-9. [DOI] [PubMed] [Google Scholar]
- Roederer E, Goldberg NH, Cohen MJ. Modification of retrograde degeneration in transected spinal axons of the lamprey by applied DC current. J. Neurosci. 1983;3:153–160. doi: 10.1523/JNEUROSCI.03-01-00153.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan M. Origins of the descending spinal projections in petromyzontid and myxinoid agnathans. J. Comp. Neurol. 1989;281:54–68. doi: 10.1002/cne.902810106. [DOI] [PubMed] [Google Scholar]
- Rovainen CM. Physiological and anatomical studies on large neurons of central nervous system of the sea lamprey (Petromyzon marinus). I. Muller and Mauthner cells. J. Neurophysiol. 1967a;30:1000–1023. doi: 10.1152/jn.1967.30.5.1000. [DOI] [PubMed] [Google Scholar]
- Rovainen CM. Physiological and anatomical studies on large neurons of central nervous system of the sea lamprey (Petromyzon marinus). II. Dorsal cells and giant interneurons. J. Neurophysiol. 1967b;30:1024–1042. doi: 10.1152/jn.1967.30.5.1024. [DOI] [PubMed] [Google Scholar]
- Rovainen CM. Regeneration of Muller and Mauthner axons after spinal transection in larval lampreys. J. Comp. Neurol. 1976;168:545–554. doi: 10.1002/cne.901680407. [DOI] [PubMed] [Google Scholar]
- Rovainen CM. Neurobiology of lampreys. Physiol. Rev. 1979;59:1007–1077. doi: 10.1152/physrev.1979.59.4.1007. [DOI] [PubMed] [Google Scholar]
- Rovainen CM, Johnson PA, Roach EA, Mankovsky JA. Projections of individual axons in lamprey spinal cord determined by tracings through serial sections. J. Comp. Neurol. 1973;149:193–201. doi: 10.1002/cne.901490205. [DOI] [PubMed] [Google Scholar]
- Ryge J, Winther O, Wienecke J, Sandelin A, Westerdahl AC, Hultborn H, Kiehn O. Transcriptional regulation of gene expression clusters in motor neurons following spinal cord injury. BMC Genomics. 2010;11:365. doi: 10.1186/1471-2164-11-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Alvarado A, Tsonis PA. Bridging the regeneration gap: genetic insights from diverse animal models. Nat. Rev. Genet. 2006;7:873–884. doi: 10.1038/nrg1923. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. Insights from a sea lamprey into the evolution of neural crest gene regulatory network. Biol. Bull. 2008;214:303–314. doi: 10.2307/25470671. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Dev. Cell. 2007;13:405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Selzer ME. Mechanisms of functional recovery and regeneration after spinal cord transection in larval sea lamprey. J. Physiol. 1978;277:395–408. doi: 10.1113/jphysiol.1978.sp012280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharman AC, Hay-Schmidt A, Holland PW. Cloning and analysis of an HMG gene from the lamprey Lampetra fluviatilis: gene duplication in vertebrate evolution. Gene. 1997;184:99–105. doi: 10.1016/s0378-1119(96)00580-x. [DOI] [PubMed] [Google Scholar]
- Shifman MI, Selzer ME. Expression of the netrin receptor UNC-5 in lamprey brain: modulation by spinal cord transection. Neurorehabil. Neural Repair. 2000a;14:49–58. doi: 10.1177/154596830001400106. [DOI] [PubMed] [Google Scholar]
- Shifman MI, Selzer ME. In situ hybridization in wholemounted lamprey spinal cord: localization of netrin mRNA expression. J. Neurosci. Methods. 2000b;104:19–25. doi: 10.1016/s0165-0270(00)00316-2. [DOI] [PubMed] [Google Scholar]
- Shifman MI, Selzer ME. Semaphorins and their receptors in lamprey CNS: cloning, phylogenetic analysis, and developmental changes during metamorphosis. J. Comp. Neurol. 2006;497:115–132. doi: 10.1002/cne.20990. [DOI] [PubMed] [Google Scholar]
- Shifman MI, Selzer ME. Differential expression of class 3 and 4 semaphorins and netrin in the lamprey spinal cord during regeneration. J. Comp. Neurol. 2007;501:631–646. doi: 10.1002/cne.21283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman MI, Zhang G, Selzer ME. Delayed death of identified reticulospinal neurons after spinal cord injury in lampreys. J. Comp. Neurol. 2008;510:269–282. doi: 10.1002/cne.21789. [DOI] [PubMed] [Google Scholar]
- Shifman MI, Yumul RE, Laramore C, Selzer ME. Expression of the repulsive guidance molecule RGM and its receptor neogenin after spinal cord injury in sea lamprey. Exp. Neurol. 2009;217:242–251. doi: 10.1016/j.expneurol.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RT. Transection of the spinal cord in developing Xenopus laevis. J. Embryol. Exp. Morphol. 1962;10:115–126. [PubMed] [Google Scholar]
- Smith J. A protocol describing the principles of cis-regulatory analysis in the sea urchin. Nat. Protoc. 2008;3:710–718. doi: 10.1038/nprot.2008.39. [DOI] [PubMed] [Google Scholar]
- Smith J, Davidson EH. Gene regulatory network sub-circuit controlling a dynamic spatial pattern of signaling in the sea urchin embryo. Proc. Natl. Acad. Sci. USA. 2008;105:20089–20094. doi: 10.1073/pnas.0806442105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Davidson EH. Regulative recovery in the sea urchin embryo and the stabilizing role of fail-safe gene network wiring. Proc. Natl. Acad. Sci. USA. 2009;106:18291–18296. doi: 10.1073/pnas.0910007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Kraemer E, Liu H, Theodoris C, Davidson E. A spatially dynamic cohort of regulatory genes in the endomesodermal gene network of the sea urchin embryo. Dev. Biol. 2008;313:863–875. doi: 10.1016/j.ydbio.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straka H, Gilland E, Baker R. Rhombomeric organization of brainstem motor neurons in larval frogs. Biol. Bull. 1998;195:220–222. doi: 10.2307/1542849. [DOI] [PubMed] [Google Scholar]
- Straka H, Baker R, Gilland E. Rhombomeric organization of vestibular pathways in larval frogs. J. Comp. Neurol. 2001;437:42–55. doi: 10.1002/cne.1268. [DOI] [PubMed] [Google Scholar]
- Straka H, Baker R, Gilland E. Preservation of segmental hindbrain organization in adult frogs. J. Comp. Neurol. 2006;494:228–245. doi: 10.1002/cne.20801. [DOI] [PubMed] [Google Scholar]
- Swain GP, Snedeker JA, Ayers J, Selzer ME. Cytoarchitecture of spinal-projecting neurons in the brain of the larval sea lamprey. J. Comp. Neurol. 1993;336:194–210. doi: 10.1002/cne.903360204. [DOI] [PubMed] [Google Scholar]
- Tanaka EM, Ferretti P. Considering the evolution of regeneration in the central nervous system. Nat. Rev. Neurosci. 2009;10:713–723. doi: 10.1038/nrn2707. [DOI] [PubMed] [Google Scholar]
- Tang F, Lao K, Surani MA. Development and applications of single-cell transcriptome analysis. Nat. Methods. 2011;8(4s):S6–S11. doi: 10.1038/nmeth.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Donkelaar HJ. Development and regenerative capacity of descending supraspinal pathways in tetrapods: a comparative approach. Adv. Anat. Embryol. Cell Biol. 2000;154:iii–ix. 1–145. doi: 10.1007/978-3-642-57125-1. [DOI] [PubMed] [Google Scholar]
- Thompson CL, Pathak SD, Jeromin A, Ng LL, MacPherson CR, Mortrud MT, Cusick A, Riley ZL, Sunkin SM, Bernard A, et al. Genomic anatomy of the hippocampus. Neuron. 2008;60:1010–1021. doi: 10.1016/j.neuron.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Tseng AS, Levin M. Tail regeneration in Xenopus laevis as a model for understanding tissue repair. J. Dent. Res. 2008;87:806–816. doi: 10.1177/154405910808700909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22:2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade N. Rare hits and heaps of misses to pay for. The New York Times. 2010 Nov 9;:D1. [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Hirano S, Murakami Y. Development of the lamprey central nervous system, with reference to vertebrate evolution. Zoolog. Sci. 2008;25:1020–1027. doi: 10.2108/zsj.25.1020. [DOI] [PubMed] [Google Scholar]
- Winden KD, Oldham MC, Mirnics K, Ebert PJ, Swan CH, Levitt P, Rubenstein JL, Horvath S, Geschwind DH. The organization of the transcriptional network in specific neuronal classes. Mol. Syst. Biol. 2009;5:291. doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MR, Cohen MJ. Synaptic regeneration in identified neurons of the lamprey spinal cords. Science. 1979;206:344–347. doi: 10.1126/science.482943. [DOI] [PubMed] [Google Scholar]
- Wood MR, Cohen MJ. Synaptic regeneration and glial reactions in the transected spinal cord of the lamprey. J. Neurocytol. 1981;10:57–79. doi: 10.1007/BF01181745. [DOI] [PubMed] [Google Scholar]
- Yeomans JS, Li L, Scott BW, Frankland PW. Tactile, acoustic and vestibular systems sum to elicit the startle reflex. Neurosci. Biobehav. Rev. 2002;26:1–11. doi: 10.1016/s0149-7634(01)00057-4. [DOI] [PubMed] [Google Scholar]
- Yin HS, Selzer ME. Axonal regeneration in lamprey spinal cord. J. Neurosci. 1983;3:1135–1144. doi: 10.1523/JNEUROSCI.03-06-01135.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HS, Mackler SA, Selzer ME. Directional specificity in the regeneration of lamprey spinal axons. Science. 1984;224:894–896. doi: 10.1126/science.6719120. [DOI] [PubMed] [Google Scholar]
- Zhang G, Selzer ME. In vivo transfection of lamprey brain neurons by gene gun delivery of DNA. Exp. Neurol. 2001;167:304–311. doi: 10.1006/exnr.2000.7584. [DOI] [PubMed] [Google Scholar]
- Zhang L, Palmer R, McClellan AD. Conditioning lesions enhance axonal regeneration of descending brain neurons in spinal-cord-transected larval lamprey. J. Comp. Neurol. 2004;478:395–404. doi: 10.1002/cne.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Jin LQ, Sul JY, Haydon PG, Selzer ME. Live imaging of regenerating lamprey spinal axons. Neurorehabil. Neural Repair. 2005;19:46–57. doi: 10.1177/1545968305274577. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Melton DA. Extreme makeover: converting one cell into another. Cell Stem Cell. 2008;3:382–388. doi: 10.1016/j.stem.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Zottoli SJ, Freemer MM. Recovery of C-starts, equilibrium and targeted feeding after whole spinal cord crush in the adult goldfish Carassius auratus. J. Exp. Biol. 2003;206:3015–3029. doi: 10.1242/jeb.00512. [DOI] [PubMed] [Google Scholar]
- Zottoli SJ, Bentley AP, Feiner DG, Hering JR, Prendergast BJ, Rieff HI. Spinal cord regeneration in adult goldfish: implications for functional recovery in vertebrates. Prog. Brain Res. 1994;103:219–228. doi: 10.1016/s0079-6123(08)61138-3. [DOI] [PubMed] [Google Scholar]