Abstract
Background
In Africa, relapsing fever borreliae are neglected vector-borne pathogens that cause mild to deadly septicemia and miscarriage. Screening vectors for the presence of borreliae currently requires technically demanding, time- and resource-consuming molecular methods. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) has recently emerged as a tool for the rapid identification of vectors and the identification of cultured borreliae. We investigated whether MALDI-TOF-MS could detect relapsing fever borreliae directly in ticks.
Methodology/Principal Findings
As a first step, a Borrelia MALDI-TOF-MS database was created to house the newly determined Mean Spectrum Projections for four Lyme disease group and ten relapsing fever group reference borreliae. MALDI-TOF-MS yielded a unique protein profile for each of the 14 tested Borrelia species, with 100% reproducibility over 12 repeats. In a second proof-of-concept step, the Borrelia database and a custom software program that subtracts the uninfected O. sonrai profile were used to detect Borrelia crocidurae in 20 Ornithodoros sonrai ticks, including eight ticks that tested positive for B. crocidurae by PCR-sequencing. A B. crocidurae-specific pattern consisting of 3405, 5071, 5898, 7041, 8580 and 9757-m/z peaks was found in all B. crocidurae-infected ticks and not found in any of the un-infected ticks. In a final blind validation step, MALDI-TOF-MS exhibited 88.9% sensitivity and 93.75% specificity for the detection of B. crocidurae in 50 O. sonrai ticks, including 18 that tested positive for B. crocidurae by PCR-sequencing. MALDI-TOF-MS took 45 minutes to be completed.
Conclusions/Significance
After the development of an appropriate database, MALDI-TOF-MS can be used to identify tick species and the presence of relapsing fever borreliae in a single assay. This work paves the way for the use of MALDI-TOF-MS for the dual identification of vectors and vectorized pathogens.
Author Summary
In Africa, relapsing fever borreliae are neglected vector-borne infections that cause mild to deadly septicemia and miscarriage. The causative relapsing fever borreliae are transmitted by the bite of soft ticks, except for Borrelia recurrentis which is transmitted by body lice. Screening vectors for these relapsing fever borreliae currently relies on time- and resource-consuming methods such as polymerase chain reaction-based method. Here, we applied matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) to the rapid detection of borreliae in ticks. As a first step, we created a Borrelia MALDI-TOF-MS database and we detected B. crocidurae in Ornithodoros sonrai ticks. As a blind validation step, the 45-minute MALDI-TOF-MS exhibited a 88.9% sensitivity and a 93.75% specificity for the detection of B. crocidurae in 50 O. sonrai ticks including 18 ticks detected positive for B. crocidurae by PCR-sequencing. These findings provide the proof-of-concept that MALDI-TOF-MS can be used to identify tick species and the presence of relapsing fever borreliae. This technique could be translated for field applications.
Introduction
The genus Borrelia is composed of bacterial pathogens responsible for relapsing fever and Lyme borreliosis [1]. Whereas the Lyme disease agents Borrelia burgdorferi [2], Borrelia garinii [3], Borrelia afzelii [4] and Borrelia valaisiana [5] are transmitted by hard ticks, the relapsing fever borreliae are transmitted by soft ticks [6], [7] and lice (in the case of Borrelia recurrentis) [8].
In Africa, four cultured species, Borrelia crocidurae, Borrelia duttonii, Borrelia recurrentis and Borrelia hispanica, and several not-yet cultured species are circulating in vector populations [1]. Screening vectors for these relapsing fever borreliae currently requires the use of technically demanding, time- and resource-consuming molecular methods [9], [10]. However, matrix-assistedlaser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) has emerged as a new, easy technique that can hasten the identification of bacteria and archaea [11], [12]. In particular, MALDI-TOF-MS has been used to identify spirochetes of the genus Leptospira [13] and Brachyspira [14], and recently cultured Borrelia spp. [15]. Also, MALDI-TOF-MS has recently been used to identify vectors [16]–[18].
In this study, we investigated whether MALDI-TOF-MS could further detect relapsing fever borreliae directly in their tick vectors. More specifically, we developed a step-by-step approach, first creating a Borrelia MALDI-TOF-MS database, then performing a proof-of-concept B. crocidurae detection in Ornithodoros sonrai ticks, and finally, blindly testing the MALDI-TOF-MS approach to detect B. crocidurae in O. sonrai ticks.
Materials and Methods
Borreliae and ticks
Fourteen Borrelia spp. were grown at 32°C in Barbour-Stoenner-Kelly-H (BSK-H) medium (Sigma, Saint Quentin Fallavier, France) supplemented with 10% heat-inactivated rabbit serum (Eurobio, Courtaboeuf, France) (Table 1). Dark-field microscopic observation was performed to ensure the absence of any contaminants and to verify the richness and viability of the culture. The identification of the growing borreliae was performed by PCR-sequencing the flagellin gene, as previously reported [19]. The Borrelia culture was centrifuged at 13,000 g for 10 min at room temperature, and the pellet was washed twice with 1 mL high performance liquid chromatography grade water (VWR International, Fontenay-sous-Bois, France) and then suspended at 104 spirochetes/mL in this water before MALDI-TOF-MS analysis.
Table 1. Borrelia species used to establish a MALDI-TOF-MS reference database.
| Species | Strain | MALDI-TOF-MS identification | Score Value |
| B. burgdorferi | B31 | B. burgdorferi | 2.34 |
| B. recurrentis | A1 | B. recurrentis | 2.39 |
| B. crocidurae | Achema | B. crocidurae | 2.45 |
| B. duttonii | Ly | B. duttonii | 2.60 |
| B. lusitaniae | Poti B2 | B. lusitaniae | 2.67 |
| B. japonica | HO14 | B. japonica | 2.65 |
| B. afzelii | PKo | B. afzelii | 2.27 |
| Borrelia sp. | CA 28 (“genomospecies 2”) | B. genomosp. | 2.36 |
| B. andersonii | 19952 | B. andersonii | 2.50 |
| B. garinii | PBi | B. garinii | 2.56 |
| B. californiensis | CA446 | B. californiensis | 2.34 |
| B. valaisiana | VS116 | B. valaisiana | 2.25 |
| B. hermsii | BH0147 | B. hermsii | 2.49 |
| B. turcica | IST7 | B. turcica | 2.15 |
Fifty ticks were collected in Senegal. Total DNA was extracted from the body of the tick by using the EZ1 DNA Tissue kit and the EZ1 apparatus (Qiagen, Courtaboeuf, France) for further PCR-sequencing-based investigations. Ticks were identified as O. sonrai (a species not registered as an endangered species) by 16S rRNA gene sequencing, as previously described [20]. The ticks were tested for the presence of B. crocidurae by glpQ gene real-time PCR using a Ct≤35 cut-off [21], and 18/50 (36%) ticks were found to be infected.
Borreliae MALDI-TOF-MS database
A 1 µL aliquot of the suspension was deposited onto a spot on an MSP 96 target polished steel micro Scout target plate (Bruker Daltonics, Wissembourg, France). After air-drying, 1.5 µL of matrix solution (a saturated solution of alpha-cyanohydroxycinnaminic acid in 50% aqueous acetonitrile containing 2.5% trifluoroacetic acid) was added, and the plate was air-dried for 15 min before being processed in the mass spectrometer. A total of 12 spots were deposited for each Borrelia strain, and this manipulation was repeated in two independent runs. On each plate, Escherichia coli DH5 alpha (Bruker Daltonics) was used as a positive control, and non-inoculated BSK-H medium and non-inoculated-matrix solution were used as negative controls. The analysis was conducted using a Microflex LT spectrometer (Bruker Daltonics), and the spectra were recorded in a linear, positive ion mode with an acceleration voltage of 20 kV. The spectra were collected as a sum of 240 shots across a spot. The preprocessing and identification steps were performed using the manufacturer's parameters. For each of the 14 Borrelia species, a consensus pattern referred to as the Mean Spectrum Projection (MSP) was obtained by the Biotyper MSP Creation Standard Method (Bruker Daltonics). To assess the reproducibility of the MALDI-TOF-MS profiling, the 14 Borrelia strains included in the database were further blindly analyzed by MALDI-TOF-MS as described above. For each strain, 12 spots were analyzed, and the 12 spectra obtained were compared with the Borrelia MSP database. The results of the pattern-matching process were expressed as an identifying score varying from 0 to 3. A score of between 3 and 1.9 indicated a species level identification; a score of 1.9–1.7 indicated a genus level identification; and a score of <1.7 was regarded as an unreliable identification [22]. The MSP spectra from Borrelia spp. were used to generate a dendogram with the MALDI Biotyper 2.0 software (Bruker Daltonics).
MALDI-TOF-MS detection of B. crocidurae in ticks
A total of 20 O. sonrai ticks, including 8 B. crocidurae-infected ticks, were studied. Each tick was placed in a 1.5 mL microcentrifuge tube, frozen at −20°C for 30 min [18], [23], rinsed once with distilled water and dried with paper. Four legs were removed with scalpels and manually homogenized in 60 µL of 70% formic acid and 60 µL of 50% acetonitrile in 1.5 mL microcentrifuge tubes using pellet pestles (Thermo Fischer Scientific, Courtaboeuf, France). All of the homogenates were centrifuged at 13,000 g for 20 s, and 1 µL of the supernatant was spotted onto a steel target plate in quadruplicate. Using in-house subtraction software, the MSP pattern of non-infected O. sonrai was removed from the pattern of infected ticks. The software normalizes the spectra comparing common peaks in infected and uninfected ticks and then generates the MSP spectra before performed the subtraction. After subtraction, the list of remaining differential masses (m/z) was compared with the B. crocidurae MSP.
Blind MALDI-TOF-MS detection of B. crocidurae in O. sonrai ticks
The 50 O. sonrai ticks were coded and blindly tested for the presence of B. crocidurae using the Borrelia database and the subtraction software described above. After blind MALDI-TOF-MS analysis, the codes were unmasked to compare the MALDI-TOF-MS assay results with the PCR-sequencing-based results. Any surprising result was double-checked by performing MALDI-TOF-MS and PCR-sequencing detection again.
Results
MALDI-TOF-MS Borrelia database
Negative controls yielded no identifiable patterns while the positive controls yielded E. coli with identification scores from 1.9–2.3. Each of the 14 Borrelia reference isolates yielded a unique reproducible protein profile combining 25 to 123 peaks (Table S1). Each profile differed from the ones in the Bruker Daltonics database (V3.1.2.0) and each profile was specific for each species of Borrelia (Table S1). Further blind test correctly identified all of the 14 Borrelia strains, with identification scores from 2.15–2.67 after incorporation of the reference spectra (Table 1).
MALDI-TOF-MS detection of B. crocidurae in O. sonrai ticks
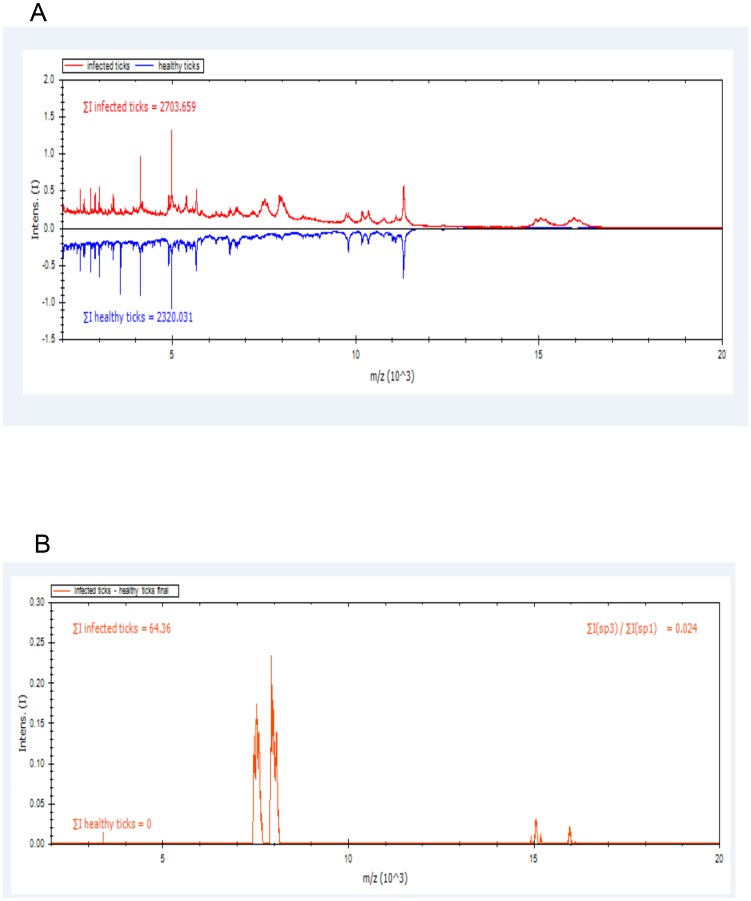
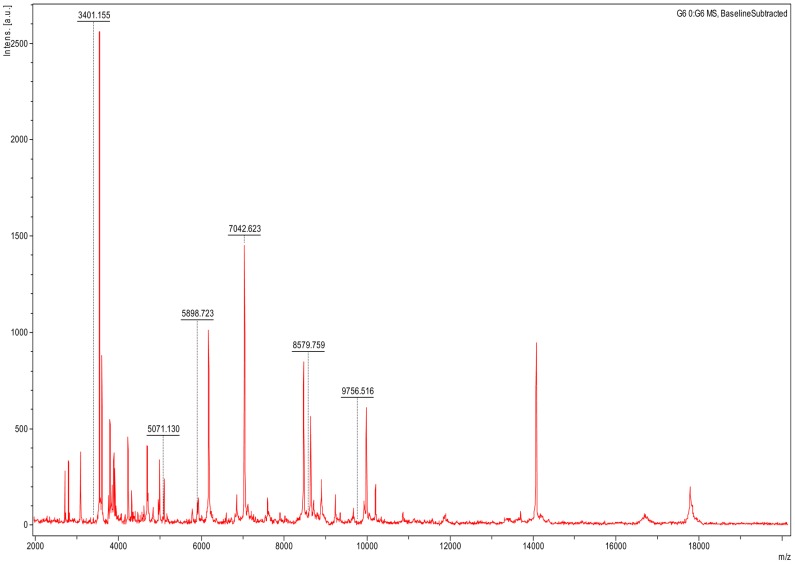
The 12 un-infected O. sonrai ticks yielded no matches, as the database we used did not contain a reference spectrum for this soft tick species; we therefore added this new reference MSP in the ticks' database being constructed in our laboratory [17]. The eight infected O. sonrai ticks consistently yielded a specific pattern of six peaks of 3405, 5071, 5898, 7041, 8580 and 9757 m/z. After subtraction of the O. sonrai MSP, none of the 12 un-infected ticks yielded such a pattern. This 6-peak pattern was found in the B. crocidurae MALDI-TOF-MS pattern (Figures 1, 2). Using this 6-peak pattern, B. crocidurae was detected in 100% of infected ticks and 0% of non-infected ticks.
Figure 1. Subtraction of infected-tick pattern minus non-infected-tick pattern by our subtraction software (1A).
Residual spectrum obtained after the subtraction of the non-infected tick pattern from the infected tick pattern (1B).
Figure 2. A 6-peak pattern for the specific detection of Borrelia crocidurae in O. sonrai ticks.
Blind detection of B. crocidurae in O. sonrai
During the blind test, 100% of the 50 tested ticks were identified as O. sonrai (after the corresponding MSP had been added to the database). Furthermore, the 6-peak B. crocidurae pattern was blindly detected in 16/18 (sensitivity, 88.9%) infected ticks and in 2/32 (6.25%) non-infected ticks (specificity of 93.75%) (Table 2). Re-testing these four ticks with both glpQ–qPCR and MALDI-TOF-MS yielded the same results as in previous experiment.
Table 2. MALDI-TOF-MS detection of B. crocidurae in 50 O. sonrai ticks collected in Senegal.
| MALDI-TOF-MS | |||
| + | − | ||
| glpQ qPCR | 18 + | 16 | 2 |
| 32 − | 2 | 30 | |
“+” denotes a positive result; “−” denotes a negative result.
Discussion
For ten years, MALDI-TOF-MS has revolutionized the routine identification of bacteria [11], [22], [24] and archaea [12], but few studies have examined its application to spirochetes, including Leptospira spp. [13] Brachyspira spp. [14] and Borreliae spp. [15]. Here, using a simple protocol consisting of only one centrifugation step, 14 Borrelia isolates yielded an interpretable, identifying protein pattern. Identification was achieved starting from only 104 Borrelia/mL, in the same range as the 105 Leptospira/mL previously reported [13]. These data completely confirm a recently published study showing that cultured Borreliae are identifiable by MALDI-TOF-MS [15]. The MSP for the 14 Borrelia isolates described herein have been deposited into our freely available Mediterranée Infection Institute website database (http://www.mediterranee-infection.com/article.php?laref=256&titre=urmsdatabase) to assist other scientists with their identification work. As the reliability of MALDI-TOF-MS bacterial identifications depends on the quality of the database [22], we carefully validated the reproducibility of the Borrelia spectra. No misidentifications were discovered when all of the Borrelia strains analyzed in this study were matched with non-borreliae spirochetes (Leptospira and Brachyspira). Further enrichments of the database and the creation of additional super spectra using several genetically/morphologically different strains of the same species will likely increase the power of MALDI-TOF-MS for the identification of borreliae. We also added an MSP for O. sonrai to the tick MALDI-TOF-MS database that we are building [17].
Using this extended database and a piece of subtraction software, MALDI-TOF-MS allowed for the blindly identification of O. sonrai ticks and detection of B. crocidurae in these ticks within 45 minutes. Interestingly, infection by B. crocidurae did not hamper the MALDI-TOF-MS identification of the infected O. sonrai. Moreover, the 6-peak signature found in B. crocidurae infected ticks is comprised of six proteins that are specifically found in B. crocidurae. Therefore, this 6-peak signature is not an unspecific pattern due to any ticks' infection or inflammatory response to Borrelia, but indeed a signature specific to B. crocidurae. Using a 96-spot plate, a total of 23 ticks could be screened in the same experiment, including controls and quadruplicate spots per tick. These data indicate that MALDI-TOF-MS can be used for the rapid, one-shot identification of ticks and tick-borne borreliae. This is the first report of dual identification of vector and vectorized pathogen at once, although a few vectors, including mosquitoes and ticks, have been previously identified by MALDI-TOF-MS [16]–[18].
Further studies are warranted to extend this concept to other pathogens and other vectors. Automation of the differential peak list interpretation would help hasten the field implementation of MALDI-TOF-MS as a first-line tool for the rapid identification of vectors and vectorized pathogens.
Supporting Information
List of peaks (m/z) detected by MALDI-TOF-MS for each of the 14 Borrelia species under study.
(XLSX)
Funding Statement
This study was supported by Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes (URMITE), IHU Méditerranée Infection, Marseille, France. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Haitham E, Raoult D, Drancourt M (2013) Relapsing fever borreliae in Africa. Am J Trop Med Hyg 89: 288–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Casjens S, Palmer N, Van Vugt R, Huang WM, Stevenson B, et al. (2000) A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi . Mol Microbiol 35: 490–516. [DOI] [PubMed] [Google Scholar]
- 3. Glockner G, Lehmann R, Romualdi A, Pradella S, Schulte-Spechtel U, et al. (2004) Comparative analysis of the Borrelia garinii genome. Nucleic Acids Res 32: 6038–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Glockner G, Schulte-Spechtel U, Schilhabel M, Felder M, Suhnel J, et al. (2006) Comparative genome analysis: selection pressure on the Borrelia vls cassettes is essential for infectivity. BMC Genomics 7: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schutzer SE, Fraser-Liggett CM, Qiu WG, Kraiczy P, Mongodin EF, et al. (2012) Whole-genome sequences of Borrelia bissettii, Borrelia valaisiana, and Borrelia spielmanii . J Bacteriol 194: 545–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vial L, Diatta G, Ba EH, Bouganali H, Durand P, et al. (2006) Incidence of tick-borne relapsing fever in West Africa: longitudinal study. Lancet 368: 37–43. [DOI] [PubMed] [Google Scholar]
- 7. Trape JF, Diatta G, Arnathau C, Bitam I, Sarih M, et al. (2013) The epidemiology and geographic distribution of relapsing fever borreliosis in West and North Africa, with a review of the Ornithodoros erraticus Complex (Acari: Ixodida). PLoS One 8: e78473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cutler SJ, Bonilla ME, Singh JR (2010) Population structure of East African relapsing fever Borrelia spp. Emerg Infect Dis 16: 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parola P, Ryelandt J, Mangold A, Mediannikov O, Guglielmone A, Raoult D (2011) Relapsing fever Borrelia in Ornithodoros ticks from Bolivia. Ann Trop Med Parasitol 105: 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parola P, Diatta G, Socolovschi C, Mediannikov O, Tall A, et al. (2011) Tick-borne relapsing fever borreliosis, rural Senegal. Emerg Infect Dis 17: 883–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bizzini A, Greub G (2010) Matrix-assisted laser desorption/ionization time-of flight mass spectrometry, a revolution in clinical microbial identification. Clin Microbiol Infect 216: 1614–1619. [DOI] [PubMed] [Google Scholar]
- 12. Dridi B, Raoult D, Drancourt M (2012) Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of Archaea: towards the universal identification of living organisms. APMIS 120: 85–91. [DOI] [PubMed] [Google Scholar]
- 13. Djelouadji Z, Roux V, Raoult D, Kodjoa A, Drancourt M (2012) Rapid MALDI-TOF mass spectrometry identification of Leptospira organisms. Vet Microbio 158: 142–146. [DOI] [PubMed] [Google Scholar]
- 14. Calderaro A, Piccolo G, Montecchini S, Buttrini M, Gorrini C, et al. (2013) MALDI-TOF MS analysis of human and animal Brachyspira species and benefits of database extension. J Proteomics 78: 273–280. [DOI] [PubMed] [Google Scholar]
- 15. Calderaro A, Gorrini C, Piccolo G, Montecchini S, Buttrini M, et al. (2014) Identification of Borrelia species after creation of an in-house MALDI-TOF MS database. PLoS One 9: e88895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoppenheit A, Murugaiyan J, Bauer B, Steuber S, Clausen P-H, et al. (2013) Identification of Tsetse (Glossina spp.) using matrix-assisted laser desorption/ionisation time of flight mass spectrometry. PLoS Negl Trop Dis 7: e2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yssouf A, Flaudrops C, Drali R, Kernif T, Socolovschi C, et al. (2013) Matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of tick vectors. J Clin Microbiol 51: 522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yssouf A, Socolovschi C, Flaudrops C, Ndiath MO, Sougoufara S, et al. (2013) Matrix-assisted laser desorption/ionization time of flight mass spectrometry: an emerging tool for the rapid identification of mosquito vectors. PLoS ONE 8: e72380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wodecka B (2011) FlaB gene as a molecular marker for distinct identification of Borrelia species in environmental samples by the PCR-restriction fragment length polymorphism method. App Environ Microbiol 77: 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Norris DE, Klompen JS, Keirans JE, Black WC (1996) Population genetics of Ixodes scapularis (Acari: Ixodidae) based on mitochondrial 16S and 12S genes. J Med Entomol 33: 78–89. [DOI] [PubMed] [Google Scholar]
- 21. Haitham E, Henry M, Diatta G, Mediannikov O, Sokhna C, et al. (2013) Multiplex real-time PCR diagnostic of relapsing fevers in Africa. PLoS Negl Trop Dis 7: e2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, et al. (2009) Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49: 543–551. [DOI] [PubMed] [Google Scholar]
- 23. Feltens R, Gorner R, Kalkhof S, Groger-Arndt H, Von Bergen M (2010) Discrimination of different species from the genus Drosophila by intact protein profiling using matrix-assisted laser desorption ionization mass spectrometry. BMC Evol Biol 10: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Bruyne K, Slabbinck B, Waegeman W, Vauterin P, De Baets B, et al. (2011) Bacterial species identification from MALDI-TOF mass spectra through data analysis and machine learning. Syst Appl Microbiol 34: 20–29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of peaks (m/z) detected by MALDI-TOF-MS for each of the 14 Borrelia species under study.
(XLSX)