Abstract
Pattern recognition receptors (PRRs) are traditionally known to recognize microbial molecules during infection to initiate inflammatory responses. However, ligands for PRRs are not exclusive to pathogens, and are abundantly produced by the resident microbiota during normal colonization. Mechanism(s) that underlie this paradox have remained unclear. Recent studies reveal that gut bacterial ligands from the microbiota signal through PRRs to promote host tissue and immune development, and protection from disease. Furthermore, evidence from both invertebrate and vertebrate models reveals that innate immune receptors are required to promote long-term colonization by the microbiota. This emerging perspective challenges current paradigms in immunology, and suggests that PRRs may have evolved, in part, to mediate the bidirectional crosstalk between microbial symbionts and their hosts.
Conventional wisdom suggests that the immune system evolved to combat infection, and that distinguishing between self and non-self molecules is a basic feature of innate immunity. As proposed by Charles Janeway, the recognition of microbial molecules, termed pathogen-associated molecular patterns (PAMPs), is critical in selectively driving immune responses to infectious agents 1. Studies identifying and characterizing host receptors that recognize specific PAMPs, named pattern recognition receptors (PRRs), have provided evidence that PRR signaling is critical in coordinating immune responses and protection against pathogens (see reviews 2-5). However, this view has been challenged by the emerging appreciation that animals harbor a diverse and complex symbiotic microbiota 6-10, which normally does not trigger inflammation. PAMPs, by definition, are universally conserved structures that are generally invariant and essential in all microorganisms. Thus, PAMP expression is not limited to pathogens, but also common to the microbiota. As such, it has been proposed that these molecules be renamed microbe-associated molecular patterns (MAMPs) 11. Furthermore, host PRRs are constantly exposed to MAMPs, in the absence of infection. These molecules are largely provided by the commensal microbiota that colonize our skin and mucosal surfaces. Despite the continuous presence of numerous MAMPs, commensal microbes usually do not elicit inflammatory responses, but rather, may contribute to various aspects of host development and enhanced immune function 12. Surprisingly, this beneficial influence is mediated, in part, by commensal stimulation of host PRRs 13.
How these molecules and receptors are able to achieve such divergent and opposing responses between pathogens and symbiosis is a frontier in our understating of innate immunity. It has been proposed that the context in which the host receives MAMP stimulation dictates the quality of the immune response. During infection, MAMP signals are received in the presence of other cues, such as cell damage caused by infection 14 and/or cytosolic detection of MAMPs 15, resulting in inflammation. During symbiosis, not only does the microbiota generally not harm host cells and MAMPs are sensed in the absence of exposed self-antigens, but it appears that some MAMPs directly promote beneficial outcomes. This review will focus on how MAMP recognition by PRRs under steady-state conditions promotes immune development, protection from disease and maintains homeostasis. The concepts presented here collectively demonstrate that PRRs may have evolved in both the invertebrate and vertebrate immune systems to communicate with commensals and maintain beneficial, symbiotic coexistence with the microbiota.
Pattern recognition in Drosophila promotes homeostasis
Extensive work using Drosophila as a model system has highlighted the important functions of PRRs in host defense, as well as homeostasis. Toll, the founding example of a PRR, was initially discovered in Drosophila melanogaster (the fruit fly) 16. However, the realization that Drosophila Toll does not directly recognize MAMPs, unlike the mammalian TLR signaling pathway, left the open question of how a suite of bacterial ligands are recognized. D. melanogaster has 13 peptidoglycan recognition protein (PGRP) genes that are alternatively spliced into 19 different proteins, one of the largest repertoires of PGRPs currently known for any organism 17. The role of Drosophila PGRPs as PRRs was discovered during the identification of upstream receptors that activate the signal transduction pathways, Toll and Imd (immune-deficiency) 18, 19, which show high similarities with the mammalian interleukin-1 (IL-1)-TLR and tumor necrosis factor (TNF) pathways 20. However, Toll does not function as a pattern recognition receptor as it does not directly recognize MAMPs 21. Instead, PGRP-SA and PGRP-SD form a complex upstream of the Toll pathway 18, 19. Upon peptidoglycan recognition (from Gram-positive bacteria), PGRP-SA triggers a protease cascade, resulting in Toll dimerization, leading to activation of the transcription factor NF-κB and production of antimicrobial peptides (AMPs) 18. Additionally, fungi and Gram-negative bacteria trigger the Imd pathway via PGRP-LC, mediating innate immune responses 19, 22, 23. Upon Imd pathway activation, NF-κB signaling results in the production of AMPs, targeting invasive pathogens during infection (Fig. 1a,b).
Figure 1. PRRs in invertebrate systems.
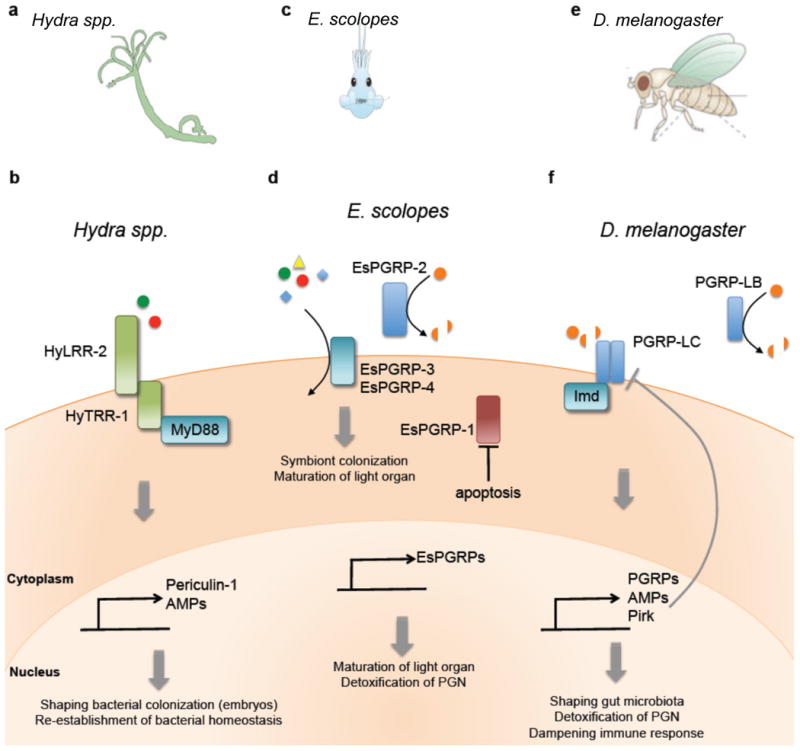
(a) Macroscopic view of Drosophilia and the midgut. (b)| PGRP-LC expression on the cell surface senses DAP-PGN (from Gram-negative bacteria), triggering the Imd pathway and the production of additional PGRPs, AMPs, Caudal and Pirk ( negative regulators of the Imd pathway). The amidase PGRP-LB, induced upon Imd activation, cleaves microbiota-derived PGN, blocking additional activation of the Imd pathway. (c) | Macroscopic view of the Hydra body plan consisting of two layers of epithelial cells, with the outermost layer exposed to bacterial symbionts. (d) | PRR signaling in Hydra is mediated by the HyLRR-2-HyTRR-1 complex, where HyLRR-2 serves as the receptor and HyTRR-1 serves as the transmembrane domain. Upon MAMP stimulation, MyD88 is recruited, triggering the production of AMPs such as periculin-1. Maternal expression of periculin-1 is responsible for shaping bacterial colonization in embryos, and re-establishing bacterial homeostasis in adults. (e) | Macroscopic view of the bobtail squid (E. scolopes) and the maturation of the light organ, induced upon colonization with V. fisheri. (f) | E. scolopes expresses four PGRPs. EsPGRP-3 and -4 function as cell surface receptors that recognize MAMPs, resulting in signaling activation. EsPGRP-2 is secreted by epithelial cells and crypts of the light organ, and detoxifies PGN via amidase activity to dampen immune responses. Loss of EsPGRP-1 upon colonization by V. fisheri results in apoptosis and loss of appendages, marking the maturation of the light organ.
Intriguingly, in addition to regulating AMP production in response to infection, the Imd pathway also shapes the response to commensal microbiota. The Imd pathway regulator, Caudal, is important in shaping the composition of gut microbiota 24. Caudal suppresses Imd-dependent expression of AMPs by blocking AMP gene promotors. Additional negative regulators of Imd, such as Pirk (Fig. 1b), sequester PGRP-LC in the cytoplasm to prevent exposure to peptidoglycan and activation of the Imd pathway, which is critical in regulating commensal populations, maintaining intestinal homeostasis and restraining overactive immune responses 25, 26.
In addition to sensing bacterial ligands and activating an inflammatory response, a subset of Drosophila PGRPs have amidase activity and participate in hydrolyzing pro-inflammatory peptidoglycan into non-immunostimulatory fragments 27, 28 (Fig. 1b). These catalytic PGRPs, such as PGRP-LB and PGRP-SC, serve to limit the availability of peptidoglycan released from symbiotic gut bacteria, preventing over-activation of the Imd pathway and dampening the immune response. In fact, deletion of PGRP-LB and PGRP-SC leads to a ten-fold increase in AMPs compared to wild-type flies 28, revealing the importance of amidase PRGPs in detoxifying the biological activity of peptidoglycan and controlling an overactive systemic immune response. While these studies highlight a role of PGRPs in maintaining gut homeostasis by controlling the immune responses to commensal bacteria, future studies are necessary to reveal how this pathway discriminates between symbiotic and pathogenic bacteria. What is clear, however, is that sensing of bacterial ligands in Drosophila is critical for host-microbial symbiosis in the absence of infection.
Hydra TLR signaling promotes bacterial colonization
As a member of the second oldest phyla, Cnidaria, Hydra spp. serve as a simple model system to study the evolutionary origins of commensalism (Fig. 1c). The intimate relationship between Hydra and specific bacterial species reveal that the primitive immune system of Hydra is able to recognize its symbionts, promoting a highly evolved partnership with its host29. Hydra lacks migratory phagocytic cells and hemolymph, rather relying on PRRs and the production of antimicrobial peptides by epithelial cells for immune protection 30. In the absence of conventional TLRs, analysis of the Hydra magnipapillata genome identified two genes with a Toll-interleukin-1 receptor (TIR) domain and transmembrane domain (HyTRR-1 and -2), each lacking leucine-rich repeats (LRR) in the extracellular region, which is typical of classical TLRs 31. Two additional genes were identified encoding for transmembrane proteins with TLR-related LRRs in the extracellular region (HyLRR-1 and -2). Signaling by the HyLRR-2 and HyTRR-1 complex following microbial pattern recognition recruits the adaptor protein, MyD88 and triggers the production of AMPs, such as periculin-1 30 (Fig. 1d). In addition to bactericidial activity, maternal expression of periculin-1 is involved in shaping the microbiome of the embryo 32. In transgenic polyps that overexpress periculin-1a, the bacterial community structure is dramatically different, with a decrease in β-Proteobacteria and an increase in α-Proteobacteria. Additionally, a significantly lower bacterial load was observed in transgenic polyps compared to control polyps, indicating that periculin-1a plays a role in both controlling and shaping bacterial colonization during Hydra development.
In addition to defense against pathogenic microbes, MyD88 signaling also contributes to host-mediated colonization by commensal bacteria. Using a transgenic MyD88 knockdown approach, the function of PRR signaling in Hydra was examined. While the overall composition of the bacterial microbiota remained unchanged, MyD88-mediated PRR signaling promotes reestablishment of bacterial homeostasis 33. Thus studies in Hydra, a primitive organism, reveal that recognition of commensal bacteria appears to be an ancient function of innate immune signaling, suggesting that PRRs have evolved to mediate host-microbe communication.
PRR signaling maintains squid-Vibrio symbiosis
The relationship between the Hawaiian bobtail squid, Euprymna scolopes, and the Gram-negative bacteria, Vibrio fisheri, is one of the most extensively studied examples of host-microbial symbiosis to date 34-36. The bioluminescent bacterium, found in seawater, colonizes the crypts of the light organ in the juvenile host as a mono-specific symbiont 37(Fig. 1e). The light organ is continually exposed to numerous microbes in the surrounding seawater, similar to the mammalian intestinal tract. Yet the squid is able to selectively and exclusively harbor a single species of bacteria. This intimate symbiosis allows the nocturnal squid to utilize light produced by V. fisheri as a counter-illumination camouflage strategy, a clever adaptation to avoid detection by predators swimming below.
A series of elegant studies have shown that shortly after hatching of the juvenile squid, the symbiont promotes colonization and beneficial co-existence with its host. MAMP signaling mediates this specificity and initiates the colonization process, promoting host light organ development 38, 39 (Fig. 1f). During the initial interaction with the squid, the bacterial symbiont induces a sequence of events to promote colonization. The juvenile light organ has a pair of ciliated appendages, which secrete mucus upon exposure to the bacterial trigger, a peptidoglycan fragment known as tracheal cytotoxin (TCT) 38. This mucus not only serves as a chemoattractant for V. fisheri, but also a substrate for growth and aggregation to seed the founding microbial population of the light organ. Following the initial colonization, the symbiont then induces a second sequence of events driving the maturation of the squid light organ by directing a switch from a permissive to a restrictive state to ensure V. fisheri colonization dominance. The bacterial triggers that drive this switch, lipopolysaccharide (LPS) and TCT, are necessary to induce morphogenesis of the light organ via apoptosis and regression of epithelial cells, leading to a loss of ciliated appendages and preventing entry of other bacteria into the developed light organ 40 (Fig. 1e, f). Therefore, the squid utilizes MAMPs as morphogens that direct normal developmental programs.
How are the symbiotic bacterial cues sensed by E. scolopes? PGRPs were implicated in mediating responses to MAMPs during symbiosis 41. To date, five EsPGRP have been identified in the bobtail squid 39, 42, although only EsPGRP-1 and EsPGRP-2 have been extensively characterized, as both are expressed in the juvenile light organ. EsPGRP-1 expression is localized within the nucleus of the ciliated epithelium of the light organ, and upon symbiont colonization, the bacterial signal TCT directs the loss of EsPGRP-1, inducing the initiation of the apoptotic pathway 43 (Fig. 1f). Apoptosis of the ciliated epithelium and the light organ appendage marks the completion of development and colonization of the squid light organ. Alternatively, EsPGRP-2 possesses N-acetyl-muramyl-L-alanine amidase activity, hydrolyzing peptidoglycan for degradation, diminishing its toxicity and pro-inflammatory properties 44 (Fig. 1f). This activity is particularly important as V. fisheri colonization occurs in high numbers and peptidoglycan is continually shed in the light organ, posing the risk of inflammatory distress. Similar to Drosophila, the EsPGRP-2 amidase that degrades peptidoglycan into non-immunostimulatory fragments implicates its role in attenuating inflammatory responses, a prerequisite for symbiosis. Thus V. fisheri has evolved to co-opt innate immunity to promote colonization of the squid, conferring benefits to both partners during mutualism. In conclusion, the collective data from the invertebrate models reveal PRRs are instrumental in host defense against infection, the recent discovery of their role during symbiosis reveals the dual function of innate immunity in responding to both pathogens and the commensal microbiota.
Maintaining tolerance in zebrafish
The zebrafish (Fig. 2a) has gained considerable attention as a model vertebrate system45, due to ease of genetic manipulation 46, 47, the ability to study germ-free conditions 48, and resemblance to mammalian physiology. Specifically, the conserved PRR system is highly similar to other vertebrates, and the zebrafish model has proven to be a useful tool to examine host-microbe interactions49. A recent study revealed a role for the zebrafish intestinal alkaline phosphatase (IAP) to dephosphorylate the immunostimulatory LPS, thereby modulating intestinal inflammation in response to commensal microbes, the primary source of LPS in the gastrointestinal tract 50 (Fig. 2b). LPS is a major component of the outer membrane of Gram-negative bacteria, found in both pathogenic and commensal bacteria alike. However, LPS recognition by TLR4 results in induction of signaling cascades that lead to activation of NF-κB and the production of pro-inflammatory cytokines. Upon bacterial colonization (or administration of exogenous LPS), IAP expression and activity is induced in a MyD88-dependent fashion 50. TLR orthologs have been identified in the zebrafish genome, with several TLR4 orthologs reported 51 and hypothesized to recognize LPS (however, it is important to note that several studies have reported a lack of LPS responsiveness in some zebrafish TLR4 paralogs 52, 53). IAP deficiency results in excessive intestinal neutrophil infiltration, a process involving functional MyD88 and TNF. Thus, commensal-derived LPS signaling via TLR4-MyD88 leads to upregulation of IAP and detoxification of LPS, and prevents inflammatory responses to the resident microbiota. Additionally, following the initial discovery in zebrafish, IAP has been shown to be involved in the maintenance of homeostasis in the bobtail squid 54 and mice 55, 56. The identification of IAP revealed yet another mechanism in which PRR signaling is crucial in preventing uncontrolled inflammation in response to commensal microbes, thus promoting colonization by the microbiota.
Figure 2. | PRRs in vertebrate systems.
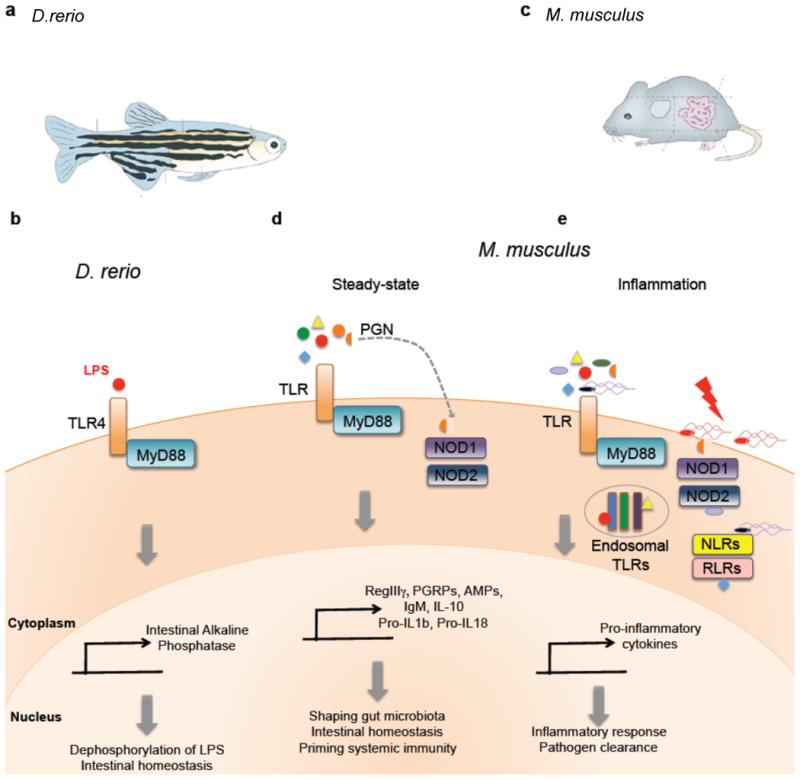
(a)| Macroscopic view of zebrafish and its intestinal tract. (b)| LPS stimulation of TLR4 in zebrafish results in MyD88 recruitment and triggers the production of intestinal alkaline phosphatase (IAP) in epithelial cells. Induction of IAP is critical in maintaining gut homeostasis through dephosphorylation of microbiota-derived LPS. (c)| Macroscopic view of mouse and its gastrointestinal tract. (d)| Under steady-state conditions, TLR and NOD1/2 signaling by microbiota-derived MAMPs results in the recruitment of MyD88 and production of AMPs (such as PGRPs, RegIIIγ and defensins) and other mediators of intestinal homeostasis. (e)| Under inflammatory conditions, pathogen-derived MAMPs trigger various TLRs, NLRs and RLRs. Invasive microbes, as well as non-invasive pathogens possessing a secretion system, are able to stimulate cytosolic and endosomal PRRs. This infectious process results in a pro-inflammatory response, leading to pathogen clearance.
PRR signaling promotes intestinal homeostasis in mice
The mammalian intestine is home to an abundant and complex consortium of bacteria that orchestrate important immune and metabolic functions within the host. Several studies in the murine model (Fig. 2c) have shown that the composition of the microbiome, as well as the spatial location of gut bacteria within the intestine, dictates the balance of tolerogenic (Fig. 2d) versus pro-inflammatory (Fig. 2e) immune responses in the gut. PRR signaling in mice appears to play a key role in both directing the spatial segregation of the microbiota, as well as shaping the composition of the commensal microbiota.
The gastrointestinal surface is coated with a layer of mucus 57, largely limiting bacterial access to the epithelium that separates the trillions of microorganisms in the gut lumen from host tissues 58 (Fig. 3a). Additionally, secretory immunoglobulin A (IgA) maintains barrier functions of the epithelium 59, 60. This physical separation of microbiota and intestinal gut mucosa is mediated by MyD88 signaling in intestinal epithelial cells 61. In the absence of MyD88, commensal bacteria gain close proximity to the intestinal surface, resulting in a 100-fold increase in mucosal-associated bacteria compared to wild-type animals. Similar to invertebrate models, MyD88 signaling results in production of AMPs in vertebrate systems as well 62. MyD88 signaling controls the production of several AMPs by specialized intestinal epithelial cells termed Paneth cells, such as RegIIIγ, a C-type lectin that targets Gram-positive bacteria (Fig. 3a) 61-63. RegIIIγ-deficient animals exhibit a defect in the spatial segregation of mucosa-associated bacteria with the microbiota penetrating the mucus layer and making intimate contact with host tissue, a phenotype similar to intestinal epithelial cell-specific MyD88-deficient mice 61. The increased bacterial burden at the intestinal surface of the small intestine results in immune activation, with elevated IgA and TH1 responses. Upon depletion of the microbiota with antibiotic treatment, the elevated IgA and TH1 responses in Myd88-deficeint mice were diminished. This suggests that commensal microbiota drives MyD88 signaling via TLR stimulation under steady-state conditions, inducing the expression of AMPs, such as RegIIIγ, to restrict bacterial colonization on intestinal surfaces and limit an immune response to resident bacteria.
Figure 3. | PRR signaling promotes immune homeostasis.
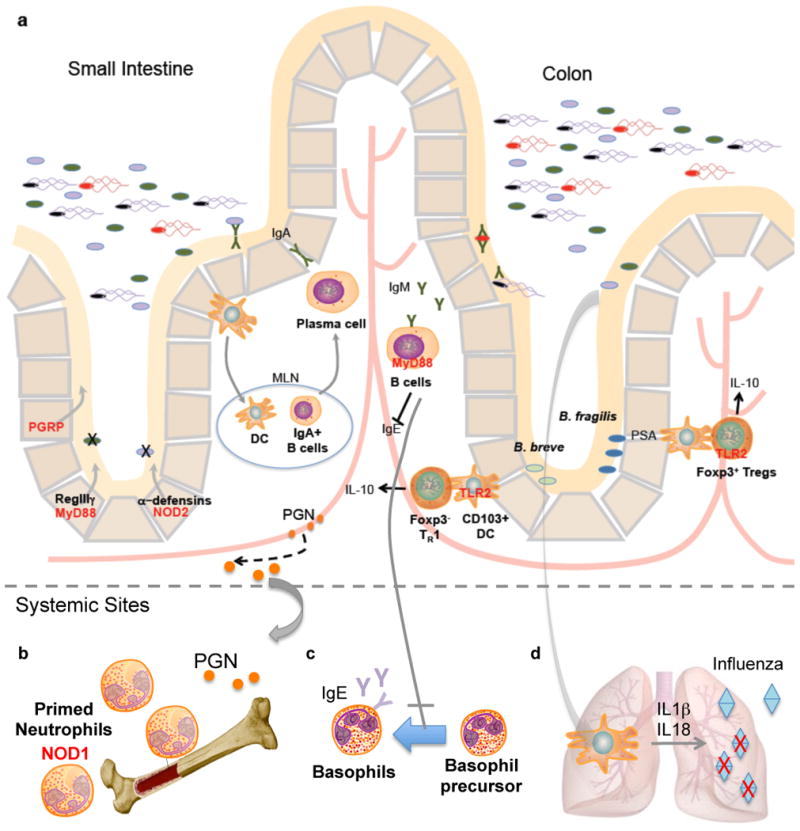
(a)| The small intestine and colon are comprised of a single layer of intestinal epithelial cells separating the abundant microbiota from host tissues. A complex mucus layer coats the epithelium, protecting the gastrointestinal tract from potential invasion. Additional protective mechanisms are present to maintain intestinal homeostasis, many of which are controlled by PRR signaling by commensal microbes. In the small intestine, AMPs such as PGRPs, RegIIIγ and defensins are induced upon PRR stimulation by commensals. In the colon, commensal microbes B. fragilis and B. breve signal through TLR2 to induce Treg cells. The MAMP polysaccharide A (PSA), expressed by B. fragilis, drives the development of IL-10-producing Foxp3+ Treg cells via TLR2. B. breve, however, induces IL-10-producing Foxp3- TR1 cells via CD103+ DCs in a TLR2 dependent manner. Additionally, B cell intrinsic MyD88 signaling promotes IgM secretion. The secretion of IgM is important in controlling systemic spread of bacteria after intestinal injury. (b)| Peptidoglycan derived from the gut microbiota is necessary to prime neutrophils in bone marrow stores in a Nod1-dependent manner. (c)| MyD88 signaling in B cells suppresses serum IgE and inhibits the differentiation of basophils in systemic sites. (d)| Commensal gut microbiota induces the production of pro-IL-1β and pro-IL-18 during steady state (signal 1). During an influenza infection in the lungs, caspase-1 mediated activation of IL-1β and IL-18 (signal 2) is critical for clearance of influenza.
In addition to controlling the spatial segregation of intestinal bacteria, PRRs play a key role in shaping the composition of the microbiota. The impact of this process is apparent in studies of inflammatory bowel disease (IBD). Crohn's disease, a form of IBD, involves an overactive immune response and impaired barrier function in the gut. Genome-wide association studies have connected polymorphisms in the Nod2 innate immune receptor to increased susceptibility to Crohn's disease 64, 65. Nod2 is a member of the NLR family of cytoplasmic proteins. It recognizes a derivative of peptidoglycan, muramyl dipeptide (MDP), found in both Gram-positive and negative bacteria. Signaling of Nod2 requires the adaptor protein Rip2k, which activates downstream signaling cascades involving NF-κB. Nod2-deficiency has been linked to impaired expression of Paneth cell α-defensins, a family of AMPs 66, 67. Accordingly, Nod2 was found to be required for the regulation of commensal microbiota in the terminal ileum, where Nod2 expression is mainly localized 68. An increase in Bacteroides and Firmicutes was observed in Nod2-deficient animals compared to wild-type littermates, revealing that Nod2 is involved not only in shaping the composition of the microbiota, but also in restricting bacterial numbers in the ileal crypts. Notably, Nod2-deficient mice are impaired in the ability to clear the pathobiont Helicobacter hepaticus. Similarly, Rip2k-deficient animals exhibit the same phenotype, and Rip2k was recently identified as a Crohn's susceptibility gene 69, corroborating this innate immune signaling pathway in shaping the commensal microbiota.
Consistent with the role of Nod2 in promoting host-microbial symbiosis, the PGRP family is involved in the regulation of commensal microbiota in mice. Mammals have four PGRPs (PGRP-1 – 4), where PGRP-1, -3, and -4 are directly bactericidal, and PGRP-2 is an amidase that hydrolyzes peptidoglycan – all of which are found at varying levels of expression in the colon70. Animals deficient in any one of the PGRPs harbor a microbiota that promote increased sensitivity to dextran sulfate sodium (DSS)-induced colitis, a mouse model of IBD 71. Indeed, germ-free animals inoculated with stool from PGRP-deficient donor mice are more sensitive to DSS colitis compared to animals that received stool from wild-type mice and display increased mortality, weight loss and colitis scores. Thus, mammalian PGRPs are important in shaping a homeostatic commensal microbiota and preventing intestinal inflammation 72.
The studies we outline involving MyD88-mediated induction of RegIIIγ, Nod2 and PGRPs highlight the role of innate immune signaling in shaping the composition and localization of the mammalian microbiota. It is important to emphasize that while this may be the result of bidirectional crosstalk between symbiotic gut bacteria and the host, there is clearly a role for pathogen detection by these immune regulators. Future studies aimed at understanding how innate immunity shapes the microbiome, and whether gut bacteria actively signal through PRRs to coordinate the immune response, will be critical. As such, the identification of Nod1 as a receptor utilized by the microbiota to induce isolated lymphoid follicles (ILFs) in the gut may provide a glimpse into a process where specific gut bacteria educate the development of the immune system via PRRs 73.
TLRs mediate host-protective immune responses
Several studies have identified a role for TLRs in mediating non-inflammatory immune responses to the microbiota, challenging the paradigm that PRRs have evolved solely to recognize and respond to pathogens. MyD88-deficient mice are more susceptible to DSS colitis, suggesting that commensal bacteria may be directly recognized by TLRs under steady-state conditions to mediate host-protective responses 74. To corroborate this notion, depletion of gut bacteria with antibiotics results in increased susceptibility to DSS; remarkably, oral feeding of LPS and lipoteichoic acid (LTA) corrects this predisposition to colitis, revealing that TLR ligands have beneficial effects on the host74. As DSS induces intestinal injury, these findings suggest that TLR signaling by the microbiota leads to maintenance of intestinal epithelial homeostasis in the absence of enteric pathogens.
Peptidoglycan, LPS and other widely conserved bacterial patterns mediate host-microbial interactions that span from Hydra to mice. It appears, however, that species-specific TLR ligands have also been evolved by the microbiota. The human commensal Bacteroides fragilis produces polysaccharide A (PSA) that directs the maturation of the mammalian immune system, specifically promoting the development and function of CD4+ T cells 75. One outcome of this process is the activation of Foxp3+ regulatory T (Treg) cells to produce interleukin 10 (IL-10), which shapes a toloregenic immune response that is protective in animal models of IBD and multiple sclerosis (MS) 76-78 (Fig. 3). Remarkably, PSA is a unique TLR2 ligand found (to our knowledge) only in the human microbiome, which orchestrates anti-inflammatory immune responses that ameliorate immune-mediated diseases. TLR2-deficient mice are not protected by PSA against colitis 79. PSA is delivered by outer membrane vesicles (OMVs) that bud from the surface of B. fragilis and are internalized by intestinal dendritic cells (DCs) 80. TLR2-deficient DCs do not promote Foxp3+ Treg responses and IL-10 production in vitro, demonstrating that specific gut bacterial molecules have evolved to promote benefits to the host via PRR signaling. The concept of interkingdom communication via innate immune receptors was recently extended to Bifidobacterium breve, a probiotic that signals through TLR2 to induce another subset of regulatory T cells termed Tr1 cells 81. While the bacterial ligand remains unknown, B. breve prevents intestinal inflammation by activating IL-10-producing Tr1 cells in the gut. The demonstration that B. fragilis and B. breve mediate beneficial adaptive immune responses via TLR signaling reveals a deep co-evolutionary symbiosis founded on a “molecular dialogue” using the PRR system.
PRRs on lymphocytes promote intestinal homeostasis
It is worth mentioning that the PRRs discussed up to this point are expressed among epithelial cells and myeloid cells, which comes to no surprise, as PRRs are classic innate immune signaling receptors (and invertebrates lack an adaptive immune system). However, several studies have highlighted the importance of TLR-MyD88 signaling among lymphocytes. In B cell-specific MyD88-deficient mice, bacteria disseminate to systemic sites, such as liver or lung, following DSS-induced colonic damage – but not in epithelial or DC-specific MyD88-deficient animals 82. Further, it has recently been appreciated that T cell subsets express functional TLRs 83. Transfer of MyD88-deficient T cells into RAG-deficient animals results in reduced intestinal inflammation 84. Conversely, while classically thought to promote immunity, it now appears that TLR signaling by T cells can restrain inflammatory responses. For example, treatment of CD4+ T cell subsets with a TLR4 agonist increases suppressive activity and enhances protection from colitis 85. Therefore, TLRs represent a dynamic signaling system that trigger various immune outcomes, and TLR signaling directly by adaptive immune cells mediates reactions in the absence of innate immune cells 86-88.
Did gut bacteria evolve to induce anti-inflammatory responses through TLR signaling solely to improve host health, or are there direct benefits to the microbe? PSA from B. fragilis enhances the anti-inflammatory function of Treg cells by signaling directly through TLR2 on CD4+ T cells 79. Modulation of Treg activity achieves long-term colonization of the gut by B. fragilis by suppressing TH17 responses directed toward the microbe. Therefore, PSA is distinct from TLR2 ligands of pathogens, which elicit inflammation, and sensing of gut bacterial molecules to mediate colonization further supports a role for TLR signaling in promoting host-microbe symbiosis. These findings imply that the host is not ‘hard-wired’ to distinguish symbionts from enteric pathogens, and specific microbial ligands have evolved to actively allow mutualism between mammals and beneficial bacteria of the microbiota.
Recognition of commensals promotes extra-intestinal immune function
While the beneficial effects of the resident microbes have been extensively studied within the gut, a more diverse role for the microbiota in modulating systemic immunity is emerging. With such abundant numbers of microbes inhabiting the mammalian gut, one can imagine the high concentrations of MAMPs that may circulate to systemic sites. Peptidoglycan from the gut microbiota is translocated into sera and bone marrow, priming systemic innate immunity by enhancing neutrophil function 89 (Fig. 3b). Specifically, peptidoglycan recognition is mediated via Nod1, which recognizes meso-diaminopimelic acid (meso-DAP) containing cell wall fragments found on Gram-negative bacteria. Treatment of mice with broad-spectrum antibiotics to deplete the microbiota, and thus circulating peptodoglycan, leads to a significant reduction in neutrophil killing of both Streptococcus pneumoniae and Staphylococcus aureus89. This defect in neutrophil function is rescued by administration of meso-DAP, which restores the innate immune response after microbiota depletion. Bone marrow derived neutrophils from Nod1-deficient mice are also defective in S. pneumoniae and S. aureus killing89. Hence, microbial products derived from the gut microbiota serve to prime systemic immune responses via Nod1 signaling to rapidly respond to pathogenic microorganisms.
The range of influence by PRR of commensals extends to protection against allergic diseases 90. Further support of systemic influences by the gut microbiota is revealed with pattern recognition of commensals by B cells, which promotes systemic immune development, resulting in decreased allergic inflammation90. Antibiotic-treated and germ-free animals display increased serum IgE and basophil frequencies, implicating a role for the microbiota in regulating TH2 responses and allergic inflammation 90. B cell-intrinsic MyD88 signaling is critical in controlling steady-state IgE levels in the serum, as well as circulating basophil populations 90 (Fig. 3c). MyD88 deficiency in B cells results in higher levels of serum IgE and increased basophil surface bound IgE frequencies 90. Mechanistically, MAMPs derived from commensal bacteria were found to limit basophil proliferation and development in the bone marrow 90. Thus, commensal-derived signals are able to direct basophil development from bone marrow precursors via MyD88 signaling, linking the gut microbiota to the regulation of hematopoiesis.
Finally, PRR signaling in the gut appears to modulate immune responses at distant mucosal surfaces. Antibiotic-treatment of mice diminishes adaptive immune responses to intranasal infection with influenza virus 91. Both CD4+ T cell and cytotoxic CD8+ T cell (CTL) responses to influenza were significantly reduced in antibiotic-treated animals together with virus-specific antibody titers, resulting in increased viral load in the respiratory tract 91. Administration of TLR ligands intrarectally restored T cell and antibody responses to influenza in the lungs of these antibiotic-treated mice, suggesting that PRR stimulation in the gastrointestinal tract is important in priming immunity at other mucosal surfaces 91. The intestinal microbiota was found to be critical in providing signals necessary for inflammasome-dependent cytokine secretion in response to influenza infection 91. Two signals are necessary for the production (signal 1) and processing (signal 2) of IL-1β and IL-18. Signal 1 is provided through TLR stimulation, resulting in the expression of pro-IL-1β and pro-IL-18. Signal 2 is mediated through inflammasome activation, leading to caspase-1 cleavage to mature IL-1β and IL-18. The commensal microbiota provides signal 1 during colonization, initially priming the immune response whereupon influenza infection (signal 2) subsequently activates the inflammasome to help clear virus from the lungs 91 (Fig. 3d). These seminal examples of recognition of molecular patterns from commensal bacteria elegantly illustrate that the microbiota co-opts PRRs to mediate beneficial outcomes.
Do hosts distinguish beneficial from harmful bacteria via PRRs?
We have discussed several examples for how the innate immunity system responds to the microbiota. However, a fundamental feature of the immune system is the critical distinction between the resident microbiota and invading pathogens, and recent data are beginning to suggest PRRs play a role on this process. The discovery of an intimate symbiosis with specific bacterial species indicates that the immune system of organisms ranging from Hydra to mammals is able to recognize its microbial partners, or alternatively, that symbiotic bacteria actively promote a highly evolved associations with their hosts. In the case of B. fragilis, PSA signals through TLR2 to promote its symbiotic association with mucosal tissue 79, a finding that implies the host is not ‘hard-wired’ to distinguish symbionts from enteric pathogens, and that specific microbial ligands have evolved to actively allow mutualism between mammals and beneficial bacteria of the microbiota. Examples revealing that the microbiota protects the host from infectious agents via PRR signaling further supports the notion that innate immunity is a mechanism of host-microbial communication. As PRRs have historically been studied in the context of infectious agents, these findings suggest that PRRs serve the dual function of sensing both pathogens and symbionts, with very different consequences for both microbes (clearance vs. symbiosis) and hosts (inflammation vs. immune homeostasis).
Conclusions
Microbes dominate as the most abundant life form on our planet, occupying almost every terrestrial, aquatic and biological ecosystem. Throughout their lives, all metazoans continuously encounter microorganisms that are essential for health or cause death. The immune system is charged with the task of distinguishing beneficial microbes from pathogens, much like it distinguishes self from non-self antigens, to coordinate appropriate immune responses. As symbiotic microbes share similar molecular patterns with pathogens, why don't we immunologically reject our microbiota during lifelong colonization? It was initially suggested that symbiotic bacteria were simply ignored by the host 92. Emerging evidence, however, reveals that certain microbes directly engage the immune system and in some cases actively shape beneficial host immune responses. Symbiosis is often achieved through microbial molecules that are sensed by PRRs, the same innate immune system that has been studied for years in responding to microbial infections. As the first eukaryotes evolved in a world inhabited by bacteria, PRRs appear to facilitate a wide range of interactions with this diverse microbial world. This new perspective suggests that simply distinguishing self from non-self is insufficient to explain the basic functions of the innate immune system, and future studies should consider how and why we tolerate our ‘microbial self’.
Acknowledgments
We are grateful to A. Khosravi, S. W. McBride, G.Sharon, Y. Lee and M. Flajnik for thoughtful comments on the manuscript, and apologize to our colleagues whose work could not be discussed due to space constraints. Work in the laboratory of the authors is supported by grants from the Burroughs Wellcome Fund, Crohn's and Colitis Foundation and the National Institutes of Health (DK078938).
References
- 1.Janeway CA., Jr Approaching the asymptote? Evolution revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 3.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 6.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 7.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hooper LV, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 10.Nyholm SV, Graf J. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol. 2012;10:815–827. doi: 10.1038/nrmicro2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackey D, McFall AJ. MAMPs and MIMPs: proposed classifications for inducers of innate immunity. Mol Microbiol. 2006;61:1365–1371. doi: 10.1111/j.1365-2958.2006.05311.x. [DOI] [PubMed] [Google Scholar]
- 12.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubinak JL, Round JL. Toll-like receptors promote mutually beneficial commensal-host interactions. PLoS Pathog. 2012;8:e1002785. doi: 10.1371/journal.ppat.1002785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 15.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 17.Werner T, et al. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97:13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 19.Gottar M, et al. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 20.Gay NJ, Keith FJ. Drosophila Toll and IL-1 receptor. Nature. 1991;351:355–356. doi: 10.1038/351355b0. [DOI] [PubMed] [Google Scholar]
- 21.Levashina EA, et al. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- 22.Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 23.Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 24.Ryu JH, et al. The homeobox gene Caudal regulates constitutive local expression of antimicrobial peptide genes in Drosophila epithelia. Mol Cell Biol. 2004;24:172–185. doi: 10.1128/MCB.24.1.172-185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleino A, et al. Pirk is a negative regulator of the Drosophila Imd pathway. J Immunol. 2008;180:5413–5422. doi: 10.4049/jimmunol.180.8.5413. [DOI] [PubMed] [Google Scholar]
- 26.Lhocine N, et al. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe. 2008;4:147–158. doi: 10.1016/j.chom.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Bischoff V, et al. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paredes JC, Welchman DP, Poidevin M, Lemaitre B. Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity. 2011;35:770–779. doi: 10.1016/j.immuni.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 29.Franzenburg S, et al. Bacterial colonization of Hydra hatchlings follows a robust temporal pattern. ISME J. 2013;7:781–790. doi: 10.1038/ismej.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bosch TC, et al. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol. 2009;33:559–569. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Kobe B, Deisenhofer J. Proteins with leucine-rich repeats. Curr Opin Struct Biol. 1995;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 32.Fraune S, et al. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci U S A. 2010;107:18067–18072. doi: 10.1073/pnas.1008573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franzenburg S, et al. MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc Natl Acad Sci U S A. 2012;109:19374–19379. doi: 10.1073/pnas.1213110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 35.McFall-Ngai M, Nyholm SV, Castillo MG. The role of the immune system in the initiation and persistence of the Euprymna scolopes--Vibrio fischeri symbiosis. Semin Immunol. 2010;22:48–53. doi: 10.1016/j.smim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McFall-Ngai M, Heath-Heckman EA, Gillette AA, Peyer SM, Harvie EA. The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol. 2012;24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 38.Koropatnick TA, et al. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 39.Goodson MS, et al. Identifying components of the NF-kappaB pathway in the beneficial Euprymna scolopes-Vibrio fischeri light organ symbiosis. Appl Environ Microbiol. 2005;71:6934–6946. doi: 10.1128/AEM.71.11.6934-6946.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foster JS, Apicella MA, McFall-Ngai MJ. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev Biol. 2000;226:242–254. doi: 10.1006/dbio.2000.9868. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Aksoy S. PGRP-LB is a maternally transmitted immune milk protein that influences symbiosis and parasitism in tsetse's offspring. Proc Natl Acad Sci U S A. 2012;109:10552–10557. doi: 10.1073/pnas.1116431109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins AJ, Schleicher TR, Rader BA, Nyholm SV. Understanding the role of host hemocytes in a squid/vibrio symbiosis using transcriptomics and proteomics. Front Immunol. 2012;3:91. doi: 10.3389/fimmu.2012.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Troll JV, et al. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell Microbiol. 2009;11:1114–1127. doi: 10.1111/j.1462-5822.2009.01315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Troll JV, et al. Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ Microbiol. 2010;12:2190–2203. doi: 10.1111/j.1462-2920.2009.02121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci U S A. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nasevicius A, Ekker SC. Effective targeted gene 'knockdown' in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 47.De Rienzo G, Gutzman JH, Sive H. Efficient shRNA-mediated inhibition of gene expression in zebrafish. Zebrafish. 2012;9:97–107. doi: 10.1089/zeb.2012.0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pham LN, Kanther M, Semova I, Rawls JF. Methods for generating and colonizing gnotobiotic zebrafish. Nat Protoc. 2008;3:1862–1875. doi: 10.1038/nprot.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stein C, Caccamo M, Laird G, Leptin M. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol. 2007;8:R251. doi: 10.1186/gb-2007-8-11-r251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meijer AH, et al. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol Immunol. 2004;40:773–783. doi: 10.1016/j.molimm.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 52.Sullivan C, et al. The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. J Immunol. 2009;183:5896–5908. doi: 10.4049/jimmunol.0803285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sepulcre MP, et al. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J Immunol. 2009;182:1836–1845. doi: 10.4049/jimmunol.0801755. [DOI] [PubMed] [Google Scholar]
- 54.Rader BA, Kremer N, Apicella MA, Goldman WE, McFall-Ngai MJ. Modulation of symbiont lipid A signaling by host alkaline phosphatases in the squid-vibrio symbiosis. MBio. 2012;3 doi: 10.1128/mBio.00093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldberg RF, et al. Intestinal alkaline phosphatase is a gut mucosal defense factor maintained by enteral nutrition. Proc Natl Acad Sci U S A. 2008;105:3551–3556. doi: 10.1073/pnas.0712140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malo MS, et al. Intestinal alkaline phosphatase preserves the normal homeostasis of gut microbiota. Gut. 2010;59:1476–1484. doi: 10.1136/gut.2010.211706. [DOI] [PubMed] [Google Scholar]
- 57.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johansson ME, Sjovall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol. 2013 doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 60.Hapfelmeier S, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 65.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 66.Wehkamp J, et al. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–1664. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wehkamp J, et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci U S A. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petnicki-Ocwieja T, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A. 2009;106:15813–15818. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu C, Xu Z, Gupta D, Dziarski R. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J Biol Chem. 2001;276:34686–34694. doi: 10.1074/jbc.M105566200. [DOI] [PubMed] [Google Scholar]
- 71.Saha S, et al. Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-gamma. Cell Host Microbe. 2010;8:147–162. doi: 10.1016/j.chom.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saha S, et al. Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-gamma. Cell Host Microbe. 2011;8:147–162. doi: 10.1016/j.chom.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bouskra D, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 74.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 75.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 76.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 77.Ochoa-Reparaz J, et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol. 2010;185:4101–4108. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- 78.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shen Y, et al. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12:509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jeon SG, et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012;8:e1002714. doi: 10.1371/journal.ppat.1002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kirkland D, et al. B cell-intrinsic MyD88 signaling prevents the lethal dissemination of commensal bacteria during colonic damage. Immunity. 2012;36:228–238. doi: 10.1016/j.immuni.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Manicassamy S, Pulendran B. Modulation of adaptive immunity with Toll-like receptors. Semin Immunol. 2009;21:185–193. doi: 10.1016/j.smim.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fukata M, et al. The myeloid differentiation factor 88 (MyD88) is required for CD4+ T cell effector function in a murine model of inflammatory bowel disease. J Immunol. 2008;180:1886–1894. doi: 10.4049/jimmunol.180.3.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Caramalho I, et al. Regulatory T cells selectively express toll-like receptors and are activated by lipopolysaccharide. J Exp Med. 2003;197:403–411. doi: 10.1084/jem.20021633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Komai-Koma M, Jones L, Ogg GS, Xu D, Liew FY. TLR2 is expressed on activated T cells as a costimulatory receptor. Proc Natl Acad Sci U S A. 2004;101:3029–3034. doi: 10.1073/pnas.0400171101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu H, Komai-Koma M, Xu D, Liew FY. Toll-like receptor 2 signaling modulates the functions of CD4+ CD25+ regulatory T cells. Proc Natl Acad Sci U S A. 2006;103:7048–7053. doi: 10.1073/pnas.0601554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sutmuller RP, et al. Toll-like receptor 2 controls expansion and function of regulatory T cells. J Clin Invest. 2006;116:485–494. doi: 10.1172/JCI25439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hill DA, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hooper LV. Do symbiotic bacteria subvert host immunity? Nat Rev Microbiol. 2009;7:367–374. doi: 10.1038/nrmicro2114. [DOI] [PubMed] [Google Scholar]


