Abstract
The HSP70 family of molecular chaperones function to maintain protein quality control and homeostasis. The major stress-induced form, HSP70 (also called HSP72 or HSPA1A) is considered an important anti-cancer drug target because it is constitutively overexpressed in a number of human cancers and promotes cancer cell survival. All HSP70 family members contain two functional domains: an N-terminal nucleotide binding domain (NBD) and a C-terminal protein substrate-binding domain (SBD); the latter is subdivided into SBDα and SBDβ subdomains. The NBD and SBD structures of the bacterial ortholog, DnaK, have been characterized, but only the isolated NBD and SBDα segments of eukaryotic HSP70 proteins have been determined. Here we report the crystal structure of the substrate-bound human HSP70-SBD to 2 angstrom resolution. The overall fold of this SBD is similar to the corresponding domain in the substrate-bound DnaK structures, confirming a similar overall architecture of the orthologous bacterial and human HSP70 proteins. However, conformational differences are observed in the peptide-HSP70-SBD complex, particularly in the loop Lα, β that bridges SBDα to SBDβ, and the loop LL,1 that connects the SBD and NBD. The interaction between the SBDα and SBDβ subdomains and the mode of substrate recognition is also different between DnaK and HSP70. This suggests that differences may exist in how different HSP70 proteins recognize their respective substrates. The high-resolution structure of the substrate-bound-HSP70-SBD complex provides a molecular platform for the rational design of small molecule compounds that preferentially target this C-terminal domain, in order to modulate human HSP70 function.
Introduction
The HSP70 family proteins represent an evolutionarily conserved group of molecular chaperones that are important for maintaining protein quality control and protein homeostasis. They were first identified more than thirty years ago in Drosophila as 70 kD proteins that were induced by heat stress or other potentially lethal stimuli [1], [2], and found to be critical for maintaining cell survival [3], [4]. Subsequently, other HSP70 family proteins were identified in both prokaryotes and eukaryotes, with many members also shown to be constitutively expressed and exhibit important housekeeping functions [5]–[8]. Among the many activities of HSP70 family proteins are the chaperoning of nascent polypeptides and unfolding of misfolded protein substrates, the facilitation of protein transport to organelles, the protection and/or dissolution of multi-protein complexes, and the targeting of some misfolded proteins for degradation [5], [6], [9]–[12]. HSP70 proteins are considered among the most conserved proteins in evolution as they are found in all kingdoms from archaebacteria to humans. While most prokaryotes have only one HSP70 gene, some gram-negative bacteria and all eukaryotes encode several HSP70 proteins. For example, Escherichia coli has three HSP70 proteins: DnaK, HscA(Hsc66) and HscC(Hsc62), Saccharomyces cerevesiae encodes at least ten family members, and Homo sapiens encodes at least eight paralogs [5]–[8], [13], [14]. Some family members are thought to serve tissue-specific or organelle-limited roles; some are constitutively expressed, and still others are stress-induced. Current evidence suggests that certain HSP70 family members may serve overlapping, or specific, functions in a cell or organism [6], [15], [16]. Thus, a better understanding of the functional diversity of the HSP70 family proteins would benefit from greater insight regarding their structure-activity relationships.
The stress inducible human protein HSP70 (also called HSPA1A/A1B, HSP70-1 and HSP72) is of particular interest because it is considered a cancer-critical survival protein [17], [18]. Unlike the closely related, but constitutively expressed HSC70 (also known as HSPA8, Hsp70-8 and HSP73) protein, HSP70 is not essential for viability, as knockout mice for HSP70 are viable and fertile [19]. Additionally, unlike HSC70, HSP70 is expressed at very low levels in unstressed normal cells but is rapidly up-regulated under a variety of stress conditions. Importantly, it is constitutively overexpressed in most human cancer cells, and its elevated expression correlates with resistance to therapy and poor prognosis [6], [13], [15], [16]. Evidence indicates that, among its cancer-supporting activities, HSP70 protects cells from apoptosis and the proteotoxic stress associated with oncoproteins and aberrant metabolism [20]–[22]. Accordingly, this molecular chaperone has emerged as an attractive therapeutic target, and several groups have focused efforts on the identification of HSP70 inhibitors for use in cancer therapy. To date, however, relatively few effective, well-characterized modulators of HSP70 activities have been reported [7], [8], [22]–[30]. In general, a lack of structural information on the human HSP70 protein has slowed the development of more effective, clinically useful inhibitors. It is expected that generating new structural information on human HSP70 should aid in the development of such modulators.
HSP70 binds to small hydrophobic stretches of amino acids in nascent or partially folded substrates; together with the actions of critical co-chaperones, it directs the substrates to a particular fate, such as re-folding or degradation. HSP70 proteins share a similar overall structure, comprised of an N-terminal nucleotide binding domain (NBD) that, on its own, exhibits modest ATPase activity, and a C-terminal peptide substrate-binding domain (SBD). The actions of the two major domains of the HSP70 proteins are allosterically regulated. In the presence of ADP, model substrate peptides such as NRLLLTG or client proteins interact with high affinity. However, when ATP is bound to the NBD, substrate binds significantly more weakly. The NBD is subdivided into four subdomains, which are partitioned into two lobes (I and II) by a central ATP/ADP binding pocket [31], [32]. The SBD is composed of a two-layered β-sandwich (SBDβ), which contains the peptide binding pocket, and an α-helical subdomain (SBDα) that functions as a lid, covering the substrate binding cleft [33], [34]. The SBDβ and SBDα subdomains are connected by the loop Lα,β, which shows limited species conservation. The NBD and SBD are bridged by a highly conserved interdomain linker (also known as loop LL,1), which has been implicated as being critically involved in modulating the allosteric regulation of HSP70 proteins [35]–[39]. The extreme C-terminal domain of HSP70 is believed to be largely unstructured, and is the docking site for some co-chaperones [6].
Among the HSP70 family proteins, E. coli DnaK has been the most extensively studied member at the structural level, though there is some structural information about the mammalian heat shock cognate (HSC70) member as well. Indeed, much of our mechanistic understanding of HSP70 structure and activity has come from analyses of E. coli DnaK. Several structures of the DnaK NBD [40] and SBD region, either alone or in complex with various client peptides [33], [34], [41]–[44], have been determined. Several near full-length DnaK structures, bound to either ADP or ATP in the presence or absence of substrates, were also more recently determined [37], [39], [45], [46]. In the past two decades, several structures of isolated NBDs of HSC70 in the absence of nucleotides and in the presence of ADP or ATP as well as the isolated SBDα subdomain of HSC70 have been determined [45], [47]–[51]. Finally, a near full-length, nucleotide-free bovine HSC70 X-ray structure (residues 1-54; PDB code 1YUW) has been reported [52].
Significantly less is known about the structure of the stress-inducible human HSP70, particularly of the SBD. To date, the following structures of human HSP70 have been determined: the nucleotide-bound or apo-form NBD [53]–[58], the isolated NBD in complex with the inhibitor VER-155008 (PDB code 4IO8) [59], and the isolated C-terminal SBDα (residues 537–610 [60]; or residues 534–615, PDB code 3LOF). No structure is available of a complete HSP70 SBD, which includes both SBDβ and SBDα, and no structure has been reported of the HSP70 SBD in complex with a substrate. Here we present the 2 Å crystal structure of the stress inducible human HSP70 (residues 386–616) bound to a hepta-peptide model substrate, NRLLLTG, used in previous DnaK protein structural analyses. This high-resolution structure represents the first of an intact eukaryotic HSP70 SBD; it provides a previously unavailable molecular platform to use in structure-function analyses and should aid in the rational design of HSP70-specific modulators that preferentially target the SBD of this important molecular chaperone.
Materials and Methods
Protein Expression and Purification
Constructs of human HSP70 (residues 1–641, 386–616, 391–615, and 395–510) were cloned into pET25 (EMD Millipore Chemicals, Inc., Billerica, MA, USA), between the NdeI and XhoI restriction sites, for expression as N-terminal His6-tagged fusion proteins in E. coli. All plasmid inserts were verified by DNA sequencing. The E. coli BL21 Star (DE3) competent cells (Invitrogen catalog number C6010-03) were transformed with the respective expression plasmids. The resulting strains were grown at 37°C in LB medium containing 50 µg/ml of carbenicillin (Sigma-Aldrich Co., St. Louis, MO, USA). At an OD600 ∼0.3–0.5, protein expression was induced with 0.5 mM IPTG, and cells were subsequently grown overnight at 25°C. Cells were collected by centrifugation and resuspended in the Bugbuster Master Mix (EMD Millipore Chemicals catalog number 71456-4), supplemented with protease inhibitors, 1 mM DTT and 20 mM imidazole. The His6-tagged fusion proteins were isolated on Ni2+-chelating resins (Ni-NTA Superflow, Qiagen catalog number 30410) by standard procedures. The purified proteins were dialyzed thoroughly against 10 mM Tris-HCl (pH 7) supplemented with 5 mM DTT at 4°C. When necessary, size-exclusion chromatography on a Superdex-200 or a Superose 6 10/300 GL (GE Healthcare) analytical column pre-equilibrated with 10 mM Tris-HCl (pH 7) and 5 mM DTT was used as a final purification step. The pure proteins were concentrated and microcentrifugated at 15,000×g for 5 min at 4°C, the concentration was determined by Bradford Protein Assay and aliquots of the soluble proteins were stored at −80°C.
Crystallization, Data Collection, and Structure Determination
We used the hanging drop vapor diffusion technique to co-crystallize the HSP70-SBD with the NRLLLTG peptide. 5 mM of NRLLLTG (Biomatik, Wilmington, Delaware, USA) was mixed with 400 µM of HSP70 protein (residues 386–616, 391–615, or 395–510) or 400 µM of Seleno-Methionine (SeMet)-derivatized HSP70 protein. The protein mixtures were preheated to 42°C for 15 minutes and gradually cooled to room temperature. The protein preparations were subsequently microcentrifugated at 15,000×g for 5 min at room temperature, and the soluble protein complexes were used to set up crystallization screens. The best crystals were obtained using a HSP70 construct containing residues 386–616 from a crystallization solution of 0.1 M Bis-Tris (pH 5.5), 0.2 M Li2SO4, with 28∼30% PEG3350 in the reservoir and 22%∼25% PEG3350 in the crystallization drop. The crystals grow to full size (0.05∼0.2 mm in three dimensions) in 2∼3 days and flash-frozen directly in liquid nitrogen. Data were collected using the X29A beamline at the National Synchrotron Light Source (Brookhaven National Laboratory). One data set each for native and SeMet-derivatized HSP70-SBD/substrate crystals were collected at wavelengths of 1.075 Å and 0.9791 Å (selenium peak), respectively, and then processed with HKL2000 [61]. The data were useful to resolution limits of 2 Å and 2.5 Å, respectively. Eight selenium sites from data collected from the SeMet-derivatized crystals were identified with the HYSS program [62] using the Phenix suite [63]. These sites were input into the Phenix/Phaser program [64] and protein phases were determined using the same dataset by the single-wavelength anomalous dispersion (SAD) method. A high quality electron density map was generated and a initial model of 323 amino acid within 16 fragments were build using Phenix/Autobuild [65] automatically. The structure was refined and manually adjusted using the high resolution native dataset by iterative cycles of refinement with Phenix/Refine [66] and model building with Coot [67] (Table 1). All the structural alignments and figures were prepared with Pymol (http://www.pymol.org).
Table 1. Data Collection and Refinement Statistics.
| SeMet-HSP70-NRLLLTG | Native HSP70-NRLLLTG | |
| Data collection | ||
| Space group | C2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 100.25, 84.80, 72.15 | 100.29, 85.28, 72.20 |
| α, β, γ (°) | 90.00, 127.20, 90.00 | 90.00, 126.97, 90.00 |
| Wavelength | 0.97910 | 1.0750 |
| Resolution (Å) | 50–2.50 (2.59–2.50) | 50–2.0 (2.07–2.00) |
| Rmerge | 8.9 (31.9) | 9.9 (24.5) |
| I/σ (I) | 20.8 (6.7) | 20.7 (8.1) |
| Data completeness (%) | 100.0 (100.0) | 99.8 (99.4) |
| Redundancy | 7.4 (7.5) | 7.5 (7.5) |
| No. reflections | 16817 | 32837 |
| Refinement | ||
| Resolution (Å) | 50–2.0 (2.07–2.00) | |
| No. reflections | 32790 | |
| Rwork/Rfree | 17.54/21.78 | |
| Molecules/a.u. | 2 | |
| No. atoms | 4105 | |
| Protein | 3448 | |
| Peptide | 99 | |
| Water | 543 | |
| B-factor (Å2) | ||
| Average | 29.59 | |
| Protein | 28.12 | |
| Peptide | 30.89 | |
| Water | 37.07 | |
| r.m.s deviations | ||
| Bond lengths (Å) | 0.0040 | |
| Bond angles (°) | 0.842 | |
| Ramachandran plot (%) | ||
| Favored regions | 99.3 | |
| Additionally allowed regions | 0.7 |
r.m.s., root-mean-square deviation. Values in parenthesis are for the highest resolution shell.
Protein Data Bank accession code
Atomic coordinates for the human HSP70 SBD structure have been deposited with the Protein Data Bank under the identifier code 4po2.
Results and Discussion
Overall Structure of the Human NRLLLTG-Bound HSP70-SBD
We set out to determine the high-resolution crystal structure of the human HSP70 SBD bound to a client peptide substrate. Towards that goal, we were able to prepare well-ordered crystals of HSP70 (aa 386–616) bound to a NRLLLTG peptide that formed in the C2 space group and diffracted to 2 Å resolution. The structure of the complex was determined with single anomalous diffraction using selenomethionine-derivatized protein and refined to 2 Å to Rwork and Rfree values of 17.54% and 21.78%, respectively, with excellent stereochemistry (Fig. 1A and Table 1).
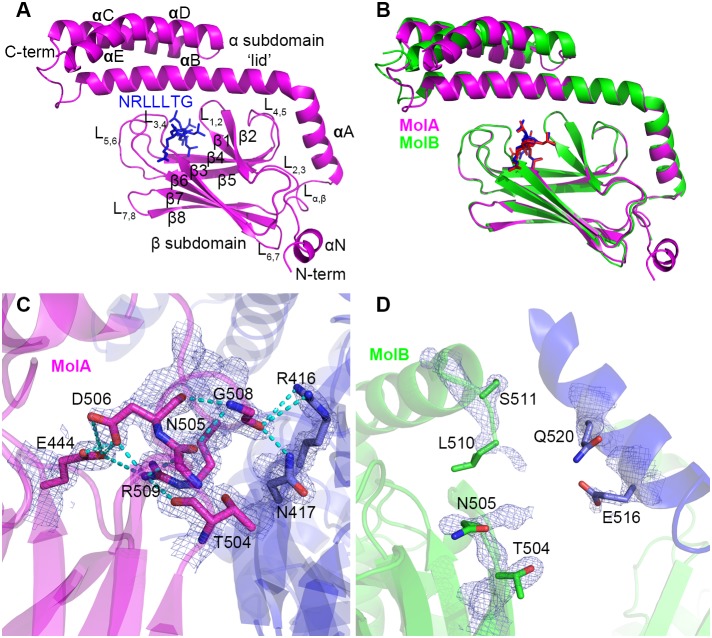
Figure 1. Overall structure of the human peptide-bound HSP70 SBD.
A. Cartoon representation of NRLLLTG-bound HSP70 SBD Molecule A. The SBD and NRLLLTG peptide are shown in magenta and blue, respectively, and all domains and secondary structural elements are labeled. B. Superposition of molecules A and B in the asymmetric unit cell. Molecules A and B are color-coded magenta and green, respectively. C and D. Hinge region between the α and β subdomains. Molecules A and B are shown in panels C and D, respectively. Interacting (MolA) or potential interacting (MolB) residues are indicated in stick and CPK coloring (oxygen in red and nitrogen in blue). The electron density maps corresponding to these residues are contoured at 1.5 (MolA) and 1.0 (MolB) sigma. Hydrogen bonds are indicated with dotted lines.
The X-ray crystal structure of NRLLLTG-bound HSP70-SBD reveals two molecules of the complex per asymmetric unit cell. Each molecule contains the characteristic SBDβ and SBDα subdomains (Fig. 1A), as previously observed in the NRLLLTG-bound DnaK-SBD [33]. Molecule A (MolA) can be traced from Asn387 to Gly613, while Molecule B (MolB) can only be traced from Asp395 to Gly613 and contains a gap in the electron density map corresponding to residues 506–509 in the loop Lα,β region. Based on sequence alignment, the human HSP70 SBD domain (residues 391–615) corresponds to residues 389–607 of DnaK, which was used previously for structural studies [33]. Since we employed residues 386–616 of human HSP70 for structure determination here, the human HSP70 SBD structure is five amino acids longer at the N-terminus than the DnaK SBD previously reported [33]. Additionally, we retained the 6-his tag at the N-terminus for crystallization. This N-terminal region of the HSP70 SBD, which represents the interdomain linker between the NBD and SBD, forms a short helix on MolA (referred to here as αN) but is disordered in MolB. These differences are likely mediated by the crystal environment since the N-terminus of MolA forms crystal contacts while the corresponding region of MolB does not. The corresponding region of intact E.coli DnaK forms a β strand [39], and the corresponding region of ADP-bound intact Geobacillus kaustophilus HTA426 DnaK (gkDnaK) forms an extended loop structure [45]; it is also distinct from a loop in the nucleotide free NBD of the bovine HSC70 structure [52]. Taking this comparison together, suggests that the interdomain linker between the NBD and SBD of human HSP70 is most likely to be inherently flexible.
Comparison of the two molecules in the asymmetrical unit cell
The overall fold of MolA and MolB in the asymmetric unit cell is essentially identical (Fig. 1B). Each monomer is composed of the SBDβ subdomain that contains the peptide-binding pocket, and an SBDα subdomain, also known as the lid. A superposition of MolA to MolB reveals that the SBDβ subdomain contains a root mean square deviation (RMSD) of 0.18 Å for 110 equivalent Cα positions. The SBDα subdomain shows more variability, particularly in the three-helix bundle of the SBDα region, with an overall RMSD of 1.7 Å for 198 equivalent Cα positions of the SBD. The latter observation is consistent with prior studies of substrate-bound DnaK structures, which indicated the existence of considerable structural variability within the SBDα region [33], [41], [42], [69].
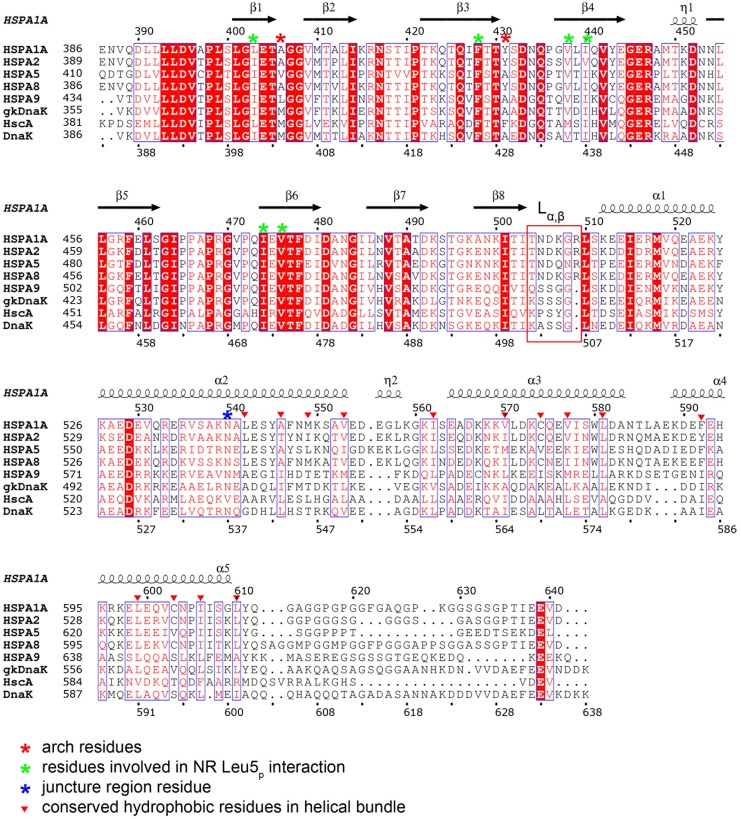
The linker region between the α and β subdomains (loop Lα,β) is of particular interest because it has been shown in the ATP-bound DnaK structures to serve as a dynamic hinge that serves to appropriately position the long helix αB of the SBDα[37], [39]. A sequence alignment of this region reveals significant differences between E. coli DnaK and human HSP70s. In DnaK, this linker region is composed of residues with small side chains (ASSGL or SSSGL), while in human HSP70 this linker region is composed of conserved amino acids with larger side chains (NDKGRL) (Fig. 2). Phylogenetic analysis shows that, unlike other regions of HSP70, this region is significantly different between prokaryotes and eukaryotes [70]. In MolA of our structure, this linker is involved in crystal packing through interactions with a loop from an adjacent molecule (Fig. 1C). Specifically, the carboxylate oxygen atom of Gly508 hydrogen bonds with arginine and asparagine residues of a symmetry-related molecule. The side chain of Arg509 also folds back to interact with both the backbone of Thr504 and Asn505, and the side chain of Asp506 and Glu444. In addition, the side chain of Asp506 forms hydrogen bonds with Glu444. All these interactions hold the linker region in a fixed position, so that the electron density map in this region is very clear. In contrast, the corresponding linker region in MolB is not observed because the adjacent molecule is more than 5 Å away and, as a consequence, the electron density is poorly defined in this region of MolB (Fig. 1D). The dramatically different conformations of the linker region in the two molecules in the asymmetric unit is consistent with the inherent flexibility of this region in human HSP70.
Figure 2. Sequence alignment of eukaryotic and bacterial HSP70 proteins.
Human HSP70 (HSPA1A or HSP72), HSPA2, HSPA5 (GRP78 or BiP), HSC70 (HSPA8 or HSP73), HSPA9 (GRP75, Mortalin-2 or MTHSP70) and E.coli DnaK (ecDnaK), E. coli HscA, Geobacillus kaustophilus HTA426 DnaK (gkDnaK) sequences are used for the alignment. Secondary structure elements and residue numbering for HSP70 is indicated above the protein sequence. The hinge region (also known as loop Lα,β) is highlighted in a red rectangular box, the arch residues are indicated with red asterisks, residues involved in NRLLLTG Leu5p substrate binding are indicated with green asterisks, and the juncture region residue Asn540 is indicated with a blue asterisk. The conserved hydrophobic residues in the helical bundle region are indicated with red triangles.
Comparison to other HSP70 SBD Structures
The human HSP70 SBD is highly homologous to the DnaK SBD (51% sequence identity in the full-length protein and 47% identity in the SBD) (Fig. 2). Not surprisingly, the overall fold of the HSP70-NRLLLTG structure closely approximates the other substrate-bound DnaK SBD structures, with the SBDβ subdomain showing the greatest degree of structural homology with an RMSD of about 0.4 Å, depending on which DnaK structure is used for the comparison (Fig. 3A). In contrast, loop Lα,β is significantly different as there is one more residue inserted in this region of the human HSP70 proteins, and, as stated previously, the side chains are longer than those in DnaK. The SBDα subdomains of the peptide-bound HSP70 and DnaK structures show greater structural variability with an average RMSD of about 1.5 Å (Fig. 3A). Importantly, the modest degree of structural variation between the SBD of HSP70 and DnaK is on par with the structural variation between the two HSP70 molecules in the asymmetric unit, which is consistent with the highly conserved structures of the SBD of both human HSP70 and bacterial DnaK. This observation is in contrast to previous observations based on partial HSC70 SBD structures, which suggested that the HSC70 SBDα region harbors considerable differences to the corresponding region of DnaK [49], [52].
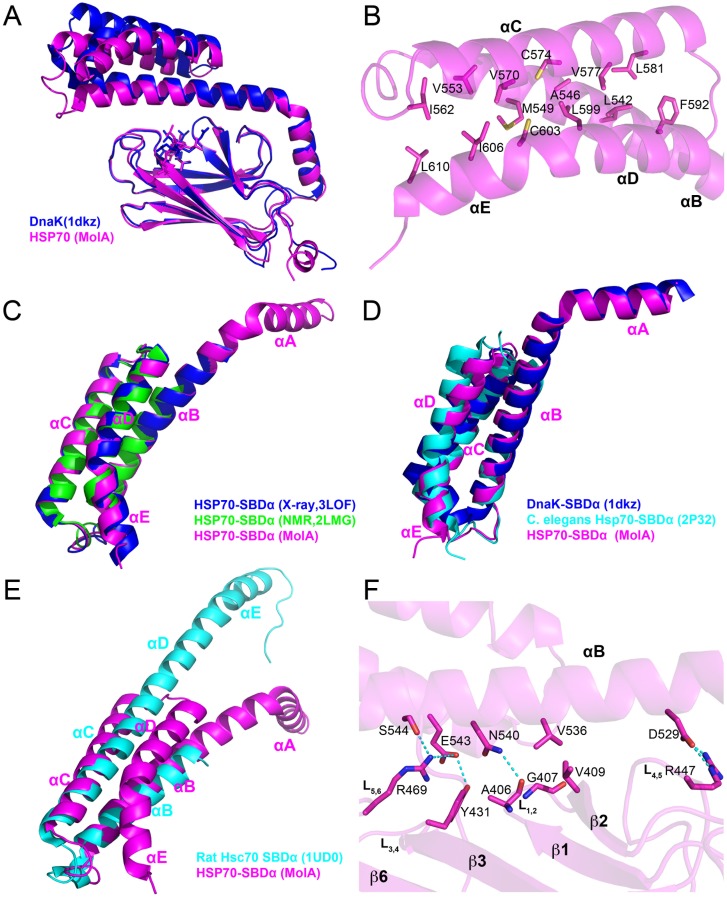
Figure 3. Comparison of the substrate-bound HSP70 SBD to other DnaK and HSC70 SBD structures.
A. Comparison of the NRLLLTG-bound HSP70 SBD to the NRLLLTG-bound DnaK SBD (PDB code 1DKZ). HSP70 and DnaK are shown in magenta and blue cartoons, respectively. The corresponding peptide substrates are shown as stick models. B. A close-up view of the hydrophobic interactions within the helical bundle region of the α subdomain. Hydrophobic residues are shown as stick models. C. Comparison of the SBDα subdomain of the human HSP70-NRLLLTG structure with the isolated three-helix bundle region of the same protein in X-ray structure (PDB code 3LOF, blue) and NMR solution structure (PDB code 2LMG, green). D, Structural alignment of the SBDα subdomain of the HSP70-NRLLLTG structure with the three-helix bundle region of C. elegans Hsp70 (PDB code 2P32, cyan) and DnaK (PDB code 1DKZ, blue). E. Superposition of the lid subdomains of human HSP70 and rat HSC70. HSP70 (MolA) and HSC70 are colored in magenta and cyan, respectively. F. A close-up view of the interaction between the SBDα and SBDβ subdomains of HSP70. Interacting residues are highlighted as sticks, and hydrogen bonds are indicated with dotted lines.
The SBDα subdomain is comprised of five α-helices (αA–αE). The helix αA connects the loop Lα,β to the αB helix, the C-terminal half of which forms an anti-parallel three-helix bundle with helices αC and αD. The αD helix is contiguous with the αE helix. The structure of HSP70 reveals that the helices within the lid are stabilized by extensive van der Waals interactions between hydrophobic residues (Fig. 3B). These hydrophobic residues are not strictly conserved (including Leu542, Ala546, Met549, Val553, Ile562, Val570, Cys574, Val577, Leu581, Phe592, Leu599, Cys603, Ile606, Leu610 in HSP70), but the corresponding positions in DnaK are always occupied by hydrophobic residues (Figs. 2). The helix αE in the HSP70-NRLLLTG structure is kinked by about 30° because of the hydrophobic interaction between this helix and the three helical bundle. This is significantly different from helix αE of crystalized DnaK (which is 1 residue shorter at the C-terminus than the human HSP70 SBD structure reported here), which is kinked by about 60°. Specifically, our findings are most consistent with the following eukaryotic HSP70 SBDα structures: the crystal structures of the human HSP70-SBDα helical bundle (residues 534–613; PDB code 3LOF; overall Cα RMSD = 0.42 Å); the solution structure of human HSP70 SBDα helical bundle (residues 537–610; PDB code 2LMG; overall Cα RMSD = 0.77Å) [60] (Fig. 3C); and the C. elegans Hsp70 SBDα helical bundle (residues 533–614; PDB code 2P32; overall Cα RMSD = 0.58 Å) [71] (Fig. 3D). Based on the comparison of these structures, it appears that eukaryotic HSP70 proteins have a different helix αE conformation than DnaK.
The previous crystal structure of the rat HSC70 lid region revealed a dramatically different organization of the helices, in which helices αC, αD and αE are combined to form a long helix [49] (Fig. 3E). Although this structure suggested that the SBDα of HSC70 is structurally distinct from that of DnaK, our structure shows that human HSP70 adopts the same SBDα fold as DnaK. The different helix organization in HSC70 might be influenced by the truncated construct used or the crystal lattice environment. This conclusion is bolstered by the analogous structures of the three isolated three-helix bundle structures from eukaryotic HSP70 discussed above. Taken together, this comparison is consistent with a structurally conserved and rigid lid region between the eukaryotic and bacterial HSP70 proteins.
The combined structural data support the notion that the SBDα subdomain is inherently more flexible than the SBDβ [33], [42], [69]. This is even true when one compares the SBDα subdomain from different molecules in the asymmetric unit of the same crystal structure, as illustrated in the comparison between MolA and MolB of HSP70 reported here (Fig. 1B). On closer inspection, it appears that the junction for this variability occurs around the center of helix αB where this helix participates in the helical bundle of the SBDα subdomain (Fig. 3F). In particular, consistent with the DnaK juncture region (536–538) [33], the juncture point for this region appears to be around residue Asn540. This residue is identical in most HSP70 proteins (except E. coli HscA) and also makes conserved interactions with residue Ala406 on the SBDβ subdomain. In this HSP70 structure, Asn540 interacts with the backbone of Ala406 (3.3 Å in MolA, 3.0 Å in MolB), which is in the L1,2 loop forming the substrate-binding groove on the SBDβ subdomain. A helical turn away, Val536, which is either a valine or isoleucine residue in HSP70 proteins, makes van der Waals contacts with Gly407 and Val409; notably, Gly407 is strictly conserved in all HSP70s and Val409 is conserved on strand β2 of the SBDβ subdomain in most HSP70 proteins, except for E. coli HscA (Fig. 2). Val536 is always placed in between Gly407 and Val409. Additional hydrogen bond interactions occur between Asp529 at the N-terminal end of helix αB and Arg447 of loop L4,5 of the SBDβ subdomain. Together, these residues are highly conserved in HSP70 proteins. These conserved interactions fix the conformation of the N-terminal half of helix αB. However, the interactions in the C-terminal part of helix αB are less conserved. For instance, in MolA, residues Glu543 and Ser544, one helical turn C-terminal to Asn540 of helix αB, hydrogen bond with Arg469 of loop L5,6 and Tyr431 of loop L3,4 of the SBDβ subdomain (Fig. 3F). In contrast, these interactions differ in MolB, with Arg469 adopting a different conformation that cannot contact Glu543 and Ser544. Since Arg469 extends to Phe547, it appears that MolB is forming a destabilizing interaction in this region. Together, this network of interactions between the N-terminal end of helix αB of the SBDα subdomain and the loops of the SBDβ subdomain serve to rigidly link this region of helix αB as well as the preceding helix αA to the SBDβ subdomain, but leaves the C-terminal part of helix αB and the associated helical bundle region of the SBDα subdomain to take on more divergent positions when compared to different HSP70 proteins. Interestingly, DnaK shows a more extensive network of interactions between residues within helix αB and the SBDβ subdomain. This is consistent with the greater degree of structural and functional plasticity of the eukaryotic HSP70 proteins relative to the DnaK proteins. We hypothesize that the less constrained SBDα subdomain of the HSP70 proteins may influence the interaction of HSP70 proteins with different co-chaperones such as CHIP [72].
Substrate Recognition by HSP70
The NRLLLTG substrate peptide is known to bind to the ADP-bound HSP70/DnaK with high affinity [33], [73]. However, upon exchange of ADP with ATP, the HSP70/DnaK protein undergoes a conformational change, switching to the lower-affinity peptide binding state [38], [73], [74]. Similar to the NRLLLTG-DnaK-SBD structure, the NRLLLTG peptide substrate binds to the SBDβ subdomain of the human HSP70 SBD (Fig. 4A). This indicates that this structure is representative of the peptide bound to the ADP form of full length HSP70. The SBDβ subdomain is made up of two layers of β sheets that interact through hydrophobic interactions (Figs. 1A and 4A). The first layer consists of strands β3, β6, β7 and β8, and the second layer contains strands β5, β4 and β1, β2 (Fig. 1B). These β-strands are connected by loops L1,2 through L6,7. The NRLLLTG peptide binding pocket is situated between loops L3,4 and L1,2, and the binding cleft is further stabilized by loops L4,5 and L5,6 through a series of hydrogen bonds and multiple van der Waals contacts (Figs. 1A, 4A and 4B).
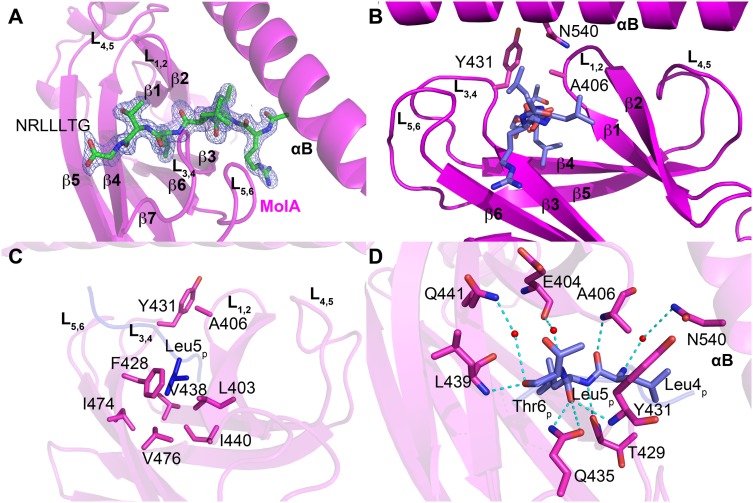
Figure 4. NRLLLTG peptide substrate binding by HSP70.
A. Omit electron density of the peptide substrate bound to the β subdomain of molecule A is contoured at 3.0 sigma. The NRLLLTG peptide is shown as a stick model in CPK coloring. B. Peptide binding site highlighting the flanking L1,2 and L3,4, and supporting L5,6 and L4,5 loops. The NRLLLTG peptide and arch residues are shown as stick models. C. Detailed view of the NRLLLTG-binding pocket in HSP70-NRLLLTG highlighting a network of van der Waals contacts with Leu5p of the NRLLLTG peptide. D. A close-up view of the interactions between Leu4p-Leu5p-Thr6p of the NRLLLTG peptide and the surrounding residues in HSP70-NRLLLTG. All interacting residues are shown as sticks and hydrogen bonds are shown as dotted lines.
The substrate-binding groove of HSP70 shows significant conservation with the corresponding groove of DnaK (and other HSP70 proteins). Highly conserved residues Leu403 (Ile401 in DnaK), Phe428, Val438, Ile440, Ile474 and Val476 at the base of the hydrophobic pocket each make van der Waals interactions with the central hydrophobic residue of the peptide substrate, Leu5p (Fig. 4C). These interactions explain the strong preference for a hydrophobic residue, and particularly leucine, in this position of HSP70 substrates. The flanking peptide residues make more minor interactions with HSP70, while the side chains of Ala406 and Tyr431 form a structurally conserved arch over the surface of the pocket and make van der Waals interactions with flanking residues Leu4p and Thr6p (methyl group) (Fig. 4D). Positions corresponding to Ala406 and Tyr431 are not conserved within DnaK but are typically hydrophobic in DnaK and other HSP70 proteins (Fig. 2). These arch residues correspond to residues Met404 and Ala429 in E. coli DnaK, Met and Phe in E. coli HscA and Ser and Ala in E. coli HscC. Taken together, the HSP70 substrate-binding site appears to be tailored for a hydrophobic residue and leucine in particular that is flanked by 2 additional residues with hydrophobic character, and this is conserved in DnaK and other HSP70 proteins.
A survey of DnaK SBD X-ray structures with the same peptide sequence (i.e. NRLLLTG) reveals that most substrate-bound structures contain Leu4p in the hydrophobic pocket [33], [42]. However, one structure (PDB code 4EZW) also has Leu5p modeled into the pocket; and another structure contains the peptide NRLILTG instead of the canonical NRLLLTG peptide, but bound in the reverse orientation with Leu3p in the hydrophobic pocket (PDB code 4EZY) [42]. Analysis of the crystal packing arrangements of several DnaK SBD/peptide structures does reveal that the crystal-packing contacts could, and likely do, influence the register of peptide binding [33], [42]. Consistent with their broad substrate specificity, we hypothesize that the HSP70 isoforms can accommodate various peptide registers, and the actual binding mode may depend on the composition of the substrate and the microenvironment.
Implications for Developing HSP70 Inhibitors
Rodina et al. (2013) recently unveiled 5 potential druggable clefts from a theoretical model of full length human HSP70, which was generated from the DnaK SBD. Combining the S-score, D-score and size of the cavity, they identified 2 sites (sites 1 and 2) in the NBD that are considered “most druggable” (Rodina et al., 2013). Furthermore, they tested a novel inhibitor called YK5 and provided evidence, in addition to computer modeling, that it binds to site 1 [30]. Intriguingly, Schlecht et al. recently cocrystallized the N-terminal NBD domain of HSP70 (residues 1–382) with the HSP70 inhibitor VER-155008 (PDB code 4IO8) [59], and in this structure the inhibitor binds to site 2 in the NBD.
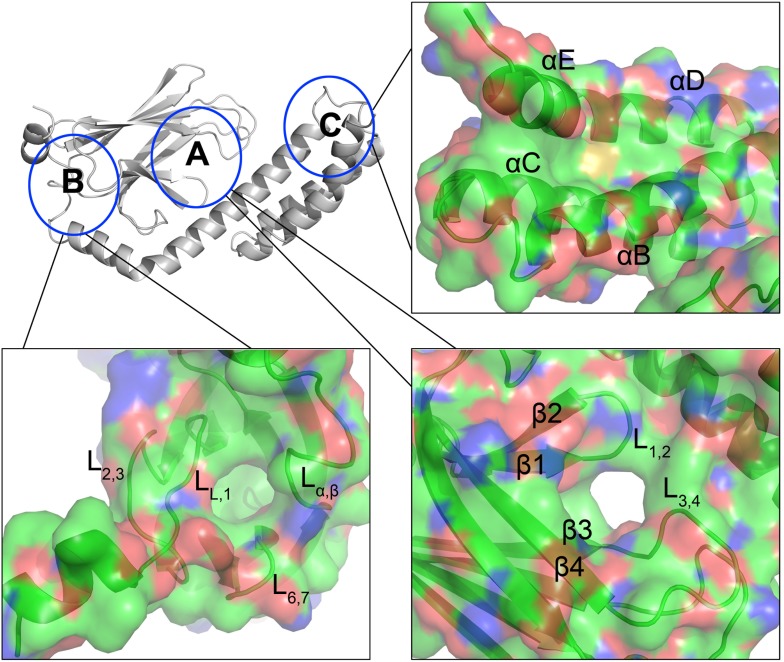
The high resolution NRLLLTG-bound HSP70-SBD structure reported here provides an opportunity to identify additional potential druggable sites within the HSP70 SBD domain. A detailed analysis of the structure reveals three potentially druggable hydrophobic pockets for the rational development of inhibitors (Fig. 5), two of which were previously noted using the theoretical model of the human SBD (Rodina et al., 2013). The first site (labeled A in Fig. 5) maps to the canonical substrate-binding site, referred to here as the Leu5p binding pocket. The second site (labeled B in Fig. 5) is distinct from the NRLLLTG-binding cleft and is supported by loops Lα,β, L2,3, L6,7 and LL,1; a similar hydrophobic binding site has been reported by Cellitti et al. (2009) [68]. A third site (labeled C in Fig. 5) is located in the helix bundle region, which is more covert and has been investigated recently [75]. Given the importance of these predicted hydrophobic pockets in orchestrating the crosstalk among the NBD, SBDβ and SBDα domains, and the interaction between the SBD domain and other co-chaperones, it is anticipated that small molecule inhibitors or inhibitory peptides that preferentially interact with one or more of these sites might negatively impact HSP70-clients interactions.
Figure 5. Putative druggable sites in the HSP70 SBD domain.
An overview of three hydrophobic pockets within the SBD domain have been circled and marked as A, B and C. Close-up of the surface area of these three sites are illustrated in CPK coloring mode, with carbon in green, oxygen in red and nitrogen in blue. Secondary structural elements within the area are labeled. The NRLLLTG substrate is removed from this figure (pocket A) to better illustrate the pocket A for comparison with the other pockets (B and C).
In summary, we report the crystal structure of the complete stress-inducible human HSP70-SBD in complex with a peptide substrate at atomic resolution. This structure provides the first substrate-bound HSP70-SBD molecular scaffold for a structure-activity based strategy to understand HSP70-substrate interactions and subsequent signaling events. Analysis of the NRLLLTG-bound HSP70-SBD structure also provides intriguing clues for potential drug discovery and development.
Acknowledgments
We thank the services provided by the University of Pennsylvania DNA Sequencing Facility at the Perelman School of Medicine, University of Pennsylvania.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Atomic coordinates for the human HSP70 SBD structure have been deposited with the Protein Data Bank under the identifier code 4po2.
Funding Statement
This work was supported by US Public Health Service National Institutes of Health Grants R01CA139319 (M.E.M.) and PO1CA114046 (J.I.L., M.E.M., D.L.G., R.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Schedl P, Artavanis-Tsakonas S, Steward R, Gehring WJ, Mirault ME, et al. (1978) Two hybrid plasmids with D. melanogaster DNA sequences complementary to mRNA coding for the major heat shock protein. Cell 14: 921–929. [DOI] [PubMed] [Google Scholar]
- 2. Ashburner M, Bonner JJ (1979) The induction of gene activity in drosophilia by heat shock. Cell 17: 241–254. [DOI] [PubMed] [Google Scholar]
- 3. Craig EA, Jacobsen K (1984) Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell 38: 841–849. [DOI] [PubMed] [Google Scholar]
- 4. Werner-Washburne M, Stone DE, Craig EA (1987) Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol 7: 2568–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mayer MP, Bukau B (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62: 670–684 10.1007/s00018-004-4464-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Daugaard M, Rohde M, Jäättelä M (2007) The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett 581: 3702–3710 10.1016/j.febslet.2007.05.039 [DOI] [PubMed] [Google Scholar]
- 7.Murphy ME (2013) The HSP70 family and cancer. Carcinogenesis 34, 1181–1188. doi:10.1093/carcin/bgt111. [DOI] [PMC free article] [PubMed]
- 8. Stricher F, Macri C, Ruff M, Muller S (2013) HSPA8/HSC70 chaperone protein: Structure, function, and chemical targeting. Autophagy 9: 1937–1954 10.4161/auto.26448 [DOI] [PubMed] [Google Scholar]
- 9. Sharma SK, De los Rios P, Christen P, Lustig A, Goloubinoff P (2010) The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat Chem Biol 6: 914–920 10.1038/nchembio.455 [DOI] [PubMed] [Google Scholar]
- 10. Munro S, Pelham HR (1987) A C-terminal signal prevents secretion of luminal ER proteins. Cell 48: 899–907. [DOI] [PubMed] [Google Scholar]
- 11. French JB, Zhao H, An S, Niessen S, Deng Y, et al. (2013) Hsp70/Hsp90 chaperone machinery is involved in the assembly of the purinosome. Proceedings of the National Academy of Sciences 110: 2528–2533 10.1073/pnas.1300173110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, et al. (1997) Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J Biol Chem 272: 9002–9010. [DOI] [PubMed] [Google Scholar]
- 13. Mosser DD, Morimoto RI (2004) Molecular chaperones and the stress of oncogenesis. Oncogene 23: 2907–2918 10.1038/sj.onc.1207529 [DOI] [PubMed] [Google Scholar]
- 14. Brocchieri L, Conway de Macario E, Macario AJL (2008) hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol Biol 8: 19 10.1186/1471-2148-8-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calderwood S, Khaleque M, Sawyer D (2006) Heat shock proteins in cancer: chaperones of tumorigenesis. Trends in biochem. Sci. 31, 164–172. [DOI] [PubMed]
- 16. Rohde M, Daugaard M, Jensen MH, Helin K, Nylandsted J, et al. (2005) Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev 19: 570–582 10.1101/gad.305405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nylandsted J, Rohde M, Brand K, Bastholm L, Elling F, et al. (2000) Selective depletion of heat shock protein 70 (Hsp70) activates a tumor-specific death program that is independent of caspases and bypasses Bcl-2. Proc Natl Acad Sci USA 97: 7871–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gyrd-Hansen M, Nylandsted J, Jäättelä M (2004) Heat shock protein 70 promotes cancer cell viability by safeguarding lysosomal integrity. Cell Cycle 3: 1484–1485. [DOI] [PubMed] [Google Scholar]
- 19. Hunt CR, Dix DJ, Sharma GG, Pandita RK, Gupta A, et al. (2004) Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol Cell Biol 24: 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Powers MV, Clarke PA, Workman P (2008) Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell 14: 250–262 10.1016/j.ccr.2008.08.002 [DOI] [PubMed] [Google Scholar]
- 21. Powers MV, Workman P (2007) Inhibitors of the heat shock response: Biology and pharmacology. FEBS Lett 581: 3758–3769 10.1016/j.febslet.2007.05.040 [DOI] [PubMed] [Google Scholar]
- 22. Leu JI-J, Pimkina J, Pandey P, Murphy ME, George DL (2011) HSP70 inhibition by the small-molecule 2-phenylethynesulfonamide impairs protein clearance pathways in tumor cells. Mol Cancer Res 9: 936–947 10.1158/1541-7786.MCR-11-0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brodsky JL, Chiosis G (2006) Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr Top Med Chem 6: 1215–1225. [DOI] [PubMed] [Google Scholar]
- 24. Patury S, Miyata Y, Gestwicki JE (2009) Pharmacological targeting of the Hsp70 chaperone. Curr Top Med Chem 9: 1337–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leu JI-J, Pimkina J, Frank A, Murphy ME, George DL (2009) A small molecule inhibitor of inducible heat shock protein 70. Mol Cell 36: 15–27 10.1016/j.molcel.2009.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Evans CG, Chang L, Gestwicki JE (2010) Heat shock protein 70 (hsp70) as an emerging drug target. J Med Chem 53: 4585–4602 10.1021/jm100054f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Powers MV, Jones K, Barillari C, Westwood I, van Montfort RLM, et al. (2010) Targeting HSP70: the second potentially druggable heat shock protein and molecular chaperone? Cell Cycle 9: 1542–1550. [DOI] [PubMed] [Google Scholar]
- 28. Rerole A-L, Gobbo J, De Thonel A, Schmitt E, Pais de Barros JP, et al. (2011) Peptides and aptamers targeting HSP70: a novel approach for anticancer chemotherapy. Cancer Res 71: 484–495 10.1158/0008-5472.CAN-10-1443 [DOI] [PubMed] [Google Scholar]
- 29. Goloudina AR, Demidov ON, Garrido C (2012) Inhibition of HSP70: a challenging anti-cancer strategy. Cancer Lett 325: 117–124 10.1016/j.canlet.2012.06.003 [DOI] [PubMed] [Google Scholar]
- 30. Rodina A, Patel PD, Kang Y, Patel Y, Baaklini I, et al. (2013) Identification of an allosteric pocket on human hsp70 reveals a mode of inhibition of this therapeutically important protein. Chem Biol 20: 1469–1480 10.1016/j.chembiol.2013.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flaherty K, DeLuca-Flaherty C, McKay D (1990) Three-dimensional structure of the ATPase fragment of a 70 K heat-shock cognate protein. Nature 346, 623–628. [DOI] [PubMed]
- 32. Flaherty KM, McKay DB, Kabsch W, Holmes KC (1991) Similarity of the three-dimensional structures of actin and the ATPase fragment of a 70-kDa heat shock cognate protein. Proc Natl Acad Sci USA 88: 5041–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, et al. (1996) Structural Analysis of Substrate Binding by the Molecular Chaperone DnaK. Science 272: 1606–1614 10.1126/science.272.5268.1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang H, Kurochkin AV, Pang Y, Hu W, Flynn GC, et al. (1998) NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction. Biochemistry 37: 7929–7940 10.1021/bi9800855 [DOI] [PubMed] [Google Scholar]
- 35. Vogel M, Mayer MP, Bukau B (2006) Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J Biol Chem 281: 38705–38711 10.1074/jbc.M609020200 [DOI] [PubMed] [Google Scholar]
- 36. Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, et al. (2007) Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell 26: 27–39 10.1016/j.molcel.2007.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kityk R, Kopp J, Sinning I, Mayer MP (2012) Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol Cell 48: 863–874 10.1016/j.molcel.2012.09.023 [DOI] [PubMed] [Google Scholar]
- 38. Zhuravleva A, Clerico EM, Gierasch LM (2012) An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 151: 1296–1307 10.1016/j.cell.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi R, Sarbeng EB, Liu Q, Le KQ, Xu X, et al. (2013) Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat Struct Mol Biol., 20, 900–907. doi:10.1038/nsmb.2583. [DOI] [PMC free article] [PubMed]
- 40. Harrison CJC, Hayer-Hartl MM, Di Liberto MM, Hartl FF, Kuriyan JJ (1997) Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276: 431–435. [DOI] [PubMed] [Google Scholar]
- 41. Liebscher M, Roujeinikova A (2009) Allosteric coupling between the lid and interdomain linker in DnaK revealed by inhibitor binding studies. J Bacteriol 191: 1456–1462 10.1128/JB.0113108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zahn M, Berthold N, Kieslich B, Knappe D, Hoffmann R, et al. (2013) Structural Studies on the Forward and Reverse Binding Modes of Peptides to the Chaperone DnaK. J Mol Biol. doi:10.1016/j.jmb.2013.03.041. [DOI] [PubMed]
- 43. Pellecchia M, Montgomery DL, Stevens SY, Vander Kooi CW, Feng HP, et al. (2000) Structural insights into substrate binding by the molecular chaperone DnaK. Nat. Struct. Biol. 7, 298–303. [DOI] [PubMed] [Google Scholar]
- 44. Stevens SY, Cai S, Pellecchia M, Zuiderweg ERP (2003) The solution structure of the bacterial HSP70 chaperone protein domain DnaK (393–507) in complex with the peptide NRLLLTG. Protein Sci 12: 2588–2596 10.1110/ps.03269103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chang Y-W, Sun Y-J, Wang C, Hsiao C-D (2008) Crystal structures of the 70-kDa heat shock proteins in domain disjoining conformation. Journal of Biological Chemistry 283: 15502–15511 10.1074/jbc.M708992200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ERP (2009) Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci USA 106: 8471–8476 10.1073/pnas.0903503106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilbanks SM, McKay DB (1998) Structural replacement of active site monovalent cations by the epsilon-amino group of lysine in the ATPase fragment of bovine Hsc70. Biochemistry 37: 7456–7462 10.1021/bi973046m [DOI] [PubMed] [Google Scholar]
- 48. Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, et al. (2001) Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291: 1553–1557 10.1126/science.291.5508.1553 [DOI] [PubMed] [Google Scholar]
- 49. Chou C-C, Forouhar F, Yeh Y-H, Shr H-L, Wang C, et al. (2003) Crystal structure of the C-terminal 10-kDa subdomain of Hsc70. Journal of Biological Chemistry 278: 30311–30316 10.1074/jbc.M304563200 [DOI] [PubMed] [Google Scholar]
- 50. Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, et al. (2007) Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell 28: 422–433 10.1016/j.molcel.2007.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Williamson DS, Borgognoni J, Clay A, Daniels Z, Dokurno P, et al. (2009) Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design. J Med Chem 52: 1510–1513 10.1021/jm801627a [DOI] [PubMed] [Google Scholar]
- 52. Jiang J, Prasad K, Lafer EM, Sousa R (2005) Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell 20: 513–524 10.1016/j.molcel.2005.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sriram M, Osipiuk J, Freeman B, Morimoto R, Joachimiak A (1997) Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain. Structure 5: 403–414 10.1016/S0969-2126(97)00197-4 [DOI] [PubMed] [Google Scholar]
- 54. Osipiuk J, Walsh MA, Freeman BC, Morimoto RI, Joachimiak A (1999) Structure of a new crystal form of human Hsp70 ATPase domain. Acta Crystallogr D Biol Crystallogr 55: 1105–1107. [DOI] [PubMed] [Google Scholar]
- 55. Wisniewska M, Karlberg T, Lehtiö L, Johansson I, Kotenyova T, et al. (2010) Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B’, and HSPA5/BiP/GRP78. PLoS ONE 5: e8625 10.1371/journal.pone.0008625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Arakawa A, Handa N, Shirouzu M, Yokoyama S (2011) Biochemical and structural studies on the high affinity of Hsp70 for ADP. Protein Sci 20: 1367–1379 10.1002/pro.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shida M, Arakawa A, Ishii R, Kishishita S, Takagi T, et al. (2010) Direct inter-subdomain interactions switch between the closed and open forms of the Hsp70 nucleotide-binding domain in the nucleotide-free state. Acta Crystallogr D Biol Crystallogr 66: 223–232 10.1107/S0907444909053979 [DOI] [PubMed] [Google Scholar]
- 58. Polier S, Dragovic Z, Hartl FU, Bracher A (2008) Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 133: 1068–1079 10.1016/j.cell.2008.05.022 [DOI] [PubMed] [Google Scholar]
- 59. Schlecht R, Scholz SR, Dahmen H, Wegener A, Sirrenberg C, et al. (2013) Functional analysis of hsp70 inhibitors. PLoS ONE 8: e78443 10.1371/journal.pone.0078443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gao X-C, Zhou C-J, Zhou Z-R, Wu M, Cao C-Y, et al. (2012) The C-terminal helices of heat shock protein 70 are essential for J-domain binding and ATPase activation. J Biol Chem 287: 6044–6052 10.1074/jbc.M111.294728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol 276: 307–326. [DOI] [PubMed] [Google Scholar]
- 62. Grosse-Kunstleve RW, Adams PD (2003) Substructure search procedures for macromolecular structures. Acta Crystallogr D Biol Crystallogr 59: 1966–1973. [DOI] [PubMed] [Google Scholar]
- 63. Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, et al. (2007) Phaser crystallographic software. urn:issn: 0021-8898 40: 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, et al. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr 64: 61–69 10.1107/S090744490705024X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, et al. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68: 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 68. Cellitti J, Zhang Z, Wang S, Wu B, Yuan H, et al. (2009) Small molecule DnaK modulators targeting the beta-domain. Chem Biol Drug Des 74: 349–357 10.1111/j.1747-0285.2009.00869.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cupp-Vickery JR, Peterson JC, Ta DT, Vickery LE (2004) Crystal structure of the molecular chaperone HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC. J Mol Biol 342: 1265–1278 10.1016/j.jmb.2004.07.025 [DOI] [PubMed] [Google Scholar]
- 70. Gupta RS, Singh B (1994) Phylogenetic analysis of 70 kD heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus. Curr Biol 4: 1104–1114. [DOI] [PubMed] [Google Scholar]
- 71. Worrall LJ, Walkinshaw MD (2007) Crystal structure of the C-terminal three-helix bundle subdomain of C. elegans Hsp70. Biochem Biophys Res Commun 357: 105–110 10.1016/j.bbrc.2007.03.107 [DOI] [PubMed] [Google Scholar]
- 72. Murata S, Chiba T, Tanaka K (2003) CHIP: a quality-control E3 ligase collaborating with molecular chaperones. Int J Biochem Cell Biol 35: 572–578. [DOI] [PubMed] [Google Scholar]
- 73. Gragerov A, Zeng L, Zhao X, Burkholder W, Gottesman ME (1994) Specificity of DnaK-peptide binding. J Mol Biol 235: 848–854 10.1006/jmbi.1994.1043 [DOI] [PubMed] [Google Scholar]
- 74. Flynn GC, Chappell TG, Rothman JE (1989) Peptide binding and release by proteins implicated as catalysts of protein assembly. Science 245: 385–390. [DOI] [PubMed] [Google Scholar]
- 75. Balaburski GM, Leu JI-J, Beeharry N, Hayik S, Andrake MD, et al. (2013) A Modified HSP70 Inhibitor Shows Broad Activity as an Anticancer Agent. Mol Cancer Res 11: 219–229 10.1158/1541-7786.MCR-12-0547-T [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Atomic coordinates for the human HSP70 SBD structure have been deposited with the Protein Data Bank under the identifier code 4po2.