Abstract
Many pathogens, particularly those that require their host for survival, have devised mechanisms to subvert the host immune response in order to survive and replicate intracellularly. Legionella pneumophila, the causative agent of Legionnaires' disease, promotes intracellular growth by translocating proteins into its host cytosol through its type IV protein secretion machinery. At least 5 of the bacterial translocated effectors interfere with the function of host cell elongation factors, blocking translation and causing the induction of a unique host cell transcriptional profile. In addition, L. pneumophila also interferes with translation initiation, by preventing cap-dependent translation in host cells. We demonstrate here that protein translation inhibition by L. pneumophila leads to a frustrated host MAP kinase response, where genes involved in the pathway are transcribed but fail to be translated due to the bacterium-induced protein synthesis inhibition. Surprisingly, few pro-inflammatory cytokines, such as IL-1α and IL-1β, bypass this inhibition and get synthesized in the presence of Legionella effectors. We show that the selective synthesis of these genes requires MyD88 signaling and takes place in both infected cells that harbor bacteria and neighboring bystander cells. Our findings offer a perspective of how host cells are able to cope with pathogen-encoded activities that disrupt normal cellular process and initiate a successful inflammatory response.
Author Summary
Translation inhibition is a common virulence mechanism used by a number of pathogens (e.g. Diphtheria Toxin, Shiga Toxin and Pseudomonas Exotoxin A). It has been a mystery how host cells mount a pathogen-specific response and clear infection under conditions where protein synthesis is blocked by pathogens. Using Legionella pneumophila as a model, a bacterium that efficiently blocks the host protein translation machinery, we show here that the innate immune system has devised a mechanism to cope with translation inhibition by selectively synthesizing proteins that are required for inflammation.
Introduction
The pathogen-associated molecular pattern (PAMP) hypothesis has been developed to explain how the innate immune system recognizes foreign microbial invaders. By this model, germline-encoded receptors recognize conserved foreign ligands associated with microbes, such as nucleic acids, lipopolysaccharide (LPS), peptidoglycan or flagellin to generate a response directed at clearing the microorganism [1], [2]. More recently, it has become clear that pattern recognition alone does not explain how multicellular organisms are able to differentiate virulent pathogens from harmless commensals and mount a response. It has been proposed that the host immune system can sense the presence of danger and respond to pathogen-encoded enzymatic activities that disrupt normal cellular processes. This mode of recognition, referred to as “effector triggered immunity” has been shown to play a significant role in pathogen clearance both in plants and mammalian cells [3], [4], [5], [6], [7], [8], [9], [10], [11], [12]. Such recognition may be sufficient to activate a host response, but because it occurs simultaneously with PAMP recognition, host cell detection of pathogens likely results from integrating the recognition of microbial patterns together with pathogen-specific activities.
Legionella pneumophila, the causative agent of Legionnaires' disease, promotes intracellular growth by translocating proteins into its host cytosol through its type IV (Dot/Icm) protein secretion machinery [13], [14], [15]. These translocated effectors serve various purposes, including recruitment of ER-derived membrane to the Legionella containing vacuole, inhibition of cell death pathways and manipulation of host lipid metabolism and regulatory pathways [16], [17], [18], [19], [20], [21]. Most importantly for the innate immune response, after contact with macrophages, the bacterium stimulates a pathogen-specific response that is the consequence of simultaneous recognition of PAMPs and pathogen-translocated proteins that results in a unique response to this microorganism [10].
Legionella pneumophila is a pathogen for a broad range of fresh water amoebae, which provide the natural environmental niche for the microorganism and the source of exposure for humans [22], [23]. After aspiration by a susceptible mammalian host, the bacterium is engulfed by alveolar macrophages in the lungs [24]. In cultured macrophages, L. pneumophila provokes signaling through various pattern-recognition receptors (PRRs), such as Toll-like receptors (TLRs) and cytosolic NOD-like receptors (NLRs) [9], [25], [26], [27], [28], [29], [30], [31]. This response is critical for clearance of the microorganism, because mouse mutants defective in these two responses succumb to lethal pneumonia [27].
Interestingly, macrophage challenge with wild type L. pneumophila (Dot/Icm+) triggers a unique transcriptional response in host cells compared to mutants that lack a functional type IV secretion system, supporting the model that there is a pathogen-specific response involved in innate immune recognition [9], [10], [11],[17],[32]. Microarray studies have identified many of these transcriptional targets as being genes controlled by the NF-κB and mitogen-associated protein kinases (MAPKs) transcriptional regulators [9], [17], including downstream dual specificity phosphatases (Dusp1 and Dusp2), stress response genes (Hsp70, Gadd45a, Egr1) and pro-inflammatory cytokines and chemokines (Il1α, Il1β, Tnfα, Il23a, Csf1, Csf2) [9], [10], [11], .
It was recently demonstrated that the pathogen-specific response to Legionella is triggered by the action of L. pneumophila translocated effectors that interfere with host protein translation [10], [11]. Disruption of the host translation machinery serves as a second signal (in concert with signaling from PRRs) to constitute the full innate immune response against Legionella pneumophila [10]. The elimination of five of these effectors is sufficient to block this response, even though it is clear that they are part of a much larger pool of translocated substrates that impinge on host protein synthesis [10], [31]. These inhibitors (the products of the lgt1, lgt2, lgt3, sidI, and sidL genes) modify eukaryotic elongation factor eEF1A and eEF1Bγ of mammalian cells and block protein synthesis both in vitro and in vivo [10], [33], [34], [35].
In addition to blocking elongation, there is evidence that suggests wild type Legionella can also inhibit cap-dependent translation initiation [36]. Recognition of pathogenic Legionella leads to ubiquitination of the mTOR pathway, which in turn suppresses the eukaryotic initiation factor 4E (eIF4E) and prevents the synthesis of various genes [36]. This mode of translation inhibition was shown to induce translational biasing of host cells towards a more pro-inflammatory state [36]. However, it is currently not clear how host cells would be able to mount an inflammatory response when protein translation is blocked by L. pneumophila both at the initiation and elongation stages [9], [17]. It is likely that pattern-recognition would play a role under conditions of intoxication, but the mechanism by which this is regulated is also unclear.
A strong pro-inflammatory cytokine response is crucial for clearance of Legionella pneumophila [9], [31]. The importance of cytokines can be seen in IL-1α, IL-12, IFN-γ and TNF knockout mouse strains that show increased susceptibility to L. pneumophila infection [31], [37]. Moreover, patients treated with TNF-α blockers are at high risk of developing severe Legionnaires' disease [38]. Given the key role that this innate immune response plays in clearance of L. pneumophila, we examined how cytokines and other immune mediators are synthesized under conditions in which the bacterium effectively blocks the host protein translation machinery. We find that although protein synthesis inhibitors induce the transcriptional response and block the translation of most genes, pro-inflammatory cytokine genes can bypass this blockade in a fashion that requires the TLR-adaptor protein MyD88.
Results
L. pneumophila translation inhibitors induce a frustrated MAPK response
We hypothesized that Legionella-induced inflammatory gene transcription will be largely inconsequential, as the transcribed genes cannot be translated to proteins due to the bacterium-derived translation inhibitors. To test this hypothesis, we examined mammalian host protein synthesis predicted to be downstream of MAPK activation following exposure to L. pneumophila.
Bone marrow-derived macrophages challenged with wild type L. pneumophila showed phosphorylation of MAPK members shortly after exposure to the bacterium, with the activation kinetics being almost identical to previous observations ([9], [39], [40]; Figure S1). In the first hour after L. pneumophila challenge, activation of JNK and P38 was independent of the Icm/Dot system, consistent with phosphorylation being the result of Tlr engagement [9]. A second wave of activation was observed beginning at two hours after challenge. This was dependent on the presence of the L. pneumophila type IV secretion system, as the levels of MAPK phosphorylation decayed when macrophages were challenged with the dotA3 mutant that lacks the Icm/Dot system (Figure S1). This two-wave activation, reflecting an early Tlr-dependent and a later L. pneumophila-specific response, mirrors our previous observations with NF-KB activation [41].
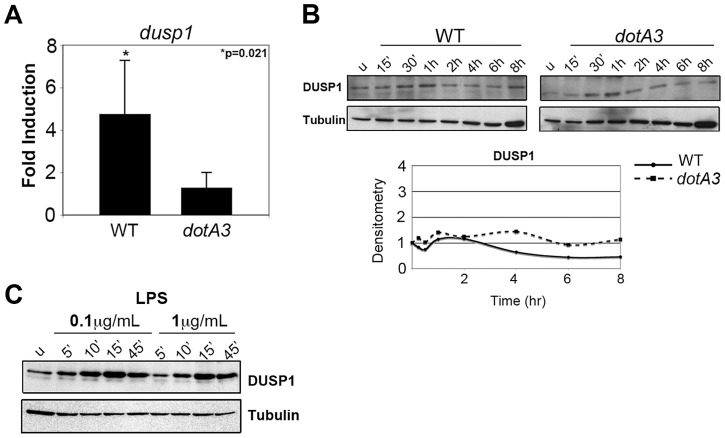
Cultured macrophages challenged with wild type L. pneumophila transcriptionally activate a number of dual specificity phosphatase (DUSP) genes, dependent on an intact Icm/Dot system [17], [40]. Bone marrow-derived macrophages challenged with wild type Legionella for 4 hrs showed significant induction of the Dusp1 transcript compared to dotA3 infection (Figure 1A). The transcriptional induction was much higher (over 60 fold) in U937 human monocytes, the cell lines in which dusp1 induction in response to wild type Legionella was first characterized (Figure S2A) [17]. This transcriptional response, however, was not accompanied by translation either in bone marrow-derived macrophages (Figure 1B) or U937 cells (Figure S2B) as DUSP-1 protein levels remained unchanged over the course of infection. Our inability to observe enhanced protein synthesis was not due to limitations with our detection system, because we observed a robust increase in DUSP-1 protein levels in response to the addition of LPS (Figure 1C). Presumably the multiple translocated substrates that inhibit translation elongation [10] frustrate the transcriptional response, preventing translation of induced genes. To test this hypothesis, we challenged cells with an L. pneumophila mutant missing five of the translation inhibitors (Δ5) to determine if the absence of these proteins could allow translation to proceed. Instead, we observed no transcriptional activation of the Dusp1 gene in response to this mutant (Figure S2A). Therefore, induction of genes in the MAPK pathway and frustration of the response are tightly coupled.
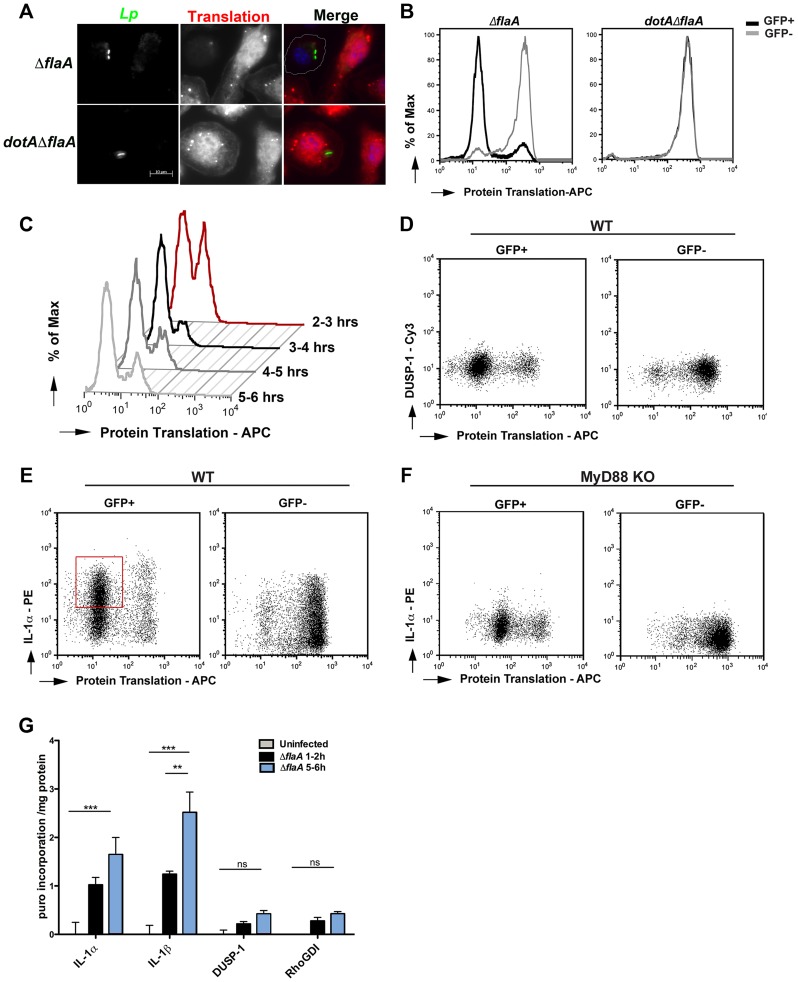
Figure 1. L. pneumophila translation inhibitors induce a frustrated MAPK response.
(A) Dusp1 transcript levels in A/J macrophages infected with wild type or dotA3 L. pneumophila for 4 hrs. Transcript levels were normalized to18S ribosomal RNA (18S) and graphed as a fold increase over uninfected controls. (B) Immunoblot analysis of DUSP-1 protein levels in A/J macrophages challenged with wild type or dotA3 strains and (C) treated with LPS (0.1 µg/mL or 1 µg/mL) for indicated time points. Data are representative of at least three independent experiments.
A subset of cytokine transcripts are selectively translated following L. pneumophila infection
We then asked if the response leading to transcriptional upregulation of pro-inflammatory cytokine genes was similarly affected by translational inhibition. To understand how cytokines are regulated in response to L. pneumophila, we measured the transcription and translation of selected pro-inflammatory cytokines in C57/Bl6 (B6) macrophages following L. pneumophila challenge. Flagellin deficient (ΔflaA) mutants were used in these experiments to avoid Caspase 1-dependent cell death downstream from NAIP5/NLRC4 recognition of flagellin by B6 macrophages [42], [43], [44].
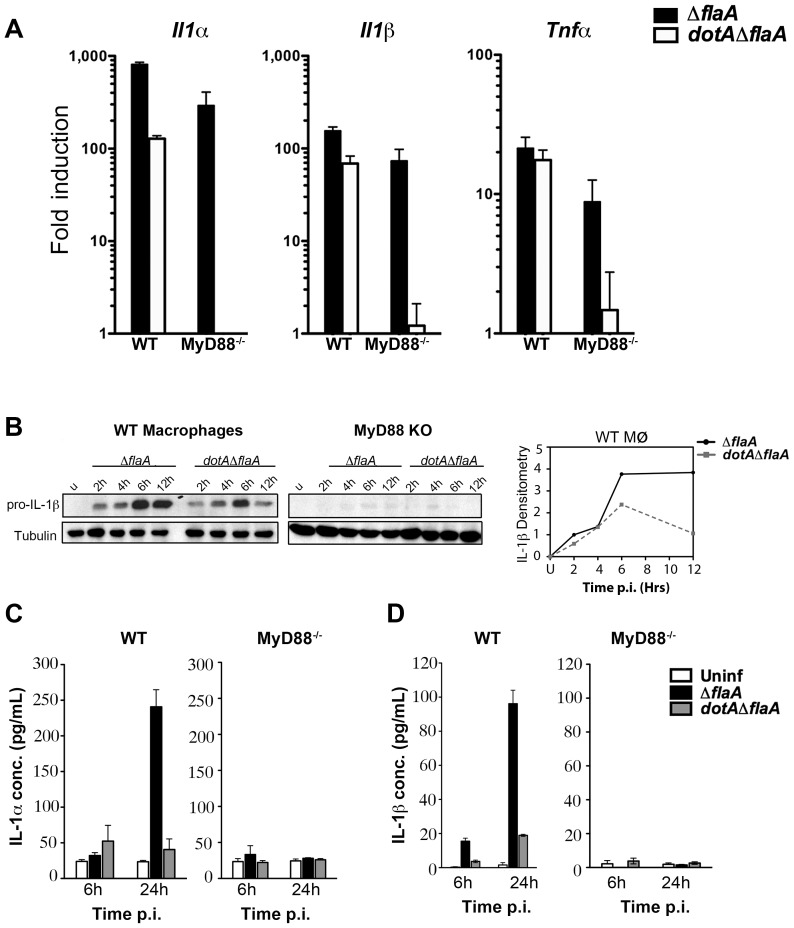
Infection of bone-marrow macrophages with virulent (Dot+) L. pneumophila induced Il1α, Il1β and Tnfα transcripts by 6 hrs post infection (Figure 2A). As previously reported, the cytokine response to Dot+ was comprised of MyD88-dependent signaling that is layered on top of MyD88-independent, effector mediated signaling (Figure 2A, black bars) [10]. The response to the avirulent, dotA3 mutants on the other hand, was mostly dependent on MyD88 signaling (Figure 2A, white bars) [9].
Figure 2. A subset of transcripts can bypass translation inhibition exerted by L. pneumophila effectors.
(A) C57BL/6 wild type and MyD88−/− macrophages were infected with indicated L. pneumophila strains at MOI-15. Cytokine and DUSP transcripts were analyzed at 6 hrs post infection by qRT-PCR. Results shown are pooled from at least four independent experiments and represent the mean fold induction and SEM of samples relative to uninfected controls. (B) Immunoblot analysis of IL-1β precursor in WT and MyD88−/− macrophages infected with virulent L. pneumophila (ΔflaA) or avirulent mutant dotAΔflaA for indicated time points. Graphs on the right show densitometry of IL-1β normalized to tubulin. (C and D) WT and MyD88−/− macrophages were challenged with indicated L. pneumophila strains for 6 and 24 hrs at MOI-15 and cytokine levels were measured in culture supernatants by ELISA. Data represent mean and SEM of samples from 3 independent experiments.
To determine if the transcribed cytokine mRNAs were efficiently translated and secreted during infection, WT and MyD88−/− macrophages were challenged with L. pneumophila and cytokine protein levels were measured by western blot and ELISA. Contrary to what we saw for DUSP-1, challenge with L. pneumophila Dot+ led to a significant increase in cell-associated pro-IL-1β levels after 4 hrs of infection (Figure 2B). IL-1α and IL-1β mature forms could also be detected in culture supernatants after 24 hrs (Figure 2C & 2D). Interestingly, challenge of macrophages with L. pneumophila dotA3 mutants accumulated pro-IL-1β transiently, with steady state levels reduced by 12 hr post infection (Figure 2B densitometry), but this was not sufficient to induce the release of mature IL-1β. This is consistent with the hypothesis that in addition to TLR signaling, Icm/Dot translocated substrates are required for persistent pro-inflammatory cytokine activation and secretion [32]. Surprisingly, cytokine translation was severely diminished in MyD88−/− macrophages in response to wild type L. pneumophila (Figure 2B) despite the presence of large amounts of transcripts (Figure 2A). Therefore, a MyD88-dependent signal appears necessary to bypass the translation block induced in response to wild type L. pneumophila.
Intracellular cytokine accumulates in infected, translation-blocked macrophages
There is no clear model for why the presence of MyD88 allowed bypass of the translation block. Engagement of MyD88 on the host cell surface may lead to selective bypass of translation inhibition on a subset of transcripts. Alternatively, translation of cytokine transcripts could largely occur in neighboring uninfected cells that have not been directly injected with L. pneumophila translocated proteins, but which have been activated by bacterial fragments liberated by infected cells. We therefore asked if the observed cytokine translation was derived from neighboring bystander cells.
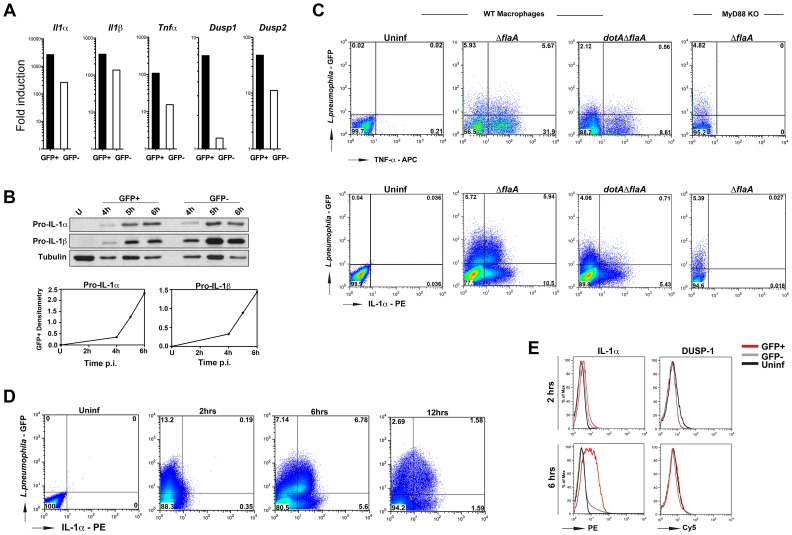
B6 macrophages were challenged with L. pneumophila-GFP strains and macrophages harboring bacteria were sorted from uninfected bystanders. Cytokine transcripts (Figure 3A) and protein levels (Figure 3B) were measured in each population by qRT-PCR and Western blots. To ensure accumulation of TNF-α protein, cells were treated with GolgiPlug (Brefeldin A; Materials and Methods) to prevent secretion of this cytokine. No such treatment was necessary for IL-1α and IL-1β, which accumulate as precursors via an alternate secretion pathway [45].
Figure 3. Cytokines are produced from both infected and bystander macrophages.
(A) B6 macrophages were challenged with ΔflaA-GFP at MOI-15 for 4 hrs and sorted by Flow Cytometry. Cytokine and Dusp transcripts were measured in both GFP+ and GFP− population by qRT-PCR. (B) B6 macrophages were infected with ΔflaA-GFP at MOI-15 for indicated times and levels of IL-1β were measured by Western blot. Bottom graphs indicate densitometry of IL-1β in GFP+ cells normalized to tubulin. (C) WT and MyD88−/− macrophages were infected with indicated L. pneumophila strains at MOI-10 and intracellular cytokine levels were measured by flow cytometry. Top panels show TNF levels at 14 hrs post infection. To prevent secretion of TNF, cells were treated with Golgiplug (Brefeldin A) for 5 hrs before samples were collected. Bottom panels show intracellular IL-1α levels at 6 hrs post infection. Data shown are representative of at least 4 independent experiments. (D) Time course analysis of intracellular IL-1α levels in ΔflaA-GFP infected macrophages and (E) Comparison of intracellular IL-1α and DUSP1 levels in B6 macrophages infected with ΔflaA-GFP for 2 and 6 hrs. Infected and uninfected (bystander) cells were gated based on GFP signal and protein levels were compared to control macrophages that were left untreated. Red lines indicate GFP+ population; grey lines indicate GFP− population and black lines show untreated macrophages.
Relative to cells that had never been exposed to bacteria, challenge of macrophages with L. pneumophila resulted in high levels of I1α, Il1β, Tnfα and Dusp2 transcripts in both infected (GFP+) and neighboring uninfected (GFP−) populations by 4 hrs post infection, (Figure 3A). Depending on the cytokine, the amount of transcription in the bystander cells varied from 10% −30% of that observed in the cells harboring bacteria. Dusp1 on the other hand, was mainly transcribed in GFP+ cells (Figure 3A). More importantly, despite the presence of the translocated protein synthesis inhibitors, macrophages harboring bacteria were able to produce high levels of pro-IL-1α and pro-IL-1β (Figure 3B, GFP+ cells). The kinetics of pro-IL-1α and pro-IL-1β production in infected cells indicated that there was enhanced accumulation of these proteins between 4–6 hrs post-infection (Figure 3B, bottom panel). We will show that during this time window, L. pneumophila translation inhibitors effectively block most protein synthesis in infected cells (below, Figure 4B).
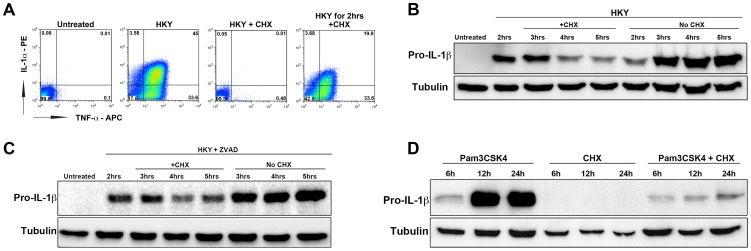
Figure 4. The elongation inhibitor cycloheximide blocks cytokine translation independent of cell death.
(A) B6 macrophages were either left untreated (1st box), treated with heat killed Yersinia at MOI = 50 to induce cytokine expression (2nd box), treated with HKY MOI = 50 and 10 µg/mL cycloheximide (CHX) (3rd box) or treated with HKY MOI = 50 for 2 hrs followed by addition of 10 µg/mL CHX (4th box). X-axis represents intracellular TNF-α levels and Y-axis represents intracellular IL-1α levels. (B) Macrophages were pre-stimulated with heat killed Yersinia at MOI = 50 for 2 hrs. Cells were then treated with either 1 µg/mL cycloheximide or left untreated (HKY alone). IL-1β protein levels were measured by Western blot at the indicated time points. (C) B6 macrophages were pre-stimulated with heat killed Yersinia at MOI = 50 in the presence of Pan-Caspase inhibitor (Z-VAD-FMK) for 2 hrs and treated with either 0.5 µg/mL cycloheximide or left untreated. IL-1β protein levels were measured by Western blot for the next 3 hrs. (D) Macrophages were treated with 2 µg/mL Pam3CSK4 for 2 hrs in the presence of Z-VAD-FMK. CHX (0.5 µg/mL) was added to cells at 2 hrs post Pam3CSK4 treatment and IL-1β protein levels were measured at 6, 12 or 24 hrs. Data is representative of at least 3 independent experiments.
The presence of persistent cytokine synthesis in infected cells was confirmed by intracellular cytokine staining. B6 WT and MyD88−/− macrophages were challenged with L. pneumophila-GFP strains and intracellular cytokine levels were measured by flow cytometry. TNF-α was produced by both macrophages bearing bacteria (GFP+) and bystander cells (GFP−) after challenge with L. pneumophila Δfla, while the dotAΔfla strain induced much lower levels of this cytokine (Figure 3C, top panel 2nd and 3rd boxes). Cells harboring bacteria (GFP+) were a significant source of IL-1α (Figure 3C, bottom panel, 2nd box). Approximately 50% of the cells harboring bacteria showed detectable accumulation of IL-1α (Figure 3C, bottom panels, 2nd box), while bacteria were associated with approximately 33% of the IL-1α-producing cells. Consistent with Figure 2B, translation of IL-1α and TNF-α were both dependent on MyD88 signaling, and accumulation of IL-1a in infected cells was dependent on the presence of the Icm/Dot translocator (Figure 3C, two rightmost boxes in each panel).
Time course analysis of intracellular IL-1α levels using flow cytometry confirmed that the highest level of IL-1α accumulation occurred between 2–6 hrs post infection in the GFP+ population (Figure 3D and 3E). During this time period, the number of cells bearing bacteria that accumulated IL-1α increased from 1% of this population to approximately 50% (Figure 3D). DUSP-1 protein levels on the other hand, remain unchanged between 2–6 hrs (Figure 3E).
Cytokine response to chemical inhibitors of protein translation
The two signals that are received by mammalian cells during L. pneumophila infection (1st signal from TLR activation and 2nd signal from protein translation inhibition) synergize to induce the full cytokine response against the bacterium. It was previously reported that pharmacological inhibitors of host protein translation induce transcription of various stress response genes and cytokines such as IL-6, IL-23, IL-α and IL-1β [10], [31]. We wanted to confirm that translation and secretion of these cytokines could always bypass translation inhibition using the protein synthesis inhibitor cycloheximide (CHX). CHX interferes with protein translation elongation by binding to the E-site of the 60S ribosomal subunit and preventing tRNA translocation [46].
Macrophages were treated with heat-killed Yersinia (HKY) to induce TLR signaling, together with 10 µg/mL of cycloheximide. Addition of the chemical inhibitor at the same time as HKY led to a complete inhibition of TNF-α and IL-1α production in bone-marrow macrophages (Figure 4A). Contrary to what we observed during L. pneumophila infection, addition of CHX dampened the signal received from TLR stimulation (Figure 4 A,B). Surprisingly, even at low concentrations of CHX that permit significant levels of protein translation (Figure S3), CHX was still able to inhibit IL-1β translation (Fig. 4B).
To rule out the possibility that the reduction in IL-1β levels during CHX treatment was due to cell death, we lowered the CHX dose to 0.5 µg/mL (inhibits less than 50% of total host protein synthesis) (Figure S3) and also incubated the cells with the apoptotic inhibitor Z-VAD-FMK (pan-caspase inhibitor). CHX was still able to inhibit IL-1β under these conditions, although it was clear that increased survival of cells was accompanied by higher accumulation of HKY-induced cytokine (Figure 4C).
MyD88-dependent stimulation of mouse macrophages in response to L. pneumophila primarily occurs via Toll-like receptor 2 (TLR2) [29]. A recent report was able to reconstruct the cytokine induction seen during L. pneumophila infection by using the synthetic TLR-2 ligand Pam3CSK4 in combination with Exotoxin A (Exo A), a toxin from Pseudomonas aeruginosa that interferes with translation elongation [31]. Based on this observation, we wanted to determine if specific activation of TLR2 is what leads to cytokine translation in the presence of protein synthesis inhibitors. Macrophages were treated with the TLR2 agonist Pam3CSK4 and pro-IL-1β levels were measured in the presence or absence of the protein synthesis inhibitor cycloheximide (CHX). Drug treatment after addition of the TLR2 agonist led to a large reduction in pro-IL-1β levels (Figure 4D). We also observed a failure to hyperstimulate pro-IL-1b in the presence of another protein elongation inhibitor, puromycin (Figure S4). Similar results were obtained when macrophages were stimulated with another TLR2 agonist lipoteichoic acid (LTA), or a TLR4 agonist LPS, followed by addition of CHX (data not shown). This indicates that the selective synthesis of cytokines may result from host cells sensing a specific mode of protein synthesis inhibition. It is also possible that the selective synthesis of pro-inflammatory cytokines is triggered by a block in translation initiation [36] instead of translation elongation, which would explain why the elongation inhibitors cycloheximide or puromycin were not able to induce the response.
Translation of IL-1 takes place in the presence of highly depressed host cell translation
We considered two possible explanations for how cytokines were translated in the presence of the bacterium-derived protein synthesis inhibitors: (1) translation inhibition by L. pneumophila is not efficient, allowing most of the cytokine to be synthesized prior to a complete block; or (2) the host preferentially translates a subset of genes after protein synthesis is shutdown by pathogens.
To distinguish between these possibilities, relative levels of protein synthesis were measured in bone-marrow macrophages at various times following L. pneumophila challenge, using an immunofluorescence readout. B6 macrophages were challenged with L. pneumophila flaA−-GFP at MOI = 10 for 2 hrs and translation of proteins was measured by incorporation of a methionine analog (L-azidohomoalanine, AHA) for an additional 4 hrs. Incorporated AHA was detected by reaction with a fluorescent-labeled phosphine reagent (phosphine-APC), which covalently links to the azido-functional group on AHA (Staudinger ligation reaction) [47].
Protein translation was significantly inhibited in macrophages harboring L. pneumophila (ΔflaA) compared to bacteria lacking the Icm/Dot system (Figure 5A). Furthermore, in macrophage cultures incubated with L. pneumophila, the macrophages harboring L. pneumophila showed selective protein synthesis interference, while the majority of the uninfected cells showed efficient incorporation of the amino acid analog (Figure 5B; compare GFP+ to GFP− population). To determine the time point at which the protein synthesis inhibitors fully shut down global protein translation, pulse-chase experiments were performed in which the methionine analog (AHA) was added for 1 hr intervals starting at 2 hrs post infection (Figure 5C). Between 2–3 hrs post-infection, approximately 40% of the cells harboring L. pneumophila were found in the population that has high levels of protein synthesis. Between 3–4 hrs post infection, we observed a major shift where almost 90% of cells harboring ΔflaA were found in the population having highly depressed protein synthesis. Later time points showed no further blockage in translation, perhaps reflecting the fact that there is a small fraction of wild type bacteria that fail to form replication compartments [48]. This population is predicted to show no significant translocation via the Icm/Dot system and should fail to inhibit protein synthesis (seen in ∼10% of the macrophage population).
Figure 5. Translation of IL-1 takes place in the presence of L. pneumophila elongation inhibitors.
(A) B6 macrophages were infected with indicated L. pneumophila strains at MOI = 10 for 2 hrs and a methionine analog, L-azidohomoalanine (AHA, 50 µM) was incorporated into newly synthesized proteins for 4 hrs. Cells were fixed, permeabilized and the incorporated analog was detected by an APC conjugated phosphine and fluorescence microscopy. Nuclei were stained with Hoechst 33342 (blue) (B) Macrophages were infected with indicated L. pneumophila strains at MOI = 10 for 2 hrs and L-azidohomoalanine (AHA, 50 µM) was added to cells for additional 4 hrs. Protein translation was quantified in infected macrophages (GFP+) and uninfected bystanders (GFP−) by staining with APC-labeled phosphine and flow cytometry. Experiment was performed three times. (C) B6 macrophages were infected with Dot+ L. pneumophila for 2 hrs and AHA was added for 1 hr intervals. Cells were fixed after each time point and protein translation was quantified by APC-conjugated phosphine and flow cytometry. Translation of DUSP1 (D) or IL-1α (E) was monitored in wild type macrophages infected with ΔflaA for 6 hrs by co-staining AHA with Cy3 conjugated DUSP1 antibody or PE-conjugated IL-1α antibody. (F) Translation of IL-1α in MyD88-deficient macrophages was determined by co-staining AHA with PE-conjugated IL-1α antibody. Dot plots show GFP+ (left column) and GFP− (right column) gated cells. (G) Kinetics of IL-1α, IL-1β, DUSP-1 and RhoGDI translation was quantified in Dot+ infected macrophages using puromycin incorporation. B6 macrophages were infected with Dot+ L. pneumophila and 10 µg/mL of puromycin was added either between 1–2 hrs or between 5–6 hrs post infection. Cells were washed, lysed and incubated on plates coated with anti- IL-1α, IL-1β, DUSP-1 and RhoGDI antibodies. Incorporation of puromycin in the indicated samples was monitored by anti-puromycin antibody and HRP-conjugated secondary antibody. Data represent absorbance at 450 normalized to total protein levels. Values for uninfected controls were subtracted from each sample to determine the increase in puromycin incorporation upon infection. Each bar represents mean and SEM of triplicate samples.
To determine if translation-blocked cells could still produce cytokines, macrophage monolayers were challenged with L. pneumophila-GFP+ and protein synthesis was monitored by addition of AHA. The cells were then probed for IL-1α accumulation by immunofluorescence and flow analysis. In the infected GFP+ population of macrophages, the majority of cells that accumulated IL-1α show evidence of an almost complete shutdown of protein translation (Figure 5E; red box). In the absence of MyD88 signaling, both the infected and uninfected populations showed little IL-1α accumulation (Figure 5F). In contrast to IL-1α, there was no DUSP-1 accumulation after L. pneumophila challenge of macrophages (Figure 5D). This confirmed our main hypothesis that there is translation of selected cytokine genes when protein synthesis is inhibited by L. pneumophila and this bypass requires MyD88 signaling.
The increase in IL-1α and IL-1β protein levels is due to enhanced protein synthesis
We have shown that pro-IL-1α and pro-IL-1β accumulate in cells that harbor L. pneumophila between 4–6 hrs post infection (Figure 3B), despite a significant block in protein translation (Figure 5C & E). To confirm that the accumulation we observe in infected cells was due to newly synthesized cytokines over the course of infection, we took advantage of another protein translation assay, SunSET. This assay uses puromycin incorporation into growing polypeptide chains to monitor active protein synthesis [49]. We modified this assay to measure the amount of puromycin incorporated into our protein of interest during a 1-hour pulse period. Accordingly, macrophages were challenged with L. pneumophila for increasing lengths of time, cells were labeled for one hour with 10 µg/mL of puromycin, lysed, and individual proteins were immobilized in assay wells using specific antibodies (Materials and Methods). An ELISA was then used to determine the amount of puromycin incorporated in the immobilized proteins (Figure 5G). Consistent with our intracellular cytokine staining and Western blot data (Figure 3), the highest levels of IL-1α and IL-1b synthesis were detected between 5–6 hrs post-infection. On the other hand, no significant puromycin incorporation was detected for DUSP-1 and RhoGDI proteins, confirming that there is selective synthesis of few genes after L. pneumophila challenge.
Translation inhibition acts in concert with TLR signaling to generate the full cytokine response
To determine the role that L. pneumophila translation inhibitors play in modulating host cytokine synthesis, we used a mutant that lacks the 5 Icm/Dot translocated substrates known to block host protein synthesis (Δ5 mutant) [10]. The level of protein synthesis was first measured using AHA incorporation (Figure 6A, B) and puromycin incorporation (Figure S5) in macrophages infected with Δ5ΔflaA-GFP+. Compared to Dot+ strain (Figure 6B & Figure S5), there was an increase in active protein translation in cells that were infected with the Δ5 mutant, although the cells showed lower levels of protein synthesis than Dot− infected cells (Figure 6B) or the uninfected population (Figure 6A; compare GFP+ to GFP− populations). This is consistent with the hypothesis that in addition to Icm/Dot translocated substrates that act on translation elongation, infection with virulent L. pneumophila also blocks translation initiation [36].
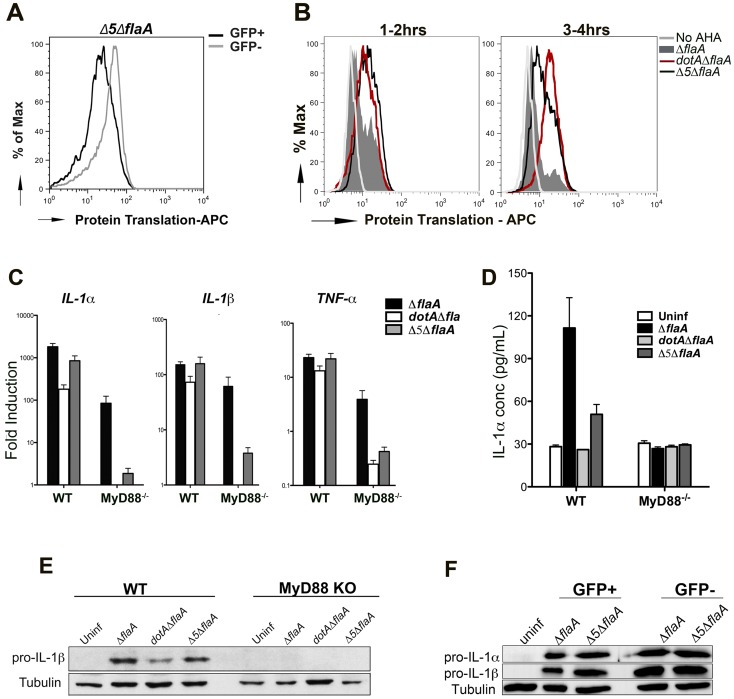
Figure 6. Production of the pro-inflammatory cytokines IL-1α and IL-1β is independent of the five translocated protein synthesis inhibitors.
(A) B6 macrophages were infected with Δ5Δfla-GFP+ and protein translation was measured between 2–6 hrs post infection by incorporation of the methionine analog AHA. Translation (incorporation of AHA) was compared between infected cells (GFP+, black line) and uninfected bystanders (GFP−, grey line). (B) B6 macrophages were infected with GFP expressing Dot+, Dot− and Δ5 strains of L. pneumophila and protein synthesis inhibition was compared between these strains by incorporation of AHA between 1–2 hrs (left graph) or 3–4 hrs (right graph). Graphs show translation in infected cells (GFP+). Cells incubated in the absence of the methionine analog (No AHA) were used as a negative control to show baseline staining. (C) WT and MyD88−/− macrophages were infected with the indicated strains at MOI-15 and cytokine transcripts were analyzed at 6 hrs post infection by qRT-PCR. Data represent the mean fold induction and SEM of samples relative to uninfected controls. (D) ELISA measurement of IL-1α secretion from WT and MyD88−/− macrophages infected with the indicated strains for 24 hrs. Data represent mean and SEM of triplicate samples. (E) Immunoblot analysis of pro-IL-1β in WT and MyD88−/− macrophages challenged with the indicated L. pneumophila strains for 6 hrs. (F) B6 macrophages were challenged with ΔflaA-GFP or Δ5Δfla at MOI-15 for 6 hrs and sorted by Flow Cytometry. Pro-IL-1β levels were measured in both GFP+ and GFP− population by westernblot.
It had been previously reported that in the absence of most known pathways of pattern recognition (MyD88−/− Nod1−/− Nod2−/− macrophages), the cytokine transcriptional response to L. pneumophila was primarily due to the presence of the translocated protein synthesis inhibitors [10], [11]. Using macrophages that are only defective for MyD88, this dependence on the translation inhibitors could be clearly observed for the Il1α, Il1β and Tnfα transcripts (Figure 6C, MyD88−/−, gray vs. black bars). Consistent with our previous data, the transcriptional response in MyD88 knockout macrophages was unproductive, with no evidence that these highly induced transcripts are translated (Figure 6E).
In the case of wild type macrophages, Dot+ and Δ5 infections induced comparable levels of MyD88-dependent cytokine transcription and translation (Figure 6C and E), and this could be observed in macrophages that were sorted by flow cytometry, as well (Figure 6F). This result is in contrast with macrophages lacking MyD88 signaling, in which it is clear that there is protein synthesis inhibitor-dependent induction of cytokine transcripts (Figure 6C), but this induction produces no apparent cytokine translation products. Interestingly, unlike what we see for dotA mutants, infection with Δ5 was still able to induce secretion of mature IL-1α at 24 hrs after infection [31], even though it was to a lesser extent than wild type (Figure 6D).
Therefore, although the protein synthesis inhibitors are responsible for the transcriptional response that occurs in the absence of pattern recognition receptors, the production of cytokine proteins associated with infections by fully virulent strains is not dependent on these translocated substrates.
MyD88-mediated bypass of translation inhibition of the il1β gene is independent of mRNA stability
Cytokine expression is regulated at various stages, including transcription, post-transcriptional processing, translation and secretion. One of the main regulatory steps for IL-1 and TNF production is their transcript stability, which is controlled by their AU-rich elements (ARE) in their 3′-noncoding regions and by various ARE-binding proteins [50], [51]. To determine if lack of cytokine translation in MyD88−/− macrophages was due to mRNA instability, the half-life of Il1β and Tnfα transcripts were compared in WT and MyD88−/− macrophages after L. pneumophila challenge. Macrophages were first infected with L. pneumophila for 2.5 hrs, actinomycin D was added to the medium to block further transcription, and the amount of transcript remaining was measured by qRT-PCR at various time points after the addition of the drug. Tnfα mRNA was highly unstable in the absence of MyD88 when compared to MyD88+/+ macrophages (Figure 7A). However, Il1β transcripts were relatively stable in the absence of MyD88, and the amount of mRNA remaining after 2 hrs was similar to control macrophages (Figure 7A). This indicates that the translation inhibition bypass of Il1β was independent of mRNA stability (Figure 7). In the case of tnfα, however, transcript stabilization via a MyD88-dependent signal may play a role in bypassing translation inhibition.
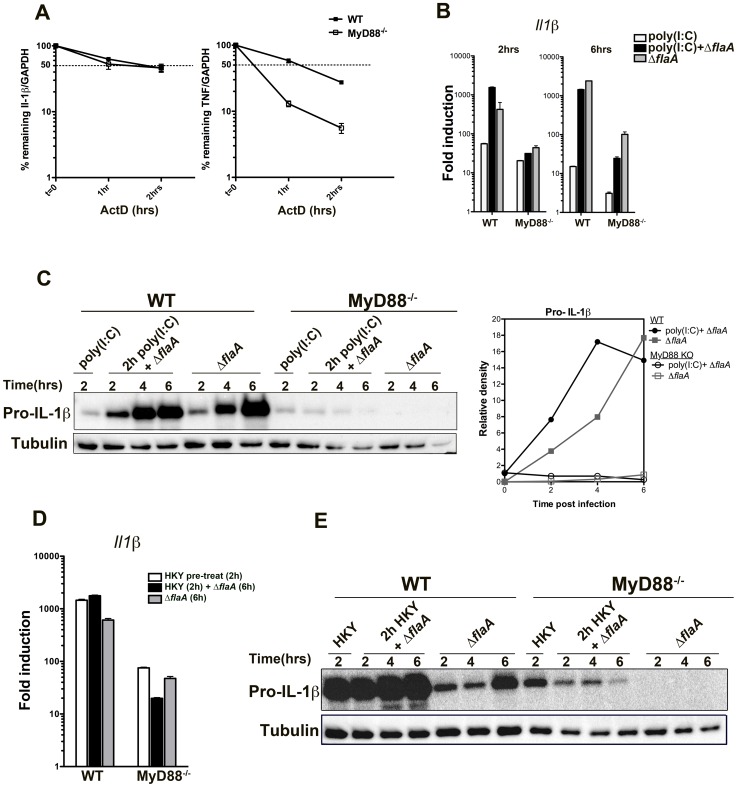
Figure 7. Stability of Il-1β mRNA or pre-activation of the NF-kB pathway is not sufficient to induce translation bypass in MyD88−/− macrophages.
(A)WT and MyD88 deficient bone-marrow macrophages were challenged with ΔflaA for 2.5 hrs and transcription was blocked by addition of 10 µg/mL Actinomycin D. The percentage of remaining Il1β and Tnfα transcripts was measured by qRT-PCR after 1 or 2 hrs post actinomycin D treatment. Data represent mean and SEM of samples relative to GAPDH. (B) WT and MyD88−/− macrophages were pre-treated with 50 µg/mL poly(I∶C) for 2 hrs to induce NF-kB activation, after which cells were either challenged with Dot+ Legionella or left untreated (poly(I∶C) only). Macrophages that were not pre-stimulated with poly(I∶C) but were challenged with Dot+ were included for comparison. RNA was collected after 2 and 6 hrs of infection and Il1β transcript levels were analyzed by qRT-PCR. (C) B6 WT and MyD88-deficient macrophages were pre-treated with poly(I∶C) for 2 hrs or left untreated. Cells were then challenged with Dot+ Legionella for additional 2, 4 and 6 hrs and production of pro-IL-1β was examined by Western blot. Densitometry of the blot is shown on the right. (D) WT and MyD88−/− macrophages were pre-treated with HKY (MOI = 100) for 2 hrs or left untreated after which cells were challenged with Dot+ L. pneumophila. Induction of Il1β transcript was determined after 6 hrs of infection. (E) Pro-IL-1β synthesis was also examined in these cells after 2, 4 and 6 hrs of challenge.
Pre-activation of NF-kB in MyD88-deficient macrophages is not sufficient to bypass translation inhibition
Another possible explanation for why few pro-inflammatory cytokines, such as IL-1α and IL-1β, bypass translation inhibition could be mRNA abundance. A recent study has shown that these cytokines are the most abundant transcripts within macrophages after challenge with L. pneumophila [36]. Although there was significant transcription of these genes in MyD88−/− macrophages, the total mRNA abundance of IL-1α, IL-1β and TNF was still significantly lower compared to wild type macrophages (Figure 2). To address if mRNA abundance plays a role in bypass of translation inhibition during Legionella infection, WT and MyD88−/− macrophages were pre-activated with the TLR3 agonist poly(I∶C) for 2 hrs to induce NF-kB signaling. We also used heat-killed Yersinia pseudotuberculosis (HKY) to activate NF-kB via TLR4.
2 hr pre-stimulation with 50 µg/mL poly(I∶C) was sufficient to trigger IL-1β transcription (Figure 7B, left graph, white bars) both in wild type and MyD88−/− macrophages. Pre-stimulation with poly(I∶C) followed by L. pneumophila infection increased Il1β transcription initially (after 2 hrs of infection) in wild type macrophages compared to cells that were untreated (Figure 7B, compare black bars with grey bars). Surprisingly, this was reversed by 6 hrs post infection and cells that were pre-treated with poly(I∶C) down regulated their Il1β transcription (Figure 7B, right graph). This was more pronounced in MyD88−/− macrophages (Figure 7B, compare black and grey bars). We observed a very similar phenomenon when cells were pre-treated with heat-killed Yersinia followed by Legionella infection (Figure 7D).
As expected, pro-IL-1β translation was robust in wild type macrophages that were pre-treated with poly(I∶C) or with HKY followed by Legionella infection (Figure 7C and 7E). Interestingly, pro-IL-1β was detected by Western blots in MyD88−/− macrophages after pre-stimulation with poly(I∶C) but the protein level was reduced when the pre-activated cells were challenged with L. pneumophila (Figure 7C). MyD88−/− macrophages that were infected with Legionella alone on the other hand, showed little detectable translation (Figure 7C) despite having higher levels of Il1β transcripts (Figure 7B, right graph, compare black and grey bars). This phenotype was more obvious during pre-treatment with HKY. 2 hr HKY pre-stimulation led to a robust pro-IL-1β translation initially, which was significantly reduced when cells were challenged with L. pneumophila (Figure 7E). Therefore, in the absence of MyD88 signaling, macrophages were unable to overcome the translation inhibition induced by Legionella pneumophila even in the presence of external stimuli.
Discussion
Inhibition of protein translation is a common virulence mechanism used by many viruses and bacteria. In this study, we showed that host cells have evolved mechanisms to cope with translation inhibition by selectively translating a subset of cytokine genes, including pro-inflammatory cytokines such as IL-1α and IL-1β in response to L. pneumophila challenge. The ability to bypass L. pneumophila translational inhibition is an important determinant of host protection, as mice defective in the IL-1α/IL-1β response and humans exposed to TNF-α inhibitors are highly susceptible to L. pneumophila infection [31], [38].
L. pneumophila challenge of bone-marrow macrophages leads to a dramatic reduction in global protein translation (Figure 5A). The bacterium interferes with protein translation both at the initiation step [36] and elongation step [10]. It has been shown previously that this inhibition triggers the transcription of various stress response genes including NF-kB- and MAPK-regulated genes, heat shock proteins and pro-inflammatory cytokines and chemokines [9], [10], [11]. We show here that L. pneumophila translocated effectors prevent the translation of these genes, resulting in a “frustrated response,” in which there is accumulation of transcripts but no increase in protein levels. A subset of cytokine genes, and potentially other genes that have not been identified yet, are insensitive to this inhibition and get translated in cells that show the highest level of protein synthesis inhibition.
We observed bypass of translational inhibition as an orderly series of events resulting in the accumulation of IL-1α in cells harboring bacteria as well as in bystanders, followed by release of the cytokine into culture supernatants. Initial transcriptional induction and translation of IL-1a occurred independently of the Icm/Dot system, and was associated with TLR-signaling, consistent with the TLR-dependent activation of the NF-κB response known to occur at early time points after L. pneumophila challenge [41]. This was followed by persistent accumulation of pro-IL-1α protein in a process that required both the presence of the Icm/Dot system and MyD88-dependent signaling, indicating collaborative signaling between the two pathways. Surprisingly, accumulation of pro-IL-1α was equally robust in both cells harboring bacteria and in bystander cells, in spite of the translocated protein synthesis inhibitors that are deposited by L. pneumophila. This observation is particularly striking, in that time points showing the strongest inhibition of protein synthesis also resulted in the fastest rate of pro-IL-1α accumulation, arguing that there is selective ribosomal loading of cytokine transcripts in intoxicated cells. In the absence of MyD88 signaling, no such bypass could be observed in either infected or uninfected cells, either because translation requires the extremely high levels of transcription that occur in the presence of MyD88, or there is pattern recognition-dependent bypass of translational inhibition. Accumulation of pro-IL-1α was then followed by its release, which required both the Icm/Dot system and MyD88.
It is conceivable that the L. pneumophila translational inhibitors could be responsible for the induction of pro-IL-1α. Arguing against this model is the fact that a strain that lacks 5 of the known translation inhibitors (Δ5) still induced considerable pro-IL-α accumulation (Figure 6E, F) [31]. The accumulation of cytokine in response to this strain could have resulted from residual translation inhibition that was observed, but it should be noted that MAPK and NF-κB activation resulting from macrophage challenge by this strain is due to a pattern recognition response and is not due to Icm/Dot signals [11]. The initial MyD88-dependent activation that occurs after contact with L. pneumophila may be amplified by unknown Icm/Dot-signals or due to inhibition of translation initiation [36]. A recent study proposed that infection of macrophages with virulent L. pneumophila strains (both Dot+ and Δ5) leads to downregulation of mTOR activity, which is sufficient to suppress cap-dependent protein translation initiation [36]. The second signal that is required for amplifying pattern-recognition could be generated from such translational suppression, and could be the trigger for induction of pro-IL-1α.
Challenge of macrophages that lack MyD88 with L. pneumophila induces cytokine gene transcription, but the transcribed genes fail to be translated. In the absence of MyD88, therefore, the protein translation inhibition takes on global dimensions. The MyD88-dependent bypass of the translation inhibition was independent of transcript stability in the case of Il1β transcript, which is a known strategy for post-transcriptional regulation of cytokines [52], [53]. This surprising result indicates that there may be a previously unrecognized MyD88-dependent signaling pathway that mediates post-transcriptional regulation of cytokine transcripts. It has previously been reported that separate transcriptional and translational signals are required for IL-1β expression [54]. Although it is not clear what these translational signals could be, it is possible that MyD88-dependent signals could result in either enhanced ribosome loading, or could regulate translation via action at the 3′ or 5′ untranslated regions. Alternatively, the role of MyD88 could be totally passive, and merely a consequence of enhancing expression of cytokine gene transcripts. Although L. pneumophila infection causes a large induction of cytokine transcription in the absence of MyD88, these levels are still lower than what is seen when pattern recognition is intact (Figure 2A). This added boost in cytokine gene transcription by MyD88 may be sufficient to push the concentrations of these transcripts above the minimum threshold necessary to support selective translation of these genes under conditions of intoxication. Consistent with this model are previous results from nanostring analysis of macrophage transcripts that are induced in response to L. pneumophila challenge [36]. In this work, it is argued that the primary determinant of translation in cells challenged with L. pneumophila is the relative abundance of a particular transcript. Translation was most likely to occur from transcripts that were the most abundantly expressed after bacterial challenge [36].
A similar phenomenon to that reported here has been observed in the model organism C. elegans upon infection with Pseudomonas aeruginosa. C. elegans intestinal cells endocytose P. aeruginosa Exotoxin A, which shuts down protein translation by inhibiting elongation factor 2 (EF2) [8]. This inhibition leads to the selective translation of ZIP-2, which is required for activation of defense pathways and pathogen clearance [7]. It was proposed that the 5′ UTR of zip-2, which contains several untranslated ORFs (uORFs), was required for the selective translation. Even so, there is no explanation for how ribosome loading and translation can selectively occur in this transcript. Interestingly, there are a few other examples from mammalian cells and yeast, where protein translation inhibition leads to selective translation of few genes that have uORFs at their 5′ UTR. The mammalian stress response transcription factor ATF4 and the yeast transcription factor GCN4 respond similarly to amino acid starvation and protein synthesis inhibition [55], [56]. The 5′ UTR or 3′ UTR of cytokines could potentially be functioning the same way to allow selective protein translation when initiation and/or elongation is blocked by Legionella pneumophila.
Interestingly, pharmacological inhibitors of host protein translation induce transcription of various stress response genes, including pro-inflammatory cytokines such as Il6, Il23, Il1α and Il1β [10], [11], [31]. Secretion of these genes can take place when the highly conserved host elongation machinery is targeted by toxins such as P. aeruginosa Exotoxin A or Corynebacterium diphtheriae encoded diptheria toxins [10], [31]. Diphtheria toxin and Pseudomonas ExoA inhibit eukaryotic elongation factor similar to the mechanism used by L. pneumophila effectors. They modify elongation factor 2 (EF2) of eukaryotic cells by ADP-ribosylation, which has been shown to trigger a strong host immune response [31], [57]. This suggests the presence of a conserved surveillance mechanism the host uses to detect and respond to inhibition of the translation elongation machinery.
A previous study has shown that the cytokine induction seen during L. pneumophila can be mimicked by the addition of P. aeruginosa Exo A in combination with the synthetic PAMP ligand Pam3Csk4 [31]. It seems likely that macrophages are able to selectively bypass the translation block of this toxin in a fashion that is similar to that described in C. elegans. Interestingly, we cannot reproduce this result using a variety of concentrations of the protein synthesis inhibitor cycloheximide, which instead reduces pro-inflammatory cytokine production in response to PAMP challenge. The mechanism by which cycloheximide inhibits protein synthesis is sufficiently different from these toxins to explain why we see differences in the innate immune response against CHX. CHX binds to the E-site of the 60S ribosomal subunit and freezes all translating ribosomes [46].The RNA/ribosome complex remains stabilized and does not dissociate, a phenomenon that may not be perceived as danger by eukaryotic cells. In contrast, both L. pneumophila and ExoA interfere with elongation factor function. There may be a set of modified elongation factors in the host cell that resist action of these effectors, or there may be a population that is sequestered from modification, allowing them to selectively act on cytokine transcripts. In either case, there must be some special property to the cytokine transcript that allows this selective utilization of these active elongation factors. Future work will focus on the nature of these transcripts that allows bypass of translation inhibition.
Materials and Methods
Ethics statement
This study was carried out in accordance with the recommendation in the Guide for Care and Use of Laboratory Animals of the National Institutes of Health. The Institutional Animal Care and Use Committee of Tufts University approved all animal procedures. Our approved protocol number is B2013-18. The animal work, which is limited to the isolation of macrophages, does not involve any procedures of infections of live animals.
Bacteria and culturing media
L. pneumophila strains Lp02 (referred to as WT) and Lp03 (referred to as dotA3) are streptomycin-resistant restriction-defective thymidine auxotrophs derived from L. pneumophila Philadelphia-1 (Lp01) (Table 1; [58]). The Δ5 and Δ5.ΔflaA strains were kindly provided by Zhao-Qing Luo (Purdue University) [10], [11], [43]. ΔflaA-GFP+, dotAΔflaA-GFP+, Δ5.ΔflaA-GFP+ carry GFP on an isopropyl-β-D-thiogalactopyranoside (IPTG)–inducible, Cm resistant plasmid (Table 1; [17], [59]). Solid medium containing buffered charcoal yeast extract (BCYE) and ACES-buffered yeast extract (AYE) broth culture medium supplemented with 100 µg/mL thymidine were used to maintain L. pneumophila strains [60], [61]. Strains containing the pGFP plasmid were maintained on BCYE plates containing 100 µg/mL thymidine and 5 µg/mL chloramphenicol and grown in AYE containing 100 µg/mL thymidine, 5 µg/mL chloramphenicol and 1 mM IPTG [17].
Table 1. Plasmids and strains used in this study.
| Plasmids/Strains | Genotype/relevant characteristics | Reference |
| pGFP | oriRSF101cmRptac::GFP+ | [17], [59] |
| pΔflaA | pSR47SΔflaA | [63] |
| WT | Lp02, Philadelphia-1, thyArpsLhsdR | [58] |
| dotA3 | Lp02 dotA03 (Lp03) | [58] |
| ΔflaA | Lp02ΔflaA (allelic exchange using pΔflaA) | [43] |
| dotA.ΔflaA | Lp03ΔflaA (allelic exchange using pΔflaA) | [63] |
| Δ5 | Missing 5 IDTS required for host protein translation inhibition | [10] |
| Δ5.ΔflaA | Lp02 background | [11] |
| ΔflaA-GFP | ΔflaA containing pGFP | This study |
| dotA.ΔflaA-GFP | dotA3.ΔflaA containing pGFP | This study |
| Δ5.ΔflaA-GFP | Δ5.ΔflaA containing pGFP | This study |
Eukaryotic cell culture
Bone marrow-derived macrophages (BMDMs) were isolated from the femurs of mice and allowed to proliferate as described [60], [61]. C57BL/6 myd88−/− femurs were kindly provided by Tanja Petnicki-Ocwieja in the laboratory of Linden Hu (Tufts Medical Center). BMDMs were differentiated for 7 days in RPMI containing 30% L-cell supernatant, 10% FBS, 2 mM L-glutamine and 1× Pen Strep (100 U/mL penicillin, 100 µg/mL streptomycin). Cells were lifted and either re-plated for experiments or quick-frozen for later use in FBS and 10% DMSO.
U937 cells (ATCC) were grown in RPMI supplemented with 10% FBS and 1 mM L- glutamine. For differentiation, cells were treated with 10 ng/ml 12-tetradecanoyl phorbol 13-acetate (TPA) for 48 hrs. For L. pneumophila infections, U937 cells were plated in fresh media without TPA and infections were carried out 12–16 hours after plating.
Immunoblotting
To evaluate protein expression in host cells, C57BL/6 bone marrow-derived macrophages were plated in medium supplemented with 200 µg/mL of thymidine. Cells were challenged with L. pneumophila at the desired MOI, subjected to centrifugation at 1000×g for 5 min and incubated at 37° for the noted time periods. Lysates were collected using 2× SDS Laemmli sample buffer (0.125 M Tric-Cl pH 6.8, 4% SDS, 20% glycerol, 10% beta-mercaptoethanol, 0.01% bromophenol blue). Proteins were electroblotted to PVDF membranes, blocked in milk and analyzed by immunoprobing.
For phospho-specific antibodies, blots were washed of all milk and incubated overnight with 1∶1000 phospho-p38 or phospho-JNK (Cell Signaling) in 5% BSA in phosphate buffered saline (PBS). Rabbit anti-DUSP1 (MKP1 V-15, Santa Cruz) and mouse anti-tubulin (sigma) antibodies were diluted to 1∶200 and 1∶7,500, respectively, in 5% milk in TBST. Goat anti-IL-1α and Goat anti-IL-1β antibodies (R&D systems, AF-400-NA & AF-401-NA) were diluted to 1∶500 in 5% milk in TBST.
Heat Killed Yersinia, LPS, Pam3CSK4, poly(I∶C) treatment
For preparation of Heat-Killed Yersinia, wild type Y. pseudotuberculosis strains were grown overnight at 26° in Luria-Bertani (LB) broth. Overnight cultures were heat killed at 60° for 30–60 min and aliquots were frozen at −80° until use.
Macrophages were stimulated with HKY (MOI = 50), LPS (Sigma, 0.1 µg/mL or 1 µg/mL) and Pam3CSK4 (Invitrogen, 2 µg/mL), poly(I∶C) (InvivoGen, 50 µg/mL) for the desired time points. Cells were washed 3× with PBS and lysed in 2× SDS Laemmli sample buffer.
Quantitative RT-PCR
RNA was extracted from mammalian cells using RNAeasy kit (Qiagen). To determine the amount of a particular transcript, a one step, RNA-to-Ct kit (Applied Biosystems) was used according to manufacturer's instructions. Primers used for transcript analysis were as follows: human Dusp1 (5′ TTTGAGGGTCACTACCAG and 3′ CCGCTTCGTAGTAGAG), mouse Dusp1 (5′GGATATGAAGCGTTTTCGGCT and 3′ ACGGACTGTCACGTCTTAGG), mouse Il1α (5′GCACCTTACACCTACCAGAGT and 3′ TGCAGGTCATTTAACCAAGTGG), mouse Il1β (5′ GCAACTGTTCCTGAACTCAACT and 3′ ATCTTTTGGGGTCCGTCAACT), mouse Tnf-α ( 5′ GCACCACCATCAAGGACTCAA and 3′ GCTTAAGTGACCTCGGAGCT), mouse 18S ribosomal RNA (5′ CGCCGCTAGAGGTGAAATTCT and 3′ GCTTTCGTAAACGGTTCTTCA).
Determination of secreted cytokines
Macrophages were plated in 24 well tissue culture plates (2.5×105 per well) and challenged with L. pneumophila for either 6 hrs or 24 hrs. Supernatants were collected from each sample and 50 µL was used for ELISA. Mouse IL-1α and IL-1β Platinum ELISA (eBiosciences) was used to measure cytokine levels according to the manufacturers manual.
Intracellular cytokine staining
Intracellular cytokine staining was performed as described before [62] with modifications. Differentiated macrophages (∼1×107cells/plate) were washed and challenged with L. pneumophila GFP+ strains in RPMI containing 200 µg/mL thymidine, 5 µg/mL Chloramphenicol and 1 mM IPTG. For measuring TNF production, infections were carried out at MOI-3 and MOI-10 for 9 hrs followed by Golgiplug incubation (BD Bioscience, 1 µL/mL) for additional 5 hrs. For IL-1α and IL-1β, infections were carried out for 6 hrs. Cells were harvested with 10 mL cold PBS, washed twice with FACS buffer (PBS+0.5%BSA+0.05%NaN), incubated with Fc Block (clone 2.4G2) for 20 minutes and fixed with 2% paraformaldehyde overnight at 4°. Macrophage were permeabilized with Perm/Wash buffer (BD Bioscience) on ice for 20 minutes and stained with Alexa Fluor 647-conjugated anti-mouse TNF (BioLegend, clone ALF 161), phycoerythrin(PE)-conjugated anti-mouse IL-1α (BioLegend, clone MP6-XT22), PE-conjugated anti-mouse IL-1β (eBioscience, clone NJTEN3), Rabbit anti-DUSP1 (MKP1 V-15, Santa Cruz) and Goat anti-Rabbit Cy5 (Invitrogen) for 40 minutes. Stained cells were analyzed by BD LSR II flow cytometer.
Translation, pulse labeling and quantification
To detect whole cell translation during defined timepoints, C57BL/6 bone marrow-derived macrophages were challenged with L. pneumophila-GFP+ at MOI-10 for 2 hrs, followed by labeling cells at various timepoints with 50 µM of L-azidohomoalanine (AHA) (Invitrogen) added to the culture medium. Cells were incubated for 1 hr (time course experiments) or for 4 hrs (IL-1α co-staining experiments) to allow incorporation of AHA into growing polypeptide chains. At the end of each incubation period, cells were washed with PBS, fixed with 4% paraformaldehyde for 15–20 min and left overnight in PBS. Incorporation of AHA was monitored by a Biotin- or APC-conjugated phosphine reagent (Pierce) and nuclei were stained with Hoechst 33342 (Molecular Probes). For flow cytometry, fixed cells were blocked with 1× BSA/PBS for 30 min at RT and 100 µM of APC-phosphine was added. Cells were incubated at 37° for 2–3 hrs and excess dye was removed by washing with 0.5% Tween-20/PBS. For IL-1α co-staining, PE conjugated anti-mouse IL-1α (BioLegend, clone MP6-XT22) was added to cells on ice for 30 min. Cells were analyzed by BD LSR II flow cytometer or by BD FACScalibur.
Puromycin incorporation (SunSET assay) and ELISA
SunSET assay was used to determine the kinetics of protein translation over time [49] with modifications. Macrophages were plated in 6 well plates and infected with ΔflaA for the desired time points. 10 µg/mL of puromycin (Sigma) was added to cells for either 15 min or 1 hr. Macrophages were washed and lysed with IP Wash/Lysis buffer (Pierce) in the presence of protease inhibitors (Roche). Lysates were incubated on ice for 20 min, centrifuged at 13,000 rpm for 5 min and the supernatants were used for ELISA. The protein concentration in each sample was determined by Bradford assay.
ELISA plates were prepared by coating 96 well Nunc MaxiSorp plates with the desired antibody. Polyclonal Goat anti-IL-1α and IL-1β (R&D systems), Rabbit anti-DUSP-1 (Santa Cruz) and Rabbit anti-RhoGDI (Santa Cruz) antibodies were diluted in Carbonate/Bicarbonate buffer (PH = 9.6) to 10 µg/mL and 100 µL was used per well. Plates were incubated overnight at 4°. The following day, plates were brought to room temperature and blocked with 0.5% BSA/PBS for 1 hr. Cell lysates that have incorporated puromycin were incubated on the ELISA plates for 2 hrs, washed with 0.05% Tween-20/PBS three times and incubated with monoclonal mouse-anti-Puromycin (12D10, Millipore) for 1 hr. Unbound antibody was washed exhaustively with 0.05% Tween-20/PBS and plates were incubated with Donkey anti-mouse-HRP secondary antibody. Unbound antibody was washed again with Tween-20/PBS four times and 100 µL of HRP substrate (TMB solution) was added to each well for 5–10 mins. The reaction was stopped by adding 100 µL of stop-solution (2N H2SO4), and absorbance was measured at 450 nm.
Supporting Information
L. pneumophila induces an early TLR-dependent and a later Icm/Dot-dependent activation of MAPK members. A/J macrophages were infected with wild type L. pneumophila or dotA3 mutant for indicated time points. Cell lysates were blotted for phosphorylated forms of JNK (p-JNK) and p38 (p-p38). Lower graphs show densitometry of p-JNK and p-p38 normalized to tubulin. Data are representative of at least three independent experiments.
(TIF)
Δ5 strains fails to induce transcription of DUSP-1 in U937 human monocytes. (A) U937 cells were challenged with wild type L. pneumophila, dotA or Δ5 mutants for 4 hrs and RNA was isolated from cells. dusp1 transcript levels were normalized to the housekeeping genes hydroxymethylbilane synthase (HMBS) and graphed as a fold increase over uninfected controls. (B) A time course analysis of DUSP1 protein levels in U937 cells infected with wild type, dotA or Δ5 strain of L. pneumophila.
(TIF)
Low concentrations of CHX allow partial protein translation to take place. B6 macrophages were either left untreated or treated with the indicated concentrations of cycloheximide for 1 hr and the methionine analog, L-azidohomoalanine (AHA, 50 µM) was incorporated into newly synthesized proteins for an additional hour. The incorporated analog was detected by an APC-conjugated phosphine and fluorescence microscopy. Bottom panel shows protein translation in macrophages that were infected with Dot+ L. pneumophila and AHA added between 5–6 hrs post infection.
(TIF)
Both cycloheximide and puromycin block IL-1β hyperstimulation. Macrophages were treated with 2 µg/mL Pam3CSK4 for 2 hrs. Cells were then treated with either cycloheximide (0.5 µg/mL) or puromycin for additional 3 hrs and pro- IL-1β protein levels were measured by western blot.
(TIF)
Δ5 mutants show partial inhibition of protein synthesis. B6 macrophages were infected with ΔflaA, dotAΔflaA and Δ5ΔflaA for 5 hrs and 10 µg/mL of puromycin was added between 5–6 hrs poi. Cells were fixed, permeabilized and incorporated puromycin was detected by anti-puromycin antibody (12D10) and fluorescence microscopy.
(TIF)
Acknowledgments
We thank Dervla Isaac, Kim Davis, Edward Geisinger and Vinay Ramabhadran for review of the text. We thank Zhao-Qing Luo for providing the Δ5 and Δ5.ΔflaA strains and Tanja Petnicki-Ocwieja for providing femurs from myD88−/− strains.
Funding Statement
RRI is an Investigator of HHMI. Work was supported by HHMI. SA is supported by an HHMI International Student Research Fellowship (http://www.hhmi.org/programs/international-student-research-fellowships), an HHMI “Med-into-Grad” training grant and a Tufts University Sackler School Dean's Fellowship (http://sackler.tufts.edu/Student-Life/Student-Awards/Deans-Fellows). ASD was supported by a Kirchstein postdoctoral fellowship F32AI084202 from NIAID and training program 5K12GM074869. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Akira S, Hemmi H (2003) Recognition of pathogen-associated molecular patterns by TLR family. Immunol Lett 85: 85–95. [DOI] [PubMed] [Google Scholar]
- 2. Medzhitov R, Janeway C Jr (2000) Innate immunity. N Engl J Med 343: 338–344. [DOI] [PubMed] [Google Scholar]
- 3. Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- 4. Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- 5. Ausubel FM (2005) Are innate immune signaling pathways in plants and animals conserved? Nat Immunol 6: 973–979. [DOI] [PubMed] [Google Scholar]
- 6. Matzinger P (2002) The danger model: a renewed sense of self. Science 296: 301–305. [DOI] [PubMed] [Google Scholar]
- 7. Dunbar TL, Yan Z, Balla KM, Smelkinson MG, Troemel ER (2012) C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe 11: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McEwan DL, Kirienko NV, Ausubel FM (2012) Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans. Cell Host Microbe 11: 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shin S, Case CL, Archer KA, Nogueira CV, Kobayashi KS, et al. (2008) Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog 4: e1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fontana MF, Banga S, Barry KC, Shen X, Tan Y, et al. (2011) Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog 7: e1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fontana MF, Shin S, Vance RE (2012) Activation of host mitogen-activated protein kinases by secreted Legionella pneumophila effectors that inhibit host protein translation. Infect Immun 80: 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tattoli I, Sorbara MT, Vuckovic D, Ling A, Soares F, et al. (2012) Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11: 563–575. [DOI] [PubMed] [Google Scholar]
- 13. Horwitz MA, Silverstein SC (1980) Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J Clin Invest 66: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vogel JP, Andrews HL, Wong SK, Isberg RR (1998) Conjugative transfer by the virulence system of Legionella pneumophila. Science 279: 873–876. [DOI] [PubMed] [Google Scholar]
- 15. Brand BC, Sadosky AB, Shuman HA (1994) The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol Microbiol 14: 797–808. [DOI] [PubMed] [Google Scholar]
- 16. Hubber A, Roy CR (2010) Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26: 261–283. [DOI] [PubMed] [Google Scholar]
- 17. Losick VP, Isberg RR (2006) NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J Exp Med 203: 2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kagan JC, Roy CR (2002) Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol 4: 945–954. [DOI] [PubMed] [Google Scholar]
- 19. Kagan JC, Stein MP, Pypaert M, Roy CR (2004) Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J Exp Med 199: 1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Banga S, Gao P, Shen X, Fiscus V, Zong WX, et al. (2007) Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc Natl Acad Sci U S A 104: 5121–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heidtman M, Chen EJ, Moy MY, Isberg RR (2009) Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol 11: 230–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rowbotham TJ (1980) Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol 33: 1179–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vance RE (2010) Immunology taught by bacteria. J Clin Immunol 30: 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nash TW, Libby DM, Horwitz MA (1984) Interaction between the legionnaires' disease bacterium (Legionella pneumophila) and human alveolar macrophages. Influence of antibody, lymphokines, and hydrocortisone. J Clin Invest 74: 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shin S (2012) Innate Immunity to Intracellular Pathogens: Lessons Learned from Legionella pneumophila. Adv Appl Microbiol 79: 43–71. [DOI] [PubMed] [Google Scholar]
- 26. Massis LM, Zamboni DS (2011) Innate immunity to legionella pneumophila. Front Microbiol 2: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Archer KA, Ader F, Kobayashi KS, Flavell RA, Roy CR (2010) Cooperation between multiple microbial pattern recognition systems is important for host protection against the intracellular pathogen Legionella pneumophila. Infect Immun 78: 2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Archer KA, Alexopoulou L, Flavell RA, Roy CR (2009) Multiple MyD88-dependent responses contribute to pulmonary clearance of Legionella pneumophila. Cell Microbiol 11: 21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Archer KA, Roy CR (2006) MyD88-dependent responses involving toll-like receptor 2 are important for protection and clearance of Legionella pneumophila in a mouse model of Legionnaires' disease. Infect Immun 74: 3325–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Newton CA, Perkins I, Widen RH, Friedman H, Klein TW (2007) Role of Toll-like receptor 9 in Legionella pneumophila-induced interleukin-12 p40 production in bone marrow-derived dendritic cells and macrophages from permissive and nonpermissive mice. Infect Immun 75: 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barry KC, Fontana MF, Portman JL, Dugan AS, Vance RE (2013) IL-1alpha Signaling Initiates the Inflammatory Response to Virulent Legionella pneumophila In Vivo. J Immunol 190: 6329–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fontana MF, Vance RE (2011) Two signal models in innate immunity. Immunol Rev 243: 26–39. [DOI] [PubMed] [Google Scholar]
- 33. Shen X, Banga S, Liu Y, Xu L, Gao P, et al. (2009) Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol 11: 911–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Belyi Y, Niggeweg R, Opitz B, Vogelsgesang M, Hippenstiel S, et al. (2006) Legionella pneumophila glucosyltransferase inhibits host elongation factor 1A. Proc Natl Acad Sci U S A 103: 16953–16958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Belyi Y, Tabakova I, Stahl M, Aktories K (2008) Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J Bacteriol 190: 3026–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ivanov SS, Roy CR (2013) Pathogen signatures activate a ubiquitination pathway that modulates the function of the metabolic checkpoint kinase mTOR. Nat Immunol 14 12: 1219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Friedman H, Yamamoto Y, Klein TW (2002) Legionella pneumophila pathogenesis and immunity. Semin Pediatr Infect Dis 13: 273–279. [DOI] [PubMed] [Google Scholar]
- 38. Li Gobbi F, Benucci M, Del Rosso A (2005) Pneumonitis caused by Legionella pneumoniae in a patient with rheumatoid arthritis treated with anti-TNF-alpha therapy (infliximab). J Clin Rheumatol 11: 119–120. [DOI] [PubMed] [Google Scholar]
- 39. Welsh CT, Summersgill JT, Miller RD (2004) Increases in c-Jun N-terminal kinase/stress-activated protein kinase and p38 activity in monocyte-derived macrophages following the uptake of Legionella pneumophila. Infect Immun 72: 1512–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Z, Dugan AS, Bloomfield G, Skelton J, Ivens A, et al. (2009) The amoebal MAP kinase response to Legionella pneumophila is regulated by DupA. Cell Host Microbe 6: 253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Losick VP, Haenssler E, Moy MY, Isberg RR (2010) LnaB: a Legionella pneumophila activator of NF-kappaB. Cell Microbiol 12: 1083–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Diez E, Lee SH, Gauthier S, Yaraghi Z, Tremblay M, et al. (2003) Birc1e is the gene within the Lgn1 locus associated with resistance to Legionella pneumophila. Nat Genet 33: 55–60. [DOI] [PubMed] [Google Scholar]
- 43. Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE (2006) Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog 2: e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wright EK, Goodart SA, Growney JD, Hadinoto V, Endrizzi MG, et al. (2003) Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr Biol 13: 27–36. [DOI] [PubMed] [Google Scholar]
- 45. Rubartelli A, Cozzolino F, Talio M, Sitia R (1990) A novel secretory pathway for interleukin-1 beta, a protein lacking a signal sequence. EMBO J 9: 1503–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, et al. (2010) Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol 6: 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR (2006) A comparative study of bioorthogonal reactions with azides. ACS Chem Biol 1: 644–648. [DOI] [PubMed] [Google Scholar]
- 48. Roy CR, Berger KH, Isberg RR (1998) Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol Microbiol 28: 663–674. [DOI] [PubMed] [Google Scholar]
- 49. Schmidt EK, Clavarino G, Ceppi M, Pierre P (2009) SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6: 275–277. [DOI] [PubMed] [Google Scholar]
- 50. Lu JY, Sadri N, Schneider RJ (2006) Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev 20: 3174–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Radwan M, Stiefvater R, Grunert T, Sharif O, Miller I, et al. (2010) Tyrosine kinase 2 controls IL-1ss production at the translational level. J Immunol 185: 3544–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shaw G, Kamen R (1986) A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46: 659–667. [DOI] [PubMed] [Google Scholar]
- 53. Palanisamy V, Jakymiw A, Van Tubergen EA, D'Silva NJ, Kirkwood KL (2012) Control of cytokine mRNA expression by RNA-binding proteins and microRNAs. J Dent Res 91: 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dinarello CA (1992) Dissociation of transcription from translation of human IL-1 beta: induction of steady state mRNA by adherence or recombinant C5a in the absence of translation. Proc Soc Exp Biol Med 200: 228–232. [DOI] [PubMed] [Google Scholar]
- 55. Hinnebusch AG (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450. [DOI] [PubMed] [Google Scholar]
- 56. Vattem KM, Wek RC (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 101: 11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jorgensen R, Wang Y, Visschedyk D, Merrill AR (2008) The nature and character of the transition state for the ADP-ribosyltransferase reaction. EMBO Rep 9: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Berger KH, Isberg RR (1993) Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7: 7–19. [DOI] [PubMed] [Google Scholar]
- 59. Solomon JM, Rupper A, Cardelli JA, Isberg RR (2000) Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect Immun 68: 2939–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Swanson MS, Isberg RR (1995) Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun 63: 3609–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Feeley JC, Gibson RJ, Gorman GW, Langford NC, Rasheed JK, et al. (1979) Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J Clin Microbiol 10: 437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Alaniz RC, Cummings LA, Bergman MA, Rassoulian-Barrett SL, Cookson BT (2006) Salmonella typhimurium coordinately regulates FliC location and reduces dendritic cell activation and antigen presentation to CD4+ T cells. J Immunol 177: 3983–3993. [DOI] [PubMed] [Google Scholar]
- 63. Creasey EA, Isberg RR (2012) The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proc Natl Acad Sci U S A 109: 3481–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
L. pneumophila induces an early TLR-dependent and a later Icm/Dot-dependent activation of MAPK members. A/J macrophages were infected with wild type L. pneumophila or dotA3 mutant for indicated time points. Cell lysates were blotted for phosphorylated forms of JNK (p-JNK) and p38 (p-p38). Lower graphs show densitometry of p-JNK and p-p38 normalized to tubulin. Data are representative of at least three independent experiments.
(TIF)
Δ5 strains fails to induce transcription of DUSP-1 in U937 human monocytes. (A) U937 cells were challenged with wild type L. pneumophila, dotA or Δ5 mutants for 4 hrs and RNA was isolated from cells. dusp1 transcript levels were normalized to the housekeeping genes hydroxymethylbilane synthase (HMBS) and graphed as a fold increase over uninfected controls. (B) A time course analysis of DUSP1 protein levels in U937 cells infected with wild type, dotA or Δ5 strain of L. pneumophila.
(TIF)
Low concentrations of CHX allow partial protein translation to take place. B6 macrophages were either left untreated or treated with the indicated concentrations of cycloheximide for 1 hr and the methionine analog, L-azidohomoalanine (AHA, 50 µM) was incorporated into newly synthesized proteins for an additional hour. The incorporated analog was detected by an APC-conjugated phosphine and fluorescence microscopy. Bottom panel shows protein translation in macrophages that were infected with Dot+ L. pneumophila and AHA added between 5–6 hrs post infection.
(TIF)
Both cycloheximide and puromycin block IL-1β hyperstimulation. Macrophages were treated with 2 µg/mL Pam3CSK4 for 2 hrs. Cells were then treated with either cycloheximide (0.5 µg/mL) or puromycin for additional 3 hrs and pro- IL-1β protein levels were measured by western blot.
(TIF)
Δ5 mutants show partial inhibition of protein synthesis. B6 macrophages were infected with ΔflaA, dotAΔflaA and Δ5ΔflaA for 5 hrs and 10 µg/mL of puromycin was added between 5–6 hrs poi. Cells were fixed, permeabilized and incorporated puromycin was detected by anti-puromycin antibody (12D10) and fluorescence microscopy.
(TIF)