Abstract
This study constructs a rat brain T 2‐weighted magnetic resonance imaging template including olfactory bulb and a compatible digital atlas. The atlas contains 624 carefully delineated brain structures based on the newest (2005) edition of rat brain atlas by Paxinos and Watson. An automated procedure, as an SPM toolbox, was introduced for spatially normalizing individual rat brains, conducting statistical analysis and visually localizing the results in the Atlas coordinate space. The brain template/atlas and the procedure were evaluated using functional images between rats with the right side middle cerebral artery occlusion (MCAO) and normal controls. The result shows that the brain region with significant signal decline in the MCAO rats was consistent with the occlusion position. Hum Brain Mapp, 2013. © 2012 Wiley Periodicals, Inc.
Keywords: SD rat brain, MRI imaging analysis, SPM, normalization, location, co‐registration
INTRODUCTION
Magnetic resonance imaging (MRI) techniques have been widely used for the investigation of human brain functions and structures in normal and diseased individuals. Most of these studies are performed on a group of subjects, and the data are analyzed voxel‐by‐voxel after their brain images are spatially normalized to a brain template, such as the MNI (Montreal Neurological Institute) template. The MNI template is an important part of a number of well known neuroimaging computer software packages, among them is SPM (Welcome Department of Cognitive Neurology, London, UK). In addition to conditioning data for voxel‐wise statistical analyses over multiple subjects, the availability of the template also makes it feasible for automated anatomical localizations and references since the MNI template is in the stereotaxic coordinate space of the Talairach atlas [Talairach and Tournoux, 1988] system. Moreover, the automatic human brain area indexing is now a common practice [Lancaster et al., 1997; Tzourio‐Mazoyer et al., 2002].
Similarly, MRI techniques have also been widely used in animal studies in the recent years. To process the animal imaging data, either existing analytic tools originally introduced for human studies are evaluated and adopted, or new ones are developed. One of such adaptation efforts is to construct the animal brain template in order to use the existing analytic tools for human studies. In the past several years, several animal brain MRI templates have been constructed for mice [Lin et al., 2003; Sawiak et al., 2009], baboons [Black et al., 2001b], macaque [Alexander et al., 2008; Black et al., 2001a], zebra finch [Poirier et al., 2008], and Sprague‐Dawley (SD) rats [Schwarz et al., 2006; Schweinhardt et al., 2003].
To our research interest in the current study, we especially note the role that SD rats played in normal and diseased brain studies [Pawela et al., 2008; Schwarz et al., 2006, 2009]. Accordingly, the SD rat brain template construction and data analysis standardization have been long a methodological topic. For example, although with limited sample size of five and all were female animals, Schweinhardt et al. built a first T 2‐weighted SD rat brain template [Schweinhardt et al., 2003]. In a later study, Schwarz et al. introduced their MRI T 2‐weighted SD rat brain template with tissue classes for pharmacological MRI studies [Schwarz et al., 2006].
We noted that these reported rat brain templates do not include olfactory bulb. Because a close association between olfactory dysfunction and cognitive impairment has been implied in some researches [Devanand et al., 2000; Peters et al., 2003; Schubert et al., 2008], the olfactory bulb is necessary not only for its own studies but also for the cognitive ones.
A brain template alone is incomplete without a brain atlas system offering detailed anatomical information. Therefore, a co‐registration step is needed in order to align the template with the atlas. In 2006, Schwarz et al. [2006] report their template together with the digital rat brain atlas derived from the fourth edition of rat brain stereotaxic coordinates built by Paxinos and Watson [1998]. In their study, the nine‐parameter affine transformation is used to co‐register the template with the atlas based on the homologous anatomical features [Schwarz et al., 2006]. However, alternative approaches might be needed to make more adequate template/atlas co‐registration using both linear and non‐linear deformation.
With the introduction above, it is desirable to make a rat brain MRI template including olfactory bulb and a compatible digital rat brain atlas available for the widely used computer package SPM. The current study is designed (1) to construct an SD rat brain template that includes olfactory bulb; (2) to create a digital rat brain atlas from the 5th edition of the Paxinos and Watson [2005]; and (3) to develop and implement an automated rat brain image analysis toolbox in the widely used SPM. To evaluate the adequacy of our template together with the digital atlas and to examine the toolbox implemented in SPM, we analyze the data from rats with the right side middle cerebral artery occlusion (MCAO) and those without. This toolbox is available by contacting the corresponding author at shanbc@ihep.ac.cn.
MATERIAL AND METHODS
Animals and Data Acquisition
Twenty‐one healthy adult SD rats (female, 9; age range, 10–11 week old; weight range, 250–300 g) were used for template construction in this study. In addition, in order to evaluate the used intracranial mask, ten healthy adult SD rats (male, 10; age range, 11–12 week old; weight range, 250–300 g) were used. Furthermore, in order to demonstrate the values and the practical use of the constructed template in voxel‐based procedure, data from additional five SD rats with the right side MCAO (male, 5; age range, 10–12 week old; weight range, 180–240 g) and five healthy SD rats (male, 5; age range, 10–12 week old; weight range, 180–240 g) were analyzed. Intraluminal occlusion of the middle cerebral artery (MCA) was accomplished using a modification of the Longa technique [Longa et al., 1989]. This surgical procedure was performed 30 min prior to the MRI acquisition.
During MRI scan, all rats were anesthetized using inhaled isoflurane/O2 (3% for induction and 1.5–2% for maintenance) and prostrated on a custom‐made holder to minimize head motion while respiration was monitored at a rate of 50 breaths per min.
T 2‐weighted data for template construction and functional data of MCAO were acquired on a 7.0 T animal MRI scanner (70/16 PharmaScan, Bruker Biospin GmbH, Germany) in Nanjing, using a 38‐mm birdcage rat brain quadrature resonator for radiofrequency transmission and receiving. The structural images were obtained with a rapid acquisition with relaxation enhancement (RARE) sequence (RARE factor = 8, repetition time (TR) = 10,493 ms, echo time (TE) = 36 ms, matrix size 256 × 256 × 90, voxel size 0.14 × 0.14 × 0.3 mm3, no slice gap). For functional cerebrum, images were obtained with an echo planar imaging (EPI) sequence (TR = 850 ms, TE = 25 ms, matrix size 128 × 128 × 15, voxel size 0.3 × 0.3 × 1 mm3, flip angle 90°, no slice gap, total of 80 volumes) and coplanar T 2‐weighted scan was also acquired in addition to the functional data. All the Bruker original images were converted to DICOM format with programs (Paravision 4.0) in the scanner. All experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by Jiang Su Animal Care and Use Committee.
T 2‐weighted data for evaluating the use of the intracranial mask were acquired on a 7 T/20 cm Bruker Biospec animal MRI scanner in Wuhan. The structural images were obtained with a RARE sequence using a volume coil for radio frequency (RF) transmission and a quadrature surface coil for signal detection (RARE factor = 4, TR = 5800 ms, TE = 40 ms, matrix size 512 × 384 × 52, voxel size 0.068 × 0.091 × 0.58 mm3, no slice gap). All the Bruker original images were converted to DICOM format with programs (Paravision 4.0) in the scanner. All experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by Hubei Animal Care and Use Committee.
Rat Brain Template Construction
Construction of the rat brain template
The rat brain template with the extracranial tissues was created using SPM2 (Welcome Department of Cognitive Neurology, London, UK). All of the 21 images included in this study were inspected and were found equally of high quality in terms of the image contrast, noise level, and resolution. The rat brain template was created recursively by registering [Ashburner and Friston, 1999; Friston et al., 1995; Zhilkin and Alexander 2004] and averaging as described by Lin et al. [2003]. The constructed template is shown in Figure 1A.
Figure 1.
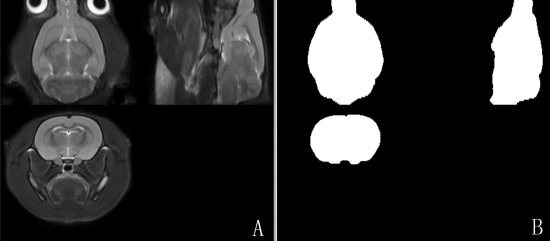
A: Axial, sagittal and coronal views of the T2‐weighted rat brain template; (B) Axial, sagittal and coronal views of the corresponding intracranial mask.
Extraction of the canonical brain and creation of an intracranial mask image
The canonical brain was extracted semi‐automatically from the template constructed above by the following two steps. Firstly, a rectangular bounding box was used to roughly remove the extracranial tissues in MRIcro [Rorden and Brett, 2000], especially the voxels of Harderian gland. Then, the intracranial portion of the image including the olfactory was extracted based on Otsu's criterion [Gonzalez et al., 2004].
In addition, an intracranial rat brain mask was formed from this canonical brain by assigning 1 for intracranial voxels and 0 for others, as shown in Figure 1B. Thus, after normalizing an individual image to the rat brain template, the intracranial tissues could be conveniently extracted.
Construction of the Digital Rat Brain Atlas From the Paxinos and Watson Atlas
First, the 2D color figures [Paxinos and Watson, 2005], from the anterior part of olfactory bulb (z bregma + 7.56 mm) to the posterior part of cerebellum (z bregma − 15.72 mm), were converted to gray‐scale figures. The background of these figures, including the structure labels and coordinate grids, was removed. Then the outline and internal contours were identified and further enhanced and smoothed by image erosion and dilation [Gonzalez et al., 2004]. Moreover, the slice‐to‐slice realignment was performed.
Each anatomical structure of the brain atlas was given a unique integer as an index. The laterality information was preserved by assigning odd/even integers for right/left corresponding structures. In this study, 624 structures could be identified in the digital atlas. By doing so, the graphical representation of the atlas was now in the form of 3D tomographic data. Then, the constructed 3D atlas image was interpolated to create a new 98‐slice image with a uniform inter‐slice separation of 0.24 mm. This final 3D atlas image was saved as SPM2 compatible Analyze 7.5 format and SPM5/SPM8 compatible NIFTI format. With this digital atlas, a structure could be queried by simply reading the voxel index inside the structure. As an example, one slice of this rat brain atlas image (Bregma −4.20 mm) [Paxinos and Watson, 2005] is given in Figure 2. Appendix A lists information on all structures of this slice including their assigned integer image intensity.
Figure 2.
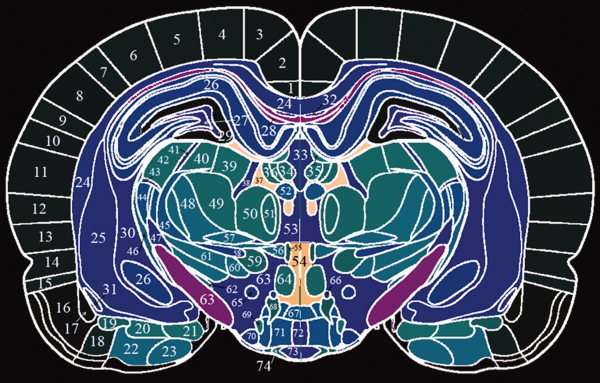
One slice of the rat brain atlas with detailed anatomical delineation. Note that the integer assigned to each VOI on the figure is not the one designated as the VOI ID. The corresponding VOI ID can be found in Appendix A. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Moreover, the 624 anatomical regions were merged into 66 functional ones according to different brain functions by an anatomist and a co‐author of this study (JHL). Among them are sensory cortex, visual cortex, and hippocampus. A new set of integer indices was assigned to these 66 functional regions. Integer indices and functionalities for these 66 regions are listed in Appendix B.
In order to co‐register the canonical brain with the prepared tomographic atlas, we created a pseudo T 2‐weighted MR image from the 3D atlas [Rubins et al., 2003]. For this purpose, the 624 regions were merged into 12 main anatomy structures, which included olfactory bulb, dorsal part of cortex, ventral part of cortex, hippocampi, corpus striatum, thalamus, hypothalamus, mesencephalon, medulla oblongata, cerebellar, corpus callosum, and ventricle. Finally, this pseudo image was further smoothed using an isotropic Gaussian kernel with a FWHM (Full Width Half Maximum) of two voxels to smooth the juncture of the neighboring segmented parts.
The fine 624 anatomical structures or the 66 functional regions can be used for localization of statistical results. And the low‐resolution the pseudo T 2‐weighted atlas with 12 regions is for co‐registration between the intracranial‐only rat brain template and the digital atlas.
Co‐Registration of the Canonical Brain With the Paxinos and Watson Rat Atlas
The pseudo MRI was chosen as the target image. The SPM intensity‐based affine transformation algorithm and subsequent non‐linear warping algorithm was employed to co‐register the canonical brain with this target atlas image [Friston et al., 2007], and the transformation matrix was saved to standardize statistical results into the Atlas coordinate space.
Data Analysis of Functional Images in SPM With the Constructed Template and Atlas
The analysis of the functional images was performed in both SPM2 and SPM5 separately. Briefly, the functional image series were realigned to each other and co‐registered to the T 2 coplanar image. They were then spatially normalized to the rat brain template via the coplanar. After removing the non‐brain tissues via the intracranial mask, the functional series were brought to the canonical space and smoothed with a Gaussian kernel of 2 × 2 × 4 mm3 FWHM.
In order to identify the difference of signals between the rats with MCAO and the healthy controls, voxel‐wise two‐sample t‐test [Friston et al., 2007] was performed accounting for global confounds with proportional scaling. Brain regions with signal reductions in MCAO rats were identified using P < 0.001 (uncorrected) for voxel intensities and 50 for cluster‐extent.
RESULTS
The Use of Constructed Rat Brain Template and Intracranial Mask Image
To illustrate the successful use of the intracranial mask in removing non‐brain tissues automatically, the dataset obtained in Wuhan was used. As this group of data was acquired using surface coil, the image intensity attenuated seriously from dorsal to ventral. Therefore, the voxels beneath the ventral side were cut off in both the rat brain template and individual images. Then the individual images were normalized to the template, and the intracranial mask was used to remove non‐brain tissues, both of which could be performed in our modified SPM. The extraction of three randomly chosen rats result is shown in Figure 3. As shown in Figure 3, the intracranial tissue could be extracted successfully by the intracranial mask.
Figure 3.
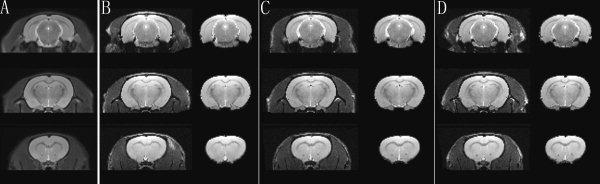
The extraction results. Panel (A) shows three planes from the average rat brain. For randomly selected individual rats 1, 2, and 3, the registered image is shown on the left and the extracted brain tissues on the right on Panels (B–D) separately.
Co‐Registration of the Canonical Brain to the Constructed Rat Brain Atlas
The co‐registration result between the canonical brain and atlas was qualitatively and quantitatively evaluated in this study. The constructed atlas is overlaid on the co‐registered canonical brain (Fig. 4). Three representative anatomical structures, hippocampus, corpus striatum, and ventricle, are selected from the atlas [Nie et al., 2010] and also overlaid on the canonical brain (Fig. 5). Moreover, the hippocampus on the right side is manually traced out on the canonical brain by an experienced rater, blinded to the atlas. Four volumetric and spatial correspondence measures, Jaccard similarity (JS) [Jaccard, 1912; Lin et al., 2003; Murugavel and Sullivan, 2009], the relative error on volume (RV), the proportions of false‐positive (FP), and false‐negative (FN) [Chupin et al., 2007; Rodionov et al., 2009], were calculated between the atlas‐derived and manual‐traced hippocampus, and between the whole brain volumes of the atlas and canonical image. The calculated results are given in Table I, and details of these measures are listed in Appendix C.
Figure 4.
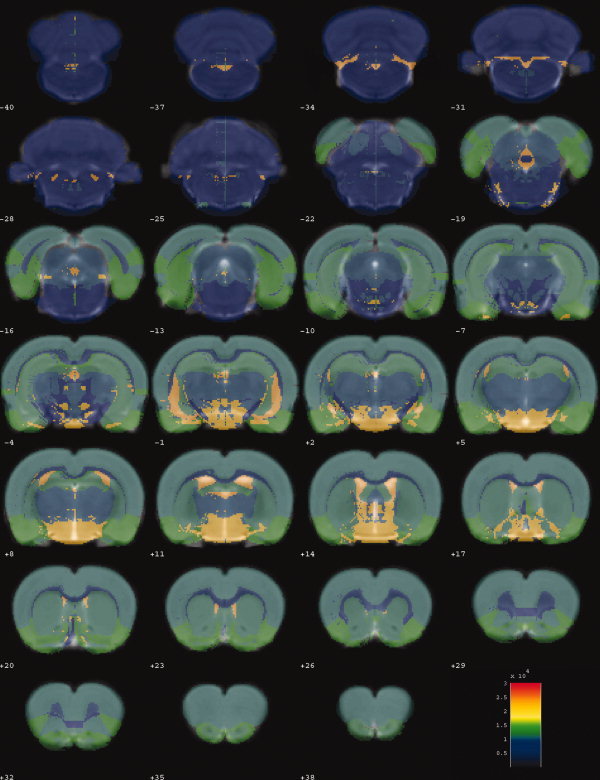
Superimposing the rat brain atlas with 12 anatomical structures on the co‐registered rat brain template. The rat brain atlas is presented with varying transparency and color scaled. The co‐registered rat brain template is presented in gray‐scale as a background. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 5.
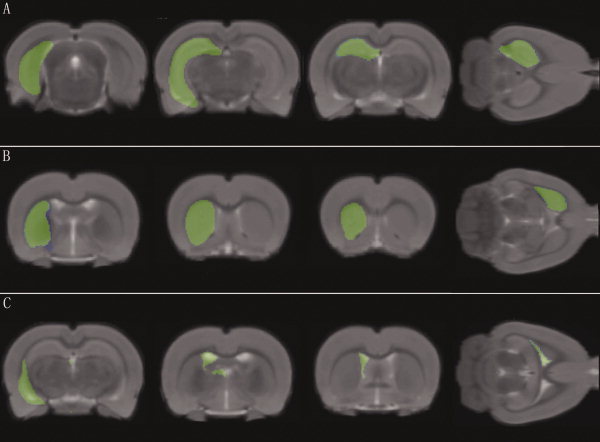
Superimposing three representative anatomical structures on the co‐registered rat brain template. The selected anatomical structures are extracted from the rat brain atlas and presented with varying transparency and color scaled. The co‐registered rat brain template is presented in gray‐scale as a background. The three selected anatomical structures are, respectively, (A) hippocampus, (B) corpus striatum, and (C) ventricle. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Table I.
Volumetric and spatial correspondence measures between the rat brain atlas and the co‐registered rat brain template
| JS (%) | RV (%) | FP (%) | FN (%) | |
|---|---|---|---|---|
| Whole brain | 93.98 | 1.83 | 3.99 | 3.63 |
| Right hippocampus | 90.56 | 8.37 | 10.29 | 9.59 |
Data Analysis of Functional Images in SPM With the Constructed Template and Atlas
The SPM2 based statistical results of the five MCAO rats comparing with the five normal controls are shown in Figure 6. Similar results were also obtained in SPM5 and will not be presented. These regions are all related to the right side MCA.
Figure 6.
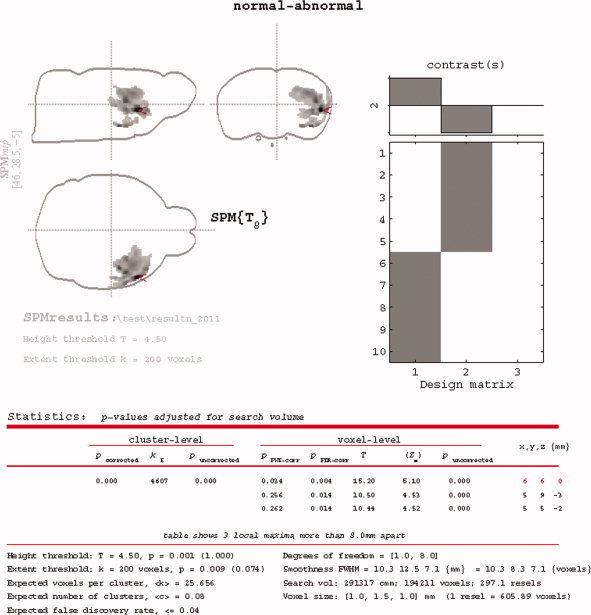
Statistical results of reduced signals in the MCAO related regions in comparison with 10 normal controls. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
DISCUSSION
In the current study, we constructed and evaluated T 2‐weighted brain template, including the whole olfactory bulb. This SD rat brain template was in the widely used Paxinos coordinate space, which is defined by Paxinos and Watson [2005]. Moreover, to make it easy to use in routine analyses, we incorporated the rat brain template together with standard atlas into the SPM package.
It was found that the spatial normalization of individual rat brains to the template space was satisfactory even when image resolutions differed from that of the data used for the template creation. As exemplified by analyzing the MCAO data, the use of this template in SPM2 or SPM5 was feasible and practical. We note that the imaging contrast differences between the rats with MCAO and normal controls might be due to the effects of stroke, such as inflammation or necrosis.
In addition, one of our primary aims was to make the removal of the extracranial tissue automated. Alternative to the successful automatic cropping method using pulse coupled neural networks (PCNN) by Murugavel and Sullivan [2009], we proposed the use of intracranial‐only mask image which was in the same coordinate space with the template. When an individual image was normalized to the template, the extracranial tissue could be simply removed by the intracranial mask. Our results showed this strategy worked satisfactorily.
The central piece of our efforts was to make the Paxinos and Watson's rat brain atlas [Paxinos and Watson, 2005] available in the coordinate space of our MRI template. We adopted the strategy of converting the atlas map into pseudo T 2‐weighted images. This allowed us to use the existing co‐registration algorithm with the option of affine only or with non‐linear warping. We also envision its general applicability in other template/atlas construction studies.
As any other electronically accessible atlas with detailed labeling for human [Talairach and Tournoux, 1988], the availability of the rat brain atlas can provide two convenient services to researchers. First, detailed structure information could be queried either automatically or interactively in statistical parametric mapping analyses, as showcased in this study (Results and Appendix B). This was very helpful for pin‐pointing the exact brain locations where the maximal group/condition differences were observed in voxel‐wise analysis, as shown in Figure 6. Second, similar as the work described by Schwarz et al. [2006], volume of interest (VOI) could now be defined straightforward, objective, and free of operator bias [Nie et al., 2010].
In demonstrating the adequacy and usefulness of our work, the functional (T 2*‐weighted) image series were spatially normalized to the constructed rat brain template via the coplanar T 2 image of each animal. To do that, we first corrected possible movements in the functional time series and co‐registered them to the coplanar T 2 data. This procedure has been common for human studies [Friston et al., 2007]. Nevertheless, if coplanar structural image is not a part of the data acquisition protocol, or not available due to any other reason, the procedure above then becomes non‐feasible. An alternative and convenient way to analyze the functional images is to make available of a functional rat brain template in the same coordinate space of the T 2‐weighted template. The template construction method and co‐registration strategy proposed in this study can be extended in establishing the functional MRI template, even the positron emission tomography (PET) and electronic computer X‐ray tomography technique (CT) templates.
As a whole‐brain unbiased technique, voxel‐based morphometry (VBM) technique has been used for characterizing regional gray/white matter concentration or volume differences. The segmentation of the structural images into grey matter (GM), white matter (WM), and cerebrospinal fluid (CSF) is prerequisite for the VBM procedure [Good et al., 2001; Keller et al., 2004]. Continuing our current work, we are in the process of establishing GM, WM, and CSF prior probability maps for rat brains and will have it reported separately.
In conclusion, we demonstrated the adequacy and practical usefulness of the constructed template in SPM environment together with the corresponding Paxinos and Watson rat brain atlas. We believe it will be helpful to streamline the neuroimaging data analyses for rat brain imaging studies from the pre‐processing stage to the reports of the statistical inference results.
APPENDIX A.
Table APPENDIX A.
Detailed information on some of the 624 structures included in the rat atlas by Paxinos and Watson and presented in Figure 2
| Index in Figure 5 | Structural name | Abbreviation | IndexL a | IndexR a |
|---|---|---|---|---|
| 1 | Retrosplenial granular cortex, b region | RSGb | 178 | 177 |
| 2 | Retrosplenial granular cortex, c region | RSGc | 156 | 155 |
| 3 | Retrosplenial dysgranular cortex | RSD | 158 | 157 |
| 4 | Senondary visual cortex, mediomedial area | V2MM | 184 | 183 |
| 5 | Senondary visual cortex, mediolateral area | V2ML | 186 | 185 |
| 6 | Parietal cortex, posterior area, dorsal part | PtPD | 180 | 179 |
| 7 | Parietal cortex, posterior area, rostral part | PtPR | 190 | 189 |
| 8 | Primary somatosensory cortex, barrel field | S1BF | 150 | 149 |
| 9 | Primary somatosensory cortex | S1 | 182 | 181 |
| 10 | Secondary auditory cortex, dorsal area | AuD | 166 | 165 |
| 11 | Primary auditory cortex | Au1 | 168 | 167 |
| 12 | Secondary auditory cortex, ventral area | AuV | 170 | 169 |
| 13 | Ectorhinal cortex | Ect | 174 | 173 |
| 14 | Perirhinal cortex | PRh | 176 | 175 |
| 15 | Lateral ectorhinal cortex | LEnt | 200 | 199 |
| 16 | Piriform cortex | Pir | 78 | 77 |
| 17 | Dorsal endopiriform nucleus | DEn | 94 | 93 |
| 18 | Amygdalopiriform transition area | APir | 208 | 207 |
| 19 | Basolateral amygdaloid nucleus, posterior part | BLP | 490 | 489 |
| 20 | Amygdalohippocampal area, posterolateral | AHiPL | 570 | 569 |
| 21 | Amygdalohippocampal area, posteromedial | AHiPM | 572 | 571 |
| 22 | Posterolateral cortical amygdaloid nucleus | PLCo | 686 | 685 |
| 23 | Posteromedial cortical amygdaloid nucleus | PMCo | 688 | 687 |
| 24 | Corpus callosum | cc | 930 | 929 |
| Cingulum | cg | |||
| Deep cerebral white matter | dcw | |||
| 25 | Lateral ventricle | LV | 934 | 933 |
| 26 | Field CA1 of the hippocampus | CA1 | 870 | 869 |
| Field CA2 of the hippocampus | CA2 | |||
| Field CA3 of the hippocampus | CA3 | |||
| Lacunosum moleculare layer of the hippocampus | LMol | |||
| Oriens layer of the hippocampus | Or | |||
| Pyramidal cell layer of the hippocampus | Py | |||
| Stratum lucidum of the hippocampus | SLu | |||
| 27 | Granular layer of the dentate gyrus | GrDG | 982 | 981 |
| Polymorph layer of the dentate gyrus | PoDG | |||
| 28 | Dorsal hippocampal commissure | dhc | 1400 | 1399 |
| Alveus of the hippocampus | alv | |||
| 29 | Molecular layer of the dentate gyrus | MoDG | 100 | 99 |
| 30 | Fimbria of the hippocampus | fi | 956 | 955 |
| 31 | Lat amygdaloid nucleus | La | 976 | 975 |
| 32 | Dorsal fornix | df | 1078 | 1077 |
| 33 | Dorsal 3rd ventricle | D3V | 934 | 933 |
| 34 | Medial habenular nucleus | MHb | 550 | 549 |
| 35 | Stria medullaris of the thalamus | sm | 605 | 604 |
| 36 | Lateral habenular nucleus, medial part | LHbM | 552 | 551 |
| Lateral habenular nucleus, lateral part | LHbL | |||
| 37 | Paraventricular thalamic nucleus | PV | 2170 | 2169 |
| 38 | Centrolateral thalamic nucleus | CL | 932 | 931 |
| 39 | Lateral posterior thalamic nucleus, mediorostral part | LPMR | 556 | 555 |
| 40 | Lateral posterior thalamic nucleus, laterorostral part | LPLR | 558 | 557 |
| 41 | Intramedullary thalamic area | IMA | 960 | 959 |
| 42 | Dorsal lateral geniculate nucleus | DLG | 560 | 559 |
| 43 | Intergeniculate leaf | IGL | 978 | 977 |
| 44 | Ventral lateral geniculate nucleus | VLG | 778 | 777 |
| 45 | Superior thalamic radiation | str | 770 | 769 |
| 46 | Optic tract | opt | 722 | 721 |
| 47 | Subgeniculate nucleus | SubG | 952 | 951 |
| 48 | Ventral posteromedial thalamic nucleus | VPM | 668 | 667 |
| Ventral posterolateral thalamic nucleus | VPL | |||
| 49 | Posterior thalamic nuclear group | Po | 554 | 553 |
| 50 | Parafascicular thalamic nucleus | PF | 546 | 545 |
| 51 | Fasciculus retroflexus | fr | 548 | 547 |
| 52 | Precommissural nucleus | PrC | 758 | 757 |
| 53 | Periventricular gray | PVG | 974 | 973 |
| 54 | 3rd ventricle | 3V | 2086 | 2085 |
| 55 | A11dopamine cells | A11 | 452 | 451 |
| 56 | Subparafascicular thalamic nucleus | SPF | 680 | 679 |
| Subparafascicular thalamic nucleus, parvicellular part | SPFPC | |||
| 57 | Ventral posterior nucleus of the thalamus, parvicellular part | VPPC | 682 | 681 |
| 58 | Medial lemniscus | ml | 866 | 865 |
| 59 | Prerubral field | PR | 514 | 513 |
| 60 | Nucleus of the fields of Forel | F | 678 | 677 |
| 61 | Zona incerta, dorsal part | ZID | 666 | 665 |
| Zona incerta, ventral part | ZIV | |||
| 62 | Subthalamic nucleus | STh | 676 | 675 |
| 63 | Internal capsule | ic | 954 | 953 |
| Cerebral peduncle | cp | |||
| 64 | Posterior hypothalamic nucleus | PH | 544 | 543 |
| 65 | Gemini hypothalamic nucleus | Gem | 874 | 873 |
| 66 | Submammillothalamic nucleus | SMT | 872 | 871 |
| 67 | Supramammillary nucleus, medial part | SuMM | 762 | 761 |
| Supramammillary nucleus, lateral part | SuML | |||
| 68 | Mammillothalamic tract | mt | 454 | 453 |
| 69 | Fornix | f | 582 | 581 |
| 70 | Lateral mammillary nucleus | LM | 876 | 875 |
| 71 | Medial mammillary nucleus, medial part | MM | 772 | 771 |
| Medial mammillary nucleus, lateral part | ML | |||
| 72 | Medial mammillary nucleus, median part | MnM | 970 | 969 |
| 73 | Mammillary recess of the 3rd ventricle | MRe | 972 | 971 |
| 74 | Arcuate hypothalamic nucleus, medial posterior part | ArcMP | 512 | 511 |
| Arcuate hypothalamic nucleus, lateroposterior part | ArcLP |
IndexL is the integer index value for the structure on the left hemisphere. And IndexR is the integer index value for the corresponding structure on the right hemisphere.
APPENDIX B.
Table APPENDIX B.
Detailed information of the 66 structures included in the rat atlas by Paxinos and Watson
| Structures | IndexL a | IndexR a | |||
|---|---|---|---|---|---|
| Cerebrum | Cerebral cortex | Olfactory bulb | 52 | 51 | |
| Limbic system | Cingulate gyrus | 54 | 53 | ||
| Retrosplenial cortex | 56 | 55 | |||
| Piriform cortex | 58 | 57 | |||
| Bed nucleus of stria terminalis | 60 | 59 | |||
| Tenia tecta | 62 | 61 | |||
| Septal area | 64 | 63 | |||
| Parietal lobe | Parietal cortex posterior area | 66 | 65 | ||
| Sensory cortex | 68 | 67 | |||
| Parietal association cortex | 70 | 69 | |||
| Frontal lobe | Prelimbic cortex | 72 | 71 | ||
| Orbital cortex | 74 | 73 | |||
| Frontal association cortex | 76 | 75 | |||
| Dorsal peduncular cortex | 78 | 77 | |||
| Frotal cortex area | 80 | 79 | |||
| Infralimbic cortex | 82 | 81 | |||
| Motor cortex | 84 | 83 | |||
| Occipital lobe | Visual cortex | 86 | 85 | ||
| Temporal lobe | Auditory cortex | 88 | 87 | ||
| Olfactory cortex | 90 | 89 | |||
| Temporal association cortex | 92 | 91 | |||
| Insular cortex | 94 | 93 | |||
| Fiber | Corpus callosum | 96 | 95 | ||
| Anterior commissure | 98 | 97 | |||
| Posterior commissure | 100 | 99 | |||
| Olfactory tract | 102 | 101 | |||
| Deep structure | Olfactory tubercle | 104 | 103 | ||
| Accumbens nucleus | 106 | 105 | |||
| Striatum | 108 | 107 | |||
| Amygdaloid body | 110 | 109 | |||
| Claustral layer | 112 | 111 | |||
| Hippocampus | 114 | 113 | |||
| Dentate gyrus | 116 | 115 | |||
| Infracerebellar | 118 | 117 | |||
| Subthalamic nucleus | 120 | 119 | |||
| Substantia nigra | 122 | 121 | |||
| Red nucleus | 124 | 123 | |||
| Interbrain | Dorsal thalamus | Midline nucleus group | 126 | 125 | |
| Anterior nucleus group | 128 | 127 | |||
| Medial nucleus group | 130 | 129 | |||
| Lateral nucleus group | 132 | 131 | |||
| Pineal gland | 134 | 133 | |||
| Hypothalamus | Preoptic region | 136 | 135 | ||
| Supraoptic region | 138 | 137 | |||
| Tuberal region | 140 | 139 | |||
| Mammillary region | 142 | 141 | |||
| Inferior colliculus | 144 | 143 | |||
| Cerebellum | Anterior lobel of cerebellum | 146 | 145 | ||
| Posterior lobel of cerebellum | 148 | 147 | |||
| Flocculonodular lobe | 150 | 149 | |||
| Cerebellar nucleus | 152 | 151 | |||
| Interstitial nucleus | 154 | 153 | |||
| Brain stem | Medulla oblongata | 156 | 155 | ||
| Pons | Basilar part of pons | 158 | 157 | ||
| Tegmentum of pons | 160 | 159 | |||
| Mesencephalon | Superior colliculus | 162 | 161 | ||
| Inferior colliculus | 164 | 163 | |||
| Tegmentum of midbrain | 166 | 165 | |||
| Cerebral peduncle | 168 | 167 | |||
| Pretectal area | 170 | 169 | |||
| Periaqueductal gray matter | 172 | 171 | |||
| Ventricle | 3rd ventricle | 174 | 173 | ||
| 4th ventricle | 176 | 175 | |||
| Blood vessel | Middle cerebral artery | 178 | 177 | ||
| Blood vessel | 180 | 179 | |||
| Internal capsule | Capsule | 182 | 181 | ||
IndexL is the integer index value for the structure on the left hemisphere. And IndexR is the integer index value for the corresponding structure on the right hemisphere.
APPENDIX C.
The following volumetric and spatial correspondence measures were calculated between the canonical brain (
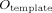 ) and each individual normalized image (
) and each individual normalized image (
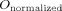 ):
):
(1) JS = Jaccard similarity as in Eq. (A1) (the optimal value is 100%);
| (A1) |
(2) RV = the relative error on volume as in Eq. (A2) (the optimal value is 0%);
| (A2) |
(3) FP = the proportions of false‐positive as in Eq. (A3);
| (A3) |
(4) FN = the proportions of false‐negative as in Eq. (A4).
| (A4) |
REFERENCES
- Alexander GE, Chen K, Aschenbrenner M, Merkley TL, Santerre‐Lemmon LE, Shamy JL, Skaggs WE, Buonocore MH, Rapp PR, Barnes CA ( 2008): Age‐related regional network of magnetic resonance imaging gray matter in the rhesus macaque. J Neurosci 28: 2710–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ ( 1999): Nonlinear spatial normalization using basis functions. Hum Brain Mapp 7: 254–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black KJ, Koller JM, Snyder AZ, Perlmutter JS ( 2001a): Template images for nonhuman primate neuroimaging. II. Macaque. NeuroImage 14: 744–748. [DOI] [PubMed] [Google Scholar]
- Black KJ, Snyder AZ, Koller JM, Gado MH, Perlmutter JS ( 2001b): Template images for nonhuman primate neuroimaging. I. Baboon. NeuroImage 14: 736–743. [DOI] [PubMed] [Google Scholar]
- Chupin M, Mukuna‐Bantumbakulu AR, Hasboun D, Bardinet E, Baillet S, Kinkingnehun S, Lemieux L, Dubois B, Garnero L ( 2007): Anatomically constrained region deformation for the automated segmentation of the hippocampus and the amygdala: Method and validation on controls and patients with Alzheimer's disease. NeuroImage 34: 996–1019. [DOI] [PubMed] [Google Scholar]
- Devanand DP, Michaels‐Marston KS, Liu X, Pelton GH, Padilla M, Marder K, Bell K, Stern Y, Mayeux R ( 2000): Olfactory deficits in patients with mild cognitive impairment predict Alzheimer's disease at follow‐up. Am J Psychiatry 157: 1399–1405. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Ashburner J, Frith CD, Pline J‐B, Heather JD, Frachowiak RSJ ( 1995a): Spatial registration and normalization of images. Hum Brain Mapp 2: 89–165. [Google Scholar]
- Friston KJ, Ashburner JT, Kiebel SJ, Nichols TE, Penny WD ( 2007): Statistical Parametric Mapping: The Analysis of Functional Brain Images. London: Academic Press. [Google Scholar]
- Gonzalez RC, Woods RE, Eddins SL ( 2004): Digital Image Processing Using MATLAB. Beijing, China: Publishing house of electronics industry. pp 305–307. [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS ( 2001): A voxel‐based morphometric study of ageing in 465 normal adult human brains. NeuroImage 14( 1 Part 1): 21–36. [DOI] [PubMed] [Google Scholar]
- Jaccard P ( 1912): The distribution of the flora in the alpine zone. New Phytol 11: 37–50. [Google Scholar]
- Keller SS, Wilke M, Wieshmann UC, Sluming VA, Roberts N ( 2004): Comparison of standard and optimized voxel‐based morphometry for analysis of brain changes associated with temporal lobe epilepsy. NeuroImage 23: 860–868. [DOI] [PubMed] [Google Scholar]
- Lancaster JL, Rainey LH, Summerlin JL ( 1997): Automated labeling of the human brain: A preliminary report on the development and evaluation of a forward transform method. Human brain mapping 5: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Chen K, Alexander GE, He J, Valla J, Galons J‐P, Hauss‐Wegrzyniak B, Reiman EM ( 2003): Construction of mouse brain MRI templates using SPM99. Modeling and control in biomedical systems. pp 113–118. [Google Scholar]
- Longa EZ, Weinstein PR, Carlson S, Cummins R ( 1989): Reversible middle cerebral artery occlusion without craniectomy in rats. Stroke 20: 84–91. [DOI] [PubMed] [Google Scholar]
- Murugavel M, Sullivan JM Jr ( 2009): Automatic cropping of MRI rat brain volumes using pulse coupled neural networks. NeuroImage 45: 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie B, Hui J, Wang L, Chai P, Gao J, Liu S, Zhang Z, Shan B, Zhao S ( 2010): Automatic method for tracing regions of interest in rat brain magnetic resonance imaging studies. J Magn Reson Imaging 32: 830–835. [DOI] [PubMed] [Google Scholar]
- Pawela CP, Hudetz AG, Ward BD, Schulte ML, Li R, Kao DS, Mauck MC, Cho YR, Neitz J, Hyde JS ( 2008): Modeling of region‐specific fMRI BOLD neurovascular response functions in rat brain reveals residual differences that correlate with the differences in regional evoked potentials. NeuroImage 41: 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C ( 1998): The Rat Brain in Stereotaxic Coordinates, 4th ed. San Diego: Academic Press. [Google Scholar]
- Paxinos G, Watson C ( 2005): The Rat Brain in Stereotaxic Coordinates, 5th ed. New York: Academic Press. [Google Scholar]
- Peters JM, Hummel T, Kratzsch T, Lotsch J, Skarke C, Frolich L ( 2003): Olfactory function in mild cognitive impairment and Alzheimer's disease: An investigation using psychophysical and electrophysiological techniques. Am J Psychiatry 160: 1995–2002. [DOI] [PubMed] [Google Scholar]
- Poirier C, Vellema M, Verhoye M, Van Meir V, Wild JM, Balthazart J, Van Der Linden A ( 2008): A three‐dimensional MRI atlas of the zebra finch brain in stereotaxic coordinates. NeuroImage 41: 1–6. [DOI] [PubMed] [Google Scholar]
- Rodionov R, Chupin M, Williams E, Hammers A, Kesavadas C, Lemieux L ( 2009): Evaluation of atlas‐based segmentation of hippocampi in healthy humans. Magn Reson Imaging 27: 1104–1109. [DOI] [PubMed] [Google Scholar]
- Rorden C, Brett M ( 2000): Stereotaxic display of brain lesions. Behav Neurol 12: 191–200. [DOI] [PubMed] [Google Scholar]
- Rubins DJ, Melega WP, Lacan G, Way B, Plenevaux A, Luxen A, Cherry SR ( 2003): Development and evaluation of an automated atlas‐based image analysis method for microPET studies of the rat brain. NeuroImage 20: 2100–2118. [DOI] [PubMed] [Google Scholar]
- Sawiak SJ, Wood NI, Williams GB, Morton AJ, Carpenter TA ( 2009): Voxel‐based morphometry in the R6/2 transgenic mouse reveals differences between genotypes not seen with manual 2D morphometry. Neurobiol Dis 33: 20–27 [DOI] [PubMed] [Google Scholar]
- Schubert CR, Carmichael LL, Murphy C, Klein BE, Klein R, Cruickshanks KJ ( 2008): Olfaction and the 5‐year incidence of cognitive impairment in an epidemiological study of older adults. J Am Geriatr Soc 56: 1517–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz AJ, Danckaert A, Reese T, Gozzi A, Paxinos G, Watson C, Merlo‐Pich EV, Bifone A ( 2006): A stereotaxic MRI template set for the rat brain with tissue class distribution maps and co‐registered anatomical atlas: Application to pharmacological MRI. NeuroImage 32: 538–550. [DOI] [PubMed] [Google Scholar]
- Schwarz AJ, Gozzi A, Bifone A ( 2009): Community structure in networks of functional connectivity: Resolving functional organization in the rat brain with pharmacological MRI. NeuroImage 47: 302–311. [DOI] [PubMed] [Google Scholar]
- Schweinhardt P, Fransson P, Olson L, Spenger C, Andersson JL ( 2003): A template for spatial normalisation of MR images of the rat brain. J Neurosci Methods 129: 105–113. [DOI] [PubMed] [Google Scholar]
- Talairach P, Tournoux J ( 1988): A Stereotactic Coplanar Atlas of the Human Brain. Stuttgart: Thieme. [Google Scholar]
- Tzourio‐Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M ( 2002): Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single‐subject brain. NeuroImage 15: 273–89. [DOI] [PubMed] [Google Scholar]
- Zhilkin P, Alexander ME ( 2004): Affine registration: A comparison of several programs. Magn Reson Imaging 22: 55–66. [DOI] [PubMed] [Google Scholar]


