Abstract
Primary central nervous system lymphoma (PCNSL) is a rare variant of non-Hodgkin lymphoma that is confined to the central nervous system. Biologic studies of PCNSL are challenging to conduct because the disease is rare and available tissue material is sparse. However, in recent years there has been progress in the understanding of PCNSL biology, largely as the result of multicenter studies using modern molecular techniques. Recent studies may improve insight into the pathogenesis of PCNSL and increase the chances of identifying prognostic factors and novel therapeutic targets. This review discusses recent advances in PCNSL biology, including immunologic and genetic risk factors, and focuses on the molecular alterations important in central nervous system lymphomagenesis.
Introduction
Primary central nervous system lymphoma (PCNSL) is a rare and aggressive form of extranodal non-Hodgkin lymphoma (NHL) involving the brain, leptomeninges, or eyes [1]. Unlike other forms of NHL, relatively little is known about the biology of PCNSL. Although insight into the biology of all forms of NHL may be achieved with studies of extraneural lymphomas, it is becoming increasingly evident that PCNSL is associated with a unique set of biologic, clinical, and therapeutic features. Recent studies have identified specific molecular alterations in PCNSL, and a “central nervous system (CNS) signature” of the disease is slowly emerging. These discoveries likely will lead to the development of targeted therapeutics in the near future. Currently, a limited number of molecular markers can be used to predict prognosis in PCNSL. The greatest challenge for researchers continues to be the paucity of archival or fresh frozen tissue available for research purposes, because this disease is commonly diagnosed by stereotactic brain biopsy and the tissue is often consumed by the diagnostic process.
Epidemiology
Although PCNSL was considered extremely rare several decades ago, its incidence increased nearly threefold between 1973 and 1984 [2]. However, recent data suggest that PCNSL incidence rates may be stabilizing or declining slightly [3,4]. This tumor accounted for 3.1% of all primary brain tumors diagnosed in the United States from 1998 to 2002 [5]. The demographics of immunocompetent and immunocompromised patients with PCNSL differ. Immunocompetent patients who develop PCNSL are predominantly older adults [1], and although PCNSL tends to affect males more than females in both groups, this difference is more pronounced in the immunocompromised population (male-to-female ratio, 7.38:1) than in the immunocompetent population (1.35:1). PCNSL incidence peaks at age 57 years in immunocompetent populations, compared with a peak range of 31 to 35 years in immunocompromised patients [6,7].
Immunologic risk factors
Congenital or acquired immunodeficiency is the only established risk factor for PCNSL. The HIV pandemic is the primary factor responsible for the increase in PCNSL incidence. In fact, persons infected with HIV have a 3600-fold higher risk of PCNSL compared with the general population [8]. HIV-related PCNSL is associated with a reduction in the number of circulating CD4+ cells, a factor linked to patient survival [9]. With the introduction of highly active antiretroviral therapy, the proportion of HIV-infected persons with CD4+ cell counts less than 50 cells/mm3 has declined, and this has correlated with a reduction in PCNSL incidence in this population.
An increased PCNSL risk has been observed in organ transplant recipients receiving immunosuppressive drugs [10] and in some autoimmune and immunomodulating diseases, as well as in patients with prior malignancies, implicating immune dysregulation as a risk factor for PCNSL [1].
Genetic risk factors
Homocysteine and folate metabolism is closely linked to DNA methylation and contributes substantially to preservation of DNA integrity. Consequently, genetic polymorphisms that influence homocysteine and folate metabolism are associated with different types of cancer, including NHL, acute leukemias, and colorectal cancer. Methionine synthase (MS) is an important enzyme in folate metabolism, and the MS missense dimorphism c.2756A > G (D919G) is associated with a lower risk of developing colorectal cancer and systemic NHL. Linnebank et al. [11] analyzed 31 cases of PCNSL and concluded that the MS c.2756A > G (G allele) also may protect against PCNSL.
Pathology and Immunopathogenesis
Immunopathologic characterization of PCNSL and tissue-based genetic studies of PCNSL have been limited by the availability of tissue for research purposes. Additionally, many patients receive corticosteroid treatment before tissue sampling. Corticosteroids promote necrosis of malignant lymphoid cells, leading to changes in tumor architecture. This often prevents pathologists from making an accurate histopathologic diagnosis and limits the amount of fresh, “untreated” tumor tissue available for molecular analysis.
PCNSL diffusely infiltrates brain parenchyma in a uniquely angiocentric pattern. The tumor is composed of immunoblasts or centroblasts with a predilection for blood vessels, resulting in lymphoid clustering around small cerebral vessels. Reactive T-cell infiltrates also are present to varying degrees either in the form of scattered lymphocytes or perivascular cuffing occurring alone or interposed between the vessel wall and malignant cells (reactive perivascular T-cell infiltrate [RPVI]), making it difficult for the pathologist to discriminate between PCNSL versus a reactive process. Relatively high percentages of activated cytotoxic T lymphocytes (CTLs) have been detected in CNS lymphomas as compared with extraneural lymphoma, yet the antitumor effect of these CTLs may be impaired by an escape mechanism common to aggressive B-cell lymphomas of the brain. In particular, CTL killing action requires HLA class I and class II molecule expression by target cells. When expression of these two molecules is lost, an efficient antitumor response cannot be mounted. One study of HLA I and HLA II expression suggests that loss of HLA expression provides a strong growth advantage for lymphoma cells and is much more common in B-cell lymphomas of the CNS than in extraneural B-cell lymphomas [12].
Diffuse large B-cell lymphoma (DLBCL) is the most common type of systemic lymphoma, comprising approximately 30% of all NHL cases worldwide. DLBCL accounts for 85% of PCNSL cases. The other 15% consists of low-grade B-cell lymphomas (mainly marginal zone B-cell lymphomas arising from the meninges), Burkitt’s lymphomas, and T-cell lymphomas. Systemic DLBCL encompasses a heterogeneous group of aggressive B-cell lymphomas [13]. These tumors arise from B cells arrested at a certain differentiation stage at which the neoplastic transformation occurs [14]. Based on gene expression profiling studies, two distinct groups of DLBCL have been described: germinal center (GC)-like and non–GC-like. The non–GC-like group can be further subdivided into activated B-cell (ABC)-like and “type 3” subtypes. These two groups (GC-like and non–GC-like) originate from distinct B-cell counterparts and employ distinct mechanisms of malignant transformation [13]. Notably, the clinical outcome for GC-like DLBCL is more favorable compared with that of non–GC-like DLBCL. Normal GC B cells have been implicated as the likely origin for GC-like DLBCL, whereas in non–GC-like DLBCL, the ABC-like subtype appears to derive from a post-GC stage such as memory B cells or plasma cells [15,16]. GC-like DLBCL tumors are marked by characteristic genetic variations that distinguish them from non–GC-like DLBCL. Somatic mutations in the variable (V) region of immunoglobulin (Ig) genes are commonly used as markers of passage through the GC, because normal pregerminal center lymphocytes harbor unmutated Ig genes. Also, the presence of intraclonal heterogeneity in Ig gene mutations is regarded as a marker of ongoing somatic mutations that occur almost exclusively in the GC and thus is used as a marker of GC origin [13,17].
In contrast to extraneural DLBCL, PCNSL is poorly understood in terms of its precise histogenetic origin and molecular pathogenesis. However, recent immunohistochemical studies and gene profiling experiments suggest that PCNSL is derived from GC B cells. Although there is no evidence of GC formation in the brain, the high levels of accumulated somatic mutations, BCL-6 gene mutations, and ongoing hypermutation (features typically observed in cells with transition through the GC) are observed in tumor tissue from immunocompetent PCNSL patients. These results suggest a GC origin for PCNSL [14,18,19]. Braaten et al. [20] investigated the histogenetic origin of PCNSL with respect to the stage of B-cell differentiation. Within a cohort of 33 immunocompetent patients with PCNSL, 79% of tumors expressed bcl-6, a protein primarily found in B lymphocytes in the GC stage of development. A zinc-finger transcriptional repressor encoded by the BCL-6 gene, bcl-6 promotes cell proliferation and blocks differentiation. Although expression of bcl-6, which is highly associated with the GC, supports the theory that the cell of origin in PCNSL is related to the GC stage of B-cell differentiation, only 19% of tumor samples in this study displayed a definite GC-like phenotype, as denoted by coexpression of two GC antigens, bcl-6 and CD10, combined with an absence of the post-GC antigens, vs38c and CD138. However, 10 additional bcl-6–positive cases were CD10 negative but lacked CD44 and vs38c as well, possibly suggesting late GC differentiation. Combining these two categories, 50% of the PCNSL cases in these series had either a definite or possible GC-like phenotype. All but one tumor expressed MUM-1, a transcriptional factor involved in differentiation from GC to plasma cells and a generally accepted marker of the ABC-like phenotype. These findings suggest a late GC or post-GC origin for the PCNSL cases studied. Furthermore, seven tumors were bcl-6–/CD10–, consistent with a non-GC immunophenotype. True plasmacytic differentiation, as evidenced by membrane staining for CD138, was not observed, and cytoplasmic staining for vs38c was distinctly rare. Taken together, these immunophenotypic findings suggest a GC or possibly late GC stage of B-cell differentiation for most cases of PCNSL (Table 1).
Table 1.
Immunohistochemistry for germinal center–related antigens
| Antigen |
Positive PCNSL
cases, n/n (%) |
Comment |
|---|---|---|
| bcl-6 | 26/33 (79) | The bcl-6 protein is a zinc-finger transcriptional repressor encoded by the BCL-6 gene |
| CD10 | 6/32 (19) | CD10 is a marker that reflects GC origin in reactive lymphoid tissue and lymphomas |
| MUM1 | 31/32 (97) | MUM1 is a late GC/early post-GC marker |
| CD44 | 11/31 (35) | CD44 is expressed most strongly by post-GC mantle zone cells |
| vs38c | 4/33 (12) | The presence of vs38c and CD138 denotes plasmacytic and/or post-GC differentiation |
| CD138 | 0/32 (0) | The presence of vs38c and CD138 denotes plasmacytic and/or post-GC differentiation |
| BCL-2 | 27/29 (93) | BCL-2 protein expression prevents cellular apoptosis and is downregulated by normal GC cells |
GC—germinal center; PCNSL—primary central nervous system lymphoma.
(From Braaten et al. [20].)
Studies of Viral Pathogenesis
The Epstein-Barr virus (EBV), a lymphotropic virus involved in malignant B-cell transformation, is involved in PCNSL pathogenesis in immunocompromised patients; PCNSL in immunocompromised patients is strongly associated with EBV infection [21]. In the immunocompromised state, chronic immune stimulation by EBV may lead to B-cell immortalization and, ultimately, malignant transformation [22]. Under natural immune conditions, proliferation of EBV-infected B cells is suppressed by normal T-cell immunity. With diminished T-cell function, EBV-infected clones may progress to malignant lymphoma [23,24]. In addition, EBV infection correlates with CNS tropism of lymphoma. The risk of CNS tumor dissemination in EBV-positive, AIDS-related systemic lymphoma is 10 times higher than in EBV-negative cases [25]. Cerebrospinal fluid (CSF) EBV titers are higher in AIDS patients with PCNSL compared with those in AIDS patients with systemic lymphomas, suggesting a distinct viral pathogenesis for PCNSL in this patient population. Meeker et al. [26] demonstrated that tumor tissue in all PCNSL cases is EBV positive, and the EBV CSF titer is widely used as a diagnostic screen in the immunocompromised patient population [27]. Conversely, EBV does not appear to play a major role in PCNSL pathogenesis in immunocompetent patients.
Del Valle et al. [28] recently reported that JC virus might play a pathogenetic role in PCNSL. Researchers identified JC virus DNA by gene amplification and immunohistochemistry in B lymphocytes and mononuclear cells in brain perivascular spaces. Of 27 PCNSL tissue samples studied, 22 (81%) were positive for JC virus DNA. Twelve patients were HIV positive, but no cases of progressive multifocal leukoencephalopathy (PML) were described in the studied cohort. Importantly, coexpression of EBV latent membrane protein (LMP1) and JC virus antigen was identified in the same cells, suggesting a possible pathogenetic role for JC virus in PCNSL development.
Although other viruses, namely human herpes simplex type 6 and type 8 and simian virus 40, have been implicated in PCNSL pathogenesis, definitive evidence regarding the causative role of these viruses is lacking [29].
Genetic Studies
Gene expression profiling studies indicate that PCNSL is a distinct biologic subtype of DLBCL. Although PCNSL can be segregated into the same three classes of DLBCL (GC B cell, activated B cell, type 3 B cell) as systemic NHL, there are critical differences. In one study involving 35 frozen tumor specimens from PCNSL patients, approximately 100 genes had at least a twofold level of differential expression between PCNSL, systemic DLBCL, and nonneoplastic brain. The gene expression signature of PCNSL includes genes involved in B-cell differentiation, proliferation, apoptosis, and cytokine signaling [30]. Additional genes associated with the unfolded protein response (UPR) pathway, which is important in the stress response crucial for tumor adaptation to glucose deprivation (X-box binding protein 1 [XBP-1]), also were identified in PCNSL [31••,32]. Proto-oncogenes MYC and PIM1 were found to be highly expressed in PCNSL, and there is evidence of somatic mutations in these genes [32].
B-cell growth factor interleukin-4 (IL-4), which may function in an autocrine or paracrine manner, was reported to be highly expressed in tumor tissue and endothelium derived from PCNSL [31••]. Identification of IL-4 in the endothelium may help explain why PCNSLs grow in an angiocentric pattern. Transcription factor STAT6 and specifically its activated form also were implicated in lymphoma pathogenesis. STAT6 is highly expressed in tumor cells and endothelium and is a mediator of IL-4–dependent gene expression. Tumors with STAT6 overexpression are associated with aggressive growth, early progression, and shorter survival in patients treated with methotrexate [31••]. Thus, STAT6 deserves further study as a potential novel biomarker for prognostic determination at the time of PCNSL diagnosis [31••,33•].
Recently, Tun et al. [34••] published an important contribution to our knowledge about the molecular biology of PCNSL. The authors performed a genome-wide analysis comparing gene expression between PCNSL and non-CNS lymphomas (nodal and extranodal). Using pathway analysis (SigPathway method), these investigators studied differential expression of multiple extracellular matrix (ECM) and adhesion-related pathways. The most significantly upregulated gene set found in their analysis was the ECM receptor pathway, suggesting that the interaction between the brain microenvironment and lymphoma cells is of great importance for PCNSL. Upregulation of two important ECM-related genes, osteopontin (SPP1) and chitinase 3–like 1 (CHI3L1), was demonstrated at the protein level. Osteopontin (SPP1, OPN) has been implicated in many cellular functions, including CNS tropism, B-cell migration and activation, and lymphoproliferation. Moreover, SPP1 overexpression is associated with more aggressive tumor behavior, greater propensity for metastatic behavior, and poor prognosis. SPP1 was found to be upregulated in other CNS diseases, including multiple sclerosis, glioblastoma, and astrocytomas [35,36]. It appears that SPP1 plays an important role in CNS disease pathogenesis. Another gene with differential expression in PCNSL versus non-CNS types is CHI3L1. This ECM-related gene plays an important role in cancer cell proliferation, differentiation, survival, migration, metastasis, and angiogenesis. High serum levels of CHI3L1 products were found in patients with metastatic cancers [37•]. This gene was found to be associated with poor prognosis in patients with breast cancer [38].
Tun et al. [34••] studied 13 PCNSL, 11 nodal DLBCL, and 19 extranodal DLBCL samples. Unique expression of ECM and adhesion-related pathways was identified only in PCNSL. Interestingly, adhesion-related genes, when analyzed individually, did not differ in their expression between PCNSL and the other two groups analyzed. However, when genes were pooled and analyzed using SigPathway methodology, they were found to be overexpressed in PCNSL. Findings from this study imply that the brain microenvironment might play a critical role in PCNSL pathogenesis. Individual genes might be responsible for different stages in the transformation of a B cell into a malignant form. SPP1 and DDR1 (ECM/adhesion gene) may be important for CNS tropism in PCNSL. CXCL13 (B-cell–attracting chemokine [BCA]) and SPP1 likely are associated with B-cell migration and SPP1, TCL1A (a gene important in lymphatic leukemias and lymphomas), and CHI3L1 with B-cell proliferation. This study was the first to present an in-depth genomic analysis differentiating PCNSL from non-CNS DLBCL and will serve as the platform for further studies, ultimately leading to identification of the true CNS signature of PCNSL.
Data regarding expression of the antiapoptotic protein survivin, platelet-derived growth factor A (PDGF-A), and PDGF receptor-α (PDGFR-α) in PCNSL were recently reported. Forty-four tumor specimens were analyzed immunohistochemically. Eighty-four percent of the samples expressed survivin, 36% PDGF-A, and 77% PDGFR-α. However, the expression of these proteins did not predict survival in this study cohort [39•]. Thus, survivin’s role in PCNSL pathology remains unclear. This protein has been identified in many human cancers, and its high expression has been associated with shorter survival and increased resistance to therapy [40]. Although the potential role of survivin and PDGF in PCNSL biology needs further study, it appears that their utility as prognostic markers may be limited.
In a study of 21 PCNSL patients, tumor specimens were analyzed for the presence of mutations in the PRDM1 gene and alterations in the expression pattern of the PRDM1 protein. This gene is important in regulating terminal B-cell differentiation into plasma cells and has been implicated as a tumor suppressor in systemic DLBCL. Direct sequencing of all coding exons of the PRDM1 gene identified mutations associated with abrogation of PRDM1 protein expression in 4 of 21 PCNSL cases (19%). The authors hypothesized that alterations in the function of this gene may lead to lymphomagenesis by disrupting terminal B-cell differentiation [41•].
Montesinos-Rongen et al. [42] investigated whether aberrant somatic hypermutation in growth regulatory genes may play a role in PCNSL development. They observed extremely high mutation frequencies for immunoglobulin H (IgH) genes in PCNSL. The investigators also found that PCNSL exhibited aberrant somatic hypermutation with involvement of four proto-oncogenes: PAX5, PIM1, c-MYC, and RhoH/TTF. These genes have a role in B-cell development and differentiation as well as in the regulation of proliferation and apoptosis [42]. Although mutation frequencies reported for IgH genes are much higher (60-fold) than mutation frequencies for PAX5, PIM1, c-MYC, and RhoH/TTF, the involvement of these four genes in PCNSL pathogenesis is potentially significant. Moreover, individual comparison of these genes in PCNSL and extraneural DLBCL revealed that their mean mutation frequencies are two- to fivefold higher in PCNSL [42]. Such high mutation frequencies may arise during a prolonged interaction of the tumor cell (or its precursor) in the GC microenvironment [19].
The p16INK4a gene is frequently inactivated by either homozygous deletion (40%–50%) or 5′-CpG hypermethylation (15%–30%) in PCNSL patients [43]. Inactivation of p14ARF and p16INK4a genes by homozygous deletion or promoter hypermethylation may represent an important step in the molecular pathogenesis of PCNSL. The p14ARF gene, for example, normally induces growth arrest and stabilizes p53 protein in the cell nucleus. Its deletion has been reported in glioblastomas and systemic NHL. p14ARF and p16INK4a are frequently co-deleted in human neoplasms; moreover, mice lacking the murine homologue of p14ARF develop a variety of tumors, including lymphomas, sarcomas, and gliomas [44–46]. In contrast, mutations in the TP53 gene have been observed in only a small proportion of PCNSL specimens.
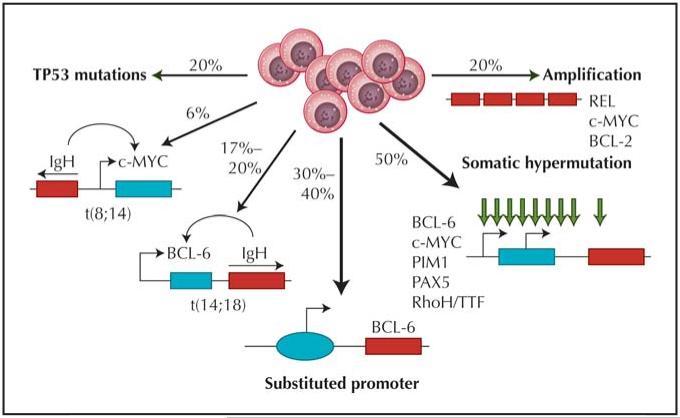
Comparative genomic hybridization has identified other potentially important genetic lesions in PCNSL. Gains have been detected on chromosome 12 and the long arms of chromosomes 1, 7, and 18. A reduced number of DNA copies was detected on chromosome 6 and the short arms of chromosomes 17 and 18 [33•]. Gain on chromosome 12 appears to be the most common chromosomal alteration, specifically in the 12q region harboring MDM2, CDK4, and GLI1 [45–47]. Figure 1 summarizes major genetic events potentially important in the pathogenesis of DLBCL, including PCNSL.
Figure 1.
Model for diffuse large B-cell lymphoma pathogenesis. Only the most common genetic lesions and their frequencies are shown. Genetic lesions more specific for primary central nervous system lymphoma (PCNSL) include somatic hypermutations as well as chromosomal translocations, especially those involving the IgH and BCL-6 genes. TP53 mutations are rare in PCNSL. (Adapted from Lossos [13].)
Specific chromosomal translocations observed frequently in PCNSL tumor specimens involve the IgH and BCL-6 genes. Such translocations are thought to originate by recombination mechanisms following double-strand DNA breaks; these breaks may be generated during class switch recombinations and somatic hypermutation of the Ig genes. Montesinos-Rongen et al. [48] have proposed that the resulting juxtaposition of some oncogenes with regulatory elements may lead to oncogene activation. BCL-6 translocations in PCNSL appear similar to translocations found in other extranodal forms of DLBCL and are thought to be pathogenetically relevant as well. In addition to the genetic alterations discussed earlier, there are differences in the expression of chemokines, chemokine receptors, cytokines, and cytokine receptors in extranodal lymphomas such as PCNSL. Compared with other extranodal lymphomas, PCNSL exhibits significantly higher expression of Th1-type cytokine IL-2 and Th2-type cytokine IL-13 [49]. It was initially believed that CXCL13 (BCA-1) was exclusively expressed in malignant lymphocytes and vascular endothelium within the CNS; however, further studies demonstrated that BCA-1 expression was not specific for CNS lymphomas and may also be found in extraneural lymphomas [49,50].
Proteomic approaches to identify possible new biomarkers of PCNSL in CSF also have been studied [51••]. In one report, investigators identified 80 CSF proteins that were differentially expressed in patients with PCNSL. One of these proteins, antithrombin III (ATIII), is typically localized selectively in tumor vasculature. The presence of ATIII in CSF was highly suggestive of PCNSL diagnosis in this study. Determining ATIII concentration by enzyme-linked immunosorbent assay was found to be significantly more accurate (> 75% sensitivity; > 98% specificity) than cytology in identifying cancer. Measuring CSF ATIII levels may potentially enhance the ability to diagnose PCNSL.
Biomarkers of prognosis
Lymphomas thought to be derived from GC cells, such as follicular lymphoma, express BCL-6, whereas lymphomas derived from naïve B cells, like chronic lymphocytic leukemia or mantle cell lymphoma, do not [52,53]. Consistent with other studies demonstrating a favorable prognostic effect of BCL-6 expression in patients with extraneural DLBCL, Braaten et al. [20] demonstrated that overexpression of BCL-6 in PCNSL patients was associated with improved survival (101 months) compared with patients whose tumors did not express BCL-6 (14.7 months). The prognostic significance of BCL-6 in CNS lymphoma was subsequently corroborated by Levy et al. [54•] and Lin et al. [55]. Another potential prognostic marker is the presence of RPVI in PCNSL. In a large multicenter retrospective series of 100 PCNSL cases, the presence of RPVI significantly correlated with a better outcome [56•]; this feature highlights the tumor microenvironment’s potential role in PCNSL pathogenesis.
Analysis of chromosomal imbalances by comparative genomic hybridization has demonstrated frequent chromosome 6q deletions in PCNSL (60% of cases) [57]. The incidence of chromosome 6q deletions is higher in PCNSL than in extraneural lymphomas. Patients with loss of heterozygosity (LOH) on 6q have a shorter survival compared with PCNSL patients without LOH on 6q. Further study consisting of deletion mapping of chromosome 6q in PCNSL disclosed a region suspected to harbor a lymphoma-related tumor suppressor gene (6q22-23) [58]. This locus is known to contain the PTPRK gene in humans. Products of this gene are important for regulating cell contact and adhesion [58]. Earlier death was observed in patients with PCNSL showing loss of PTPRK expression compared with those maintaining PTPRK expression; however, this correlation was not statistically significant [58].
In preliminary gene expression studies in PCNSL, Rubenstein et al. [30,31••] demonstrated that differential expression of genes may distinguish between patients with long-term versus short-term survival. Ongoing gene expression profiling studies may yield other markers with specific clinical relevance. For example, as noted previously, STAT6 was implicated as a possible marker of poor prognosis in a subset of patients with PCNSL receiving methotrexate therapy [31••,33•]. Table 2 summarizes selected important genetic events in the biology of PCNSL and their possible prognostic and therapeutic implications.
Table 2.
Selected genetic factors in primary central nervous system lymphoma and their biologic and prognostic significance
| Gene | Biologic effects | Possible clinical significance |
|---|---|---|
| ECM family: SPP1 and CHI3L1 |
CNS tropism, B-cell migration, lymphoproliferation, cell differentiation, migration, angiogenesis, and metastasis |
Aggressive tumor growth, high metastatic potential, poor prognosis |
| STAT6 | Expressed in tumor tissue and endothelium | Overexpression associated with aggressive tumor growth, decreased survival in patients treated with methotrexate |
| PRDM1 | Tumor suppressor gene, regulation of B-cell differentiation | Mutations associated with lymphomagenesis |
| p14 ARF | Induction of cell growth arrest and stabilization of p53 protein |
Deletion or promoter methylation can induce tumor growth |
| PTPRK | Regulation of cell contact and adhesion | Shorter survival in patients with loss of expression |
| BCL-6 | Transcriptional repressor, oncogene, malignant transformation of germinal center B cells |
Overexpression associated with favorable prognosis |
CNS—central nervous system; ECM—extracellular matrix.
Possible therapeutic implications
Based on a better understanding of PCNSL biology, investigators are now in a position to investigate potential therapeutic targets in this distinct form of NHL. For example, elevated expression of regulators of the UPR signaling pathway might suggest a potential therapeutic role for proteosome inhibitors. Expression of IL-4 and several IL-4–dependent genes, including XBP-1, by tumor vasculature as well as by tumor cells in CNS lymphomas also suggests that inhibitors of these molecules may have therapeutic value. Other possible targets include multiple ECM and adhesion-related pathways. It is hoped that the identification of these potential therapeutic targets in PCNSL will lead to future clinical trials of novel agents capable of inhibiting or modulating these pathways.
Conclusions
PCNSL, a rare form of extranodal NHL, is typically represented by a DLBCL that is confined to the nervous system. Our understanding of PCNSL biology has improved over the past few years. Molecular profiling is now possible although not yet widely available. Genetic studies demonstrate that CNS lymphomas exhibit a distinct gene expression profile compared with nodal lymphomas of the same histologic type. Emerging data also indicate that CNS lymphomas are derived from a distinct cell of origin and exhibit a unique immunophenotype. The importance of the brain microenvironment in the malignant transformation of B cells has been implicated. Efforts continue to identify novel tumor biomarkers to possibly facilitate noninvasive diagnosis and identify patients with a better prognosis.
A better biologic understanding of PCNSL will enable the development of customized treatment approaches for this patient population. Until recently, biologic studies of PCNSL were lacking, mainly because of the rarity of the disease and the paucity of available research specimens. However, multicenter collaboration has resulted in the first gene expression profile study in PCNSL, and other studies regarding biomarkers of prognosis have been published recently. Although there is still a large gap between our understanding of other forms of extranodal B-cell lymphomas and PCNSL, it is anticipated that this gap will close in the coming years as multicenter collaboration, tissue preservation methods, and molecular techniques are improved and refined.
Footnotes
Disclosures
Dr. Batchelor has been a consultant for Enzon Pharmaceuticals. Dr. Mrugala has been on the speakers’ bureaus of Schering-Plough and UCB Pharma. No other potential conflicts of interest relevant to this article were reported.
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Pfannl R, Harris FJ. Pathology of primary central nervous system lymphoma and related conditions. In: Batchelor TT, editor. Lymphoma of the Nervous System. Butterworth-Heinemann; Boston: 2004. pp. 29–41. [Google Scholar]
- 2.Eby NL, Grufferman S, Flannelly CM, et al. Increasing incidence of primary brain lymphoma in the US. Cancer. 1988;62:2461–2465. doi: 10.1002/1097-0142(19881201)62:11<2461::aid-cncr2820621135>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 3.O’Neill BP. Epidemiology of PCNSL. In: Batchelor TT, editor. Lymphoma of the Nervous System. Butterworth-Heinemann; Boston: 2004. pp. 43–51. [Google Scholar]
- 4.Kadan-Lottick NS, Skluzacek MC, Gurney JG. Decreasing incidence rates of primary central nervous system lymphoma. Cancer. 2002;95:193–202. doi: 10.1002/cncr.10643. [DOI] [PubMed] [Google Scholar]
- 5.Primary Brain Tumors in the United States 1998–2002. Central Brain Tumor Registry of the United States; Chicago: CBTRUS, Statistical Report; pp. 2005–2006. [Google Scholar]
- 6.Fine HA, Mayer RJ. Primary central nervous system lymphoma. Ann Intern Med. 1993;119:1093–1104. doi: 10.7326/0003-4819-119-11-199312010-00007. [DOI] [PubMed] [Google Scholar]
- 7.Schabet M. Epidemiology of primary CNS lymphoma. J Neurooncol. 1999;43:199–201. doi: 10.1023/a:1006290032052. [DOI] [PubMed] [Google Scholar]
- 8.Coté TR, Manns A, Hardy CR, et al. Epidemiology of brain lymphoma among people with or without acquired immunodeficiency syndrome. AIDS/Cancer Study Group. J Natl Cancer Inst. 1996;88:675–679. doi: 10.1093/jnci/88.10.675. [DOI] [PubMed] [Google Scholar]
- 9.Bashir R, Chamberlain M, Ruby E, Hochberg FH. T-cell infiltration of primary CNS lymphoma. Neurology. 1996;46:440–444. doi: 10.1212/wnl.46.2.440. [DOI] [PubMed] [Google Scholar]
- 10.Ferry J, Harris N. Pathology of post-transplant lymphoproliferative disorders. In: Solez K, Racusen LC, Billingham ME, editors. Pathology and Rejection Diagnosis in Solid Organ Transplantation. Marcel Dekker; New York: 1994. pp. 277–301. [Google Scholar]
- 11.Linnebank M, Schmidt S, Kölsch H, et al. The methionine synthase polymorphism D919G alters susceptibility to primary central nervous system lymphoma. Br J Cancer. 2004;90:1969–1971. doi: 10.1038/sj.bjc.6601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riemersma SA, Oudejans JJ, Vonk MJ, et al. High numbers of tumour-infiltrating activated cytotoxic T lymphocytes, and frequent loss of HLA class I and II expression, are features of aggressive B cell lymphomas of the brain and testis. J Pathol. 2005;206:328–336. doi: 10.1002/path.1783. [DOI] [PubMed] [Google Scholar]
- 13.Lossos IS. Molecular pathogenesis of diffuse large B-cell lymphoma. J Clin Oncol. 2005;23:6351–6357. doi: 10.1200/JCO.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Thompsett AR, Ellison DW, Stevenson FK, Zhu D. V(H) gene sequences from primary central nervous system lymphomas indicate derivation from highly mutated germinal center B cells with ongoing mutational activity. Blood. 1999;94:1738–1746. [PubMed] [Google Scholar]
- 15.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 16.Rosenwald A, Wright G, Chan WC, et al. Lymphoma/Leukemia Molecular Profiling Project: The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 17.Lossos IS, Alizadeh AA, Eisen MB, et al. Ongoing immunoglobulin somatic mutation in germinal center B cell-like but not in activated B cell-like diffuse large cell lymphomas. Proc Natl Acad Sci U S A. 2000;97:10209–10213. doi: 10.1073/pnas.180316097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larocca LM, Capello D, Rinelli A, et al. The molecular and phenotypic profile of primary central nervous system lymphoma identifies distinct categories of the disease and is consistent with histogenetic derivation from germinal center-related B cells. Blood. 1998;92:1011–1019. [PubMed] [Google Scholar]
- 19.Montesinos-Rongen M, Küppers R, Schlüter D, et al. Primary central nervous system lymphomas are derived from germinalcenter B cells and show a preferential usage of the V4-34 gene segment. Am J Pathol. 1999;155:2077–2086. doi: 10.1016/S0002-9440(10)65526-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braaten KM, Betensky RA, de Leval L, et al. BCL-6 expression predicts improved survival in patients with primary central nervous system lymphoma. Clin Cancer Res. 2003;9:1063–1069. [PubMed] [Google Scholar]
- 21.Roychowdhury S, Peng R, Baiocchi RA, et al. Experimental treatment of Epstein-Barr virus-associated primary central nervous system lymphoma. Cancer Res. 2003;63:965–971. [PubMed] [Google Scholar]
- 22.Tanner JE, Alfieri C. The Epstein-Barr virus and post-transplant lymphoproliferative disease: interplay of immunosuppression, EBV, and the immune system in disease pathogenesis. Transpl Infect Dis. 2001;3:60–69. doi: 10.1034/j.1399-3062.2001.003002060.x. [DOI] [PubMed] [Google Scholar]
- 23.Thomas JA, Crawford DH, Burke M. Clinicopathologic implications of Epstein-Barr virus related B cell lymphoma in immunocompromised patients. J Clin Pathol. 1995;48:287–290. doi: 10.1136/jcp.48.4.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penn I. The role of immunosuppression in lymphoma formation. Springer Semin Immunopathol. 1998;20:343–355. doi: 10.1007/BF00838048. [DOI] [PubMed] [Google Scholar]
- 25.Cingolani A, Gastaldi R, Fassone L, et al. Epstein-Barr virus infection is predictive of CNS involvement in systemic AIDS-related non-Hodgkin’s lymphomas. J Clin Oncol. 2000;18:3325–3330. doi: 10.1200/JCO.2000.18.19.3325. [DOI] [PubMed] [Google Scholar]
- 26.Meeker TC, Shiramizu B, Kaplan L, et al. Evidence for molecular subtypes of HIV-associated lymphoma: division into peripheral monoclonal, polyclonal and central nervous system lymphoma. AIDS. 1991;5:669–674. [PubMed] [Google Scholar]
- 27.Rubin N, Chamberlain M. Primary central nervous system lymphoma in acquired immune deficiency syndrome. In: Batchelor TT, editor. Lymphoma of the Nervous System. Butterworth-Heinemann; Boston: 2004. pp. 97–103. [Google Scholar]
- 28.Del Valle L, Enam S, Lara C, et al. Primary central nervous system lymphoma expressing the human neurotropic polyomavirus, JC virus, genome. J Virol. 2004;78:3462–3469. doi: 10.1128/JVI.78.7.3462-3469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montesinos-Rongen M, Besleaga R, Heinsohn S, et al. Absence of simian virus 40 DNA sequences in primary central nervous system lymphoma in HIV-negative patients. Virchows Arch. 2004;444:436–438. doi: 10.1007/s00428-004-1001-9. [DOI] [PubMed] [Google Scholar]
- 30.Rubenstein JL, Shen A, Fridlyand J, et al. Gene expression profile analysis of primary CNS lymphoma: class distinction and outcome prediction. Presented at the Eighth Annual Meeting of the Society for Neuro-Oncology; Keystone, CO. Nov, 2003. pp. 13–16. [Google Scholar]
- 31••.Rubenstein JL, Fridlyand J, Shen A, et al. Gene expression and angiotropism in primary CNS lymphoma. Blood. 2006;107:3716–3723. doi: 10.1182/blood-2005-03-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comprehensive analysis of novel molecular features specific for PCNSL with possible therapeutic and prognostic implications.
- 32.Park HR, Tomida A, Sato S, et al. Effect on tumor cells of blocking survival response to glucose deprivation. J Natl Cancer Inst. 2004;96:1300–1310. doi: 10.1093/jnci/djh243. [DOI] [PubMed] [Google Scholar]
- 33•.Kadoch C, Treseler P, Rubenstein JL. Molecular pathogenesis of primary central nervous system lymphoma. Neurosurg Focus. 2006;21:E1. doi: 10.3171/foc.2006.21.5.2. [DOI] [PubMed] [Google Scholar]
- The authors discuss the current knowledge of the molecular pathogenesis of CNS lymphomas and review the recent advances in gene expression profile analysis and identification of novel prognostic biomarkers.
- 34••.Tun HW, Personett D, Baskerville KA, et al. Pathway analysis of primary central nervous system lymphoma. Blood. 2008;111:3200–3210. doi: 10.1182/blood-2007-10-119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The authors describe a bioinformatics approach to genetic testing in PCNSL. The gene expression signature discovered in this study may represent a true “CNS signature” of PCNSL.
- 35.El-Tanani MK, Campbell FC, Kurisetty V, et al. The regulation and role of osteopontin in malignant transformation and cancer. Cytokine Growth Factor Rev. 2006;17:463–474. doi: 10.1016/j.cytogfr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 36.Chabas D, Baranzini SE, Mitchell D, et al. The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science. 2001;294:1731–1735. doi: 10.1126/science.1062960. [DOI] [PubMed] [Google Scholar]
- 37•.Johansen JS, Jensen BV, Roslind A, Price PA. Is YKL-40 a new therapeutic target in cancer? Expert Opin Ther Targets. 2007;11:219–234. doi: 10.1517/14728222.11.2.219. [DOI] [PubMed] [Google Scholar]
- The authors hypothesize that inhibition of YKL-40 by monoclonal antibodies either directly or toward its receptor may be as efficient a cancer therapy as monoclonal antibodies against HER2, HER1, vascular endothelial growth factor, and CD20.
- 38.Kim SH, Das K, Noreen S, et al. Prognostic implications of immunohistochemically detected YKL-40 expression in breast cancer. World J Surg Oncol. 2007;5:17. doi: 10.1186/1477-7819-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.Karabatsou K, Pal P, Dodd S, et al. Expression of survivin, platelet-derived growth factor A (PDGF-A) and PDGF receptor alpha in primary central nervous system lymphoma. J Neurooncol. 2006;79:171–179. doi: 10.1007/s11060-005-9102-0. [DOI] [PubMed] [Google Scholar]
- This study investigated expression of the antiapoptotic protein survivin, PDGF-A, and PDGFR-α in PCNSL. Prognostic importance was not confirmed.
- 40•.Lossos IS, Czerwinski DK, Alizadeh AA, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 41•.Courts C, Montesinos-Rongen M, Brunn A, et al. Recurrent inactivation of the PRDM1 gene in primary central nervous system lymphoma. J Neuropathol Exp Neurol. 2008;67:720–727. doi: 10.1097/NEN.0b013e31817dd02d. [DOI] [PubMed] [Google Scholar]
- This study investigated the role of PRDM1 in PCNSL. The authors determined that PRDM1 may be a tumor suppressor in some PCNSLs and may contribute to lymphomagenesis by impairing terminal differentiation.
- 42.Montesinos-Rongen M, Van Roost D, Schaller C, et al. Primary diffuse large B-cell lymphomas of the central nervous system are targeted by aberrant somatic hypermutation. Blood. 2004;103:1869–1875. doi: 10.1182/blood-2003-05-1465. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura M, Sakaki T, Hashimoto H, et al. Frequent alterations of the p14(ARF) and p16(INK4a) genes in primary central nervous system lymphomas. Cancer Res. 2001;61:6335–6339. [PubMed] [Google Scholar]
- 44.Kamijo T, Bodner S, van de Kamp E, et al. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–2222. [PubMed] [Google Scholar]
- 45.Rubenstein JL, Treseler P, O’Brien JM. Pathology and genetics of primary central nervous system and intraocular lymphoma. Hematol Oncol Clin North Am. 2005;19:705–717. vii. doi: 10.1016/j.hoc.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura M, Shimada K, Ishida E, Konishi N. Histopathology, pathogenesis and molecular genetics in primary central nervous system lymphomas. Histol Histopathol. 2004;19:211–219. doi: 10.14670/HH-19.211. [DOI] [PubMed] [Google Scholar]
- 47.Harada K, Nishizaki T, Kubota H, et al. Distinct primary central nervous system lymphoma defined by comparative genomic hybridization and laser scanning cytometry. Cancer Genet Cytogenet. 2001;125:147–150. doi: 10.1016/s0165-4608(00)00377-0. [DOI] [PubMed] [Google Scholar]
- 48.Montesinos-Rongen M, Zühlke-Jenisch R, Gesk S, et al. Interphase cytogenetic analysis of lymphoma-associated chromosomal breakpoints in primary diffuse large B-cell lymphomas of the central nervous system. J Neuropathol Exp Neurol. 2002;61:926–933. doi: 10.1093/jnen/61.10.926. [DOI] [PubMed] [Google Scholar]
- 49.Fujii A, Ohshima K, Hamasaki M, et al. Differential expression of chemokines, chemokine receptors, cytokines and cytokine receptors in diffuse large B cell malignant lymphoma. Int J Oncol. 2004;24:529–538. [PubMed] [Google Scholar]
- 50.Smith JR, Braziel RM, Paoletti S, et al. Expression of B-cell-attracting chemokine 1 (CXCL13) by malignant lymphocytes and vascular endothelium in primary central nervous system lymphoma. Blood. 2003;101:815–821. doi: 10.1182/blood-2002-05-1576. [DOI] [PubMed] [Google Scholar]
- 51••.Roy S, Josephson SA, Fridlyand J, et al. Protein biomarker identification in the CSF of patients with CNS lymphoma. J Clin Oncol. 2008;26:96–105. doi: 10.1200/JCO.2007.12.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- This is the first study to attempt proteomic analysis of CSF in patients with PCNSL. The authors reported that CSF yields individual biomarkers with greater sensitivity than CSF cytology for identifying cancer.
- 52.Falini B, Fizzotti M, Pileri S, et al. Bcl-6 protein expression in normal and neoplastic lymphoid tissues. Ann Oncol. 1997;8:101–104. Suppl 2. [PubMed] [Google Scholar]
- 53.Dogan A, Bagdi E, Munson P, Isaacson PG. CD10 and BCL-6 expression in paraffin sections of normal lymphoid tissue and B-cell lymphomas. Am J Surg Pathol. 2000;24:846–852. doi: 10.1097/00000478-200006000-00010. [DOI] [PubMed] [Google Scholar]
- 54•.Levy O, Deangelis LM, Filippa DA, et al. Bcl-6 predicts improved prognosis in primary central nervous system lymphoma. Cancer. 2008;112:151–156. doi: 10.1002/cncr.23149. [DOI] [PubMed] [Google Scholar]
- This study confirmed and supplemented earlier data indicating that Bcl-6 expression is associated with a better prognosis in patients with PCNSL.
- 55.Lin CH, Kuo KT, Chuang SS, et al. Comparison of the expression and prognostic significance of differentiation markers between diffuse large B-cell lymphoma of central nervous system origin and peripheral nodal origin. Clin Cancer Res. 2006;12:1152–1156. doi: 10.1158/1078-0432.CCR-05-1699. [DOI] [PubMed] [Google Scholar]
- 56•.Ponzoni M, Berger F, Chassagne-Clement C, et al. International Extranodal Lymphoma Study Group: Reactive perivascular T-cell infiltrate predicts survival in primary central nervous system B-cell lymphomas. Br J Haematol. 2007;138:316–323. doi: 10.1111/j.1365-2141.2007.06661.x. [DOI] [PubMed] [Google Scholar]
- The authors identified a novel marker of prognosis in PCNSL that can be assessed easily and routinely at diagnosis on histopathologic specimens.
- 57.Weber T, Weber RG, Kaulich K, et al. Characteristic chromosomal imbalances in primary central nervous system lymphomas of the diffuse large B-cell type. Brain Pathol. 2000;10:73–84. doi: 10.1111/j.1750-3639.2000.tb00244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakamura M, Kishi M, Sakaki T, et al. Novel tumor suppressor loci on 6q22-23 in primary central nervous system lymphomas. Cancer Res. 2003;63:737–741. [PubMed] [Google Scholar]