Abstract
We have analyzed the available genome and transcriptome resources from the coelacanth in order to characterize genes involved in adaptive immunity. Two highly distinctive IgW-encoding loci have been identified that exhibit a unique genomic organization, including a multiplicity of tandemly repeated constant region exons. The overall organization of the IgW loci precludes typical heavy chain class switching. A locus encoding IgM could not be identified either computationally or by using several different experimental strategies. Four distinct sets of genes encoding Ig light chains were identified. This includes a variant sigma-type Ig light chain previously identified only in cartilaginous fishes and which is now provisionally denoted sigma-2. Genes encoding α/β and γ/δ T-cell receptors, and CD3, CD4 and CD8 co-receptors also were characterized. Ig heavy chain variable region genes and TCR components are interspersed within the TCR α/δ locus; this organization previously was reported only in tetrapods and raises questions regarding evolution and functional cooption of genes encoding variable regions. The composition, organization and syntenic conservation of the major histocompatibility complex locus have been characterized. We also identified large numbers of genes encoding cytokines and their receptors, and other genes associated with adaptive immunity. In terms of sequence identity and organization, the adaptive immune genes of the coelacanth more closely resemble orthologous genes in tetrapods than those in teleost fishes, consistent with current phylogenomic interpretations. Overall, the work reported described herein highlights the complexity inherent in the coelacanth genome and provides a rich catalog of immune genes for future investigations.
Keywords: Coelacanth, adaptive immunity, immune receptor, immunoglobulin, T-cell receptor, variable region, V(D)J rearrangement, cytokine, cluster of differentiation
Introduction
The lobe-finned vertebrates (Sarcopterygii) evolved from a stem lineage of bony fishes ∼400 million years ago and are comprised, collectively, of fish with fleshy fins as well as the land vertebrates, or tetrapods. Coelacanths (Latimeria) and lungfishes are the only surviving fishes within the Sarcopterygii, and are placed, phylogenetically, in a critically informative position between the ray-finned fishes and tetrapods. Most recently, the assembled genome sequence from the African coelacanth, Latimeria chalumnae, has been reported, and this has provided entrez into studying genes involving numerous aspects of vertebrate biology, notably the evolutionary transition from aquatic to terrestrial environments (Amemiya et al. 2013). Whereas the coelacanth is undeniably a fish, phylogenetic analyses most often indicate a closer relationship to tetrapods at the molecular level. Herein, we highlight those genes encoding components of its anticipatory or “adaptive” immune system. A separate companion paper on the coelacanth’s “innate” immune repertoire can be found elsewhere in this issue (Boudinot et al. 2014).
The B-lymphocytes of vertebrates such as mammals, utilize segmental V(D)J genetic recombination, somatic hypermutation, and other somatic mechanisms to generate, hypothetically, upwards of 1014 antibody specificities in its immunoglobulin genes (Fanning et al. 1996). However, the genomic organization, gene content, as well as the ratio of functional genes to nonfunctional pseudogenes among immunoglobulin (Ig) loci, have undergone notable changes during vertebrate evolution (Das et al. 2012). This characteristic of the Ig genes seems to be largely true for the analogous receptors on the T-lymphocytes, the T-cell receptor (TCR) gene families. Accordingly, studies of the genomic structure and organization of vertebrate Ig and TCR genes and functionally associated genes such as Rag and Aicda, which are integral to the generation of diversity, provide valuable insight into the genetic mechanisms and evolutionary divergence of adaptive immune recognition systems. Further, in the context of antigen recognition by TCR, CD4 (cluster of differentiation molecule 4) dimerizes and binds to the α2 and β2 domains of major histocompatibility complex (MHC) class II molecules (Huang et al. 1997;Wu et al. 1997), thereby serving as a TCR co-receptor. Similarly, cytotoxic T-cells utilize CD8 as a co-receptor, which, together, interact with MHC class I molecules during antigen presentation to T-cells. The identification of all subsets of TCR, key T-cell markers such as CD3, CD4, CD8, CD28, CD40L, and a great number of cytokines and chemokines in teleost fishes, suggests that so-called T helper cells Th1, Th2 and Th17 and the regulatory counterpart, Treg, were present prior to the emergence of tetrapods (Reyes-Cerpa et al. 2012).
Up to this point, descriptive immunological studies in the coelacanth, an endangered species, had been hampered by the lack of fresh material for examination. The only reported papers were from two decades ago. One of these used a genomic lambda library of coelacanth DNA to describe a mosaic genomic organization of immunoglobulin heavy chain gene segments, a hybrid structure between the "cluster" organization in cartilaginous fishes and the "translocon" organization in mammals (Amemiya et al. 1993). The other used a genomic lambda library and RT-PCR to isolate several partial sequences of coelacanth class I genes of the MHC and document gene structure and evolutionary relationships (Betz et al. 1994). Thus, the availability of the genome assembly from an African coelacanth, a bacterial artificial chromosome (BAC) library from the closely-related Indonesian coelacanth, L. menadoensis (Danke et al. 2004), and limited transcriptomic assemblies from both species, enabled us to conduct an initial survey for genes encoding immunoglobulin superfamilies involved in adaptive immunity, as well as several other genes whose proteins are known to be associated intimately with the adaptive immune system. We show that the coelacanth possesses, to a large degree, genes for requisite canonical immune molecules as would be expected for a typical vertebrate species, and further highlight major distinctions between the coelacanth genes and those of other vertebrate taxa.
Methods
Identification and Analysis of Genes of the Adaptive Immune System
The conserved nature of most of the key genes of the adaptive immune system together with the intrinsically slow rate of molecular evolution of coelacanth coding sequences (Amemiya et al. 2013), allowed for easy identification via database searches employing commonly-used search tools. The query sequences included Ig heavy and light chains, TCR (α, β, γ, δ), MHC (class I, class II), various interleukins, recombination activating genes (Rag1, Rag2), CD molecules, and activation-induced cytidine deaminase (Aicda). Available databases included the genome assembly (GenBank AFYH00000000.1) and an automated annotation of the L. chalumnae scaffolds (available on the Ensembl site: ensembl.org). All genomic scaffolds described in this report use GenBank or Ensembl nomenclature: JHxxxxxx or AFYHxxxxxxxx, respectively, for scaffold ID, and the Ensembl ENSLACGxxxxxxxxxxx for protein ID (where x’s denote a unique numerical identifier). The “JH” prefix of the scaffolds is not to be confused with the JH gene segments of IgH. Findings from the genomic surveys were validated using a composite testis+liver transcriptome assembly from L. menadoensis (NCBI GAPS00000000.1), or respective assemblies from the transcriptomes of the individual tissues (Pallavicini et al. 2013). A muscle transcriptome assembly from L. chalumnae also was available (unpublished); however, this resource only provided limited numbers of hits to genes of the immune system and only was used sparingly and then largely for the purposes of confirmation. Details regarding the coelacanth sequence datasets and the very high sequence identity between the two coelacanth species (∼99.7% across coding sequences) have been described (Amemiya et al. 2013). For certain gene families (e.g., interleukins, CD molecules) keyword searches on the Ensembl annotated assembly were used to extract pertinent genes en masse; each candidate then was manually validated via BLASTX. Where necessary, scaffolds were downloaded into Vector NTI sequence analysis software (Invitrogen) and further analyzed with regard to gene composition and organization. Phylogenetic analyses employed standard alignment programs as well as those in the MEGA5 package (Kumar et al. 2008). Calculation of % similarity and % identity used a locally installed version of Matrix Global Alignment Tool (Campanella et al. 2003).
Analysis of Ig Heavy Chains
Prior to initiation of the coelacanth genome project, a ≥7X coverage Latimeria menadoensis BAC library (Danke et al. 2004) was screened using a variety of Latimeria and lungfish VH and CH probes (Amemiya et al. 1993;Ota et al. 2003a). Resultant clones were validated as IgH-hybridizing and restriction fingerprinted using an automated system (Fjell et al. 2003). Five clones were strategically selected based on their restriction mapping patterns, and sequenced to 10X coverage by the Joint Genome Institute (Walnut Creek) using Sanger sequencing on ABI 3730xl instruments (Crow et al. 2012). Phred and Phrap were used for sequence editing and assembly (Gordon et al. 1998). Manual annotation was facilitated using Vector NTI software (Invitrogen). The L. menadoensis BAC library and a 100X coverage lambda genomic library from the same specimen (unpublished) were screened exhaustively and systematically with several other VH, JH and CH probes (including degenerate oligonucleotides against highly conserved transmembrane regions of vertebrate Cµ) in order to identify any other IgH-containing clones that escaped initial detection. In addition, various robust PCR strategies were employed with Latimeria genomic DNA to amplify any putative Cµ-containing fragments (Supplementary Table 10). Attempts to isolate divergent VH fragments employed PCR primers targeting CDR1 and FR3, which previously had been successful at amplifying VH sequences from genomic DNA of teleost fishes and lungfish (Turchin and Hsu 1996). L. chalumnae IgH-containing scaffolds were downloaded and annotated manually.
Analysis of TCRs
L. chalumnae genomic TCR scaffolds were identified and downloaded, and manually annotated. The TCR V, D and J gene segments were detected by sequence homology to corresponding gene segments from other species and by identifying the corresponding recombination signal sequences (RSSs). V gene segments were designated as Vα, Vβ, Vδ, Vγ or VH based on overall percentage identity with known variable region sequences.
Results and Discussion
Ig Heavy Chains
Comparative studies of immunoglobulins in numerous species of chondrichthyans, teleost fishes, amphibians and reptiles have facilitated efforts to understand the nature of diverse antibody production (Danilova et al. 2005;Anderson et al. 1999;Saha et al. 2005;Rast et al. 1998). In mammals, the arrangement of the IgH locus is a “translocon” type, wherein multiple variable heavy chain (VH) segments are linked distantly to diversity (DH), joining (JH) and CH domains [(V)n-(D)n-(J)n-(C)n]. This translocon type arrangement also is conserved with limited variations in teleost fish, wherein C genes encode three distinct classes (IgM, IgD and IgZ/T) as compared to as many as five major classes (IgM, IgD, IgG, IgE and IgA) in mammals. In contrast, the most basal lineages of jawed fishes such as sharks and rays (elasmobranchs) possess IgH loci (IgM, IgW, IgNAR) of distinct "cluster" type arrangement comprised of repeated units of (V-Dn-J-C) or slight variants thereof, sometimes with germline-fused segments (Rast et al. 1989;Flajnik 2002). A third and highly divergent IgH organization evolved within the avian lineage and consists of a single functional VH gene that undergoes gene conversion to generate antibody diversity (Reynaud et al. 1989). Outside of the invariant and close proximity (∼ 190 nt) of VH to DH segments in the coelacanth genome (Amemiya et al. 1993), nothing was known about its immunoglobulin loci prior to the acquisition of its genome sequence.
Characterization of IgH isotypes in L. menadoensis from BAC clones
A ≥7X coverage BAC library generated from the Indonesian coelacanth, L. menadoensis, was screened via colony hybridization using a mixed coelacanth VH probe and 50 positive clones initially were identified. Screening with a horn shark Cμ probe identified 20 clones (Supplementary Table 1). All VH+, CH+ and VH+CH+ clones were restriction fingerprinted using Internet Contig Explorer (iCE) (Fjell et al. 2003) and assembled into two contigs (Supplementary Fig. 1). Clones 189I9 (177 kb), 58E24 (183 kb), 130A21 (159 kb), 206D14 (167 kb) and 217L16 (167 kb) were selected strategically and sequenced to high draft coverage (10X) via Sanger shotgun sequencing. Overlaps between clones 189I9 and 58E24 (Fig. 1A), and between clones 130A21 and 206D14 (Contig 2) were confirmed (Fig. 1B); 217L16 did not show any appreciable overlaps with the other clones even though it had been placed in Contig 2 via automated restriction fingerprinting.
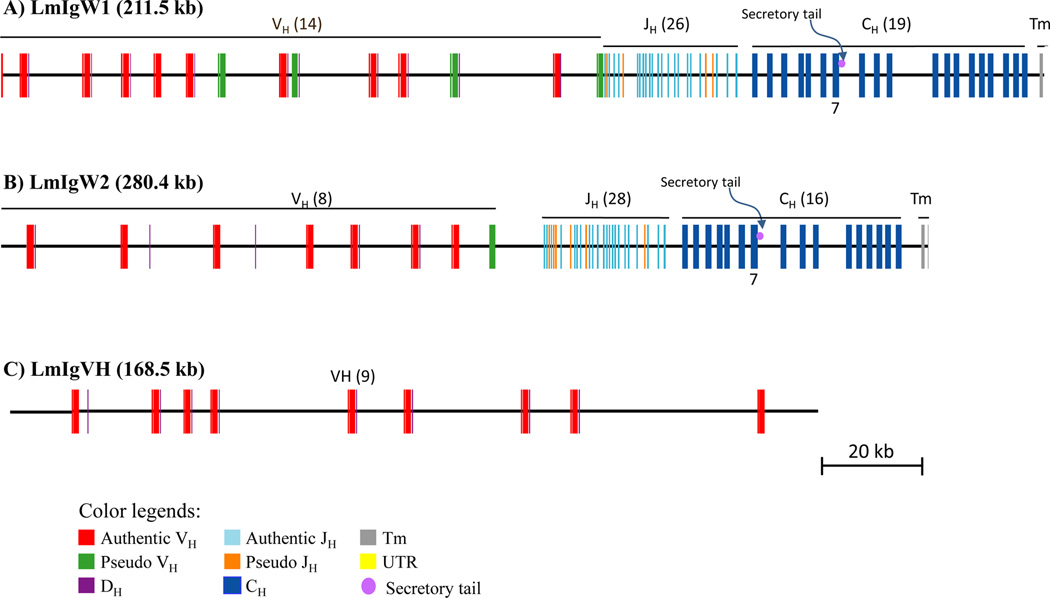
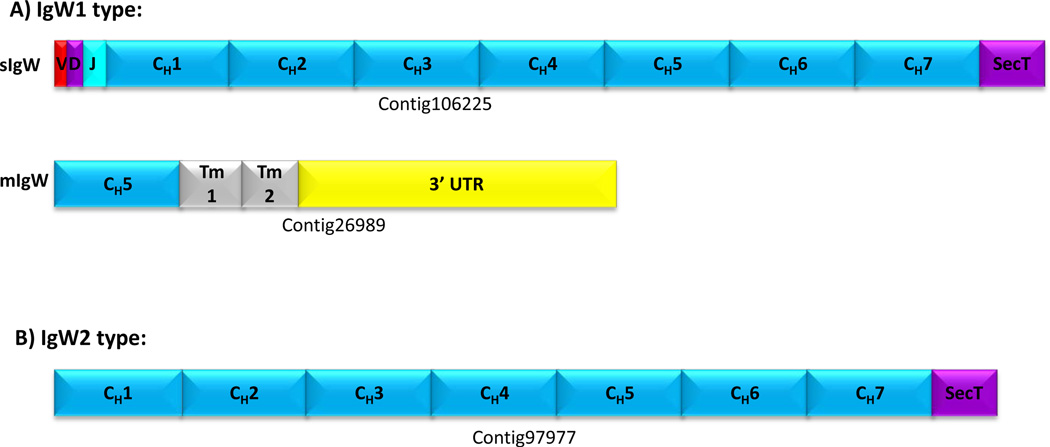
Figure 1. Immunoglobulin heavy chain genome organization in Latimeria menadoensis.
Overlapping BAC clones encompassing the immunoglobulin heavy chain loci of L. menadoensis were isolated and sequenced. Locus 1, designated IgW1 (BAC clones 58E24 and 189I9; GenBank JX848736.1) contains 19 constant region exons (A), whereas locus 2, designated IgW2 (BAC clones 130A21 and 206D14; GenBank JX840472) encodes 16 constant region exons (B). Both IgH loci are comparatively large and organized in a different pattern in which V and D segments are in close (∼190 bp) V-D linkage and contain consensus RSSs. The respective genes appear to be most similar in overall sequence relatedness to IgW from the African lungfish and cartilaginous fishes. Both IgW isotypes encode a secretory tail at the end of the first seven C domains and possess two transmembrane exons at the end of last C domain. A separate BAC clone, 217L16, was identified in the same large restriction fingerprint set as were the clones encompassing the IgW2 locus (Supp. Fig. 1); however, it contains only VH and DH segments; no JH segments or CH domains were identified.
VH, JH and CH elements readily were identified via motif searches. DH elements were identified by inference using the positions of flanking RSS sequences. BLAST analyses using authentic IgM sequences as queries showed that neither contig contained an IgM-type heavy chain. Two distinct gene loci consisting of VH, DH, JH and CH elements were identified (Fig. 1); however, their CH segments are clearly not of the Cμ type (Supplementary Table 2) and most are related closely to the IgW type reported in cartilaginous fish (Rumfelt et al. 2004;Harding et al. 1990) and lungfish (Ota et al. 2003b). Based on their genomic structures and notable sequence differences, the two contigs represent distinct loci and clearly are not allelic forms. A fifth VHDH-containing clone, 217L16, (Fig. 1C) lacks the downstream JH and CH exons and does not show sequence overlap with the two contigs. Overall, the gene segments are organized in an intrinsically different pattern than observed in other vertebrate species, wherein individual V and D gene segments are paired and in close proximity (Amemiya et al. 1993), with a large track of J segments and several C region exons further downstream: [(VH-DH)n-(JH)n-(CH)16/19]. The internal duplication of CH exons is reminiscent of the IgD-encoding locus of pufferfish (Saha et al. 2004) (Supplementary Fig. 2). The patterns of organization of the loci preclude typical heavy chain class switching, although alternative splicing ostensibly could produce different isoforms of IgW.
IgW1 is predicted to contain 19 C domains (Fig. 1A), whereas IgW2 is predicted to contain 16 C domains (Fig. 1B). Each locus encodes two transmembrane domains and a termination codon followed by a 3′ untranslated region; a region that is predicted to represent a secretory tail (SecT) (Figure 1 and Suppl Fig. 2) is just downstream of CH7. Assuming that the primary transcript consists of seven CH domains (see below), these observations are consistent with predicted secretory and membrane-bound forms of the two IgW molecules for Latimeria. The evolutionary scenario and usage of the other CH domains largely are unknown at this time. These other downstream exons appear to be completely in-frame, devoid of stop codons and possess predicted splice donor/acceptor sites.
A total of 31 VH genes have been identified in the five sequenced BACs; of these, one is a partial sequence and five are pseudogenes (lacking one or more characteristic features such as an octamer and/or a TATA-box or possess stop codons and/or frame shifts in their coding sequences). All putatively functional VH sequences possess upstream regulatory sequences (an octamer that is separated from a TATA-box by 18 bp), a leader peptide sequence split by an intron with consensus splice sites, a reading frame that can be divided readily into framework regions (FRs) and complementarity determining regions (CDRs), and a 3′ RSS with a typical 23 bp spacer (Supplementary Table 3). Most of the VH genes span 291 to 300 nucleotides (97–100 amino acid residues from FR1 through FR3), typical for VH genes of other vertebrates. All DH segments exhibit conserved upstream and downstream RSSs as reported previously for L. chalumnae (Amemiya et al. 1993).
Characterization of IgH isotypes from L. chalumnae whole genome sequence
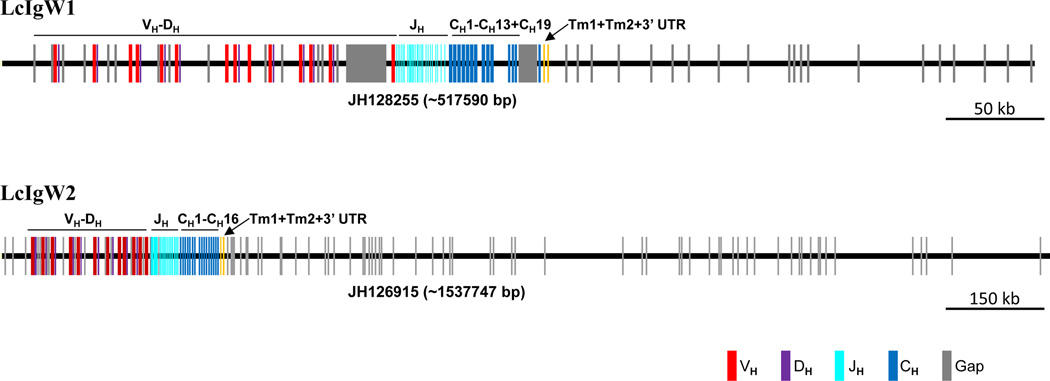
The scaffolds corresponding to the sequenced L. menadoensis Ig heavy chain loci were identified from the assembled L. chalumnae genome. Two separate extended scaffolds, JH128255 (517,590 bp) and JH126915 (1,537,747 bp), were identified and annotated (Fig. 2); these correspond unequivocally to IgW1 and IgW2, respectively. The IgW loci in the two species of coelacanth exhibit high overall concordance and sequence identity (> 99%), excluding problematic sequence stretches from both loci (Fig. 3). Based on our annotations, the Lc scaffolds extend the IgH regions ∼65 kb and ∼106 kb upstream of the BAC-based Lm-IgW1 and Lm-IgW2 loci, respectively, and primarily contain VH and DH segments. Fourteen additional scaffolds containing from one to several VH genes were identified in L. chalumnae; none of these contain a CH domain (Table 1). Of the 66 VH distinct genes thus far identified, at least 13 represent pseudogenes (Supplementary Table 4). The IgH scaffolds of L. chalumnae and corresponding sequence analysis of selected L. menadoensis IgH-containing BAC clones are consistent with the existence of two distinct IgW loci in the coelacanth. Of note, the downstream boundary of IgW2 extends to the TCRα locus (discussed below).
Figure 2. Immunoglobulin heavy chain genome organization in Latimeria chalumnae.
Genomic scaffolds containing the two extended IgH loci were downloaded and annotated. Analyses of these L. chalumnae scaffolds confirm that they also possess two IgW heavy chain loci, which correspond unequivocally to L. menadoensis IgW1 and IgW2. Orthologous IgW1 and IgW2 loci between the two species are highly similar (>99% identical across alignable regions). A locus encoding a heavy chain recognizable as Cµ was not identified either from bioinformatics searches or via direct hybridization and PCR screening strategies.
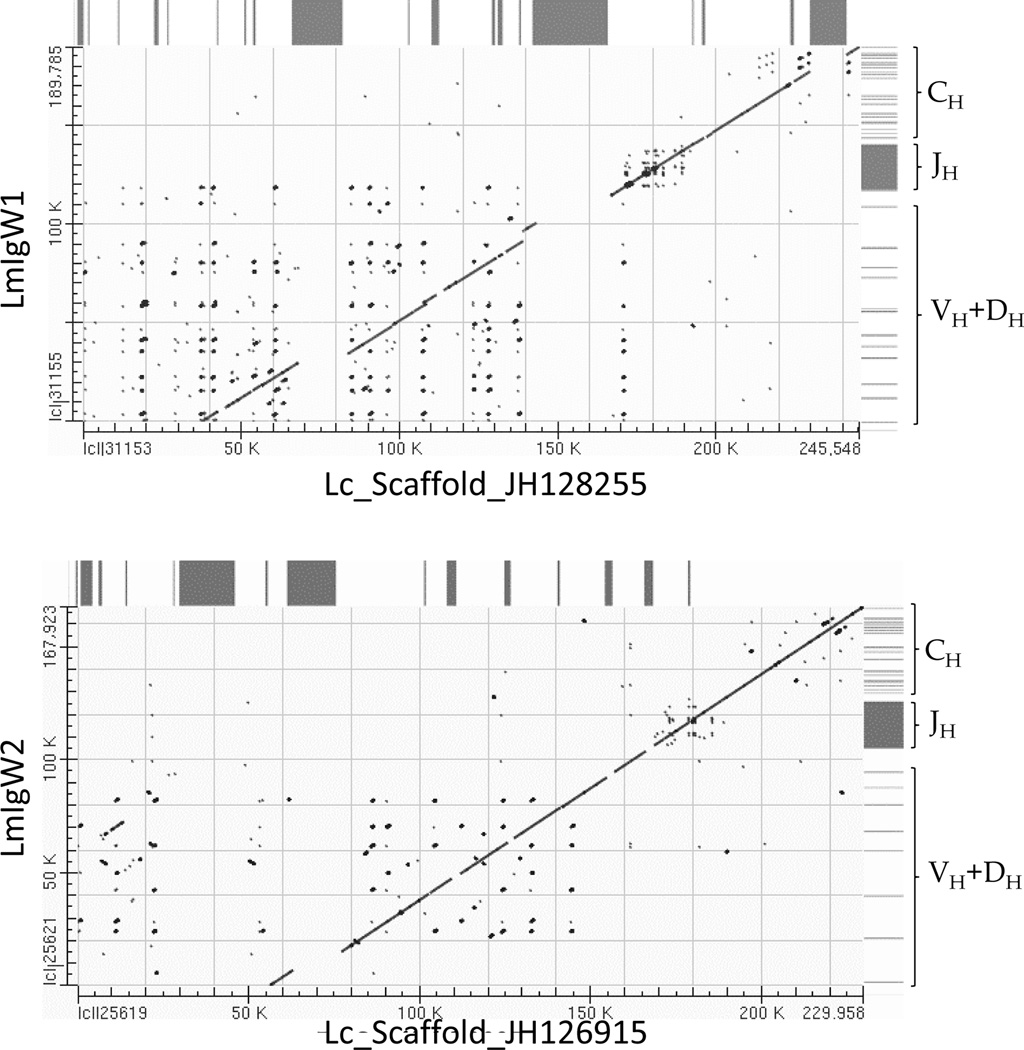
Figure 3. Concordance of L. chalumnae and L. menadoensis genomic sequences encoding two IgW heavy chain loci.
The dot-plots are graphical depictions of MEGABLAST alignments of orthologous IgW heavy chain genomic regions from the two coelacanth species. Horizontal axes represent L. chalumnae scaffolds and vertical axes represent L. menadoensis assemblies based on overlapping BAC clones. Boxes on the right side of the plots denote relative positions of coding sequences in the respective IgW heavy chain loci within the BAC assemblies. Diagonal lines indicate strong concordance between the IgW heavy chain loci of both species. Orthologous regions were highly similar (99% identical across alignable regions). The tracts that are not aligned (gaps in the diagonal line) are largely accounted for by runs of N’s in the L. chalumnae genomic scaffold and are depicted above the plot by black boxes.
Table 1.
IgVH-containing scaffolds in L. chalumnae. The two predominant scaffolds that were analyzed contained the IgW loci (IgW1, IgW2). Other scaffolds contained many VH elements, but their relationship to these other two loci are unclear. Some of the VH elements in these other scaffolds also contained downstream DH elements.
| Receptor | Scaffold | Length (bp) | Notes |
|---|---|---|---|
| IgW1 | JH128255 | 517,590 | Contains 14 VH-DH, JH and CW, cluster-translocon, (VHDH)n-JHn-CW |
| IgH | JH130781 | 102,095 | Contains 5 VH genes |
| IgH | JH129820 | 195,380 | Contains 14 VH genes, 6 of which are pseudogenes |
| IgH | JH135971 | 10,639 | Contains 1 VH gene |
| IgW2 | JH126915 | 1,537,747 | Contains 12 VH-DH, JH and CW, cluster-translocon, (VHDH)n-JHn-CW |
| IgH | JH132402 | 21,611 | Contains 2 VH genes |
| IgH | JH128757 | 372,416 | Contains 2 VH genes |
| IgH | JH135868 | 5,841 | Contains 1 VH gene |
| IgH | JH130737 | 105,473 | Contains 7 VH genes |
| IgH | JH136021 | 5,567 | Contains 1 VH gene |
| IgH | JH135110 | 7,584 | Contains 1 VH gene |
| IgH | JH132703 | 17,134 | Contains 1 VH gene |
| IgH | JH132971 | 14,983 | Contains 1 VH gene |
| IgH | JH133119 | 14106 | Contains 1 VH gene |
| IgH | JH135509 | 6690 | Contains 1 VH gene |
| IgH | JH133573 | 11916 | Contains 2 VH genes |
VH repertoire in Latimeria
Multiple alignments of the deduced amino acid sequences of VH gene segments identified in this study indicate that the coelacanth VH germline repertoire is largely comparable to those characterized in other jawed vertebrates (Supplementary Fig. 3). The conserved GKGLEW and YYCAR motifs along with other canonical residues underscore the overall conservation of vertebrate VH sequences in vertebrate phylogeny. Both VH and VH pseudogenes from the two IgW loci are in the same transcriptional orientation; no observable inverted sequences have been detected.
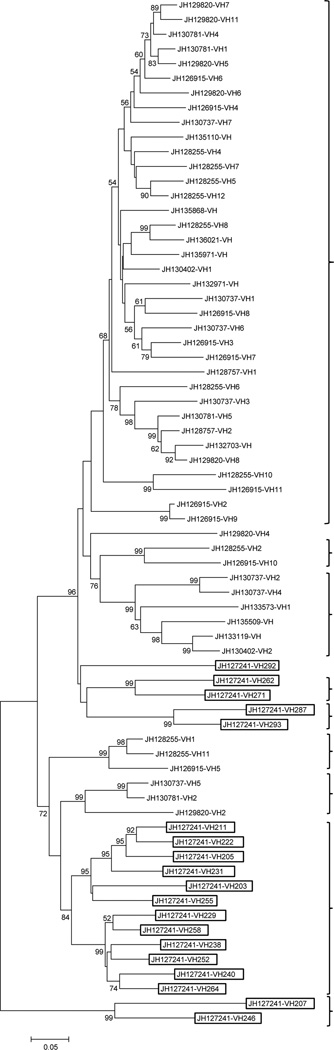
VH gene families are defined on the basis of percent nucleotide sequence identity. Sequences that are greater than 80% identical are categorized as representing a single family. At least five distinct phylogenetic lineages of VH gene families have been identified in vertebrates (Andersson and Matsunaga 1995;Ota and Nei 1994) , although the number of VH gene families can vary widely among species. Mice and humans possess 14 and seven families, respectively (Tomlinson et al. 1992;Tutter et al. 1991). Rainbow trout and channel catfish possess 11 and six VH gene families, respectively (Roman et al. 1996;Tutter et al. 1991;Ghaffari and Lobb 1991;Warr et al. 1991), whereas the horn shark, a cartilaginous fish, possesses only two VH gene families (Hinds-Frey et al. 1993). Based on our analysis, at least nine VH families can be recognized in coelacanth. In addition, several unique VHs were delineated that cannot be ascribed to any specific gene families. A phylogenetic tree indicating the relationships of 70 VH genes identified in the L. chalumnae genome is presented in Fig. 4; pseudogenes and VH genes containing ambiguous sequences were eliminated from the comparison. Nineteen of the VH genes are located in a TCRαδ locus (see below).
Figure 4. Phylogenetic analysis of VH segments in coelacanth.
The relationships of VH genes were inferred using the Neighbor-Joining method. All ambiguous positions were removed for each sequence pair. The analysis involved 70 amino acid sequences; % bootstrap replicates are given on the tree. Coelacanth VH genes fall into nine bona fide VH families as indicated by brackets. VH elements that were identified within the TCR locus are in boxes.
Analysis of IgH-transcripts
Although the L. menadoensis transcriptome assembly was produced from non-haematopoietic tissues (liver and testis), a small percentage (0.7%) of the ∼ 13,000 annotated genes correspond to immune system processes (Gene Ontology term 0002376). This dataset was used to identify multiple hits encompassing IgW transcripts; these represent both IgW1 and IgW2 molecules although none were found to be full-length rearranged molecules (Fig. 5). One IgW1 transcript (contig106265) is rearranged with a unique V-D segment (truncated), a known J segment and seven C domains (CH1 to CH7) followed by a secretory tail composed of 20 amino acid residues. A second, truncated IgW1 transcript (contig26989) was identified that lacks a V region but contained CH5 spliced directly to the TM-encoding exons (located downstream of the 12 additional CH domains, Fig. 1A) followed by a 3’ untranslated region. This likely represents a membrane form of IgW. A similar feature is seen in teleost IgM whereby the transmembrane domain is spliced directly to a CH3 domain (Saha et al. 2005;Bengten et al. 1992;Hansen et al. 1994). The genomic sequence for the secretory tail is located at the 3′ end of the CH7 domain in both IgW1 and IgW2 loci (Fig. 1). The IgW genes that were identified in coelacanth structurally resemble IgW described previously in lungfish and cartilaginous fish, although a characteristic two-domain form (Ota et al. 2003b;Harding et al. 1990) has yet to be identified in the transcriptome. The finding of multiple, internally repeated CH domains (Suppl Fig. 2) also is curious and it will be interesting to determine whether or not any of these other domains are utilized, and in what context. This may be challenging given the difficulty in procuring any coelacanth tissue, let alone a hematopoietic source; however, one alternative may be to use a surrogate in vitro B-cell system to assess the functionality.
Figure 5. IgW transcript validation in coelacanth.
IgW1 and IgW2 heavy chain transcripts were identified in the L. menadoensis RNAseq combined assembly, despite the dataset being generated from non-lymphoid (liver and testes) sources. (A) A rearranged IgW1 heavy chain transcript that includes a partial VD region, a known J-segment and seven constant domains (CH1 to CH7) followed by a secretory tail (upper panel). Another IgW1 heavy chain transcript that lacks the variable region and most of the constant region but contains a CH5 domain spliced to transmembrane domains followed by its 3’ UTR, suggests it derives from a membrane isoform of IgW. (B) The IgW2 heavy chain transcript lacks a variable domain but possess an intact constant region (7 domains) in which its secretory tail was attached at the 3’ end of CH7 domain. Three other constant region transcripts were identified in the muscle transcriptome of L. chalumnae (not shown).
Lack of IgM in Latimeria
Ig heavy chain that is encoded by IgM has been reported in all vertebrates characterized thus far and is considered to be essential for the initial phase of thean IgM gene constant region could not be identified, even though orthologs of most of the major genes involved in the adaptive immune system of jawed vertebrates are present. Moreover, L. menadoensis genomic BAC and λ libraries were screened exhaustively using numerous strategies and a variety of probes but no Cμ like sequences were identified. Additionally, PCR primers (Turchin and Hsu 1996) that amplified VH from teleost fish, lungfish, amphibians, reptiles, and mammals produced fragments that fell into two distinct groups. One set consisted of bona fide IgW VH elements. Sequencing of VH elements from the second set, which we surmised may be embedded in a different heavy chain locus, instead were found in the TCR α/δ locus, described below. Furthermore, numerous additional degenerate primers that were designed to amplify Cμ sequences, based on published sequence data, failed to identify a Cμ homolog. No traces of Cµ were found in the RNA-seq data of coelacanth although as stated above, transcripts encoding both IgW1 and IgW2 heavy chains were identified (Fig. 5). The apparent lack of genes encoding IgM heavy chain is unexpected although it is known that the codfish and its close relatives apparently have lost major components (MHC class II) of their immune systems (Star et al. 2011). The evolutionary relationships of Ig heavy chains, including IgD (to which the coelacanth IgW shows a relationship), will be addressed elsewhere. The lack of IgM in the coelacanth raises questions as to whether an IgW molecule supplants classical IgM in a manner analogous to the compensatory modifications seen in the codfish with respect to the function of MHC class II (Malmstrom et al. 2013).
Ig Light Chains
Multiple immunoglobulin light chain isotypes have been identified in all vertebrates studied to date, with the exception of birds, bats and snakes, in which only a single light chain has been described (Lundqvist et al. 2006;Gambon-Deza et al. 2012;Magadan-Mompo et al. 2013). In tetrapods, IgL can be classified into three distinct groups: kappa (κ), lambda (λ), and sigma (σ) (Criscitiello and Flajnik 2007). However, the σ isotype was thought to have been lost in all lineages after the divergence of amphibians (Das et al. 2012). A close examination of VL based on its phylogenetic relationships, CDR lengths and RSS orientation, recognized four ancestral VL clades that were maintained throughout the vertebrates (Criscitiello and Flajnik 2007). A distinct variant of the σ isotype, which was named σ-cart (for cartilaginous fish), has been identified only in the shark (Criscitiello and Flajnik 2007). The organization of the light chain loci among the vertebrates is not as definitive or diagnostic as for the heavy chain loci and can consist of cluster-type, translocon-type or perhaps other variations (Hsu and Criscitiello 2006).
Coelacanth IgL
Homology searches with various vertebrate immunoglobulin light chain amino acid sequences have identified a large number of IgL genes in the African coelacanth genome. Following previous classification schemes (Edholm et al. 2011;Criscitiello and Flajnik 2007) most coelacanth IgL genes can be separated into four groups based on the amino acid sequence, i.e., sigma-cart (cartilaginous fish type I/NS5), sigma (fish L2), kappa (cartilaginous fish type III/NS4, fish L1/L3/F/G, Xenopus rho) and lambda (cartilaginous fish type II/NS3, Xenopus type III) (fig. 6) and overall are in agreement with the generally accepted classification scheme (Criscitiello and Flajnik 2007).
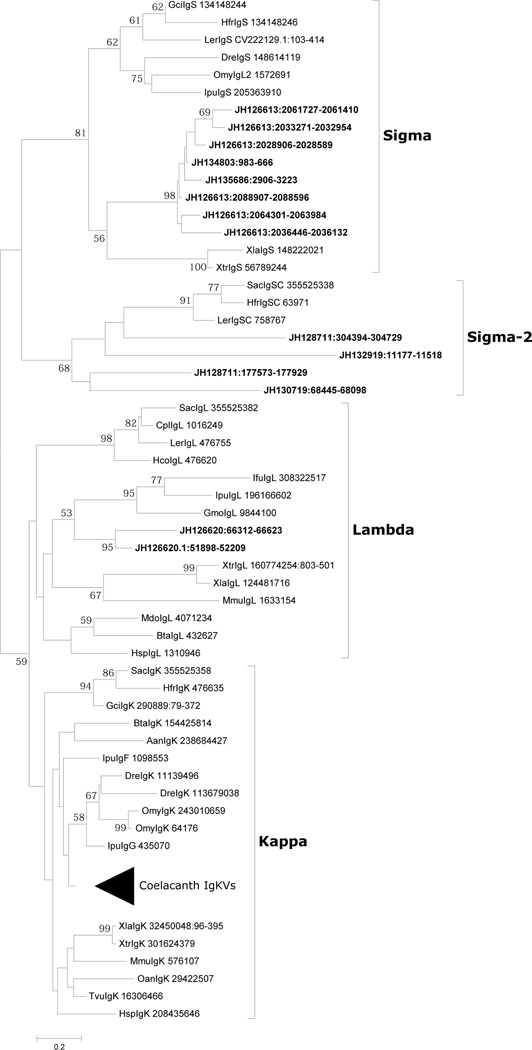
Figure 6. Phylogenetic analysis of VL segments in coelacanth.
The relationships of VL genes were inferred using the Neighbor-Joining method. All positions containing gaps and missing data were eliminated; % bootstrap replicates are given on the tree. A total of 66 positions of framework region were represented in the final dataset. The Lc sequences are denoted by ‘JHxxxxxx’, followed by their positions on the scaffolds. Sequence identification numbers (GI) of all other taxa are mentioned after the species name. The light chain classes are given to the right of the tree and are in complete accordance with branching topology. The Sigma-2 designation is the former Sigma-cart subclass that had previously been found only in cartilaginous fishes.
The coelacanth genome encodes IgL genes of the sigma-cart type, which we provisionally denote sigma-2. These loci are in a cluster-type pattern of organization and four clusters are found in three different scaffolds (JH130719, JH128711 and JH132919), where V and J gene segments are germline-joined. An extra C region exon, which shows 94% identity at the nucleotide level with other C regions, is also observed in the scaffolds scaffold JH130719. It is uncertain if this distinctive C gene exon is expressed together with the VJ gene of the neighboring complete cluster or if it represents a pseudogene. Given the notable numbers of ambiguous regions (due to assembly gaps), it is possible that an additional VJ gene segment(s), which may be associated with a C gene segment, is present. CDR1 and CDR2 of V gene segments encode 13 and 11 amino acids, respectively, a characteristic feature of the sigma-cart IgLs of cartilaginous fish. Certain IgLs have insertions in their FRs. One scaffold (JH132380) contains a single C region, of which the amino acid sequence is similar to that in JH130719. The ortholog of this C region was identified in the Indonesian coelacanth transcriptome (testis: comp76432_c0_seq1) but the 5’ region preceding the C region showed little sequence homology with VJ region. The C gene segment of scaffold JH132919 is unusual in that its exon structure and 3′ end are predicted to be encoded by separate exons, somewhat different from that seen in other sigma-cart type IgL genes. Sigma cart initially was found only in elasmobranchs (Hikima et al. 2011;Sun et al. 2012), hence its presence in the coelacanth implies a wider distribution than initially thought.
The sigmatype of IgL in coelacanth was detected in three scaffolds: JH126613, which contains 3(V-J)-3V–J-C; JH134803, which contains V-J; and JH135686, which contains V-J and a V pseudogene segment. Most J gene segments, with the exception of those most proximal to the C gene segment, may represent pseudo gene segments, as they contain an in-frame internal termination codon. The possibility remains that scaffolds JH134803 (8271 bp) and JH135686 (6455 bp) are located within JH126613 (3,167,360 bp). VL and JL gene segments are flanked by RSSs with 12 bp and 23 bp spacers, respectively. The CDR1 and CDR2 of V gene segments encode 10–11 and 12 amino acid residues, respectively, and are equivalent to those of cartilaginous and fishes. A YGxG (or PxYGxGFS) motif located at the CDR2-FR3 boundary region is conserved among most of the V gene segments of coelacanth IgL of the sigma type. Genes encoding Kcnv2 (potassium channel, subfamily V, member 2), Kank3 (KN motif and ankyrin repeat domains 3), Angpt14 (angiopoietin-like 4) and Rab11b (member of RAS oncogene family) downstream of C gene segments map to JH126613 (ENSLACG00000017294, ENSLACG00000017209, ENSLACG00000017018 and ENSLACG00000016974, respectively); these same genes also map downstream of the Ig sigma locus (XB-GENE-5806081) on Xenopus scaffold GL173022.1.
Igκ is present in all jawed vertebrates except birds and includes the amphibian rho-type light chain. The most extensive IgL gene family in coelacanth is of the kappa-type and encoded in three scaffolds: JH128084 (580,075 bp), JH129712 (214,159 bp) and JH130074 (167,776 bp). Four V gene segments, four J gene segments and one C segment are encoded in JH130074 in the same transcriptional orientation. A large number of VL gene elements without JL elements and CL segments have been identified in JH128084 and JH129712. The gene encoding ribose 5-phosphate isomerase A (RPIA) is located downstream of C gene segments in JH130074; close linkage of RPIA to the kappa locus is a tetrapod condition (Edholm et al. 2011). The gene encoding succinate-CoA ligase alpha subunit is upstream of V gene segments in JH128084 indicating that the 5’ end of IgL kappa loci likely is encoded in this scaffold. In addition to the above, three V gene segments were identified in scaffolds JH131133 (80,170 bp) and JH131467 (58,214 bp), and a single Vκ gene segment was identified in scaffolds JH130471 (127,935 bp), JH132852 (16,005 bp), JH133287 (13,169 bp), AFYH01278842 (5496 bp) and AFYH01285422 (1902 bp). Scaffolds JH128084, JH129712 and JH130074 and other short scaffolds could ostensibly be part of a longer contig. Some V gene segments detected in the aforementioned scaffolds are apparent pseudogenes, as defined by internal truncation, termination codons and/or frame-shift mutations.
Igλ constitutes the only IgL isotype in avians and was considered missing in fishes until its identification in channel catfish, Atlantic cod, and rainbow trout (Edholm et al. 2009). IgLλ in coelacanth maps to scaffold JH126620, which contains four V gene segments, two J gene segments and one gene segment in a translocon-type gene organization. The genes for car15 (ENSLACG00000004420) and dgcr2 (ENSLACG00000005606) map downstream of the λ locus. Orthologous genes are located near the Ig type III locus in Xenopus tropicalis and near Igλ chain loci in human (chromosome 22q11) and in mouse (chromosome 16). CDR1 and CDR2 of the coelacanth V gene segments contain 13 and 11 amino acid residues, respectively and the length of CDR2 is longer generally than those of other vertebrate Igλ V gene segments but similar to some Xenopus Igλ V gene segments.
Other Ig-like genes
In addition to the bona fide IgH and IgL genes discussed above, there are a few other genes encoding Ig domains that were detected in several scaffolds (JH127746, JH132194, JH132693, JH134408 and JH129664) that could not be assigned confidently to a specific gene family or Ig class (Supplementary Fig. 4). JH127746 encodes a VJ-VJ-C configuration whereby its C terminal end is encoded by five exons. JH132194 and JH132693 encode a VJ and a C gene segment; however, placement of the 3’ end of the C gene segment in this case is uncertain. JH134408 and JH129664 encode one VJ gene segment, the nature of which also is unclear (Supplementary Table 5). The functionality of these genes remains to be determined. BLAST comparisons of the respective V and C domains do not reveal anything strongly resembling IgM or any other IgH, IgL or TCR-type genes.
T-cell receptor
T-cell receptors (TCRs) are expressed on the surface of T-lymphocytes that recognize antigens presented by MHC and induce a series of intracellular signaling cascades, although it is not clear yet whether the γ/δ chains, which are found only in 5% of T-cells, are MHC-restricted. These signaling cascades regulate T-cell development, homeostasis, activation, acquisition of effector functions and apoptosis (Okkenhaug et al. 2004;Lin and Weiss 2001). All jawed vertebrates thus far characterized possess four different types of TCR-chains: α, β, δ and γ. T-lymphocytes are characterized as either being αβ or γδ. In addition, a divergent TCR, TCRµ, which has some features resembling a recently described TCRδ isoform in sharks, has been described in marsupials and a monotreme (Parra et al. 2007;Wang et al. 2011). The T-cell receptor genes, like those of immunoglobulins, consist of V, D and J segments and a C region.
As expected, genes encoding the four basic types of TCR (α, β, γ, δ) have been identified in the coelacanth genome (described below). These were validated by partial cDNA sequences from both L. menadoensis and L. chalumnae (Suppl. Fig. 7). A phylogenetic tree of the C regions of the four TCR types of the coelacanth confirmed that each type is grouped with the expected clades (Supplementary Fig. 8). No genes encoding a conspicuous TCRμ were identified.
Coelacanth TCR α/δ
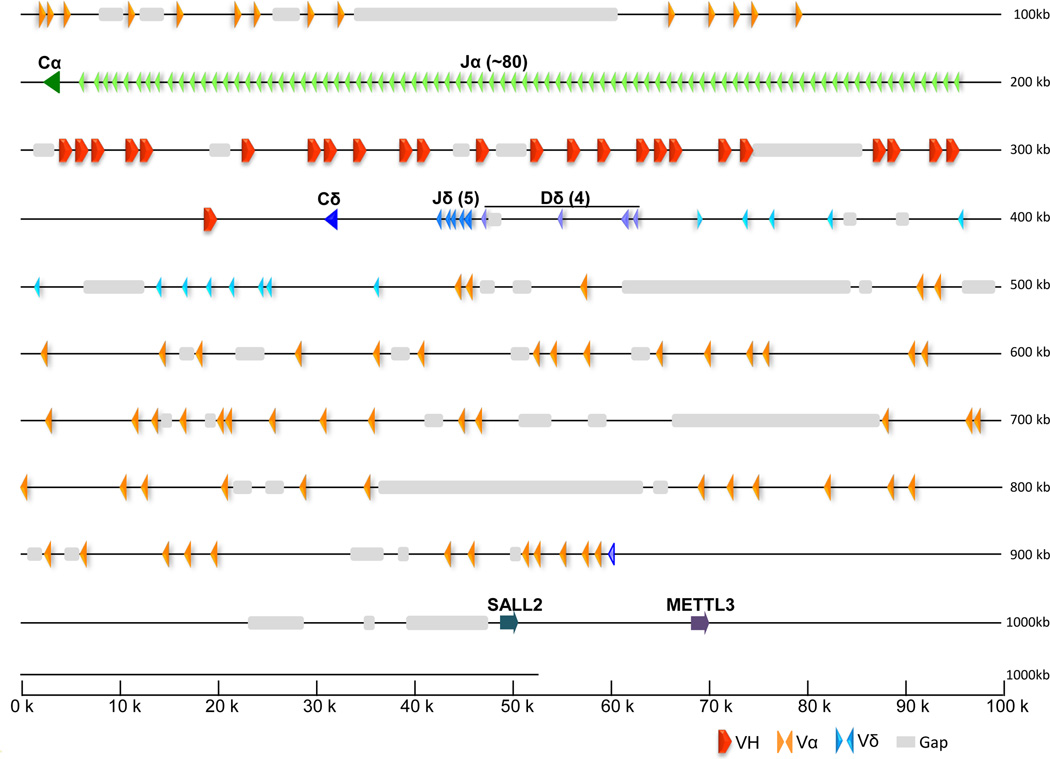
Scaffold JH127241 contains the extended TCR α/δ region and scaffold JH126915 contains primarily genes of the TCR-α locus; whether or not these two scaffolds are localized to the same chromosomal region has not been determined definitively. As in all other tetrapods examined to date, TCR-α locus is embedded with the genes encoding the TCR-δ chains (Fig. 7). The TCR α/δ region encompasses a track encoding 25 VH genes; these genes are nearly indistinguishable from those encoded at the IgW loci except they are not associated with DH segments. The VHs are located between the TCRα and TCRδ genes. VH genes also were reported at the TCRδ locus in the frog; however, there was no evidence for trans-locus somatic recombination between the loci despite the fact that both loci contain multiple VH gene segments (Parra et al. 2010). Analogous to the casein gene in Xenopus, VH genes also were found to be embedded in the TCRα/δ locus of the platypus and opossum (Parra et al. 2009). In marked contrast, several cDNAs that contained IgM or IgW V segments were rearranged with other gene segments of TCRδ and α in nurse shark (Criscitiello et al. 2010). At this point it is not known whether or not the coelacanth uses these VH gene segments in the context of TCRα/δ genes as no transcripts encoding VH gene segments could be identified in the available transcriptome databases. Fourteen Vα gene segments are located upstream and in the opposite transcriptional orientation as Cα. Another 58 Vα gene segments are in reverse orientation. A total of 80 Jα genes, which are in the same transcriptional orientation as Cα, have been identified. The large number of Jα elements exceeds that reported in frog (Parra et al. 2010), human and mice (Giudicelli et al. 2005). Only one TCRδ gene encodes Vδ in the same transcriptional orientation as Cδ and at least five Jδ elements have been identified. Sal-like protein 2 (SALL-2) and Methyl-transferase-like 3 (METTL3) delimit one end of the TCRα/δ locus in coelacanth, similar to the situation in birds and mammals.
Figure 7. Physical map and annotation of T-cell receptor α/δ locus (TCR locus 1).
Scaffold JH127241 contains the coelacanth TCR α/δ locus. This locus contains genes for both TCR-α and TCR-δ in a typical arrangement; however, the locus also contains 25 V gene segments that are very closely relatedto those VHs encoded in the IgW loci. Transcriptional orientation is indicated by the direction of the arrowheads for each segment. Syntenic genes shown in gray are those conserved with mammalian TCRα/δ locus: methyl-transferase like 3 (METTL3), zinc finger protein (SALL2).
The overall organization of the coelacanth TCRα/δ locus as depicted in scaffold JH127241 is highly conserved with those of amphibians, birds and mammals, suggesting that it is a very stable genomic region. VH genes embedded in the TCRα/δ locus (previously named VHd) have been reported in different lineages of jawed vertebrates. In the amphibian, Xenopus tropicalis, the VHδ genes were only found expressed in TCRδ chains (Parra et al. 2010). The organization of TCRα/δ locus in birds varies among lineages. In the zebra finch the TCRα/δ locus contains VHδ, similar to the amphibians (Parra and Miller 2012). However in galliformes and anseriformes, the TCRα/δ locus does not contain any VH genes; instead the VHδ genes have been translocated to a separate chromosome creating a second TCRδ locus (Parra et al JI 2012). Among the mammalian lineages only the monotremes (the platypus) contains a single VHδ in the TCRα/δ locus (Parra et al. 2012); monotremes and marsupials have an additional TCR locus (TCRm) which contains VH-like and Cδ-like genes (Parra et al. 2007;Wang et al. 2011). These rearrangements suggest that VH-TCRδ genes are mobile and prone to translocation. In cartilaginous fish (nurse shark) there is also evidence that TCRδ chains use VH genes. NAR-TCR is a three domain receptor composed by double rearrangement of two V genes (VH and Vd) expressed with TCR Cδ (Criscitiello et al. 2006)(. In addition, shark VH (IgM and IgW) have been found rearranged with TCRα/δ D and J gene segments and then spliced to either Cα or Cδ TCR constant regions (Criscitiello et al 2010). While no transcripts have been found, its highly intact and conserved VH elements might imply that the coelacanth also undergoes similar deployment of the VH-TCRδ chains seen in mammalian TCRµ, shark NAR-TCR and VH-TCR transrearrangements, and the VHδ-TCRδ chains in frogs and birds. These results are consistent with a selective pressure to maintain T cells that are capable of direct antigen binding.
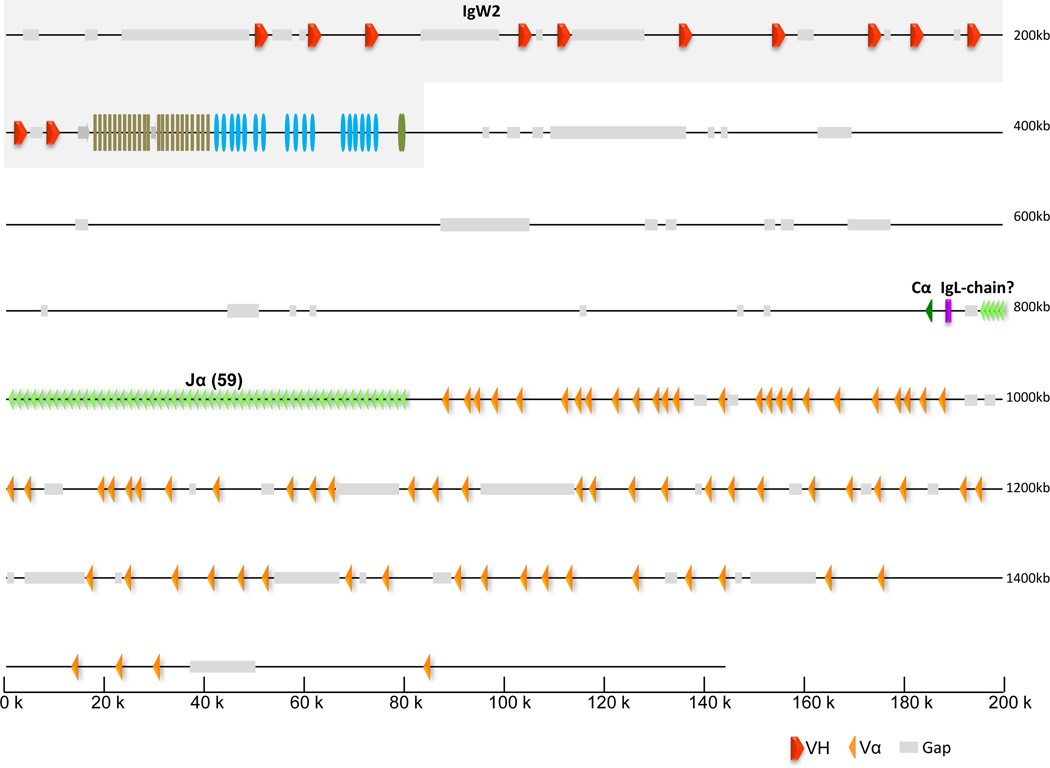
The 2nd TCRα-containing scaffold (JH126915) is unlike those reported in other species. The IgW2 immunoglobulin heavy chain locus maps to the same scaffold but is in opposite transcriptional orientation as the respective TCRα genes (Fig. 8). The TCRα region consists of 74 Vα and 59 Jα segments followed by a single Cα domain. All of the TCRα components are in the same transcriptional orientation in this second TCRα scaffold. The linkage of TCRα with an IgH locus and the interdigitation of putatively functional VH segments with TCRα/δ, raises speculation about the genomic origins of immunoglobulin domains and their possible cooption to new immune functions. A short sequence segment, which is located between Cα and the first Jα, shows limited homology to the C region of kappa light chains in cattle and the secreted form of IgH rainbow trout. Although this fragment is an obvious member of Ig-superfamily, its relationship is unclear..
Figure 8. Physical map and annotation of T-cell receptor α locus (TCR locus 2) and tight linkage with IgW2.
Scaffold JH126915 was annotated and shown to contain both the IgW2 locus (see Fig. 2) as well as TCRα. TCRα components are in reverse orientation with respect to IgW2. Unlike the components in TCRα/δ-containing scaffold (Fig. 7), all TCRα components are in the same transcriptional orientation. There are 74 Vα and 59 Jα segments followed by a single constant domain (Cα). Transcriptional orientation is demonstrated by the direction of the arrowhead for each segment. The chromosomal relationship of this region to the TCRα/δ locus (Fig. 7) is unknown.
Coelacanth TCRβ
Gene segments encoding coelacanth TCRβ were found in seven scaffolds. JH127253 (1,055,683 bp) contains 84 V gene segments, at least one D gene segment, 29 J gene segments and a C gene segment in one orientation and six V gene segments in the opposite orientation downstream of the C gene segment. JH134430 (10,111bp) and JH134555 (8,965 bp) contain five and two V gene segments, respectively. The other scaffolds, JH137264 (3,510 bp), AFYH01288189 (1,308 bp), AFYH01289906 (1,164 bp) and AFYH01290638 (1,118 bp) each contain only a single V gene segment. Some of V gene segments apparently are pseudogenes with internal frameshift and/or nonsense mutations. In addition, several segments only are partially characterized and contain bad or marginal sequence stretches. Nonetheless, numerous, potentially functional V gene segments as well as J gene segments can be identified. In marked contrast to TCRα, coelacanth TCRβ is encoded at a single locus represented by the scaffold JH127253. It is assumed that the remaining six scaffolds either map to the middle of scaffold JH127253 or encode orphan genes. In addition, orthologs of CLCN1 (chloride channel voltage-sensitive 1; ENSLACG00000013923), FAM131B (family with sequence similarity 131 member B; ENSLACG00000013702), EPHA1 (EPH receptor A1; ENSLACG00000013051), NOBOX (NOBOX oogenesis homeobox; ENSLACG00000012815) and ARHGEF5 (Rho guanine nucleotide exchange factor (GEF) 5; ENSLACG00000012495) are localized on the 5’ side of scaffold JH127253 and orthologs of EPHB6 (EPH receptor B6; ENSLACG00000005719), KEL (Kell blood group), metallo-endopeptidase; ENSLACG00000002852), NECAP1 (NECAP endocytosis associated 1, i.e., adaptin ear-binding coat-associated protein 1; ENSLACG00000001735) are localized to the 3’ side of scaffold JH127253. The close linkage of CLCN1, FAM131B, EPHA1, NOBOX, ARHGEF35, EPHB6, and KEL to the TCR-β locus has been established in other vertebrates. Specifically, these genes are localized to the 3’ side of the TCRB locus on human chromosome 7q34–35, indicating local chromosomal rearrangement, such as inversion, occurred during vertebrate evolution.
Coelacanth TCRγ
TCRγ genes in the coelacanth can be detected among seven scaffolds: JH127368 (947,744 bp) containing four V gene segments; JH128975 (325,176 bp) containing 11 V gene segments, a pseudo V gene segment, 18 J gene segments and a C gene segment; JH132947 (15,197 bp) containing two V gene segments; JH134594 (8,858 bp) containing two V gene segments and a pseudo V gene segment; JH133588 (11,855 bp) containing two V gene segments and a partial V gene segment; AFYH01286773 (1,488 bp) containing a partial V gene segment and AFYH01287628 (1,377 bp) containing a V gene segment. The 5’ region of the coelacanth TCRγ locus likely is encoded by JH127368 as it contains non-TCRγ genes such as DNAH5 (dynein, axonemal, heavy chain 5, ENSLACG00000002629), whereas the 3’ region of the coelacanth TCRγ locus is encoded by JH128975. As in other vertebrates, V and J gene segments are associated with RSSs containing 23 bp and 12 bp internal spacers, respectively.
Major Histocompatibility Complex genes
MHC proteins are integral molecules in adaptive immunity and their genes provide one of the best examples of balancing selection in vertebrates. Unlike Ig and TCR, genes of the MHC do not undergo genomic rearrangement. MHC proteins present peptide fragments of processed intra-cellular antigens to CD4+ or CD8+ T-cells. MHC I is composed of MHC class I alpha and the invariant beta-2-microglobulin subunits. MHC II is composed of MHC class II alpha and beta subunits. The number and diversity of genes encoding MHC I alpha subunit and MHC II genes are related directly to the potential repertoire of peptide antigens that can be recognized. MHC I and II molecules are linked in many vertebrates; but in teleost fishes, MHC I and II genes are localized to separate genomic regions (Flajnik and DuPasquier 2008). The syntenic relationship of MHC regions provides compelling evidence for two rounds of whole genome duplications occurring at an early stage in vertebrate evolution (Kasahara 1997).
Genes encoding MHC I alpha, beta-2-microglobulin (β2M), MHC II alpha and beta have been identified in homology searches of the African coelacanth genome database. Only the first exon of β2M was detected in the genome database (JH127334: 422278–422347); however, complete transcripts (i.e., comp30430_c0_seq1 of testis transcript) were identified in the Indonesian coelacanth transcriptome (Pallavicini et al. 2013), underscoring the lack of contiguity in some regions of the genome assembly. Nevertheless, at least 29 MHC I alpha, nine MHC II alpha and 12 MHC II beta genes can be recognized in the coelacanth genome database, including a few apparent pseudogenes (Table 2). Notably, the MHC loci are polymorphic and it is expected that some of the sequences could be allelic to each other. In several instances, multiple MHC genes are in close proximity, i.e., six and five MHC I alpha genes in scaffold JH127214 and scaffold JH129212, respectively, and four MHC II alpha and four MHC class II beta genes in scaffold JH128941. However, others are in separate scaffolds, many of which represent extended chromosomal regions (Table 2). Some MHC I alpha and MHC II genes can be localized to scaffolds that contain homologs of COL11A2, RXRB, and SLC39A7 (in scaffold JH128993.1) or homologs of DAXX, SYNGAP1, PHF1, KIFC1, ZBTB9, CUTA, PFDN6, RGL2, TAPBP, WDR46, RPS18, RING1, VPS52, HSD17B8, PSMB8, TAP1, PSMB9 and BRD2 (in scaffold JH127214). Many of these also are found in the MHC region in human and Xenopus (Flajnik and DuPasquier 2008), although the overall degree of chromosomal synteny in the MHC regions is not entirely clear at this point because of the fragmented nature of the current assembly. Furthermore, some genes such as those of complement components (eg., C2, C4A/B, CFB) present in MHC class III region (supplementary table 7), locating between MHC class I and MHC class II region in higher vertebrates, have been reported in the coelacanth genome paper (Amemiya et al. 2013).
Table 2.
Major Histocompatibility Complex genes in Latimeria chalumnae
| (1) MHC class Iα | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Scaffold | Scaffold Size (bp) | Location in the scaffold | |||||||
| Exon 1 | Exon 2 | Exon 2’ | Exon 3 | Exon 4 | Exon 5 | Exon 6 | Exon 7 | ||
| JH126818 | 1794345 | 462259–462307 | 464491–464754 | 465827–466102 | 468353–468628 | 471820–471939 | |||
| JH127214 | 1088405 | 566140–566089 | 565118–564849 | 564207–563929 | 563178–562903 | 562043–561945 | 561360–561328 | 558233–558216 | |
| JH127214 | 1088405 | 612101–612038 | 607138–606869 | 603863–603588 | 601791–601513 | ||||
| JH127214 | 1088405 | 656018–655955 | 654809–654549 | 650086–649810* | 647527–647250* | ||||
| JH127214 | 1088405 | 669524–669262 | |||||||
| JH127214 | 1088405 | 701062–700793* | 699468–699193 | 697298–697020 | |||||
| JH127214 | 1088405 | 865322–865385 | 908130–908405 | 910781–911059 | 912512–912622 | 915120–915137 | 915300–915349 | ||
| JH128073 | 586066 | 153687–153418 | 140330–140055 | 136998–136720 | |||||
| JH128073 | 586066 | 248515–248455* | 212796–212551 | 208340–208308 | 204621–204342 | 202272–201994 | |||
| JH128073 | 586066 | 261470–261724 | 263698–263973 | 266037–266309 | |||||
| JH128073 | 586066 | 273989–274267 | |||||||
| JH128472 | 453462 | 411589–411843 | 414650–414925 | 417963–418241 | |||||
| JH128993 | 322248 | 294360–294088 | 290524–290249 | ||||||
| JH128993 | 322248 | 319591–319319 | 313902–313627 | 311453–311175 | 310305–310189 | 309835–309803* | |||
| JH129212 | 282836 | 167686–167958 | 169382–169657 | 170218–170496 | |||||
| JH129212 | 282836 | 179381–179429 | 181502–181768 | 183016–183291 | 183813–184091 | 184640–184762 | 185229–185261 | 186092–186132 | |
| JH129212 | 282836 | 216314–216586 | 218963–219235 | 221388–221666 | 222528–222644 | 228168–228208 | |||
| JH129212 | 282836 | 248010–248056* | 250273–250545 | 252072–252347 | 253179–253457 | 254026–254139 | 254509–254541 | 255208–255248 | |
| JH129212 | 282836 | 274491–274763 | 277827–278102 | 280159–280437 | 280986–281099 | 281452–281484 | 282156–282196 | ||
| JH129714 | 208590 | 99789–99514 | |||||||
| JH130167 | 156402 | 147202–146933 | |||||||
| JH130808 | 100269 | 43099–43353 | 46245–46520 | 49557–49835 | |||||
| JH130808 | 100269 | 86006–86260 | 88855–89130 | 92486–92764 | |||||
| JH130480 | 126838 | 78758–79027 | 81551–81826 | 83466–83744 | |||||
| JH130480 | 126838 | 96595–96864 | 123365–123640 | 125312–125590 | |||||
| JH130480 | 126838 | 120592–120861 | |||||||
| JH130646 | 112270 | 108385–108322 | 106744–106475* | ||||||
| JH130654 | 112651 | 15338–15084 | 13108–12833 | 6175–5913* | |||||
| JH130654 | 112651 | 81589–81861 | 82741–83016 | 84873–85149* | |||||
| JH132002 | 46392 | 12919–13179 | 15721–15999 | ||||||
| JH132348 | 23389 | 850–743 | |||||||
| JH134769 | 8345 | 3708–3962 | 6586–6861 | ||||||
| JH134901 | 8035 | 3539–3285 | |||||||
| AFYH01279416 | 5177 | 3322–3068 | 1050–775 | ||||||
| AFYH01283960 | 2885 | 1545–1291* | |||||||
| AFYH01287256 | 1426 | 952–1227 | |||||||
| (2) MHC class IIα | ||||||
|---|---|---|---|---|---|---|
| Scaffold | Scaffold Size (bp) | Location in the scaffold | ||||
| Exon 1 | Exon 2 | Exon 3 | Exon 4 | Exon 5 | ||
| JH128941 | 332998 | 3322–3325 | ||||
| JH128941 | 332998 | 62062–62143 | 64994–65251 | 74161–74442 | 75274–75412 | 77767–77770 |
| JH128941 | 332998 | 151751–151832 | 155539–155790 | 176762–176765 | ||
| JH128941 | 332998 | 325931–326069 | 328282–328285 | |||
| JH131877 | 48675 | 41269–41517 | 41959–42240 | 44448–44586 | 46714–46717 | |
| JH132119 | 43233 | 3707–3845 | ||||
| JH133334 | 12986 | 5895–6146 | 8990–9271 | 12390–12528 | ||
| JH133683 | 11598 | 5404–5332 | 4970–4722 | 4281–4000 | ||
| JH134128 | 10105 | 3916–4197 | 8942–9080 | |||
| JH135774 | 6059 | 4200–3949 | ||||
| AFYH01281632 | 4068 | 221–140 | ||||
| AFYH01282120 | 3832 | 2036–2287 | ||||
| AFYH01284662 | 2301 | (2301)-2166 | 142–4 | |||
| AFYH01288882 | 1242 | (1242)-1169 | 806–558 | 142-(1) | ||
| AFYH01285476 | 1880 | 1239–1164 | 775–527 | 50-(1) | ||
| (3) MHC class IIβ | |||||||
|---|---|---|---|---|---|---|---|
| Scaffold | Scaffold Size (bp) | Location in the scaffold | |||||
| Exon 1 | Exon 2 | Exon 3 | Exon 4 | Exon 5 | Exon 6 | ||
| JH127214.1 | 1088405 | 310508–310224 | |||||
| JH128941.1 | 332998 | 11339–11438 | 24130–24402 | 29823–30104 | 31326–31439 | 32932–32969 | |
| JH128941.1 | 332998 | 105250–105349 | 107185–107457 | 109707–109988 | 111265–111378 | 112188–112220 | 112971–112996 |
| JH128941.1 | 332998 | 186058–186157 | 187393–187665 | 190179–190460 | 196080–196193 | 199838–199870 | |
| JH128941.1 | 332998 | 300189–300093 | 296851–296582 | 293251–292973 | 290843–290730 | 289298–289266 | 289162–289137 |
| JH128993.1 | 322248 | 45498–45244 | 41707–41429 | 39296–39183 | |||
| JH131812.1 | 49899 | 6788–6675 | 3740–3708 | 3585–3560 | |||
| JH131877.1 | 48675 | 11390–11121 | 6266–5988 | 3886–3773 | 2329–2297 | 2193–2168 | |
| JH132119.1 | 43233 | 41828–41876 | |||||
| JH132855.1 | 15952 | 2699–2977 | 4132–4245 | 5389–5421 | 5525–5550 | ||
| JH132855.1 | 15952 | 15510–15478 | 15374–15349 | ||||
| JH133922.1 | 10792 | 3829–3730 | |||||
| JH134191.1 | 9945 | (9945)-9744 | 6004–5891 | 5123–5091 | 4984–4959 | ||
| JH134411.1 | 9293 | 6864–6583 | 4538–4425 | 2434–2402 | 2279–2254 | ||
| JH134772.1 | 8384 | 535–634 | 5207–5479 | ||||
| JH135383.1 | 6924 | 979–1251 | 5209–5490 | ||||
| AFYH01279633.1 | 5076 | 1402–1130 | |||||
| AFYH01283994.1 | 2863 | 840–568 | |||||
| AFYH01284697.1 | 2290 | 419–691 | |||||
| AFYH01287635.1 | 1377 | (1377)-1215 | |||||
| AFYH01288041.1 | 1323 | 681–794 | |||||
| AFYH01289521.1 | 1191 | 778–877 | |||||
an exon containing apparent defects, such as a termination codon in-frame or frameshift mutation.
Our data on MHC class I genes largely corroborates a previous report (Betz et al. 1994) that identified sequences of L. chalumnae MHC I genes. The analysis of the MHC genes, with reference to their polymorphism, coalescence and evolution, should now be possible given these data and that of a recently published report of genome sequences of additional coelacanth specimens (Nikaido et al. 2013).
Recombination Activating Genes
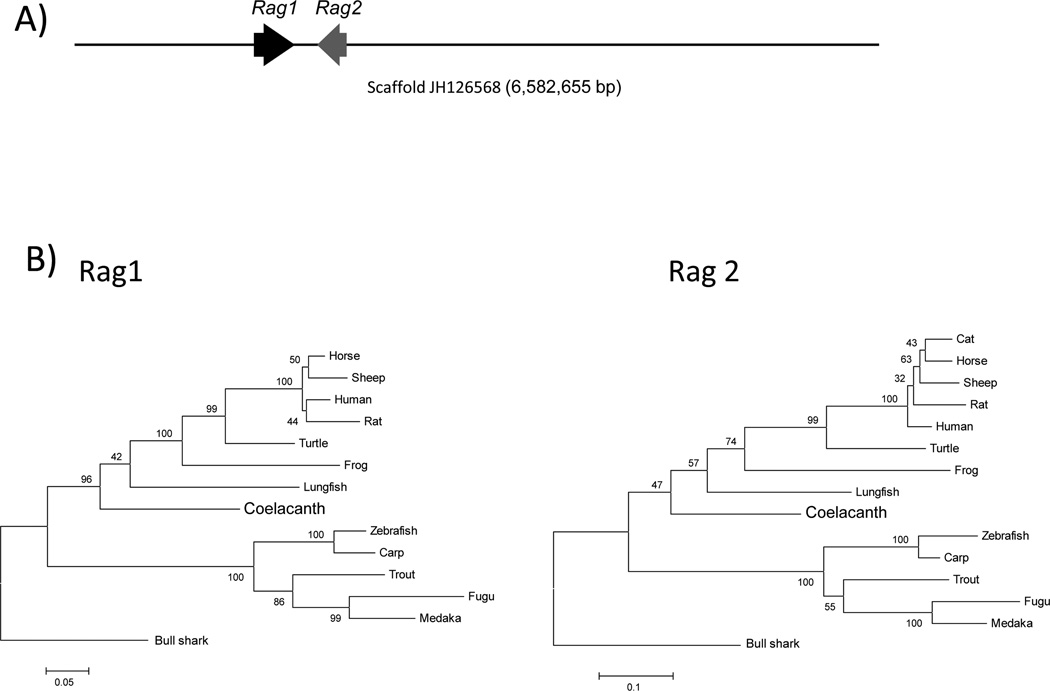
Arguably, one of the most important events in the evolution of the adaptive immune system was the integration of the Rag genes via a transposon-based insertion event into the genome of a common ancestor of deuterostomes (Fugmann et al. 2006). Rag1 and Rag2, which mediate V(D)J recombination, are imperative for both the somatic generation of Ig and TCR, and ultimately, for the maturation of B- and T- lymphocytes. The genomic and transcriptomic databases of the Latimeria were searched using the partial sequences of previously identified coelacanth Rag1 and Rag2 (Brinkmann et al. 2004). The coelacanth Rag1 and Rag2 genes were localized to a 6.58 megabase scaffold (JH126568). Both genes consist of a single exon, unlike teleost fishes, and are in opposite transcriptional orientation (Fig. 9A). Coelacanth Rag1 is predicted to consist of 1058 amino acids, whereas Rag2 consists of 522 amino acids. The distance between the two genes is 10.6 kb, which is shorter than in human (15 kb) but longer than in zebrafish (2.6 kb) and trout (2.4 kb) (Hansen and Kaattari 1996;Willett et al. 1997). Of greater significance, long-range synteny is evident over the 17 genes flanking the Rag locus of coelacanth and tetrapods, compared with only 2 conserved flanking genes between coelacanth and bony fish (Brinkmann et al. 2004). These results suggest that the Rag genes have been highly conserved during sarcopterygian evolution in terms of both their gene organization and extended genomic milieu.
Figure 9. Analysis of coelacanth Rag genes.
(A) Both Rag1 and Rag2 are located on genomic scaffold JH126568 (6,582,655 bp) at positions 121275–124451 and 135133–136701, respectively, and are oriented in a head-to-head manner. Each Rag gene is composed of a single exon. Only the first 150 kp of the large scaffold is illustrated. (B) Phylogenetic analysis of amino acid sequences of Rag1 (left) and Rag2 (right) by Maximum Likelihood method. The trees are rooted with bull shark Rag1 and Rag2; the % bootstrap replicates are indicated on the tree. The topology of the tree is consistent with known phylogeny, though strong bootstrap support is lacking for some of the nodes. GenBank accession numbers for all sequences used for (Rag1; Rag2): horse (NP_001243830; XP_001488023), rat (NP_445920; NP_001093998), sheep (XP_004016460; XP_004016461), lungfish (AAS75810; AAS75812), turtle (ACJ48241; AF369089), frog (ABS00344; AAI29720), zebrafish (NP_71464; NP_571460), fugu (AAD20561; AAD20562), trout (NP_001118209; AAB18138), carp (AAX16495; AAX16496), medaka (XP_004070148; XM_004069726), human (NP_00439; NP_000527) and cat (XP_003993217; XP_004001473).
The lack of lymphopoietic tissues in the RNAseq analyses limits the capacity to identify actively transcribed Rag genes. We only could identify three short transcripts (contig3388, contig89407 and contig10412) aligning to different locations within the Rag1 gene from the L. menadoensis liver transcriptome (Pallavicini et al. 2013) (Supplementary Fig. 5). No Rag2 transcripts were identified.
Rag1 and 2 sequences are frequently used for phylogenetic analyses due to their ubiquity in all jawed vertebrate taxa and to evolutionary behavior that is not outwardly affected by differences in molecular evolutionary rates (Brinkmann et al. 2004;Cramer et al. 2011). The degree to which these sequences are conserved is shown in Supplementary Table 6, which lists percent similarities and percent identities between coelacanth Rag1 and Rag2 versus those of various vertebrate taxa, and demonstrates that the Rag proteins are moderately, but not highly conserved. Phylogenetic trees constructed from the same amino acid sequences were used to assess the interrelationships of Rag1 and Rag2 among vertebrates (Fig. 9B) and more-or-less corroborate established phylogenetic relationships, although not always with high bootstrap support.
Activation-induced cytidine deaminase
Activation-induced cytidine deaminase (Aicda, AID) is currently thought to be the master regulator of secondary antibody diversification through the initiation of three separate Ig diversification processes: somatic hypermutation, gene conversion, and class switch recombination. Somatic hypermutation involves a programmed mutational process affecting the V regions of Ig genes, whereas gene conversion is involved in partially templated replacement of portions of V regions of genes (Longerich et al. 2008). Both processes are mediated by AID and diversify the antibody repertoire. In contrast, class switch recombination does not alter the specificity of the antibody but supplants the C region of the Ig heavy chain and, consequently, its effector function (Kataoka et al. 1980). Class switch recombination (CSR) appears at the time of the emergence of the amphibians and is conserved in all tetrapods. It is absent in teleost fish (Stavnezer and Amemiya 2004), although, paradoxically, teleost AID protein has been shown to undergo CSR catalytic activity in in vitro assays despite the fact that teleosts lack genomic loci amenable to CSR (Wakae et al. 2006;Barreto et al. 2005). The coelacanth AID gene is encompassed in two overlapping scaffolds: scaffold JH127875 (∼656 kb) and scaffold JH135912 (3,785 bp) as depicted in Supplementary Fig. 6A. The predicted coding sequence is 582 bp (183 amino acids). The first few amino acids at the amino terminus are somewhat uncertain because of compromised sequence stretches at the 5' end that confound annotation; however, the predicted protein contains the most important functional portions of the AID protein, including its catalytic domain and carboxy-terminal region, which are essential for the CSR activity (Ichikawa et al. 2006). Although the C-terminus clearly has the NLS motif the coelacanth C-terminus is distinct from any other AID in having that 10–20 residue extension. The alignment of the coelacanth AID to representative AID molecules from other species is given in Supplementary Fig. 6B and shows high overall identity. The IgW loci of coelacanth do not possess cognate switch regions within and around their constant region, thereby precluding classical CSR; however, from an evolutionary standpoint it will be curious to assess the biochemical capabilities of coelacanth AID in a surrogate assay system.
Cluster of differentiation (CD) molecules
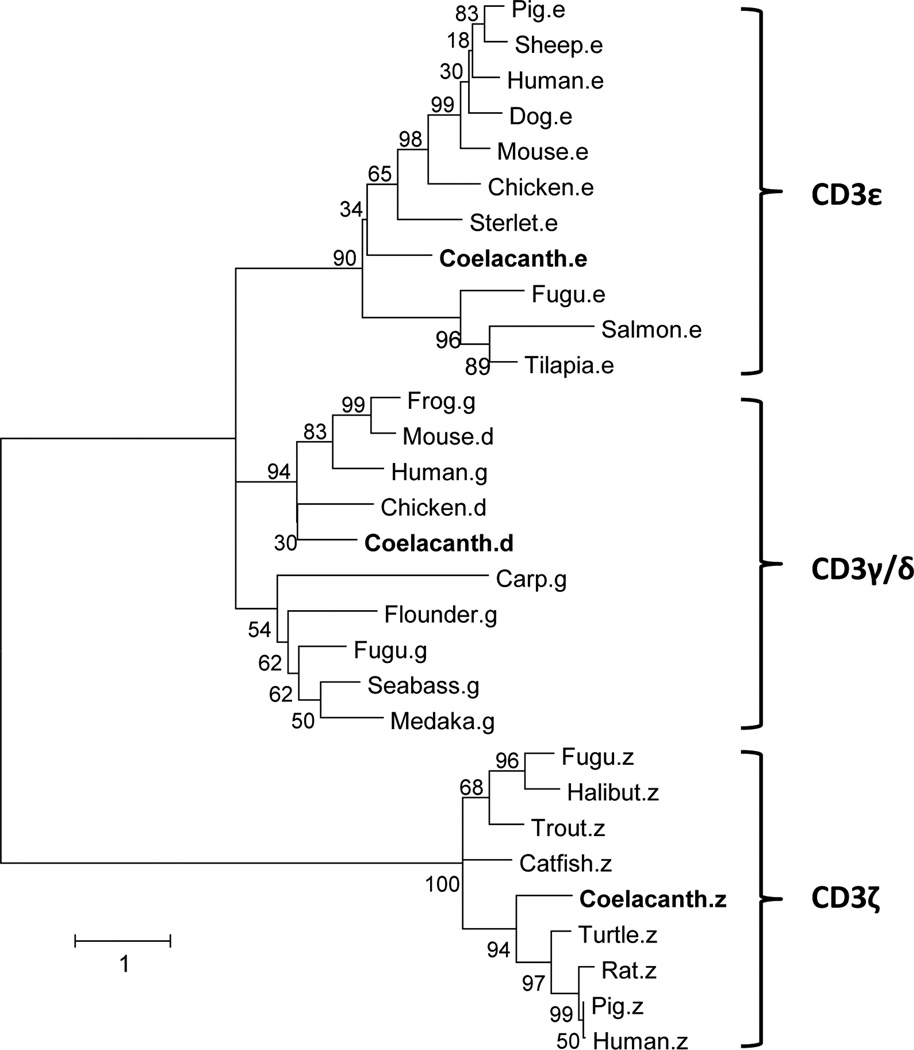
T-cells are classified into subsets based on their functionality and expression of distinct surface receptors. In mammals, the protein encoded by CD3-epsilon (CD3ε), together with CD3-gamma (Cd3γ), CD3-delta (CD3δ) and CD3-zeta (CD3ζ) and the TCR-α/β and -γ/δ heterodimers, form the TCR-CD3 complex. The CD3 components largely are responsible for antigen ligation events with intracellular signaling leading to the activation of the T-cell. CD3γ, CD3δ and CD3ε chains, each of which contain a single extracellular Ig domain, are closely related. However, in chickens, amphibians and fish, the CD3γ and CD3δ subunits are replaced by a CD3γ/δ subunit (Ropars et al. 2002;Bernot and Auffray 1991;Dzialo and Cooper 1997;Araki et al. 2005;Park et al. 2005). It is inferred that separate mammalian CD3γ and CD3δ molecules were derived from a tandem gene duplication. In coelacanth, three CD3 chains, which are orthologous to CD3ɛ, CD3γδ and CD3ζ, have been identified in both the genome assembly and transcriptome datasets. The scaffold, JH126582, contains both CD3γ/δ and CD3ε genes at nucleotide positions 2908204–2913094 and 2923833–2931815, respectively. CD3ζ is located on scaffold JH128766 between 158334 and 168776. The complete cDNA sequences of CD3 gamma-delta (Contig 43331) and zeta (Contig 96288) have been identified in the liver transcriptome dataset of L. menadoensis. A phylogenetic analysis of amino acid sequences of CD3ɛ, CD3γ and CD3γ/δ has been performed (Fig. 10). All three chains of CD3 from coelacanth are distinct from the corresponding sequences from other fishes and, in terms of sequence homology, group together with the corresponding molecules found in avians and mammals.
Figure 10. Phylogenetic relationships of CD3.
The phylogenetic tree was generated via Maximum Likelihood method. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial trees for the heuristic search were obtained automatically by applying Neighbor-Joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with the superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 30 predicted amino acid sequences. There were a total of 234 positions in the final dataset. GenBank accession numbers used in this analysis: carp.γ (DQ340867), fugu.γ (AB166800), seabass. γ (FN667954), medaka. γ (XM_004076021), flounder. γ (AB044573), human. γ (NP_000064), mouse.δ (NP_038515), frog. δ (XP_508789), chicken. δ (NP_990843), sterlet.ɛ (AJ242941), salmon. ɛ (GU180241), fugu. ɛ (AB166798), pig. ɛ (AY323829), mouse. ɛ (BC145926), dog. ɛ (M55410), sheep. ɛ (S53077), chicken. ɛ (EU779493), human. ɛ (BC049847), tilapia. ɛ (XP_003449345), Fugu.ζ (XM_003966619), Catfish. ζ (FJ809774), Halibut. ζ (FJ769820), trout. ζ (NM_001165113), human. ζ (AAA60394), rat. ζ (NP_740770), turtle. ζ (ADP21384), pig. ζ (NP_000725).
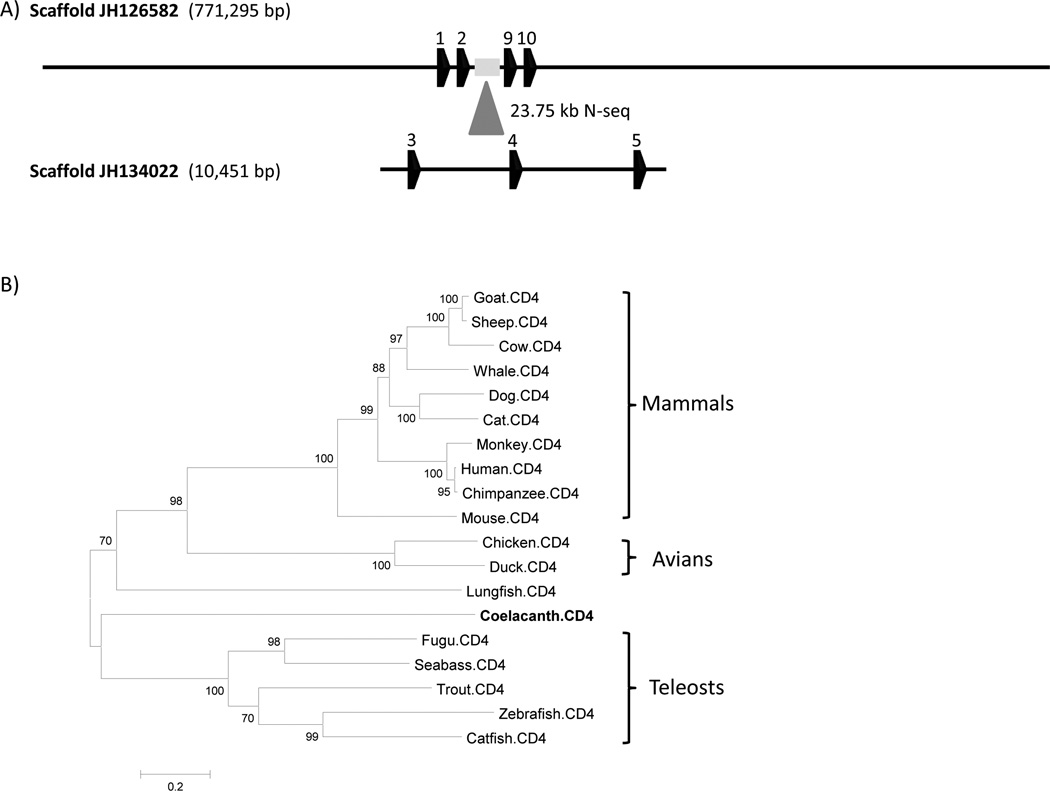
CD4, a single chain transmembrane glycoprotein, is expressed by helper T-cells and is a co-receptor with TCR in MHC II-mediated antigen recognition. CD4 has a fundamental role in thymocyte selection during development. In the context of antigen recognition by TCR, CD4 dimerizes and binds to the α2 and β2 domains of MHC class II molecules (Wu et al. 1997;Huang et al. 1997), acting as a TCR co-receptor. CD4 is composed of four Ig domains, a transmembrane region and a cytoplasmic tail that contains the canonical CXC motif involved in the interaction of CD4 with p56LCK, which is required for signal 1 of T-cell activation. A CD4 ortholog was identified in the L. menadoensis transcriptome dataset (Supplementary Fig. 9). Key functional motifs that potentially could be involved in the regulation of CD4 transcription also were identified. A large scaffold (JH126582, 771 kb) containing CD4 was identified in the L. chalumnae genome (Fig. 11A). However, many of the exons could not be identified owing to a 27 kb assembly gap (position 304318–356023 bp). This scaffold contains exon 1 (5’ UTR), exon 2 (5’ UTR and leader peptide), exon 9 and exon 10 (3’ UTR), based on the CD4 molecules found in human (Ansari-Lari et al. 1996), chicken (Koskinen et al. 2002) and other fish species, including zebrafish (unpublished data). A more extensive search led to the identification of a 10.4 kb scaffold (JH134022), which consists of the “missing” exons 3, 4 and 5. A phylogenetic analysis was carried out that included lungfish CD4 (Fig. 11B); however, the resolution of the coelacanth and lungfish branches was poor due to overtly long branches.
Figure 11. Coelacanth CD4 annotation and phylogenetic analysis.
(A) Two scaffolds, JH126582 and JH10451, contain 4 and 3 exons, respectively. Scaffold JH134022, which contains exon 3, 4 and 5, maps to the assembly gap (∼ 23.75 kb) of scaffold JH126582. (B) The unrooted phylogenetic tree was inferred by using the Maximum Likelihood method. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 19 CD4 amino acid sequences, with a total of 535 positions in the final dataset. The resolution of the coelacanth and lungfish branches is poor due to the deep branch lengths. GenBank accession numbers of CD4s: Zebrafish (NP_001128568), fugu (NP_001072091), trout (NP_001118011), catfish (ABD93355), seabass (CAO98731), chicken (NP_989980), duck (AF378701), chimpanzee (NP_001009043), human (NP_000607), mouse (NP_038516), dog (NP_001003252), cow (NP_001096695), cat (NP_001009250), monkey (CAA51752), sheep (NP_001123374), whale (NP_001267583), goat (ACG76115).
CD8 is a membrane bound glycoprotein found on cytotoxic T-cells that consists of either CD8αα homodimers or CD8αβ heterodimers. Both chains are composed of a single Ig domain linked to the membrane by a segment of extended polypeptide chain. Both CD8α and CD8β have been identified in most jawed vertebrates (Suetake et al. 2007;Nagarajan et al. 2004;Moore et al. 2005), and both genes also have been identified in the coelacanth genome assembly within the same locus (JH128706) at a distance of ∼ 84 kb (Fig. 12). The CD8β gene consists of 9 exons and the spans ∼49 kb. For CD8α, four exons were predicted over a span of 25 kb that includes ∼16 kb of poorly assembled sequence. However, it was not possible to identify sequences corresponding to a transmembrane and cytoplasmic tail region of CD8α because of a sequence gap downstream of exon 4. Both coelacanth CD8α and CD8β show strong similarities to corresponding molecules of other vertebrates. A partial CD8α sequence (contig40330) has been identified in the liver transcriptome, but no expressed CD8β was identified.
Figure 12. Annotation of the scaffold containing CD8 genes.
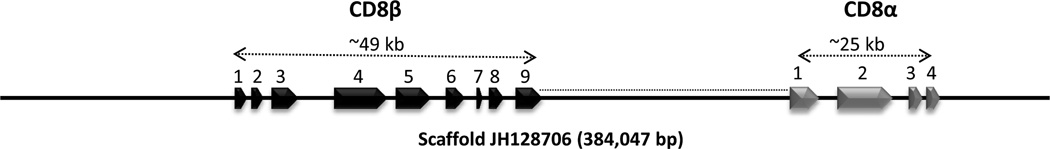
Genes for both CD8α and CD8β are located on JH128706, with an intergenic distance of ∼ 84 kb. Four exons of CD8α span ∼ 25 kb, including a stretch of ∼16 kb of poorly assembled sequence. The transmembrane and cytoplasmic tail could not be definitively identified owing to a sequence gap just downstream of exon 4. The CD8β gene consists of 9 exons that span ∼ 49 kb.
Use of text searches on the Ensembl annotated assembly uncovered numerous other CD molecules (Supplementary Table 8). Manual BLAST searches on these molecules were used to validate that these were orthologous to their mammalian counterparts. The T-cell-specific surface glycoprotein, CD28, is located on JH127402 (430316–43931). CD9, which associates with CD3, CD4, CD5, CD29 and CD44, also was found in coelacanth. A gene encoding CD40 (costimulatory molecule involved in antigen presentation and class switching) was not identified. However, CD40L, also known as CD154 and a key member of TNF superfamily expressed on activated T-cells and is found on scaffold JH126623 (1333194–1340474). CD40L is comprised of 3 exons and the translated amino acid residues show significant identity (∼40%) with CD40L of turtle (data not shown). CD45, known as leukocyte common antigen, was found on two scaffolds, JH127371 and JH126742. Key mammalian CD molecules, such as stem cell markers CD34, CD31 and CD117 have not been identified in the coelacanth genome, however, without better search tools and a more complete genome, it is, as yet, difficult to definitely state that they (including CD40) are truly absent.
Cytokines
Cytokines are small signaling molecules secreted by specific cells of the immune system that mediate signals between cells. Cytokines are transcribed from many cell types as needed in the course of an immune response. They are critical to the development and functioning of both the innate and adaptive immune responses, and modulate the responses in an autocrine or paracrine manner upon binding to their corresponding receptors (Zhu et al. 2013). Cytokines can be divided into interferons (IFNs), interleukins (ILs), tumor necrosis factors (TNFs), colony stimulating factors (CSF), and chemokines (Savan and Sakai 2006).
A large number of ILs orthologous to those in mammals have been identified in the coelacanth genome (Fig. 13), however, others such as IL-2, IL-4, IL-5, IL-6, IL-7, IL-9, IL-15, and IL-21, which play crucial roles in the adaptive immunity of mammals, were not detected in this study. As is the case with many other genes, it is unclear whether they are absent from the genome or are not being detected because of substantial sequence divergence and/or sequencing-assembly issues. However, the cognate receptors for some of these (e.g., IL-2, IL-6, IL-7 and IL-21) definitively have been identified in the genome (Supplementary Table 9), making the latter explanation more plausible. STAT-6, a member of STAT family transcription factors that play a central role in exerting IL4-mediated biological responses, has been identified (JH126563: 4604284–4647669 bp). The genes encoding IL-1β and IL-18 have been discussed in detail in a companion paper (Boudinot et al. 2014).
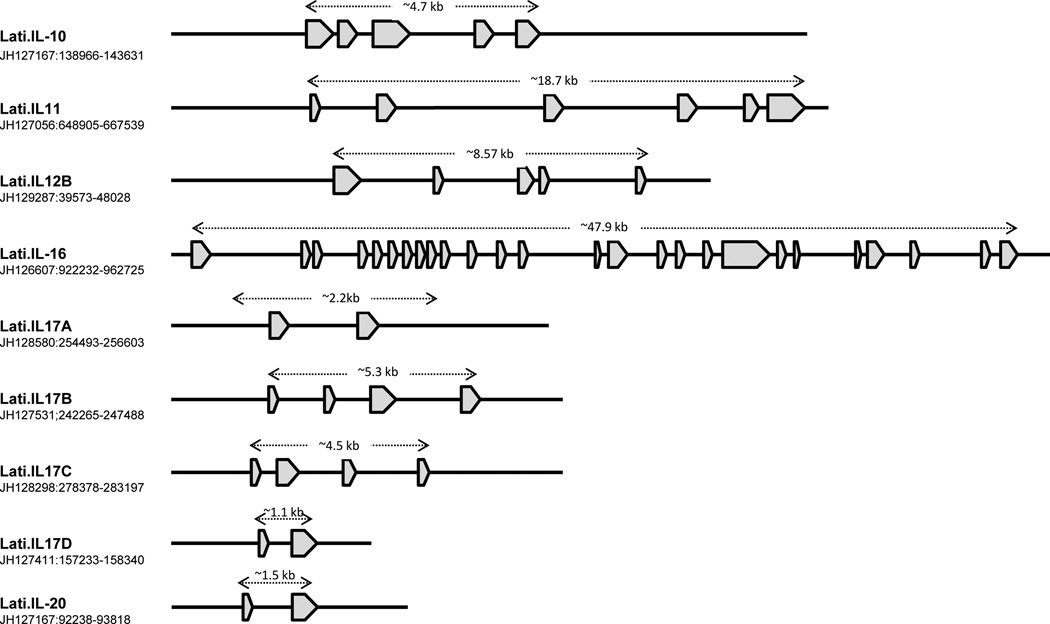
Figure 13. Annotation of the scaffolds encoding coelacanth interleukin genes.
Genes encoding interleukins were identified from the coelacanth annotated assembly at the Ensembl. Coding sequences were retrieved from their corresponding scaffolds and imported into Vector NTI sequence analysis software (Invitrogen). GENESCAN (Burge and Karlin 1997) and BLASTX programs were used, respectively, to predict the exon-intron boundaries and determine amino acid alignments to other vertebrates.
Interleukin-10 is an anti-inflammatory cytokine capable of inhibiting synthesis of pro- inflammatory cytokines such as IFN-γ, IL-2, IL-3, TNFα and GM-CSF, which are made by cells such as macrophages and regulatory T-cells. In mammals, IL-10 regulates growth and/or differentiation of B-cells, NK cells, cytotoxic and helper T-cells, mast cells, granulocytes, dendritic cells, keratinocytes, and endothelial cells (Moore et al. 2001), and also stimulates certain Th2 cells, mast cells and B-cells. The coelacanth IL-10 gene, is comprised of five exons (like that of human) and codes for a protein of 184 amino acid residues (Fig. 13). Coelacanth IL-10 has highest identity with IL-10 of green anole (52%), western clawed frog (50%), chicken (46%) and bottlenose dolphin (46%). The IL-10 family of interleukins also contains IL-20, which is present on the same scaffold (JH127167) as the gene for IL-10, localized ∼45 kb upstream and in opposite transcriptional orientation. Although human IL-20 is composed of five exons, only two exons were identified in coelacanth. This partial amino acid sequence has the highest overall identity (63%) to that of the gray short-tailed opossum.
IL-11, a stromal cell-derived member of the IL-6-type cytokine family, shares its receptor and signal transduction partially with IL-6. IL-11 functions in a wide range of hematopoietic and non-hematopoietic systems and supports the growth of plasmacytoma and hybridoma cells. IL-11 in coelacanth encodes six exons, as opposed to the human ortholog, which consists of five exons (Figure 13). Although the homologous regions of exon 1 and 6 have not been established in any other animals, exons 2–5 are highly conserved with IL-11 genes of other animals, e.g., zebra finch (66%), chicken (62%), western painted turtle (60%) and western clawed frog (57%). In addition, two IL-11 receptors (IL-11Rα and IL-11Rβ) also have been identified in coelacanth (Supplementary Table 9).
IL-12 is produced by activated macrophages and dendritic cells, stimulates the production of IFN-γ, induces the differentiation of Th cells to become Th1 cells (Heufler et al. 1996) and enhances the cytolytic functions of cytotoxic T-cells and NK cells. An IL-12 ortholog, which is comprised of exons, has been identified in coelacanth (Fig. 13) and is shown to exhibit moderate identity to rock pigeon (40%), chicken (40%), western painted turtle (40%) and peregrine falcon (39%). IL-10 production by Th1 cells requires an IL-12-induced STAT4 transcription factor (Saraiva et al. 2009), which also has been identified in coelacanth (JH128710 at the position of 208614–275761 bp).
IL-16 is involved with adaptive immunity. It is highly sensitive to mitogens phytohemaglutinnin and ConA and stimulates T-lymphocyte proliferation and activation in pufferfish (Wen et al. 2006). An IL-16 homolog, which consists of 26 exons (Fig. 13), has been detected in coelacanth. The predicted IL-16 encodes 1,592 amino acids and exhibits significant homology to that of the green sea turtle (45%), chicken (43%) and mallard (43%).
In human, the interleukin 17 family includes six members, IL-17A, IL-17B, IL-17C, IL-17D, IL-17E/IL-25, and IL-17F, which are produced by multiple cell types. At least four IL-17 genes (IL-17A, IL-17B, IL-17C and IL-17D) (Fig. 13), as well as genes for three of the corresponding receptors have been identified in coelacanth (Supplementary Table 9). Both Il-17 and their receptors show closer phylogenetic relationships to orthologous forms in tetrapods than to the teleost orthologs (tree not shown).
In addition to interleukins, the coelacanth genome also contains many other orthologs to mammalian homolog cytokine genes and their receptors. For example, the gene for Transforming growth factor beta (TGF-β) has been identified in one scaffold (JH126565:1664929–1738138), whereas those for TGF-β receptors are present in two other scaffolds (JH126740:103899–129604 and JH128485:346074–379745). Macrophage migration inhibitory factor (MIF), which is one of the important regulators of innate immunity, is located in scaffold JH126570. A tumor necrosis factor receptor superfamily member 1B (TNF-1β) is found on scaffold JH126880 and discussed at length in the coelacanth innate immune paper (this issue). Lastly, we have identified genes for a large number of putative cytokines via our data mining efforts (Suppl. Table 9); these will be described and characterized in a separate report.
Conclusions
The recent genome sequencing of the coelacanth has provided unique insights into its biology and evolutionary position (Amemiya et al. 2013). The availability of the genome and transcriptome assemblies as well as BAC resources, has allowed characterization of genes and gene families encoding the coelacanth immunome. The coelacanth genome encodes large numbers of immune receptors of the Ig superfamily, including Igs, TCRs, MHC, TCR co-receptors, as well as immune regulatory molecules, differentiation antigens, and presumptive additional immune multigene families that cannot be placed definitively. The numbers of surveyed genes is far from exhaustive, though we have focused on the most relevant ones to adaptive immunity in this paper. Most of the phylogenetic analyses of gene trees support a placement of coelacanth between the teleost fishes and the tetrapods, with coelacanth having an overall higher affinity with tetrapods. Certain findings stand out and will require further investigation including the chimeric gene organization of the IgH loci, lack of IgM (long considered a conditio sine qua non for the adaptive immune response), the presence of two different loci for IgW, the multiplicity of constant domains in its IgW loci, the close proximity of TCRα and IgW loci, the evolutionary relationships of IgW with IgD, and interdigitation of VH genes within the α/δ TCR locus. The adaptive immunome of the coelacanth is certainly as complex as that for any vertebrate thus far studied and this is reflected in its genome complexity
Supplementary Material
Acknowledgments
The authors thank the coelacanth genome sequencing consortium for access to the sequence resources prior to the publication of the landmark coelacanth genome paper. This work was supported, in part, by National Science Foundation grants IOS-0321461 & MCB-0719558 (to C.T.A.), and National Institutes of Health grants HL66728 (to E. Rubin and J.-F.C.), AI23338 & AI57559 (to GWL) and RR14085 & GM090049 (to C.T.A.), and The US Geological Survey (JH). Any use of trade names is for descriptive purposes only and does not imply endorsement by the U.S. Government. We thank Marco Gerdol and Mark Robinson for help with bioinformatics, Gail Mueller for help with screening and PCR experiments for Cμ, and Giuseppe Scapigliati, Martin Flajnik and Louis Du Pasquier for early discussions of the data.
Reference List
- Amemiya CT, Alfoldi J, Lee AP, Fan S, Philippe H, Maccallum I, Braasch I, Manousaki T, Schneider I, Rohner N, Organ C, Chalopin D, Smith JJ, Robinson M, Dorrington RA, Gerdol M, Aken B, Biscotti MA, Barucca M, Baurain D, Berlin AM, Blatch GL, Buonocore F, Burmester T, Campbell MS, Canapa A, Cannon JP, Christoffels A, De MG, Edkins AL, Fan L, Fausto AM, Feiner N, Forconi M, Gamieldien J, Gnerre S, Gnirke A, Goldstone JV, Haerty W, Hahn ME, Hesse U, Hoffmann S, Johnson J, Karchner SI, Kuraku S, Lara M, Levin JZ, Litman GW, Mauceli E, Miyake T, Mueller MG, Nelson DR, Nitsche A, Olmo E, Ota T, Pallavicini A, Panji S, Picone B, Ponting CP, Prohaska SJ, Przybylski D, Saha NR, Ravi V, Ribeiro FJ, Sauka-Spengler T, Scapigliati G, Searle SM, Sharpe T, Simakov O, Stadler PF, Stegeman JJ, Sumiyama K, Tabbaa D, Tafer H, Turner-Maier J, van HP, White S, Williams L, Yandell M, Brinkmann H, Volff JN, Tabin CJ, Shubin N, Schartl M, Jaffe DB, Postlethwait JH, Venkatesh B, DiPalma F, Lander ES, Meyer A, Lindblad-Toh K. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya CT, Ohta Y, Litman RT, Rast JP, Haire RN, Litman GW. VH gene organization in a relict species, the coelacanth Latimeria chalumnae: evolutionary implications. Proc Natl Acad Sci U S A. 1993;90:6661–6665. doi: 10.1073/pnas.90.14.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MK, Strong SJ, Litman RT, Luer CA, Amemiya CT, Rast JP, Litman GW. A long form of the skate IgX gene exhibits a striking resemblance to the new shark IgW and IgNARC genes. Immunogenetics. 1999;49:56–67. doi: 10.1007/s002510050463. [DOI] [PubMed] [Google Scholar]
- Andersson E, Matsunaga T. Evolution of immunoglobulin heavy chain variable region genes: a VH family can last for 150–200 million years or longer. Immunogenetics. 1995;41:18–28. doi: 10.1007/BF00188428. [DOI] [PubMed] [Google Scholar]
- Ansari-Lari MA, Muzny DM, Lu J, Lu F, Lilley CE, Spanos S, Malley T, Gibbs RA. A gene-rich cluster between the CD4 and triosephosphate isomerase genes at human chromosome 12p13. Genome Res. 1996;6:314–326. doi: 10.1101/gr.6.4.314. [DOI] [PubMed] [Google Scholar]
- Araki K, Suetake H, Kikuchi K, Suzuki Y. Characterization and expression analysis of CD3varepsilon and CD3gamma/delta in fugu, Takifugu rubripes. Immunogenetics. 2005;57:158–163. doi: 10.1007/s00251-005-0772-8. [DOI] [PubMed] [Google Scholar]
- Barreto VM, Pan-Hammarstrom Q, Zhao Y, Hammarstrom L, Misulovin Z, Nussenzweig MC. AID from bony fish catalyzes class switch recombination. J Exp Med. 2005;202:733–738. doi: 10.1084/jem.20051378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengten E, Leanderson T, Pilstrom L. Immunoglobulin heavy chain cDNA from the teleost Atlantic cod (Gadus morhua L.): nucleotide sequences of secretory and membrane form show an unusual splicing pattern. Eur J Immunol. 1992;22:294. doi: 10.1002/eji.1830220145. [DOI] [PubMed] [Google Scholar]
- Bernot A, Auffray C. Primary structure and ontogeny of an avian CD3 transcript. Proc Natl Acad Sci U S A. 1991;88:2550–2554. doi: 10.1073/pnas.88.6.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz UA, Mayer WE, Klein J. Major histocompatibility complex class I genes of the coelacanth Latimeria chalumnae. Proc Natl Acad Sci U S A. 1994;91:11065–11069. doi: 10.1073/pnas.91.23.11065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudinot P, Zou J, Ota T, Buonocore F, Scapigliati G, Canapa A, Cannon J, Litman GW, Hansen AJ. A tetrapod-like repertoire of innate immune receptors and effectors for coelacanths: an emphasis on antiviral immunity. Journal of Experimental Zoology, part B, Molecular and Developmental Evolution. 2014 doi: 10.1002/jez.b.22559. Ref Type: In Press. [DOI] [PubMed] [Google Scholar]
- Brinkmann H, Venkatesh B, Brenner S, Meyer A. Nuclear protein-coding genes support lungfish and not the coelacanth as the closest living relatives of land vertebrates. Proc Natl Acad Sci U S A. 2004;101:4900–4905. doi: 10.1073/pnas.0400609101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Campanella JJ, Bitincka L, Smalley J. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics. 2003;4:29. doi: 10.1186/1471-2105-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer CA, Bonatto SL, Reis RE. Molecular phylogeny of the Neoplecostominae and Hypoptopomatinae (Siluriformes: Loricariidae) using multiple genes. Mol Phylogenet Evol. 2011;59:43–52. doi: 10.1016/j.ympev.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Criscitiello MF, Flajnik MF. Four primordial immunoglobulin light chain isotypes, including lambda and kappa, identified in the most primitive living jawed vertebrates. Eur J Immunol. 2007;37:2683–2694. doi: 10.1002/eji.200737263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscitiello MF, Ohta Y, Saltis M, McKinney EC, Flajnik MF. Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. J Immunol. 2010;184:6950–6960. doi: 10.4049/jimmunol.0902774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscitiello MF, Saltis M, Flajnik MF. An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc Natl Acad Sci U S A. 2006;103:5036–5041. doi: 10.1073/pnas.0507074103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow KD, Smith CD, Cheng JF, Wagner GP, Amemiya CT. An independent genome duplication inferred from Hox paralogs in the American paddlefish--a representative basal ray-finned fish and important comparative reference. Genome Biol Evol. 2012;4:937–953. doi: 10.1093/gbe/evs067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Bussmann J, Jekosch K, Steiner LA. The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nat Immunol. 2005;6:295–302. doi: 10.1038/ni1166. [DOI] [PubMed] [Google Scholar]
- Danke J, Miyake T, Powers T, Schein J, Shin H, Bosdet I, Erdmann M, Caldwell R, Amemiya CT. Genome resource for the Indonesian coelacanth, Latimeria menadoensis. J Exp Zoolog A Comp Exp Biol. 2004;301:228–234. doi: 10.1002/jez.a.20024. [DOI] [PubMed] [Google Scholar]
- Das S, Hirano M, Tako R, McCallister C, Nikolaidis N. Evolutionary genomics of immunoglobulin-encoding Loci in vertebrates. Curr Genomics. 2012;13:95–102. doi: 10.2174/138920212799860652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzialo RC, Cooper MD. An amphibian CD3 homologue of the mammalian CD3 gamma and delta genes. Eur J Immunol. 1997;27:1640–1647. doi: 10.1002/eji.1830270708. [DOI] [PubMed] [Google Scholar]
- Edholm ES, Wilson M, Bengten E. Immunoglobulin light (IgL) chains in ectothermic vertebrates. Dev Comp Immunol. 2011;35:906–915. doi: 10.1016/j.dci.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Edholm ES, Wilson M, Sahoo M, Miller NW, Pilstrom L, Wermenstam NE, Bengten E. Identification of Igsigma and Iglambda in channel catfish, Ictalurus punctatus, and Iglambda in Atlantic cod, Gadus morhua. Immunogenetics. 2009;61:353–370. doi: 10.1007/s00251-009-0365-z. [DOI] [PubMed] [Google Scholar]
- Fanning LJ, Connor AM, Wu GE. Development of the immunoglobulin repertoire. Clin Immunol Immunopathol. 1996;79:1–14. doi: 10.1006/clin.1996.0044. [DOI] [PubMed] [Google Scholar]
- Fjell CD, Bosdet I, Schein JE, Jones SJ, Marra MA. Internet Contig Explorer (iCE)--a tool for visualizing clone fingerprint maps. Genome Res. 2003;13:1244–1249. doi: 10.1101/gr.819303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol. 2002;2:688–698. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]
- Flajnik MF, DuPasquier LD. Evolution of the Immune System. In: WE Paul., editor. Fundamental Immunology. Lippincott Williams & Wilkins, Wolters Kluwer; 2008. pp. 56–124. [Google Scholar]
- Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci U S A. 2006;103:3728–3733. doi: 10.1073/pnas.0509720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambon-Deza F, Sanchez-Espinel C, Mirete-Bachiller S, Magadan-Mompo S. Snakes antibodies. Dev Comp Immunol. 2012;38:1–9. doi: 10.1016/j.dci.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Ghaffari SH, Lobb CJ. Heavy chain variable region gene families evolved early in phylogeny. Ig complexity in fish. J Immunol. 1991;146:1037–1046. [PubMed] [Google Scholar]
- Giudicelli V, Chaume D, Lefranc MP. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33:D256–D261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Hansen J, Leong JA, Kaattari S. Complete nucleotide sequence of a rainbow trout cDNA encoding a membrane-bound form of immunoglobulin heavy chain. Mol Immunol. 1994;31:499–501. doi: 10.1016/0161-5890(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Hansen JD, Kaattari SL. The recombination activating gene 2 (RAG2) of the rainbow trout Oncorhynchus mykiss. Immunogenetics. 1996;44:203–211. doi: 10.1007/BF02602586. [DOI] [PubMed] [Google Scholar]
- Harding FA, Amemiya CT, Litman RT, Cohen N, Litman GW. Two distinct immunoglobulin heavy chain isotypes in a primitive, cartilaginous fish, Raja erinacea. Nucleic Acids Res. 1990;18:6369–6376. doi: 10.1093/nar/18.21.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heufler C, Koch F, Stanzl U, Topar G, Wysocka M, Trinchieri G, Enk A, Steinman RM, Romani N, Schuler G. Interleukin-12 is produced by dendritic cells and mediates T helper 1 development as well as interferon-gamma production by T helper 1 cells. Eur J Immunol. 1996;26:659–668. doi: 10.1002/eji.1830260323. [DOI] [PubMed] [Google Scholar]
- Hikima J, Jung TS, Aoki T. Immunoglobulin genes and their transcriptional control in teleosts. Dev Comp Immunol. 2011;35:924–936. doi: 10.1016/j.dci.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Hinds-Frey KR, Nishikata H, Litman RT, Litman GW. Somatic variation precedes extensive diversification of germline sequences and combinatorial joining in the evolution of immunoglobulin heavy chain diversity. J Exp Med. 1993;178:815–824. doi: 10.1084/jem.178.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu E, Criscitiello MF. Diverse immunoglobulin light chain organizations in fish retain potential to revise B cell receptor specificities. J Immunol. 2006;177:2452–2462. doi: 10.4049/jimmunol.177.4.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Yachou A, Fleury S, Hendrickson WA, Sekaly RP. Analysis of the contact sites on the CD4 molecule with class II MHC molecule: co-ligand versus co-receptor function. J Immunol. 1997;158:216–225. [PubMed] [Google Scholar]
- Ichikawa HT, Sowden MP, Torelli AT, Bachl J, Huang P, Dance GS, Marr SH, Robert J, Wedekind JE, Smith HC, Bottaro A. Structural phylogenetic analysis of activation-induced deaminase function. J Immunol. 2006;177:355–361. doi: 10.4049/jimmunol.177.1.355. [DOI] [PubMed] [Google Scholar]
- Kasahara M. New insights into the genomic organization and origin of the major histocompatibility complex: role of chromosomal (genome) duplication in the emergence of the adaptive immune system. Hereditas. 1997;127:59–65. doi: 10.1111/j.1601-5223.1997.t01-1-00059.x. [DOI] [PubMed] [Google Scholar]
- Kataoka T, Kawakami T, Takahashi N, Honjo T. Rearrangement of immunoglobulin gamma 1-chain gene and mechanism for heavy-chain class switch. Proc Natl Acad Sci U S A. 1980;77:919–923. doi: 10.1073/pnas.77.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen R, Salomonsen J, Tregaskes CA, Young JR, Goodchild M, Bumstead N, Vainio O. The chicken CD4 gene has remained conserved in evolution. Immunogenetics. 2002;54:520–525. doi: 10.1007/s00251-002-0490-4. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Weiss A. T cell receptor signalling. J Cell Sci. 2001;114:243–244. doi: 10.1242/jcs.114.2.243. [DOI] [PubMed] [Google Scholar]
- Longerich S, Orelli BJ, Martin RW, Bishop DK, Storb U. Brca1 in immunoglobulin gene conversion and somatic hypermutation. DNA Repair (Amst) 2008;7:253–266. doi: 10.1016/j.dnarep.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist ML, Middleton DL, Radford C, Warr GW, Magor KE. Immunoglobulins of the non-galliform birds: antibody expression and repertoire in the duck. Dev Comp Immunol. 2006;30:93–100. doi: 10.1016/j.dci.2005.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magadan-Mompo S, Zimmerman AM, Sanchez-Espinel C, Gambon-Deza F. Immunoglobulin light chains in medaka (Oryzias latipes) Immunogenetics. 2013;65:387–396. doi: 10.1007/s00251-013-0678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom M, Jentoft S, Gregers TF, Jakobsen KS. Unraveling the Evolution of the Atlantic Cod's (Gadus morhua L.) Alternative Immune Strategy. PLoS One. 2013;8:e74004. doi: 10.1371/journal.pone.0074004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KW, de Waal MR, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- Moore LJ, Somamoto T, Lie KK, Dijkstra JM, Hordvik I. Characterisation of salmon and trout CD8alpha and CD8beta. Mol Immunol. 2005;42:1225–1234. doi: 10.1016/j.molimm.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Nagarajan UM, O'Connell C, Rank RG. Molecular characterization of guinea - pig (Cavia porcellus) CD8alpha and CD8beta cDNA. Tissue Antigens. 2004;63:184–189. doi: 10.1111/j.1399-0039.2004.00155.x. [DOI] [PubMed] [Google Scholar]
- Nikaido M, Noguchi H, Nishihara H, Toyoda A, Suzuki Y, Kajitani R, Suzuki H, Okuno M, Aibara M, Ngatunga BP, Mzighani SI, Kalombo HW, Masengi KW, Tuda J, Nogami S, Maeda R, Iwata M, Abe Y, Fujimura K, Okabe M, Amano T, Maeno A, Shiroishi T, Itoh T, Sugano S, Kohara Y, Fujiyama A, Okada N. Coelacanth genomes reveal signatures for evolutionary transition from water to land. Genome Res. 2013;23:1740–1748. doi: 10.1101/gr.158105.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug K, Bilancio A, Emery JL, Vanhaesebroeck B. Phosphoinositide 3-kinase in T cell activation and survival. Biochem Soc Trans. 2004;32:332–335. doi: 10.1042/bst0320332. [DOI] [PubMed] [Google Scholar]
- Ota T, Nei M. Divergent evolution and evolution by the birth-and-death process in the immunoglobulin VH gene family. Mol Biol Evol. 1994;11:469–482. doi: 10.1093/oxfordjournals.molbev.a040127. [DOI] [PubMed] [Google Scholar]
- Ota T, Rast JP, Litman GW, Amemiya CT. Lineage-restricted retention of a primitive immunoglobulin heavy chain isotype within the Dipnoi reveals an evolutionary paradox. Proc Natl Acad Sci U S A. 2003a;100:2501–2506. doi: 10.1073/pnas.0538029100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Rast JP, Litman GW, Amemiya CT. Lineage-restricted retention of a primitive immunoglobulin heavy chain isotype within the Dipnoi reveals an evolutionary paradox. Proc Natl Acad Sci U S A. 2003b;100:2501–2506. doi: 10.1073/pnas.0538029100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallavicini A, Canapa A, Barucca M, Alf LJ, Biscotti MA, Buonocore F, De MG, Di PF, Fausto AM, Forconi M, Gerdol M, Makapedua DM, Turner-Meier J, Olmo E, Scapigliati G. Analysis of the transcriptome of the Indonesian coelacanth Latimeria menadoensis. BMC Genomics. 2013;14:538. doi: 10.1186/1471-2164-14-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CI, Hirono I, Aoki T. Molecular characterization of the Japanese flounder, Paralichthys olivaceus, CD3epsilon and evolution of the CD3 cluster. Dev Comp Immunol. 2005;29:123–133. doi: 10.1016/j.dci.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Parra ZE, Baker ML, Lopez AM, Trujillo J, Volpe JM, Miller RD. TCR mu recombination and transcription relative to the conventional TCR during postnatal development in opossums. J Immunol. 2009;182:154–163. doi: 10.4049/jimmunol.182.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra ZE, Baker ML, Schwarz RS, Deakin JE, Lindblad-Toh K, Miller RD. A unique T cell receptor discovered in marsupials. Proc Natl Acad Sci U S A. 2007;104:9776–9781. doi: 10.1073/pnas.0609106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra ZE, Lillie M, Miller RD. A model for the evolution of the mammalian t-cell receptor alpha/delta and mu loci based on evidence from the duckbill Platypus. Mol Biol Evol. 2012;29:3205–3214. doi: 10.1093/molbev/mss128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra ZE, Miller RD. Comparative analysis of the chicken TCRalpha/delta locus. Immunogenetics. 2012;64:641–645. doi: 10.1007/s00251-012-0621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra ZE, Ohta Y, Criscitiello MF, Flajnik MF, Miller RD. The dynamic TCRdelta: TCRdelta chains in the amphibian Xenopus tropicalis utilize antibody-like V genes. Eur J Immunol. 2010;40:2319–2329. doi: 10.1002/eji.201040515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast JP, Anderson MK, Litman GW. The structure and organization of immunoglobulin genes in lower vertebrates. In: T Honjo, FW Alt, TH Rabbitts., editors. Immunoglobulin Genes. London: Academic Press; 1989. pp. 315–341. [Google Scholar]
- Rast JP, Amemiya CT, Litman RT, Strong SJ, Litman GW. Distinct patterns of IgH structure and organization in a divergent lineage of chrondrichthyan fishes. Immunogenetics. 1998;47:234–245. doi: 10.1007/s002510050353. [DOI] [PubMed] [Google Scholar]
- Reyes-Cerpa S, Maisey K, Reyes-Lopez F, Toro-Ascuy D, Sandion AM, Imarai M. Fish Cytokines and Immune Response. Türker Hakan., editor. New Advances and Contributions to Fish Biology. Intech. 2012:3–57. [Google Scholar]
- Reynaud CA, Dahan A, Anquez V, Weill JC. Somatic hyperconversion diversifies the single Vh gene of the chicken with a high incidence in the D region. Cell. 1989;59:171–183. doi: 10.1016/0092-8674(89)90879-9. [DOI] [PubMed] [Google Scholar]
- Roman T, Andersson E, Bengten E, Hansen J, Kaattari S, Pilstrom L, Charlemagne J, Matsunaga T. Unified nomenclature of Ig VH genes in rainbow trout (Oncorhynchus mykiss): definition of eleven VH families. Immunogenetics. 1996;43:325–326. doi: 10.1007/BF02441003. [DOI] [PubMed] [Google Scholar]
- Ropars A, Bautz AM, Dournon C. Sequencing and expression of the CD3 gamma/delta mRNA in Pleurodeles waltl (urodele amphibian) Immunogenetics. 2002;54:130–138. doi: 10.1007/s00251-002-0437-9. [DOI] [PubMed] [Google Scholar]
- Rumfelt LL, Lohr RL, Dooley H, Flajnik MF. Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol. 2004;5:8. doi: 10.1186/1471-2172-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha NR, Suetake H, Kikuchi K, Suzuki Y. Fugu immunoglobulin D: a highly unusual gene with unprecedented duplications in its constant region. Immunogenetics. 2004;56:438–447. doi: 10.1007/s00251-004-0693-y. [DOI] [PubMed] [Google Scholar]
- Saha NR, Suetake H, Suzuki Y. Analysis and characterization of the expression of the secretory and membrane forms of IgM heavy chains in the pufferfish, Takifugu rubripes. Mol Immunol. 2005;42:113–124. doi: 10.1016/j.molimm.2004.06.034. [DOI] [PubMed] [Google Scholar]
- Saraiva M, Christensen JR, Veldhoen M, Murphy TL, Murphy KM, O'Garra A. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity. 2009;31:209–219. doi: 10.1016/j.immuni.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savan R, Sakai M. Genomics of fish cytokines. Comp Biochem Physiol Part D Genomics Proteomics. 2006;1:89–101. doi: 10.1016/j.cbd.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrom M, Gregers TF, Rounge TB, Paulsen J, Solbakken MH, Sharma A, Wetten OF, Lanzen A, Winer R, Knight J, Vogel JH, Aken B, Andersen O, Lagesen K, Tooming-Klunderud A, Edvardsen RB, Tina KG, Espelund M, Nepal C, Previti C, Karlsen BO, Moum T, Skage M, Berg PR, Gjoen T, Kuhl H, Thorsen J, Malde K, Reinhardt R, Du L, Johansen SD, Searle S, Lien S, Nilsen F, Jonassen I, Omholt SW, Stenseth NC, Jakobsen KS. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011;477:207–210. doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin Immunol. 2004;16:257–275. doi: 10.1016/j.smim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Suetake H, Araki K, Akatsu K, Somamoto T, Dijkstra JM, Yoshiura Y, Kikuchi K, Suzuki Y. Genomic organization and expression of CD8alpha and CD8beta genes in fugu Takifugu rubripes. Fish Shellfish Immunol. 2007;23:1107–1118. doi: 10.1016/j.fsi.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liu Z, Ren L, Wei Z, Wang P, Li N, Zhao Y. Immunoglobulin genes and diversity: what we have learned from domestic animals. J Anim Sci Biotechnol. 2012;3:18. doi: 10.1186/2049-1891-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IM, Walter G, Marks JD, Llewelyn MB, Winter G. The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J Mol Biol. 1992;227:776–798. doi: 10.1016/0022-2836(92)90223-7. [DOI] [PubMed] [Google Scholar]
- Turchin A, Hsu E. The generation of antibody diversity in the turtle. J Immunol. 1996;156:3797–3805. [PubMed] [Google Scholar]
- Tutter A, Brodeur P, Shlomchik M, Riblet R. Structure, map position, and evolution of two newly diverged mouse Ig VH gene families. J Immunol. 1991;147:3215–3223. [PubMed] [Google Scholar]
- Wakae K, Magor BG, Saunders H, Nagaoka H, Kawamura A, Kinoshita K, Honjo T, Muramatsu M. Evolution of class switch recombination function in fish activation-induced cytidine deaminase, AID. Int Immunol. 2006;18:41–47. doi: 10.1093/intimm/dxh347. [DOI] [PubMed] [Google Scholar]
- Wang X, Parra ZE, Miller RD. Platypus TCRmu provides insight into the origins and evolution of a uniquely mammalian TCR locus. J Immunol. 2011;187:5246–5254. doi: 10.4049/jimmunol.1101113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr GW, Middleton DL, Miller NW, Clem LW, Wilson MR. An additional family of VH sequences in the channel catfish. Eur J Immunogenet. 1991;18:393–397. doi: 10.1111/j.1744-313x.1991.tb00038.x. [DOI] [PubMed] [Google Scholar]
- Wen Y, Shao JZ, Xiang LX, Fang W. Cloning, characterization and expression analysis of two Tetraodon nigroviridis interleukin-16 isoform genes. Comp Biochem Physiol B Biochem Mol Biol. 2006;144:159–166. doi: 10.1016/j.cbpb.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Willett CE, Cherry JJ, Steiner LA. Characterization and expression of the recombination activating genes (rag1 and rag2) of zebrafish. Immunogenetics. 1997;45:394–404. doi: 10.1007/s002510050221. [DOI] [PubMed] [Google Scholar]
- Wu H, Kwong PD, Hendrickson WA. Dimeric association and segmental variability in the structure of human CD4. Nature. 1997;387:527–530. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- Zhu LY, Nie L, Zhu G, Xiang LX, Shao JZ. Advances in research of fish immune-relevant genes: a comparative overview of innate and adaptive immunity in teleosts. Dev Comp Immunol. 2013;39:39–62. doi: 10.1016/j.dci.2012.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.