Background: Molecular mechanisms explaining stress-induced leaky scanning translation initiation regulation are poorly understood.
Results: We demonstrate this relation and identify a post-translational modification on eIF1 that is stress responsive.
Conclusion: Leaky scanning mediated by eIF1 phosphorylation is a stress-regulated process.
Significance: Stress-induced global translation initiation can be regulated by means of leaky scanning as well as re-initiation.
Keywords: Gene Expression, Post-translational Modification (PTM), Ribosome, Translation Control, Translation Initiation Factor, AUG Fidelity, AIRAP, Arsenite, Leaky Scanning, Upstream Open Reading Frame
Abstract
Initial steps in protein synthesis are highly regulated processes as they define the reading frame of the translation machinery. Eukaryotic translation initiation is a process facilitated by numerous factors (eIFs), aimed to form a “scanning” mechanism toward the initiation codon. Translation initiation of the main open reading frame (ORF) in an mRNA transcript has been reported to be regulated by upstream open reading frames (uORFs) in a manner of re-initiation. This mode of regulation is governed by the phosphorylation status of eIF2α and controlled by cellular stresses. Another mode of translational initiation regulation is leaky scanning, and this regulatory process has not been extensively studied. We have identified arsenite-inducible regulatory particle-associated protein (AIRAP) transcript to be translationally induced during arsenite stress conditions. AIRAP transcript contains a single uORF in a poor-kozak context. AIRAP translation induction is governed by means of leaky scanning and not re-initiation. This induction of AIRAP is solely dependent on eIF1 and the uORF kozak context. We show that eIF1 is phosphorylated under specific conditions that induce protein misfolding and have biochemically characterized this site of phosphorylation. Our data indicate that leaky scanning like re-initiation is responsive to stress conditions and that leaky scanning can induce ORF translation by bypassing poor kozak context of a single uORF transcript.
Introduction
Initiation of translation at a designated start site is a fundamental process required to obtain a correct proteome. In eukaryotes, the 43 S pre-initiation complex (PIC)2 is composed of the 40 S ribosomal subunit, several initiation factors, and the eIF2-GTP-Met-tRNA ternary complex (TC, see Refs. 1, 2). Upon assembly near the 5′ cap site of an mRNA transcript, the PIC will initiate scanning along the transcript using the anticodon of the Met-tRNA to identify the AUG initiation site (3, 4). Factors known to influence recognition efficiency and subsequent translational initiation can be split into factors in cis (mRNA sequences) or trans (PIC components). Factors in cis are the AUG surrounding sequence (Ref. 5, also known as the kozak sequence), the distance of the AUG site from the 5′ cap (6), and near cognate sequences surrounded by optimal kozak sequences (7). Factors in trans are several initiation subunits (8–11) and TC availability (12). Upon start site recognition by the scanning PIC, the eIF1 factor is ejected from the PIC allowing the release of Pi from the hydrolyzed GTP within the TC (8, 13). Upon eIF1 release from the PIC, the “open” conformation of the PIC is converted into a “closed” conformation (14), thus shifting the PIC to downstream events that lead to the formation of the 80 S initiation complex (9).
Translational control by means of regulating initiation have been reported to occur by two distinct mechanisms, re-initiation involving TC availability (12) and leaky scanning (3) involving eIF1 (8). The model explaining re-initiation regulation predicts that the mRNA transcript has at least two upstream open reading frames (uORF) in the 5′-untranslated region (UTR), and that TC availability is limiting (for a review see Ref. 9). Limiting TC availability is achieved by eIF2α phosphorylation leading to reduction in eIF2-GTP levels (15). This mode of regulation has been best characterized in the case of Gcn4 (12) and its mammalian functional homologue ATF4 (16, 17). Regulation, molecular requirements, and the cellular mechanisms that sense various environmental signals that lead to eIF2α phosphorylation have been well characterized (18, 19). Human ATF4 contains a short 9nt uORF followed by a second 98nt uORF that overlaps out-of-frame with the downstream ATF4 CDS. Conditions that do not limit TC levels enable uORF1 translation, efficient re-initiation of a 43 S complex on the second ORF thereby bypassing the CDS AUG of ATF4. Upon eIF2α phosphorylation that limits TC availability, inefficient re-initiation of a 43 S complex occurs prior to the second uORF2, thus enabling the scanning complex to arrive at the CDS AUG of ATF4 and re-initiate translation on the CDS only upon TC-limiting conditions.
Leaky scanning has been studied with respect to the capability to initiate translation either from near cognate AUGs (20) or sub-optimal kozak sequences surrounding an AUG (5). In this type of regulation, eIF1 has been shown both genetically (21) and biochemically to be the central factor in respect to AUG fidelity, and therefore, eIF1 is considered as the “gate keeper” in this process (22). Leaky scanning has been recently shown to serve as an auto-regulatory mechanism in the translation of eIF1 itself and that of eIF5 (23, 24); however additional examples are limited (for example see Ref. 25).
In this report, we show that the transcript of an arsenite-inducible regulatory particle-associated protein (AIRAP, see Ref. 26) is translationally regulated by means of leaky scanning. We further identify eIF1 as a key component in this arsenite-induced translational regulation. Upon arsenite treatment eIF1 is phosphorylated, AUG fidelity enhanced, leading to leaky scanning across the inhibitory uORF present in AIRAP 5′-UTR. By identifying a stress-dependent phosphorylation event on eIF1, we make the molecular connection between arsenite-induced leaky scanning and eIF1 activity, which in the case of AIRAP translational induction would initiate proteasomal modulation that is required during early stages of protein misfolding condition (26).
EXPERIMENTAL PROCEDURES
Reporter Gene Construction
The 5′-end of mouse AIRAP cDNA was recovered by 5′ RACE from mRNA of arsenite-treated mouse fibroblasts and was ligated between the HindIII-BamHI sites of pEGFP-N1 (Clontech Laboratories, Inc.), Point mutations described in the text were introduced into the AIRAP cDNA using overlapping PCR. The eIF1 activity reporter was constructed by changing the sequence surrounding the AUG in the pSEAP2 control reporter (Clontech) to eIF1 responsive and kozak sequences as previously described for the eIF1 luciferase reporter (23). SEAP activity was performed as previously described (27).
Cell Culture and Cell Transfection
293T cells, mouse embryonic fibroblasts (MEFs) were maintained in DMEM supplemented with 10% fetal bovine serum. Unless otherwise indicated, cells were treated for 4 h with 0.4 μm thapsigargin (586005, Calbiochem) or 1 h 25 μm sodium arsenite (S7400, Sigma). Unless indicated, 293T cells were transfected with 3 μg of plasmid DNA per 60-mm dish using the calcium-phosphate precipitation method. MEF cells were transfected with 6 μg of plasmid DNA per 60-mm dish using turbofect (R0531; Thermo Scientific).
Cell Metabolic Labeling and Lysis
Transfected 293T cells were starved for 15 min in methionine/cysteine-free DMEM containing 10% dialyzed fetal calf serum, and labeled for 15 min with 200 μCi/ml35S-Translabel (ICN) in the same medium. Cells were lysed in TNH buffer (20 mm HEPES-7.9, 100 mm NaCl, 1% Triton X-100, 1 mm EDTA, 1.5 mm MgCl2, 1 mm DTT, and protease inhibitors) immediately after labeling and clarified at 20,000 × g for 10 min. The labeled protein was immunoprecipitated with the indicated antibody and resolved by 12% SDS-PAGE.
Antibodies
AIRAP and GFP antisera for immunoblots were a kind gift from David Ron and have been previously described (26). P97 and eIF1 antiserum for immunoblots and immunoprecipitations was produced by immunizing rabbits against the full-length rat P97 or human eIF1 (Covance). Alternatively eIF1 antiserum and SEAP were used from a commercial source (Abcam 2B9 and Santa Cruz Biotechnology 28904).
Two-dimensional Gel Electrophoresis
293T cells were lysed in two-dimensional lysis buffer (20 mm HEPES-7.9, 10% Chaps, 1 mm DTT, protease, and phosphatase inhibitors, unless indicated otherwise). First dimension protein separation was performed using 3–10 NL pH gradients (IPG strips, 163-2002; Bio-Rad). For detection of endogenous eIF1, 150 μg of total protein in a final volume of 120 μl were loaded per strip. For detection of ectopic eIF1, only 5 μg of protein was loaded per strip to maximize resolution and eliminate detection of endogenous eIF1. Isoelectric focusing (IEF) was conducted using the PROTEAN system (Bio-Rad) applying 16,000 volt hours as indicated by the manufacturer. Second dimension SDS-PAGE separation was achieved using 15% acrylamide gels.
Polysome Profiles
Untreated and arsenite-treated 293T cells were incubated with cycloheximide (100 μg/ml) for 5 min to arrest polyribosome migration. Cells were lysed in polysome extraction buffer (0.3% Triton X-100, 0.3% deoxycholate, 1 mm DTT, RNase inhibitor, and protease inhibitors). Thereafter, 0.5 ml of each cell lysate was loaded onto a sucrose gradient (10–50% w/v, 140 mm KCl, 20mm Tris-HCl (pH 7.5), and 5 mm MgCl2). After centrifugation (SorvallTH641 rotor at 41,000 rpm for 2.5 h at 4 °C), the gradient UV profile (A254) of ribosomal content was obtained using a TELEDYNE ISCO UA6.
RNA Extraction, Reverse Transcription, and Quantitative Real-time PCR
RNA was extracted from whole-cell lysates, using the RNA easy kit (Qiagen 217004). RNA was converted into cDNA using the Verso cDNA kit (Thermo Scientific, AB-1453/A). RT-qPCR was performed using Stratagene Mx3000P device and SYBR Premix Ex TaqII reaction mix (TaKaRa). Primers for mouse b-globin, human GAPDH, and mouse AIRAP were self-designed, and primers for GFP evaluation were obtained from Primer design.
RESULTS
Translational Regulation of AIRAP
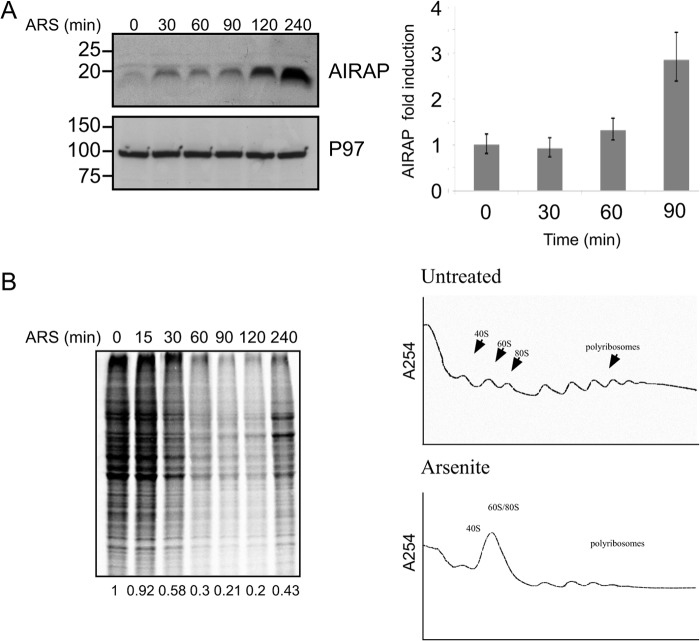
Airap is transcriptionally induced during arsenite stress (26, 28), conditions previously reported to cause translational inhibition (16, 29), we therefore wanted to understand the molecular events that enable AIRAP translation under these conditions. Time course analysis of AIRAP mRNA and protein levels indicated a minor but consistent amount of Airap induction that could be detected as soon as a 30-min post-arsenite treatment (Fig. 1A), at times when translational repression is observed as seen by metabolic label decreased incorporation and polysomal profiles analyses (Fig. 1B). Interestingly, the increased protein level detected at 30 min is not caused by changes in the transcript levels, as shown by the lack of mRNA elevation at this time point (Fig. 1A, right panel). Transcription induction is detected at 90 min, which is followed kinetically by the increase in protein levels at 120 and 240 min. This result attributes the early induction in Airap levels to translational regulation, while the later stage induction is also due to the increase in transcript levels and is consistent with the observed translational recovery.
FIGURE 1.
Translational induction of AIRAP. A, immunoblot of AIRAP and p97 (used as a loading control) in mouse embryonic fibroblasts (MEFs) pretreated with arsenite (25 μm) for the indicated time course. AIRAP mRNA levels from the same cells were quantified and normalized to a housekeeping gene using qPCR, and fold induction is represented in the right panel. Quantitation and statistical evaluations were obtained from three independent experiments. Comparison of Time 0 with Time 90 showed a statistical significance of t test p < 0.001. B, left panel, metabolic labeling performed in MEFs pretreated with arsenite for the indicated time course. Cell lysates were resolved by SDS-PAGE, and the autoradiogram is presented. Right panel, polysome profiles from untreated and arsenite-treated cells showing the accumulation of ribosomal subunits and monosomes in the arsenite-treated cells.
uORF-mediated AIRAP Translation Induction
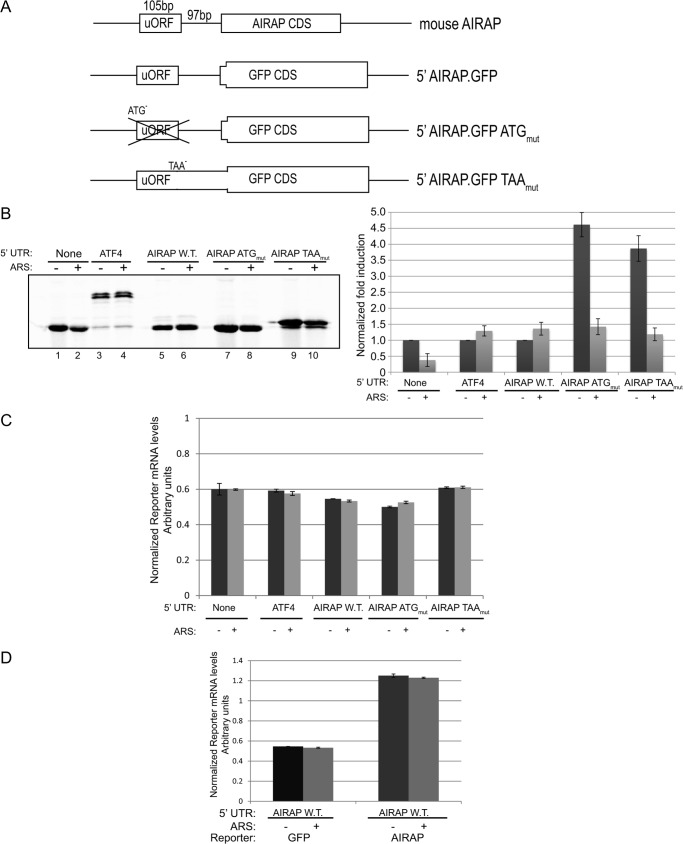
To eliminate any transcriptional-mediated AIRAP induction, we constructed several reporter plasmids consisting of a heterologous CMV promoter with the 5′-UTR of the mouse AIRAP genomic region fused to a GFP reporter (Fig. 2A). To evaluate reporter translational rates during arsenite conditions that induce translation inhibition, we performed metabolic label incorporation during the last 15 min of the arsenite treatment. Cell lysates were immunoprecipitated for GFP to determine translational rates during the pulse period (see “Experimental Procedures”). As seen in Fig. 2B, translational repression could be observed in the control reporter (without the AIRAP 5′-UTR) upon arsenite treatment; however, AIRAP 5′-UTR insertion enabled to induce reporter translation under otherwise translational repressing conditions (Fig. 2B). Based on these results, we conclude that the 5′-UTR of AIRAP harbors elements that enable translational induction upon arsenite stress conditions. Close evaluation of the mouse AIRAP 5′-UTR indicates a single uORF, the presence of which is also conserved in humans (data not shown). uORFs have long been implicated in translational regulation mediated by eIF2α phosphorylation by means of re-initiation regulation (12). However, this regulation requires the presence of at least two uORFs in the 5′-UTR (9). To establish a system for studying the impact of a 5′-UTR cis acting element on Airap translation, we inserted a set of AIRAP 5′-UTR reporters into a GFP plasmid (Fig. 2A). This set of reporters included the wild-type 5′ AIRAP UTR (5′ AIRAP.GFP), a point mutation eliminating the uORF ATG (5′ AIRAP.GFP ATGmut), and a point mutation in the uORF stop codon thus leading to an in-frame fusion with the downstream GFP reporter (5′ AIRAP.GFP TAAmut). As controls, we used a reporter that did not contain the AIRAP 5′-UTR and a positive control consisting of the ATF4 5′-UTR reporter previously shown to be translationally regulated by eIF2α phosphorylation (16). To eliminate any effect of the 5′-UTR mutations or cellular conditions on the reporter transcript levels, quantification of translated labeled reporter products were normalized to the transcripts levels to eliminate possible transcriptional effects (see “Experimental Procedures”). A representative evaluation of the transcript levels indicated no significant changes in response to arsenite of 5′-UTR mutations (Fig. 2C). Also, any effect of the GFP CDS on the transcript was also evaluated by replacing the GFP CDS with that of AIRAP, indicating no changes were observed in response to arsenite treatment (Fig. 2D). Introduction of the aforementioned reporters into cells and pulse labeling during control and arsenite treatment, enabled to evaluate the contribution of the 5′-UTR uORF to AIRAP translational regulation. As seen in Fig. 2B, reduction in translation is observed during arsenite treatment when the AIRAP 5′-UTR is absent (Fig. 2B, lanes 1 and 2), consistent with the general translational repression upon arsenite treatment (Fig. 1B). ATF4 5′-UTR was sufficient to eliminate any such repression and further induced a modest translational induction (Fig. 2B, lanes 3 and 4), consistent with previous reports of modest translational induction of the ATF4.GFP reporter (16). As with the ATF4-positive control, AIRAP 5′-UTR was sufficient to eliminate any translational repression upon arsenite treatment and further enabled a modest translational induction (Fig. 2B, lanes 5 and 6). The AIRAP.GFP ATGmut reporter indicated translational repression upon arsenite treatment (Fig. 2B) and was 4.5-fold higher compared with the AIRAP wild type 5′-UTR reporter (Fig. 2B, lanes 5 and 7). This result indicates the necessity of the uORF for induced Airap translation during arsenite treatment (Fig. 2B, compare lanes 5 and 6 with lanes 7 and 8). Furthermore, the induced basal activity in the AIRAP.GFP ATGmut reporter indicates an active repressing activity of the uORF toward basal translation (Fig. 2B, lanes 5 and 7). The reporter with a uORF stop codon elimination (5′ AIRAP.GFP TAAmut reporter) represents translation products that are initiated from the AUG uORF as well as the AUG of GFP. We noted that the reporter expressed from this construct appeared in two molecular weights (Fig. 2B, lanes 9 and 10). The major species represented in the below quantifications had a higher molecular weight (HMW) due to the extended in-frame 5′ sequence that is being translated upstream from the GFP AUG (as the stop codon has been eliminated). The minor LMW species is due to a minority of ribosomes that bypass the uORF AUG and initiate translation from the downstream in-frame AUG present in the GFP reporter. We noted that while the overall translation during arsenite was reduced in the AIRAP TAAmut reporter (Fig. 2B, compare lanes 9 and 10), the internal ratio between the lower and higher molecular weight reporter species was increased 3-fold during arsenite treatment. The importance of this ratio fold will be further discussed. Basal differences between AIRAP.GFP ATGmut and AIRAP TAAmut (lanes 7 and 9) may be explained by their different AUG contexts (see below).
FIGURE 2.
uORF mediates AIRAP translation induction. A, organization of the 5′-UTR of the mouse AIRAP transcript and the derivative 5′ AIRAP-GFP reporters. Exons 1 and 2 of the mouse AIRAP (encoding the 5′-UTR and the initiating amino acids of AIRAP CDS) were fused in-frame to the GFP CDS and are indicated in the GFP reporter. B, autoradiogram of radiolabeled proteins after a brief labeling pulse of untreated and pretreated cells. Cells were transfected with the indicated GFP reporter and subjected to a GFP IP, resolved by SDS-PAGE. Untreated wild-type reporter quantities were set as 1 for each reporter and relative quantities are shown. All quantifications were normalized to GFP mRNA transcript levels shown in panel B. Quantitation and statistical evaluations are presented from three independent experiments. Both ATF4 and AIRAP WT reporters showed a statistical significance of t test p < 0.001 upon comparing to the non-harboring 5′-UTR GFP reporter arsenite response. C, transcript levels from cells transfected with the indicated reporter and arsenite treated as indicated were quantified and normalized to a housekeeping gene using qPCR. Quantitation and statistical evaluations were obtained from three independent experiments with error bars representing the S.D. D, RT-PCR analysis of two reporters containing the 5'-UTR of AIRAP and the GFP CDS or AIRAP CDS. The GAPDH housekeeping gene was used as an internal loading control for total mRNA. Both reporters do not show any substantial changes and a role for mRNA stability in response to a brief arsenite treatment. The same conditions were used to measure translational induction; 60 min, 50 mm).
AIRAP Translational Regulation by Means of Leaky Scanning
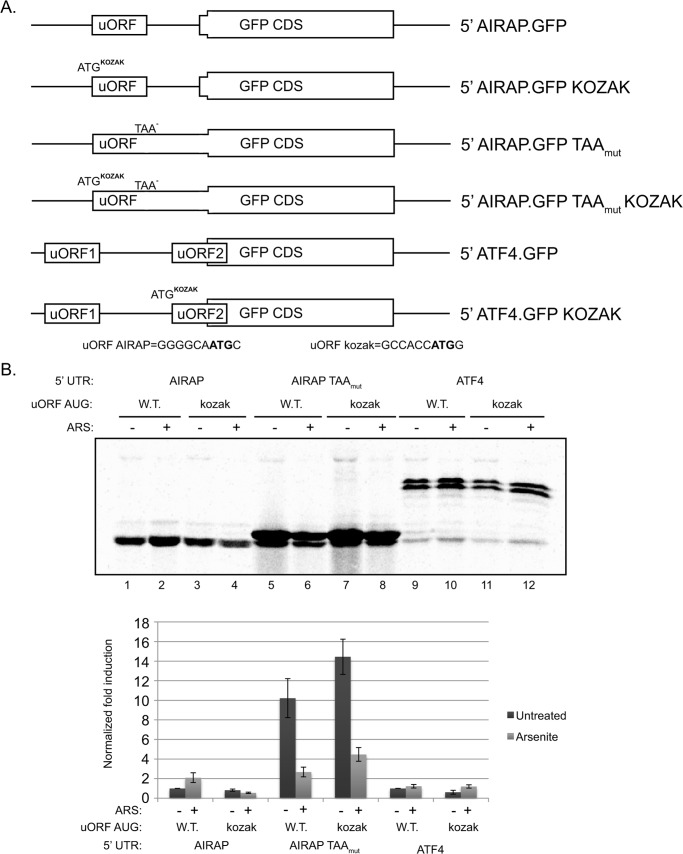
Our results thus far indicate that AIRAP is regulated at a translational level, with importance of a singular uORF in mediating this regulation. The re-initiation model requires the presence of at least two uORFs (as exemplified in the case of ATF4), this is due to the slow recruitment of a new TC needed to re-initiate translation of the downstream uORF. A slow reloading of TC enables to bypass the second inhibitory uORF only during eIF2α phosphorylation conditions (16). Furthermore, translation from the second uORF of ATF4 was shown not to be induced during eIF2α phosphorylation, thus ruling out re-initiation as a mechanism regulating AIRAP translational induction during arsenite conditions (9, 12, 16). A second mechanism known to regulate translational initiation is leaky scanning (3, 8). This mode of regulation involves the ability of a scanning ribosome to bypass a putative AUG initiation-site (20). In metazoan the optimal context for translation initiation was found to be GCC(A/G)CCAUGG with the two critical positions within the sequence being a purine at the −3 and a G at the +4 positions (relative to the A of the AUG codon, which is designated +1, Ref. 5). When examining the AIRAP 5′-UTR we noted a conserved non-optimal sequence (GGGGCAAUGC) present in the uORF AUG, while the ORF AUG is in optimal context. To evaluate translational regulation of AIRAP by means of leaky scanning and to address the role of a non-optimal uORF AUG context in AIRAP translational regulation, we modified the uORF AUG context of AIRAP to an optimal sequence. The mutation was performed in the 5′ AIRAP.GFP and 5′ AIRAP.GFP TAAmut reporters (Fig. 3A). We also noted that the ATF4 reporter previously shown to be dependent on eIF2α phosphorylation (16) contains a non-optimal kozak sequence in the AUG of uORF2, and therefore a similar mutation in this reporter was also constructed (ATF4.GFP kozak; Fig. 3A).
FIGURE 3.
AIRAP translation regulation is mediated by means of leaky scanning. A, diagram presentation of the reporters used. B, autoradiogram of radiolabeled proteins after a brief labeling pulse of untreated and arsenite-pretreated 293T cells. Cells were transfected with the indicated reporter and subjected to a GFP IP resolved by SDS-PAGE. Untreated 5′ AIRAP WT reporter quantities were set as 1, and relative quantities are shown below. All quantifications were normalized to GFP mRNA transcript levels. Quantitation and statistical evaluations are present from three independent experiments.
As noted in Fig. 3B, kozak optimization of the AIRAP uORF abolished the translational induction observed during arsenite treatment (Fig. 3B, lanes 1 and 2 versus lanes 3 and 4). Furthermore, the results obtained using the AIRAP.GFP TAAmut reporter show the enhancement of the uORF translation upon kozak optimization (Fig. 3B, lanes 5 and 7). Basal repression is observed upon kozak optimization in the ATF4.GFP reporter (Fig. 3B, lanes 9 and 11), however no reduction in translational induction is noted upon arsenite treatment (Fig. 3B, lanes 9 and 10 versus lanes 11 and 12). This situation is unlike the observed reduction noted in AIRAP 5′-UTR (Fig. 3B, lanes 1 and 2 versus lanes 3 and 4), indicating the different mechanisms of translational regulation in these two 5′-UTRs.
eIF1 Regulates AIRAP Translational Induction
A key player enabling scanning 43 S ribosomes to discriminate between various potential initiation sites is eIF1, thus eIF1 is a key component in leaky scanning regulation (8). In fact, one of the best characterized regulations of leaky scanning by eIF1 is the translation of eIF1 itself that is initiated from an AUG located within a poor-kozak context (23). Translational regulation of AIRAP by means of leaky scanning (Fig. 3) is predicted therefore to be highly responsive to eIF1. Using the 5′ AIRAP.GFP and 5′ AIRAP.GFP TAAmut reporters, we evaluated the effect of eIF1 overexpression on AIRAP translation. eIF1 overexpression has been previously shown to increase the bypass of the 43 S complex from non-optimal AUG initiation sites (23). Therefore, in the case of the AIRAP 5′-UTR, overexpression of eIF1 is predicted to increase the bypass over the non-optimal AUG kozak context of the uORF and enable leaky scanning toward the downstream kozak optimal ORF. As seen in Fig. 4A, overexpression of eIF1 increased the translation from the downstream ORF (Fig. 4A, lanes 1 and 2). This induction was entirely dependent on the sub-optimal AUG kozak context of the uORF, as we could not observe any induction upon eIF1 overexpression in the AIRAP.GFP kozak reporter (Fig. 4A, lanes 3 and 4) a scenario resembling that of arsenite induction (Fig. 3). To evaluate eIF1's role in leaky scanning regulation of AIRAP translation, we utilized the AIRAP.GFP TAAmut reporter. As noted previously (Figs. 2 and 3), the HMW and LMW species of the translated GFP, correspond to the uORF and ORF initiation sites, respectively. Overexpression of eIF1 induced the translation of the LMW GFP (i.e. ORF translation) while reducing the translation of the HMW GFP (i.e. uORF translation, Fig. 4A, lanes 5 and 6). This regulation was once again entirely dependent on the sub-optimal kozak sequence of the uORF AUG, as no LMW GFP was induced upon eIF1 expression in this reporter context (AIRAP.GFP TAAmut Kozak; Fig. 4A, lanes 7 and 8). We further confirmed the ability of eIF1 to induce the translation of endogenous AIRAP. To this end, cells were transfected with eIF1, and AIRAP levels were monitored. Using arsenite treatment as a positive control for Airap induction, we noted the ability of eIF1 overexpression to induce by 2-fold the endogenous levels of AIRAP even in the absence of arsenite (Fig. 4B). The higher induction levels observed during arsenite are also an effect of transcriptional induction (Fig. 1A).
FIGURE 4.
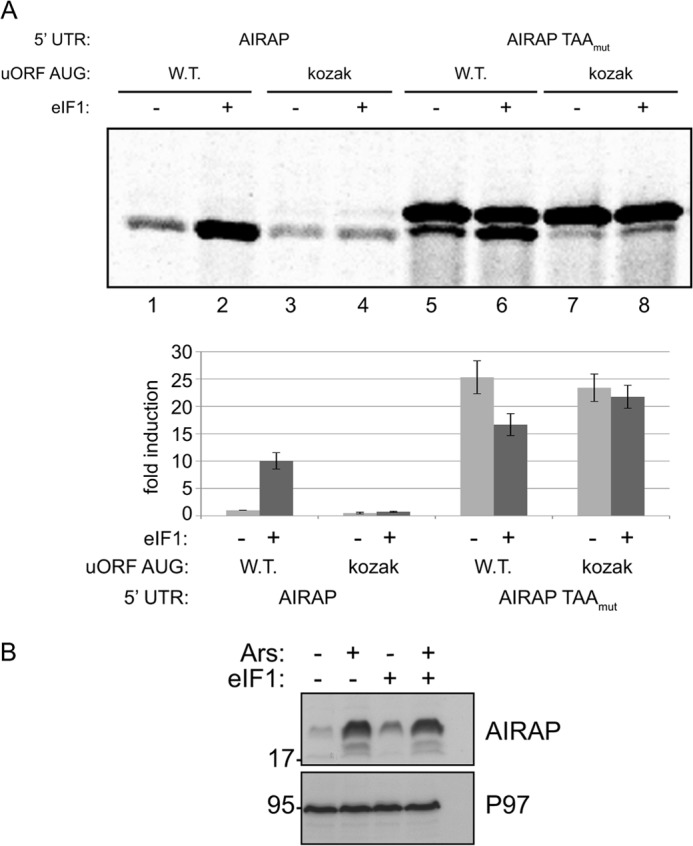
eIF1 regulates AIRAP translational induction. A, autoradiogram of radiolabeled proteins after a brief labeling pulse of293T cells. The cells were transfected with the indicated AIRAP reporter and with an eIF1 expressing plasmid. Quantification of the GFP or HMW GFP (AIRAP TAAmut reporter) were normalized to GFP mRNA transcript levels. Quantitation and statistical evaluations are presented from three independent experiments. B, MEF cells were transfected with an empty vector or an eIF1-expressing plasmid and were either untreated or arsenite treated as indicated. Immunoblots revealing the AIRAP and p97 content (used as a loading control) are presented.
Arsenite-induced eIF1 Phosphorylation and Function
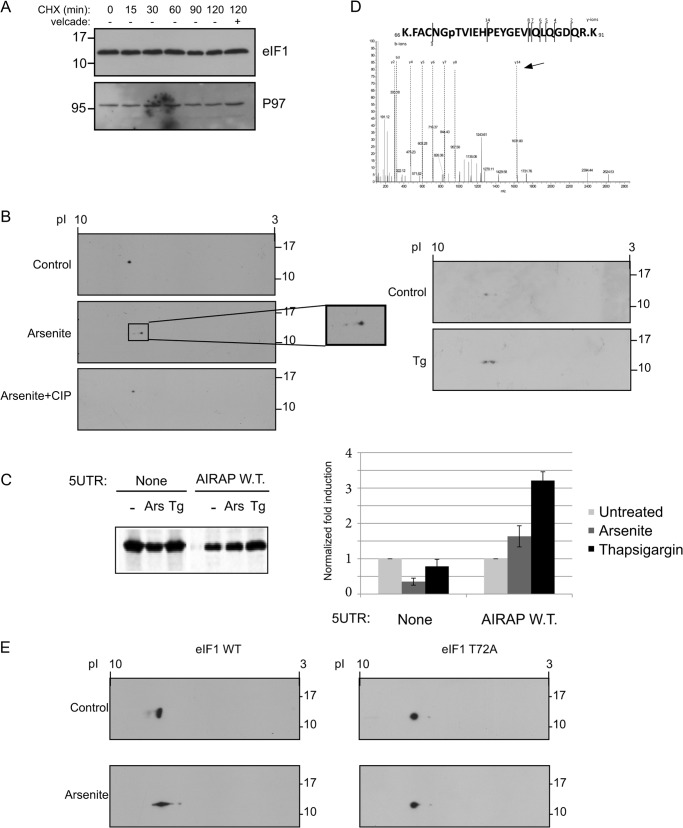
Arsenite-induced AIRAP translation by means of leaky scanning in a mechanism resembling eIF1 overexpression (Figs. 3 and 4). We hypothesized that arsenite may regulate eIF1 levels or activity, thereby inducing AIRAP translation by means of leaky scanning. Previous reports indicated the role of AIRAP as a proteasome regulator induced during UPS-impaired conditions (26). However, we did not find any indication of elevated levels of eIF1 during arsenite treatment (data not shown). Furthermore, evaluation of eIF1 half-life indicated the protein to be long-lived at least in respect to the time line used in our experiments. This conclusion is observed by the lack of reduction in eIF1 levels upon cyclohexamide chase experiments (Fig. 5A). Therefore, an auto-regulatory network of eIF1 on its own translation (23), is probably not useful for immediate leaky scanning responses due to the protein's long half-life. We therefore evaluated if eIF1 may be regulated by means of a post-translational modification. To this end, we performed two-dimensional gel analysis on eIF1 extracted from control and arsenite-treated cells. Upon arsenite treatment, we noted a decrease in the protein pI, but no increase in eIF1 MW (Fig. 5B). As this pattern of modification can correlate with a phosphorylation modification, we analyzed the protein shift upon arsenite treatment by incubating the lysate extract with alkaline phosphatase. As noted in Fig. 5B, the arsenite-induced pI shift was completely abolished upon alkaline phosphatase treatment, implying an arsenite-induced phosphorylation event on eIF1. Further analysis of the phosphorylation modification, indicated thapsigargin was also able to induce eIF1 phosphorylation (Fig. 5B), consistent with the ability of thapsigargin to induce AIRAP reporter translation (Fig. 5C). The phosphorylation site of the endogenous eIF1 was identified by means of mass spectrometry, implying Thr-72 is the arsenite-induced phosphorylation site (Fig. 5D). To confirm the MS analysis, eIF1 T72A mutation abolished the phosphorylation of eIF1 upon arsenite treatment, as indicated by the lack of pI shift of the mutated eIF1 (Fig. 5E), thus confirming Thr-72 phosphorylation as a site of modification in eIF1 under arsenite treatment.
FIGURE 5.
Arsenite-induced eIF1 phosphorylation. A, eIF1 protein half-life was determined by incubating cells in the presence of 10 μg/ml cycloheximide (CHX) for the indicated time course, after which lysates were immunoblotted for eIF1 and P97 as a control for a long lived protein. Where indicated, Velcade (10 μg/ml) was added to evaluate proteasomal-dependent degradation. B, following a two-dimensional gel separation of 293T cell lysates (treated with arsenite or thapsigargin as indicated), immunoblots toward the endogenous eIF1 are presented. Where indicated (+CIP), arsenite-treated lysate was also incubated with alkaline phosphatase to evaluate if the pI shift is due to a phosphorylation event. C, autoradiogram of radiolabeled proteins obtained from 293T cells after a brief labeling pulse performed on cells that were treated as indicated. Cells were transfected with the indicated GFP reporter, subjected to a GFP IP, and content was resolved by SDS-PAGE. Untreated quantities were set as 1 for each reporter and relative quantities are shown. All quantifications were normalized to GFP mRNA transcript levels. Quantitation and statistical evaluations are present from three independent experiments. Both arsenite and thapsigargin treatments showed a statistical significant response with t test p < 0.001 upon comparing to the non-harboring 5′-UTR GFP reporter. D, fragmented MS/MS showing the only m/z fragment with a +80 shift. The presence of the y14 ion (labeled by arrow) indicates Tyr-79 is not the phosphorylation site leaving only Thr-72 as the possible site of phosphorylation. E, 24 h following the indicated eIF1 transfection, cells were treated with arsenite, and lysates were subjected to a two-dimensional gel separation analysis. Immunoblots revealing the exogenous eIF1 are presented.
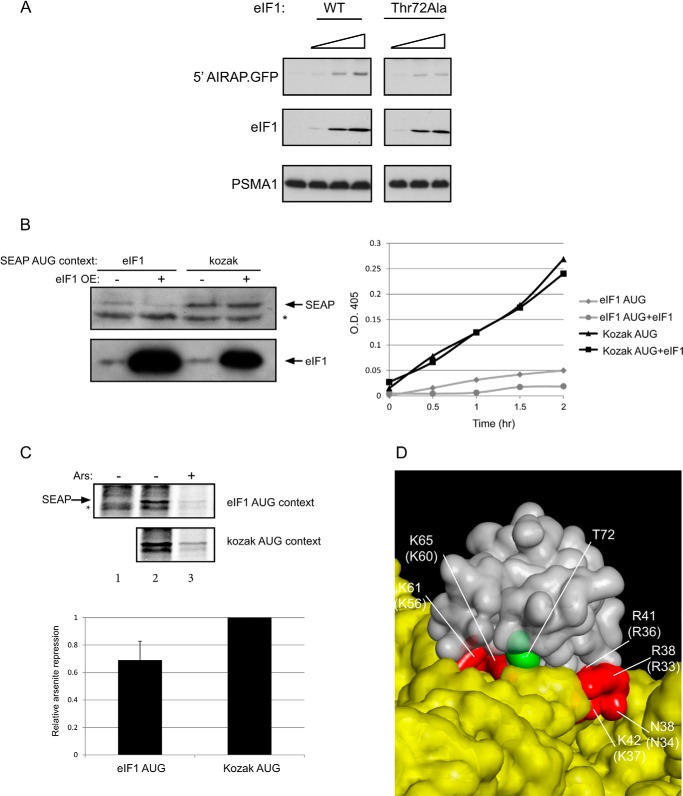
Since arsenite induced eIF1 phosphorylation (Fig. 5) and increased leaky scanning fidelity (Figs. 2 and 3) in the AIRAP transcript, we wanted to address the impact of arsenite-induced phosphorylation on eIF1 activity. Using the AIRAP reporter as a means to determine eIF1 activity (Fig. 4) indicated a dose-dependent response of the reporter toward eIF1 levels (Fig. 6A). Comparing the ability to induce AIRAP translation of the eIF1 T72A overexpression and the WT eIF1, indicated a reduction in the mutant eIF1 (T72A) ability to induce AIRAP translation (Fig. 6A). This result is consistent with a role of eIF1 phosphorylation in promoting AIRAP translation. eIF1 activity (i.e. determining start site fidelity) was also shown to regulate its own translation (23); therefore, we monitored the translational rate of eIF1 under normal and arsenite conditions. Since eIF1 does not contain any cysteine or methionine in the mature processed polypeptide (the N-terminal Met is processed), we could not label eIF1 itself. Thus, as previously shown (23), we constructed an eIF1 reporter (SEAP) in an eIF1 and a kozak AUG context. Both immunoblots and reporter activity indicated the ability of eIF1 overexpression to reduce reporter levels in an eIF1 AUG context-dependent manner (Fig. 6B). Using both these reporters, we performed pulse labeling under control and arsenite conditions and evaluated the amount of the reporter translated. Because arsenite is a strong inducer of eIF2α phosphorylation, we observed severe translational repression under arsenite conditions in both reporters; yet the eIF1 AUG context reporter consistently showed a higher degree of repression compared with the kozak AUG context reporter (Fig. 6C). Thus, we conclude that arsenite can increase start codon fidelity in an eIF1-dependent manner.
FIGURE 6.
Arsenite and eIF1 activity. A, 5′ AIRAP-GFP reporter was transfected alongside increasing amounts of the indicated eIF1 expression vector (25, 100, and 250 ng). 24 h post-transfection, cells were directly evaluated for GFP, eIF1, and PSMA1 content by immunoblot (IB). PSMA1 served as a loading control. B, SEAP reporter was introduced into 293 cells with eIF1 as indicated. SEAP levels were determined by immunoblot (left) or assayed kinetically for the reporter activity (right). * indicates a nonspecific band. C, non-transfected cells (lane 1) or transfected cells with the indicated SEAP reporter were untreated (lane 2) or arsenite-treated (lane 3) metabolically labeled, and immunoprecipitated against SEAP. * indicates a nonspecific band. Quantitation of three independent experiments is shown. Label incorporation decrease upon arsenite treatment in the kozak AUG reporter was set to 1, and relative increase in eIF1 AUG context repression is shown. D, presented is a surface structure of the human eIF1 (gray) and the rabbit 18 S RNA (yellow). The reported eIF1 residues contacting the 18 S (red) original positions in the yeast eIF1 are indicated in brackets. The phosphorylation site within eIF1 (Thr-72) is labeled in green. The figure is based on the published 4KZY PDB structural coordinates (33).
DISCUSSION
Our results exemplify a translational initiation regulation by means of leaky scanning on a transcript that is not encoding a PIC subunit. The molecular mechanisms governing this regulation include the presence of a single uORF that contains a sub-optimal kozak sequence. Increasing initiation fidelity enables the bypass of the uORF inhibitory role as a decoy start site and initiation of translation from the downstream ORF that harbors an optimal kozak AUG. Thus, AIRAP is an example of a transcript whose translational induction is brought upon by increasing fidelity as opposed to the reduction in translation that has been reported thus far (23). In the case of AIRAP, increased translation can be viewed as part of a cellular response that is required to increase proteasomal processing during certain protein misfolding conditions (26).
While our results confirm the ability of overexpressed eIF1 to increase AUG fidelity and mimetic arsenite-induced AIRAP translation (Fig. 4), we could not observe any changes in eIF1 levels during arsenite treatment. In addition, our findings also point to the fact that the estimated half-life of eIF1 is not short (Fig. 5A); thus, a mechanism to regulate AUG fidelity by quantitative means (i.e. eIF1 levels) is not be expected to produce short term oscillations or to be responsible for an immediate response regulating translation fidelity initiation. Several point mutations in eIF1 (Sui mutants) have previously demonstrated the capability to regulate AUG fidelity (20); therefore, our identified phosphorylation site in eIF1 (Thr-72) would be a suitable candidate to modulate AUG fidelity in a qualitative form and may be suitable to quickly respond to environmental conditions. Based on the previous mutants that increase AUG fidelity (21, 22), we would expect the phosphorylation of Thr-72 to reduce eIF1 release from the PIC in the context of a sub-optimal initiation site. The crystal structure of the human eIF1 (30) places Thr-72 at the center of a basic surface partially overlapping with a region previously termed KR surface, a surface involved in eIF3 and 40 S binding (31, 32). This is further evident from the structures of the 40 S-eIF1 complex (33, 34), as the aligned conserved phosphorylation site is clearly located in the close proximity of the eIF1–18 S interface (Fig. 6D). This interaction surface has also given rise to several Sui mutations (35) (Fig. 6D). Thus, Thr-72 phosphorylation may affect interactions with PIC components, thereby influencing AUG fidelity. Arsenite and thapsigargin (Fig. 5) modifications may not be limited to eIF1 phosphorylation and may include additional components in the PIC; thereby changing the interaction properties of eIF1 with the PIC. While our biochemical results indicated Thr-72 as an arsenite-induced phosphorylation site (Fig. 5E), an eIF1 T72D mutant introduced into cells (putatively a phosphomimetic mutant), did not enhance Airap induction compared with the wild type eIF1 (data not shown). This lack of hyperactivity in eIF1 may reflect the inability of the above mutant to mimic phosphorylation or may indicate the necessity of additional events in other PIC components to modulate the eIF1-PIC interactions during arsenite conditions. An eIF1 T72A mutant did demonstrate a reduction in eIF1 capacity to regulate leaky scanning fidelity (Fig. 6A), consistent with the role of eIF1 phosphorylation in AUG fidelity and Airap translational induction.
Several aspects remain to be revealed in the ability to regulate initiation fidelity by means of eIF1 phosphorylation. As stress conditions reported here are also known to regulate the integrated stress response (19), a possible cross-talk between leaky scanning (eIF1) phosphorylation and re-initiation (eIF2α) phosphorylation remains to be explored. Another point is how extensive is the effect of eIF1 phosphorylation on the proteome content. The presence of single uORFs in 5′-UTRs of the human genome is abundant. This situation resembles that of the AIRAP transcript studied in this report. It remains to be determined how many of these single uORF contain non-optimal kozak sequences, but it is clear that eIF1 influence on AUG fidelity may have large implications in the ability to regulate leaky scanning across these uORFs, as recently exemplified in the case of CHOP (25). Future analysis and experiments should enable to reveal the extent of eIF1 and its phosphorylation influence on leaky scanning as a means for regulating translational initiation at the proteome level.
Acknowledgments
We thank David Ron for the indicated antibodies and ATF4-GFP reporter, Ivaylo Ivanov for the eIF1 expression plasmid, Nick Ingolia and Jon Lorsch for fruitful discussions and suggestions during this project, and Steven Blais, NYU protein analysis facility for mass spectrometry phosphorylation analysis.
This work was supported by the John Hopkins-Technion grant foundation and the Israeli Science Foundation Grant 168/12.
- PIC
- pre-initiation complex
- ORF
- open reading frame
- uORF
- upstream open reading frame
- AIRAP
- arsenite-inducible regulatory particle-associated protein
- TC
- ternary complex
- UTR
- untranslated region
- HMW
- high molecular weight.
REFERENCES
- 1. Algire M. A., Maag D., Savio P., Acker M. G., Tarun S. Z., Jr., Sachs A. B., Asano K., Nielsen K. H., Olsen D. S., Phan L., Hinnebusch A. G., Lorsch J. R. (2002) Development and characterization of a reconstituted yeast translation initiation system. Rna 8, 382–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chaudhuri J., Chowdhury D., Maitra U. (1999) Distinct functions of eukaryotic translation initiation factors eIF1A and eIF3 in the formation of the 40 S ribosomal preinitiation complex. J. Biol. Chem. 274, 17975–17980 [DOI] [PubMed] [Google Scholar]
- 3. Pestova T. V., Kolupaeva V. G. (2002) The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16, 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Unbehaun A., Borukhov S. I., Hellen C. U., Pestova T. V. (2004) Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 18, 3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kozak M. (1991) Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266, 19867–19870 [PubMed] [Google Scholar]
- 6. Elfakess R., Sinvani H., Haimov O., Svitkin Y., Sonenberg N., Dikstein R. (2011) Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res. 39, 7598–7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zitomer R. S., Walthall D. A., Rymond B. C., Hollenberg C. P. (1984) Saccharomyces cerevisiae ribosomes recognize non-AUG initiation codons. Mol. Cell. Biol. 4, 1191–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheung Y. N., Maag D., Mitchell S. F., Fekete C. A., Algire M. A., Takacs J. E., Shirokikh N., Pestova T., Lorsch J. R., Hinnebusch A. G. (2007) Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 21, 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jackson R. J., Hellen C. U., Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nature Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lorsch J. R., Dever T. E. (2010) Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. J. Biol. Chem. 285, 21203–21207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nanda J. S., Cheung Y. N., Takacs J. E., Martin-Marcos P., Saini A. K., Hinnebusch A. G., Lorsch J. R. (2009) eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol. 394, 268–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hinnebusch A. G. (1993) Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2. Mol. Microbiol. 10, 215–223 [DOI] [PubMed] [Google Scholar]
- 13. Algire M. A., Maag D., Lorsch J. R. (2005) Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell 20, 251–262 [DOI] [PubMed] [Google Scholar]
- 14. Passmore L. A., Schmeing T. M., Maag D., Applefield D. J., Acker M. G., Algire M. A., Lorsch J. R., Ramakrishnan V. (2007) The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell 26, 41–50 [DOI] [PubMed] [Google Scholar]
- 15. Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., Hinnebusch A. G. (1992) Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68, 585–596 [DOI] [PubMed] [Google Scholar]
- 16. Lu P. D., Harding H. P., Ron D. (2004) Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vattem K. M., Wek R. C. (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brostrom C. O., Brostrom M. A. (1998) Regulation of translational initiation during cellular responses to stress. Progr. Nucleic Acid Res. Mol. Biol. 58, 79–125 [DOI] [PubMed] [Google Scholar]
- 19. Mathews M., Sonenberg N., Hershey J. W. B. (2007) Translational Control in Biology and Medicine, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20. Yoon H. J., Donahue T. F. (1992) The suil suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNA(iMet) recognition of the start codon. Mol. Cell. Biol. 12, 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Valásek L., Nielsen K. H., Zhang F., Fekete C. A., Hinnebusch A. G. (2004) Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell. Biol. 24, 9437–9455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saini A. K., Nanda J. S., Lorsch J. R., Hinnebusch A. G. (2010) Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNA(i)(Met) binding to the ribosome. Genes Dev. 24, 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ivanov I. P., Loughran G., Sachs M. S., Atkins J. F. (2010) Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc. Natl. Acad. Sci. U. S. A. 107, 18056–18060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loughran G., Sachs M. S., Atkins J. F., Ivanov I. P. (2012) Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 40, 2898–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Palam L. R., Baird T. D., Wek R. C. (2011) Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 286, 10939–10949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stanhill A., Haynes C. M., Zhang Y., Min G., Steele M. C., Kalinina J., Martinez E., Pickart C. M., Kong X. P., Ron D. (2006) An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol. Cell 23, 875–885 [DOI] [PubMed] [Google Scholar]
- 27. Glinka T., Alter J., Braunstein I., Tzach L., Wei Sheng C., Geifman S., Edelmann M. J., Kessler B. M., Stanhill A. (2014) Signal-peptide-mediated translocation is regulated by a p97-AIRAPL complex. Biochem. J. 457, 253–261 [DOI] [PubMed] [Google Scholar]
- 28. Sok J., Calfon M., Lu J., Lichtlen P., Clark S. G., Ron D. (2001) Arsenite-inducible RNA-associated protein (AIRAP) protects cells from arsenite toxicity. Cell Stress Chaperones 6, 6–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McEwen E., Kedersha N., Song B., Scheuner D., Gilks N., Han A., Chen J. J., Anderson P., Kaufman R. J. (2005) Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280, 16925–16933 [DOI] [PubMed] [Google Scholar]
- 30. Fletcher C. M., Pestova T. V., Hellen C. U., Wagner G. (1999) Structure and interactions of the translation initiation factor eIF1. EMBO J. 18, 2631–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lomakin I. B., Kolupaeva V. G., Marintchev A., Wagner G., Pestova T. V. (2003) Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 17, 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reibarkh M., Yamamoto Y., Singh C. R., del Rio F., Fahmy A., Lee B., Luna R. E., Ii M., Wagner G., Asano K. (2008) Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J. Biol. Chem. 283, 1094–1103 [DOI] [PubMed] [Google Scholar]
- 33. Lomakin I. B., Steitz T. A. (2013) The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature 500, 307–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rabl J., Leibundgut M., Ataide S. F., Haag A., Ban N. (2011) Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331, 730–736 [DOI] [PubMed] [Google Scholar]
- 35. Martin-Marcos P., Nanda J., Luna R. E., Wagner G., Lorsch J. R., Hinnebusch A. G. (2013) beta-Hairpin Loop of Eukaryotic Initiation Factor 1 (eIF1) Mediates 40 S Ribosome Binding to Regulate Initiator tRNAMet Recruitment and Accuracy of AUG Selection in vivo. J. Biol. Chem. 288, 27546–27562 [DOI] [PMC free article] [PubMed] [Google Scholar]