Background: CHD5 is a tumor suppressor and putative chromatin remodeling ATPase, although its activity is unknown.
Results: We found that CHD5 is a unique remodeling enzyme that partially unwraps the nucleosome.
Conclusion: CHD5 makes nucleosomal DNA accessible without nucleosome repositioning.
Significance: Our discovery adds to the diversity of known remodeling activities.
Keywords: ATPase, Chromatin Remodeling, Neuroblastoma, Nucleosome, Transcription Factor, CHD5
Abstract
Although mutations or deletions of chromodomain helicase DNA-binding protein 5 (CHD5) have been linked to cancer and implicate CHD5 in tumor suppression, the ATP-dependent activity of CHD5 is currently unknown. In this study, we discovered that CHD5 is a chromatin remodeling factor with a unique enzymatic activity. CHD5 can expose nucleosomal DNA at one or two discrete positions in the nucleosome. The exposure of the nucleosomal DNA by CHD5 is dependent on ATP hydrolysis, but continued ATP hydrolysis is not required to maintain the nucleosomes in their remodeled state. The activity of CHD5 is distinct from other related chromatin remodeling ATPases, such as ACF and BRG1, and does not lead to complete disruption or destabilization of the nucleosome. Rather, CHD5 likely initiates remodeling in a manner similar to that of other remodeling factors but does not significantly reposition the nucleosome. While the related factor CHD4 shows strong ATPase activity, it does not unwrap nucleosomes as efficiently as CHD5. Our findings add to the growing evidence that chromatin remodeling ATPases have diverse roles in modulating chromatin structure.
Introduction
Chromodomain helicase DNA-binding protein 5 (CHD5)2 2 is a member of a subgroup of SNF2-like ATP-dependent motor proteins that contain both a conserved SNF2-like motor domain and tandem chromodomains (1–3). CHD5 is also a key tumor suppressor that is frequently lost in a variety of cancers characterized by deletions of chromosome 1p36.3 (4–6). In addition to deletion, loss of CHD5 expression by promoter CpG-hypermethylation occurs in some cancers (7, 8). In neuroblastoma tumors, loss of CHD5 expression from both deletion of 1p36.3 and promoter silencing is correlated with a poor prognosis, and neuroblastoma cells that express CHD5 are more responsive to treatment than those that have lost CHD5 expression (9).
Despite the link between CHD5 and tumor suppression, the role of CHD5 in cells remains poorly understood. CHD5 appears closely related to CHD4 (also known as Mi-2β), a reported nucleosome sliding factor that is a subunit of the Nucleosome Remodeling and Deacetylase (NuRD) complex (10–13). In humans, CHD4 appears to be widely expressed (14) and, despite the sequence similarity with CHD5, does not appear to compensate for the loss of CHD5 in CHD5-deficient tumors. Rat CHD5 has been shown to interact with some subunits of the NuRD complex, suggesting that CHD5 may also play a role in the regulation of transcription (15). A recent genome-wide study of CHD5 has found that CHD5 binds to promoters near transcription start sites, providing further evidence that CHD5 may function as a transcriptional regulator (16).
Despite the links between CHD5 and tumor progression, it is not known what effect the activity of CHD5 has on chromatin structure. ATP-dependent chromatin remodeling enzymes act on chromatin in different ways (reviewed in Refs. 17). For example, chromatin assembly is mediated by the ISWI and CHD1 ATPases, while nucleosome disruption has been linked to SWI/SNF factors, such as the mammalian BRG1 and BRM ATPases. In addition, histone exchange is carried out by the INO80-family of remodelers. The precise mechanisms by which ATP hydrolysis is coupled to chromatin remodeling is not entirely understood. One proposed mechanism for another chromatin remodeling activity, nucleosome repositioning, is a loop propagation model (18–22). In this model, ∼30 bp of the linker DNA is fed into the nucleosome, which causes a small section of DNA to loop-out. This loop is then propagated around the nucleosome, and once the loop reaches the other end, the nucleosome would have been repositioned by 30 bp. Nucleosome repositioning allows for DNA within the nucleosome to move to the linker region between nucleosomes, thereby becoming more accessible to DNA-binding proteins.
In our current study, we analyzed the biochemical activity of CHD5 and found it remodels chromatin in a way that is distinct from that of other, well-studied chromatin-remodeling factors. CHD5 loops-out or unwraps the nucleosomal DNA at one or two internal sites located at 40 to 50 bp from the ends of the nucleosome, without repositioning the nucleosomes. CHD5 appears to have low processivity and remains bound to the remodeled nucleosomes. This suggests that CHD5 exposes nucleosomal DNA through stable looping out of the DNA. We also found that while the related family member CHD4 is a robust chromatin remodeling ATPase, it does not appear to efficiently unwrap nucleosomes as observed for CHD5. The finding that CHD5 is a unique remodeler is consistent with the evidence that loss of CHD5 expression causes defects in cell proliferation, and may shed light on the reason why other remodeling factors are unable to compensate for the absence of CHD5.
EXPERIMENTAL PROCEDURES
Protein Purification
The human wild-type CHD5, mut CHD5, and BRG1 proteins were expressed as FLAG-tagged fusions using the Bac-to-Bac baculovirus expression system (Invitrogen), as previously described for other ATPases (23, 24). Mutant CHD5 is identical to the wild-type protein except it contains a 2 amino acid substitution of D847A and E848A. Recombinant Drosophila ACF, NAP1, and native core histones were purified as described (25). Protein concentrations were estimated using A280 absorbance and SDS-PAGE followed by Coomassie Blue staining, with standards.
HaeIII Accessibility Assay
The restriction enzyme accessibility assay using HaeIII was performed essentially as previously described (26). Briefly, plasmid DNA (pGIE-0) was assembled into chromatin by salt dialysis, using purified Drosophila core histones. The average number of nucleosomes per array was estimated at 12, based on the patterns of partial digestion using MNase titration and agarose gel analysis (data not shown). The remodeling reactions were performed in Reaction Buffer (20 mm Tris-acetate, pH 7.9, 50 mm potassium acetate, 10 mm magnesium acetate, 1 mm DTT). Either 3 mm ATP or UTP (as a minus ATP control) was included, where indicated. Each reaction also contained 0.7 μg of chromatin (DNA), 0.5 μl HaeIII (New England Biolabs), and 1 μg of CHD5 (wild-type or mutant), 1 μg of ACF, or 2 μg of BRG1. Reactions proceeded at 30 °C for 2 h, were stopped with Stop Buffer (20 mm EDTA, 1% SDS, 0.2 m NaCl, 0.25 mg/ml glycogen, 0.06 mg/ml proteinase K), phenol extracted, and EtOH precipitated. The purified DNA was resolved on a 1.5% agarose gel and visualized by ethidium bromide staining.
Micrococcal Nuclease Sensitivity Assay
For the MNase digestion assays, plasmid-assembled chromatin (1 μg DNA) was incubated with CHD5 (0.3 μm), ACF (0.17 μm), or BRG1 (0.45 μm) in Reaction Buffer with 3 mm ATP or UTP (as a minus ATP control) for 20 min at 30 °C. CaCl2 was added (1 mm final), and the reactions digested with MNase (Worthington) at room temperature for 4 min. The reactions were stopped with Stop Buffer, and the DNA was phenol extracted, EtOH precipitated, and analyzed on an agarose gel with EtBr staining. The DNA marker used was a 123-bp ladder (Invitrogen).
For the glycerol gradient assays, the remodeling reactions containing CHD5 and plasmid-assembled chromatin were scaled almost 2-fold and digested with MNase for 10 min. The reactions were then stopped by adding EDTA (to 10 mm final concentration) and loaded onto a 5-ml linear gradient consisting of 10 to 40% glycerol in Reaction Buffer plus 10 mm EDTA. The gradients were centrifuged at 171,500 × g for 16 h at 4 °C. Twenty fractions of 250 μl each were collected from the top of the gradient. The DNA in each fraction was phenol extracted, EtOH precipitated, resolved on a 3% agarose gel, and visualized by EtBr staining. The DNA marker used was a 20-bp ladder (Fermentas).
Purified mononucleosomes for remodeling assays (50 ng of DNA) were incubated with CHD5 (10 nm) in Reaction Buffer with ATP or UTP for 20 min at 30 °C. CaCl2 was added (1 mm final concentration), followed by MNase (final concentrations of 0.05, 0.1, 0.25, 0.5, 1 unit/μl), and the reactions digested for 4 min at RT. For the time course assays, the ratios of CHD5 to nucleosomes, and the time points are indicated. Following digestion, the reactions were stopped with Stop Buffer and the DNA precipitated and analyzed by PAGE and SYBR green staining.
Restriction Enzyme Accessibility of Mononucleosomes
A 157-bp EcoRI-RsaI fragment containing a Xenopus 5S rDNA sequence was PCR amplified from the plasmid pXP-10 (27). The fragment was gel purified and assembled into mononucleosomes by salt dialysis, briefly digested with MNase, and purified on a glycerol gradient. The mononucleosomes (20 ng DNA) were incubated with the indicated remodeling factors (0.3 μg of each for the reactions with Cac8I or 0.6 μg of each for the reactions with EaeI and EcoRV) in Reaction Buffer for 10 min at 30 °C. Cac8I (5 units/ml), EaeI (1.5 units/ml) or EcoRV (10 units/ml) (New England Biolabs) was added, and the reactions incubated at 37 °C for an additional 1 h. ATP or UTP (3 mm final) was included for the Cac8I and EaeI digestions. For the EcoRV digestion, ATP or AMP-PNP (2 mm final) was included. In the EaeI reactions that contained Nap1 or plasmid DNA, 1 μg of Nap1 or 100 ng of pGIE-0 plasmid was included at the beginning of the reactions. DNA fragments were resolved by PAGE and visualized by SYBR green staining.
ATPase Assays of CHD4 and CHD5
The ATPase assays were carried out in 20 μl of Reaction Buffer with 0.3 mm ATP, a trace amount of [γ-32P]ATP and WT CHD4, WT CHD5, or mut CHD5 (22 nm final concentration). Plasmid chromatin (100 ng DNA) was included where indicated. The reactions were incubated at 30 °C for 1 h. One microliter of each reaction was spotted on a PEI cellulose plate (EMD Millipore), and the samples were resolved by thin-layer chromatography with 1 m formic acid, 0.5 m LiCl for 1 h. The plates were dried and quantitated with a phosphorimager (Bio-Rad). Each experiment was performed in triplicate, and the mean and standard deviation for each set were determined and displayed in a graph with GraphPad Prism software.
Nucleosome Sliding Assay
The nucleosome sliding assay was performed as described with slight modification (28). A 359-bp restriction fragment digested from pBSK-359 was assembled into mononucleosomes by salt jump, as previously described (28). Mononucleomes (50 ng DNA) were incubated for 30 min at 37 °C with ACF (20 nm) or CHD5 (120 nm) in a buffer consisting of 10 mm Tris-HCl, pH 7.8, 50 mm NaCl, 3 mm MgCl2, and 1 mm 2-mercaptoethanol. ATP or AMP-PNP was included (3 mm), where indicated. EDTA (0.1 mm) and glycerol (6%) were added, and the reactions resolved on a 4.5% polyacrylamide/0.5× TBE gel and visualized by SYBR green staining.
Mononucleosome Mobility Shift Assay
Mononucleosomes (25 ng DNA) used in the micrococcoal nuclease sensitivity assay described above were incubated for 5 min with increasing amounts of wild-type or mut CHD5 (0, 80 ng, 120 ng, 200 ng) in Reaction Buffer supplemented with 12% glycerol and 3 mm ATP or AMP-PNP. The reactions were resolved on an 8% polyacrylamide/0.5× TBE gel and visualized by SYBR green staining.
RESULTS
Chromatin Remodeling by CHD5 Increases Accessibility of Internal Nucleosome Sites
To study the chromatin remodeling activity of CHD5 and compare its activity with other SNF2-like family ATPases, we purified wild-type human CHD5 from baculovirus-infected insect cells (Fig. 1A) along with human BRG1 and Drosophila ACF, which contains the ISWI ATPase, as controls. We first tested whether CHD5 is able to remodel chromatin by using an assay that relies on the accessibility of restriction sites in a nucleosome array to a specific restriction enzyme (26, 29–32). Like ACF and BRG1, wild-type CHD5 leads to an increase in the accessibility of plasmid chromatin to the restriction enzyme HaeIII in an ATP-dependent manner (Fig. 1B). Thus, using a general assay for chromatin remodeling activity, we find that CHD5 is a chromatin remodeling enzyme that is able to modulate the accessibility of chromatin to DNA-modifying enzymes. However, this assay does not distinguish the different types of remodeling activity.
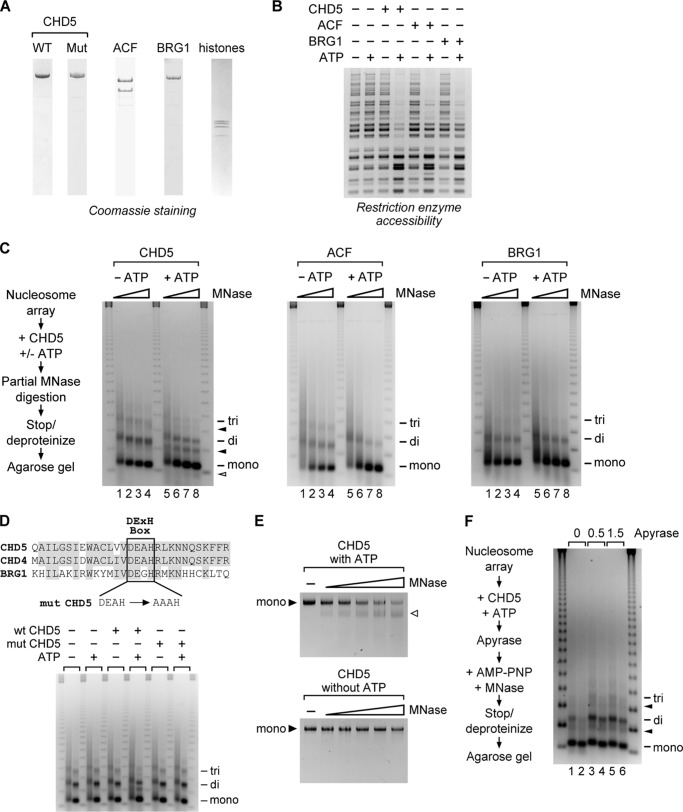
FIGURE 1.
CHD5 is a chromatin remodeling enzyme that increases accessibility of internal nucleosomal DNA sites. A, purification of wild-type CHD5, mut CHD5, ACF, BRG1. B, restriction enzyme accessibility assay using HaeIII and plasmid-assembled chromatin incubated with CHD5, ACF, or BRG1 in the presence or absence of ATP. DNA fragments were purified and resolved by agarose gel electrophoresis. C, plasmid chromatin was incubated with the indicated remodeling factors and partially digested with increasing amounts of MNase, and the DNA fragments were resolved by agarose gel electrophoresis. DNA fragments corresponding to mono-, di-, and trinucleosomes are indicated. DNA fragments migrating between canonical nucleosome fragments are indicated with solid arrowheads, while fragments migrating faster than mononucleosomes are indicated with the open arrowhead. D, top, alignment of the conserved DExH region from human CHD5, CHD4, and BRG1. Bot, partial MNase digestion assay of plasmid chromatin incubated with either wild-type CHD5 or mut CHD5 (DExH to AAxH mutation). The reactions were divided and partially digested with either a low (left lane) or high (right lane) concentration of MNase. DNA fragments corresponding to mono-, di-, and trinucleosomes are indicated. E, purified mononucleosomes were incubated with CHD5 in the presence or absence of ATP and digested with an increasing concentration of MNase. Following digestion, the DNA was purified, resolved by PAGE, and visualized by SYBR green staining. DNA corresponding to full-length mononucleosomal DNA is indicated as mono. Fragments migrating faster than mononucleosomes are indicated by the open arrowhead. F, left, CHD5 was incubated with plasmid chromatin and ATP. The remaining ATP in the reactions was depleted by incubation with apyrase. A non-hydrolyzable ATP analog (AMP-PNP) was then added, and the chromatin partially digested by MNase. DNA fragments migrating between canonical nucleosome fragments are indicated with solid arrowheads.
To further characterize the remodeling activity of CHD5, we analyzed the effects of CHD5 on the structure of a nucleosome array. In this assay, plasmid chromatin was incubated with CHD5 followed by partial digestion with micrococcal nuclease (MNase). Because MNase preferentially cuts DNA at protein-free regions, such as the linker region between nucleosomes, changes in the accessibility of DNA to MNase reflect changes in the structure of the nucleosome array (33). As a control, we first incubated the nucleosome array with CHD5 in the absence of ATP and then performed a partial MNase digestion. We analyzed the digested DNA fragments by agarose gel electrophoresis and observed the characteristic nucleosome ladder representing mono-, di-, and trinucleosomes (Fig. 1C, lanes 1–4), which are also observed with ACF and BRG1 in the absence of ATP (Fig. 1C). We then incubated the nucleosomes with CHD5 and ATP followed by digestion with MNase and observed the appearance of additional nucleosomal DNA species, which migrate at distinct positions between the mononucleosomal and dinucleosomal fragments and between the dinucleosomal and trinucleosomal fragments (Fig. 1C, lanes 5–8). Incubation with higher amounts of MNase leads to the generation of a DNA fragment shorter than mononucleosomal DNA (Fig. 1C, lanes 7–8). Based on the pattern of migration of molecular weight markers, we estimate that these DNA species are ∼40 to 50 base pairs (bp) smaller than the canonical nucleosomal DNA fragments. These intermediate size DNA fragments do not appear when samples are incubated with ACF or BRG1 in the presence of ATP (Fig. 1C). To confirm that an intact ATPase domain is required for the remodeling activity of CHD5, we tested a mutant version of CHD5 (mut CHD5) that carries a two amino acid substitution in the conserved DExH box (Fig. 1D, top). The distinct pattern of nucleosomal DNA fragments is not detected when the nucleosome array is incubated with mut CHD5 (Fig. 1D, bot) and confirms our finding that the observed remodeling activity of CHD5 is ATP-dependent.
To determine whether CHD5 is capable of increasing MNase accessibility of individual nucleosomes, we investigated the chromatin remodeling activity of CHD5 on mononucleosomes. We purified mononucleosomes by glycerol gradient sedimentation of MNase-digested plasmid chromatin. The purified mononucleosomes were then incubated with CHD5 in the presence of increasing amounts of MNase. In reactions containing CHD5 and ATP, MNase digests the mononucleosomal DNA at an internal site ∼50 bp from its end (Fig. 1E). Digestion of the mononucleosomal DNA is observed even in samples that are incubated with low concentrations of MNase. In contrast, no digestion is detected when mononucleosomes are incubated with CHD5 in the absence of ATP. Together, our results suggest that the altered patterns of MNase digestion observed in the experiments in which nucleosomal arrays are incubated with CHD5 and ATP are due to cleavage of the nucleosomal DNA and not due to protection of extranucleosomal linker DNA by bound CHD5.
We considered two possible mechanisms by which the activity of CHD5 can lead to an increase in the accessibility of the nucleosomal DNA to MNase. First, CHD5 might disrupt histone:DNA contacts (e.g. by looping or unwrapping), leading to the exposure of regions of the nucleosomal DNA to the nuclease. Alternatively, CHD5 could push or slide nucleosomes by 40 to 50 bp off the ends of the DNA, allowing MNase to digest the exposed region. To determine whether CHD5 slides nucleosomes off the ends of DNA, we took advantage of the MNase assays that use plasmid chromatin as substrates. Plasmid chromatin has no free DNA ends off which nucleosomes could slide. Free ends are only generated upon incubation with MNase. Therefore, to eliminate the possibility that the observed remodeling activity of CHD5 requires free DNA ends, we allowed CHD5 to remodel plasmid chromatin in the presence of ATP and then stopped the remodeling activity by depleting the remaining ATP in the reactions with apyrase (Fig. 1F), followed by the addition of an excess of a non-hydrolyzable ATP analog (AMP-PNP). We used two concentrations of apyrase and both were sufficient to completely hydrolyze all of the ATP in parallel reactions containing trace amounts of [32P]ATP (data not shown). We then performed the MNase digestion and found that removal of ATP by apyrase before MNase digestion does not inhibit the formation of the intermediate size nucleosomal DNA species (Fig. 1F, lanes 3–6). This result shows that, once CHD5 remodels nucleosomes, continued ATP hydrolysis is no longer required. Together, our results suggest that the activity of CHD5 leads to an increase in accessibility of the nucleosomal DNA at an internal site and in a manner not observed for other remodeling factors like ACF and BRG1.
CHD5 Can Remodel Two Sites within Nucleosomes
To better understand the nature of the nucleosomes remodeled by CHD5, we resolved CHD5-remodeled nucleosomes digested with MNase by glycerol gradient sedimentation. Following MNase digestion and sedimentation of the samples incubated with CHD5 in the absence of ATP, we observe a large amount of DNA fragments corresponding to mononucleosomes (∼160 bp) and dinucleosomes (∼320 bp), which sediment to two distinct positions in the gradient (Fig. 2A). After sedimentation of samples incubated with CHD5 in the presence of ATP, we detect mononucleosomal and dinucleosomal DNA in the same fractions as observed for the samples incubated in the absence of ATP (Fig. 2B). Importantly, we find that the nucleosomes containing shorter DNA fragments co-sediment with intact nucleosomes. The nucleosomes containing DNA of ∼115 bp co-sediment with canonical mononucleosomes, whereas the nucleosomes containing DNA of ∼270 bp co-sediment with canonical dinucleosomes. Thus, the extra bands observed in nucleosome arrays and plasmid chromatin incubated with CHD5 and ATP and digested with MNase represent nucleosomes containing a decrease in the amount of nucleosomal DNA and not an increase in the length of linker DNA.
FIGURE 2.
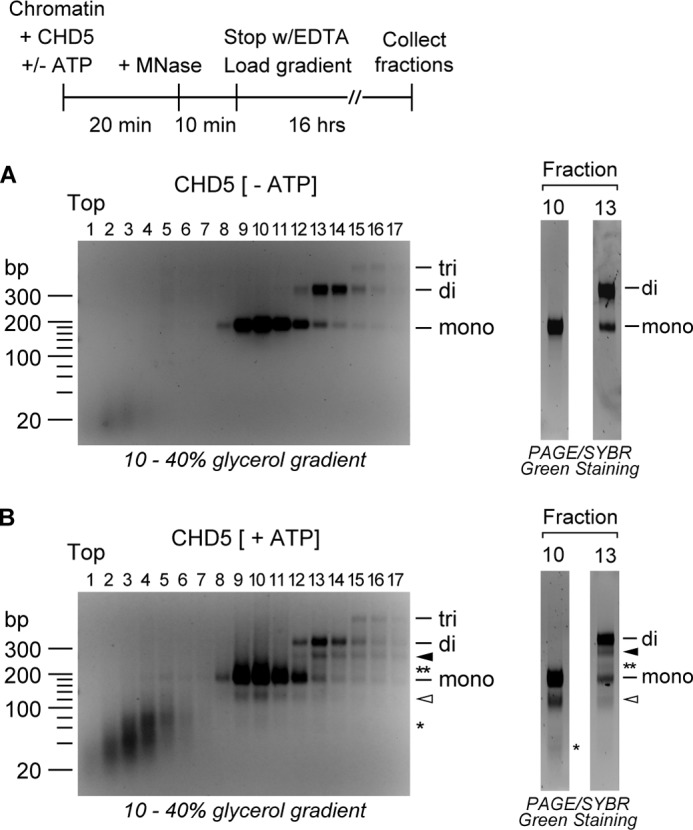
CHD5 increases accessibility of nucleosomal DNA at two sites without significant changes in nucleosome mass. Chromatin was incubated with CHD5 in the absence (A) or presence (B) of ATP, digested with MNase, and loaded onto a 10% to 40% linear glycerol gradient. Following sedimentation, fractions were collected, and the DNA in each fraction was purified and resolved by agarose gel electrophoresis. To better visualize the DNA fragments, peak fractions were resolved by PAGE and visualized by SYBR green staining. DNA fragments corresponding to mono-, di-, and trinucleosomes are indicated. DNA fragments derived from mononucleosomes digested at one internal site are indicated with the open arrowhead and those digested at two sites are indicated with the asterisk. DNA fragments from dinucleosomes digested at one internal site are indicated with the solid arrowhead, and fragments from dinucleosomes digested at two sites are indicated with the double asterisk.
The glycerol gradient fractions containing mononucleosomes and the shorter ∼115 bp species also contain a small amount of a DNA fragment of ∼70 bp (Fig. 2B, right). Similarly, we detect two shorter DNA fragments (of ∼270 bp and ∼220 bp) in the same fractions that contain dinucleosomes. These fragments are not observed in the samples incubated with CHD5 in the absence of ATP (Fig. 2A, right). The presence of two distinct species co-sedimenting with canonical mononucleosomes or dinucleosomes suggests that CHD5 is able to increase the accessibility of the DNA to MNase at one or two sites within the nucleosome.
The co-fractionation of canonical dinucleosomes with dinucleosomes containing shorter nucleosomal DNA is consistent with the hypothesis that the activity of CHD5 activity leads to the exposure of internal sites within the nucleosomes but not to nucleosome sliding. The sliding of nucleosomes off the ends of the DNA and subsequent cleavage of exposed DNA by MNase could yield mononucleosomes containing shorter DNA of various lengths but not dinucleosomes containing shorter DNA. In addition, CHD5 activity leads to an increase in the overall digestion of the nucleosome array and yields larger amounts of mononucleosomes (see Figs. 1C and 2). This implies that CHD5 is not “pushing” two nucleosomes together (e.g. Ref. 34), which would lead to a decrease in the amounts of mononucleosomes observed after MNase digestion.
Incubation of plasmid chromatin with CHD5 in the presence of ATP also generates DNA fragments that sediment near the top of the gradient (i.e. Fig. 2B, fractions 1–6). These DNA fragments are likely free-DNA ranging in size from 40 to 80 bp, based on molecular weight markers. A more prominent band, corresponding to a DNA fragment with a molecular weight of ∼70, can be detected in fractions 3 to 5. We suspect that this fragment represents the shortest DNA that is able to remain weakly bound to the nucleosome but can also dissociate from the histones. We also detect the accumulation of small DNA fragments of ∼50 bp that may correspond to the DNA that is cleaved off the nucleosome by MNase. Significantly less free-DNA was observed in the samples incubated with CHD5 in the absence of ATP (Fig. 2A).
Unwrapping of DNA by CHD5 Is Limited to Discrete Positions of the Nucleosome
Our studies show that the chromatin remodeling activity of CHD5 leads to the unwrapping of the DNA at more than one site within the nucleosome. We next investigated whether the CHD5-mediated unwrapping of the nucleosomal DNA extends through the dyad. Unwrapping of DNA through the dyad would be consistent with a loop propagation model of chromatin remodeling (18–22). To determine whether CHD5 increases the accessibility of nucleosomal DNA near the dyad, we reconstituted mononucleosomes in vitro, using a defined DNA sequence that contains the natural Xenopus 5S rDNA nucleosome positioning sequence (35, 36) with various restriction sites. We then removed all free and extranucleosomal DNA through digestion with MNase and purified the reconstituted mononucleosomes by glycerol gradient sedimentation. Next, we incubated the mononucleosomes with CHD5 and a restriction enzyme and determined whether the activity of CHD5 leads to an increase in the accessibility of its respective restriction site. When mononucleosomes were incubated with Cac81, which cleaves at the dyad, we did not observe a significant increase in the digestion of DNA treated with CHD5, ACF, or BRG1 in the presence of ATP (Fig. 3A), as compared with mononucleosomes treated with the same remodeling factors in the absence of ATP. These results indicate that the activity of CHD5 does not lead to an increase in the accessibility of this site, suggesting that CHD5 does not unwrap the nucleosomal DNA at the dyad. In contrast, when the mononucleosomes were digested with EcoRV, which cleaves at a site ∼44 bp from one end of the nucleosomal DNA, we observed a significant increase in the digestion of the DNA in the samples incubated with CHD5 in the presence of ATP (Fig. 3B), as compared with the samples treated with CHD5 in the absence of ATP. Digestion of the nucleosomal DNA with EcoRV was not affected when mononucleosomes were incubated with ACF or BRG1, in the presence or absence of ATP, suggesting the activity of CHD5 is not shared with these remodeling enzymes.
FIGURE 3.
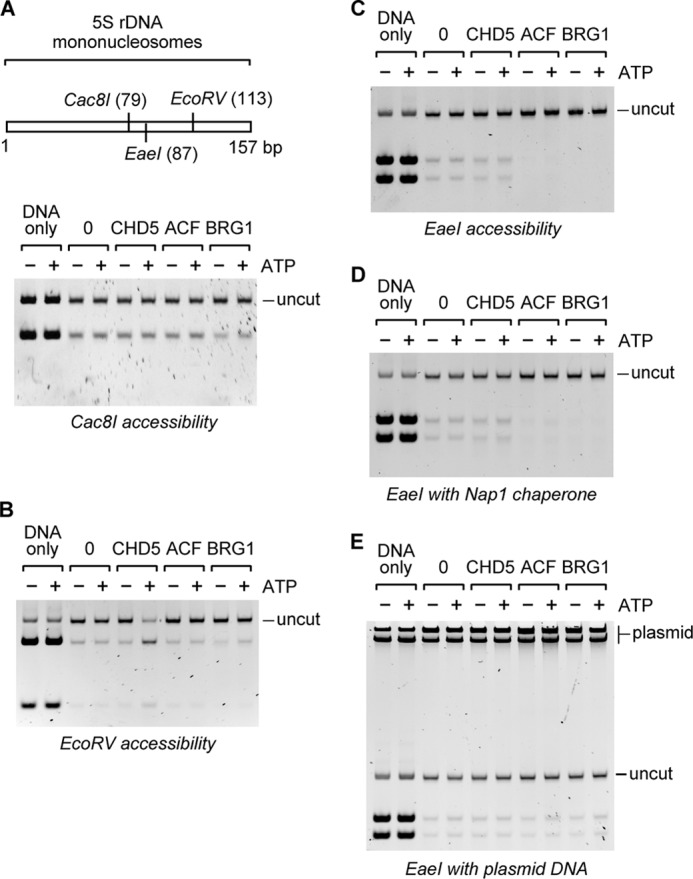
CHD5 increases accessibility of mononucleosomal DNA at specific sites. Mononucleosomes lacking extranucleosomal DNA were reconstituted on a natural 5 S rDNA positioning sequence and incubated with CHD5 in the presence of a restriction enzyme. A, schematic of the mononucleosome DNA with restriction sites and their positions indicated in parentheses. Mononucleosomes were incubated with CHD5 and digested with Cac8I, which cleaves at the dyad, or (B) with EcoRV, which cleaves at a single site at ∼44 bp from one end, or (C) with EaeI, which cleaves at a single site ∼70 bp from one end. To aid in the capture of potentially released histones, the histone chaperone NAP1 (D) or plasmid DNA (indicated as plasmid at the top of the gel) (E) was included in the EaeI digestion reactions. DNA corresponding to full-length mononucleosomal DNA is indicated as uncut. Samples incubated in the absence of remodeling factors are indicated by 0. DNA-only refers to samples with naked DNA.
The same mononucleosome accessibility assay was performed with the restriction enzyme EaeI, which cleaves at a site ∼70 bp from one end (i.e. between the EcoRV and Cac8I sites). None of the tested remodeling factors led to a significant increase in digestion with EaeI (Fig. 3C), confirming that the activity of CHD5 is limited to specific sites within the nucleosome. We repeated the EaeI digestion in the presence of either the histone chaperone NAP1 (Fig. 3D), which has been shown to aid nucleosome disruption (37), or plasmid DNA (Fig. 3E), which would capture any dissociated histones and prevent them from rebinding to the 5 S rDNA template. In neither case did we observe an increase in accessibility of the EaeI site of mononucleosomes treated with CHD5 in the presence of ATP. A slight ATP-dependent increase in the accessibility of all three restriction sites was observed in samples incubated with BRG1 in the presence of ATP, which is consistent with reports that BRG1 exhibits weak nucleosome disruption activity on mononucleosomes (18). We should note that the digestion of naked DNA by Cac8I, EcoRV, and EaeI is not affected by incubation with CHD5, ACF, or BRG1 (data not shown). Together, our data suggest that CHD5-mediated unwrapping of nucleosomal DNA is limited to regions positioned at 40 to 50 bp from the entry or exit sites of the nucleosome and does not extend through the dyad. Our results also indicate that the remodeling activity of CHD5 does not result in complete nucleosome disruption.
CHD5 Has Increased Affinity for Remodeled Nucleosomes
We have shown that continuous ATP hydrolysis by CHD5 is not required to maintain nucleosomes in a remodeled state (Fig. 1D). This raises the possibility that nucleosomes remodeled by CHD5 are stable and have low mobility. To investigate this, we used a 359 bp restriction fragment and assembled mononucleosomes, which were then resolved by native polyacrylamide gel electrophoresis (PAGE). Based on the migration of the mononucleosomes, we observe four major nucleosomes species, which were previously shown to represent the four preferred nucleosome positions (28). We then incubated the assembled mononucleosomes with CHD5 or ACF and examined whether any significant repositioning of nucleosomes occurred. In the presence of ATP, ACF repositions the nucleosomes resulting in an increase in two of the four nucleosome positions. In contrast, CHD5 did not exhibit any detectable nucleosome repositioning in our assays. Thus, while we cannot completely exclude the possibility that CHD5 confers some degree of nucleosome mobility, our results show that the remodeling activity of CHD5 does not lead to significant mononucleosome repositioning. This is consistent with our previous finding (Fig. 1F) that the increase in MNase accessibility of plasmid chromatin by CHD5 is not due to the sliding of nucleosomes off of free DNA ends.
One possible explanation for the fact that CHD5 increases nucleosomal DNA accessibility without promoting significant dissociation or repositioning of mononucleosomes is that CHD5 unwraps nucleosomes yet remains bound to the them. To investigate this, we performed a gel mobility shift assay, in which we analyzed the binding of wild-type or mut CHD5 to purified mononucleosomes by PAGE. Using this assay, we observed a shift in the migration of mononucleosomes only in reactions containing wild-type CHD5 and ATP (Fig. 4B). Mononucleosome binding is nearly undetectable in reactions containing wild-type CHD5 and the non-hydrolyzable AMP-PNP, or in reactions containing mut CHD5. These results suggest that, in the presence of ATP, CHD5 remodels nucleosomes and remains bound to them. We should note that in sliding assays such as the one shown in Fig. 4A, we also observe a mobility shift of mononucleosomes when we increase the concentration of CHD5 beyond the one shown in Fig. 4A (data not shown). We would also like to note that while the binding of CHD5 to mononucleosomes is enhanced in the presence of ATP, we cannot be certain that the shift in mobility of mononucleosomes is solely due to CHD5-binding. It is possible that some of the observed mobility shift is due to alterations of nucleosome structure caused by the remodeling activity of CHD5. Further, while the affinity of CHD5 for nucleosomes is increased in the presence of ATP, CHD5 binding may not be required to maintain the remodeled state.
FIGURE 4.
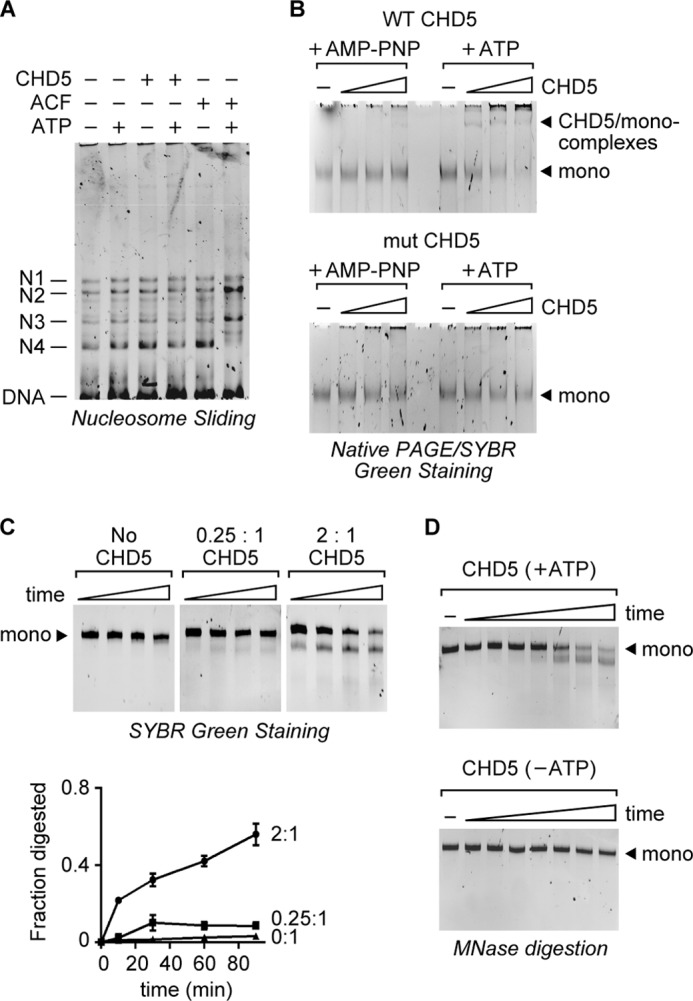
CHD5 does not readily dissociate from remodeled mononucleosomes. A, nucleosome sliding assay using mononucleosomes assembled onto a 359 bp restriction fragment. The four major nucleosome positions are labeled as N1–4 (28). CHD5 (125 nm) or ACF (20 nm) were incubated with the mononucleosomes in the presence of ATP or AMP-PNP (as a minus ATP control). The reactions were then analyzed by native PAGE and SYBR green staining. B, gel mobility shift assays were performed using increasing amounts of wild-type CHD5 or mut CHD5 incubated with purified mononucleosomes in the presence of ATP or non-hydrolyzable AMP-PNP. The reactions were then resolved by native PAGE and visualized by SYBR green staining. C, sub-stoichiometric amounts of CHD5 do not lead to remodeling of all available nucleosomes over time. Purified mononucleosomes were incubated with CHD5 at the indicated ratios and in the presence of ATP and MNase. The reactions were stopped at various times and the DNA resolved by PAGE (top). Quantification of the results is shown and is based on the fraction of digested mononucleosomal DNA in each lane (bot). Values shown are mean and S.D. (n = 3). D, time course of MNase digestion of mononucleosomes remodeled by excess CHD5 in the presence or absence of ATP. The time points are (from left to right): 0, 15 s, 30 s, 1 min, 2 min, 10 min, 20 min, 30 min. The initial remodeling rate was estimated (kcat = 0.16 ± 0.03 min -1).
The increase in affinity for nucleosomes in the presence of ATP raises the possibility that CHD5 has low processivity and does not readily move from one nucleosome to another. To investigate this, we analyzed whether CHD5 has a tendency to persist on remodeled nucleosomes If so, then reactions containing a sub-stoichiometric amount of CHD5 relative to the amount of nucleosomes should yield a limited number of remodeled nucleosomes that do not significantly increase over time. On the other hand, if CHD5 quickly dissociates following remodeling and is free to bind and remodel other nucleosomes, then the number of remodeled nucleosomes should steadily increase over time. To determine the processivity of CHD5, mononucleosomes were incubated with CHD5 in the presence of ATP and MNase for increasing amounts of time. After the reactions were stopped, the DNA fragments were analyzed by PAGE. We used CHD5:nucleosome ratios of ∼2:1 and 0.25:1, and at both ratios, we observed remodeling of mononucleosomes within 10 min, as indicated by the generation of a DNA species of ∼115 bp (Fig. 4D). The number of nucleosomes that were digested by MNase increased slightly between 10 and 30 min, but appeared to remain constant after 30 min in the reactions containing sub-stoichiometric amounts of CHD5 (e.g. 0.25:1 ratio). Between 60 and 90 min, we detected some loss of full-length mono-nucleosomal DNA in all reactions, including those lacking CHD5. This is likely due to nonspecific digestion of the DNA by MNase. These results indicate that, once CHD5 remodels nucleosomes, it does not rapidly dissociate and continue remodeling other nucleosomes. The observed persistence of CHD5 on remodeled nucleosomes digested with MNase also suggests that the prolonged interaction of CHD5 with the nucleosomes is not dependent on the presence of a full nucleosomal DNA fragment. Our findings are consistent with a genome-wide binding study (16) that found that CHD5 is enriched within a narrow region DNA surrounding transcription start sites of its genomic targets. This implies that CHD5 has a tendency to stably bind to nucleosomes within promoter regions. It is possible that CHD5 found at the promoter regions functions to increase the accessibility of nucleosomal DNA to DNA-binding factors.
Previous studies have suggested that nucleosomes exist in a dynamic state in which spontaneous unwrapping or peeling of the DNA from a nucleosome occurs at a low frequency (38, 39). If true, it is likely that CHD5 acts either to catalyze the unwrapping or to stabilize the unwrapped state. Under our experimental conditions, we observe very little MNase digestion of mononucleosomes in the absence of CHD5 (Fig. 4C) or in the presence of CHD5 without ATP (Fig. 1E). To better analyze the rate of nucleosome accessibility catalyzed by CHD5 and ATP, we extended the time course experiment to include early time points. In reactions containing excess CHD5 and ATP, we observe MNase digestion within 30 s, and the digestion increases throughout the 30 min time course. In contrast, we observe only a small amount of MNase digestion in the reactions lacking ATP. These results show that under the conditions in which CHD5 rapidly increases nucleosomal DNA accessibility, spontaneous nucleosome unwrapping is not readily detected. Using the results of this time-course and replicate assays, we were able to estimate the rate of remodeling by CHD5 in the presence of ATP. We estimate the initial kcat at 0.16 ± 0.03 min−1 for CHD5 on mononucleosomes. This rate is consistent with the remodeling rates reported for other ATP-dependent remodeling enzymes examined under similar conditions (40, 41).
CHD4 Is Less Robust Than CHD5 at Nucleosome Unwrapping
Several reports have shown that loss of CHD5 is linked to cell-proliferation defects and cancer (4–8) in cells that still express CHD4, indicating that CHD4 is unable to fully compensate for the absence of CHD5. To compare the remodeling activities of CHD4 and CHD5, we purified recombinant human CHD4 using the same purification strategy described above for CHD5. We then analyzed the specific activities of CHD4 and CHD5 using a standard ATPase assay. We measured the amount of ATP hydrolyzed by wild-type CHD5, wild-type CHD4, or mut CHD5 in the presence or absence of plasmid chromatin. We found that wild-type CHD4 and wild-type CHD5 have comparable levels of ATPase activity, which are stimulated by the presence of chromatin, with CHD4 having a slightly higher specific activity than CHD5 (Fig. 5A). We then compared the chromatin remodeling activities of CHD4 and CHD5 using the restriction enzyme accessibility assay and plasmid chromatin as the substrate. As observed with CHD5, ACF, and BRG1, CHD4 is able to stimulate digestion of the plasmid chromatin by the restriction enzyme in an ATP-dependent manner (Figs. 5B and 1B), confirming that our purified CHD4 is active. We then incubated CHD4 and CHD5 with plasmid chromatin and performed a partial MNase digestion using similar conditions to the experiments performed with CHD5, ACF, and BRG1 shown in Fig. 1A. Unlike CHD5, CHD4 does not lead to significant MNase digestion at internal sites within the nucleosome (Fig. 5C). The lack of clear unwrapping activity observed CHD4 cannot be explained by differences in the specific activity between CHD4 and CHD5, as CHD4 is as active as CHD5 in the ATPase and restriction enzyme accessibility assays. We cannot rule out the possibility that CHD4 has latent unwrapping activity that is stimulated under different conditions.
FIGURE 5.
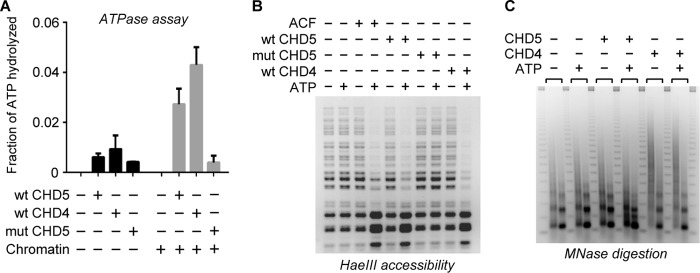
CHD4 shows minimal nucleosome unwrapping activity. A, recombinant human CHD4 was purified and the ATPase activities of CHD4, CHD5, and mut CHD5 (22 nm) were measured in the presence or absence of plasmid chromatin (100 ng). The fraction of ATP hydrolyzed (300 nm initial concentration) was determined and graphed relative to the initial amount of hydrolyzed ATP. Error bars represent S.D. (n = 3). B, restriction enzyme accessibility assay using plasmid chromatin and HaeIII. The plasmid chromatin contains 15 HaeIII cleavage sites that are only be partially digested by HaeIII in the absence of remodeling activity. CHD4, CHD5, or mut CHD5 were included where indicated. C, plasmid chromatin was incubated with CHD4 or CHD5 in the presence or absence of ATP, and the reactions digested with either a low (left-hand lane) or high (right-hand lane) concentration of MNase.
DISCUSSION
The link between the loss of CHD5 expression and progression of neuroblastoma suggests that CHD5 plays an essential and unique role in neurons. We have analyzed the chromatin remodeling activity of CHD5 and found it increases DNA accessibility within the nucleosome through a DNA unwrapping mechanism. This type of unwrapping activity has not been observed for other chromatin remodeling enzymes and adds to the diversity of known chromatin remodeling activities, which include chromatin assembly, nucleosome disruption, histone exchange and nucleosome sliding (reviewed in Ref. 17). Based on our data, we believe that CHD5 acts to stably unwrap nucleosomal DNA at sites positioned ∼40 to 50 bp from the entry/exit points of the nucleosome, without the requirement for complete nucleosome displacement or disruption. We predict that CHD5 unwraps the nucleosomal DNA by transiently altering the histone:DNA contacts of the DNA entering the nucleosomes until a stably unwrapped site is formed. Previous studies on the dynamic nature of nucleosomes have suggested that nucleosomal DNA can transiently unwrap, particularly where the DNA enters and exits the nucleosome (38, 39). Thus, CHD5 could serve to catalyze and stabilize this spontaneous unwrapping of nucleosomal DNA that occurs at a low frequency in the absence of CHD5.
The precise mechanism of nucleosome unwrapping by CHD5 is not known, yet it is possible that CHD5 initiates the unwrapping of nucleosomes in a manner similar to other chromatin remodeling factors that have been shown to reposition nucleosomes via the propagation of a transient loop starting at 50 bp from the entry side of the nucleosome (18–22). However, CHD5 does not appear to propagate the unwrapped loop around the nucleosome and has higher affinity for remodeled nucleosomes. The mechanism by which CHD5 accomplishes this distinct remodeling activity is not currently known. The CHD3/4/5 subgroup of the CHD family of chromatin remodeling enzymes is characterized by the presence of tandem chromodomains and PHD fingers, both of which are histone binding motifs (2, 16, 42). Whether these motifs play a direct role in the remodeling activity of CHD5 is not clear, although it is likely they help target CHD5 to chromatin and may serve to regulate the activity of CHD5 until it encounters a nucleosome.
The stable unwrapping of sites within the nucleosome by CHD5 possibly serves to enhance the exposure or accessibility of both the nucleosomal DNA and the underlying histones to DNA-binding or modifying enzymes and to histone-modifying enzymes, respectively. The reports that CHD5 interacts with transcription factors (15), and it is enriched at promoter regions (16) are consistent with our model that CHD5 remodels nucleosomes to expose nucleosomal DNA at promoters. CHD5 could accomplish this by unwrapping DNA to create longer stretches of linker DNA or by potentially flipping out nucleosomes from higher order nucleosome arrays. It is important for future investigations to determine the impact of cofactors on the remodeling activity of CHD5. If, like CHD4, CHD5 is part of a NuRD-like complex (15), its cofactors may regulate the activity of CHD5 or aid in targeting CHD5 to specific chromatin domains. At the least, the unwrapping activity of CHD5 should facilitate the binding of DNA-binding cofactors, which could mask the accessibility of the nucleosomal DNA in the experiments we have reported.
Overall, our study adds to the increasing evidence that cells evolved multiple, distinct ATP-dependent mechanisms to alter chromatin structure. Our cells contain numerous ATP-dependent remodeling proteins. The fact that mutations or deletions of many remodeling factors are linked to human diseases suggests they have non-overlapping roles. This may be partially due to spatial and temporal differences in expression of some remodeling factors, differences in cofactor association, or differences in their remodeling activities.
Acknowledgments
We thank P. Szajner and A. D'Andrea for critical reading of the manuscript. We thank S. Shikov for cloning assistance with the mut CHD5. We thank C. Wu for providing the pBSK-395 plasmid. We also thank J. Kadonaga for helpful discussions of this work prior to publication.
This work was supported by the Dana-Farber Cancer Institute, the William F. Milton Fund, and the Claudia Adams Barr Program.
- CHD5
- chromodomain helicase DNA-binding protein 5
- ACF
- ATP-utilizing chromatin assembly and remodeling factor
- BRG1
- BRM/SWI2-related gene 1
- NuRD
- nucleosome remodeling and deacetylase
- ISWI
- imitation SWI
- NAP1
- nucleosome assembly protein 1
- PHD
- plant homeodomain
- SNF2
- sucrose non-fermenting 2
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate.
REFERENCES
- 1. Eisen J. A., Sweder K. S., Hanawalt P. C. (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23, 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flaus A., Martin D. M., Barton G. J., Owen-Hughes T. (2006) Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34, 2887–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gorbalenya A. E., Koonin E. V. (1993) Helicases: amino acid sequence comparisons and structure-function relationship. Curr. Opin. Struct. Biol. 3, 419–429 [Google Scholar]
- 4. Bagchi A., Papazoglu C., Wu Y., Capurso D., Brodt M., Francis D., Bredel M., Vogel H., Mills A. A. (2007) CHD5 is a tumor suppressor at human 1p36. Cell 128, 459–475 [DOI] [PubMed] [Google Scholar]
- 5. Fujita T., Igarashi J., Okawa E. R., Gotoh T., Manne J., Kolla V., Kim J., Zhao H., Pawel B. R., London W. B., Maris J. M., White P. S., Brodeur G. M. (2008) CHD5, a tumor suppressor gene deleted from 1p36.31 in neuroblastomas. J. Natl. Cancer Inst. 100, 940–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang L., He S., Tu Y., Ji P., Zong J., Zhang J., Feng F., Zhao J., Gao G., Zhang Y. (2013) Downregulation of chromatin remodeling factor CHD5 is associated with a poor prognosis in human glioma. J. Clin. Neurosci. 20, 958–963 [DOI] [PubMed] [Google Scholar]
- 7. Gorringe K. L., Choong D. Y., Williams L. H., Ramakrishna M., Sridhar A., Qiu W., Bearfoot J. L., Campbell I. G. (2008) Mutation and methylation analysis of the chromodomain-helicase-DNA binding 5 gene in ovarian cancer. Neoplasia 10, 1253–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mulero-Navarro S., Esteller M. (2008) Chromatin remodeling factor CHD5 is silenced by promoter CpG island hypermethylation in human cancer. Epigenetics 3, 210–215 [DOI] [PubMed] [Google Scholar]
- 9. Garcia I., Mayol G., Rodríguez E., Suñol M., Gershon T. R., Ríos J., Cheung N. K., Kieran M. W., George R. E., Perez-Atayde A. R., Casala C., Galván P., de Torres C., Mora J., Lavarino C. (2010) Expression of the neuron-specific protein CHD5 is an independent marker of outcome in neuroblastoma. Mol. Cancer 9, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tong J. K., Hassig C. A., Schnitzler G. R., Kingston R. E., Schreiber S. L. (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395, 917–921 [DOI] [PubMed] [Google Scholar]
- 11. Wade P. A., Jones P. L., Vermaak D., Wolffe A. P. (1998) A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 8, 843–846 [DOI] [PubMed] [Google Scholar]
- 12. Xue Y., Wong J., Moreno G. T., Young M. K., Côté J., Wang W. (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2, 851–861 [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y., LeRoy G., Seelig H. P., Lane W. S., Reinberg D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279–289 [DOI] [PubMed] [Google Scholar]
- 14. Ramsköld D., Wang E. T., Burge C. B., Sandberg R. (2009) An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 5, e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Potts R. C., Zhang P., Wurster A. L., Precht P., Mughal M. R., Wood W. H., 3rd, Zhang Y., Becker K. G., Mattson M. P., Pazin M. J. (2011) CHD5, a brain-specific paralog of Mi2 chromatin remodeling enzymes, regulates expression of neuronal genes. PloS one 6, e24515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paul S., Kuo A., Schalch T., Vogel H., Joshua-Tor L., McCombie W. R., Gozani O., Hammell M., Mills A. A. (2013) Chd5 requires PHD-mediated histone 3 binding for tumor suppression. Cell Reports 3, 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clapier C. R., Cairns B. R. (2009) The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 [DOI] [PubMed] [Google Scholar]
- 18. Dechassa M. L., Sabri A., Pondugula S., Kassabov S. R., Chatterjee N., Kladde M. P., Bartholomew B. (2010) SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell 38, 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kassabov S. R., Zhang B., Persinger J., Bartholomew B. (2003) SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell 11, 391–403 [DOI] [PubMed] [Google Scholar]
- 20. Saha A., Wittmeyer J., Cairns B. R. (2005) Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat. Struct. Mol. Biol. 12, 747–755 [DOI] [PubMed] [Google Scholar]
- 21. Strohner R., Wachsmuth M., Dachauer K., Mazurkiewicz J., Hochstatter J., Rippe K., Längst G. (2005) A 'loop recapture' mechanism for ACF-dependent nucleosome remodeling. Nat. Struct. Mol. Biol. 12, 683–690 [DOI] [PubMed] [Google Scholar]
- 22. Zofall M., Persinger J., Kassabov S. R., Bartholomew B. (2006) Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 13, 339–346 [DOI] [PubMed] [Google Scholar]
- 23. Rattner B. P., Yusufzai T., Kadonaga J. T. (2009) HMGN proteins act in opposition to ATP-dependent chromatin remodeling factors to restrict nucleosome mobility. Mol. Cell 34, 620–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yusufzai T., Kadonaga J. T. (2008) HARP is an ATP-driven annealing helicase. Science 322, 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fyodorov D. V., Kadonaga J. T. (2003) Chromatin assembly in vitro with purified recombinant ACF and NAP-1. Methods Enzymol. 371, 499–515 [DOI] [PubMed] [Google Scholar]
- 26. Alexiadis V., Kadonaga J. T. (2002) Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 16, 2767–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wolffe A. P., Jordan E., Brown D. D. (1986) A bacteriophage RNA polymerase transcribes through a Xenopus 5S RNA gene transcription complex without disrupting it. Cell 44, 381–389 [DOI] [PubMed] [Google Scholar]
- 28. Hamiche A., Sandaltzopoulos R., Gdula D. A., Wu C. (1999) ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97, 833–842 [DOI] [PubMed] [Google Scholar]
- 29. Almer A., Rudolph H., Hinnen A., Hörz W. (1986) Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5, 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boyer L. A., Logie C., Bonte E., Becker P. B., Wade P. A., Wolffe A. P., Wu C., Imbalzano A. N., Peterson C. L. (2000) Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem. 275, 18864–18870 [DOI] [PubMed] [Google Scholar]
- 31. Fan H. Y., He X., Kingston R. E., Narlikar G. J. (2003) Distinct strategies to make nucleosomal DNA accessible. Mol. Cell 11, 1311–1322 [DOI] [PubMed] [Google Scholar]
- 32. Varga-Weisz P. D., Wilm M., Bonte E., Dumas K., Mann M., Becker P. B. (1997) Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388, 598–602 [DOI] [PubMed] [Google Scholar]
- 33. Noll M., Kornberg R. D. (1977) Action of micrococcal nuclease on chromatin and the location of histone H1. J. Mol. Biol. 109, 393–404 [DOI] [PubMed] [Google Scholar]
- 34. Engeholm M., de Jager M., Flaus A., Brenk R., van Noort J., Owen-Hughes T. (2009) Nucleosomes can invade DNA territories occupied by their neighbors. Nat. Struct. Mol. Biol. 16, 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Simpson R. T., Stafford D. W. (1983) Structural features of a phased nucleosome core particle. Proc. Natl. Acad. Sci. U. S. A. 80, 51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Young D., Carroll D. (1983) Regular arrangement of nucleosomes on 5S rRNA genes in Xenopus laevis. Mol. Cell. Biol. 3, 720–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lorch Y., Maier-Davis B., Kornberg R. D. (2006) Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl. Acad. Sci. U. S. A. 103, 3090–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li G., Widom J. (2004) Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 11, 763–769 [DOI] [PubMed] [Google Scholar]
- 39. Polach K. J., Widom J. (1995) Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 254, 130–149 [DOI] [PubMed] [Google Scholar]
- 40. Cho I., Tsai P. F., Lake R. J., Basheer A., Fan H. Y. (2013) ATP-dependent chromatin remodeling by Cockayne syndrome protein B and NAP1-like histone chaperones is required for efficient transcription-coupled DNA repair. PLoS genetics 9, e1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang J. G., Madrid T. S., Sevastopoulos E., Narlikar G. J. (2006) The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat. Struct. Mol. Biol. 13, 1078–1083 [DOI] [PubMed] [Google Scholar]
- 42. Oliver S. S., Musselman C. A., Srinivasan R., Svaren J. P., Kutateladze T. G., Denu J. M. (2012) Multivalent recognition of histone tails by the PHD fingers of CHD5. Biochemistry 51, 6534–6544 [DOI] [PMC free article] [PubMed] [Google Scholar]