Background: The function of KAP1 is regulated by multiple posttranslational modifications during DNA damage response.
Results: A previously unidentified arginine-rich motif (ARM) of RNF4 regulates the phosphorylation-induced, SUMO-dependent recruitment and degradation of KAP1.
Conclusion: The ARM of RNF4 enhances SIM-SUMO-dependent recruitment.
Significance: The bimodular recognition by RNF4 could be critical for fine-tuning substrate selection during DNA damage response and other stress conditions.
Keywords: DNA Damage Response, Phosphorylation, Protein-Protein Interaction, SUMOylation, Ubiquitylation (Ubiquitination), ATM, KAP1, RNF4, STUbL, Arginine-rich Motif
Abstract
Krüppel-associated box domain-associated protein 1 (KAP1) is a universal transcriptional corepressor that undergoes multiple posttranslational modifications (PTMs), including SUMOylation and Ser-824 phosphorylation. However, the functional interplay of KAP1 PTMs in regulating KAP1 turnover during DNA damage response remains unclear. To decipher the role and cross-talk of multiple KAP1 PTMs, we show here that DNA double strand break-induced KAP1 Ser-824 phosphorylation promoted the recruitment of small ubiquitin-like modifier (SUMO)-targeted ubiquitin E3 ligase, ring finger protein 4 (RNF4), and subsequent RNF4-mediated, SUMO-dependent degradation. Besides the SUMO interacting motif (SIM), a previously unrecognized, but evolutionarily conserved, arginine-rich motif (ARM) in RNF4 acts as a novel recognition motif for selective target recruitment. Results from combined mutagenesis and computational modeling studies suggest that RNF4 utilizes concerted bimodular recognition, namely SIM for Lys-676 SUMOylation and ARM for Ser(P)-824 of simultaneously phosphorylated and SUMOylated KAP1 (Ser(P)-824-SUMO-KAP1). Furthermore, we proved that arginines 73 and 74 within the ARM of RNF4 are required for efficient recruitment to KAP1 or accelerated degradation of promyelocytic leukemia protein (PML) under stress. In parallel, results of bimolecular fluorescence complementation assays validated the role of the ARM in recognizing Ser(P)-824 in living cells. Taken together, we establish that the ARM is required for RNF4 to efficiently target Ser(P)-824-SUMO-KAP1, conferring ubiquitin Lys-48-mediated proteasomal degradation in the context of double strand breaks. The conservation of such a motif may possibly explain the requirement for timely substrate selectivity determination among a myriad of SUMOylated proteins under stress conditions. Thus, the ARM dynamically regulates the SIM-dependent recruitment of targets to RNF4, which could be critical to dynamically fine-tune the abundance of Ser(P)-824-SUMO-KAP1 and, potentially, other SUMOylated proteins during DNA damage response.
Introduction
Protein posttranslational modification (PTM)4 plays a critical role in modulating the stability, localization, and behavior of a protein for signaling. By covalently attaching small proteins to specific amino acid residues or by changing the structure or the chemical nature of the amino acids, the diversity of protein function can be largely expanded. Most PTMs are processed rapidly and reversibly, thereby serving as a regulatory switch of proteins involved in stress signaling pathways (1–3). Among them, phosphorylation has long been known as a central regulatory signal in response to various physiological stimuli, such as growth factors, replication stress, and DNA damage. Recently, emerging roles of other types of PTMs, including ubiquitylation and small ubiquitin-like modifications (SUMOylation) have also been described in a wide variety of (patho)physiological conditions. Multiple PTMs can occur sequentially or simultaneously on a specific protein substrate and result in cooperative or antagonizing effects in determining the fate of the protein. It is becoming increasingly clear that multiple PTMs converge upon DNA double strand breaks (DSBs) to initiate temporal and spatial coordination of the DNA damage checkpoint and repair response (3, 4).
Krüppel-associated box domain-associated protein 1 (KAP1), also known as TRIM28/TIF1β, a transcriptional corepressor and chromatin-remodeling protein, undergoes multiple PTMs, including DSB-induced phosphorylation at Ser-824 (5–8). We and others have reported that KAP1 is SUMOylated at several lysine residues located in and around the PHD/bromodomains (9, 10). Moreover, the SUMOylation of KAP1 represses the transcription of pro-apoptosis and pro-cell cycle arrest genes, including p21, BAX, GADD45α, NOXA, and PUMA, during DNA damage response (DDR) (7, 8, 10). The transcriptional repression of KAP1 target genes is mediated through the recruitment of HP1, SETDB1, the Mi-2α·NuRD complex, and histone deacetylases via its PHD and bromodomain (6, 12–15). In contrast, Ser-824 phosphorylation of KAP1 (Ser(P)-824-KAP1) promotes the repair of DSBs within heterochromatin by inducing the chromatin to decondense (6, 17). Furthermore, we have reported that the reduction of SUMOylated KAP1 (SUMO-KAP1) induced by KAP1 Ser-824 phosphorylation resulted in the transcriptional derepression of KAP1-targeted genes during DDR (7, 8, 10). Both phosphorylation and SUMOylation are critical PTMs for proper DDR, although little is known about the molecular interplay employed by KAP1 Ser-824 phosphorylation to regulate the abundance of SUMO-KAP1 molecules in the context of DNA damage. Therefore, we sought to decipher how the cross-talk of different types of PTMs regulates the fate of KAP1 in the context of DSB induction.
The conventional thought that the SUMOylation and ubiquitylation systems operate independently or antagonistically has changed with recent discoveries of SUMO-targeted ubiquitin ligase (STUbL) (reviewed in Ref. 18). The identification of Slx8-Rfp, MIP1, Slx5-Slx8, and ring finger protein 4 (RNF4) suggests that SUMOylation and ubiquitylation can work cooperatively (19–22). Therefore, STUbLs belong to a new class of ubiquitin E3 ligases that are recruited to SUMO-conjugated targeting proteins, resulting in ubiquitylation and subsequent degradation (23). Among them, RNF4 is the only human homolog of Slx8-Rfp. The most conspicuous structural feature of RNF4 or STUbLs is the tandem SUMO-interacting motifs (SIMs) that have specific consensus sequences to interact with the SUMO or SUMO-like domains of their ubiquitylation targets (23). Although published reports have contributed extensively to our understanding of how STUbLs recruit substrates and their role in connecting the SUMO and ubiquitin pathways (24–29), the exact mechanistic bases by which to regulate the SIM-dependent recruitment of SUMOylated targets to RNF4 for timely degradation during DDR remain to be determined.
Given the short half-life of the KAP1 Ser-824 phosphorylation signal (5–8) and the involvement of DSBs-induced phosphorylation in regulating both SUMOylation and ubiquitylation systems (4), we speculated that RNF4 could have a key role in regulating Ser-824-phosphorylated KAP1 abundance. Here, we show that RNF4 engages an active process to accelerate the turnover of Ser-824-phosphoylated, Lys-676-SUMOylated KAP1 through a previously unrecognized arginine-rich motif (ARM). Our study further suggests that RNF4 utilizes bimodular recognition by both SIM and ARM to dynamically fine-tune its target selection.
EXPERIMENTAL PROCEDURES
Cloning and Construction of Plasmids
Human RNF4 was tagged with Myc by cloning it into pCMV-Tag3B after standard RT-PCR of cDNA that was reverse-transcribed from total human RNA and amplified with RNF4-specific primer pairs (5′-GACCGTGGATCCATGAGTACAAGAAAGCGT-3′ (forward) and 5′-CCTGGAGAATTCTCATATATAAATGGGGTG-3′ (reverse). All Myc-RNF4 and HA-PML point mutations or deletions were generated from the Myc-RNF4 and HA-PML templates using a site-directed mutagenesis kit (Clontech). FLAG-RNF4, FLAG-ligase-dead RNF4, and FLAG-RNF4(Δ41–63) were provided by Dr. Ronald Hay. SFB-RNF4 was provided by Dr. Junjie Chen. All SFB-RNF4 deletion mutants were generated by the sequential PCR method and verified by sequencing. For the in vitro binding assay, SFB-RNF4 constructs containing different deletions were used as templates, and RNF4 was cloned into pT7CFE1 (Thermo Scientific) for in vitro translation. For bimolecular fluorescence complementation (BiFC) assays, RNF4 was cloned into the pBiFC-CC155 vector for fusion with C-terminal enhanced cyan fluorescent protein (ECFP) at the C terminus of RNF4 (30). FLAG-KAP1(WT) and the mutants were used as templates to generate short-form constructs. The full-length or PHD-bromodomain (aa 625–835) of KAP1 was then cloned into the pBiFC-VN173 vector for fusion with N-terminal venus at the C terminus of KAP1 (30).
Cell Cultures and Reagents
HEK293, HeLa, and U2OS cells were maintained (37 °C, 5% CO2) in DMEM supplemented with 10% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin. MCF7 cells were maintained in the same medium supplemented with recombinant human insulin (0.01 mg/ml). HEK293/shRNF4, MCF7/shKAP1, and MCF7/shRNF4 cells were cultured in HEK293 or MCF7 medium with puromycin (2 μg/ml). The proteasome inhibitor MG132 was obtained from Calbiochem and used at 5 or 10 μm in this study. Cycloheximide (CHX) was obtained from Sigma-Aldrich and used at 100 μg/ml to inhibit protein synthesis.
Western Blot Analyses and Antibodies
Whole cell lysates were prepared by lysing cells with Laemmli sample buffer supplemented with Complete protease inhibitor mixture (Roche) and PhosphoSTOP phosphatase inhibitor mixture (Roche). Equal amounts of whole cell extracts were first separated by SDS-PAGE and then Western blotted and probed with antibodies. The antibodies used on the Western blots were FLAG (M2, Sigma-Aldrich), KAP1 (Bethyl and a gift from Dr. David Schultz), Ser(P)-824-KAP1 (Bethyl), HA (Covance), β-actin (Millipore), RNF4 (Abnova and a gift from Drs. Ronald Hay and Jorma Palvimo (31)), tubulin (D-10), Ser(P)-1981-ATM, His, ubiquitin, EGFP and Myc (Santa Cruz Biotechnology), ATM (GeneTex), and SUMO-2 (Novus). Blots were visualized by enhanced chemiluminescence (ECL-Plus, GE Biosciences) using a Versadoc 3000 imaging system (Bio-Rad). Densitometric data were obtained and analyzed with Quantity One software (Bio-Rad), which provides a 4.8-order linear range of tracing. The Western blot analyses shown are representative of two to four independent experiments.
Immunoprecipitation of KAP1 and Coimmunoprecipitation Assay
FLAG-KAP1(WT) or its mutants were cotransfected with an EGFP-SUMO-1 expression construct into HEK293 cells using Lipofectamine 2000. Whole cell lysates were prepared by lysing the cells as described previously (8). Anti-FLAG (for KAP1) or other antibodies (1 μl) were mixed with whole cell lysate (1 mg), and samples were rotated (4 °C, 2 h). Protein A/G Plus-agarose (Santa Cruz Biotechnology) was added to bind and elute precipitated proteins. Eluates were examined on Western blots probed with the appropriate antibodies. To pull down SFB-RNF4, cells were lysed with NETN buffer (0.5% Nonidet P-40, 20 mm Tris-HCl (pH 8.0), 100 mm NaCl, and 1 mm EDTA) and mixed with S protein-agarose (Novagen), and then we analyzed the eluates on Western blots probed with the appropriate antibodies.
Production of Lentivirus and Lentiviral Transduction
The lentiviral vectors pLKO.1-shRNF4, pΔ8.7, and pVSV-G were constructed and used to produce lentivirus in HEK293FT cells, as described previously (32). Viral supernatant was collected, pooled, concentrated, and stored as described previously (32). For lentiviral infection, HEK293 and MCF7 cells were plated 1 day prior to infection and cultured overnight to ∼50% confluency. The culture medium was then aspirated, and fresh medium was added that contained concentrated lentiviruses with an empty vector, an shRNA against human RNF4, or a random sequence (as a control). Cells and viruses were incubated for 24 h in the presence of 8 μg/ml Polybrene. All studies used mixed populations of transduced cells.
Transfection
Lipofectamine 2000 (Invitrogen) was used for the plasmid DNA transfections according to the protocol of the manufacturer. The transfected cells were analyzed 48 h after transfection.
Irradiation (IR)
A Shepherd Mark I cesium 137 γ irradiator was used to irradiate cells at a fixed dose rate of 1–2 Gy/min.
Immunofluorescence Staining and Imaging
Cells were irradiated, washed with PBS, and fixed (30 min, −20 °C) in ice-cold methanol and ethanol (1:1, v/v) at the end of the desired, post-IR incubation period. For pre-extraction, after the PBS wash, cells were pre-extracted using CSK buffer (10 mm PIPES (pH 6.8), 100 mm NaCl, 1.5 mm MgCl2, 300 mm sucrose, and 0.5% Triton X-100) (5 min, on ice), washed with CSK buffer and PBS, and fixed in 4% formaldehyde (15 min, 4 °C). Cells were blocked in blocking buffer (PBS containing 2% BSA) and then incubated with the appropriate primary antibody. Antibodies used were Ser(P)-824-KAP1 (Bethyl, 1:500), RNF4 (Abnova, 1:200), γ-H2AX (Millipore, 1:500), and FK2 and Lys-48-Ub (Millipore, 1:1000) in incubation buffer (PBS containing 2% BSA). Cells were then washed with PBS-Tween 20 and incubated with a fluorescent secondary antibody (Alexa Fluor 488 goat anti-mouse or Alexa Fluor 568 goat anti-rabbit, Invitrogen). Cells were then washed with PBS-Tween 20 and mounted on slides using ProLong Gold antifade reagent with DAPI (Invitrogen). Slides were viewed using a ×60 objective on an inverted IX81 microscope or a ×100 objective on an AX70 fluorescence microscope. Images were collected and processed using Image-Pro 6.3 software.
Construction of a Molecular Docking Model
The final conformation was chosen from the top 2000 docked conformations. These were ranked on the basis of the geometrical, hydrophobic, and electrostatic complementarities of the molecular surface and by considering the biochemical evidence reported here. In consideration of the flexibility of the SUMO C-terminal loop region from aa residues 94–97 and the NMR structure of KAP1 missing aa residues 813–824, these two regions were modeled by using the loop search method in the Biopolymer module implemented in the SYBYL 6.8 (TRIPOS, St. Louis, MO) package. In the modeled structure, an isopeptide bond was assigned between SUMO Gly-97 and KAP1 Lys-676, and the Ser(P)-824 of KAP1 was constructed and located close to the ARM motif in RNF4. Then, the resultant structure was minimized stepwise with the Assisted Model Building with Energy Refinement (AMBER) force field in the SYBYL package to remove possible steric hindrances and to obtain a lower energy conformation. Subsequently, a 1-ns molecular dynamics simulation was carried out using the AMBER package (version 10.0) to sample a more reasonable conformation.
Chromatin Fractionation
Chromatin was fractionated using a modified version of a previous protocol (33). Approximately 1 × 107 cells were resuspended in 1 ml of buffer A (10 mm HEPES (pH 7.9), 10 mm KCl, 1.5 mm MgCl2, 340 mm sucrose, 10% glycerol, and 1 mm DTT) supplemented with protease/phosphatase inhibitors. Triton X-100 was added to a final concentration 0.1%, and the cells were incubated for 5 min on ice. Nuclei were collected by centrifugation at low speed (4 min, 1300 × g, 4 °C) and then lysed in buffer B (3 mm EDTA, 0.2 mm EGTA, and 1 mm DTT) that was supplemented with protease/phosphatase inhibitors. The insoluble chromatin fraction was pelleted by centrifugation (4 min, 1700 × g, 4 °C) and then resuspended in 1× Laemmli sample buffer. Samples were boiled, separated by SDS-PAGE, and analyzed on Western blots.
In-cell Ubiquitylation Assay
HEK293 cells were cotransfected with FLAG-KAP1, Myc-SUMO-2, and His-Myc-Ub-WT or the −K48R or −K63R mutants. MG132 (5 μm) was added 8 h before lysing the cells with 6 m guanidine-HCl (6 m guanidine-HCl, 0.1 m NaH2PO4, and 10 mm imidazole). Cell lysates were then processed as described under “Immunoprecipitation of KAP1 and Coimmunoprecipitation Assay,” but nickel-nitrilotriacetic acid-agarose beads (Sigma-Aldrich) were used to pull down His-Ub substrates. Beads were washed with wash buffer (50 mm Na3PO4 (pH 8.0), 0.3 m NaCl, and 10 mm imidazole), and proteins were eluted by adding Laemmli sample buffer. Eluted proteins were separated by SDS-PAGE and analyzed on Western blots.
In Vitro Binding Assay
HEK293 cells were cotransfected with FLAG-KAP1 or FLAG-PML with Myc-SUMO-2. MG132 (5 μm) was added 30 min prior to exposure to Dox (5 μm) or ATO (2 μm) for 1 h. FLAG-KAP1 or FLAG-PML was pulled down as described under “Immunoprecipitation of KAP1 and Coimmunoprecipitation Assay,” and the protein was eluted by 3× FLAG peptide (150 ng/μl). The eluted SUMO-KAP1 or SUMO-PML protein was incubated with RNF4 protein generated by a one-step human coupled in vitro translation kit (Thermo Scientific). The reaction was carried out by incubating the proteins in Tris-HCl (50 mm (pH 7.5)), NaCl (250 mm), Nonidet P-40 (0.1%), and glycerol (5%) for 1 h at 22 °C. Anti-FLAG M2-agarose beads (Sigma-Aldrich) were added to pull down FLAG-KAP1 or FLAG-PML, and the beads were washed with the reaction buffer (increased NaCl to 1 m). The proteins were eluted by Laemmli sample buffer and subjected to Western blot analysis.
Statistical Analysis
Error bars represent mean ± S.D. Unpaired Student's t test, assuming equal variances, was performed using Microsoft Excel to determine significant differences between groups. p < 0.05 was considered to be statistically significant.
RESULTS
RNF4 Accelerates the DSBs-induced Decrease of Steady-state Ser(P)-824-SUMO-KAP1 Abundance
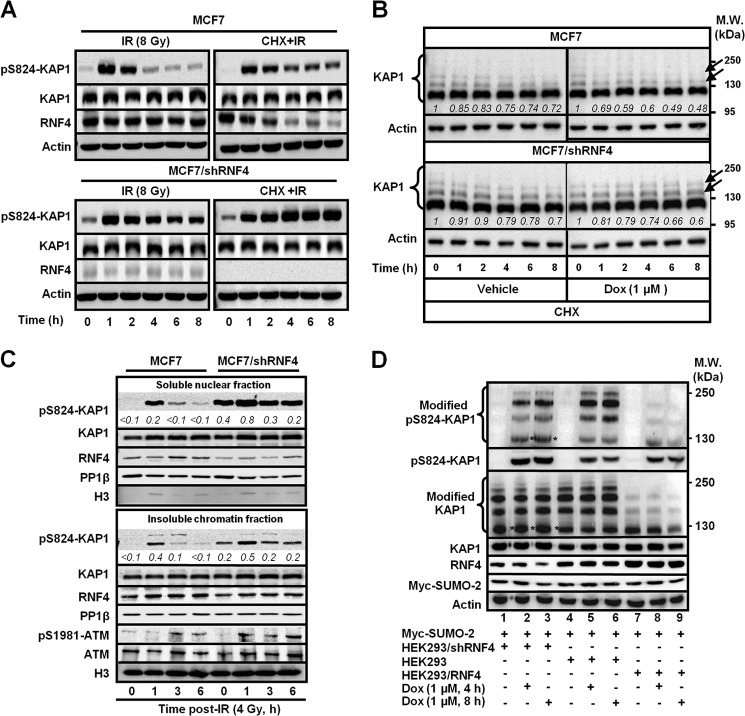
We have shown previously that KAP1 is SUMOylated and that genotoxic stress by IR or Dox induces a rapid and transient phosphorylation at Ser-824 of KAP1, which causes a reduction of SUMO-KAP1 (7, 8, 10). However, the mechanism to regulate the steady-state abundance of Ser(P)-824-KAP1 or Ser(P)-824-SUMO-KAP1 is still unclear. In this report, we aimed to explore the contribution of each KAP1 PTM, namely phosphorylation, SUMOylation, and ubiquitylation, to the temporal regulation of KAP1 turnover. RNF4 is the first known human STUbL to ubiquitylate SUMOylated proteins for degradation (34, 35), and SUMO-KAP1 was identified as a RNF4 substrate through a global proteomic survey (36). Therefore, we sought to examine whether RNF4 is responsible for regulating Ser(P)-824-KAP1 abundance during the course of genotoxic stress. To achieve this goal, MCF7 cells that expressed an shRNA targeting RNF4 were established (MCF7/shRNF4). Both MCF7 and MCF7/shRNF4 cells were exposed to IR, together with CHX, which inhibits protein biosynthesis, to determine the effect of RNF4 on the level of endogenous Ser(P)-824-KAP1. We found that IR induced a transient appearance of Ser(P)-824-KAP1 in the absence of CHX (Fig. 1A, top left panel) and that there was a correlation between decreased RNF4 and increased Ser(P)-824-KAP1 signals at later time points in the CHX-treated, IR-exposed MCF7 cells compared with those in the vehicle-treated, IR-exposed counterparts (Fig. 1A, top right panel). Knockdown of RNF4 stabilized the Ser(P)-824-KAP1 signal (Fig. 1A, bottom panels). Next, both MCF7 and MCF7/shRNF4 cells were treated with Dox in the presence or absence of CHX. Although the levels of unmodified KAP1 remained largely unaffected upon combined Dox and CHX treatment, Dox caused a time-dependent decrease of the endogenous high molecular weight KAP1 species (Fig. 1B, arrows, top right panel). Knockdown of RNF4 preserved these slowly migrating KAP1 species (Fig. 1B, arrows, bottom right panel).
FIGURE 1.
DNA damage induces an RNF4-dependent decrease of KAP1 abundance. A, RNF4 regulates the turnover of endogenous Ser(P)-824-KAP1 (pS824-KAP1) after IR exposure. MCF7 and MCF7/shRNF4 cells were exposed to IR (8 Gy) and allowed to recover for the indicated times. CHX (100 μg/ml) was used to inhibit protein biosynthesis. Equal amounts of total lysates were subjected to Western blot analyses with antibodies as indicated. B, Dox triggers a faster turnover of endogenous PTM-modified KAP1. Endogenous KAP1 in MCF7 and MCF7/shRNF4 cells was probed after treatment with vehicle or Dox (1 μm) in the presence of CHX (100 μg/ml) for the indicated times. Arrows indicate the slowly migrating KAP1 species. Italic numbers represent the levels of slowly migrating species of KAP1 relative to the levels of total KAP1. The respective slowly migrating species level in individual cells prior to each treatment was set as 1. MW, molecular weight. C, RNF4 regulates the chromatin-associated Ser(P)-824-KAP1 level. Nuclear extracts from MCF7 and MCF7/shRNF4 cells were fractionated and probed with the indicated antibodies. Italic numbers represent the normalized Ser(P)-824-KAP1 levels relative to total KAP1. H3, histone 3. D, differential levels of RNF4 correlate inversely with the level of endogenous KAP1. HEK293/shRNF4-, HEK293-, and HEK293/RNF4-overexpressing cells were transfected with Myc-SUMO-2 and then treated with Dox for the indicated times. Western blot analyses were performed as described in A. A–D, one representative Western blot analysis is shown (n = 3).
Next, nuclear extracts from MCF7 cells were fractionated into soluble nucleoplasm and insoluble chromatin-bound fractions, and Ser(P)-824-KAP1 abundance was assessed. Knockdown of RNF4 increased both the soluble and insoluble, chromatin-associated Ser(P)-824-KAP1 in irradiated MCF7 cells (Fig. 1C), suggesting that RNF4 targets KAP1 at both euchromatic and heterochromatic regions. In contrast, we did not observe significant changes of ATM, Ser(P)-1981-ATM (kinase for KAP1), or PP1β (phosphatase for KAP1), factors that could potentially affect Ser(P)-824-KAP1 levels. Lastly, we engineered HEK293 cells with distinct RNF4 levels and determined whether the effect of RNF4 on the endogenous levels of Ser-824-phosphorylated or SUMO-KAP1 is only limited to MCF7 cells. HEK293, HEK293-expressing shRNF4 (HEK293/shRNF4), and HEK293 cells overexpressing RNF4 (HEK293/RNF4) were treated with Dox. Because the steady-state abundance of SUMO-KAP1 species only represented <3% relative to the total KAP1 signals (Fig. 1B), we separated modified KAP1 from unmodified KAP1 for a longer exposure to enhance the visibility of modified KAP1 in Fig. 1D and the remaining figures. Lower abundances of higher molecular weight-modified Ser(P)-824-KAP1 and modified KAP1 species were observed in Dox-treated HEK293/RNF4 cells (Fig. 1D, first and third panels, lane 8 versus lane 5 and lane 9 versus lane 6), whereas lower molecular weight-modified Ser(P)-824-KAP1 and modified KAP1 species accumulated in Dox-treated, HEK293/shRNF4 cells (Fig. 1D, first and third panels, asterisks). On the basis of these observations, we rationalized that RNF4 regulates the steady-state abundance of simultaneously Ser-824-phsphorylated and SUMOylated KAP1 (Ser(P)-824-SUMO-KAP1) in response to DSBs.
KAP1 Ser-824 Phosphorylation Promotes RNF4-mediated Turnover
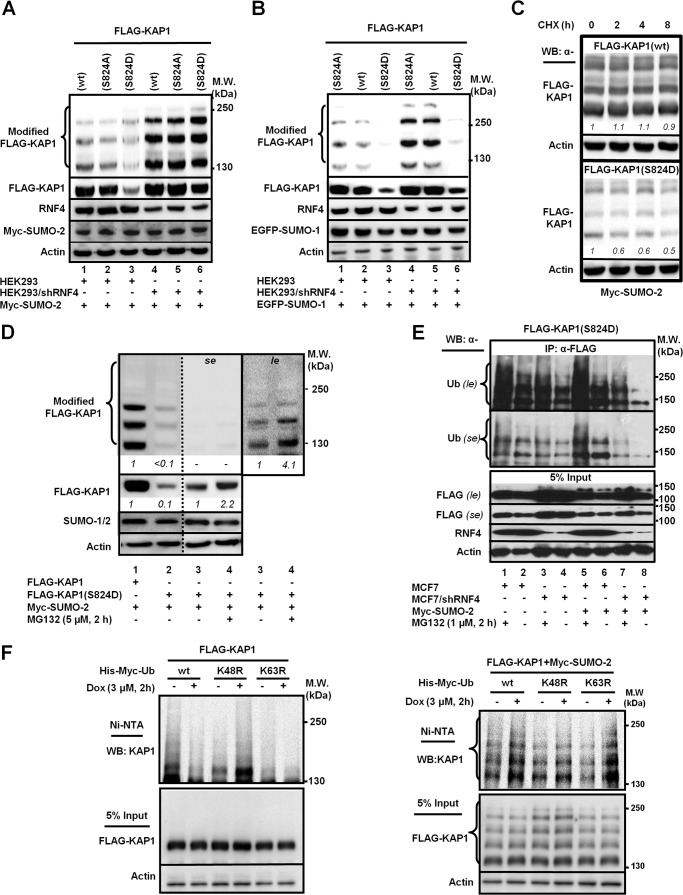
Next, to determine whether KAP1 phosphorylation at Ser-824 affects the turnover of SUMO-KAP1, we examined the turnover of a KAP1 mutant that was Ser-824 phosphorylation-defective (Ser-824 to alanine, S824A) and a KAP1 mutant that was Ser-824-phosphomimic (Ser-824 to aspartate, S824D) in the presence of Myc-SUMO-2 or EGFP-SUMO-1. Overexpression of Myc-SUMO-2 (Fig. 2A) or EGFP-SUMO-1 (Fig. 2B) alone markedly decreased the abundance of modified and unmodified KAP1(S824D). Consistently, knockdown of RNF4 caused the accumulation of polySUMO-KAP1(S824D) more over that of polySUMO-KAP1(WT or S824A) in cells transfected with Myc-SUMO-2 (Fig. 2A), suggesting that Ser-824-phosphorylation accelerates KAP1 turnover upon SUMOylation. Because the effect on the abundance of KAP1(S824D) was more noticeable in cells transfected with SUMO-2 than EGFP-SUMO-1, the remaining studies reported here were mainly performed with cotransfection of SUMO-2. In the presence of CHX, polySUMO-KAP1(WT) was more stable than polySUMO-KAP1(S824D) (Fig. 2C), suggesting that the mimicking phosphorylation at KAP1 Ser-824 destabilized KAP1. Treatment with the proteasome inhibitor MG132 over the last 2 h of incubation partially restored the levels of KAP1(S824D) and SUMO-2-KAP1(S824D) (Fig. 2D, lanes 3 and 4 in se and le), supporting that RNF4 targeted SUMO-2-modified KAP1(S824D) for degradation. Coimmunoprecipitation followed by Western blot analyses revealed that knocking down RNF4 reduced the level of ubiquitylated KAP1(S824D) (Fig. 2E, first and second panels, lane 8 versus lane 6) and that this effect was SUMO-2-dependent (Fig. 2E, first and second panels, lane 8 versus lane 4). In contrast, the addition of MG132 for the final 2 h increased the Ub-modified KAP1(S824D) species, consistent with the degradation of ubiquitylated KAP1 being proteasome-dependent (Fig. 2E, first and second panels, lane 1 versus lane 2 and lane 5 versus lane 6). Overexpression of Myc-SUMO-2 increased the Ub-KAP1(S824D) signal (Fig. 2E, first and second panels, lane 5 versus lane 1). Lastly, cell-based ubiquitylation assays showed that, in the absence of SUMO-2, only marginal ubiquitylation was detected on KAP1 (Fig. 2F, left panel). In contrast, when SUMO-2 was present, a marked increase of ubiquitylation of KAP1 was detected in the cells exposed to Dox (Fig. 2F, right panel), suggesting that KAP1 underwent DNA damage-induced, SUMO-dependent ubiquitylation. In addition, DNA damage-induced, SUMO-dependent ubiquitylation was attenuated only when Lys-48, but not Lys-63 of Ub was mutated (Fig. 2F, right panel, fourth lane). These results suggest that KAP1 is subjected to SUMO-dependent polyubiquitylation through Lys-48 linkage, leading to proteasomal degradation in response to DNA damage.
FIGURE 2.
The KAP1 phosphomimic S824D mutation promotes SUMO-2-dependent KAP1 ubiquitylation and turnover. A and B, phosphomimic S824D promotes KAP1 degradation in the presence of Myc-SUMO-2. HEK293 and HEK293/shRNF4 cells were transfected with FLAG-KAP1(WT, S824A, or S824D) and Myc-SUMO-2 (A) or EGFP-SUMO-1 (B). RNF4 knockdown increased the steady-state levels of KAP1. MW, molecular weight. C, the turnover of SUMO-2-modified KAP1(S824D) was more rapid than that of KAP1(WT). HEK293 cells were transfected with FLAG-KAP1(WT or S824D) and treated with CHX (100 μg/ml) for the indicated times. WB, Western blot. D, MG132 increases the levels of unmodified and SUMO-2-modified KAP1-S824D. Transfected HEK293 cells were treated with MG132 for 2 h prior to cell harvesting. The relative levels (italic numbers) of SUMO-modified and SUMO-unmodified KAP1 are shown after normalization to actin. The respective levels of SUMO-modified and SUMO-unmodified FLAG-KAP1 in lanes 1 and 3 were set to 1. se, short exposure; le, longer exposure. E, RNF4 enhances the phospho-SUMO-dependent ubiquitylation of KAP1. MCF7 and MCF7/shRNF4 cells were transfected with FLAG-KAP1(S824D) and Myc-SUMO-2 and then treated with MG132. FLAG-KAP1(S824D) was immunoprecipitated (IP) by an anti-FLAG antibody and blotted with an anti-Ub antibody. Input controls were detected with appropriate antibodies. F, DSBs induce KAP1 SUMO-dependent, Lys-48-linked-polyubiquitylation. Transfected HEK293 cells were pretreated with MG132, and Dox was added 2 h prior to cell lysis. His-tagged ubiquitylated proteins were pulled down using nickel-nitrilotriacetic acid (NTA)-agarose beads. Ubiquitylated KAP1 was detected by anti-KAP1 antibody. Input controls (5%) were blotted with the appropriate antibodies. A—F, one representative Western blot is shown (n = 3).
SUMOylation of KAP1 at Lys-676 is critical for Ser-824 Phosphorylation to Promote KAP1 Ubiquitylation and Degradation
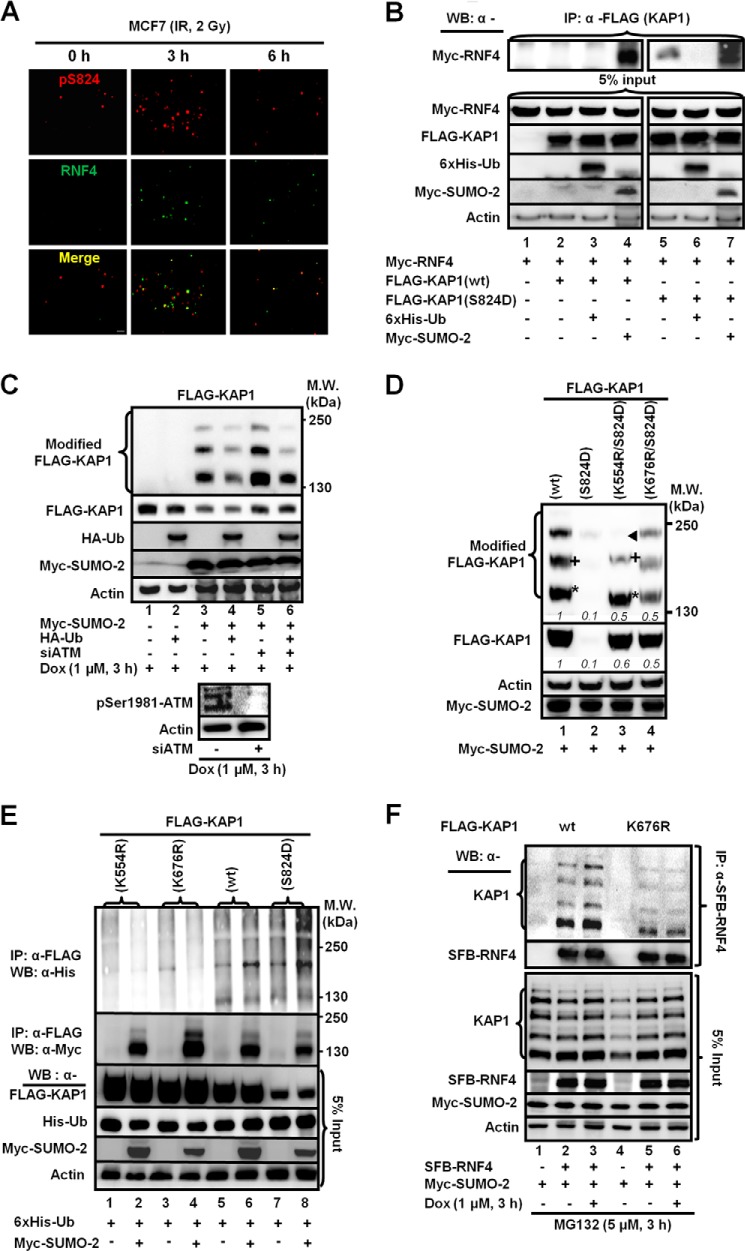
One possible mechanism accounting for RNF4 to reduce Ser(P)-824-SUMO-KAP1 abundance is that the DSB-induced KAP1 Ser-824 phosphorylation enhances the interaction of KAP1 with RNF4. To characterize the spatiotemporal recruitment of RNF4 to KAP1, we collected images that localized the Ser(P)-824 signal and RNF4 in the cell prior to and after IR. We observed a partial colocalization of the RNF4 foci with Ser(P)-824 foci and then an increase (3 h) followed by a decrease (6 h), in the number of RNF4 and Ser(P)-824 foci in MCF7 cells post-IR (Fig. 3A). Next, coimmunoprecipitation assays of whole cell extracts from HEK293 cells that were cotransfected with RNF4 and KAP1(WT) or KAP1(S824D) showed that KAP1(S824D) pulled down substantially more RNF4 than KAP1(WT) (Fig. 3B, lane 5 versus lane 1). Furthermore, SUMO-2 stimulated the interaction of RNF4 with both KAP1(WT) and KAP1(S824D) (Fig. 3B, lanes 4 and 7). To ascertain the role of Ser-824 phosphorylation in promoting the turnover of SUMO-KAP1 in response to DSBs, we knocked down ATM using siRNA (Fig. 3C, bottom panels). The level of modified KAP1 was then assessed in the presence or absence of exogenous Ub (Fig. 3C, first panel). Overexpression of Ub reduced the levels of modified KAP1, presumably SUMO-modified (Fig. 3C, first and second panels, lanes 4 and 6). But in Dox-treated cells that had the ATM kinase knocked down, both modified and unmodified KAP1 accumulated (Fig. 3C, first and second panels, lanes 5 and 6).
FIGURE 3.
The interaction between KAP1 and RNF4 requires phosphorylation of KAP1 Ser-824 and SUMOylation of KAP1 Lys-676. A, Ser(P)-824-KAP1 (pS824) and RNF4 colocalize partially in MCF7 cells. MCF7 cells were fixed at the indicated times post-IR. Immunofluorescent staining is shown using antibodies against Ser(P)-824-KAP1 (red) and RNF4 (green). Scale bar = 1 μm. B, the S824D phosphomimic facilitates KAP1-RNF4 interaction. Shown are immunoprecipitates (IP) from HEK293 cells transfected with FLAG-KAP1(WT or S824D), Myc-RNF4, His6-Ub, and Myc-SUMO-2 using an anti-FLAG antibody. WB, Western blot. C, knocking down ATM slows the turnover of KAP1. HEK293 cells were cotransfected with FLAG-KAP1(WT), His6-Ub, and Myc-SUMO-2, followed by a second transfection with siATM and treatment with Dox 2 h prior to cell lysis. MW, molecular weight. D, Lys-554 and Lys-676 are critical for SUMO-2-dependent turnover of KAP1(S824D). HEK293 cells were transfected with the indicated constructs and analyzed on Western blots. The asterisk, +, and ◀ indicate SUMO-KAP1 species with different migration speeds or abundances, as discussed in the text. The relative protein levels shown in italic numbers were normalized to actin, and the relative levels in cells cotransfected with FLAG-KAP1(WT) were set to 1. E, Ser-824-phosphorylation and Lys-554/Lys-676 SUMOylation promote KAP1 ubiquitylation. HEK293 cells were transfected with the indicated constructs. Anti-FLAG immunoprecipitation was performed and analyzed on Western blots using an anti-His (Ub) or anti-Myc (SUMO-2) antibody. F, KAP1 Lys-676 SUMOylation is required for the interaction of KAP1 with RNF4. HEK293 cells were cotransfected with SFB-RNF4, FLAG-KAP1(WT), or FLAG-KAP(K676R) and Myc-SUMO-2 and then treated with Dox in the presence of MG132. S beads were used to pull down SFB-RNF4. B–F, one representative Western blot is shown (n = 3).
Because KAP1 contains multiple SUMOylation sites (9, 10), we wished to determine the SUMOylation of which lysine residue(s) was required for the RNF4-KAP1 binding that occurs in response to DNA damage. We tested SUMOylation mutants that had Lys-to-Arg substitutions at all the potential SUMOylation sites in KAP1(WT) and KAP1(S824D). Among all mutants examined, we found that only the K554R and K676R substitution increased the levels of KAP1(S824D) in the presence of Myc-SUMO-2 (Fig. 3D). Notably, the K554R substitution rendered modified KAP1(S824D) species with different migration patterns compared with KAP1(WT) and KAP1(K676R/S824D) (Fig. 3D, asterisk, +, and ◀), whereas the K676R mutation mainly affected the respective abundance. Next, HEK293 cells cotransfected with FLAG-KAP1(WT, S824D, K554R, or K676R), His6-Ub, and/or Myc-SUMO-2 were subjected to immunoprecipitation to determine the effect of the K554R and K676R substitutions on the ubiquitylation of KAP1. Less ubiquitylated KAP1 was present in precipitates from KAP1(K554R)- and KAP1(K676R)-expressing cells compared with those of KAP1(WT) and KAP1(S824D) (Fig. 3E, first panel). The levels of ubiquitylated KAP1(K554R, K676R, WT, and S824D) were correlated inversely with the corresponding level of unmodified KAP1. However, SUMO-KAP1(K676R) bound less efficiently to RNF4 compared with SUMO-KAP1(WT), which has an enhanced interaction with RNF4 in Dox-treated cells (Fig. 3F). Collectively, we postulated that the K676R substitution impaired the SUMOylation that is responsible for RNF4 to ubiquitylate KAP1.
Molecular Modeling of RNF4 and Ser(P)-824·SUMO·KAP1 Complex
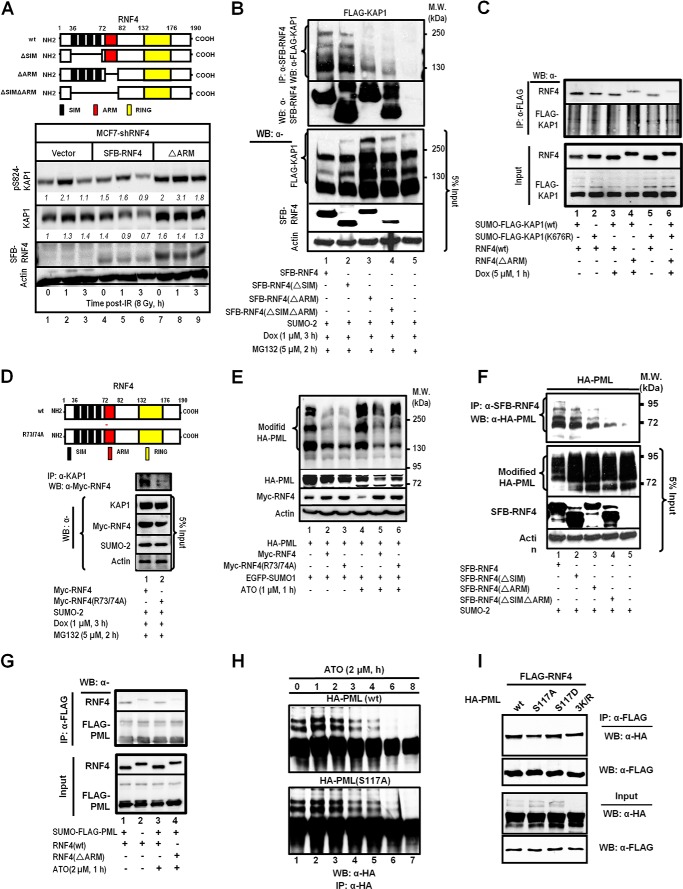
RNF4 has four tandem SIMs (Fig. 4A), which preferentially recognize and interact with poly-SUMO chains on its target proteins (35). We used coimmunoprecipitation assays to determine whether endogenous KAP1 formed complexes with RNF4(WT), ligase-dead RNF4 (RNF4cs) (20, 34, 35), or RNF4 that has the SIM2 and SIM3 deleted (aa residues 41–63, Δ41–63; Fig. 4B, top panel; Ref.35). Although endogenous poly-SUMO-KAP1 species were readily detected in the anti-RNF4(WT) (Fig. 4B, bottom panel, lanes 2 and 3), these modified species were more abundant in the anti-RNF4cs immunoprecipitates (Fig. 4B, bottom panel, lanes 5 and 6). Notably, the amount of modified KAP1 species pulled down by RNF4 decreased in Dox-treated cells (Fig. 4B, bottom panel, lane 3 versus lane 2 and lane 6 versus lane 5). The deletion of aa residues 41–63 of RNF4 reduced the amount of poly-SUMO-KAP1 pulled down (Fig. 4B, bottom panel (se), lanes 8 and 9). However, a longer exposure revealed that RNF4(Δ41–63) was still able to bind to oligo-SUMO-2-KAP1 but to a much lesser extent (Fig. 4B, bottom panel (le), lanes 8 and 9, arrows). Therefore, RNF4 could still interact with oligo-SUMO-KAP1, but the interaction was less efficient. As expected and similar to RNF4cs, overexpressing RNF4(Δ41–63) increased the SUMO-KAP1 level (Fig. 4C), consistent with the notion that functional RNF4 decreases the SUMO-KAP1 abundance.
FIGURE 4.
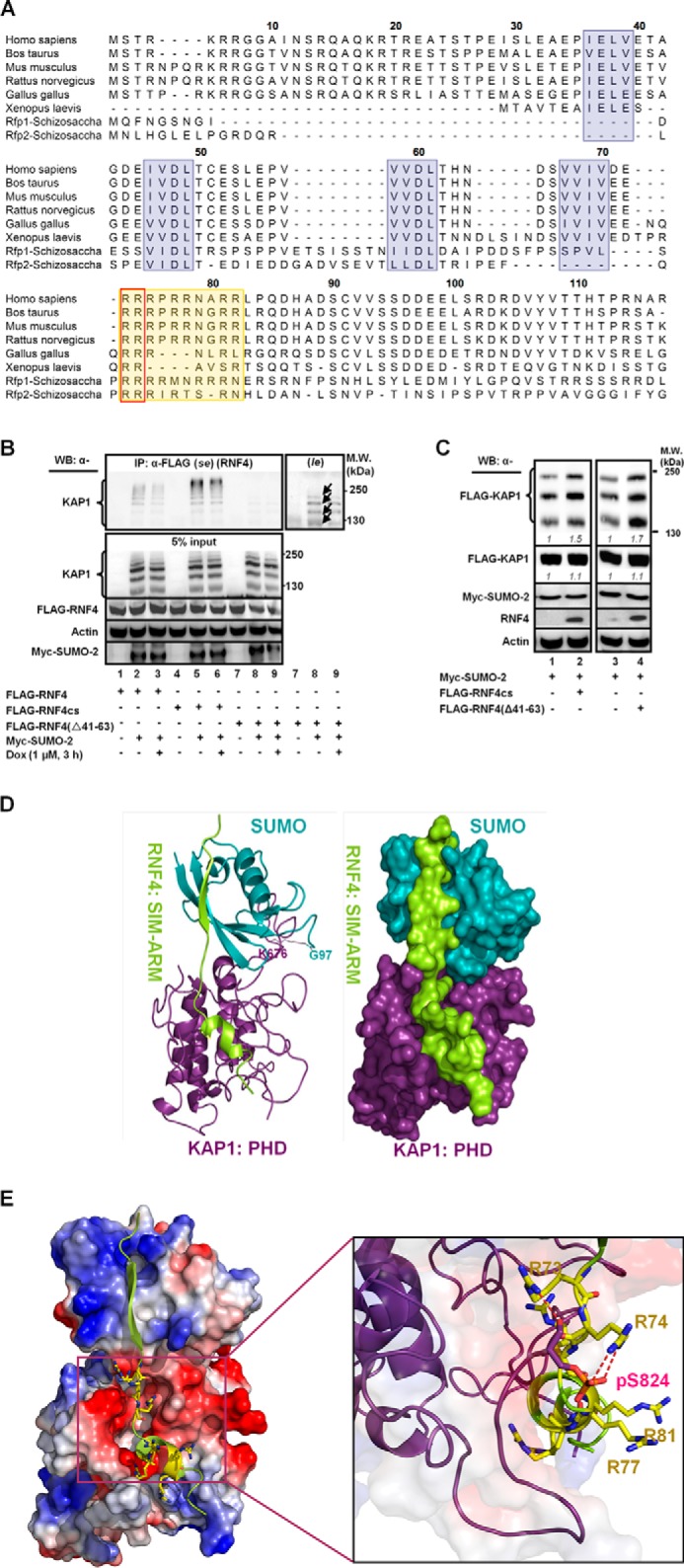
The ARM and SIMs are important motifs for RNF4 to selectively target its substrates. A, the conserved SIM and ARM domains are shaded. SIMs are blue, the ARM is orange, and Arg-73/74 are boxed. B, HEK293 cells were transfected with Myc-SUMO-2 and FLAG-RNF4(WT, cs, or Δ41–63) and treated with Dox. Proteins were immunoprecipitated (IP) with an anti-FLAG antibody to analyze the FLAG-RNF4-associated complex. Endogenous KAP1 and levels of input control were visualized using the appropriate antibodies. se, short exposure; le, long exposure. WB, Western blot; MW, molecular weight. C, overexpressing the ligase-dead RNF4 (RNF4cs) or RNF4 with the SIMs deleted (RNF4(Δ41–63)) causes the accumulation of SUMO-KAP1. HEK293 cells were transfected with FLAG-KAP1, Myc-SUMO-2, and FLAG-RNF4cs or RNF4(Δ41–63) and then treated with Dox for 3 h. The relative levels of SUMO-KAP1 and FLAG-KAP1, after they were normalized to actin, are shown in italics. The relative levels in the control cells cotransfected with vector were set to 1. D, ribbon structure of the complex formed by the interaction of SUMO-KAP1 with the fourth SIM and the ARM of RNF4. SUMO is colored cyan, KAP1 is purple, and RNF4 is yellow (left panel). The isopeptide bond formed between the Gly-97 of SUMO and Lys-676 of KAP1 is shown in line. The representative surface of the complex is shown in the right panel using the same color schemes as in the left panel. E, a molecular docking model for the RNF4·Ser(P)-824·SUMO·KAP1 complex. Shown is the positioning of the SIM and the ARM on the electrostatic surface of Ser(P)-824-SUMO-KAP1 (red, negatively charged; blue, positively charged). The residues in the ARM of RNF4 are represented by sticks (left panel). The magnified region shows the interaction of Ser(P)-824 of KAP1 with the arginines in the ARM of RNF4. The interacting residues are represented by sticks. Negatively charged groups are red, and positively charged groups are blue. Hydrogen bonds are indicated by dotted red lines (right panel). B and C, one representative Western blot is shown (n = 3).
We noticed that the sequence (aa residues 73–82, designated as ARM) immediately following the fourth SIM is rich in Arg and that this ARM is conserved throughout evolution (Fig. 4A). The internal deletion of the ARM (Fig. 4B, top panel) impaired the interaction of SUMO-KAP1 with RNF4 (Fig. 4D, lane 3). Together with the results shown in Figs. 2, A–D, 3F, and 4B, we rationalized that ARM synergizes with the SIMs to recruit SUMO-KAP1 upon Ser-824 phosphorylation. To examine this possibility, we modeled the interaction between the fourth SIM and ARM of RNF4 and Ser(P)-824-K676-SUMO-KAP1 (Fig. 4D). Stepwise, we first used the RNF4 aa sequence of the fourth SIM motif and its adjacent ARM to search the Protein Data Bank (PDB), but no highly homologous sequences were identified. Then, on the basis of the known crystal structures of SUMO-1·SIM complexes (PDB codes 1Z5S, 2ASQ, and 1WYW), we predicted that the RNF4 fourth SIM and its adjacent aa residues 71–83 (Fig. 4A), analogous to RanGAP1, tend to form a common “sheet-loop-helix” structure when bound to their partners. In the sheet-loop-helix structure, the SIM interacts with SUMO by adopting an anti-parallel β sheet conformation, whereas aa residues 71–83 fold into a short α helix that is linked to the SIM by a flexible loop. We surmised that the interaction of SUMO (KAP1) with SIM-ARM (RNF4) could be modeled on the basis of the specific structure of the SUMO-SIM complex with relatively high accuracy. Among three SUMO-1·SIM complexes that we identified (PDB codes 1Z5S, 1WYW, and 2ASQ), the PDB 1Z5S complex contains NUP358 (an E3 ligase), has longer SIM flanking sequences, and is more functionally relevant to RNF4 than UBC9 (an E2 ligase) and SUMO-1-conjugated RanGAP1. As such, the SUMO-1·Nup358 complex was chosen as the template to model the SUMO·SIM-ARM of the SUMO-conjugated KAP1-RNF4 complex.
After modeling the SUMO·(RNF4)SIM-ARM complex, we connected SUMO to Lys-676 of KAP1 (PDB code 2RO1) to establish (KAP1)PHD-SUMO-Lys-676. ZDOCK (37) was used to predict the optimal interactions between the modeled (KAP1)PHD·SUMO·Lys-676 (designated as the receptor) and (RNF4)SIM-ARM complex (designated as the ligand). The top 2000 docked conformations, on the basis of the ZRANK, were further filtered by the criterion that Lys-676 was located at the interface between SUMO and KAP1. Then, the “Loop Search” module implemented in the SYBYL 6.8 package (TRIPOS) was used to model a loop corresponding to a linear aa sequence from Gln-94 to Gly-97 of SUMO that formed the isopeptide bond between the carboxyl group of the SUMO terminal Gly-97 and the side chain amino group of Lys-676 of KAP1. Likewise, the Loop Search strategy was used to build the missing loop containing the KAP1 Pro-813 to Ser-824 residues, which are missing in the KAP1 PHD NMR structure (PDB code 2RO1). KAP1 Ser-824 was modeled to approach the ARM of RNF4. Finally, the complete model of SUMO-conjugated and phosphorylated KAP1 complexed with the fourth SIM and its adjacent ARM of RNF4 was obtained by adding the phosphate group to the side chain hydroxyl of Ser-824 and making structural refinements. In the modeled structure, the phosphorylated Ser-824 of KAP1 was constructed and located close to the ARM of RNF4. Subsequently, the resulting structure was minimized using the AMBER force field in the SYBYL package to remove possible steric hindrances and to obtain a lower energy conformation. Lastly, the 1-ns molecular dynamics simulation was run using the AMBER package (version 10.0) to sample and identify the most stable conformation.
A model consisting of KAP1 SUMO-modified at Lys-676 and phosphorylated Ser-824, interacting with the fourth SIM and the adjacent ARM of RNF4, is shown in Fig. 4D. This model further predicts that the SUMO group of Ser(P)-824-K676-SUMO-KAP1 and the fourth SIM of RNF4 form a conserved β-β antiparallel interaction (38, 39) and that the fourth SIM extends along the groove between the PHD region and C-terminal tail of KAP1. Analyses on the electrostatic surface of the predicted complex revealed that the negatively charged KAP1 surface was surrounded by the positively charged RNF4 ARM, suggesting electrostatic interactions (Fig. 4E, left panel). KAP1 Ser-824 phosphorylation presumably increases the negative charge of this binding surface. Arg-73 and Arg-74 in RNF4, predictably, formed hydrogen bonds with Ser(P)-824 of KAP1 (Fig. 4E, right panel). Two hydrogen bonds formed between Arg-74 and Ser(P)-824 through the positively charged guanido-nitrogen atoms of Arg-74 and the negatively charged oxygen atoms of the phosphate group of Ser(P)-824. The distance of the two hydrogen bonds is 2.8 and 2.9 Å (Fig. 4E, dotted red lines), respectively. Moreover, the positively charged guanido-nitrogen atom of Arg-73 and the main chain carboxyl oxygen atom of Ser(P)-824 formed another strong hydrogen bond within a 2.9 Å-distance (Fig. 4E, dotted red line).
The ARM of RNF4 Promotes the Interaction of RNF4 with KAP1 and PML
Two sets of experiments were performed to test this model. First, to ascertain the role of the ARM in modulating the levels of Ser(P)-824-KAP1, MCF7/shRNF4 cells were complemented with a control vector or cDNAs that encode an RNF4 (SFB-RNF4) or an RNF4 lacking the ARM domain (SFB-RNF4(ΔARM)) (Fig. 5A, top panel). Clearly, the deletion of the ARM from RNF4 increased the levels of Ser(P)-824-KAP1 prior to and post-IR (Fig. 5A, bottom panel, lanes 7-9). Next, overexpression of the RNF4 with internal deletion of the ARM or SIM (Fig. 5A, top panel) increased the abundance of basal modified KAP1 (Fig. 5B, bottom panel, lanes 2–4). Moreover, the internal deletion of the ARM or both the SIM and the ARM noticeably weakened the binding with phospho-SUMO-KAP1 (Fig. 5B, top panel, lanes 3 and 4). In vitro interaction studies between KAP1 and RNF4 also showed decreased binding when KAP1 Lys-676 was mutated and the RNF4 ARM was deleted (Fig. 5C). Lastly, substituting Ala for Arg-73/74 (Fig. 5D, top panel) decreased the level of RNF4 in the anti-KAP1 immunoprecipitates (Fig. 5D, bottom panel, lane 2). Together, these observations support the role of the ARM in promoting the interaction of RNF4 with KAP1 and subsequent degradation in Dox-treated cells. Next, this observation was expanded further to hypothesize that the ARM also promotes the RNF4-mediated degradation of Ser-117-phosphorylated SUMO-PML, which is induced by ATO. ATO increased PML SUMOylation (Fig. 5E, lane 4 versus lane 1). It has been reported that RNF4 accelerates the destabilization of PML in cells treated with ATO (40). Although that overexpression of RNF4 decreased the abundance of both unmodified and modified PML (Fig. 5F, lanes 2 and 5), R73A/R74A substitutions partially prevented RNF4 from reducing SUMO-PML and unmodified PML in ATO-treated cells (Fig. 5E, lane 6 versus lane 5). Likewise, we observed that the deletion of the ARM impaired the ability of RNF4 to bind SUMO-PML and that overexpression of RNF4(ΔARM) resulted in increased levels of SUMO-PML in the immunoprecipitates (Fig. 5F). Deletion of the ARM also decreased the in vitro binding between RNF4 and SUMO-PML (Fig. 5G). Moreover, ATO/CHK2 reportedly phosphorylates PML at Ser-117 (41), and a PML S117A substitution reduced the amount of PML degradation induced by ATO (Fig. 5H, bottom panel). As expected, PML(S117D) showed an enhanced interaction with RNF4. The S117A substitution, similar to 3K/R (K65R, K160R, K490R) substitutions, decreased PML binding to RNF4 but without affecting its SUMOylation as 3K/R (Fig. 5I, first and third panels). Together, these results indicate that the aa 73–82 domain of RNF4 is a binding motif that recognizes both phospho-KAP1 and phospho-PML.
FIGURE 5.
The interaction of the ARM of RNF4 with Ser(P)-824-KAP1 or Ser(P)-117-PML plays critical roles in target regulation. A, schematics of RNF4 and its mutants showing the deletion of the fourth SIM or the ARM (top panel). Deleting the ARM prevents the degradation of Ser(P)-824-KAP1 protein (bottom panel). MCF7/shRNF4 cells were transfected with an empty vector, shRNA resistant-RNF4-WT, or shRNA-resistant RNF4(ΔARM), irradiated (8 Gy), and allowed to recover for the indicated times. Endogenous Ser(P)-824-KAP1 and KAP1 levels were detected on Western blot analyses of whole cell lysates. Italic numbers indicate the relative quantity of Ser(P)-824-KAP1 and KAP1 after normalization to actin. Untreated MCF7/shRNF4 cells were set to 1. RING, really interesting new gene. B, the ARM is critical for the interaction between RNF4 and KAP1 that is induced by Dox. HEK293 cells were transfected as indicated and treated with Dox and MG132. SFB-RNF4 was pulled down using S beads, and KAP1 was detected on Western blot (WB) analyses using an anti-FLAG antibody. MW, molecular weight; IP, immunoprecipitation. C, in vitro binding between SUMO-KAP1 and RNF4. SUMO-KAP1 pulled down from HEK293 cells transfected with FLAG-KAP1(WT or K676R) and Myc-SUMO-2 and treated with Dox (5 μm, 1 h) was incubated with in vitro-translated RNF4, and the interaction was detected. D, R73A/R74A substitutions in the ARM of RNF4 reduce the efficiency of RNF4 to interact with KAP1 in cells treated with Dox. E, R73/74A substitutions in the ARM of RNF4 reduce the ability of RNF4 to down-regulate the levels of PML in ATO-treated cells. F, the ARM is also important for the targeting of PML by RNF4. HEK293 cells were transfected as indicated, and SFB-RNF4 was pulled down using S beads. PML was detected using an anti-HA antibody. G, in vitro binding between SUMO-PML and RNF4. SUMO-PML pulled down from HEK293 cells transfected with FLAG-PML and Myc-SUMO-2 and treated with ATO (2 μm, 1h) was incubated with in vitro-translated RNF4, and the interaction was detected. H, the PML S117A mutation reduces PML degradation induced by ATO. HA-PML(WT or S117A) was transfected in HEK293 cells, and then the cells were exposed to ATO for the indicated time periods. PML was immunoprecipitated and immunoblotted using an anti-HA antibody. I, the PML phosphomimic S117D enhances the interaction of RNF4 with PML, whereas the S117A substitution decreases RNF4 binding to PML. HEK293 cells were cotransfected with FLAG-RNF4 and HA-PML(WT, S117A, S117D, or 3K/R). Proteins were immunoprecipitated using anti-FLAG antibody to FLAG-RNF4, and then precipitates were blotted with anti-HA antibody to detect HA-PML. 3K/R are the K65R, K160R, and K490R substitutions of PML that prevent PML SUMOylation. A–I, one representative Western blot is shown (n = 3).
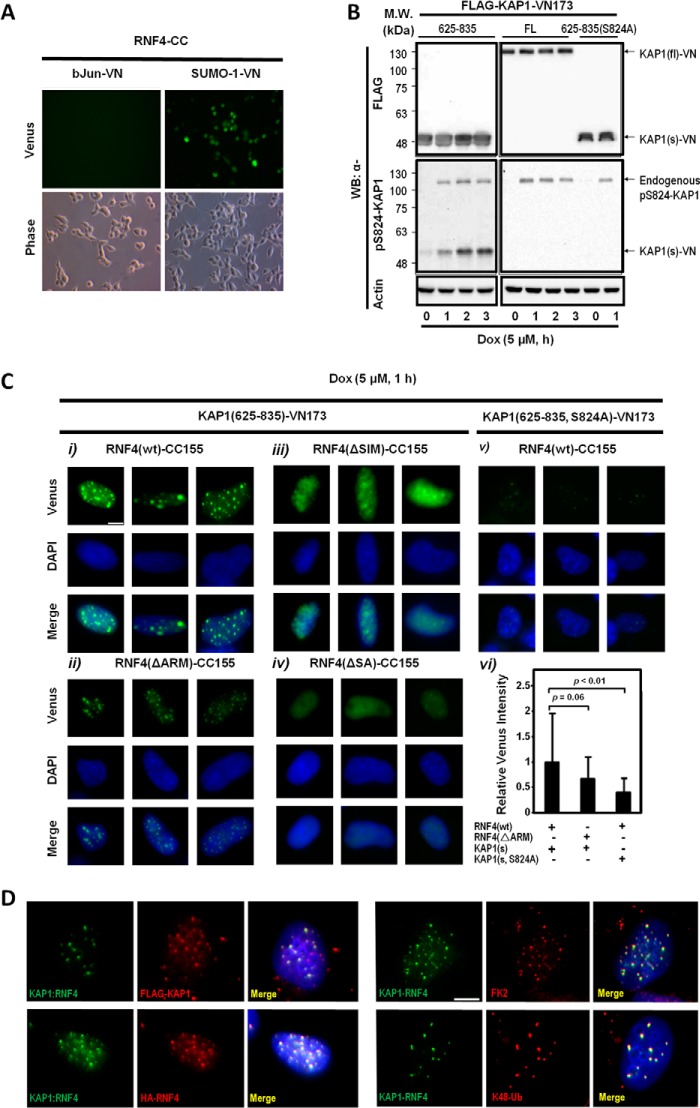
Validation of the ARM-promoted RNF4-KAP1 Interaction in Living Cells
The second set of experiments was to independently validate the role of the ARM in the recruitment of RNF4 to Ser-824-phosphorylated KAP1 in living cells by using BiFC assays to visualize the protein-protein interactions (42). To achieve this goal, the full-length (fl) or the PHD-bromodomains (aa residues 625–835 [s, short form]) of KAP1 were engineered into a BiFC construct that harbors the N-terminal fragment of venus (KAP1-VN173). RNF4(WT) and various RNF4(ΔSIM, ΔARM, ΔSA) mutants were cloned into another pBiFC that encodes the C-terminal fragment of enhanced cyan fluorescent protein (ECFP) (RNF4-CC155). Green fluorescence was detected in cells cotransfected with RNF4-CC155 and SUMO-1-VN173 but not bJun-VN, serving as control (Fig. 6A). Western blots analyses revealed, unexpectedly, that KAP1(fl)-VN173 could not be phosphorylated at Ser-824 upon Dox treatment (Fig. 6B, center right panel). The lack of phosphorylation was likely due to the steric hindrance caused by the attached VN173, preventing ATM from accessing the Ser-824 residue. The capacity of KAP1(s)-VN173 to be phosphorylated by Dox treatment (Fig. 6B, center left panel) and SUMOylated by SUMO-2 was not affected (data not shown). As such, we used KAP1(s)-VN173 and its corresponding phosphorylation-defective mutant, KAP1(s, S824A)-VN173, for following studies.
FIGURE 6.
BiFC assays validate Ser-824 phosphorylation-dependent interaction in living cells. A, interaction of RNF4-CC155 with SUMO-1-VN173. HeLa cells were transfected with the indicated constructs. Venus (green fluorescence) was observed only in cells transfected with SUMO-1-VN173 and RNF4-CC155, but not bJun-VN173, which served as a negative control. B, KAP1(fl)-VN173 cannot be phosphorylated. The fl or short form (s, 625–835) of KAP1 was transfected into HeLa cells. Cells were exposed to Dox (5 μm) for the indicated time periods prior to harvesting, and Ser-824-phosphorylated KAP1 (pS824-KAP1) was visualized using an anti-Ser(P)-824-KAP1 antibody. MW, molecular weight; WB, Western blot. C, BiFC assays reveal that RNF4-CC155 interacts with KAP1(s)-VN-173 in living cells. HeLa cells were first cotransfected with KAP1(s: aa 625–835, WT, or S824A)-VN173 and RNF4(WT)-CC155 or mutants, and then the cells were treated with MG132 and Dox for 1 h at 48 h after transfection. Cells were then pre-extracted, fixed, and examined under a fluorescence microscope (i–v) (green (venus), RNF4-CC155:KAP1(s)-VN173 interaction foci; blue (DAPI), nucleus). Nuclei (n = 1000) were analyzed for the intensity of venus, and the bar graph is shown in vi. Error bars show mean ± S.D. Scale bar = 5 μm. D, Ub colocalizes with interaction foci of RNF4(WT)-CC155 and KAP1(s)-VN173. Left panel, U2OS cells were transfected with KAP1(s)-VN173 and RNF4(WT)-CC155, and then treated with MG132 (10 μm) and exposed to IR (4 Gy, 1 h) prior to imaging analyses. Cells were pre-extracted, fixed, and stained with the indicated antibodies. Venus, RNF4(WT)-CC155:KAP1(s)-VN173 interaction foci; red: indicated protein visualized by the antibody against the tag; DAPI, nucleus. Right panel, BiFC assays were performed in U2OS cells as described in C. Cells were exposed to IR (4 Gy, 1 h), pre-extracted, fixed, and immunostained with the indicated antibodies. Green (venus), RNF4-CC155:KAP1(s)-VN173 interaction foci; red, the indicated antibody; blue (DAPI, nucleus). Scale bar = 5 μm.
In Dox-treated, pre-extracted nuclei, we observed that the green fluorescence, resulting from the interaction of RNF4(WT)-CC155 with KAP1(s)-VN173, formed focus-like structures (Fig. 6C, i). On average, 10–30 RNF4:KAP1 foci were observed in each nucleus. The specificity of these RNF4-KAP1 foci was, at least in part, confirmed by the colocalization analyses with either FLAG-KAP1(s)-VN173 or HA-RNF4(WT)-CC155 (Fig. 6D, left panel). By comparing the interaction between KAP1(s)-VN173 and RNF4(WT)-CC155 and its various mutants, we observed that RNF4(ΔARM)-CC155 still formed interaction foci with KAP1(s)-VN173 in the nucleus, albeit with lower green fluorescence intensity (Fig. 6C, ii). The observation of RNF4(ΔARM)-CC155-KAP1(s)-VN173 interaction foci in the nucleus also suggested that the internal deletion of ARM did not affect RNF4 nuclear localization. Notably, nuclei with RNF4(ΔSIM)-CC155 had less discrete interacting foci in a diffused background (Fig. 6C, iii). As expected, the RNF4(ΔSA)-CC155 mutant showed no interaction with KAP1(s)-VN173 (Fig. 6C, iv). Moreover, the S824A substitution clearly dampened the fluorescence intensity of the RNF4(WT)-CC155:KAP1(s)-VN173 interaction foci (Fig. 6C, v). Together, these observations suggest that, although these SIMs are a crucial motif for RNF4 to recognize SUMO-KAP1, the ARM further enhances their interaction through phosphorylation at Ser-824 of KAP1 (Fig. 6C, vi). Lastly, we observed that Lys-48-linked Ub colocalized with RNF4(WT)-CC155:KAP1(s)-VN173 interaction foci (Fig. 6D, bottom right panel). In contrast, a partial colocalization of total Ub (stained by FK2; Fig. 6D, top right panel) with the interaction foci was noted. This is consistent with observations that the Lys-48, but not Lys-63, linkage of Ub attached to KAP1 was initiated and extended by RNF4 (Fig. 2F).
DISCUSSION
In this study, we established unambiguously that KAP1 is a target of RNF4 and that DSB-induced Ser-824 phosphorylation promotes the simultaneous recruitment of Ser-824-phosphorylated and SUMOylated KAP1 to RNF4. It is noteworthy that KAP1 has been identified as a putative RNF4 substrate in a previous proteomic study (36). Given that KAP1 is readily SUMOylated at multiple Lys residues close to the C terminus (10), we propose that ATM-mediated Ser-824-phosphorylation plays a key role in its RNF4-mediated dynamic turnover in the context of genotoxic stress. To our knowledge, the findings presented here are the first to indicate that a previously uncharacterized ARM, together with the adjacent fourth SIM of RNF4, accelerates the simultaneous turnover of Ser-824-phosphorylated, Lys-676-SUMOylated KAP1, presumably in part via Lys-48-linked ubiquitylation and proteasome-dependent degradation.
Recent studies have indicated an important role for RNF4 or RNF4-like proteins in several cellular functions, including ATO-induced degradation of SUMOylated promyelocytic leukemia protein (PML) (31, 34, 35), phorbol 12-myristate 13-acetate (PMA)/MAPK-stimulated degradation of SUMOylated polyomavirus enhancer activator 3 (PEA3) (43), heat shock-triggered degradation of SUMOylated poly(ADP-ribose) polymerase 1 (PARP-1) (44), hypoxia-activated degradation of SUMOylated hypoxia-inducible factor 2α (HIF-2α) (45), and degradation of SUMOylated Rta viral transcription factor during the lytic cycle of EBV (46). Moreover, Kaposi sarcoma-associated herpesvirus K-Rta resembles RNF4 to degrade SUMOylated PML nuclear bodies against host immune response and to facilitate viral reactivation (47). Here we report that RNF4 is the STUbL for KAP1 in the context of DNA damage. Notably, the RNF4-targeted degradation of SUMOylated proteins, instead of altering protein-protein interaction via deSUMOylation, suggests that both SUMO conjugates and their unconjugated forms require to be entirely dislodged. The exact roles for individual SUMOylation for many DDR proteins, including KAP1, and their RNF4-mediated degradation during DDR are still largely unclear.
So far, more than 300 targets for mammalian RNF4 have been identified (36). However, the process for the spatial and temporal selection of specific substrates by RNF4, and STUbLs in general, is not well understood. Our data indicate that phosphorylation at Ser-824 of KAP1 and Ser-117 of PML promotes their respective association with RNF4 in the context of stress conditions. Several lines of evidence indicate that the interaction by the SIMs of RNF4 alone, although sufficient, was not able to provide maximal binding of RNF4 with KAP1 (Figs. 4B and 5B). This is not surprising because of the typical low affinity for SIM-SUMO interactions (38, 39). The ARM-Ser(P)-824 or ARM-Ser(P)-117 recognition could potentially synergize with SIM-SUMO interaction (40, 48) for the timely regulation of the abundance of SUMO-modified KAP1 or PML. Our results further uncovered that the Arg-73 and Arg-74 located at the ARM of RNF4 are essential for the accelerated degradation KAP1 and PML under stress. There are four SIMs located at the N terminus of RNF4 (34, 35, 49). Both Tatham et al. (35) and Keusekotten et al. (56) have reported that the second and third SIMs of RNF4 were critical for SUMO polymer binding, whereas the first SIM did not contribute much and the fourth SIM appeared to have a limited capacity for interaction with SUMO polymers. However, when the fourth SIM domain and its adjacent region, including the ARM, were deleted, approximately 50% of the SUMO binding capacity was lost (35), which is consistent with our results here showing that the SIM (fourth)-SUMO interaction synergizes with the ARM upon target phosphorylation.
We propose that RNF4 utilizes bimodular recognition, namely through conserved SIMs and the ARM to exert dynamic and adaptable responses to cellular stresses. The proposed bimodular recognition mechanism may represent a general paradigm aimed at stimulating the turnover of SUMO-conjugated proteins under cellular stress. In support, Cremona et al. (50) have determined that 27% of the proteins examined so far that are SUMO-modified in response to DNA damage are also phosphorylated. To prove the concept, the Arg-73- and Arg-74-accelerated degradation of simultaneously phosphorylated and SUMOylated protein was expanded to PML upon treatment with ATO (Fig. 5E). As additional DDR proteins, such as 53BP1, BRCA1, BLM1, RAP80, HERC2, XRCC4, and replication protein A (RPA) (28, 51–55) are SUMOylated, an analogous degradation by RNF4 is worth investigating. Moreover, our observations are in agreement with the broad and general function of SIM-SUMO interaction in recruiting RNF4 targets. We theorize that the identified ARM-Ser(P)-824 or Ser(P)-117 interaction will synergize the low-affinity SIM-SUMO interaction to enable RNF4 to specifically recognize SUMOylated KAP1 or PML with a high avidity, leading to ubiquitylation and subsequent proteasome-mediated degradation upon external stimulation. This model could also account for our observation of the ability of EGFP-SUMO-1 to stimulate KAP1(S824D) turnover. It is possible that the ARM also allows RNF4 to ubiquitylate targets with mono-SUMOylation on single or multiple distinct Lys residues upon its phosphorylation. However, much still remains to be solved, and further research will be necessary. For example, what is the role of other arginines embedded within the ARM? Does the spacing between phosphorylation and SUMOylation sites located at the target protein affect the recruitment of RNF4?
KAP1 plays a key role in transcriptional repression and chromatin remodeling. Accordingly, the fact that DSBs cause a RNF4-mediated decline of Ser(P)-824-SUMO-KAP1 could constitute a means to balance the transcriptional repressive function of SUMO-KAP1 (7, 8, 10) with the chromatin decondensation function of Ser(P)-824-KAP1 (6, 17). Our findings also highlight the role of concerted KAP1 PTMs in modulating KAP1 function. Previously, we and others have shown that the balance of ATM activation and phosphatases PP1/PP2/PP4 impacts the Ser(P)-824-KAP1 level (6–8, 57, 58). KAP1 SUMOylation and deSUMOylation have been demonstrated to regulate checkpoint activation and chromatin relaxation (7, 9, 10, 16, 17). Our results reported here suggest that the respective Ser-824 phosphorylation and Lys-676 SUMOylation are linked to the recruitment of RNF4 and, presumably, KAP1 function. Although not demonstrated directly, it appears that ATM phosphorylates Ser-824 of already SUMOylated KAP1 (7, 10) or that PP1β-mediates the SUMOylation of KAP1 after Ser-824 is phosphorylated (7, 8, 10). However, we cannot exclude the possibility that other unidentified Ub E3 ligases or deubiquitinases are also involved in regulating KAP1 turnover during DDR. It is worth noting that the DSB-induced KAP1 Ser-824-phosphorylation also promotes the dissociation of CHD3 from SUMO groups at Lys-779/804 in KAP1 to relax chromatin (17). We speculate that the release of CHD3 may permit the recruitment of RNF4 to SUMO-Lys-676-KAP1 for subsequent ubiquitylation and turnover, thus enabling the relaxation of compacted chromatin.
In summary, our observations strengthen and redefine the dynamic interaction between RNF4 and KAP1, which could be essential for proper DDR by acting in several pathways, including transcriptional derepression of KAP1-targeted pro-apoptosis and pro-arrest genes and chromatin decondensation. We propose that evolutionarily conserved RNF4 ARM and DSB-induced KAP1 Ser-824 phosphorylation concertedly promote the interaction of Ser(P)-824-SUMO-KAP1 with RNF4 to accelerate its turnover during DDR. Given the widespread involvement of SUMOylation and phosphorylation in DDR and that Kap1-null and Rnf4-null are both embryonic lethal (11, 24), the proposed bimodular recognition could affect a broader spectrum of biological processes. With these caveats, the ARM-dependent accelerated degradation of Ser(P)-824-SUMO-KAP1 or Ser(P)-117-SUMO-PML by RNF4, to our knowledge, represents the first example demonstrating that the substrate selection by RNF4 can be fine-tuned through the ARM.
Acknowledgments
We thank Drs. David Schultz, Ronald Hay, and Jorma Palvimo and the RNAi consortium at Academia Sinica for reagents and advice. We also thank Sonya Liu and Lawrance Chung for technical assistance, members of the Ann laboratory for discussions, and Dr. Margaret Morgan for editing.
This work was supported, in whole or in part, by National Institute of Health Grants R01DE10742 and R01DE14183 (to D. K. A.). This work was also supported by National Science Council Grants 99-2321-B-001-010 and 98-2321-B-001-012 (to H. M.S.), by National 973 Program Grant 2011CB510102 (to K. K. L.), and by National Natural Science Foundation Grant 81270622 (to K. K. L.).
- PTM
- posttranslational modification
- SUMO
- small ubiquitin-like modifier
- DSB
- double strand break
- DDR
- DNA damage response
- STUbL
- small ubiquitin-like modifier-targeted ubiquitin ligase
- SIM
- small ubiquitin-like modifier-interacting motif
- ARM
- arginine-rich motif
- BiFC
- bimolecular fluorescence complementation
- aa
- amino acids
- CHX
- cycloheximide
- IR
- irradiation
- Gy
- gray
- Ub
- ubiquitin
- Dox
- doxorubicin
- ATO
- arsenic trioxide
- fl
- full-length
- PML
- promyelocytic leukemia protein
- SFB
- S tag/FLAG/streptavidin-binding peptide
- PHD
- plant homeodomain
- ATM
- ataxia telangiectasia mutated
- EGFP
- enhanced green fluorescent protein
- AMBER
- Assisted Model Building with Energy Refinement.
REFERENCES
- 1. Seo J., Lee K.-J. (2004) Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. J. Biochem. Mol. Biol. 37, 35–44 [DOI] [PubMed] [Google Scholar]
- 2. Walsh C. T., Garneau-Tsodikova S., Gatto G. J. (2005) Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 44, 7342–7372 [DOI] [PubMed] [Google Scholar]
- 3. Pinder J. B., Attwood K. M., Dellaire G. (2013) Reading, writing, and repair: the role of ubiquitin and the ubiquitin-like proteins in DNA damage signaling and repair. Front. Genet. 4, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ciccia A., Elledge S. J. (2010) The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. White D. E., Negorev D., Peng H., Ivanov A. V., Maul G. G., Rauscher F. J., 3rd. (2006) KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res. 66, 11594–11599 [DOI] [PubMed] [Google Scholar]
- 6. Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D. C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. (2006) Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 8, 870–876 [DOI] [PubMed] [Google Scholar]
- 7. Li X., Lee Y. K., Jeng J. C., Yen Y., Schultz D. C., Shih H. M., Ann D. K. (2007) Role for KAP1 serine 824 phosphorylation and SUMOylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J. Biol. Chem. 282, 36177–36189 [DOI] [PubMed] [Google Scholar]
- 8. Li X., Lin H. H., Chen H., Xu X., Shih H. M., Ann D. K. (2010) SUMOylation of the transcriptional co-repressor KAP1 is regulated by the serine and threonine phosphatase PP1. Sci. Signal. 3, ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ivanov A. V., Peng H., Yurchenko V., Yap K. L., Negorev D. G., Schultz D. C., Psulkowski E., Fredericks W. J., White D. E., Maul G. G., Sadofsky M. J., Zhou M. M., Rauscher F. J., 3rd. (2007) PHD domain-mediated E3 ligase activity directs intramolecular SUMOylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee Y.-K., Thomas S. N., Yang A. J., Ann D. K. (2007) Doxorubicin Down-regulates Kruppel-associated Box domain-associated protein 1 SUMOylation that relieves its transcription repression on p21WAF1/CIP1 in breast cancer MCF-7 Cells. J. Biol. Chem. 282, 1595–1606 [DOI] [PubMed] [Google Scholar]
- 11. Cammas F., Mark M., Dollé P., Dierich A., Chambon P., Losson R. (2000) Mice lacking the transcriptional corepressor TIF1β are defective in early postimplantation development. Development 127, 2955–2963 [DOI] [PubMed] [Google Scholar]
- 12. Nielsen A. L., Ortiz J. A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R. (1999) Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 18, 6385–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ryan R. F., Schultz D. C., Ayyanathan K., Singh P. B., Friedman J. R., Fredericks W. J., Rauscher F. J., 3rd. (1999) KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell Biol. 19, 4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schultz D. C., Friedman J. R., Rauscher F. J., 3rd. (2001) Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev. 15, 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schultz D. C., Ayyanathan K., Negorev D., Maul G. G., Rauscher F. J., 3rd. (2002) SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16, 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garvin A. J., Densham R. M., Blair-Reid S. A., Pratt K. M., Stone H. R., Weekes D., Lawrence K. J., Morris J. R. (2013) The deSUMOylase SENP7 promotes chromatin relaxation for homologous recombination DNA repair. EMBO Rep. 10.1038/ncb1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goodarzi A. A., Kurka T., Jeggo P. A. (2011) KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 18, 831–839 [DOI] [PubMed] [Google Scholar]
- 18. Bologna S., Ferrari S. (2013) It takes two to tango: ubiquitin and SUMO in the DNA damage response. Front. Genet. 4, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prudden J., Pebernard S., Raffa G., Slavin D. A., Perry J. J., Tainer J. A., McGowan C. H., Boddy M. N. (2007) SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26, 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun H., Leverson J., Hunter T. (2007) Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4053–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Uzunova K., Göttsche K., Miteva M., Weisshaar S. R., Glanemann C., Schnellhardt M., Niessen M., Scheel H., Hofmann K., Johnson E. S., Praefcke G. J., Dohmen R. J. (2007) Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282, 34167–34175 [DOI] [PubMed] [Google Scholar]
- 22. Xie Y., Rubenstein E. M., Matt T., Hochstrasser M. (2010) SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor. Genes Dev. 24, 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perry J. J., Tainer J. A., Boddy M. N. (2008) A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 33, 201–208 [DOI] [PubMed] [Google Scholar]
- 24. Vyas R., Kumar R., Clermont F., Helfricht A., Kalev P., Sotiropoulou P., Hendriks I. A., Radaelli E., Hochepied T., Blanpain C., Sablina A., van Attikum H., Olsen J. V., Jochemsen A. G., Vertegaal A. C., Marine J. C. (2013) RNF4 is required for DNA double-strand break repair in vivo. Cell Death Differ. 20, 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Poulsen S. L., Hansen R. K., Wagner S. A., van Cuijk L., van Belle G. J., Streicher W., Wikström M., Choudhary C., Houtsmuller A. B., Marteijn J. A., Bekker-Jensen S., Mailand N. (2013) RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J. Cell Biol. 201, 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yin Y., Seifert A., Chua J. S., Maure J.-F., Golebiowski F., Hay R. T. (2012) SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26, 1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luo K., Zhang H., Wang L., Yuan J., Lou Z. (2012) SUMOylation of MDC1 is important for proper DNA damage response. EMBO J. 31, 3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guzzo C. M., Berndsen C. E., Zhu J., Gupta V., Datta A., Greenberg R. A., Wolberger C., Matunis M. J. (2012) RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci. Signal. 5, ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galanty Y., Belotserkovskaya R., Coates J., Jackson S. P. (2012) RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26, 1179–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shyu Y. J., Liu H., Deng X., Hu C.-D. (2006) Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques 40, 61–66 [DOI] [PubMed] [Google Scholar]
- 31. Häkli M., Karvonen U., Jänne O. A., Palvimo J. J. (2005) SUMO-1 promotes association of SNURF (RNF4) with PML nuclear bodies. Exp. Cell Res. 304, 224–233 [DOI] [PubMed] [Google Scholar]
- 32. Nguyen H.-V., Chen J.-L., Zhong J., Kim K.-J., Crandall E. D., Borok Z., Chen Y., Ann D. K. (2006) SUMOylation attenuates sensitivity toward hypoxia- or desferroxamine-induced injury by modulating adaptive responses in salivary epithelial cells. Am. J. Pathol. 168, 1452–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Méndez J., Stillman B. (2000) Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de Thé H. (2008) Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10, 547–555 [DOI] [PubMed] [Google Scholar]
- 35. Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. (2008) RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10, 538–546 [DOI] [PubMed] [Google Scholar]
- 36. Bruderer R., Tatham M. H., Plechanovova A., Matic I., Garg A. K., Hay R. T. (2011) Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 12, 142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen R., Li L., Weng Z. (2003) ZDOCK: an initial-stage protein-docking algorithm. Proteins 52, 80–87 [DOI] [PubMed] [Google Scholar]
- 38. Baba D., Maita N., Jee J. G., Uchimura Y., Saitoh H., Sugasawa K., Hanaoka F., Tochio H., Hiroaki H., Shirakawa M. (2005) Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 435, 979–982 [DOI] [PubMed] [Google Scholar]
- 39. Song J., Zhang Z., Hu W., Chen Y. (2005) Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 280, 40122–40129 [DOI] [PubMed] [Google Scholar]
- 40. Geoffroy M. C., Jaffray E. G., Walker K. J., Hay R. T. (2010) Arsenic-induced, SUMO-dependent Recruitment of RNF4 into PML nuclear bodies. Mol. Biol. Cell 21, 4227–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang S., Kuo C., Bisi J. E., Kim M. K. (2002) PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2. Nat. Cell Biol. 4, 865–870 [DOI] [PubMed] [Google Scholar]
- 42. Hu C.-D., Chinenov Y., Kerppola T. K. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 [DOI] [PubMed] [Google Scholar]
- 43. Guo B., Sharrocks A. D. (2009) Extracellular signal-regulated kinase mitogen-activated protein kinase signaling initiates a dynamic interplay between SUMOylation and ubiquitination to regulate the activity of the transcriptional activator PEA3. Mol. Cell Biol. 29, 3204–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Martin N., Schwamborn K., Schreiber V., Werner A., Guillier C., Zhang X. D., Bischof O., Seeler J. S., Dejean A. (2009) PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. EMBO J. 28, 3534–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Hagen M., Overmeer R. M., Abolvardi S. S., Vertegaal A. C. (2010) RNF4 and VHL regulate the proteasomal degradation of SUMO-conjugated hypoxia-inducible factor-2α. Nucleic Acids Res. 38, 1922–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang Y.-C., Yoshikai Y., Hsu S.-W., Saitoh H., Chang L.-K. (2013) Role of RNF4 in the ubiquitination of Rta of Epstein-Barr virus. J. Biol. Chem. 288, 12866–12879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Izumiya Y., Kobayashi K., Kim K. Y., Pochampalli M., Izumiya C., Shevchenko B., Wang D.-H., Huerta S. B., Martinez A., Campbell M., Kung H.-J. (2013) Kaposi's sarcoma-associated herpesvirus K-Rta exhibits SUMO-targeting ubiquitin ligase (STUbL) like activity and is essential for viral reactivation. PLoS Pathog. 9, e1003506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liew C. W., Sun H., Hunter T., Day C. L. (2010) RING domain dimerization is essential for RNF4 function. Biochem. J. 431, 23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun H., Leverson J. D., Hunter T. (2007) Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cremona C. A., Sarangi P., Yang Y., Hang L. E., Rahman S., Zhao X. (2012) Extensive DNA damage-induced SUMOylation contributes to replication and repair and acts in addition to the Mec1 checkpoint. Mol. Cell 45, 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dou H., Huang C., Singh M., Carpenter P. B., Yeh E. T. (2010) Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol. Cell 39, 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K. M., Jackson S. P. (2009) Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Morris J. R., Boutell C., Keppler M., Densham R., Weekes D., Alamshah A., Butler L., Galanty Y., Pangon L., Kiuchi T., Ng T., Solomon E. (2009) The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462, 886–890 [DOI] [PubMed] [Google Scholar]
- 54. Ouyang K. J., Woo L. L., Zhu J., Huo D., Matunis M. J., Ellis N. A. (2009) SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol. 7, e1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Danielsen J. R., Povlsen L. K., Villumsen B. H., Streicher W., Nilsson J., Wikström M., Bekker-Jensen S., Mailand N. (2012) DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J. Cell Biol. 197, 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Keusekotten K., Bade V. N., Meyer-Teschendorf K., Sriramachandran A. M., Fischer-Schrader K., Krause A., Horst C., Schwarz G., Hofmann K., Dohmen R. J., Praefcke G. J. (2014) Multivalent interactions of the SUMO-interaction motifs in RING finger protein 4 determine the specificity for chains of the SUMO. Biochem. J. 457, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee D.-H., Goodarzi A. A., Adelmant G. O., Pan Y., Jeggo P. A., Marto J. A., Chowdhury D. (2012) Phosphoproteomic analysis reveals that PP4 dephosphorylates KAP-1 impacting the DNA damage response. EMBO J. 31, 2403–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu J., Xu L., Zhong J., Liao J., Li J., Xu X. (2012) Protein phosphatase PP4 is involved in NHEJ-mediated repair of DNA double-strand breaks. Cell Cycle 11, 2643–2649 [DOI] [PubMed] [Google Scholar]